Abstract
Despite their wide applications in soluble macromolecules, optical tweezers have rarely been used to characterize the dynamics of membrane proteins, mainly due to the lack of model membranes compatible with optical trapping. Here, we examined optical trapping and mechanical properties of two potential model membranes, giant and small unilamellar vesicles (GUVs and SUVs, respectively) for studies of membrane protein dynamics. We found that optical tweezers can stably trap GUVs containing iodixanol with controlled membrane tension. The trapped GUVs with high membrane tension can serve as a force sensor to accurately detect reversible folding of a DNA hairpin or membrane binding of synaptotagmin-1 C2AB domain attached to the GUV. We also observed that SUVs are rigid enough to resist large pulling forces and are suitable for detecting protein conformational changes induced by force. Our methodologies may facilitate single-molecule manipulation studies of membrane proteins using optical tweezers.
Significance
Numerous biological processes on membranes involve complex protein-protein and protein-membrane interactions that are further regulated by mechanical forces. These interactions are difficult to study using traditional experimental approaches due to ensemble averaging or lack of mechanical force. As a step to manipulate single membrane proteins using optical tweezers, we tested the optical trapping of GUVs and SUVs and examined their mechanical properties. We found that both could serve as potential model membranes to study dynamics of membranes, membrane proteins, or protein-membrane interactions in the presence of force with high spatiotemporal resolution by optical tweezers.
Introduction
Numerous biological processes occurring on membranes involve complex protein-protein and protein-membrane interactions that are further regulated by mechanical forces. These processes include membrane protein folding (1, 2, 3), membrane fusion or lipid exchange (4, 5, 6), immune responses (7), mechanosensation or mechanotransduction (8, 9, 10), and cell growth, migration, and differentiation (11,12). The molecular interactions involved in these processes are difficult to study using traditional experimental approaches based on a large number of molecules due to ensemble averaging or lack of mechanical force (5,6,9,10,13). Single-molecule force spectroscopy, including atomic force microscopy (AFM), optical tweezers, and magnetic tweezers, has widely been applied to study the dynamics of soluble proteins (14, 15, 16, 17, 18, 19). However, applications of the methodology into membrane proteins are very limited. AFM can image membranes, apply force to membrane proteins, and probe protein dynamics (20). Consequently, AFM has long been used to study membrane protein folding by pulling single proteins out of surface-supported lipid bilayers (3,21). AFM generally uses large and stiff fabricated cantilevers as force probes, which lead to high spatial resolution but low force resolution compared with magnetic or optical tweezers (15,22). In addition, the underlying surfaces may perturb the structure and dynamics of membranes or embedded membrane proteins, leading to reduced lateral diffusion of lipids or proteins (23, 24, 25, 26). Magnetic tweezers have been successfully applied to detect stepwise association and dissociation of transmembrane helices of rhomboid protease GlpG or β2-adrenergic receptor in bicelles, and, recently, unfolding of GlpG in small unilamellar vesicles (SUVs) (2,27). So far, reversible protein folding has not been observed in an authentic membrane environment under an equilibrium condition, except for small regions of transmembrane helices, which prevents measurements of folding energy for larger domains of membrane proteins, including the insertion energy of a single transmembrane helix. This calls for improved single-molecule manipulation approaches for studying membrane protein dynamics. Compared with AFM and magnetic tweezers, optical tweezers are more widely used to study dynamics of soluble proteins, including the unidirectional movement of molecular motors and folding dynamics of proteins or protein complexes (14,19,28), partly due to the extremely high precision of optical tweezers for measurements of distance (∼0.2 nm) and force (∼0.01 pN) with high temporal resolution (∼10 μs) (29). In contrast, optical tweezers are also least used to investigate membrane proteins, especially their folding dynamics, partly due to lack of proper model membranes for suspension in optical traps to pull membrane proteins.
Giant unilamellar vesicles (GUVs) and SUVs are common model membranes to study membrane proteins in bulk (30). Integral proteins in both GUV and SUV membranes are fully mobile (31). Aspirated on the tips of micropipettes, GUVs have been utilized as membrane reservoirs to pull long membrane tethers or tubules with controllable diameters or curvatures with optical tweezers (32, 33, 34). These membrane tethers not only are used to measure membrane tension and bending stiffnesses (34,35) but also serve as substrates to test many proteins that bind to membranes in a curvature-dependent manner or deform the membranes upon their binding (36). Optical tweezers have been applied to trap micron-sized GUVs in many applications. They were used to probe the mechanical properties of lipid bilayers (37), sort GUVs with different properties, fuse GUVs, or assemble GUVs into artificial cell networks (38). However, the optical trapping was weak (39) due to the small difference in the refractive indices (RI) of GUVs and water, making it unlikely to directly pull membrane proteins reconstituted onto the trapped GUVs. Furthermore, reconstitution of integral membrane proteins into GUV membranes is generally challenging, as there have been no general methods for reliable protein reconstitution (40). SUVs are popular model membranes for membrane protein studies, partly because reconstitution of membrane proteins onto SUVs is generally easier. However, with a diameter ranging from 20 to 100 nm, SUVs are invisible by conventional optical microscopy and cannot be directly trapped to withstand high pulling force (39). Taking this evidence together, it remains challenging to pull single macromolecules on membranes using optical tweezers.
As a step to study membrane protein dynamics using optical tweezers, we developed methods to pull macromolecules attached to the membranes of GUVs and SUVs to measure the dynamics of proteins and/or membranes with high resolution. We validated our methods using well-studied DNA hairpins and synaptotagmin-1. Our work may facilitate potential applications of both model membranes to studies of integral or peripheral membrane proteins using optical tweezers.
Materials and methods
Lipids
All lipids were purchased from Avanti Polar Lipids (Alabaster, AL, USA), including 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS), 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-4′,5′-bisphosphate) (PI(4,5)P2), 1,2-dioleoyl-sn-glycero-3-phospho-L-serine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl) (NBD-DOPS), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine-rhodamine-B-sulfonyl) (rhodamine-DOPE), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[biotinyl(polyethylene glycol)-2000] (biotin-DSPE), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[4-(p-maleimidophenyl)]butyramide (MPB-DOPE).
DNA handles
A total of four DNA handles was used in the different experiments. All had the same length of 2260 bp and dual digoxigenin labels at one end but different overhang oligonucleotides and/or labels (biotin or thiol group) at the other end. These DNA handles were made by polymerase chain reaction (PCR) using λDNA cl857 Sam7 (Promega, Madison, WI, USA, D1501) as a template and a forward primer containing two digoxigenin labels at the 5′ end. Four reverse primers contained either the overhangs and/or biotin or thiol labels at the 5′ end. The DNA handle used in Fig. 2 A had an overhang DNA hairpin sequence of biotin-5′-TTTGAGTCAA-CGTCTGGATC-CTGTTTTCAG-GATCCAGACG-TTGACTCTTT-(spacer), while the left DNA handle in Fig. 5 A contained an overhang sequence 5′-CTCGCCAACG-TACATACAAC-TGTACGCCCTC-(spacer) that hybridizes to the 5′ region of the DNA hairpin. Here the 18-atom hexa-ethylene glycol spacer connected the overhang sequences to the remaining part of the PCR primers at the 3′ end but prevented polymerase extension to the overhang regions during PCR. All primers were synthesized by Integrated DNA Technologies (Coralville, IA, USA).
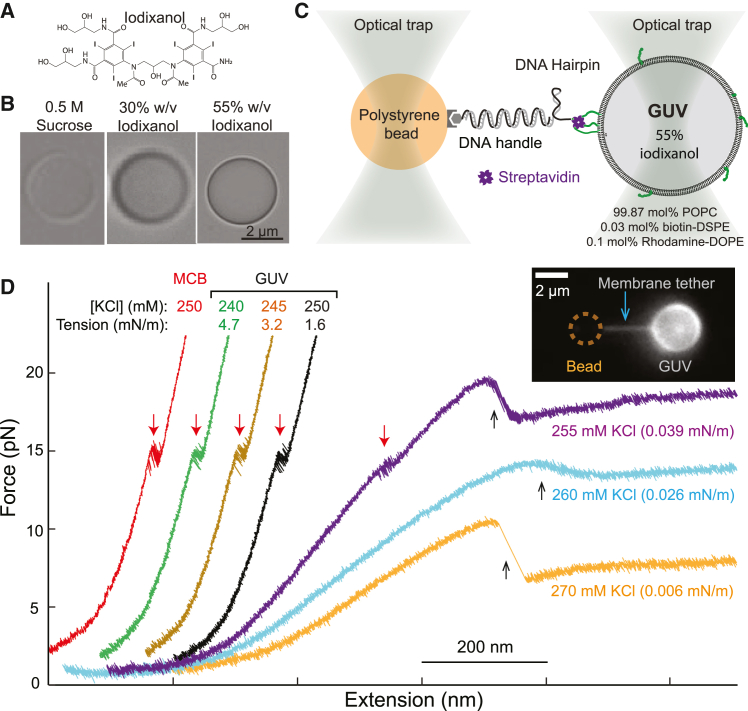
Figure 2.
Trapping iodixanol-containing GUVs for single-molecule manipulation. (A) Molecular formula of iodixanol. (B) Bright-field images of optically trapped GUVs containing 0.5 M sucrose only, and 30% or 55% iodixanol. (C) Schematic diagram of the experimental setup to pull a single DNA hairpin attached to the optical trapped GUV. (D) Force-extension curves (FECs) obtained by pulling the DNA hairpin attached to the MCB or the same GUV containing 55% iodixanol but with different membrane tensions in the buffers containing different concentrations of KCl ([KCl]). The three FECs on the left well overlap the FEC corresponding to 250 mM KCl but are successively shifted to the left for clarity. Red and black arrows indicate reversible unfolding/refolding transition of the DNA hairpin and abrupt formation of membrane tethers, respectively. The inset shows the fluorescence image of a membrane tether pulled out of the optically trapped GUV. The [KCl]-dependent results were repeatable and observed with more than eight GUVs from different batches of GUV preparations.
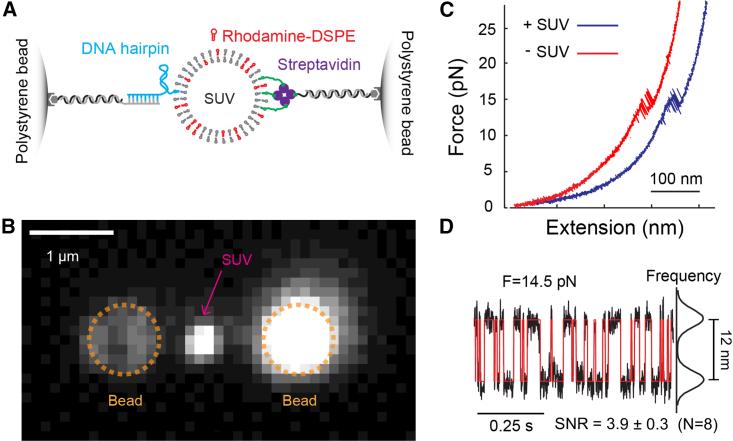
Figure 5.
Folding and unfolding transition of the DNA hairpin detected on the surface of a single SUV tethered between two polystyrene beads. (A) Schematic diagram of the experimental setup to pull the DNA hairpin conjugated to a single lipid in the tethered SUV via one of the DNA handles. The other DNA handle was directly attached to the SUV lipids through biotin-streptavidin interactions. The SUV contained 98.47 mol % POPC, 0.5 mol % rhodamine-DOPE, 1 mol % DNA hairpin-labeled DOPE, and 0.03 mol % biotin-DSPE. (B) Bright-field fluorescence image of a single rhodamine-labeled SUV tethered between two optical trapped polystyrene beads. Note that untethered SUVs bound specifically to the right bead containing an excess of free biotinylated DNA handles. (C) FEC obtained by pulling the DNA hairpin to high force in the presence of the SUV (+SUV as depicted in A) or in the absence of SUV (−SUV) by directly attaching a biotinylated DNA hairpin molecule to the DNA handle on the right. (D) Time-dependent extension trajectory at constant mean force showing reversible unfolding/refolding of the DNA hairpin.
Oligo-DOPE conjugation
The DNA hairpin-labeled DOPE lipids (oligo-DOPE, Fig. 5 A) were made by conjugating thiol-labeled oligonucleotide to the maleimide-labeled DOPE lipids (41). The oligonucleotide with a 3′ thiol group has the following sequence:
5′-GAGGGCGTAC-AGTTGTATGT-ACGTTGGCGA-GTTGAGTCAA-CGTCTGGATC-CTGTTTTCAG-GATCCAGACG-TTGACTCT-SH
The lyophilized oligonucleotide was dissolved in the buffer containing 20 mM Tris (pH 7.4), 250 mM KCl, 55 mM glucose (buffer A), plus 20 mM tris(2-carboxyethyl)phosphine (TCEP) for a 4 mM stock solution. The maleimide-labeled lipids MPB-DOPE dissolved in chloroform were dried in a clear glass vial first in nitrogen flow for 5 min and then in a vacuum desiccator for 1 h. Before lipid labeling, the stock oligonucleotide solution was diluted to 0.8 mM with buffer A plus 2.5% (w/v) n-octyl-β-D-glucoside (OG) and added to the glass vial with the dried lipid film with an MPB-DOPE to oligonucleotide molar ratio of 10:1. The solution was gently vortexed at room temperature for 4 h to complete the maleimide-thiol reaction. Finally, 2-mercaptoethanol was added to the mixture to a final concentration of 40 mM to quench all the unreacted MPB-PE. The oligonucleotide-labeled DOPE was aliquoted and stored at −80°C before use.
SUV preparation
SUVs were made for direct use (Fig. 5 A) or preparation of the membrane-coated silica beads (MCBs) (Figs. 1 and 2 D) or VAMP2-anchored GUVs (Fig. 1 D). Three types of SUVs were prepared that contained either pure lipids, oligo-DOPE, or VAMP2. Different lipids (except for the oligo-DOPE) were mixed in chloroform and dried to form lipid films as described in the preceding section. buffer A was then added to hydrate the lipids to make a solution with a total lipid concentration of 5 mg/mL. The cloudy vesicle solution was sonicated with a water bath sonicator for 30 min until the solution became clear. These SUVs were ready for use. For SUVs containing oligo-DOPE, Triton X-100 (Thermo Fisher Scientific, Waltham, MA, USA, 28314) was added to the SUV solution to a final concentration of 8 mM and incubated at room temperature with gentle agitation for 10 min. Oligo-DOPE was then added to the SUV solution to 1 mol % total lipid concentration and further incubated at room temperature for 1 h. Triton X-100 was removed by adding 40 mg of Bio-beads (Bio-Rad Laboratories, Hercules, CA, USA, 1523920) per 100 μL SUV solution and then nutating at 4°C overnight. VAMP2-anchored SUVs were prepared as previously described (42). In brief, the purified Alexa Fluor 647-labeled VAMP2 in 1.5% (w/v) OG, 140 mM KCl, and 25 mM HEPES (pH 7.4) was added to the dried lipids for a total lipid concentration of 5 mg/mL and a protein/lipid molar ratio of 1:1000. The mixture was vortexed for 15 min at room temperature, then diluted by the buffer containing 140 mM KCl and 25 mM HEPES (pH 7.4) for a final OG concentration of 0.33% (w/v). OG was removed by dialyzing the liposome solution in the same buffer using Slide-A-Lyzer Dialysis Cassettes (20 kDa cutoff) (Thermo Fisher Scientific, 66003) for 2 days at 4°C with a buffer change every 16 h. All SUVs were harvested, stored at 4°C, and used within 3 weeks.
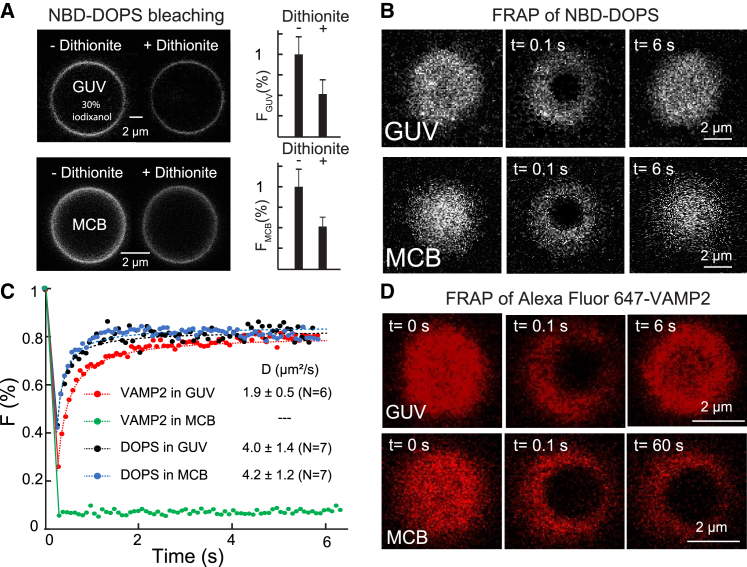
Figure 1.
Representative transmembrane protein is immobilized in the supported bilayer but fully mobile in the GUV membrane. (A) Confocal fluorescence images of the lipids in the same GUV membrane or membrane-coated bead (MCB) before and after NBD bleaching by dithionite treatment (left), with their normalized average fluorescence intensities shown in the right panel. The error bar indicates the standard deviation. (B) Confocal fluorescence images of the same GUV or MCB taken before (t = 0) and after photobleaching at the indicated time. (C) Fluorescence intensities as a function of time after photobleaching (symbols) and their best fits (dashed curves) to determine the diffusion coefficients of lipids or VAMP2 as indicated. The intensities were normalized by the corresponding intensities just before photobleaching. (D) Fluorescence images of Alexa Fluor 647-labeled VAMP2 in the GUV or MCB taken before (t = 0) and after photobleaching. The GUV or MCB membranes used in the FRAP experiments contained 99.65 mol % POPC, 0.25 mol % NBD-DOPS, and 0.1 mol % Alexa Fluor 647-labeled VAMP2. All GUVs encapsulated 30% (w/v) iodixanol.
Syt1 C2AB preparation and VAMP2 labeling
The sequences and purification of the Syt1 C2AB construct and the full-length VAMP2 with single cysteine mutation Q36C were previously described (13,43). In brief, the Syt1 C2AB construct contained an Avi-tag at its N-terminus and a unique cysteine at its C-terminus. The C2AB domain and the thiol-containing DNA handle were crosslinked as previously described (13). VAMP2 and Alexa Fluor 647 maleimide (Thermo Fisher Scientific, A20347) were mixed with a molar ratio of 1:3 in the presence of 1 mM TCEP and incubated at room temperature for 1 h. Dithiothreitol was then added to the mixture to a final concentration of 5 mM to quench unreacted maleimide. Free dye was removed by Micro Bio-Spin 6 columns (Bio-Rad Laboratories, 7326222).
Preparation of membrane-coated beads
MCBs were prepared as described elsewhere in detail (13). In brief, 100 μL of prewashed silica beads (Bangs Laboratories, Fishers, IN, USA, SS04002 and SS05003) with a diameter of 2.06 μm (for the pulling experiment) or 6 μm (for the fluorescence recovery after photobleaching (FRAP) experiment) were added into the corresponding 500 μL of SUV solution containing 1 mg/mL lipids. SUVs spontaneously bound to and collapsed on the surfaces of silica beads to form supported bilayers. The bead solution was vortexed at 1500 rpm at 37°C using Thermal Mixer C (Eppendorf, Hamburg, Germany) for 1 h to complete the membrane-coating process. MCBs were separated from the excessive liposomes by centrifuging the bead solution at 500 × g at room temperature for 1 min to precipitate the beads and then removing the supernatant. The beads were washed three times by adding 1 mL of buffer A, resuspending the beads, and centrifugation. The MCBs were stored in 100 μL of buffer A at 4°C and used within 1 week.
GUV preparation
GUVs containing sucrose only or iodixanol (≤30% w/v) were generated by the electroformation method (30). Twenty microliters of lipids with a final total lipid concentration of 5 mg/mL in chloroform was deposited onto platinum electrodes in small drops (∼0.5 μL per drop). The lipids were dried in the vacuum desiccator for 1 h to form lipid films on the electrodes. The electrodes were then gently immersed in a plastic tube with a buffer containing either 0.5 M sucrose, 1 M sucrose, or the iodixanol solution containing 30% (w/v) iodixanol, 0.43 M sucrose, and 5 mM HEPES (pH 7.4). For the GUVs containing Alexa Fluor 647 VAMP2, 40 μL of SUV solution containing 2 mg/mL lipids was deposited onto platinum electrodes in small drops (∼0.5 μL per drop). The SUV solution was dried first in the fume hood for 15 min and then in the vacuum desiccator for 1 h to form lipid films on the electrodes. The iodixanol solution was then used to immerse the electrodes. An alternating current with a sine wave (function/arbitrary waveform generators, SDG2042X; SIGLENT, Shenzhen, China) was applied to the platinum electrodes with a peak-to-peak voltage of 2.3 V and frequency of 10 Hz for 4 h. The GUVs were harvested, stored at 4°C, and used within 1 week.
The GUVs containing over 30% iodixanol were made by an alternative inverted-emulsion method (44,45) because of the poor yield of the GUVs generated by the electroformation. A total of 0.4 μmol lipids was mixed in chloroform and dried in a clean glass vial. Next, 400 μL of liquid paraffin was added to the dried lipids before incubation at 50°C for 1 h to dissolve the lipids, which yielded a solution of 1 mM lipids in paraffin. Two hundred microliters of the solution was gently deposited on top of 500 μL of buffer that eventually remains outside the GUVs (outside buffer) in a 1.5 mL centrifuge tube and incubated at room temperature overnight until the interface between the oil and aqueous phases became flat, where a monolayer of lipids formed. The outside buffer contained 20 mM Tris (pH 7.4), 55 mM glucose, and 250 mM KCl (Fig. 2 C) or 200 mM KCl (Fig. 4 A), depending upon the applications. Twenty microliters of inside buffer to be encapsulated into the GUVs, i.e., 55% (w/v) iodixanol, 5 mM HEPES (pH 7.4), and 0.355 M sucrose (Fig. 2 C) or 0.21 M sucrose (Fig. 4 A), was added to the remaining 200 μL of lipid solution in paraffin and sonicated for 5 min to prepare the inverted-emulsion solution. This emulsion was added on top of the lipid solution in paraffin above the aqueous solution in the centrifuge tube. The mixture was then centrifuged at 1000 × g for 5 min to allow water droplets in the emulsion to pass through the lipid monolayer into the bottom aqueous solution to form GUVs. The bottom GUV solution was collected and stored at 4°C before use.
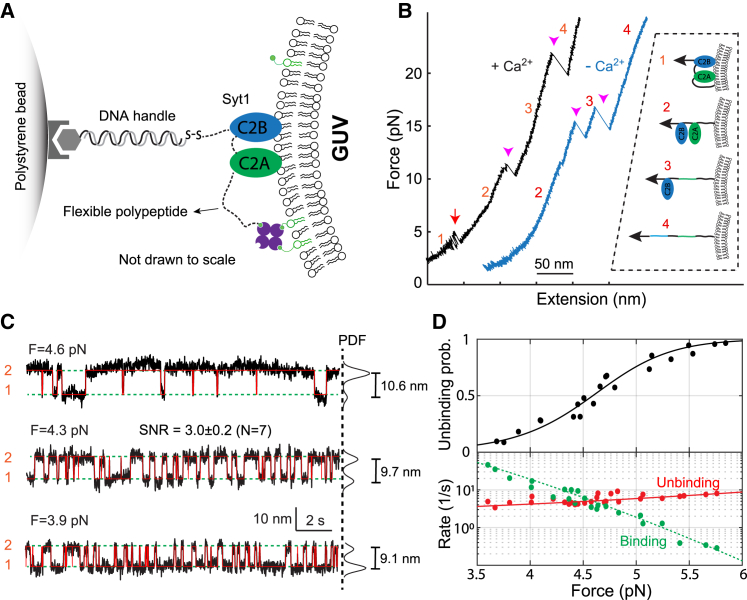
Figure 4.
Dynamic membrane binding of Syt1 C2AB domain detected on the surface of the optically trapped GUV. (A) Schematic diagram of the experimental setup. The N-terminus of Syt1 C2AB domain was attached to the GUV membrane through a flexible polypeptide linker and pulled from its C-terminus via the 2260 bp DNA handle. The GUV contains 55% iodixanol in the lumen and 84.97 mol % POPC, 10 mol % DOPS, 5 mol % PI(4,5)P2, and 0.03 mol % biotin-DSPE in the membrane. (B) FECs obtained in the presence 100 μM Ca2+ (+Ca2+) or absence Ca2+ (−Ca2+). The red arrow denotes reversible membrane binding and unbinding of Syt1 C2AB domain, and magenta arrowheads indicate the unfolding of the C2A and C2B domains. Red numbers label the four states associated with different FEC regions as depicted in the inset (13). (C) Time-dependent extension trajectories (black) and their idealized transitions derived from hidden-Markov modeling (red) showing reversible Syt1 C2AB binding to and unbinding from the GUV membrane at constant trap separation or mean force. On the right are probability density functions (PDF) of the extension, which yield the indicated extension changes and the average SNRs. The trajectories were mean-filtered to 100 Hz and shown. (D) Force-dependent unbinding probabilities (symbols in the top panel) and transition rates (symbols in the bottom panel) and their best model fits (solid and dashed curves). The fitting revealed the energy and kinetics of the C2AB binding at zero force (13).
Confocal fluorescence imaging and FRAP
All images were acquired by the laser scanning confocal microscope model SP8 (Leica, Wetzlar, Germany) equipped with LCS software and a 63× oil immersion objective at a scan speed of 1800 Hz or a frame rate of 13.04 per second. Samples were imaged in glass-bottom dishes (D35-14-1.5-U; Matsunami, Gunma, Japan), coated with β-casein (Sigma-Aldrich, Burlington, MA). The stock solutions of the GUVs or MCBs containing Alexa Fluor 647 VAMP2 and NDB-DOPS were diluted 3- or 10-fold with buffer A and added to the glass-bottom dishes. For the FRAP experiments, the excitation wavelength (λex) and emission wavelength (λem) were chosen as follows: λex = 488 nm, λem = 492–547 nm for NBD-DOPS and λex = 647 nm, λem = 650–695 nm for Alexa Fluor 647-VAMP2. Photobleaching was achieved by scanning the membrane region with a 2 μm diameter at the top of the GUV or MCB 1–3 times with maximum power of the corresponding excitation laser (20 mW for the 488 nm laser and 30 mW for the 647 nm laser) combined with the maximum power of a 405 nm laser (50 mW). Fluorescence recovery was monitored at 2%–10% of the maximum excitation laser power with the 405 nm laser off. Time-dependent average fluorescence intensities (Fig. 1C) were calculated from 6–7 FRAP experiments on different GUVs or MCBs and fitted by a modified Bessel function (46) using a script written in MATLAB, yielding the diffusion time τ. The diffusion coefficient (D) was computed based on the formula D = r2/(4τ) with r = 1 μm. To test the lamellarity of the lipid membranes, GUVs or MCBs were treated with 10 mM sodium dithionite (final concentration) for 10 min to bleach NBD-DOPS in the outer leaflet of the membrane. The averaged fluorescent intensities of GUVs/MCBs (N = 100–120) were measured before and after the dithionite treatment.
Estimations of the GUV membrane tension
Suppose a GUV has a radius and buffers of osmolarity and inside and outside the GUV, respectively, in a hypotonic solution with . The osmolarity difference generates an osmotic pressure
| (1) |
in terms of Van't Hoff's law, where is the molar gas constant and the absolute temperature. This osmotic pressure, in turn, generates membrane tension in the GUV membrane . Based on the Young-Laplace equation (47),
| (2) |
The membrane tension can then be solved from Eqs. (1) and (2) as
| (3) |
The membrane tension is also related to the area increase of the GUV membrane, i.e.,
| (4) |
where is the membrane area of the GUV in the hypotonic solution and is the elastic modulus of the GUV membrane (48). Here is the increase in the membrane area when the GUV is transferred from an isotonic solution to the hypotonic solution, which causes water to enter the GUV. This leads to the corresponding small increases in the GUV diameter () with
| (5) |
The osmolarity of the solution inside the GUV decreases when the GUV is transferred from the isotonic solution used to prepare the GUV (with osmolarity ) to the hypotonic solution (47), i.e.,
| (6) |
Substituting Eq. (6) into Eqs. (3) and (4) and equating the right sides of the latter two equations, one has
| (7) |
Substituting Eq. (7) into Eq. (4), we derive the equilibrium GUV membrane tension in the hypotonic solution as
| (8) |
The membrane tension can also be calculated using the measured equilibrium force of the membrane tether () (34), i.e.,
| (9) |
where is the membrane bending rigidity. The corresponding radius of the membrane tether can be calculated as
| (10) |
In our estimations for membrane tension (Fig. 2 D), we chose = 220 mN/m (47,48), = 23 kBT = 94 pN × nm (35,49), and = 2.5 kJ/mol. In the case of 255 mM, 260 mM, and 270 mM [KCl], the GUV membrane tension was calculated based on the measured equilibrium force of the membrane tether using Eq. (9). The GUV membrane tension at 240 mM, 245 mM, and 250 mM [KCl] was computed based on Eq. (8) using an osmolarity value = 852 mOsm for the concentrated iodixanol solution inside the GUV. This value was derived from the membrane tension of the GUV in 255 mM [KCl] again using Eq. (8). In all our derivations, the small intrinsic curvature of the GUV membrane induced by the asymmetric salt concentrations on both sides of the membrane (35) was neglected.
Dual-trap high-resolution optical tweezers
The dual-trap optical tweezers were home-built as described elsewhere in detail (50). In brief, a single laser beam of 1064 nm from a solid-state laser (Spectra-Physics, Utrecht, the Netherlands, J20I-BL-106C) was collimated, expanded, and split into two orthogonally polarized laser beams. One of the laser beams was reflected by a mirror attached to a piezoelectrically controlled stage that could turn in two axes, which was used to accurately move the corresponding optical trap in the sample plane. The two beams were further combined, expanded, and finally focused by a water-immersion 60× objective with a numerical aperture of 1.2 (Olympus, Tokyo, Japan) to form two optical traps. The outgoing laser beams were collimated by a second identical objective, split by polarization, and projected onto two position-sensitive detectors (Pacific Silicon Sensor, Westlake Village, CA, USA) to detect bead movements through back-focal plane interferometry. The trap stiffness was determined by the Brownian motions of the trapped beads or GUVs (51). To this end, the displacement of the bead or GUV in the trap was recorded at 80 kHz for over 3 s. The displacement trajectory was evenly divided into 128 regions, and a Fourier transformation of each region was performed to calculate its power spectrum density. The average of all power spectrum densities was computed and fit with a Lorentzian distribution , where is the frequency, the Boltzmann constant, = 300 K the temperature, and and are two fitting parameters. The trap stiffness was derived from the corner frequency, i.e., , with the drag coefficient of the trapped GUV or bead. The drag coefficient was calculated based on the GUV or bead radius or . The radii of the trapped GUVs or beads were determined by their images (Fig. S1). A customized microfluidic chamber with three parallel flow channels was used to deliver beads through the top and bottom channels to the central channel, where optical trapping occurred (52).
Single-molecule experiments
All pulling experiments were performed using the dual-trap high-resolution optical tweezers as previously described (16,52,53). In brief, ∼500 ng of DNA handles with biotin (Fig. 5 A), the overhang DNA hairpin (Fig. 2 B), or the Syt1 C2AB domain (Fig. 4 A) were mixed with a streptavidin solution with streptavidin/DNA handle molar ratio of 100:1 in a final volume of 5 μL and incubated at room temperature for 15 min. An aliquot of the mixture containing 1–10 ng of DNA was mixed with 10 μL of anti-digoxigenin antibody-coated polystyrene beads 2.1 μm in diameter (Spherotech), incubated at room temperature for 15 min, and diluted in 1 mL of buffer A. An aliquot of stock GUV or MCB solution was diluted by 10- to 20-fold or 1000-fold, respectively, in 1 mL of buffer A. Subsequently, the 1 mL DNA-bound bead solution and GUV or MCB solution were injected into the top and bottom channels in a home-made microfluidic chamber filled with buffer A with an oxygen scavenging system containing 55 mM glucose, 0.02 unit/mL glucose oxidase (Sigma-Aldrich), and 0.06 unit/mL catalase (Sigma-Aldrich). For the SUV pulling experiment, 10 μL of anti-digoxigenin antibody-coated polystyrene beads was mixed with 1 μL of 20 ng/μL DNA handle containing an overhang oligonucleotide and 9 μL of 1 mg/mL SUVs containing oligo-DOPE and incubated at room temperature for 15 min. The beads were then diluted in 1 mL of buffer A and injected into the bottom channel. A single anti-digoxigenin bead from the top channel and a single MCB, GUV, or anti-digoxigenin bead from the bottom channel were separately trapped and brought close to form a single protein (or lipid)-DNA tether. The tether was pulled or relaxed by moving one optical trap relative to the other fixed trap at a speed of 10 nm/s.
Results and discussion
Representative integral membrane protein is mobile on GUVs but not on supported bilayers
We have recently adopted MCBs to study membrane-binding affinity and kinetics of the C2 domains in synaptotagmin-1 (Syt1) and extend synaptotagmins using optical tweezers (6,13). In principle, integral membrane proteins can be reconstituted into the supported bilayers and similarly pulled in a direction perpendicular to the membrane surface to study their dynamics. However, like in other supported bilayers (23), the integral membrane proteins might suffer from nonspecific interactions with the underlying glass surfaces. This motivated us to examine the lateral mobility of integral membrane proteins in the lipid bilayers coated on silica beads using FRAP. We chose VAMP2, an SNARE protein of 116 amino acids in length with a single C-terminal transmembrane domain, as a representative for integral membrane proteins (5). We labeled VAMP2 with the Alexa Fluor 647 dye and reconstituted the protein into the bilayer on the surface of a silica bead 6 μm in diameter. For comparison, we also reconstituted the dye-labeled proteins into GUV membranes. Both GUV and supported membranes also contained dye-labeled lipid NBD-DOPS. First, we examined the unilamellarity of both membranes. We treated the MCBs and GUVs with dithionite that specifically quenches the NBD dyes labeled on the lipids in the outer leaflets of the membranes. Comparing bead or GUV images before and after dithionite treatment, we found that their fluorescence intensities decreased by ∼50% (Fig. 1 A), indicating unilamellar membranes coated on bead surfaces as well as in the GUVs. Next, we tested the mobility of NBD-DOPS in the membranes using FRAP. After photobleaching NBD in a small region (∼2 μm in diameter) on the top of GUV or MCB (26), the fluorescence in the region quickly recovered within 6 s with comparable recovery rates for the lipids on both GUV and MCB (Fig. 1, B and C), suggesting that the lipids are fully mobile. Similar diffusion coefficients of NPD-DOPS in both membranes were derived from the time-dependent fluorescence intensities (∼4 μm2/s, Fig. 1 C). While both diffusion coefficients fall in the ranges of previous measurements (2–9 μm2/s) (25,26), the approximately equal diffusion coefficients of DOPS in both membranes contrast with previous measurements for DOPE obtained by us and others, which show at least twofold slower lipid diffusion in the supported bilayer than in the free-standing membranes (13,25,26). The diffusion of negatively charged DOPS may be less hindered by the negatively charged silica surface than the neutral DOPE, contributing to the higher diffusion coefficient of DOPS than that of DOPE. Finally, we tested VAMP2 mobility in the membrane of GUV or MCB using FRAP (Fig. 1 D). The resultant diffusion coefficient of VAMP2 in the GUV (∼2 μm2/s, Fig. 1 C) is close to the previous measurement for another SNARE protein, syntaxin-1, which also contains a single C-terminal transmembrane (26), confirming rapid diffusion of transmembrane proteins in GUV membranes (Video S1). In contrast, no fluorescence recovery was observed for VAMP2 in the supported bilayer even 30 min after photobleaching (Fig. 1, C and D; Video S2). Thus, the VAMP2 proteins were immobilized on the bead surface. Combined with previous results (23,24,54), our experiments revealed an intrinsic drawback of the supported bilayers as a model membrane to study integral membrane proteins using optical tweezers, despite its success in studies of protein-membrane interactions with mobile lipids (6,13). We thus turned to GUVs and SUVs as potential model membranes to pull macromolecules on membranes.
Real-time confocal fluorescence images of the Alexa Fluor 647-labeled VAMP2 in the GUV membrane after photobleaching.
Real-time confocal fluorescence images of the Alexa Fluor 647-labeled VAMP2 in the lipid bilayer coated on a silica bead 6 μm in diameter after photobleaching.
Optical tweezers stably trap GUVs containing iodixanol
To trap GUVs for pulling macromolecules, we planned to increase the GUV trapping strength characterized by the stiffness of the optical trap. Given the size of a micron-sized object and the laser trapping power (typically a few hundred milliwatts), the trap stiffness increases with the RI of the object relative to that of water (RI = 1.33) (55). Therefore, we encapsulated solutions with different RIs inside GUVs and measured their trapping stiffness based on their Brownian motion in the optical trap with a fixed trapping laser power (51). All GUV membranes contained 99.87 mol % POPC, 0.03 mol % biotin-DSPE, and 0.1 mol % rhodamine-DOPE. The buffers outside the GUVs contained 20 mM Tris (pH 7.4), 55 mM glucose, and different concentrations of potassium chloride to balance the osmotic pressure of the GUVs. We first tested GUVs encapsulating 0.5 M sucrose (RI = 1.36) or 1 M sucrose (RI = 1.38), as they were used in previous trapping experiments (37, 38, 39). We obtained average trap stiffnesses of 0.025 ± 0.005 (mean ± standard deviation) pN/nm and 0.045 ± 0.006 pN/nm for the GUVs with 0.5 M sucrose and 1 M sucrose, respectively (Table 1). To compare GUV trapping, we specifically tested GUVs with a diameter in the range of 2.5–3.5 μm, although GUVs with 1.5–10 μm diameter could conveniently be trapped. The GUV traps were rather weak compared with the average trap stiffnesses of 0.162 pN/nm and 0.244 pN/nm for MCBs (RI = 1.45) and polystyrene beads (RI = 1.57), respectively, with diameters of ∼2 μm. Thus, despite being widely used in GUV preparation, sucrose does not significantly enhance GUV trapping due to its low refractive index.
Table 1.
Trapping stiffnesses of GUVs containing sucrose or iodixanol in different concentrations, membrane-coated beads, and polystyrene beads
| GUV |
GUV |
GUV |
GUV |
Membrane-coated bead |
Polystyrene bead |
|
|---|---|---|---|---|---|---|
| 0.5 M sucrose | 1 M sucrose | 30% iodixanol | 55% iodixanol | |||
| Diameter (μm) | 2.5–3.5 | 2.5–3.5 | 2.5–3.5 | 2.5–3.5 | 2.06 | 2.17 |
| Trapping stiffness (pN/nm) | 0.025 ± 0.005 (N = 15) | 0.045 ± 0.006 (N = 16) | 0.083 ± 0.005 (N = 21) | 0.113 ± 0.006 (N = 22) | 0.162 ± 0.006 (N = 29) | 0.244 ± 0.006 (N = 29) |
The number N in parentheses represents the number of GUVs or beads tested. The errors indicate the standard error of the mean.
To promote GUV trapping, we added iodixanol, also known as OptiPrep (Fig. 2 A), inside the GUV. The iodixanol solution has widely been used as a medium for density gradient centrifugation and as a radiocontrast agent in diagnostic imaging because of its high density and low osmolarity, viscosity, and toxicity (56,57). Recently, it has also gained applications in optical imaging due to its high refractive index and low absorbance for visible or infrared light (58). The typical 60% iodixanol stock solution has a high reflective index of 1.43, close to that of silica. The low absorbance is important for GUV trapping, because it minimizes laser heating due to the extremely high laser power density in optical traps (∼10 MW/cm2) (50,59). We prepared two batches of GUVs, one containing 30% (w/v) iodixanol and 0.43 M sucrose and the other, 55% (w/v) iodixanol and 0.355 M sucrose. Here sucrose was used to adjust both solutions to approximately equal osmolarity. All GUVs appeared spherical and could be readily imaged and trapped (Fig. 2 B). Due to their high refractive index, these GUVs exhibited significantly higher contrast than those containing sucrose only. The trapping stiffnesses for GUVs containing 30% and 55% iodixanol were 0.083 pN/nm and 0.113 pN/nm, respectively (Table 1). The latter was close to that of MCBs (0.162 pN/nm) but about half of the stiffness of polystyrene beads (0.244 pN/nm). Besides its high refractive index, iodixanol has another advantage over sucrose for GUV trapping due to the low osmolarity of iodixanol. To balance the osmotic pressure of the GUV containing 1 M sucrose, a high concentration of KCl (up to 560 mM) must be added in the solution outside the GUV, which tends to interfere with the structures and dynamics of the proteins in the solution. In contrast, the GUVs containing 55% iodixanol could be stably trapped in solutions containing as low as 100 mM KCl, which allows the testing of protein dynamics under a more physiological condition. In conclusion, GUVs containing ≥30% iodixanol could be stably trapped by optical tweezers to potentially detect conformational changes of macromolecules on membranes.
Pulling single DNA hairpins attached to trapped GUVs
To examine whether the trapped GUVs could further serve as a force and displacement sensor to directly measure the dynamics of macromolecules, we investigated the folding and unfolding dynamics of a DNA hairpin attached to the GUV containing 55% iodixanol. The DNA hairpin had a stem of 20 bp and a thymidine tetraloop (Fig. 2 C). It was directly tethered to the GUV at one end and to the 2.1 μm anti-digoxigenin antibody-coated polystyrene bead at the other end via a 2260 bp DNA handle. As a force and displacement sensor (60), the GUV needs to be sufficiently rigid to minimize their deformation induced by the pulling force and thermal fluctuations of the membrane. Thus, we controlled the membrane tension of the GUV by changing the concentration of potassium chloride ([KCl]) in the buffer outside the GUV. As [KCl] decreased below 270 mM, both the osmotic pressure and the GUV membrane tension increased in a predictable manner (see materials and methods) (47).
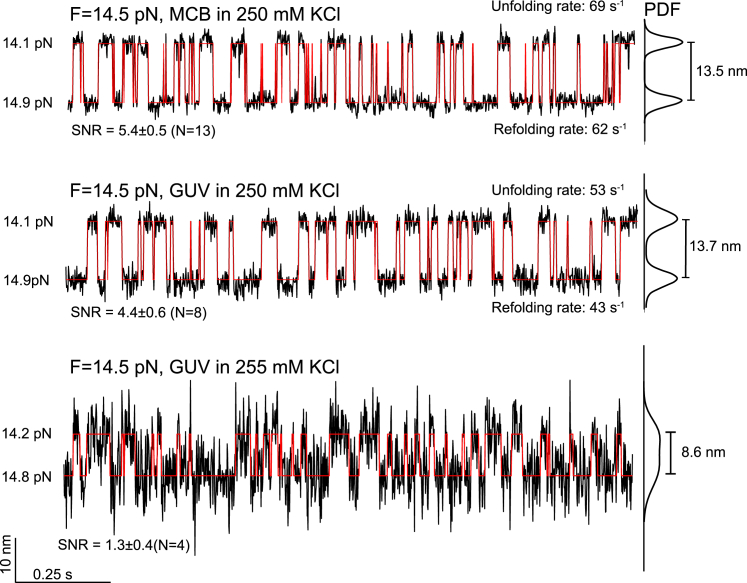
To test the effect of the GUV deformation on the single-molecule manipulation experiment, we pulled the DNA hairpin on the same GUV but adjusted its membrane tension by varying [KCl] from 240 mM to 270 mM using a microfluidic system (51). The DNA hairpin was being pulled by moving one trap away from the other fixed trap at a speed of 10 nm/s. At a high membrane tension with low [KCl] at 240 mM, 245 mM, and 250 mM, the resultant force-extension curves (FECs) were nearly identical, showing clear folding and unfolding transitions of the DNA hairpin at an equilibrium force of ∼14.5 pN (Fig. 2 D). In addition, all three FECs overlapped the FEC obtained by replacing the GUV with the MCB. Extension trajectories at a constant trap separation or mean force of 14.5 pN also revealed approximately equal extension changes and close folding and unfolding rates (Fig. 3, top and middle traces). The signal/noise ratio (SNR) detected on the GUV (4.4) was slightly lower than that on the MCB (5.4). These comparisons demonstrate that, at high membrane tension, the GUV is suited to pulling macromolecules on membranes and detecting their conformational transitions with high resolution. This conclusion implies that the GUV was relatively rigid and minimally deformed in response to a high pulling force. Consistent with this derivation, no significant GUV deformation was observed from the images of GUVs subjected to up to 40 pN pulling force (Fig. S1).
Figure 3.
Time-dependent extension trajectories at constant mean force showing reversible unfolding and refolding of the DNA hairpin attached to either the MCB in 250 mM KCl (top), or the GUV in 250 mM KCl (middle) or 255 mM KCl (bottom). The trajectories were mean-filtered to 1000 Hz and shown with their idealized transitions (red lines) derived from hidden-Markov modeling. The DNA hairpin transitions at constant trap separation were accompanied by small force fluctuations. The mean force (F) indicated is the mean of the two average forces corresponding to the folded and unfolded states labeled on the left. On the right are the probability density functions (PDF) of the extensions, which yield the indicated extension changes and the average signal/noise ratios (SNR) from a number (N) of independent measurements. The errors indicate the standard deviation.
Theoretical analyses corroborated the negligible GUV deformation induced by the pulling force under our experimental conditions with high GUV membrane tension. Cell or GUV membranes have been used as force probes based on membrane deformation, whose force constant was estimated to be twofold that of the membrane tension (61). Based on the membrane tension measured by membrane tether pulling described in the forthcoming section and the relative [KCl], we derived membrane tensions in the range of 4.7–1.6 pN/nm for the GUV in 240–250 mM KCl (Fig. 2 D, see materials and methods), with the corresponding GUV elongation of 2–6 nm in the presence of 20 pN pulling force. This contribution to the absolute extension was negligible compared to the ∼741 nm extension of the 2260 bp DNA handle at the same force. For the DNA hairpin transition measured in 250 mM KCl (Fig. 3), the GUV deformation dampened the extension change by ∼0.2 nm, which is significantly smaller than the measured 13.7 nm extension change. In conclusion, these calculations corroborated our experimental observations that GUV containing high concentrations of iodixanol with high membrane tensions can be used as a force probe to accurately measure the dynamics of macromolecules on membrane surfaces.
Pulling membrane tethers from trapped GUVs
In contrast, the FECs obtained at lower [KCl] significantly deviated from those described above. At 255 mM KCl, the FEC tilted to higher extension at a force below 18 pN (Fig. 2 D, purple), indicating significant GUV elongation along the pulling direction, which contributed to the extra extension compared to the extension measured using MCBs at the same force. Although the DNA hairpin transition still equilibrated at ∼14.5 pN, the extension change decreased to 8.6 nm, with the corresponding SNR decreasing to 1.3 (Fig. 3, bottom trace). Further pulling led to a sudden extension increase and accompanying force decrease (Fig. 2 D, purple FEC). Continued pulling only slowly increased the force as extension significantly increased. The sudden extension increase and the subsequent approximate force plateau indicate that a membrane tether or nanotubule was being pulled out of the GUV, as confirmed by fluorescence imaging (Fig. 2 D, inset). Our observations are consistent with previous experimental results and theoretical analyses based on membrane mechanics (32,62). As [KCl] was further reduced to 260 mM or 270 mM, the approximate plateau force of the membrane tether decreased with the corresponding decrease in membrane tension, again consistent with previous results (34). Quantitative relationships have been established among the plateau force, the radius of the membrane tether, and the membrane tension and bending rigidity (Eqs. 9 and 10). Thus, we derived the membrane tensions of the GUV in the three concentrations of potassium chloride (Fig. 2 D) and the radii of the associated membrane tethers (35 nm, 42 nm, and 86 nm at [KCl] of 255 mM, 260 mM, and 270 mM, respectively). Membrane tethers are widely observed in cells and play important roles in information and material transfer within or between cells (63). They are generated by pulling force and/or various proteins that bind to membranes to sense or generate membrane curvature (33,64). Thus, the trapped GUVs with low membrane tensions can be used to pull membrane tethers to probe the mechanical properties of membranes or curvature-dependent protein binding and membrane remodeling.
Protein-GUV membrane binding
Next, we asked whether the trapped GUV could be applied to study protein-membrane interactions, using the C2AB domain of synaptotagmin-1 (Syt1) as our model system. Anchored on synaptic vesicles, Syt1 binds to the presynaptic plasma membrane via the C2AB domain in the presence of Ca2+, thereby mediating Ca2+-triggered fusion of synaptic vesicles with the plasma membrane (4). We previously measured the membrane-binding energy and kinetics of Syt1 C2AB using MCBs and optical tweezers (13). Therefore, we repeated the experiment by replacing MCBs with GUVs containing 55% iodixanol. We attached the N-terminus of the Syt1 C2AB domain to the GUV membrane through a flexible polypeptide linker and pulled it from its C-terminus via the 2260 bp DNA handle (Fig. 4 A). In the presence of 100 μM Ca2+ in the solution, the FEC shows reversible membrane binding and unbinding at ∼4.5 pN, followed by sequential unfolding of the C2A and C2B domains at higher force (Fig. 4 B). The membrane binding was Ca2+ dependent, as the binding signal disappeared when Ca2+ was omitted in the solution. At constant trap separations, the force-dependent C2AB binding and unbinding transitions were clearly seen in the extension trajectories (Fig. 4 C). Detailed analyses of these trajectories based on hidden-Markov modeling revealed the unbinding probabilities and transition rates as a function of force (52,65) (Fig. 4 D). The nonlinear fitting of these data yielded the C2AB membrane unbinding energy of 9.5 (±0.1) kBT. These observations, including the average equilibrium force, the extension change, and the unbinding energy, are consistent with our previous measurements using MCBs (13). These comparisons indicate that the iodixanol-containing GUVs can be used to study the dynamics of membrane proteins in optical tweezers force spectroscopy. Compared with MCBs, the transmembrane proteins in GUV membranes are fully mobile and free from perturbation by the underlying glass surface. In addition, various macromolecules, small molecules, and buffers can be added to the relatively large interior space of GUVs, which may facilitate studies of many membrane proteins.
SUV as a model membrane to manipulate macromolecules
Despite the potential advantages of GUVs to manipulate macromolecules on membranes, it is often challenging to reconstitute transmembrane proteins into GUV membranes. In contrast, it is relatively easier to reconstitute proteins into SUV membranes with well-established protocols (40). However, with a diameter in the range of 20–100 nm, SUVs are generally too small to be stably trapped for pulling macromolecules (39). Therefore, we suspended single SUVs between two polystyrene beads using two DNA handles (Fig. 5 A). SUVs have been used to pull transmembrane proteins parallel to membranes (2). Here, we pulled the DNA hairpin attached to the SUV to mimic pulling membrane proteins in a direction perpendicular to the membrane. The experiment was designed to test how SUV deformation may affect the dynamics of macromolecules detected by optical tweezers. Wide-field fluorescence imaging of the SUV containing rhodamine-DOPE confirmed that a single SUV was being tethered between two beads (Fig. 5 B). The FEC of the SUV-DNA tether revealed the characteristic DNA hairpin unfolding and refolding transition similar to the transition of the hairpin directly attached to the bead without the SUV (Fig. 5 C). The DNA hairpin transition again exhibited an equilibrium force of 14.5 pN (Fig. 5 D), suggesting that a single DNA hairpin and SUV were tethered between two beads. A smaller extension change (12 nm vs. 13.5 nm, compare with Fig. 2 A, top trajectory) and SNR (3.9 vs. 5.4) were expected, because longer DNA handles used here slightly dampened the extension change detected by the beads (66). Therefore, conformational transitions could be accurately measured on the surfaces of SUVs. The observation implied that SUVs are relatively rigid and minimally deform in response to the pulling force. This derivation is consistent with the large force constants of the SUVs in the range of 15–32 pN/nm detected by AFM (67). Using these values, the estimated SUV elongation in the presence of 20 pN was less than 1.3 nm, and the extension change of the SUV during the DNA hairpin transition was 0.05 nm. In conclusion, SUVs may serve as a model membrane to study the dynamics of macromolecules using optical tweezers.
Conclusions
Optical tweezers have widely been used to study the dynamics of soluble proteins due to their high resolution and dynamic ranges of measurements for force, extension, and time. As a step to apply optical tweezers to membrane proteins, model membranes compatible for optical trapping and single-molecule manipulation are required. In addition, the mechanical properties of the model membranes should be examined. We found that iodixanol could be encapsulated inside GUVs to enhance their refractive index, thereby enabling their stable trapping. The trapped GUVs could serve as a model membrane to study the dynamics of membranes, proteins, and protein-membrane interactions. With proteins on two trapped GUVs, it is possible to investigate their transmembrane binding. The membrane tension of the trapped GUVs was conveniently regulated by the osmolarity of the buffer outside the GUV, which was facilitated by the microfluidic system used in optical tweezers. We found that GUVs with high membrane tensions were rigid enough to resist significant deformation due to high pulling force, thereby allowing accurate measurements of the extension changes associated with macromolecular conformational transitions around membranes. Under low membrane tension, membrane tethers could be pulled from the trapped GUVs, which could serve as model membranes with tunable curvatures to study curvature-sensitive membrane-binding proteins. Membrane tethers have previously been pulled from GUVs aspirated on the tip of micropipettes using optical tweezers (33). Our approach does not require micromanipulators and other devices required to control micropipettes. In addition, the optically trapped GUVs introduce fewer measurement noises than the aspirated GUVs due to stage drift (59). However, our method offers less accurate control in membrane tension than the aspiration approach. Finally, we validated the use of GUVs and SUVs as model membranes in single-molecule manipulation based on optical tweezers with relatively simple model macromolecules, the DNA hairpin, and the Syt1 C2AB domain. Further experiments are needed to apply the methodologies to membrane proteins, including multi-span transmembrane proteins or protein complexes.
Author contributions
Y.W., H.J., and Y.Z. designed the experiments; Y.W., A.K., and H.J. performed the experiments; Y.W., A.K., and Y.Z. analyzed the data; Y.W. and Y.Z. wrote the paper.
Acknowledgments
This work is supported by the National Institute of General Medical Sciences of the National Institutes of Health grants R35 GM131714, R01GM093341, and R01GM120193 to Y.Z. The authors declare no competing financial interests.
Editor: Rumiana Dimova.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.11.2884.
Supporting material
References
- 1.Popot J.L., Engelman D.M. Helical membrane protein folding, stability, and evolution. Annu. Rev. Biochem. 2000;69:881–922. doi: 10.1146/annurev.biochem.69.1.881. [DOI] [PubMed] [Google Scholar]
- 2.Choi H.K., Min D., et al. Yoon T.Y. Watching helical membrane proteins fold reveals a common N-to-C-terminal folding pathway. Science. 2019;366:1150–1156. doi: 10.1126/science.aaw8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu H., Siewny M.G.W., et al. Perkins T.T. Hidden dynamics in the unfolding of individual bacteriorhodopsin proteins. Science. 2017;355:945–949. doi: 10.1126/science.aah7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sudhof T.C., Rothman J.E. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y.L., Hughson F.M. Chaperoning SNARE folding and assembly. Annu. Rev. Biochem. 2021;90:581–603. doi: 10.1146/annurev-biochem-081820-103615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ge J., Bian X., Zhang Y.L., et al. Stepwise membrane binding of extended synaptotagmins revealed by optical tweezers. Nat. Chem. Biol. 2021 doi: 10.21203/rs.3.rs-523346/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huse M. Mechanical forces in the immune system. Nat. Rev. Immunol. 2017;17:679–690. doi: 10.1038/nri.2017.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin P., Jan L.Y., Jan Y.N. Mechanosensitive ion channels: structural features relevant to mechanotransduction mechanisms. Annu. Rev. Neurosci. 2020;43:207–229. doi: 10.1146/annurev-neuro-070918-050509. [DOI] [PubMed] [Google Scholar]
- 9.Zheng W., Holt J.R. The mechanosensory transduction machinery in inner ear hair cells. Annu. Rev. Biophys. 2021;50:31–51. doi: 10.1146/annurev-biophys-062420-081842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eyckmans J., Boudou T., et al. Chen C.S. A Hitchhiker's guide to mechanobiology. Dev. Cell. 2011;21:35–47. doi: 10.1016/j.devcel.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Helvert S., Storm C., Friedl P. Mechanoreciprocity in cell migration. Nat. Cell Biol. 2018;20:8–20. doi: 10.1038/s41556-017-0012-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Belly H., Stubb A., et al. Chalut K.J. Membrane tension gates erk-mediated regulation of pluripotent cell fate. Cell Stem Cell. 2021;28:273. doi: 10.1016/j.stem.2020.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma L., Cai Y., et al. Zhang Y.L. Single-molecule force spectroscopy of protein-membrane interactions. eLife. 2017;6:e30493. doi: 10.7554/eLife.30493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cecconi C., Shank E.A., et al. Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 15.Stigler J., Ziegler F., et al. Rief M. The complex folding network of single calmodulin molecules. Science. 2011;334:512–516. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- 16.Gao Y., Zorman S., et al. Zhang Y.L. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 2012;337:1340–1343. doi: 10.1126/science.1224492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.del Rio A., Perez-Jimenez R., et al. Sheetz M.P. Stretching single Talin rod molecules activates Vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao M.X., Goult B.T., et al. Yan J. Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci. Rep. 2014;4:4610. doi: 10.1038/srep04610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bustamante C., Alexander L., et al. Kaiser C.M. Single-molecule studies of protein folding with optical tweezers. Annu. Rev. Biochem. 2020;89:443–470. doi: 10.1146/annurev-biochem-013118-111442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin Y.C., Guo Y.R., et al. Scheuring S. Force-induced conformational changes in PIEZO1. Nature. 2019;573:230–234. doi: 10.1038/s41586-019-1499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oesterhelt F., Oesterhelt D., et al. Muller D.J. Unfolding pathways of individual bacteriorhodopsins. Science. 2000;288:143–146. doi: 10.1126/science.288.5463.143. [DOI] [PubMed] [Google Scholar]
- 22.Junker J.P., Ziegler F., Rief M. Ligand-dependent equilibrium fluctuations of single Calmodulin molecules. Science. 2009;323:633–637. doi: 10.1126/science.1166191. [DOI] [PubMed] [Google Scholar]
- 23.Wagner M.L., Tamm L.K. Tethered polymer-supported planar lipid bilayers for reconstitution of integral membrane proteins: silane-polyethyleneglycol-lipid as a cushion and covalent linker. Biophys. J. 2000;79:1400–1414. doi: 10.1016/S0006-3495(00)76392-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mashaghi S., van Oijen A.M. A versatile approach to the generation of fluid supported lipid bilayers and its applications. Biotechnol. Bioeng. 2014;111:2076–2081. doi: 10.1002/bit.25273. [DOI] [PubMed] [Google Scholar]
- 25.Beckers D., Urbancic D., Sezgin E. Impact of nanoscale hindrances on the relationship between lipid packing and diffusion in model membranes. J. Phys. Chem. B. 2020;124:1487–1494. doi: 10.1021/acs.jpcb.0c00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pincet F., Adrien V., et al. Tareste D. FRAP to characterize molecular diffusion and interaction in various membrane environments. PLoS One. 2016;11:e0158457. doi: 10.1371/journal.pone.0158457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Min D., Jefferson R.E., et al. Yoon T.Y. Mapping the energy landscape for second-stage folding of a single membrane protein. Nat. Chem. Biol. 2015;11:981–987. doi: 10.1038/nchembio.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avellaneda M.J., Franke K.B., et al. Tans S.J. Processive extrusion of polypeptide loops by a Hsp100 disaggregase. Nature. 2020;578:317–320. doi: 10.1038/s41586-020-1964-y. [DOI] [PubMed] [Google Scholar]
- 29.Sudhakar S., Abdosamadi M.K., et al. Schaffer E. Germanium nanospheres for ultraresolution picotensiometry of kinesin motors. Science. 2021;371:eabd9944. doi: 10.1126/science.abd9944. [DOI] [PubMed] [Google Scholar]
- 30.Angelova M.I., Dimitrov D.S. Liposome electroformation. Faraday Discuss. Chem. Soc. 1986;81:303–311. [Google Scholar]
- 31.Ramadurai S., Holt A., et al. Poolman B. Lateral diffusion of membrane proteins. J. Am. Chem. Soc. 2009;131:12650–12656. doi: 10.1021/ja902853g. [DOI] [PubMed] [Google Scholar]
- 32.Derenyi I., Julicher F., Prost J. Formation and interaction of membrane tubes. Phys. Rev. Lett. 2002;88:238101. doi: 10.1103/PhysRevLett.88.238101. [DOI] [PubMed] [Google Scholar]
- 33.Roux A., Koster G., et al. Bassereau P. Membrane curvature controls dynamin polymerization. Proc. Natl. Acad. Sci. USA. 2010;107:4141–4146. doi: 10.1073/pnas.0913734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roux A. The physics of membrane tubes: soft templates for studying cellular membranes. Soft Matter. 2013;9:6726–6736. [Google Scholar]
- 35.Karimi M., Steinkuhler J., et al. Dimova R. Asymmetric ionic conditions generate large membrane curvatures. Nano Lett. 2018;18:7816–7821. doi: 10.1021/acs.nanolett.8b03584. [DOI] [PubMed] [Google Scholar]
- 36.Shen H., Pirruccello M., De Camilli P. SnapShot: membrane curvature sensors and generators. Cell. 2012;150:1300. doi: 10.1016/j.cell.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solmaz M.E., Biswas R., et al. Povinelli M.L. Optical stretching of giant unilamellar vesicles with an integrated dual-beam optical trap. Biomed. Opt. Express. 2012;3:2419–2427. doi: 10.1364/BOE.3.002419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolognesi G., Friddin M.S., et al. Elani Y. Sculpting and fusing biomimetic vesicle networks using optical tweezers. Nat. Commun. 2018;9:1882. doi: 10.1038/s41467-018-04282-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bendix P.M., Oddershede L.B. Expanding the optical trapping range of lipid vesicles to the nanoscale. Nano Lett. 2011;11:5431–5437. doi: 10.1021/nl203200g. [DOI] [PubMed] [Google Scholar]
- 40.Jorgensen I.L., Kemmer G.C., Pomorski T.G. Membrane protein reconstitution into giant unilamellar vesicles: a review on current techniques. Eur. Biophys. J. 2017;46:103–119. doi: 10.1007/s00249-016-1155-9. [DOI] [PubMed] [Google Scholar]
- 41.Yang Y., Wang J., et al. Lin C.X. Self-assembly of size-controlled liposomes on DNA nanotemplates. Nat. Chem. 2016;8:476–483. doi: 10.1038/nchem.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tareste D., Shen J., et al. Rothman J.E. SNAREpin/Munc18 promotes adhesion and fusion of large vesicles to giant membranes. Proc. Natl. Acad. Sci. USA. 2008;105:2380–2385. doi: 10.1073/pnas.0712125105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma L., Rebane A.A., et al. Zhang Y.L. Munc18-1-regulated stage-wise SNARE assembly underlying synaptic exocytosis. eLife. 2015;4:e09580. doi: 10.7554/eLife.09580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pautot S., Frisken B.J., Weitz D.A. Production of unilamellar vesicles using an inverted emulsion. Langmuir. 2003;19:2870–2879. [Google Scholar]
- 45.Litschel T., Schwille P. Protein reconstitution inside giant unilamellar vesicles. Annu. Rev. Biophys. 2021;50:525–548. doi: 10.1146/annurev-biophys-100620-114132. [DOI] [PubMed] [Google Scholar]
- 46.Lopez A., Dupou L., et al. Tocanne J.F. Fluorescence recovery after photobleaching (FRAP) experiments under conditions of uniform disk illumination. Critical comparison of analytical solutions, and a new mathematical method for calculation of diffusion coefficient. D. Biophys. J. 1988;53:963–970. doi: 10.1016/S0006-3495(88)83177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alam Shibly S.U., Ghatak C., et al. Yamazaki M. Experimental estimation of membrane tension induced by osmotic pressure. Biophys. J. 2016;111:2190–2201. doi: 10.1016/j.bpj.2016.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rawicz W., Olbrich K.C., et al. Evans E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys. J. 2000;79:328–339. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dimova R. Recent developments in the field of bending rigidity measurements on membranes. Adv. Colloid Interfac. 2014;208:225–234. doi: 10.1016/j.cis.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 50.Sirinakis G., Ren Y.X., et al. Zhang Y.L. Combined and versatile high-resolution optical tweezers and single-molecule fluorescence microscopy. Rev. Sci. Instrum. 2012;83:093708. doi: 10.1063/1.4752190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y.L., Sirinakis G., et al. Gao Y. DNA translocation of ATP-dependent chromatin remodelling factors revealed by high-resolution optical tweezers. Methods Enzymol. 2012;513:3–28. doi: 10.1016/B978-0-12-391938-0.00001-X. [DOI] [PubMed] [Google Scholar]
- 52.Jiao J.Y., Rebane A.A., et al. Zhang Y.L. Single-molecule protein folding experiments using high-resolution optical tweezers. Methods Mol. Biol. 2017;1486:357–390. doi: 10.1007/978-1-4939-6421-5_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma L., Kang Y., et al. Zhang Y. α-SNAP enhances SNARE zippering by stabilizing the SNARE four-helix bundle. Cell Rep. 2016;15:531–539. doi: 10.1016/j.celrep.2016.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diaz A.J., Albertorio F., et al. Cremer P.S. Double cushions preserve transmembrane protein mobility in supported bilayer systems. Langmuir. 2008;24:6820–6826. doi: 10.1021/la800018d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Svoboda K., Block S.M. Biological applications of optical forces. Annu. Rev. Biophys. Biomol. Struct. 1994;23:247–285. doi: 10.1146/annurev.bb.23.060194.001335. [DOI] [PubMed] [Google Scholar]
- 56.Ford T., Graham J., Rickwood D. Iodixanol—a nonionic isosmotic centrifugation medium for the formation of self-generated gradients. Anal. Biochem. 1994;220:360–366. doi: 10.1006/abio.1994.1350. [DOI] [PubMed] [Google Scholar]
- 57.Svaland M.G., Haider T., et al. Hals P.A. Human pharmacokinetics of iodixanol. Invest Radiol. 1992;27:130–133. doi: 10.1097/00004424-199202000-00007. [DOI] [PubMed] [Google Scholar]
- 58.Boothe T., Hilbert L., et al. Rink J.C. A tunable refractive index matching medium for live imaging cells, tissues and model organisms. eLife. 2017;6:e27240. doi: 10.7554/eLife.27240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moffitt J.R., Chemla Y.R., et al. Bustamante C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc. Natl. Acad. Sci. USA. 2006;103:9006–9011. doi: 10.1073/pnas.0603342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gittes F., Schmidt C.F. Interference model for back-focal-plane displacement detection in optical tweezers. Opt. Lett. 1998;23:7–9. doi: 10.1364/ol.23.000007. [DOI] [PubMed] [Google Scholar]
- 61.Evans E., Ritchie K., Merkel R. Sensitive force technique to probe molecular adhesion and structural linkages at biological interfaces. Biophys. J. 1995;68:2580–2587. doi: 10.1016/S0006-3495(95)80441-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koster G., Cacciuto A., et al. Dogterom M. Force barriers for membrane tube formation. Phys. Rev. Lett. 2005;94:068101. doi: 10.1103/PhysRevLett.94.068101. [DOI] [PubMed] [Google Scholar]
- 63.Davis D.M., Sowinski S. Membrane nanotubes: dynamic long-distance connections between animal cells. Nat. Rev. Mol. Cell Biol. 2008;9:431–436. doi: 10.1038/nrm2399. [DOI] [PubMed] [Google Scholar]
- 64.Sorre B., Callan-Jones A., et al. Roux A. Nature of curvature coupling of amphiphysin with membranes depends on its bound density. Proc. Natl. Acad. Sci. USA. 2012;109:173–178. doi: 10.1073/pnas.1103594108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y.L., Jiao J., Rebane A.A. Hidden Markov modeling with detailed balance and its application to single protein folding. Biophys. J. 2016;111:2110–2124. doi: 10.1016/j.bpj.2016.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Greenleaf W.J., Woodside M.T., et al. Block S.M. Passive all-optical force clamp for high-resolution laser trapping. Phys. Rev. Lett. 2005;95:2081021–2081024. doi: 10.1103/PhysRevLett.95.208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vorselen D., MacKintosh F.C., et al. Wuite G.J.L. Competition between bending and internal pressure governs the mechanics of fluid nanovesicles. ACS Nano. 2017;11:2628–2636. doi: 10.1021/acsnano.6b07302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Real-time confocal fluorescence images of the Alexa Fluor 647-labeled VAMP2 in the GUV membrane after photobleaching.
Real-time confocal fluorescence images of the Alexa Fluor 647-labeled VAMP2 in the lipid bilayer coated on a silica bead 6 μm in diameter after photobleaching.