Graphical abstract
Keywords: Soluble epoxide hydrolases, Structure–function relationship, Tunnel network, Protein engineering
Abbreviations: sEHs, soluble epoxide hydrolases; msEH, Mus musculus soluble epoxide hydrolase; hsEH, Homo sapiens soluble epoxide hydrolase; StEH1, Solanum tuberosum soluble epoxide hydrolase; VrEH2, Vigna radiata soluble epoxide hydrolase; TrEH, Trichoderma reesei soluble epoxide hydrolase; bmEH, Bacillus megaterium soluble epoxide hydrolase; CH65-EH, soluble epoxide hydrolase from an unknown source, sampled in hot springs in China; Sibe-EH, soluble epoxide hydrolase from an unknown source, sampled in hot springs in Russia
Highlights
-
•
Lineage of a sEH determines its overall flexibility and tunnels architecture.
-
•
sEHs flexibility and tunnel network regulate substrate positioning.
-
•
Extensive tunnel network can be substantial for hydrolytic specificity.
-
•
sEHs predominantly utilize one tunnel or both of them.
-
•
Rearrangements in protein tunnel network can be caused by fragment insertions.
Abstract
Enzymes with buried active sites maintain their catalytic function via a single tunnel or tunnel network. In this study we analyzed the functionality of soluble epoxide hydrolases (sEHs) tunnel network, by comparing the overall enzyme structure with the tunnel’s shape and size. sEHs were divided into three groups based on their structure and the tunnel usage. The obtained results were compared with known substrate preferences of the studied enzymes, as well as reported in our other work evolutionary analyses data. The tunnel network architecture corresponded well with the evolutionary lineage of the source organism and large differences between enzymes were observed from long fragments insertions. This strategy can be used during protein re-engineering process for large changes introduction, whereas tunnel modification can be applied for fine-tuning of enzyme.
1. Introduction
Enzymes are proteins that facilitate catalytic reactions in their active site. In most known enzymes, the active site is buried inside their structure [1], and connected with the environment by tunnels. These tunnels enable and regulate not only substrate entrance and product release, but also ensure specific conditions within the active site for the reaction to occur. For example, cytochrome CYP3A4 structure is equipped with a tunnel called an aqueduct, which is used only for water molecules transport [2]. The aqueduct is regulated by R375 side chain, which is able to rotate and switch between different conformations, thus allowing a fine control of the presence of water molecules in the active site cavity [3]. Therefore, tunnels may have a regulatory mechanism, known as molecular gates that can form via one or more amino acids, often bulky or charged residues, able to rotate their side chain, hence, controlling access to the active site [4]. Despite controlling transport of substrates, products, and additional solvent molecules or even ions, gates in tunnels are also capable of controlling and synchronizing the reaction, or protecting the active site from poisoning [[5], [6]].
Since tunnels are involved in the functioning of enzymes with buried active sites, it may seem that they are evolutionary conserved structural features, which has been supported by reports [[7], [8], [9]]. It is known that the most conserved structural feature of an enzyme is its active site [10]. Additionally, the amino acids forming the protein core are also more conserved [[11], [12]] and tend to evolve slower than the surface residues, excluding those involved in protein–protein and/or protein–ligand interactions [[13], [14]]. However, in our other study [15] we elucidate that in the case of the soluble epoxide hydrolases (sEHs) most of their tunnels should be considered as variable structural features with only one exception - the tunnel located at the border between the main and cap domains. These counterintuitive findings has inspired the investigation of the structure–function relationship of sEHs in more detail.
Epoxide hydrolases (EHs) have been subjects of several structural and genome analyses. Heikinheimo et al. [16] provided four requirements, to distinguish epoxide hydrolases from other α/β-hydrolase fold members. A structure is an α/β-hydrolase fold member, when it fulfils at least two of them and maintains the sequence order of the catalytic triad. The four epoxide hydrolases features are as follows: i) the sequence order of the catalytic triad is nucleophile-acid-histidine, with the nucleophile on the canonical strand β5; ii) the “catalytic elbow” on top of the β5 strand with a sequence pattern that is often Gly-X-Nuc-X-Gly; iii) the structure starts from strand β3 and is at least five strands long, including the cross-over connection at the nucleophile (strands 43567); iv) a long loop at the end of β7 strand allows the side chains of the triad to form a hydrogen bond. Barth et al. [17] systematically compared known EHs based on their sequences, structures and biochemical properties. They identified three conserved and three variable regions mixed together within the protein’s sequence: i) the highly variable N-terminal region, which is absent in plant and most bacterial EHs, while in mammalian and insect microsomal EHs this region act as a membrane anchor; ii) the conserved first half of the α/β-hydrolase core domain; iii) the variable NC-loop, which starts directly after the β6 strand and ends before the first cap domain helix, linking the N-terminal part of the core domain with the cap domain; iv) the conserved mostly helical cap domain; v) a variable cap-loop inserted between helix α3 and α4 of the cap domain, and vi) the conserved C-terminal half of the core domain consisting of two β-strands and two α-helices. A comprehensive genome analysis by van Loo et al. [18] supports the work by Barth et al. [17]. They screened various genomic databases for EHs of the α/β-hydrolase family and divided them into 8 groups from a phylogenetic tree. Thus, identifying the following: i) sequences with proteobacterial origin and proteins with N-terminal signal peptides related to association with membranes (group 1); ii) sequences of bacterial, archaeal and eukaryotic origins, and even from multicellular organisms that have N-terminal extensions of unknown function (group 2); iii) sequence of mostly putative EHs from actinobacteria, β-proteobacteria and fungi (group 3); iv) sequence of both EHs and haloalkane dehalogenases (group 4); v) sequences of mammalian, bacterial and fungal microsomal EHs and the insect juvenile hormone EHs (group 5); vi) sequence of both fluoroacetate dehalogenases and EHs with the charge-relay aspartate located at the loop after β6 strand position (group 6); vii) sequences of EHs similar to those from group 6 with the first conserved ring-opening tyrosine and charge-relay aspartate located at different positions (group 7), and viii) sequence of a large number of known plant and mammalian EHs, including the mammalian sEHs (group 8). Unfortunately, structures of certain group members remain unknown.
Herein, we analyzed the available crystal structures of sEHs and performed a detailed analysis of their tunnel network. We focused on functional tunnels, i.e., tunnels in which we identified pathways of water molecules leading to/from the active site. We used water molecules as a molecular probe that enabled the investigation of the protein intramolecular voids and provided insights into the protein internal architecture. Thus, we were able to describe the structural basis of the tunnel network of sEHs. The available information on the sEHs substrate preferences were analyzed and combined with the data of the shape and size of their tunnel network. This study provides insight into the relationship between the structure of enzyme, usage of tunnels, and substrate preferences.
2. Materials and methods
2.1. Structure selection and preparation for analysis
Eight crystal structures of sEHs were downloaded from the Protein Data Bank (PDB) database [19]. The selected structure represent different clades of animals (Mus musculus (msEH, PDB ID: 1cqz [20]) and Homo sapiens (hsEH, PDB ID: 1s8o [21])), plants (Solanum tuberosum (StEH1, PDB ID: 2cjp [22]) and Vigna radiata (VrEH2, PDB ID: 5xm6 [23])), fungi (Trichoderma reesei (TrEH, PDB ID: 5uro [24])), and bacteria (Bacillus megaterium (bmEH, PDB ID: 4nzz [25])), as well as two thermophilic enzymes collected from hot springs in Russia (Sibe-EH, PDB ID: 5ng7 [26]) and China (CH65-EH, PDB ID: 5nfq [26]) from an unknown source. Incomplete structures with missing structural information regarding the position of amino acids were discarded (structures were collected in December 2019). Such structures may introduce bias into the results of the water molecules flow analysis. Additional ligands and ions were manually removed, as well as the N-terminal phosphatase domain of msEH and hsEH.
2.2. Multiple structure alignment
The selected sEHs structures, comprising only of the selected EH domains, were submitted to the mTM-align webserver [27]. The structural alignment was carried out using default parameters. The obtained alignment was viewed and processed by SeaView [28].
2.3. Molecular dynamics simulations
The H++ server [29] was used to protonate the analyzed structures using standard parameters at reported optimal pH for the enzyme activity (Supplementary Table 1). Additionally, counterions were added in the structures. Water molecules were placed using the combination of 3D-RISM theory [30] and Placevent algorithm [31]. Water molecules were added to fill the internal cavities and pockets of the proteins’ structures [32]. The Amber14 tLEaP package [33] was used to immerse the models in a truncated octahedral box with 10 Å radius of TIP3P water molecules and the ff14SB force field [34] was used for the parametrization of each system. PMEMD CUDA package of AMBER 14 software was used to run a set of 50 ns MD simulations of selected EHs. To improve conformation sampling, the starting geometry for each system was kept but the initial vectors were randomly assigned. The minimization procedure consisted of 2000 steps, involving 1000 steepest descent steps followed by 1000 steps of conjugate gradient energy minimization, with decreasing constraints on the protein backbone (500, 125, and 25 kcal × mol−1 × Å−2) and a final minimization with no constraints of conjugate gradient energy minimization. Next, gradual heating was performed from 0 K to 300 K over 20 ps using a Langevin thermostat with a collision frequency of 1.0 ps−1 in periodic boundary conditions with constant volume. Equilibration stage was conducted using the periodic boundary conditions with constant pressure for the time stated in Supplementary Table 1 with 1 fs time step using Langevin dynamics with a frequency collision of 1 ps−1 to maintain temperature. Production stage was conducted for 50 ns with a 2 fs time step using Langevin dynamics with a collision frequency of 1 ps−1 to maintain constant temperature. Long-range electrostatic interactions were modelled using the particle mesh Ewald method with a non-bonded cut-off of 10 Å and SHAKE algorithm. The coordinates were saved at 1 ps intervals. The number of added water molecules and ions is shown in Supplementary Table 1.
2.4. Water path analysis
AQUA-DUCT software [35] version 1.0 was used to trace paths of all water molecules that were found within a defined distance from the center of masses of atoms in the catalytic center (as listed in Supplementary Table 2). The Scope was defined as an interior of the convex hull of C-alpha atoms of the protein. Each water molecule path was cut to fit to the protein surface (auto_barber set to protein). All inlets were then clustered using the barber method with cutting sphere correction to the van der Waals radius of the closest atom (auto_barber_tovdw set to True).
3. Results
For the purpose of water molecules flow analysis we selected only crystal structures, which were unique and complete i.e. no information was missing about the position of a particular residue. In the case of repeated structures in PDB database, the apo structure and/or the one with the best resolution was selected. It was considered that incomplete or low resolution structures may introduce bias into the results. Eight sEHs structures were chosen that represent the clades of animals (Mus musculus (msEH) and Homo sapiens (hsEH)), plants (Solanum tuberosum (StEH1) and Vigna radiata (VrEH2)), fungi (Trichoderma reesei (TrEH)), and bacteria (Bacillus megaterium (bmEH)), as well as two thermophilic enzymes collected from hot springs in Russia (Sibe-EH) and China (CH65-EH) from an unknown source (structures were collected in December 2019). The obtained sEHs structures were compared using mTM-align web server [27] for multiple protein structure alignment (MSTA) analysis (Fig. 1). Then, the functional tunnels were identified using AQUA-DUCT [35] to compare the transport pathways. The functional tunnels were defined as those that display in which pathways of water molecules leading to/from the active site were identified. In this study we examined if using only crystal structures of the available sEHs and the information about the usage of tunnels, could the obtained results of such analyses be convergent with those of the evolutionary studies based on multiple sequences of EHs. Finally, we combined all data to investigate and evaluate the structural basis of the tunnel network of sEHs.
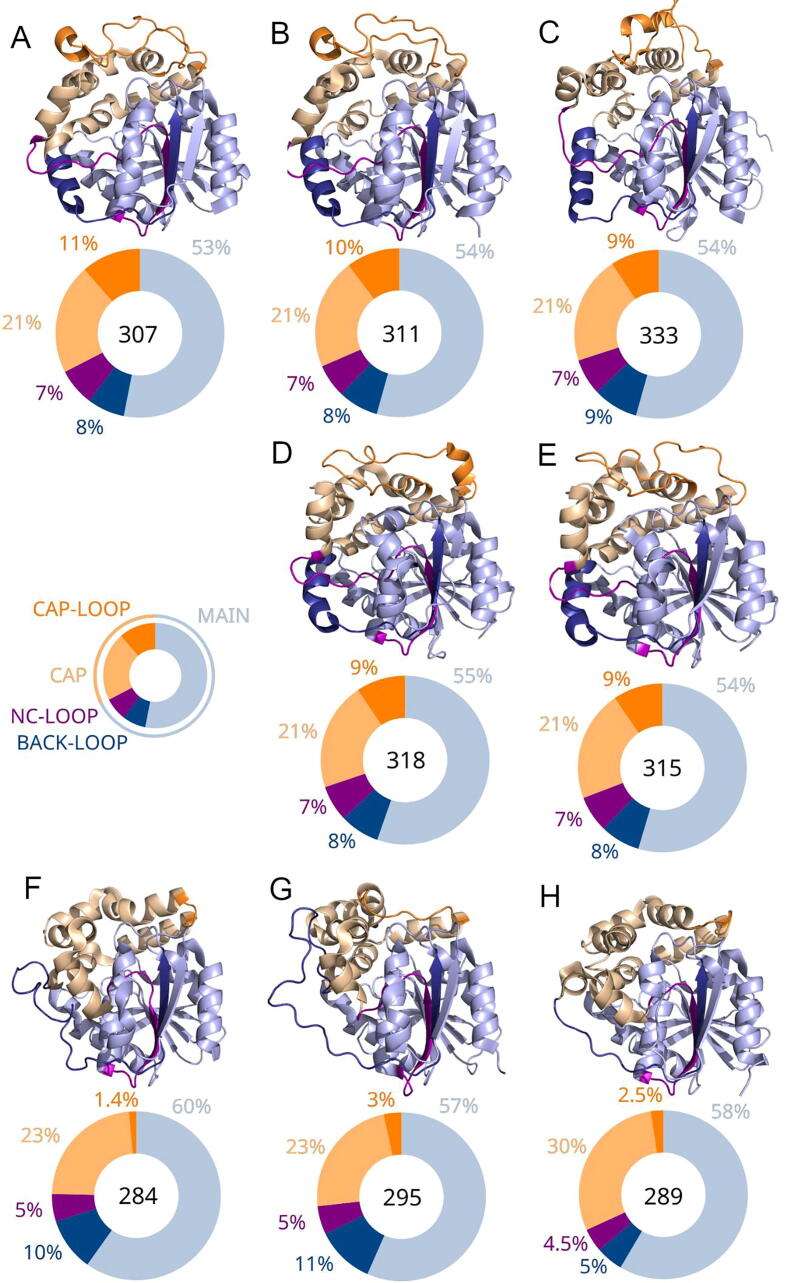
Fig. 1.
Crystal structures of selected soluble epoxide hydrolases (sEHs) and a pie chart representing the size of particular compartments. A) Mus musculus sEH (msEH), B) Homo sapiens sEH (hsEH), C) Trichoderma reesei sEH (TrEH), D) Solanum tuberosum sEH (StEH1), E) Vigna radiata sEH (VrEH2), F) Bacillus megaterium sEH (bmEH), and thermophilic G) CH65-EH, and H) Sibe-EH from an unknown organism. The data herein represents the data shown in Supplementary Table 3. The proteins are shown as cartoons. Pie charts under each protein structure show the share of particular compartments in the overall structure, while the number inside the pie chart stands for the number of amino acids comprising of the whole soluble epoxide hydrolase structure.
3.1. Structure comparison
To perform the structural comparison, we analyzed the structural features of sEHs using quantitative and qualitative descriptors (length, location/position). Nomenclature from the work of Barth et al. [17] was employed. The lengths of structural compartments were determined, such as the active site, cap and main domains, cap-loop and NC-loop, as well as some additional compartments (Supplementary Table 3, Fig. 1, and Fig. 2). All analyzed structures consisted of the main and cap domains, which were characteristic of the α/β-hydrolase fold. The additional N-terminal phosphatase domain - which is a known mammalian sEHs feature - was excluded from the structural analysis. The length of the analyzed structures varied from 284 (bmEH) to 333 amino acids (TrEH). The number of main domain amino acids varied from 197 (Sibe-EH) to 233 (TrEH), while the number of cap domain amino acids was correlated with the length of the cap-loop. The shortest cap domain and cap-loop were found in bmEH, with 70 and 4 amino acids, respectively, and the longest in msEH structure, with 100 and 35 amino acids, respectively. The length of NC-loop connecting the cap and main domains was similar in almost all analyzed structures (22 amino acids), except for the thermophilic enzymes (Sibe-EH with 13, and CH65-EH with 16 amino acids) and bmEH (15 amino acids). To provide a precise description of the structural differences between sEHs another loop connecting the cap domain with the main domain, which is referred to as the back-loop in this study, was distinguished. The back-loop was defined as a loop between the αD helix of the cap domain and β7 strand of the main domain together with β7 strand (dark blue on Fig. 1, Fig. 2). Length of the back-loop varied the most between two thermophilic sEHs: the back-loop of Sibe-EH consists of 15, while in the case of CH65-EH it consisted of 33 amino acids.
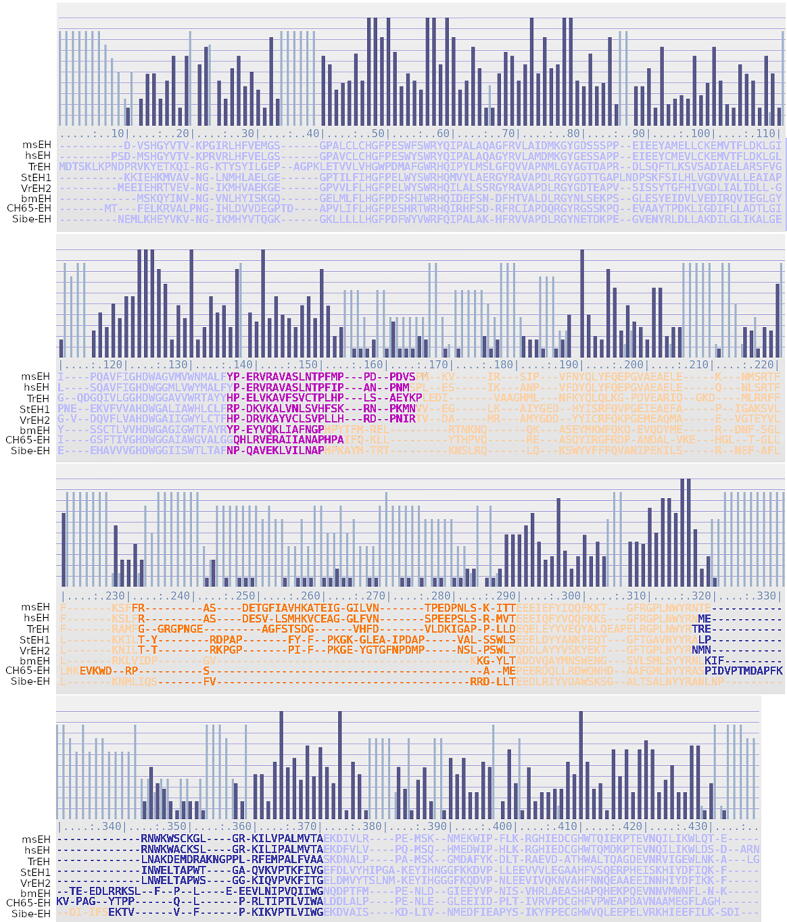
Fig. 2.
Multiple Protein Structures Alignment (MSTA) of selected soluble epoxide hydrolases (sEHs). The proteins’ sequences are color-coded (cap-loop – dark orange, cap – orange, NC-loop – violet, back-loop – dark blue, main domain – lilac). The dark blue bars indicate regions of higher structural similarity, while the light blue bars indicate regions of lower structural similarity. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
MSTA of selected sEHs structures provided by the mTM-align web server were analyzed (Fig. 2 and Supplementary Fig. 1). Thus, regions of higher structural similarity were identified as those that display greater differences. The regions of high sequence similarity corresponded mostly to the main domain. The part of the main domain that connects with the cap domain by the NC-loop showed high structural similarity. The NC-loop region also showed high similarity, however, it is clearly the NC-loop of the group IIb enzymes differed from the other analyzed sEHs structures. Hence, the cap domain region was less similar with two exceptions (alignment positions 190–206, and 288–320, Fig. 2) that corresponded to the α-helical regions between the NC-loop and cap-loop, and between the cap-loop and back-loop. These α-helices formed two layers of the cap domain (Fig. 2). The cap-loop and back-loop regions displayed high structural differences. Additionally, the main domain region connected with the cap domain by the back-loop displayed higher structural similarity.
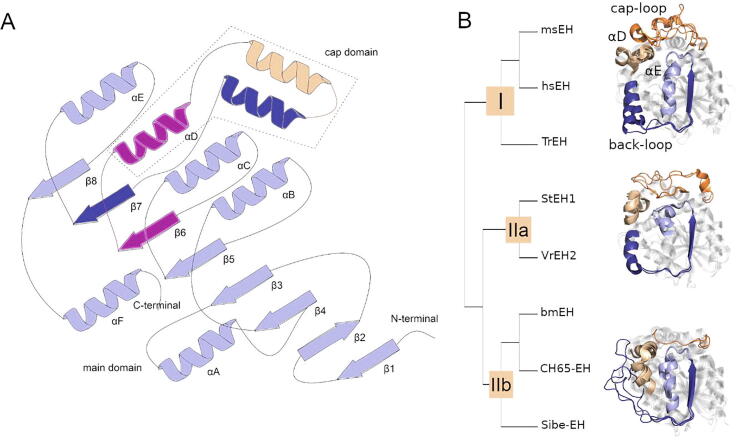
The β-barrel shape of the EHs main domain was observed in all analyzed structures, as well as the location of the cap domain. The active site was located in the buried cavity between the cap and main domains. In order to determine the structural factors that characterize for specific sEHs, the sequences and structures were aligned using mTM-align webserver. This enabled separation of structures into three groups (Fig. 3). Surprisingly, both mammalian sEHs were grouped with fungal TrEH (group I), while the second group consisted of plant StEH1 and VrEH2 (group IIa), and the third was bacterial bmEH and thermophilic EHs (group IIb). Structures were grouped together that displayed some common unique features. Enzymes from group I had relatively long cap-loops. The back-loop of TrEH was longer than that of msEH and hsEH, and slightly shifted away from the main domain. The α-helix located after the back-loop, αE helix, was parallel to the adjacent β-strands, β7, and β8 (secondary structure was derived from the work of Barth et al. [17], Supplementary Table 3 and Supplementary Table 4). Enzymes from group IIa had relatively long cap-loops which were positioned closer to the αD and αE helices compared to group I enzymes. Additionally, in contrast to group I enzymes, their αE helix rotated towards the αD helix region adjacent the NC-loop. Enzymes from group IIb had relatively short cap-loops, and the longest back-loops. Interestingly, the part of the back-loop closest to the cap domain was unfolded, whereas enzymes from groups I and IIa formed an α-helix. The αD helix was close to the αE helix. The αE helix had similar orientation as in group I enzymes’ structure. The main domain structure was similar in shape in all analyzed structures.
Fig. 3.
Structural similarity analysis of soluble epoxide hydrolases (sEHs). A) Schematic representation of sEHs structure. The nomenclature used was in accordance with [36]. B) Cladogram of analyzed sEHs structures. The proteins’ structures are shown as cartoons and for clarity only the most unique regions are shown.
3.2. Water molecules transport analysis
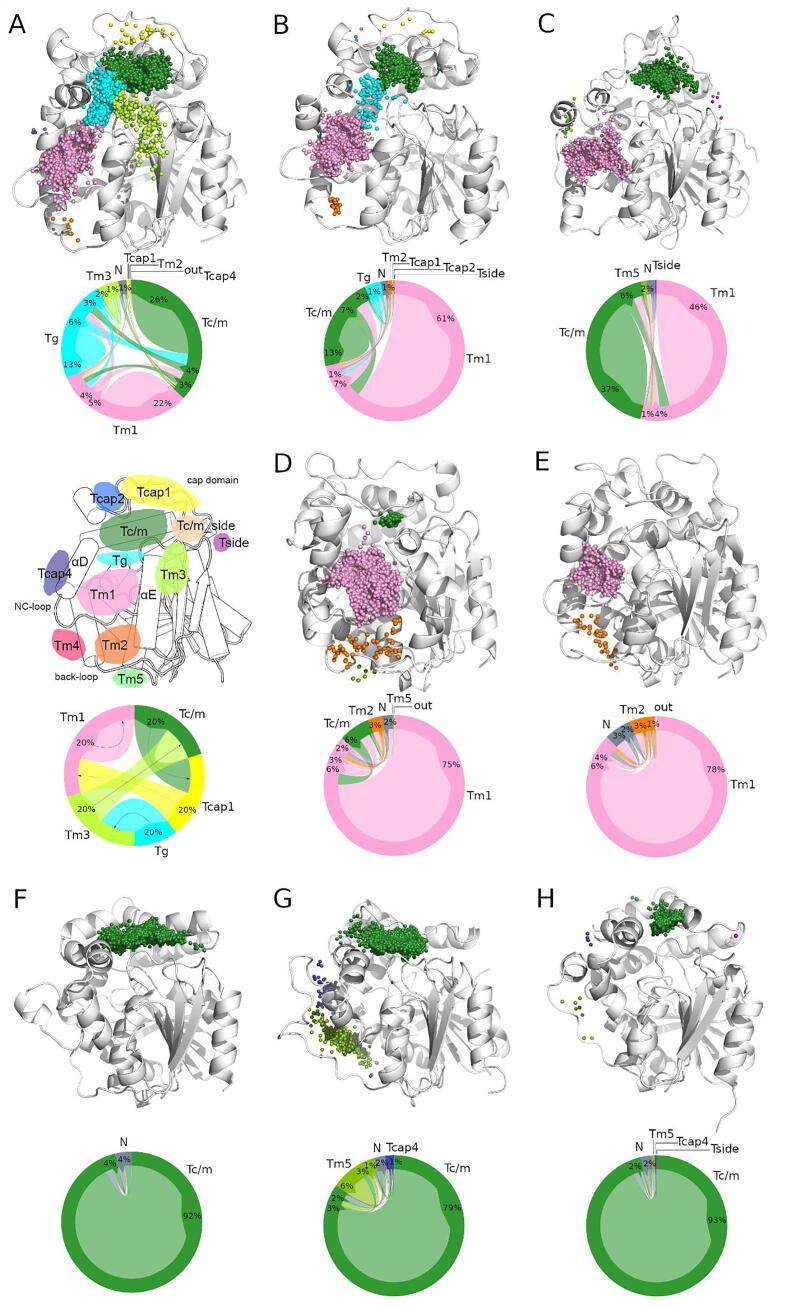
The main aim of our study was to describe the structural basis of sEHs tunnel network with focus on access to the active site and the implication of structural differences on the dynamics of the analyzed proteins. In order to conduct such analysis, five repetitions of molecular dynamics (MD) simulations were conducted (total of 250 ns per system) to mimic the conformational changes that occur in physiological conditions to a protein in solution. Then, the potential transport pathways were investigated, using water molecules as a molecular probe. During explicit solvent MD simulations, the protein was immersed in solvent (such as water) molecules that were able to penetrate the protein’s interior. Given the number of identified water molecules which entered the protein’s active site, provides the rate of exchange between the enzyme’s interior and environment. Analysis of these movements provided detailed information on the tunnel network as well as their usage, while maintaining the required simulation time at a relatively low level. In our previous report, we showed that 50 ns of a Solanum tuberosum sEHs was enough to sample rare events of water molecules entering a particular tunnel [37]. The water molecules transport analysis was facilitated by the AQUA-DUCT software and was used exclusively to trace pathways of water molecules that entered the active site of analyzed EHs. The utilization of such workflow allowed the observation of changes to the protein’s structure, which, for example, opens or closes a particular pathway leading to the active site. The obtained results gave insight into such pathways in three regions of sEHs structure - the main domain, cap domain, and border between these domains. In order to further clarify these results, the observed pathways were associated with tunnels. Therefore, the identified tunnels leading to/from the active site were marked, according to their localization; Tm - main domain, Tcap - cap domain, and Tc/m - border between those domains (Fig. 4). The regions in which water molecules entered and/or left the protein interior are shown as small balls (so-called inlets) in Fig. 4. The inlets were then clustered to represent tunnels entries reported elsewhere [15]. It should be noted that not all tunnels were represented in other structures.
Fig. 4.
Identified entries/exits of selected epoxide hydrolases (EHs) and the intramolecular flow plot. The intramolecular flow plot (also known as the migration flow plot) depicts the flow of water molecules through particular tunnels. The outer ring represents the size of the tunnel, while the size of the inner part of the plot (called here flow) represents a particular transport pathway by direction (shown in the legend by small arrows). In the sample plot five flows are shown: Tc/m to Tcap1, Tcap1 to Tm1, Tg to Tm3, Tm3 to Tc/m, Tm3 to Tc/m, and Tm1 to Tm1. The ‘out’ flow stands for the pathways that do not belong to specific clusters, while ‘N’ flow stands for the pathways that started and/or ended within the protein structure. The figure represents data presented in Supplementary Table 6. A) Mus musculus EH (msEH), B) Homo sapiens EH (hsEH), C) Trichoderma reesei EH (TrEH), D) Solanum tuberosum EH (StEH1), E) Vigna radiata EH (VrEH2), F) Bacillus megaterium EH (bmEH), and thermophilic G) CH65-EH, and H) Sibe-EH from an unknown organism. The proteins are shown as cartoons, and the entries/exits are marked as small balls (so-called inlets). For picture clarity only the epoxide hydrolase domain of msEH and hsEH structures are shown.
Comparison of all analyzed structures revealed only two tunnels that were predominantly utilized by water molecules (above 30% of all identified inlets, Supplementary Table 5): Tc/m tunnel located at the border between the main and cap domain, and Tm1 tunnel located in the main domain. Moreover, we identified other tunnels, such as Tm2, Tm3, Tm4, Tm5, Tg, and Tside in the main domain, and another tunnel located between these two domains, namely Tc/m_side, and Tcap1, Tcap2, and Tcap4 in the cap domain; those tunnels were, however, rarely used by water molecules. Information regarding the predominant tunnel allowed the determination of three different patterns of tunnel usage of sEHs: i) both Tc/m and Tm1 tunnels are predominantly used (msEH, hsEH, and TrEH); ii) only Tm1 tunnel was predominantly used (StEH1, and VrEH2), and iii) only Tc/m tunnel was predominantly used (bmEH, CH65-EH, and Sibe-EH) (Supplementary Table 5, Fig. 4). Interestingly, the observed patterns corresponded to structural analysis described above. In mammalian and fungal sEHs two tunnels were predominantly used by water molecules - Tm1 and Tc/m. In contrast to TrEH, mammalian sEHs additionally utilized Tg and Tm3 tunnels, as well as Tcap1. In the case of plant sEHs, Tm1 tunnel was utilized by 92% of water molecules entering the active sites. Additionally, plant EHs employed several other tunnels, such as Tc/m, Tm2, and Tm5. Finally, in both thermophilic enzymes, CH65-EH, and Sibe-EH, Tc/m tunnel was used by the vast majority (78% and 98%, respectively) of water molecules entering the active site cavity. Both enzymes also used additional tunnels located in the main domain and the border between the main and cap domains. Similarly, bmEH utilizes mostly Tc/m tunnel; however, in a single MD simulation several water molecules employed other tunnels. VrEH2 enzyme was the only sEH unable to utilize Tc/m tunnel, instead the Tc/m_side tunnel was used. The number of inlets per simulation nanosecond was examined to determine the water molecules’ flux. bmEH, which utilizes only one tunnel, was found as the most ‘open’ structure (96 inlets/ns), while VrEH2 was the most ‘closed’ structure (only 7 inlets/ns). The inlets/ns values were similar for hsEH, msEH, StEH1, and TrEH (40–45 inlets/ns) (Supplementary Table 5). Therefore, the number of functional tunnels does not reflect the water molecules flow through the enzyme’s active site.
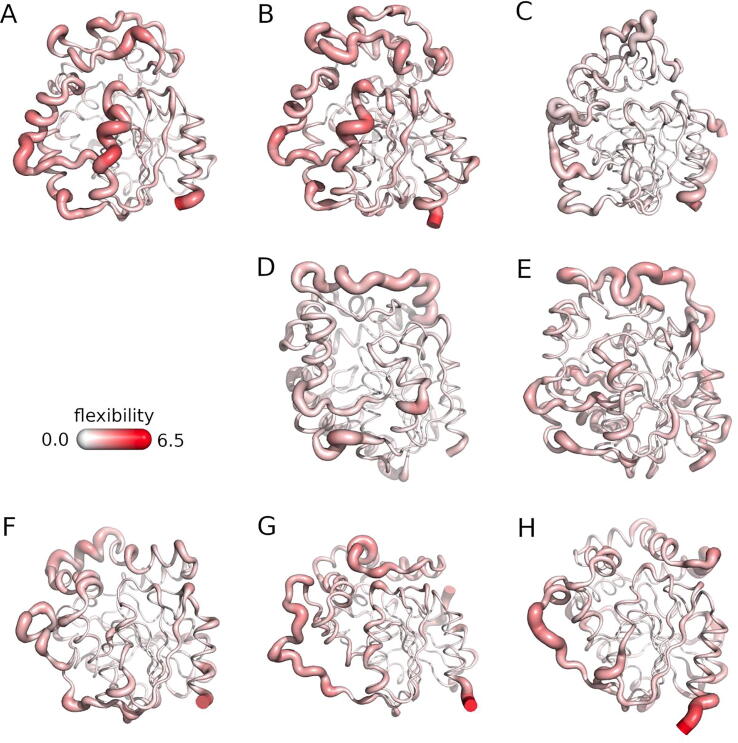
To complement the small-molecules transport analysis, we investigated the most flexible regions of the selected sEHs (according to RMSF data from MD simulations). The obtained results showed that both mammalian sEHs most flexible regions were αD and αE helices, and the cap-loop region, whereas TrEH only the αD helix and part of the cap-loop were identified (Fig. 5). Moreover, the accumulated movements of these regions were weaker than in case of mammalian sEHs. Similarly, the cap-loop and αD helix were the most flexible regions in plant sEHs, with little movement observed in the αE helix. Finally, in the case of bacterial and thermophilic sEHs, the most flexible regions were the back-loop, cap-loop, and part of the αE helix. Mammalian and thermophilic sEHs showed the greatest overall flexibility.
Fig. 5.
The overall flexibility of the selected soluble epoxide hydrolases. A) Mus musculus EH (msEH), B) Homo sapiens EH (hsEH), C) Trichoderma reesei EH (TrEH), D) Solanum tuberosum EH (StEH1), E) Vigna radiata EH (VrEH2), F) Bacillus megaterium EH (bmEH), and thermophilic G) CH65-EH, and H) Sibe-EH from an unknown source organism. The proteins are shown as springs, with the thin white lines indicating lower flexibility, and thicker reddish lines - higher flexibility.
4. Discussion
To date, little is known about EHs, however, several studies have been conducted to investigate their structural features [[15], [16], [17], [18]]. sEHs belong to the α/β-hydrolases family that display a modular structure with a central catalytic domain, the main domain, formed by eight superhelically twisted β-strands [[17], [36], [38]]. This superfamily can tolerate large insertions to the scaffold without losing their catalytic activity [39]. The most important modification of the fold is the insertion after the β6 strand, which forms the cap domain. This domain has great impact on substrate recognition and catalysis [[40], [41], [42]]. The cap-loop covers the active site, and thus, limits the pathways of substrate and products transport to a specific tunnel. Tunnel locations can, therefore, be constructed as a natural consequence of the active site positioning between both domains. Tunnels have been identified passing through the main and cap domains as well as the interdomain space. The cap domain is connected with the main domain by two flexible loops acting as hinges, namely NC-loop and back-loop. The NC-loop is considered to participate in substrate binding by defining the binding pocket and regulating the access to the active site [43]. Therefore, the tunnel network of sEHs may be regulated through a set of structural features: i) intramolecular voids in the main domain, ii) intramolecular voids in the cap domain, iii) hinge loops connecting both domains. Since the NC- and back-loops act as hinges they can regulate the tunnel network either by positioning the cap domain on the main domain, while affecting the entrances/exits of neighboring tunnels. In this study we confirmed the findings of Barth et al. and Bauer et al. regarding the modular structure of EHs [[17], [38]]. Analysis of the MSTA suggested that sEHs consisted of several modules (compartments), including the main and cap domains, as well as the NC-loop, cap-loop, and back-loop. Moreover, it was found that the main domain regions and mostly helical region of the cap domain displayed a high level of structural similarity, whereas the NC-loop, cap-loop, and the back-loop regions display dissimilarity (Fig. 2). These findings also suggest that the most dissimilar regions are more prone to modifications.
In our other study [15], sEHs were employed as a sample system in order to investigate the evolution of tunnels. It was determined that most tunnels should be considered as variable structural features of proteins. Tc/m tunnel was found to be the only exception, located between the cap and main domains. We proposed that insertion of the cap domain defined the buried active site cavity and the tunnel linking it with the environment. Such structural arrangement was preserved in most of the EHs which supports the hypothesis regarding the origin of the positioning of the active site between both domains. However, according to other reports [[15], [21], [37], [44], [45]], this was not the only pathway leading to the active site. Other tunnels were located in the cap and main domains, as well as between those domains. It should be noted that when predominant tunnels were used for substrate and/or product transport, the rarely used tunnels should not be neglected because they could be used, for example, for water molecules transport during the hydrolysis step.
In this study, we focused on the functionality of the sEHs tunnel network. Functional tunnels were defined as those which were used by water molecules to reach the active site cavity. A relationship was found between the protein structure and the shape and size of its tunnel network. Hence, despite overall structural similarity, the sEHs structures were divided into three groups, based on their structure and tunnel usage (Fig. 3, Fig. 4). The results of the structural compartments and tunnels usage analyses suggested a close evolutionary relationship of the proteins, which were consider unprecedented. Notably, the obtained results were based on a relatively low number of structures that represented different clades. Nonetheless, the sampling of the EHs family is only fragmentary. However, our results were in good agreement with that of Barth et al. [17] based not only on tunnel usage, but on the analysis of multiple sequences of EHs. Moreover, the presented data were in-line with the theory that animals and fungi were more closely related than animals and plants [46].
Mammalian (hsEH and msEH) and fungal (TrEH) structures were assigned to group I. Members of this group shared common features such as relatively long back-loop and cap-loop. Enzymes in this group primarily utilize two main tunnels - Tc/m, and Tm1. In all sEHs from the group I, T/cm tunnel was found conserved [15]. This was also the case for Tm1 tunnel, but only in the case of msEH [15]. The results of the structure flexibility analysis (Fig. 5) showed significant differences between sEHs that represent mammalian and fungal families. Mammalian sEHs were more flexible, and the regions with high RMSF values surround Tc/m, Tg, and Tm1 tunnels’ entries/exits regions. Furthermore, during MD simulations we observed that those regions merged and created a long gorge. In contrast, these regions were quite rigid for TrEH structure, and consequently both tunnels clearly separated. Such differences had substantial implications on the substrate preferences, which will be discussed below.
Plant sEHs (StEH1 and VrEH2) were assigned to group IIa. The interaction between the cap and main domains was much tighter, thus Tc/m tunnel was narrower relative to other analyzed sEHs. A subtle rearrangement of the αD helix region adjacent to the NC-loop caused narrowing of the Tc/m tunnel’s mouth and dramatically limited the tunnel usage (StEH1) or closed it permanently (VrEH2). Moreover, access to the active site through the cap domain was also nearly completely blocked. Similar to mammalian and fungal sEHs, plant sEHs structures had relatively long cap-loop and back-loop, however, the enzymes predominantly utilize the Tm1 tunnel, which was identified as a variable feature in StEH1 structure [15]. The flexibility analysis results of plant sEHs showed that the most flexible regions were distant to the tunnel entries and, therefore, the conformational changes were only limited to slight effect on for catalytic efficiency (if any).
Finally, bacterial (bmEH), and thermophilic enzymes from an unknown organism (CH65-EH and Sibe-EH) were assigned to group IIb. Members of this group had relatively short cap-loops and longest back-loops. They mainly utilized the Tc/m tunnel. Their αD helix was close to the αE helix, which caused narrowing of the tunnel mouths located in the main domain, namely Tm1, Tg, and Tm2. As a result the location of other tunnels on the other side of the back-loop, namely Tcap4 and Tm5, they could be opened. However, since the vast majority of water molecules were transported via the Tc/m tunnel, the role of the other tunnels for substrates/products transportation can be neglected. This observation supported the hypothetical origin of sEHs via insertion resulting in active site positioning between cap and main domains. Surprisingly, in the case of IIb group enzymes the Tc/m tunnel was found to be a variable feature [15]. This could be due a small number of residues lining the walls of the tunnel, which was significantly shorter in comparison to Tc/m tunnels in other sEHs. It was shown that the cap-loop, back-loop, and part of the αE helix were the most flexible regions. The results suggested that the movement of the back-loop may cause opening of the neighboring tunnels, such as Tcap4 and Tm5, which were commonly used by the enzymes.
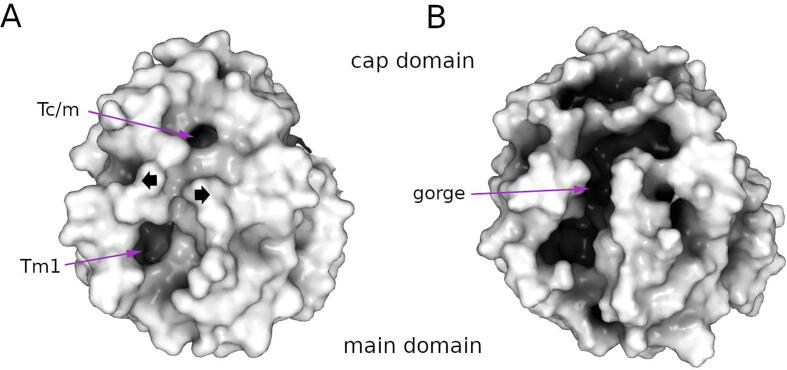
Barth et al. [17] found a correlation between the length of the NC-loop and cap-loop and the type of catalyzed substrates and evolutionary lineage of the source organism. Their results indicated that sEHs of eukaryotes had long cap-loops and medium-sized NC-loops, as well as being more active towards aliphatic epoxides, while participating in fatty acid metabolism. It must be noted that the function of an enzyme and often its name reflects substrate preference based on a very limited data set. Therefore, the functional names given to enzymes early in their investigation can bias a whole field. Additionally, the overall knowledge on the functions and applications of EHs in humans and other organisms is also limited. Known compounds synthesized by and/or tested on all analyzed sEHs are shown at Supplementary Figs. 2-8. Mammalian EHs are involved in the xenobiotic metabolism and the degradation of endogenously derived epoxy fatty acids [[47], [48]], as well as hydrolysis of trans-epoxy alcohol 1 (skin-related allylic epoxide) to RSR triol-3, which is the most abundant triol isomer in human and porcine epidermis [49]. Plant EHs are involved in the biosynthesis of essential aliphatic cuticular compounds [50], detoxification of epoxy fatty acids in seeds [51], and conversion of the epoxides that accumulate during stress into less reactive compounds [52]. EHs in plants are also involved in the defence system, where their activity can be enhanced by water deprivation, wounding or during virus infection [[53], [54], [55]]. For example, NtEH-1 gene encoding an EHs product of the Nicotiana tabacum L. is induced in the presence of the tobacco mosaic virus (TMV) [[54], [56]]. Also, not all plants metabolize the epoxy fatty acids in their seeds. Large amounts of fatty acids in the form of triacylglycerols are used as sources of energy and biosynthetic intermediates [57]. Epoxy fatty acids are common storage lipids in seeds of certain species, such as Astraceae which store about 70% of their lipids in this form [[57], [58]]. This may be the origin of the observed structural rearrangement discussed previously allowing mammalian EHs to transform long-chained epoxy fatty acids. Summerer et al. [59] highlighted a substantial difference in the catalysis of 9,10-epoxystearic acid between mammalian (from rat liver) and plant (from soybean) sEHs. They found that although the reaction catalyzed by the plant sEH was highly enantioselective towards (R)-configured carbon, the mammalian sEH catalysis involved non-enantioselective hydrolysis. Hence, the different binding mode may be related to the structural features which enable positioning of the epoxide ring. This conclusion was in agreement with that of Pineau et al. [60] in which different inhibition patterns were observed between the plant (Arabidopsis thaliana) and mammalian EHs. The presented data showed the observed differences may stem from different substrate preferences between these enzymes, and combined with the work of Mowbray et al. [22], showed that Solanum tuberosum EH may be very efficient in metabolizing substrates with aliphatic substituents of the epoxide ring. The presented tunnel network analysis sheds light on its potential mechanism. The sEHs flexibility analysis suggested a plausible mechanism of substrate/product transport, instead of the cap domain movement, which could facilitate large substrate access, or highly improbable passage of long-chained substrate entering through one tunnel and leaving by another. Additionally, the secondary structure elements surrounding the two main tunnels could move away, merging Tc/m, Tm1 and Tg tunnels into one long gorge (Fig. 6), which could encompass even long-chained substrates. In the case of F497 residue in hsEH, which is located between Tc/m and Tm1 tunnels, two different orientations in the crystal structure were detected [44]. Due to phenylalanine side chain bulky character, it operates as a molecular gate controlling access through the gorge and promoting proper positioning of the epoxide ring, or closing the tunnel to create a hydrophobic environment for the reaction to occur. This gate may provide constraints affecting a particular substrate preferences. Indeed, the mammalian sEHs have not been used in industry due to their limitations of accepting other epoxide substrates and difficulties in engineering their regiospecificity [61].
Fig. 6.
Opening of the gorge in mouse soluble epoxide hydrolase (msEH): A) the entrance to the active site in the crystal structure (PDB ID: 1cqz), and B) during molecular dynamic simulation. The protein structure is shown as white surface.
Furthermore, plant sEHs predominantly utilize only one funnel-shape tunnel. In our case, the size of the tunnel’s mouth and its funnel shape facilitated substrate access to the active site. Thus, in contrast to the long expandable tunnel of mammalian sEHs (which were created by merging of Tc/m, Tm1, and Tg tunnels), Tm1 remained open for a wide range of substrates without causing steric hindrances. Additionally, Tm1 tunnel was capable of transporting the substrates and product, whereas the side tunnels transported water molecules. Therefore, the active site cavity and its surroundings were easily modified and such enzymes were often used in industry. Several reports on StEH1 highlight the high potential of plant EHs as regioselective catalysts [[62], [63], [64], [65]]. Notably, plant EHs are usually highly regio- and/or stereoselective [[22], [66], [67], [68]] towards specific substrates. This issue was carefully analyzed both experimentally and theoretically in StEH1, VrEH2 and Phaseolus vulgaris PvEH3 [[23], [69], [70], [71], [72], [73]]. Reports have shown that modification of the stereo- and regioselectivity of the enzymes is related to the direction of the attack of water molecules and stabilization of particular transition states [72]. Moreover, modifications of the NC-loop, located near the entrance to the Tm1 tunnel, may also enhance the enantioselectivity of PvEH3 [73]. The aforementioned properties can be easily modified in plant EHs with a funnel-shaped entrance to the active site pocket than in mammalian EHs with occluded active site.
Bacterial sEHs have the shortest cap- and NC-loops among all analyzed EHs, which may be related to several bacterial EHs that accept small substrates such as styrene oxide, and mono- and disubstituted epoxides [17]. The main tunnel identified in bacterial structures is short and well-defined, which prevents conversion of long-chained substrates. The short and well-defined tunnel makes such enzymes an easy system for future modification and applicability in industrial processes.
As previously mentioned, a relationship was observed between the enzyme’s structure and its overall flexibility and substrate preferences. Due to the limited number of analyzed structures it was hypothesizes that in the case of sEHs, divergent evolution occurred. All analyzed enzymes belonged to the same α/β-hydrolase superfamily and shared the same fold, however, they present different substrate preferences profiles, which implied the presence of a common ancestor. Indeed, in the case of other enzymes related to the same superfamily the ancestral protein was identified [74], and displayed enhanced thermal stability and higher specific activity than the extant enzymes. Therefore, bacteria, the most primitive group of living organisms, would present the least complicated mechanism. Due to EHs being required to transport both the substrate and water molecule to the active site cavity, a multi-purpose tunnel or a tunnel network may be needed, in which the substrate could be transported by one tunnel, and the water molecule by another. Our results suggested that bmEH utilized only one tunnel to maintain transport of these two reagents, as well as the reaction product. The Tc/m tunnel was identified in almost all analyzed sEHs (excluding VrEH2), and was considered to be evolutionarily preserved due to its location within the intramolecular voids between the main and cap domains. Other analyzed sEHs utilized predominantly at least one tunnel (Tc/m, Tm1, or both), whereas additional tunnels were rarely used. This may suggest that other sEHs - mammalian, plant and fungal - were more specialized relative to the bacterial bmEH, as they functioned through separate transport water molecules from the transport of substrate and/or products. In our study sEHs of multicellular organisms were divided into two groups - mammalian and fungal (group I, Fig. 3), and plant (group IIa, Fig. 3). Structures of mammalian and fungal sEHs although seemed similar, they display a different pattern of overall flexibility. Two mammalian sEHs displayed high flexibility of the cap domain, as well as the αD and αE helices, and in TrEH only the cap-loop and αD helix were slightly flexible. Therefore, the substrates and/or products in the case of mammalian sEHs could possibly be transported via the widest and always open tunnel, such as the large gorge formed by merging of Tc/m, Tm1, and Tg tunnels. In the case of TrEH, two separated tunnel entries were observed (Tc/m and Tm1), therefore, it could suggest that the substrate entry and product release occurred via different tunnels. Water molecules may enter the active site cavity by a side tunnel. In the case of another α/β-hydrolase superfamily member, dehalogenases, a strategy of separating the substrate entry and product release pathways resulted in the most active dehalogenase identified to date [75]. Moreover, mammalian sEHs are bifunctional enzymes, a product of gene fusion event [[76], [77]] with an N-terminal domain exhibiting phosphatase activity, and the C-terminal domain being an actual EH [78]. Additionally, sEHs in plants and other multicellular organisms evolve independently. We speculated that insertion resulting in cap domain formation was the starting point of the specialization of the EHs. The cap domain covered the active site pocket, and thus enabled precise control of the conditions of the enzymatic reaction. In mammalian and fungal enzymes, the ancestral Tc/m tunnel was preserved and insertions resulted in higher flexibility of the enzyme structure and more complex tunnel network, whereas in plants, the inserts had closed (partially or fully) origin Tc/m tunnel and a new predominant tunnel located in main domain overcharge the substrate/products transportation. Furthermore, the sequences of the plant EHs were divided into two clades – EH1 and EH2 [56]. Moreover, it was shown that the main differences between those clades were located in the cap domain [79]. Among all examined sEHs, mammalian sEHs were subjected to the most precise and rigorous control. The most complicated tunnel network was observed, as well as extensive water exchange between all potential pathways and long range conformational changes capable of merging or separating the particular tunnels.
Our study also explored expansive strategies employed for protein re-engineering. Several approaches have been previously proposed to fine-tune enzyme’s activity and/or selectivity through the introduction of additional tunnel or modification of an existing one. Reetz and Kotik’s groups focused on Aspergillus niger EH existing tunnel leading to the active site and targeted the tunnel-lining and adjacent residues for mutagenesis [[80], [81], [82], [83], [84], [85]]. Thus, they obtained highly enantioselective variants which were useful for producing enantiopure terminal epoxides containing various side chains. A more advanced approach was shown by Kong et al. who introduced an additional tunnel in bmEH using targeted mutagenesis to unblock the steric hindrance in the active pocket [25]. This resulted in the formation of an EH with unusual (R)-enantioselectivity and much higher activity toward α-naphthyl glycidyl ether. Brezovsky et al. furthered this work by engineering a de novo tunnel in a haloalkane dehalogenase LinB, an enzyme closely related to EHs and shared very similar structural features [75]. They opened a novel tunnel by modifications of three residues W140A/F143L/I211L resulting in surface perforation and tunnel opening. The successful tunnel engineering strategies also supported the surface perforation model proposed in our other study on sEHs [15], which described the evolution of tunnels. The presented model suggested that tunnels appeared through even a single-point mutation promoting the formation of two adjacent cavities or permanently opening of an existing cavity. Our study suggested that the tunnel network was also vulnerable to more dramatic modifications such as large fragment indels (insertions/deletions), as depicted the cap domain formation. The MSTA data suggested that the longer region of the cap-loop identified in sEHs from group I and IIa stem from insertion, as well as the unfolded region of the back-loop of sEHs from group IIb. A strategy based on longer fragment insertion may lead to more complex modifications of the enzyme’s activity and/or selectivity, however, such results will be difficult to predict, while the previously described approach based on cavity perforation and tunnel modification could be used for enzyme fine-tuning.
5. Conclusions
This paper is an extension of our other work, in which the evolution of tunnels is studied. We found that tunnels are mostly variable structural features of proteins and a surface perforation model was proposed to describe the mechanism of tunnel appearance. Additionally, interconnection of the protein structure, shape and size of its tunnel network and the substrate preferences are explored. Moreover, our results suggest that tunnels may appear not only due to a single-point mutation, but also by more dramatic structural modifications such as large fragments indels. Finally, sEHs were divided into three groups based on their structure, usage of tunnels, and substrate preferences, indicating that these features are mutually connected.
Funding
This work was funded by the National Science Centre, Poland, grant no DEC-2013/10/E/NZ1/00649. Publication supported by the Own Scholarship Fund of the Silesian University of Technology in the year 2019/2020 (Grant No 919/RN2/RR4/2019).
CRediT authorship contribution statement
Karolina Mitusińska: Conceptualization, Data curation, Funding acquisition, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Piotr Wojsa: Data curation, Funding acquisition, Investigation, Methodology, Software, Validation, Visualization. Maria Bzówka: Data curation, Funding acquisition. Agata Raczyńska: Data curation, Funding acquisition. Weronika Bagrowska: Data curation, Funding acquisition, Visualization. Aleksandra Samol: Data curation, Funding acquisition. Patryk Kapica: Data curation. Artur Góra: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.10.042.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Pravda L., Berka K., Svobodová Vařeková R., Sehnal D., Banáš P., Laskowski R.A., et al. Anatomy of enzyme channels. BMC Bioinf. 2014;15(1) doi: 10.1186/s12859-014-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mancini G., Zazza C., Soares C.M. F429 regulation of tunnels in cytochrome P450 2B4: a top down study of multiple molecular dynamics simulations. PLoS ONE. 2015;10(9) doi: 10.1371/journal.pone.0137075. e0137075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fishelovitch D., Shaik S., Wolfson H.J., Nussinov R. How does the reductase help to regulate the catalytic cycle of cytochrome P450 3A4 using the conserved water channel? J Phys Chem B. 2010;114(17):5964–5970. doi: 10.1021/jp101894k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gora A., Brezovsky J., Damborsky J. Gates of enzymes. Chem Rev. 2013;113(8):5871–5923. doi: 10.1021/cr300384w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marques S.M., Daniel L., Buryska T., Prokop Z., Brezovsky J., Damborsky J. Enzyme tunnels and gates as relevant targets in drug design. Med Res Rev. 2017;37(5):1095–1139. doi: 10.1002/med.21430. [DOI] [PubMed] [Google Scholar]
- 6.Kingsley L.J., Lill M.A. Substrate tunnels in enzymes: Structure-function relationships and computational methodology. Proteins Struct Funct Bioinforma. 2015;83(4):599–611. doi: 10.1002/prot.24772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J., Raushel F.M. Perforation of the tunnel wall in carbamoyl phosphate synthetase derails the passage of ammonia between sequential active sites. Biochemistry. 2004;43:5334–5340. doi: 10.1021/bi049945+. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura A., Yao M., Chimnaronk S., Sakai N., Tanaka I. Ammonia channel couples glutaminase with transamidase reactions in GatCAB. Science (80-) 2006;312(5782):1954–1958. doi: 10.1126/science.1127156. [DOI] [PubMed] [Google Scholar]
- 9.Thangapandian S., John S., Lee Y., Arulalapperumal V., Lee K.W., Gaetano C. Molecular modeling study on tunnel behavior in different histone deacetylase isoforms. PLoS ONE. 2012;7(11) doi: 10.1371/journal.pone.0049327. e49327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franzosa E.A., Xia Y. Structural determinants of protein evolution are context-sensitive at the residue level. Mol Biol Evol. 2009;26(10):2387–2395. doi: 10.1093/molbev/msp146. [DOI] [PubMed] [Google Scholar]
- 11.Tseng Y.Y., Liang J. Estimation of amino acid residue substitution rates at local spatial regions and application in protein function inference: a Bayesian Monte Carlo approach. Mol Biol Evol. 2006;23:421–436. doi: 10.1093/molbev/msj048. [DOI] [PubMed] [Google Scholar]
- 12.Ramsey D.C., Scherrer M.P., Zhou T., Wilke C.O. The relationship between relative solvent accessibility and evolutionary rate in protein evolution. Genetics. 2011;188:479–488. doi: 10.1534/genetics.111.128025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J.-R., Liao B.-Y., Zhuang S.-M., Zhang J. Protein misinteraction avoidance causes highly expressed proteins to evolve slowly. Proc Natl Acad Sci. 2012;109(14):E831–E840. doi: 10.1073/pnas.1117408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Echave J., Spielman S.J., Wilke C.O. Causes of evolutionary rate variation among protein sites. Nat Rev Genet. 2016;17(2):109–121. doi: 10.1038/nrg.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bzówka M., Mitusińska K., Raczyńska A., Skalski T., Samol A., Bagrowska W., et al. Evolution of tunnels in α/β-hydrolases fold proteins – what can we learn from studying epoxide hydrolases? bioRxiv. 2021 doi: 10.1101/2021.12.08.471815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heikinheimo P., Goldman A., Jeffries C.y., Ollis D.L. Of barn owls and bankers: a lush variety of α/β hydrolases. Structure. 1999;7(6):R141–R146. doi: 10.1016/S0969-2126(99)80079-3. [DOI] [PubMed] [Google Scholar]
- 17.Barth S., Fischer M., Schmid R.D., Pleiss J. Sequence and structure of epoxide hydrolases: a systematic analysis. Proteins Struct Funct Bioinforma. 2004;55(4):846–855. doi: 10.1002/prot.20013. [DOI] [PubMed] [Google Scholar]
- 18.van Loo B., Kingma J., Arand M., Wubbolts M.G., Janssen D.B. Diversity and biocatalytic potential of epoxide hydrolases identified by genome analysis. Appl Environ Microbiol. 2006;72(4):2905–2917. doi: 10.1128/AEM.72.4.2905-2917.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berman H.M., Battistuz T., Bhat T.N., Bluhm W.F., Bourne P.E., Burkhardt K., et al. The protein data bank. Acta Crystallogr D Biol Crystallogr. 2002;58(6):899–907. doi: 10.1107/S0907444902003451. [DOI] [PubMed] [Google Scholar]
- 20.Argiriadi M.A., Morisseau C., Hammock B.D., Christianson D.W. Detoxification of environmental mutagens and carcinogens: Structure, mechanism, and evolution of liver epoxide hydrolase. Proc Natl Acad Sci. 1999;96(19):10637–10642. doi: 10.1073/pnas.96.19.10637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez G.A., Morisseau C., Hammock B.D., Christianson D.W. Structure of human epoxide hydrolase reveals mechanistic inferences on bifunctional catalysis in epoxide and phosphate ester hydrolysis. Biochemistry. 2004;43(16):4716–4723. doi: 10.1021/bi036189j. [DOI] [PubMed] [Google Scholar]
- 22.Mowbray S.L., Elfström L.T., Ahlgren K.M., Andersson C.E., Widersten M. X-ray structure of potato epoxide hydrolase sheds light on substrate specificity in plant enzymes. Protein Sci. 2006;15(7):1628–1637. doi: 10.1110/ps.051792106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F.-L., Kong X.-D., Chen Q.i., Zheng Y.-C., Xu Q., Chen F.-F., et al. Regioselectivity engineering of epoxide hydrolase: near-perfect enantioconvergence through a single site mutation. ACS Catal. 2018;8(9):8314–8317. doi: 10.1021/acscatal.8b02622. [DOI] [Google Scholar]
- 24.Wilson C., De Oliveira G.S., Adriani P.P., Chambergo F.S., Dias M.V.B. Structure of a soluble epoxide hydrolase identified in Trichoderma reesei. Biochim Biophys Acta - Proteins Proteomics. 2017;1865(8):1039–1045. doi: 10.1016/j.bbapap.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Kong X.-D., Yuan S., Li L., Chen S., Xu J.-H., Zhou J. Engineering of an epoxide hydrolase for efficient bioresolution of bulky pharmaco substrates. Proc Natl Acad Sci U S A. 2014;111(44):15717–15722. doi: 10.1073/pnas.1404915111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferrandi E.E., Sayer C., De Rose S.A., Guazzelli E., Marchesi C., Saneei V., et al. New thermophilic α/β Class epoxide hydrolases found in metagenomes from hot environments. Front Bioeng Biotechnol. 2018;6 doi: 10.3389/fbioe.2018.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong R., Peng Z., Zhang Y., Yang J. mTM-align: an algorithm for fast and accurate multiple protein structure alignment. Bioinformatics. 2018;34:1719–1725. doi: 10.1093/bioinformatics/btx828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gouy M., Guindon S., Gascuel O. Sea view version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27(2):221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 29.Anandakrishnan R., Aguilar B., Onufriev A.V. 3.0: Automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 2012;40(W1):W537–W541. doi: 10.1093/nar/gks375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luchko T., Gusarov S., Roe D.R., Simmerling C., Case D.A., Tuszynski J., et al. Three-dimensional molecular theory of solvation coupled with molecular dynamics in amber. J Chem Theory Comput. 2010;6(3):607–624. doi: 10.1021/ct900460m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sindhikara D.J., Yoshida N., Hirata F. Placevent: an algorithm for prediction of explicit solvent atom distribution-application to HIV-1 protease and F-ATP synthase. J Comput Chem. 2012;33(18):1536–1543. doi: 10.1002/jcc.22984. [DOI] [PubMed] [Google Scholar]
- 32.Mitusińska K., Raczyńska A., Bzówka M., Bagrowska W., Góra A. Applications of water molecules for analysis of macromolecule properties. Comput Struct Biotechnol J. 2020;18:355–365. doi: 10.1016/j.csbj.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Case D.A., Babin V., Berryman J.T., Betz R.M., Cai Q., Cerutti D.S., et al. Univ Calif; 2014. AMBER14. [Google Scholar]
- 34.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11(8):3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magdziarz T., Mitusińska K., Bzówka M., Raczyńska A., Stańczak A., Banas M., et al. AQUA-DUCT 1.0: structural and functional analysis of macromolecules from an intramolecular voids perspective. Bioinformatics. 2020;36:2599–2601. doi: 10.1093/bioinformatics/btz946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ollis D.L., Cheah E., Cygler M., Dijkstra B., Frolow F., Franken S.M., et al. The α/β hydrolase fold. Protein Eng Des Sel. 1992;5(3):197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 37.Mitusińska K., Magdziarz T., Bzówka M., Stańczak A., Gora A. Exploring solanum tuberosum epoxide hydrolase internal architecture by water molecules tracking. Biomolecules. 2018;8:143. doi: 10.3390/biom8040143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bauer T.L., Buchholz P.C.F., Pleiss J. The modular structure of α/β-hydrolases. FEBS J. 2020;287(5):1035–1053. doi: 10.1111/febs.15071. [DOI] [PubMed] [Google Scholar]
- 39.Jochens H., Hesseler M., Stiba K., Padhi S.K., Kazlauskas R.J., Bornscheuer U.T. Protein Engineering of α/β-Hydrolase Fold Enzymes. ChemBioChem. 2011;12(10):1508–1517. doi: 10.1002/cbic.201000771. [DOI] [PubMed] [Google Scholar]
- 40.Miled N., Bussetta C., De caro A., Rivière M., Berti L., Canaan S. Importance of the lid and cap domains for the catalytic activity of gastric lipases. Comp Biochem Physiol Part B Biochem Mol Biol. 2003;136(1):131–138. doi: 10.1016/S1096-4959(03)00183-0. [DOI] [PubMed] [Google Scholar]
- 41.Dugi K.A., Dichek H.L., Talley G.D., Brewer H.B., Santamarina-Fojo S. Human lipoprotein lipase: the loop covering the catalytic site is essential for interaction with lipid substrates. J Biol Chem. 1992;267(35):25086–25091. [PubMed] [Google Scholar]
- 42.Li C., Hu B.-C., Wen Z., Hu D., Liu Y.-Y., Chu Q., et al. Greatly enhancing the enantioselectivity of PvEH2, a Phaseolus vulgaris epoxide hydrolase, towards racemic 1,2-epoxyhexane via replacing its partial cap-loop. Int J Biol Macromol. 2020;156:225–232. doi: 10.1016/j.ijbiomac.2020.04.071. [DOI] [PubMed] [Google Scholar]
- 43.Li B., Yang G., Wu L., Feng Y., Oberer M. Role of the NC-loop in catalytic activity and stability in lipase from Fervidobacterium changbaicum. PLoS ONE. 2012;7(10) doi: 10.1371/journal.pone.0046881. e46881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bzówka M., Mitusińska K., Hopko K., Góra A. Computational insights into the known inhibitors of human soluble epoxide hydrolase. Drug Discov Today. 2021;26(8):1914–1921. doi: 10.1016/j.drudis.2021.05.017. [DOI] [PubMed] [Google Scholar]
- 45.Gomez G.A. Human soluble epoxide hydrolase: structural basis of inhibition by 4-(3-cyclohexylureido)-carboxylic acids. Protein Sci. 2006;15(1):58–64. doi: 10.1110/ps.051720206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simpson A.G.B., Roger A.J. The real “kingdoms” of eukaryotes. Curr Biol. 2004;14(17):R693–R696. doi: 10.1016/j.cub.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 47.Zeldin D.C., Wei S.Z., Falck J.R., Hammock B.D., Snapper J.R., Capdevila J.H. Metabolism of epoxyeicosatrienoic acids by cytosolic epoxide hydrolase: substrate structural determinants of asymmetric catalysis. Arch Biochem Biophys. 1995;316(1):443–451. doi: 10.1006/abbi.1995.1059. [DOI] [PubMed] [Google Scholar]
- 48.Fretland A.J., Omiecinski C.J. Epoxide hydrolases: biochemistry and molecular biology. Chem Biol Interact. 2000;129(1-2):41–59. doi: 10.1016/S0009-2797(00)00197-6. [DOI] [PubMed] [Google Scholar]
- 49.Yamanashi H., Boeglin W.E., Morisseau C., Davis R.W., Sulikowski G.A., Hammock B.D., et al. Catalytic activities of mammalian epoxide hydrolases with cis and trans fatty acid epoxides relevant to skin barrier function. J Lipid Res. 2018;59(4):684–695. doi: 10.1194/jlr.M082701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blee E., Schuber F. Biosynthesis of cutin monomers: involvement of a lipoxygenase/peroxygenase pathway. Plant J. 1993;4(1):113–123. doi: 10.1046/j.1365-313X.1993.04010113.x. [DOI] [Google Scholar]
- 51.Arahira M., Nong V.H., Udaka K., Fukazawa C. Purification, molecular cloning and ethylene-inducible expression of a soluble-type epoxide hydrolase from soybean (Glycine max[L.] Merr.) Eur J Biochem. 2000;267:2649–2657. doi: 10.1046/j.1432-1327.2000.01276.x. [DOI] [PubMed] [Google Scholar]
- 52.Murray G.I., Paterson P.J., Weaver R.J., Ewen S.W.B., Melvin W.T., Burke M.D. The expression of cytochrome P-450, epoxide hydrolase, and glutathione s-transferase in hepatocellular carcinoma. Cancer. 1993;71:36–43. doi: 10.1002/1097-0142(19930101)71:1<36::AID-CNCR2820710107>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 53.Kiyosue T., Beetham J.K., Pinot F., Hammock B.D., Yamaguchi-Shinozaki K., Shinozaki K. Characterization of an Arabidopsis cDNA for a soluble epoxide hydrolase gene that is inducible by auxin and water stress. Plant J. 1994;6(2):259–269. doi: 10.1046/j.1365-313X.1994.6020259.x. [DOI] [PubMed] [Google Scholar]
- 54.Guo A., Durner J., Klessig D.F. Characterization of a tobacco epoxide hydrolase gene induced during the resistance response to TMV. Plant J. 1998;15 doi: 10.1046/j.1365-313X.1998.00241.x. [DOI] [PubMed] [Google Scholar]
- 55.Gomi K., Yamamato H., Akimitsu K. Epoxide hydrolase: a mRNA induced by the fungal pathogen Alternaria alternata on rough lemon (Citrus jambhiri Lush) Plant Mol Biol. 2003;53(1/2):189–199. doi: 10.1023/B:PLAN.0000009287.95682.24. [DOI] [PubMed] [Google Scholar]
- 56.Huang F.-C., Schwab W. Molecular characterization of NbEH1 and NbEH2, two epoxide hydrolases from Nicotiana benthamiana. Phytochemistry. 2013;90:6–15. doi: 10.1016/j.phytochem.2013.02.020. [DOI] [PubMed] [Google Scholar]
- 57.Stark A., Lundholm A.-K., Meijer J. Comparison of fatty acid epoxide hydrolase activity in seeds from different plant species. Phytochemistry. 1995;38(1):31–33. doi: 10.1016/0031-9422(94)00646-B. [DOI] [Google Scholar]
- 58.Badami R., Patil K. Structure and occurrence of unusual fatty acids in minor seed oils. Prog Lipid Res. 1980;19(3-4):119–153. doi: 10.1016/0163-7827(80)90002-8. [DOI] [PubMed] [Google Scholar]
- 59.Summerer S., Hanano A., Utsumi S., Arand M., Schuber F., Blée E. Stereochemical features of the hydrolysis of 9,10-epoxystearic acid catalysed by plant and mammalian epoxide hydrolases. Biochem J. 2002;366(2):471–480. doi: 10.1042/BJ20011778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pineau E., Xu L., Renault H., Trolet A., Navrot N., Ullmann P., et al. Arabidopsis thaliana EPOXIDE HYDROLASE1 (AtEH1) is a cytosolic epoxide hydrolase involved in the synthesis of poly-hydroxylated cutin monomers. New Phytol. 2017;215(1):173–186. doi: 10.1111/nph.14590. [DOI] [PubMed] [Google Scholar]
- 61.Decker M., Arand M., Cronin A. Mammalian epoxide hydrolases in xenobiotic metabolism and signalling. Arch Toxicol. 2009;83(4):297–318. doi: 10.1007/s00204-009-0416-0. [DOI] [PubMed] [Google Scholar]
- 62.Thomaeus A., Naworyta A., Mowbray S.L., Widersten M. Removal of distal protein-water hydrogen bonds in a plant epoxide hydrolase increases catalytic turnover but decreases thermostability. Protein Sci. 2008;17(7):1275–1284. doi: 10.1110/ps.034173.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lindberg D., Ahmad S., Widersten M. Mutations in salt-bridging residues at the interface of the core and lid domains of epoxide hydrolase StEH1 affect regioselectivity, protein stability and hysteresis. Arch Biochem Biophys. 2010;495(2):165–173. doi: 10.1016/j.abb.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 64.Carlsson Å.J., Bauer P., Ma H., Widersten M. Obtaining optical purity for product diols in enzyme-catalyzed epoxide hydrolysis: contributions from changes in both enantio- and regioselectivity. Biochemistry. 2012;51(38):7627–7637. doi: 10.1021/bi3007725. [DOI] [PubMed] [Google Scholar]
- 65.Bauer P., Carlsson Å.J., Amrein B.A., Dobritzsch D., Widersten M., Kamerlin S.C.L. Conformational diversity and enantioconvergence in potato epoxide hydrolase 1. Org Biomol Chem. 2016;14(24):5639–5651. doi: 10.1039/C6OB00060F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang C., Li C., Zhu X., Liu Y., Zhao J., Wu M. Highly regio- and enantio-selective hydrolysis of two racemic epoxides by GmEH3, a novel epoxide hydrolase from Glycine max. Int J Biol Macromol. 2020;164:2795–2803. doi: 10.1016/j.ijbiomac.2020.08.011. [DOI] [PubMed] [Google Scholar]
- 67.Hu B.-C., Hu D., Li C., Xu X.-F., Wen Z., Wu M.-C. Near-perfect kinetic resolution of racemic p-chlorostyrene oxide by SlEH1, a novel epoxide hydrolase from Solanum lycopersicum with extremely high enantioselectivity. Int J Biol Macromol. 2020;147:1213–1220. doi: 10.1016/j.ijbiomac.2019.10.091. [DOI] [PubMed] [Google Scholar]
- 68.Blée E., Schuber F. Regio- and enantioselectivity of soybean fatty acid epoxide hydrolase. J Biol Chem. 1992;267(17):11881–11887. doi: 10.1016/S0021-9258(19)49780-9. [DOI] [PubMed] [Google Scholar]
- 69.Elfström L.T., Widersten M. Catalysis of potato epoxide hydrolase, StEH1. Biochem J. 2005;390:633–640. doi: 10.1042/BJ20050526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carlsson J.Å., Bauer P., Dobritzsch D., Kamerlin S.C.L., Widersten M. Epoxide hydrolysis as a model system for understanding flux through a branched reaction scheme. IUCrJ. 2018;5(3):269–282. doi: 10.1107/S2052252518003573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carlsson J.Å., Bauer P., Dobritzsch D., Nilsson M., Kamerlin S.C.L., Widersten M. Laboratory-evolved enzymes provide snapshots of the development of enantioconvergence in enzyme-catalyzed epoxide hydrolysis. ChemBioChem. 2016;17(18):1693–1697. doi: 10.1002/cbic.201600330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lind M.E.S., Himo F. Quantum chemical modeling of enantioconvergency in soluble epoxide hydrolase. ACS Catal. 2016;6(12):8145–8155. doi: 10.1021/acscatal.6b01562. [DOI] [Google Scholar]
- 73.Zhang C., Liu Y., Li C., Xu Y., Su Y., Li J., et al. Significant improvement in catalytic activity and enantioselectivity of a Phaseolus vulgaris epoxide hydrolase, PvEH3, towards ortho-cresyl glycidyl ether based on the semi-rational design. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-58693-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Babkova P., Sebestova E., Brezovsky J., Chaloupkova R., Damborsky J. Ancestral haloalkane dehalogenases show robustness and unique substrate specificity. ChemBioChem. 2017;18(14):1448–1456. doi: 10.1002/cbic.201700197. [DOI] [PubMed] [Google Scholar]
- 75.Brezovsky J., Babkova P., Degtjarik O., Fortova A., Gora A., Iermak I., et al. Engineering a de Novo Transport Tunnel. ACS Catal. 2016;6(11):7597–7610. doi: 10.1021/acscatal.6b02081. [DOI] [Google Scholar]
- 76.Harris T.R., Aronov P.A., Hammock B.D. Soluble epoxide hydrolase homologs in Strongylocentrotus purpuratus suggest a gene duplication event and subsequent divergence. DNA Cell Biol. 2008;27(9):467–477. doi: 10.1089/dna.2008.0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Harris T.R., Hammock B.D. Soluble epoxide hydrolase: gene structure, expression and deletion. Gene. 2013;526(2):61–74. doi: 10.1016/j.gene.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cronin A., Mowbray S., Durk H., Homburg S., Fleming I., Fisslthaler B., et al. The N-terminal domain of mammalian soluble epoxide hydrolase is a phosphatase. Proc Natl Acad Sci. 2003;100(4):1552–1557. doi: 10.1073/pnas.0437829100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wijekoon C.P., Goodwin P.H., Valliani M., Hsiang T. The role of a putative peroxisomal-targeted epoxide hydrolase of Nicotiana benthamiana in interactions with Colletotrichum destructivum, C. orbiculare or Pseudomonas syringae pv. tabaci. Plant Sci. 2011;181(2):177–187. doi: 10.1016/j.plantsci.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 80.Reetz M.T., Torre C., Eipper A., Lohmer R., Hermes M., Brunner B., et al. Enhancing the enantioselectivity of an epoxide hydrolase by directed evolution. Org Lett. 2004;6(2):177–180. doi: 10.1021/ol035898m10.1021/ol035898m.s001. [DOI] [PubMed] [Google Scholar]
- 81.Reetz M.T., Bocola M., Wang L.-W., Sanchis J., Cronin A., Arand M., et al. Directed evolution of an enantioselective epoxide hydrolase: uncovering the source of enantioselectivity at each evolutionary stage. J Am Chem Soc. 2009;131(21):7334–7343. doi: 10.1021/ja809673d. [DOI] [PubMed] [Google Scholar]
- 82.Reetz M.T., Zheng H. Manipulating the expression rate and enantioselectivity of an epoxide hydrolase by using directed evolution. ChemBioChem. 2011;12(10):1529–1535. doi: 10.1002/cbic.201100078. [DOI] [PubMed] [Google Scholar]
- 83.Reetz M.T., Wang L.-W., Bocola M. Directed evolution of enantioselective enzymes: Iterative cycles of CASTing for probing protein-sequence space. Angew Chemie - Int Ed. 2006;45(8):1236–1241. doi: 10.1002/anie.200502746. [DOI] [PubMed] [Google Scholar]
- 84.Kotik M., Štěpánek V., Kyslík P., Marešová H. Cloning of an epoxide hydrolase-encoding gene from Aspergillus niger M200, overexpression in E. coli, and modification of activity and enantioselectivity of the enzyme by protein engineering. J Biotechnol. 2007;132(1):8–15. doi: 10.1016/j.jbiotec.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 85.Kotik M., Archelas A., Faměrová V., Oubrechtová P., Křen V. Laboratory evolution of an epoxide hydrolase – towards an enantioconvergent biocatalyst. J Biotechnol. 2011;156(1):1–10. doi: 10.1016/j.jbiotec.2011.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.