Summary
Background
HIV/SIV-associated periodontal disease (gingivitis/periodontitis) (PD) represents a major comorbidity affecting people living with HIV (PLWH) on combination anti-retroviral therapy (cART). PD is characterized by chronic inflammation and dysbiosis. Nevertheless, the molecular mechanisms and use of feasible therapeutic strategies to reduce/reverse inflammation and dysbiosis remain understudied and unaddressed.
Methods
Employing a systems biology approach, we report molecular, metabolome and microbiome changes underlying PD and its modulation by phytocannabinoids [delta-9-tetrahydrocannabinol (Δ9-THC)] in uninfected and SIV-infected rhesus macaques (RMs) untreated (VEH-untreated/SIV) or treated with vehicle (VEH/SIV) or Δ9-THC (THC/SIV).
Findings
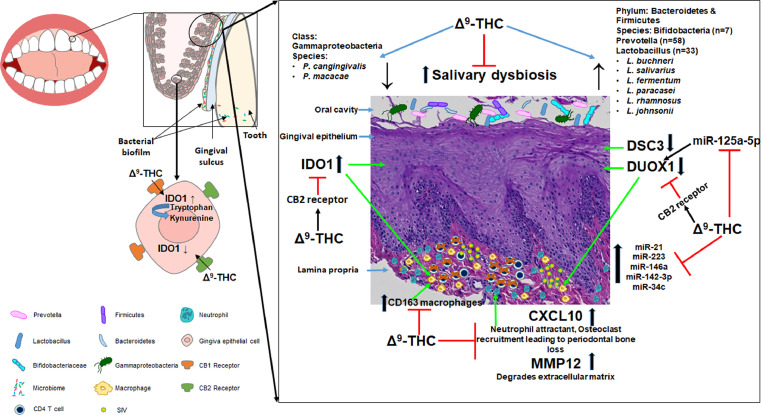
VEH- untreated/SIV but not THC/SIV RMs showed significant enrichment of genes linked to anti-viral defense, interferon-β, NFκB, RIG-1, and JAK-STAT signaling. We focused on the anti-microbial DUOX1 and immune activation marker IDO1 that were reciprocally regulated in the gingiva of VEH-untreated/SIV RMs. Both proteins localized to the gingival epithelium and CD163+ macrophages, and showed differential expression in the gingiva of THC/SIV and VEH/SIV RMs. Additionally, inflammation-associated miR-21, miR-142–3p, miR-223, and miR-125a-5p showed significantly higher expression in the gingiva of VEH/SIV RMs. In human primary gingival epithelial cells, miR-125a-5p post-transcriptionally downregulated DUOX1 and THC inhibited IDO1 protein expression through a cannabinoid receptor-2 mediated mechanism. Interestingly, THC/SIV RMs showed relatively reduced plasma levels of kynurenine, kynurenate, and the neurotoxic quinolinate compared to VEH/SIV RMs at 5 months post SIV infection (MPI). Most importantly, THC blocked HIV/SIV-induced depletion of Firmicutes and Bacteroidetes, and reduced Gammaproteobacteria abundance in saliva. Reduced IDO1 protein expression was associated with significantly (p<0.05) higher abundance of Prevotella, Lactobacillus (L. salivarius, L. buchneri, L. fermentum, L. paracasei, L. rhamnosus, L. johnsonii) and Bifidobacteria and reduced abundance of the pathogenic Porphyromonas cangingivalis and Porphyromonas macacae at 5MPI.
Interpretation
The data provides deeper insights into the molecular mechanisms underlying HIV/SIV-induced PD and more importantly, the anti-inflammatory and anti-dysbiotic properties of THC in the oral cavity. Overall, these translational findings suggest that phytocannabinoids may help reduce gingival/systemic inflammation, salivary dysbiosis and potentially metabolic disease/syndrome in PLWH on cART and those with no access to cART or do not suppress the virus under cART.
Funding
Research reported in this publication was supported by the National Institutes of Health Award Numbers R01DA052845 (MM and SNB), R01DA050169 (MM and CO), R01DA042524 and R56DE026930 (MM), and P51OD011104 and P51OD011133. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Keywords: THC, SIV, Rhesus macaque, Gingival inflammation, miR-125a-5p, DUOX1, IDO1, Lactobacilli, Prevotella, Bifidobacteria
Research in context.
Evidence before this study
HIV-associated periodontal diseases (PD), together with other oral infections, are serious comorbidities that affect people living with HIV on combination anti-retroviral therapy (cART). Despite viral suppression by cART, HIV-induced immunosuppression may facilitate the development of chronic periodontal disease and other oral infections. The ensuing chronic inflammation can induce salivary dysbiosis, which can disrupt the oral epithelial barrier leading to translocation of oral microbial and microbial by-products resulting in systemic immune activation and inflammation-driven comorbidities. The pathogenesis of HIV-associated periodontal diseases is an understudied area, especially, with regards to its molecular mechanisms. Further and more importantly, the impact of feasible therapeutic strategies to reverse gingival inflammation and dysbiosis in the context of HIV infection remains unknown and unexplored.
Added value of this study
Here, we demonstrate the molecular mechanisms by which HIV/SIV induces gingival mucosal dysfunction and how long-term controlled daily dosing of delta-9-tetrahydrocannabinol (THC) to cART-naive chronically SIV-infected rhesus macaques (RMs) successfully reversed gingival inflammation without producing any adverse effects. More importantly, we show that chronic THC can effectively suppress activation of the Indoleamine-2,3-Dioxygenase-1 (IDO1) pathway via a cannabinoid receptor 2 mediated mechanism. The suppressive effects of THC on IDO1 expression were systemic and not restricted to the gingiva as metabolomic profiling revealed significantly reduced plasma concentrations of the IDO1 catalyzed metabolite kynurenine and its downstream metabolites kynurenate and quinolinic acid. Finally, chronic THC successfully ameliorated HIV/SIV induced salivary dysbiosis of Firmicutes and Bacteroidetes, and reduced Gammaproteobacteria abundance in saliva. Further, chronic THC preserved the relative abundance of the commensal Prevotella, Lactobacillus and Bifidobacteria and reduced the relative abundance of the periodontal pathogens Porphyromonas cangingivalis and Porphyromonas macacae. These findings for the first time suggest that cannabinoids can exert a probiotic-like effect by suppressing inflammation.
Implications of all the available evidence
The induction and maintenance of IDO1 protein expression has been reported as an important triggering event in maintaining a persistent inflammatory state in progressive HIV/SIV infection. Accordingly, our findings highlight the potential of a feasible cannabinoid-based IDO1 inhibition strategy to mitigate/reduce not only chronic inflammation in both HIV and non-HIV patients but also metabolic disease/syndrome as IDO1 inhibition has been shown to increase insulin sensitivity, decrease endotoxemia, and chronic inflammation, and positively regulate lipid metabolism in liver and adipose tissues. In addition, by inhibiting oral inflammation, cannabinoids may reduce or prevent major alterations in the oral microbial populations. These translational findings provide novel insights into the mechanism of action of phytocannabinoids like THC and its potential for ameliorating HIV/SIV-induced PD and salivary dysbiosis.
Alt-text: Unlabelled box
Introduction
With the introduction of modern combination antiretroviral therapy (cART), people living with HIV (PLWH) can now achieve a near-normal life expectancy, provided cART is started early in infection. Nevertheless, PLWH are at increased risk for HIV infection-associated comorbidities such as periodontitis, salivary gland adenitis, etc. that affect the oral cavity.1, 2, 3 These are thought to be the adverse consequence of a persistent inflammatory state that is not fully resolved by cART despite successful suppression of plasma viremia.
Periodontitis is a chronic inflammatory condition caused by a complex interaction between the microbial biofilm and host immune responses that begins as a clinically manageable gingivitis. When left untreated, gingivitis can cause the gums to separate from the teeth leading to injury to periodontal connective tissue and bone and, in severe cases, tooth loss. Bacterial pathogens including Porphyromonas gingivalis, Fusobacterium nucleatum, Prevotella intermedia, and various other bacterial species in the subgingival biofilm or dental plaque have been identified as critical etiological agents that drive gingival/periodontal inflammation and tooth loss. While periodontal disease affects about 46% of the adult population, HIV infection can act as a disease modifier as it is frequently associated with the occurrence of severe necrotizing gingivitis, linear gingival erythema, and worsening of existing periodontitis.1, 2, 3 More importantly, periodontal disease has been linked to increased risk for systemic illnesses, such as cardiovascular disease, diabetes, and cancer4,5, all of which are significant comorbidities affecting PLWH.
Despite periodontal disease being a serious oral comorbidity affecting PLWH, the underlying molecular changes and mechanisms associated with its pathogenesis remain unknown. From a research perspective, simian immunodeficiency virus (SIV)-infected rhesus macaques (RMs) develop oral lesions such as gingivitis6,7 and serves as a suitable model for studying HIV/SIV-induced periodontal disease. This model, in addition to uncovering the molecular changes longitudinally, also provides an unparalleled platform to test the effects of feasible and safe pharmacological strategies to inhibit damaging gingival/periodontal inflammation. HIV/SIV-induced chronic oral inflammation, and the consequent oral dysbiosis can synergistically increase the risk for developing periodontitis and salivary gland disease. Previously, we showed that chronic administration of delta-9-tetrahydrocannabinol (Δ9-THC) [FDA approved (Dronabinol; Marinol®) potent anti-inflammatory cannabinoid8 for stimulating appetite and weight gain9,10 and found effective for symptom relief in inflammatory bowel disease patients11, 12, 13, 14 to SIV-infected RMs suppressed intestinal proinflammatory miRNA/gene expression, T-cell activation/exhaustion, and lymph node fibrosis.15,16 More recently, we showed that chronic THC administration protected minor salivary gland acini by inhibiting the expression of proinflammatory genes and preserving the expression of AGR2, WFDC2, and TSC22D3.6
It is important to note that there is growing interest within the dental community for more research to explore the potential beneficial effects of non-smoking related use of cannabinoids on the oral commensal bacteria and periodontal disease.17 Accordingly, based on our recently published findings in the intestine16 and oral mucosa6, we hypothesized that long-term low dose THC may reduce gingival immune activation and prevent salivary dysbiosis in chronic HIV/SIV infection. Here, we provide significant new knowledge on gene and microRNA expression in the gingiva using the chronically SIV-infected rhesus macaque model. We further characterized dual oxidase-1 (DUOX1) and indoleamine 2,3-dioxygenase-1 (IDO1) protein expression in gingival tissues for a couple of reasons. First, DUOX1 produces hydrogen peroxide (H2O2) that is utilized by lactoperoxidase to convert thiocyanate to the anti-bacterial/anti-microbial hypothiocyanite in mucosal surfaces. Second, IDO1 is an important anti-microbial enzyme that functions to limit tryptophan availability to oral pathogenic bacteria by converting tryptophan to kynurenine and its persistent upregulation has been reported to trigger and maintain a persistent inflammatory state in progressive HIV/SIV infection18. Accordingly, we further investigated the regulation and functional significance of both proteins using immunofluorescence, miRNA/endocannabinoid receptor-mediated mechanisms, metabolomic and metagenomic profiling. Briefly, chronic THC administration maintained the expression of DUOX1 and blocked the upregulation of the immune activation marker IDO1 in the gingiva. Reduced IDO1 gene and protein expression was associated with markedly reduced plasma levels of kynurenine and the neurotoxic quinolinate. Using primary gingival epithelial cells, we provide a mechanism by which miR-125a-5p directly targets and downregulates DUOX1 in the gingiva. More importantly, we demonstrate that THC downregulates interferon-gamma (IFNγ) induced IDO1 expression in primary human gingival epithelial cells mainly via a cannabinoid receptor-2 mechanism. Finally, suppression of IDO1 upregulation in THC/SIV RMs was associated with significantly reduced alterations in salivary microbiota composition and, more importantly, maintenance of commensal Prevotella, Lactobacillus, and Bifidobacteria species.
Methods
Animal care, ethics and experimental procedures
All experiments using rhesus macaques were approved by the Tulane Institutional Animal Care and Use Committee (Protocol No-P0308). The Tulane National Primate Research Center (TNPRC) is an Association for Assessment and Accreditation of Laboratory Animal Care International accredited facility (AAALAC #000,594). The NIH Office of Laboratory Animal Welfare assurance number for the TNPRC is A3071–01. All clinical procedures, including administration of anesthesia and analgesics, were carried out under the direction of a laboratory animal veterinarian. Animals were anesthetized with ketamine hydrochloride for blood collection procedures. Animals were pre-anesthetized with ketamine hydrochloride, acepromazine, and glycopyrrolate, intubated and maintained on a mixture of isoflurane and oxygen. All possible measures were taken to minimize the discomfort of all the animals used in this study. Tulane University complies with NIH policy on animal welfare, the Animal Welfare Act, and all other applicable federal, state and local laws.
Animal model and experimental design
A total of forty-four age and weight-matched Mamu-A0∗1−/B08−/B17− specific-pathogen-free (free of SIV, D retrovirus, STLV and Herpes B) male Indian rhesus macaques were randomly assigned to four experimental groups (Table 1). Out of the five group 1 vehicle-untreated chronically SIV-infected (VEH-untreated/SIV) male Indian RMs, all were used for RNA-seq, while four were used for global microRNA expression studies (Table 1). An additional eight age and weight-matched RMs (Table 1) were randomly assigned to two groups. One group (n = 4; Group 2) received twice daily injections of vehicle (VEH/SIV) (1:1:18 of emulphor: alcohol: saline) and second (n = 4; Group 3) received twice-daily injections of Δ9-THC (THC/SIV) beginning four weeks prior to SIV infection until 6 months post-SIV infection. In group 2, two animals each were used for both RNA-seq and microRNA expression studies. In group 3, three animals each were used for both RNA-seq and microRNA expression studies. All eight animals in groups 2 and 3 were used for salivary microbiome studies. Out of the six uninfected control (Group 4) RMs (Table 1) five and four animals were used for RNA-seq and global microRNA expression studies, respectively. A separate set of twenty-five male Indian RMs comprising four VEH/SIV and five THC/SIV previously published16 and sixteen uninfected control macaques were used to quantify plasma tryptophan metabolites (kynurenine, kynurenate and quinolinic acid).
Table 1.
Animal IDs, SIV inoculum, duration of infection and viral loads in vehicle or delta-9-tetrahydrocannabinol (Δ9-THC) treated chronic SIV-infected and uninfected rhesus macaques.
| Animal ID | SIV Inoculum | Duration of Infection | Plasma viral loads 106/mL | Gingival viral loads 106/mg RNA | Opportunistic Infections |
|---|---|---|---|---|---|
| Chronic SIV-Infected and Vehicle Untreated (Group 1) for microRNA, gene expression and Immunofluorescence studies | |||||
| IN30%,* | SIVmac239 | 246 | NA | 6.2 | Gingivitis |
| LB06#,*,%,* | SIVmac239 | 120 | 0.7 | 2.7 | ND |
| LB49#,*,%,* | SIVmac239 | 120 | 3.6 | 4.3 | ND |
| LF29#,*,%,* | SIVmac239 | 120 | 1.6 | 8.3 | ND |
| KM98#,*,%,* | SIVmac239 | 120 | NA | 0.11 | ND |
| Chronic SIV-Infected and Vehicle treated (Group 2) for microRNA, gene expression and Immunofluorescence studies | |||||
| JH47#,%,*,,^ | SIVmac251 | 180 | 2 | 2.4 | ND |
| JR36%,^ | SIVmac251 | 180 | 0.5 | 0.05 | ND |
| JD66#,%,*,^ | SIVmac251 | 180 | 0.04 | 2.9 | ND |
| IV95^ | SIVmac251 | 180 | 0.04 | NA | Gingivitis |
| Chronic SIV-Infected and Δ9-THC treated (Group 3) for microRNA, gene expression and Immunofluorescence studies | |||||
| JI45#,%,*,^ | SIVmac251 | 180 | 3 | 1.8 | ND |
| JT80#,%,*,^ | SIVmac251 | 180 | 1 | 0.58 | ND |
| IV90#,%,*,^ | SIVmac251 | 180 | 0.02 | ND | ND |
| JC85^ | SIVmac251 | 180 | 0.02 | NA | ND |
| Uninfected Controls (Group 4) for microRNA and gene expression studies | |||||
| KK15* | NA | NA | NA | NA | NA |
| JT95* | NA | NA | NA | NA | NA |
| JB84#,* | NA | NA | NA | NA | NA |
| JB61#,* | NA | NA | NA | NA | NA |
| II28#,* | NA | NA | NA | NA | NA |
| IG78# | NA | NA | NA | NA | NA |
NA- Not available, ND- None detected.
Denotes animals used for gene expression (RNA-seq) studies.
Denotes animals used for microRNA expression studies.
Denotes animals used for DUOX1 and IDO1 immunofluorescence studies.
Denotes animals used for microbiome studies.
Chronic administration of VEH (group 2) or Δ9-THC (group 3) was initiated four weeks before SIV infection at 0.18 mg/kg as used in previous studies.15,16,19,20 This dose of Δ9-THC was found to eliminate responding in a complex operant behavioral task in almost all animals20. All macaques were infected intravenously with 100TCID50 dose of the CCR5 tropic SIVmac251 or SIVmac239. Beginning the day of SIV infection, the THC dose was increased for each subject to 0.32 mg/kg, over a period of approximately two weeks when responding was no longer affected by 0.18 mg/kg on a daily basis (i.e., tolerance developed), and maintained for the duration of the study. The optimization of the THC dosing in RMs accounts for the development of tolerance during the initial period of administration. Because in previously published studies19,20 this dose of THC showed protection, the same dose was used in this study. SIV levels in plasma and gingiva were quantified using the TaqMan One-Step Real-time RT-qPCR assay that targeted the LTR gene.15,16 At necropsy, gingival tissue segments were collected in RNAlater (Thermo Fisher Scientific) and Z-fix for total RNA extraction and embedding in paraffin blocks.
Global microRNA (miRNA) profiling
Micro-RNA expression profiling was performed using TaqMan OpenArray Human MicroRNA panels (Thermo Fisher Scientific) as described previously.15,16
RNA-seq library construction, clustering and sequencing
Transcriptome profiling by RNA-seq and data analysis were performed by Novogene (Sacramento, CA). cDNA library construction and sequencing were performed by Novogene Co. Ltd, Beijing, CA (http://www.novogene.cn/). For library construction, ∼3 μg of total RNA from each sample was used to enrich mRNA with the poly-T oligo-attached magnetic beads. The purified mRNA was then randomly cleaved into small fragments using NEBNEXT RNA fragmentation buffer (NEB, MA) following the manufacturer's instructions. First-strand cDNA was synthesized using random hexamers and M-MuLV Reverse Transcriptase (RNase H-). Second-strand cDNA synthesis by nick translation was subsequently performed using E coli DNA polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. The final cDNA library was prepared after a round of purification, terminal repair, A-tailing, ligation of sequencing adapters, size selection and PCR enrichment. The cDNA fragments of preferentially 250−300 bp in length were selected using with AMPure XP system (Beckman Coulter, Beverly, U.S.A.) and PCR amplified using Phusion High-Fidelity DNA polymerase (NEB, Beijing, China), Universal PCR primers and Index (X) Primer. PCR products were then purified using AMPure XP system (Beckman Coulter, U.S.A.), and library quality was assessed on the Agilent Bioanalyzer 2100 system (Agilent Technologies, U.S.A.). The clustering of the index-coded samples was performed on a cBot Cluster Generation System, using TruSeq SR Cluster Kit v3-cBot-HS (Illumina), according to the manufacturer's instructions. After cluster generation, the library preparations were sequenced on an Illumina Novaseq 6000 platform and 150 bp paired end reads were generated.
Cloning of 3′-UTR of DUOX1 mRNA and Dual-GLO luciferase reporter gene assay
The 3′ UTR of the rhesus macaque DUOX1 mRNA contains a single predicted miR-125a-5p binding site (TargetScan 7.2).15,16 Accordingly, two separate short 34 nucleotide sequences containing the predicted miR-125a-5p site on the 3′ UTRs of DUOX1 (5′- AUGGUCAGUCACCAGGUAUGGGCUCAGGGACCU-3′) were synthesized (IDTDNA Technologies Inc., IA) for cloning into the pmirGLO Dual-Luciferase vector (Promega Corp, Madison, WI). Two separate oligonucleotides with the miR-125a-5p binding site deleted (n = 7 nucleotides) on the 3′ UTR sequence of DUOX1 (5′- AUGGUCAGUCACCAGGUAUGGGCCCU −3′) were also synthesized to serve as a negative control. Cloning of the pmirGLO vector and luciferase reporter assays was performed as described previously.15,16 Experiments were performed using HEK293 cells (ATCC, Manassas, VA) in 6 replicates and repeated thrice.
Immunofluorescence for cellular localization of IDO1 and DUOX1 in gingival tissues
Immunofluorescence studies for the detection of DUOX1 (1 in 100 dilution) (Abcam, Cat No: ab227584), IDO1 (1 in 100 dilution) (Sigma Life Sciences, Cat No: HPA023072), CB1R (1 in 50 dilution) (Abcam, Cat No: ab2373) and CB2R (1 in 50 dilution) (Abcam, Cat No: ab3560) was performed as described previously15. Expression of DUOX1 and IDO1 protein by macrophages was confirmed using the CD163 (1 in 50 dilution) (Biorad, Cat No: MCA1853).
miR-125a-5p overexpression studies
To determine the impact of miR-125a-5p on DUOX1, we overexpressed FAM-labeled locked nucleic conjugated miR-125a-5p mimics (Qiagen Inc) in primary human gingival epithelial cells pooled (HGEPp) (CELLnTEC Advanced Cell Systems AG, 3014 Bern, Switzerland) as DUOX1 protein expression was intense in the gingival epithelium. HGEPp cells were first cultured in CnT Prime medium (CnT-PR, CELLnTEC Advanced Cell Systems AG, Bern, Switzerland) in 8 well chamber slides (Thermo-Fisher Scientific Waltham, MA USA) at 37 °C in a humidified atmosphere with 5% CO2. At 95% confluency, CnT-Prime medium was replaced with CnT-Prime epithelial 2D differentiation medium (CnT-PR) and cultured for seven days to induce cellular differentiation. On day 8 of culture, cells were transfected with two different concentrations (20 and 30 nM) of FAM-LNA-miR-125a-5p or FAM-LNA-negative control mimic using the Lipojet transfection reagent (Signagen, DE). Cells were fixed with 2% paraformaldehyde at 96 h post-transfection and immunostained with DUOX1 and later with DAPI for nuclear localization.
Endocannabinoid control of IDO1 expression in primary human gingival epithelial cells
To identify an endocannabinoid mechanism regulating IDO1 expression, HGEPp cells (cultured as described in the previous section) were treated with either DMSO or 3 µM THC only or challenged subsequently with 50 U of recombinant human interferon-gamma (rIFN-γ) and incubated for 18 h. In separate experiments, HGEPp cells were preincubated with the 10 µM cannabinoid receptor 1 inverse agonist AM251 (Tocris Bioscience, Minneapolis, MN) or the cannabinoid receptor 2 antagonist AM630 (Tocris Bioscience, Minneapolis, MN) alone or in combination for 1 h followed by 3 µM THC treatment for 1 h and then challenged with 50 U of rIFN-γ and incubated for 18 h. At the end of the incubation period, cells were fixed with 2% paraformaldehyde in PBS for IDO1 immunofluorescence staining.
Quantitative image analysis of gingival sections
Briefly, two slides containing gingival tissue sections from each animal were stained with antibodies specific for DUOX1, IDO1 and CD163. No differences in staining intensity were detected between slides for each macaque. A total of ten bright-field sections for each of the eleven rhesus macaques (Groups 1, 2 and 3) scanned using a Zeiss LSM700 confocal microscope (Carl ZEISS Microscopy, LLC) at 20X objective was imported as digital images into HALO software (Indica Labs) for image quantitation analysis. Since whole/intact gingival tissues (containing all regions, namely epithelial, lamina propria, submucosal, muscular and serosal) were used for mRNA and microRNA profiling, we decided to use the area quantification module available on HALO v3.2 (Indica Labs) to quantify DUOX1 and IDO1 (green signal/Alexa-488) and CD163 (red signal/Alexa-568) fluorescence from the entire gingival tissue section. In this new computational method, the artificial intelligence driven software identifies all cells that express DUOX1 or IDO1 in green, CD163 in red and nuclei in blue and also categorizes the cells based on predefined fluorescence intensity levels. Specifically, the HALO software normalizes the threshold across all images and enables quantification of the number of cells, and relative intensity of fluorescence per cell, of single channel fluorescence (green or red) corresponding to the expression of DUOX1, IDO1 and CD163 that was intensely expressed in the gingival epithelial cells and lamina propria resident mononuclear cells. The output values (total area and average positive intensity) were used to calculate the total DUOX1, IDO1 and CD163 fluorescent intensity/tissue area. The data were graphed using Prism v8 software (GraphPad software).
Quantification of kynurenine and its metabolites in plasma
Sample preparation was performed at Metabolon Inc (Morrisville, NC). Briefly, individual samples were subjected to methanol extraction and then split into aliquots for analysis by ultrahigh performance liquid chromatography/mass spectrometry (UHPLC/MS). The global biochemical profiling analysis comprises four unique arms consisting of reverse phase chromatography positive ionization methods optimized for hydrophilic compounds (LC/MS Pos Polar) and hydrophobic compounds (LC/MS Pos Lipid), reverse phase chromatography with negative ionization conditions (LC/MS Neg), as well as a HILIC chromatography method coupled to negative (LC/MS Polar). All of the methods alternated between full-scan MS and data-dependent MSn scans. The scan range varied slightly between methods but generally covered 70–1000 m/z.
Metabolites were identified by automated comparison of the ion features in the experimental samples to a reference library of chemical standard entries that includes retention time, molecular weight (m/z), preferred adducts, and in-source fragments, as well as, associated MS spectra and curated by visual inspection for quality control using software developed at Metabolon. Identification of known chemical entities was based on comparison to metabolomic library entries of purified standards.
Quantitation of mucosal viral loads
Total RNA samples from all SIV-infected animals were subjected to a quantitative real-time TaqMan One-step RT-qPCR analysis to determine the viral load in plasma and gingival tissue. Briefly, primers and probes specific to the SIV LTR sequence were designed and used in the real-time TaqMan PCR assay. Probes were conjugated with a fluorescent reporter dye (FAM) at the 5′ end and a quencher dye at the 3′ end. Fluorescence signal was detected with a Quantstudio 5 Real-Time PCR system (Thermo Fisher). Data were captured and analyzed with QuantStudio Design & Analysis Software (Thermo Fisher). Viral copy number was determined by plotting CT values obtained from gingival samples against a standard curve (y= −3.32x + 40.4) (r2=0.999) generated with in vitro transcribed RNA representing known viral copy numbers.
Metagenomic sequencing of salivary microbiota
Saliva was collected using weck-cel sponges before and at 5-months post-SIV infection. DNA was isolated from the weck-cel sponge using the cell-free DNA purification kit (Norgen Biotek Corp, ON, Canada) following the manufacturer's protocol. Shotgun metagenomics sequencing and data analysis was performed by LC Sciences, Houston, Texas. DNA library was constructed using TruSeq Nano DNA LT Library Preparation Kit (FC-121–4001) and following the manufacture's protocol. Briefly, DNA was fragmented by dsDNA Fragmentase (NEB, M0348S) by incubating at 37 °C for 30 min. Library construction begins with fragmented genomic DNA (gDNA). Blunt-end DNA fragments are generated using a combination of fill-in reactions and exonuclease activity, and narrowly size selected with sample purification beads. An A-base is then added to the blunt ends of each strand, preparing them for ligation to the indexed adapters. Each adapter contains a T-base overhang for ligating the adapter to the A-tailed fragmented DNA. These adapters contain the full complement of sequencing primer hybridization sites for single, paired-end, and indexed reads. Single-index adapters were ligated to the fragments and the ligated products were amplified with reduced-bias PCR. Quality control analysis and quantification of the DNA library were performed using agarose gel and KAPA Library Quantification Kits. Paired-end 2 × 150 sequencing was performed on Illumina's NovaSeq sequencing system.
Data analysis and availability
For RNA-seq analysis, raw reads were first processed through perl scripts to remove reads containing adapter or -N or with a base quality score lower than 20. At the same time, the Q20, Q30 and GC contents of the clean data were calculated. The clean reads were aligned to the genome assembly of Macaca mulatta 10 (https://www.ncbi.nlm.nih.gov/genome/215?genome_assembly_id=468623) using TopHat2,21 and read numbers mapped to each gene were calculated using the HTSeq program22. The fragments per kilobase of transcript sequence per million base pairs of each gene were determined by the length of the gene and read counts mapped to this gene. Differential expression analyses of VEH or THC-treated SIV-infected RMs and control groups were performed using the DESeq R package (1.18.0).23 Genes with a P-value <0.05 and |log2 fold change| > 0.585 were defined as differentially expressed.
QuantStudio™ run files from all groups were imported into ExpressionSuite software v1.0.2 (Thermo Fisher) and simultaneously analyzed using the uninfected control group as the calibrator to obtain relative gene expression values. MiRNA expression data were normalized using the global normalization method. In all experiments, the CT upper limit was set to 28. A p-value of less than 0.05 (<0.05) was considered significant. OpenArray TaqMan miRNA (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181541) and Novogene RNA-seq data (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181487) have been deposited with GEO. Metagenomics data (https://dataview.ncbi.nlm.nih.gov/object/26651245) has been submitted to the SRA.
DUOX1, IDO1 and CD163 immunofluorescence image and RT-qPCR (DUOX1 and miR-125a-5p) data were analyzed using nonparametric Mann Whitney “U” test available on GraphPad using Prism software (v5). Shapiro-Wilk test (GraphPad Prism) was used to test for normality. While DUOX1 was found to be normally distributed (no significant p values), one group each for IDO1 (THC/SIV) and CD163 (Uninfected controls) had significant P values suggesting that the data were not normally distributed. Therefore, to maintain uniformity, we used the non-parametric Mann-Whitney test to compare statistical significance for all three datasets. Firefly/Renilla ratios and in vitro endocannabinoid regulation of IDO1 assays were analyzed using one-way ANOVA and post hoc analysis using Tukey's multiple comparison test.
For metabolite data analysis, after log transformation and imputation of missing values, if any, with the minimum observed value for each compound, Welch's two-sample t-Test was used as significance test to identify biochemicals that differed significantly (p < 0.05) between experimental groups. Imputation for replacing missing values in the metabolite data analysis was performed by replacing the missing values with its observed minimum (performed after batch normalization). Imputing with the minimum was chosen based on Metabolom's internal simulation studies comparing this to other methods with regards to the Type I error and power for the two-sample t-test. No imputation was performed for RNA-seq, microbiome and microRNA data analysis.
For metagenomic sequencing, raw sequencing reads were processed to obtain valid reads for further analysis. First, sequencing adapters were removed from sequencing reads using cutadapt v1.9. Secondly, low-quality reads were trimmed by fqtrim v0.94 using a sliding-window algorithm. Thirdly, reads were aligned to the host genome using bowtie2 to remove host contamination. Once quality-filtered reads were obtained, they were de novo assembled to construct the metagenome for each sample by IDBA-UD. All coding regions (CDS) of metagenomic contigs were predicted by MetaGeneMark v3.26. CDS sequences of all samples were clustered by CD-HIT v4.6.1 to obtain unigenes. Unigene abundance for a certain sample was estimated by TPM based on the number of aligned reads by bowtie2 v2.2.0 [2]. The lowest common ancestor taxonomy of unigenes were obtained by aligning them against the NCBI NR database by DIAMOND v 0.7.12. To determine the abundance profile of unigenes, differential analysis was carried out at each taxonomic level by Fisher's exact test. Unigenes with a P-value <0.05 and |log2 fold change| > 0.585 were defined as differentially expressed.
Although hypothesis-driven, due to the small sample size, particularly in the THC/SIV group (n = 3), we have reported genes, microRNA, plasma kynurenine pathway metabolites, and bacterial taxa that showed statistical significance at the level of p<0.05 without applying multiple comparison testing (False discovery rate). It is important to note that significant changes in mRNA expression (p and q values <0.05) do not always guarantee concurrent changes in protein expression. Similarly, differential miRNA expression does not provide information about their functional significance. For these reasons, we used the dual-labeled immunofluorescence technique to confirm changes in protein expression of three genes of interest (DUOX1, IDO1 and CD163) in the same gingival tissues. In addition, we used the RT-qPCR assay to confirm miR-125a-5p expression (OpenArray data) and then followed up with dual luciferase and miRNA overexpression assays to validate DUOX1 as a direct target of miR-125a-5p.
The funding agency (NIH) had played no role in the study design, data collection, data analyses, interpretation, or writing of the manuscript.
Results
Plasma viral loads, CD4+ and CD8+ T cell status, and oral histopathology
All SIV-infected RMs (groups 1, 2, and 3) had a wide range (0.02 × 106 to 3.6 × 106/mL) of viral RNA copies in the plasma during chronic SIV infection (Table 1). Plasma viral loads for animal IN30 were not available. However, animal IN30 and most other SIV-infected RMs had substantial viral loads in the gingiva (0.05 × 106 to 8.3 × 106/mg total RNA). Gingival viral loads were not available for animals IV95 and JC85, and no SIV RNA was detected in the gingiva of animal IV90 (Table 1). Longitudinal peripheral blood viral loads, CD4+ and CD8+ T cell dynamics from the four VEH/SIV (JH47, JR36, JD66, IV95) and THC/SIV (JI45, JT80, IV90, JC85) RMs were recently published16. Marked depletion of CD4+ T cells in the intestine and peripheral blood was detected in these eight animals16. CD4+ T cell depletion was accompanied by a concomitant increase in CD8+ T cell percentages16. No difference in plasma and gingival viral RNA copies were detected between the two groups16. Histopathological evidence of mild to moderate gingivitis was confirmed in one VEH/SIV and one VEH-untreated RMs (Table 1).
Genes associated with anti-viral defense, type I interferon signaling, JAK-STAT, and kynurenine pathway are markedly upregulated in the gingiva of VEH-untreated/SIV RMs
As periodontal disease is a significant comorbidity affecting HIV-infected individuals on cART,1, 2, 3 we performed RNA-seq to better understand its molecular pathogenesis and its modulation by cannabinoids, in particular, THC. Compared to uninfected controls, 206 genes were found to be significantly (p<0.05) upregulated and differentially expressed in the gingiva of VEH-untreated/SIV RMs. Using DAVID (NIAID) (Species: Macaca mulatta), we successfully annotated 147 genes (Figure 1a). Further, gene enrichment analysis using DAVID showed differential enrichment of biological functions associated with Influenza A infection (n = 12) (p = 5.3 × 10−7), 2′−5′-Oligoadenylate synthetase activity (n = 3) (p = 4.3 × 10−4), antigen processing and presentation (n = 7) (p = 4.6 × 10−5), positive regulation of interferon-beta production (n = 4) (p = 4.0 × 10−4), RIG-1-like receptor signaling pathway (n = 5) (p = 3.4 × 10−3), cell-adhesion molecules (n = 7) (p = 1.9 × 10−3), inflammatory bowel disease (n = 6) (p = 3.0 × 10−4), lipoprotein metabolic process (n = 3) (p = 7.2 × 10−3), JAK-STAT signaling pathway (n = 6) (p = 9.0 × 10−3) and positive regulation of NF-κB transcription factor activity (n = 5) (p = 9.0 × 10−3) (Figure 1b & c). Supplementary Table 1 lists the 50 key differentially upregulated genes linked to gingival immune activation in VEH-untreated SIV-infected RMs. Notable differentially upregulated genes with critical roles in the pathogenesis of HIV infection and periodontal disease (Figure 1a) included IDO1 (a marker of immune activation),24,25 IL21R (proliferation of T, B, and NK cells),26 CXCL10 (neutrophil migration and periodontal bone loss)27, 28, 29, MMP12 (gingival extracellular matrix degradation),30,31 IRF1 & IRF7 (innate and adaptive immune response)32, VCAM1 (leukocyte-endothelial adhesion/migration),33 LYN (inflammatory response to bacterial LPS),33 TLR2 (immune response to bacterial lipoproteins and lipopeptides),34 TRIM5 and SAMHD1 (HIV/SIV restriction), ALOX5AP (Leukotriene synthesis), GZMA and GZMH (chronic inflammation and antiviral response), KLK6 (inflammation), CD163 (anti-inflammatory macrophages) and GSDMD (bactericidal activity).35,36
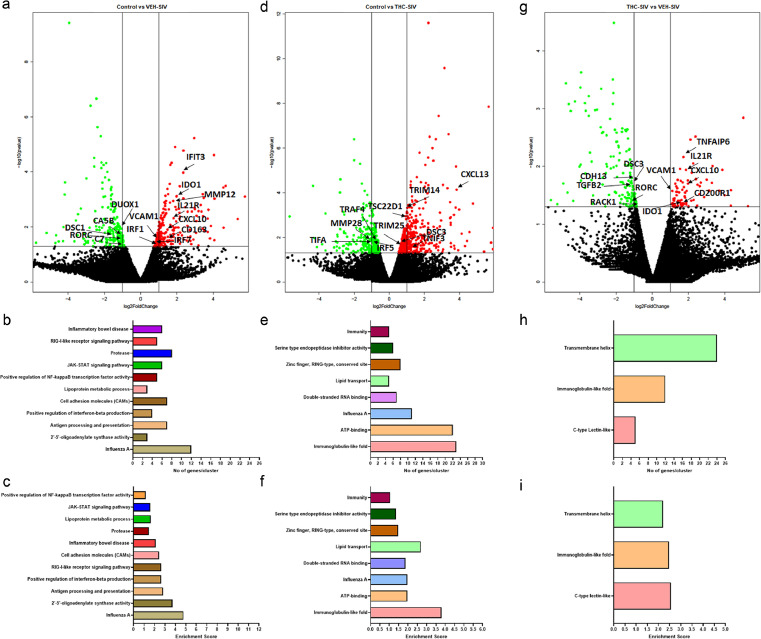
Figure 1.
Long-term low dose THC administration inhibits proinflammatory gene expression in gingiva of chronically SIV-infected RMs. Volcano plot shows the relationship between fold-change (X-axis) and statistical significance (Y-axis) of differentially expressed mRNAs in VEH/SIV (a) and THC/SIV (d) RMs relative to controls and in VEH/SIV relative to THC/SIV RMs (g). The vertical lines in (a, d, g) correspond to 2.0-fold up and down, respectively, and the horizontal line represents p ≤ 0.05. The negative log of statistical significance (p-value) (base 10) is plotted on the Y-axis, and the log of the fold change base (base 2) is plotted on the X-axis. Notable differentially expressed mRNAs are shown in the volcano plots. DAVID functional annotation cluster analysis of upregulated genes in VEH/SIV (b, c) and THC/SIV RMs (e, f) relative to controls and in VEH/SIV relative to THC/SIV RMs (h, i). Number of genes represented in each cluster in VEH/SIV (b) and THC/SIV (e) relative to controls and in VEH/SIV (h) relative to THC/SIV RMs. Top clusters of genes upregulated in VEH/SIV (c) and THC/SIV (f) relative to controls and in VEH/SIV (i) relative to THC/SIV RMs.
In contrast, greater number of genes (n = 346) were significantly upregulated in gingiva of THC/SIV RMs compared to uninfected controls (Figure 1d). Out of these, 260 genes were annotated. This represented a 1.76-fold increase (260 vs 147) in the total number of upregulated genes in THC/SIV compared to VEH-untreated/SIV RMs. Gene enrichment analysis using DAVID showed differential enrichment (Figure 1e & f) of biological functions involved in Immunoglobulin-like fold (n = 23) (p = 9.8 × 10−5), lipid transport (n = 5) (p = 1.7 × 10−3), ATP-binding (n = 22) (p = 4.1 × 10−3), Influenza A infection (n = 11) (p = 4.0 × 10−4), double-stranded RNA binding (n = 7) (p = 1.1 × 10−4), lipid transport (n = 5) (p = 1.7 × 10−3), zinc finger, RING-type, conserved site (n = 8) (p = 7.5 × 10−3), serine‑type endopeptidase inhibitor activity (n = 6) (p = 4.6 × 10−3) and immunity (n = 5) (p = 2.2 × 10−2). Although a few functional annotation clusters overlapped with VEH-untreated/SIV RMs (Figure 1b & c), fewer genes associated with proinflammatory pathways comprised the clusters detected in THC/SIV RMs, demonstrating the potent anti-inflammatory effects of long-term cannabinoid administration in the gingiva in untreated HIV/SIV infection. Interestingly, even with a sample size of n = 3, we detected a greater number of upregulated genes in gingival tissue of THC/SIV RMs. We want to emphasize that similar to our study, sample sizes consisting of three rhesus or cynomolgus macaques per group were used in numerous previously published high impact studies to investigate the pathogenesis of viral diseases (SARS-CoV-2, HIV),37, 38, 39 asthma,40 LPS mediated inflammation41 and tuberculosis.42 Noteworthy genes that were significantly upregulated in gingiva of THC/SIV RMs included CXCL13 (anti-microbial chemokine and B cell chemoattractant),43 EPAS1 and HIF1A (both associated with metabolic adaptation to inflammation-induced hypoxia for cell survival),44 TSC22D1 (TGF-β stimulated transcriptional repressor), complement 2 (C2), RNF31 (anti-inflammatory), SLFN11 (anti-viral protein), DSC3 (desmosome formation and epithelial barrier integrity) and TNFSF10 (proapoptotic)45 (Figure 1d and Supplementary Table 2). Supplementary Table 2 lists the top 50 differentially upregulated genes in the gingiva of THC/SIV RMs.
Anti-microbial DUOX1 and epithelial barrier associated DSC3 are significantly downregulated in the gingiva of VEH-untreated/SIV RMs
Compared to uninfected controls, 240 and 472 genes were downregulated in the gingiva of VEH-untreated/SIV and THC/SIV RMs, respectively. Out of these, 150 and 362 genes were successfully annotated in the VEH-untreated/SIV and THC/SIV groups, respectively. DUOX1, a member of the NADPH oxidase family associated with anti-microbial function at mucosal surfaces46,47 was significantly downregulated in the gingiva of VEH-untreated/SIV RMs. Other down-regulated genes included ALDH3A1 (aldehyde detoxification), C7 (membrane attack complex), DSC3 (desmosome formation and epithelial barrier integrity), MAOA (xenobiotic metabolism) and PTGR1 (inactivation of the chemotactic factor, leukotriene B4). In THC/SIV RMs, at least five notable genes with well-established functions in interferon induction and inflammation (IRF5), extracellular matrix degradation [MMP28, CTSK, CTSV], and NFκB and JNK activation (TRAF4 and TIFA) were found to be significantly downregulated in the gingiva (Figure 1d). Supplementary Tables 3 and 4 list the top 50 differentially downregulated genes in the gingiva of VEH-untreated/SIV and THC/SIV RMs, respectively.
The anti-inflammatory effects of THC in the gingiva became clearer when gene expression was directly compared between VEH-untreated/SIV and THC/SIV RMs at the terminal time point (Figure 1g). Consistent with the presence of a persistent inflammatory state in the gingiva, VEH-untreated/SIV RMs showed significantly higher expression of inflammation-associated genes (ADA2, ALOX5AP, CSF3R, IDO1,24,25 IL21R,26 CXCL10,27, 28, 29 CXCR6, GADD4B, KLK6, KLRD1, LTB, MAMU-DRB1, MRC1, SEMA7A, SLA, STAB1, TIMP1, VCAM133 and TNFAIP6 (Figure 1g and Supplementary Table 5) and reduced expression of oral epithelial barrier regulating genes (CASP14, CDH13, DSC3, ITGA3, ITGA6, KRT10, LAMB4) and those associated with antimicrobial function and negative regulation of inflammation (HP, PARD3B, PDGFC, TRIM35, TGFβ2) (Figure 1g and Supplementary Table 6). Gene enrichment analysis using DAVID showed differential enrichment (Figure 1h & i) of biological functions involved in C-type lectin-like (n = 5) (p = 8.5 × 10−5), immunoglobulin-like-fold (n = 11) (p = 2.7 × 10−6), and transmembrane helix (n = 21) (p = 2.8 × 10−3).
Chronic THC administration reciprocally regulated DUOX1 and IDO1 protein expression in the gingiva of chronic SIV-infected RMs
The interesting finding on the significant differential expression of the anti-microbial DUOX1 and IDO1 gene expression and its beneficial modulation by THC in the gingiva of SIV-infected RMs prompted us to identify specific gingival cell types that contributed to its differential protein expression. We focused on these two genes for the following reasons. First, DUOX1 is a specialized reactive-oxygen species (ROS)-generating enzyme expressed in epithelial tissues46. Second, the hydrogen peroxide (H2O2) generated by DUOX1 permeates into the mucosal surface, where it is utilized by lactoperoxidase to convert thiocyanate to the anti-bacterial/anti-microbial hypothiocyanate.46 Similarly, IDO1 is an important anti-microbial enzyme activated by proinflammatory cytokines such as interferon-gamma (IFN-γ), lipopolysaccharide etc., that catalyzes the conversion of tryptophan to kynurenine, thereby limiting availability of this essential amino acid to the oral pathogenic bacteria.25
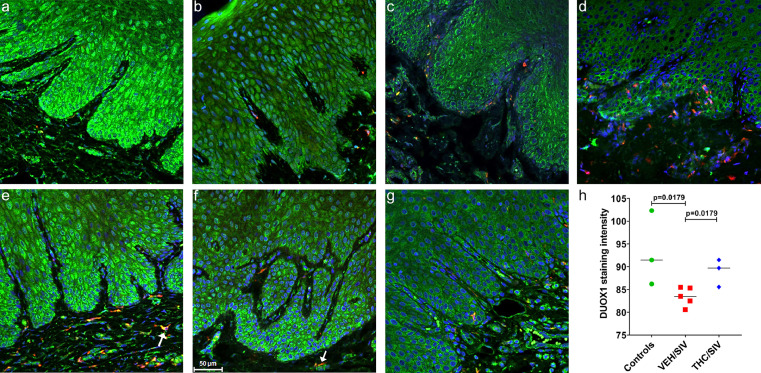
In agreement with RNA-seq data (Figure 1a), DUOX1 staining intensity (green staining) was considerably weaker in gingiva of VEH-untreated/SIV (Figure 2b, c, d) compared to uninfected controls (Figure 2a) and THC/SIV RMs (Figure 2e, f, g), confirming marked downregulation of DUOX1 protein expression in the gingiva during chronic HIV/SIV infection. In contrast, DUOX1 staining intensity was brighter and more robust in the gingiva of the uninfected control (Figure 2a) and THC/SIV RMs (Figure 2e, f, g). Although DUOX1 protein expression was detected in several cell types, the stratified squamous epithelium showed the strongest expression. This is evident in Figure 2a showing considerably stronger DUOX1 staining intensity in the gingival epithelium of uninfected control (Figure 2a) and THC/SIV (Figure 2e, f, g) compared to VEH-untreated/SIV (Figure 2b, c, d) RMs. Further, quantitative image analysis confirmed the significant (p<0.05) reduction of DUOX1 protein expression in the gingiva of VEH-untreated/SIV compared to uninfected controls and THC/SIV RMs (Figure 2h). Furthermore, DUOX1 protein expression in the gingiva of THC/SIV RMs was comparable to uninfected controls (Figure 2h). Several CD163 positive macrophages present in the gingival lamina propria also expressed DUOX1 (yellow staining; white arrows in Figure 2e, f).
Figure 2.
Chronic THC administration preserved DUOX1 protein expression in the gingiva of chronically SIV-infected RMs. Gingival tissues of uninfected control (a), VEH/SIV (b, c, d) and THC/SIV RMs (e, f, g) were immunostained for DUOX1 (green), CD163 (red) and Topro3 for nuclear staining (blue). Note the significantly decreased DUOX1 (b, c, d) staining in the gingival epithelium of VEH/SIV RMs. In contrast, DUOX1 (e, f, g) staining is intense in the gingival epithelium of the THC/SIV RMs. Representative immunofluorescence images were captured using a Zeiss confocal microscope at 20X magnification. Yellow staining (e, f) indicates colocalization of DUOX1 to CD163+ macrophages (white arrow). Quantitation of DUOX1 (h) signal intensity was performed using Halo software. Differences in DUOX1 signal intensity between groups were analyzed using Mann Whitney “U” test employing the Prism v5 software (GraphPad software). A p value of <0.05 was considered significant.
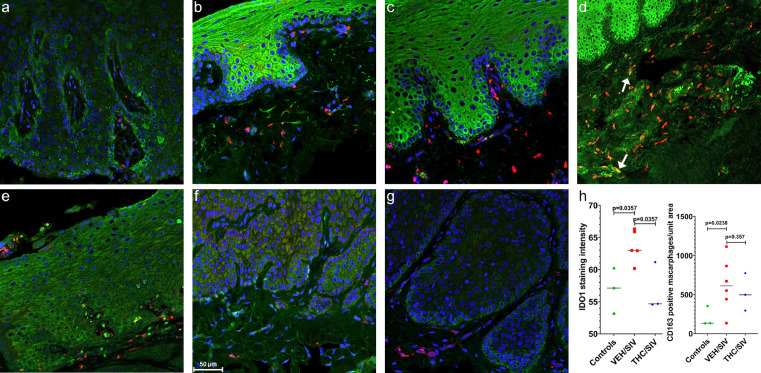
The increased mRNA expression of IDO1 in the gingiva of VEH-untreated/SIV macaques was equally intriguing as it is induced by inflammatory stimuli and its upregulation indirectly suggests the presence of mucosal inflammation/immune activation and microbial overgrowth/dysbiosis25. Similar to DUOX1 protein expression, expression of IDO1 (Figure 3b, c, d) was detected in the gingival epithelium and CD163+ macrophages in the gingival lamina propria. However, unlike DUOX1, expression of IDO1 protein was significantly (p<0.05) reduced in the gingival epithelium of uninfected control (Figure 3a) and THC/SIV (Figure 3e, f, g) RMs compared to VEH/SIV RMs (Figure 3b, c, d). These findings agree with the RNA-seq data showing increased IDO1 mRNA expression in VEH-untreated/SIV relative to both uninfected control and THC/SIV RMs (Figure 1a, g). Consistent with the RNA-seq data, significantly greater number of CD163 expressing macrophages were detected in the gingival lamina propria of VEH-untreated/SIV RMs (Figure 3b, c, d). This is clearly evident in Figure 3d (10X magnification), showing numerous CD163+ macrophages in the gingival lamina propria. Image quantitation confirmed significantly elevated IDO1 protein expression and increased numbers of CD163+ macrophages in the gingiva of VEH-untreated/SIV RMs (Figure 3h) relative to uninfected controls. On the other hand, the number of CD163+ cells was reduced in the gingiva of THC/SIV RMs compared to VEH/SIV RMs (Figure 3h). Finally, we want to point out that even though all three genes did not pass the false discovery rate (adjusted p-value), consistent with RNA-seq data, we detected significant differences in DUOX1, IDO1, and CD163 protein expression between treatment groups.
Figure 3.
Chronic THC administration reduced HIV/SIV induced IDO1 protein upregulation in the gingiva of chronically SIV-infected RMs. Gingival tissues of uninfected control (a), VEH/SIV (b, c, d) and THC/SIV RMs (e, f, g) were immunostained for IDO1 (green), CD163 (red) and Topro3 for nuclear staining (blue). Note the significantly decreased IDO1 staining in the gingival epithelium of uninfected control (a) and THC/SIV RMs (e, f, g). In contrast, IDO1 staining is significantly intense in the gingival epithelium of the VEH/SIV RMs (b, c, d). Representative immunofluorescence images were captured using a Zeiss confocal microscope at 20X magnification except (d) (10X magnification). Yellow staining (d) indicates colocalization of IDO1 to CD163+ macrophages (white arrow). Quantitation of IDO1 signal intensity and total number of CD163+ cells in the gingival lamina propria (h) was performed using Halo software. Differences in IDO1 and CD163 signal intensity between groups were analyzed using Mann Whitney “U” test employing the Prism v5 software (GraphPad software). A p-value of <0.05 was considered significant.
MiR-125a-5p post-transcriptionally regulates DUOX1 in primary gingival epithelial cells
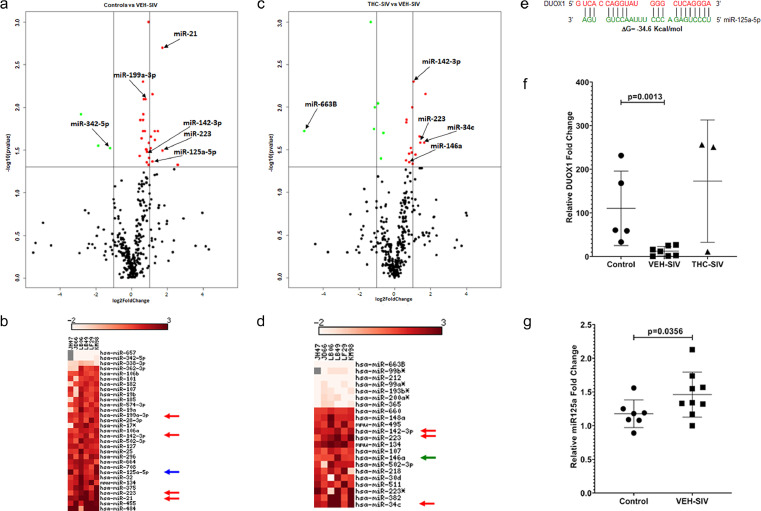
To determine the post-transcriptional mechanisms regulating DUOX1 and IDO1 expression, we profiled miRNA expression in the same gingival samples from VEH-untreated/SIV (n = 6) (Figure 4a, b) and THC/SIV RMs (n = 3) (Figure 4c, d) and identified 31 (28-up and 3-down) differentially expressed miRNAs relative to uninfected controls (n = 4). In terms of magnitude, miR-199a-3p, miR-142–3p, miR-21, and miR-223 showed the highest upregulation in the gingiva (∼3.7–11 fold-black and red arrows in Figure 4a, b, respectively). More importantly, miR-199a-3p, miR-142–3p, miR-223 and miR-21 have been confirmed to show consistent upregulation in human periodontal lesions48, 49, 50. Interestingly, no differentially expressed miRNAs (at p<0.05) were identified when comparing THC/SIV RMs to uninfected controls. Nevertheless, when comparing VEH-untreated/SIV and THC/SIV RMs, 22 miRNAs (15-up and 7-down) were differentially expressed in the gingiva of THC/SIV RMs. (Figure 4c, d). Similar to RNA-seq findings, long-term THC administration significantly downregulated the expression of several miRNAs such as miR-142–3p, miR-21, miR-223 (red arrows in Figure 4d) and the LPS induced miR-146a (green arrow in Figure 4d) that have been well associated with inflammation in periodontal disease48, 49, 50 (black arrows in Figure 4a, c).
Figure 4.
Effect of THC on miRNA expression in the gingiva of chronically SIV-infected RMs. Volcano plot shows the relationship between fold-change (X-axis) and statistical significance (Y-axis) of differentially expressed miRNAs in VEH/SIV (a) and THC/SIV (c) RMs relative to controls. The vertical lines in (a, c) correspond to 2.0-fold change up and down, respectively, and the horizontal line represents p ≤ 0.05. The negative log of statistical significance (p-value) (base 10) is plotted on the Y-axis, and the log of the fold change base (base 2) is plotted on the X-axis in a & c. The location of miRNAs of interest is denoted with arrows in a & c. The heat map shows all differentially expressed (p ≤ 0.05) miRNAs in the gingiva of VEH/SIV relative to uninfected controls (b) and THC/SIV (d) RMs. MiRNA species originating from the opposite arm of the precursor are denoted with an asterisk (*). Red, blue and green arrows (b, d) indicate inflammation-associated miRNAs differentially upregulated in VEH/SIV RMs. MiRNA-mRNA duplex showing a single miR-125a-5b binding site (seed nucleotide region) on the RM DUOX1 (e) mRNA 3′ UTR that is conserved in the human, chimpanzee and RM. RT-qPCR validation of DUOX1 mRNA (f) and miR-125a-5p (g) expression in gingiva of VEH/SIV, THC/SIV relative to uninfected control RMs.
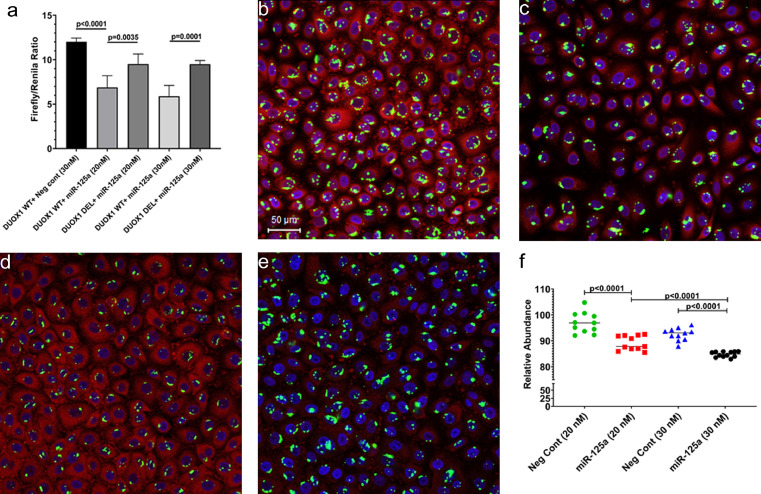
We next performed a bioinformatics analysis using the TargetScan 7.2 algorithm51 to identify miRNAs upregulated in the gingiva that are predicted to directly target DUOX1. Perfect miRNA seed nucleotide matches (miRNA nucleotide positions 2–7) were identified for miR-125a-5p on the 3′ mRNA UTR of DUOX1 (conserved in human, chimp and RM) that had a minimum free energy of −34.6 kcal/mol (Figure 4e). Moreover, miR-125a-5p dysregulation has been confirmed in periodontal disease52 (blue arrow in Figure 4b). Using RT-qPCR, we further confirmed significant downregulation of DUOX1 (Figure 4f) and upregulation of its targeting miR-125a-5p in the gingiva of VEH/SIV RMs (Figure 4g). As shown in Figure 5a, transfection of HEK293 cells with both 20 and 30 nM LNA-conjugated miR-125a-5p mimic significantly reduced firefly/renilla ratios suggesting that miR-125a-5p can regulate DUOX1 expression by directly binding to its 3′ UTR and exerting post-transcriptional repression. Next, we overexpressed FAM-labeled LNA-conjugated miR-125a-5p mimics in differentiated primary human gingival epithelial cells (HGEPp) to determine its effect on DUOX1 protein expression. Unlike western blotting, the immunofluorescence approach allowed the quantification of DUOX1 protein levels exclusively in cells that were successfully transfected with the FAM-labeled miR-125a-5p mimic (green fluorescing cells) and concurrently ensured that DUOX1 protein expression was not quantified in miR-125a-5p untransfected cells. Note the successful transfection (>95%) of HGEPp cells with the negative control [Figure 5b (20 nM) & Figure 5d (40 nM)] and miR-125a-5p [Figure 5c (20 nM) & Figure 5e (40 nM)] based on the presence of green fluorescence. Confirming the results of the luciferase reporter assay (Figure 5a), miR-125a-5p overexpression significantly decreased DUOX1 protein expression (Figure 5c, e, f) in HGEPp cells in a dose-dependent manner compared to cells transfected with the negative control mimic (Figure 5b, d, f), thus providing a potential miRNA mediated post-transcriptional mechanism regulating gingival immune/inflammatory responses in chronic HIV/SIV infection. Moreover, significant reduction in DUOX1 protein expression was successfully achieved with a low physiological dose (20 nM) of miR-125a-5p mimic (Figure 5c, f).
Figure 5.
DUOX1 is a direct target of miR-125a-5p. Luciferase reporter vectors containing a single highly conserved miR-125a-5p (a) binding site (seed nucleotide region) on the rhesus macaque DUOX1 mRNA 3′ UTR or the corresponding construct with the binding sites deleted were co-transfected into HEK293 cells with 20 nM or 30 nM miR-125a-5p or negative control mimic. Firefly and Renilla luciferase activities were detected using the Dual-Glo luciferase assay system 96 h after transfection. Luciferase reporter assays were performed thrice in six replicate wells (a). Representative immunofluorescence images showing the expression of DUOX1 (red) protein at 96 h post transfection of primary human gingival epithelial (HGEPp) cells in triplicate wells with 20 nM (b, c) and 30 nM (d, e) LNA-conjugated FAM-labeled negative control (b, d) or miR-125a-5p (green) (c, e) mimics and images were quantitated (f). Topro3 was used for nuclear staining (blue). Firefly/Renilla ratios and immunofluorescence data were analyzed using one-way ANOVA followed by Tukey's multiple comparison post hoc test. A p-value of <0.05 was considered significant.
THC downregulates IDO1 protein expression in primary gingival epithelial cells through a cannabinoid receptor 2-dependent mechanism
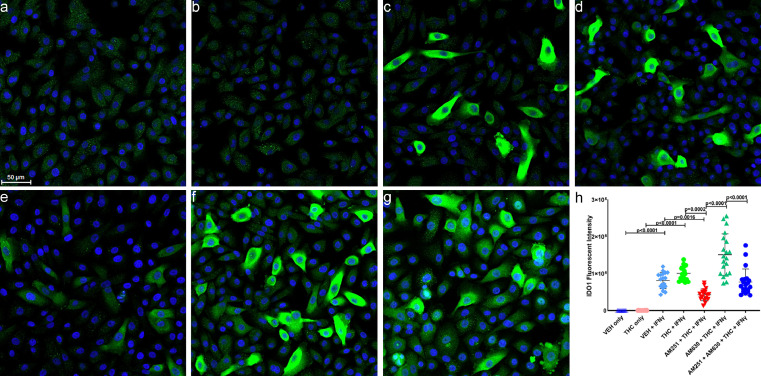
In contrast to DUOX1, none of the downregulated miRNAs detected in the gingiva of VEH-untreated/SIV RMs had predicted binding site/s on the 3′ UTR of RM IDO1 mRNA. Therefore, we sought to determine the endocannabinoid mechanisms, in particular, the role of cannabinoid receptors 1 (CB1R) and 2 (CB2R) in mediating the suppressive effects of THC. IDO1 protein expression was very weak and barely detecTable 16 h post-treatment with only vehicle (VEH; DMSO) (Figure 6a), or 3 µM THC (Figure 6b). Out of a range of THC concetrations (1, 2, 3, 4 and 5 µM) tested, 3 µM was found to be the highest dose that was well tolerated by cells (no cell detachment/death) after 16 h treatment. However, treatment with 50 units of human recombinant interferon-gamma (rIFNγ) significantly (p<0.0001) increased IDO1 protein expression in about 30–40% of the cells (Figure 6c). In contrast to that observed in gingival tissues of THC/SIV RMs (Figure 3e, Figure 3f and Figure 3g), pretreatment of HGEPp cells in vitro with 3 µM THC slightly increased the intensity of IFN-γ-induced IDO1 protein expression (Figure 6d, h). We next performed immunofluorescence to determine if differences in CB1R and CB2R expression patterns contributed to the inability of THC to suppress rIFN-γ-induced IDO1 protein expression. As shown in Supplementary Figure 1a, b, both CB1R and CB2R were abundantly expressed on in vitro differentiated HGEPp cells in the presence of rIFN-γ. As a next logical step, we preincubated HGEPp cells with 10 µM of the CB1R antagonist or inverse agonist AM251 (blocks signaling via CB1R) or the CB2R antagonist AM630 (both from Tocris Bioscience, Minneapolis, MN) or both together for 1 h followed by 3 µM THC and then IFN-γ treatments (50 units) to determine whether blocking either receptor separately or together will facilitate THC's ability to suppress rIFNγ induced IDO1 protein expression. We performed dose optimizations using 2, 5, and 10 µM of CB1R and CB2R antagonists and found 10 µM to be the most effective dose in modulating rIFN-γ-induced IDO1 protein expression, respectively. It is clear from Figure 6e that blockade of CB1R with AM251 significantly (p<0.0001) increased the ability of THC to decrease rIFN-γ induced IDO1 protein expression compared to cells treated with VEH/rIFN-γ (p = 0.0016) (Figure 6c) and THC/rIFNγ (p = 0.0002) (Figure 6d). In contrast, inhibition of CB2R with AM630 substantially increased IDO1 protein expression (Figure 6f) exceeding that observed with VEH/rIFN-γ (p = 0.0016) (Figure 6c) and THC/rIFN-γ (p = 0.0002) (Figure 6d). Inhibiting both CB1R and CB2R significantly decreased rIFN-γ-induced IDO1 protein expression (Figure 6g) compared to cells treated with AM630/THC/rIFN-γ (Figure 6f, h) and almost to the level observed with VEH/rIFN-γ treated wells (Figure 6c, h).
Figure 6.
THC downregulates rIFN-γ-induced IDO1 protein expression in HGEPp cells via a cannabinoid receptor 2 mediated mechanism. HGEPp cells were treated with VEH (DMSO) (a), THC (b), VEH + rIFN-γ (c), THC + rIFN-γ (d), AM251 + THC + rIFN-γ (e), AM630 + THC + rIFN-γ (f), AM251 + AM630 + THC+ rIFN-γ (g). Cells were fixed after 18 h and the expression of IDO1 (green) protein along with nuclear staining using Topro3 (blue) was performed and images were captured using Zeiss confocal microscope, and quantitated (h). Experiments were performed in triplicate wells using 3 µM of THC, 10 µM of AM251/AM630 and 50 U of rIFN-γ (50 U), and repeated thrice. Immunofluorescence data were analyzed using one-way ANOVA followed by Tukey's multiple comparison post hoc test. A p-value of <0.05 was considered significant.
Long-term low dose THC administration reduced plasma kynurenine, kynurenate and quinolinate levels in chronically SIV-infected RMs
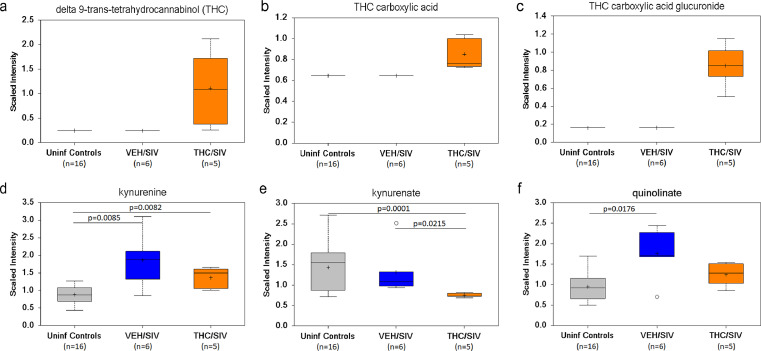
Although THC inhibition of IDO1 mRNA and protein expression is restricted to the gingiva, owing to the ubiquitous expression of CB1R and CB2R and other receptors with affinity for THC8, we predicted THC to exert suppressive effects on IDO1 and its metabolites systemically. As IDO1 catalyzes the conversion of tryptophan to kynurenine, the first step in the kynurenine pathway, we hypothesized that by reducing IDO1 protein upregulation (potentially systemically) THC will reduce plasma kynurenine, and downstream metabolites kynurenate and quinolinate levels. To test this hypothesis, we included plasma samples archived from a previously published16 cohort of five THC/SIV (IA83, IH69, HI09, IA04, JB82) and four VEH/SIV (IH96, HV48, IN24, JC81), and sixteen uninfected control RMs and quantified tryptophan metabolites using the world-class services of Metabolon Inc (Morrisville, NC). Two VEH/SIV (JH47, IV95) and one THC/SIV (IV90) (Table 1) from the current study were also included for a total of six VEH/SIV and five THC/SIV RMs in both groups. All six VEH/SIV RMs received vehicle treatments for six months. Unfortunately, animal IH69 from the THC/SIV group met Metabolon's threshold of the definition of an outlier as this macaque was found to be an outlier for 40% of the metabolites. As a result, the total number of THC/SIV RMs used for metabolomic profiling was reduced to five. The presence of THC (Figure 7a), its metabolite, THC carboxylic acid (Figure 7b) and the secreted form THC carboxylic acid glucuronide (Figure 7c) was confirmed in the plasma of THC/SIV RMs. Interestingly, plasma kynurenine levels were significantly higher in VEH/SIV and THC/SIV RMs compared to uninfected controls (Figure 7d). Nevertheless, plasma kynurenine levels were relatively lower in THC/SIV RMs compared to VEH/SIV RMs (Figure 7d). Further, plasma kynurenate levels were significantly (p<0.05) reduced in THC/SIV compared to both VEH/SIV and uninfected control RMs (Figure 7e). Furthermore, plasma levels of the neurotoxic quinolinate were significantly (p<0.05) elevated in VEH/SIV but not in THC/SIV RMs relative to uinfected control RMs (Figure 7f). These findings confirm that the anti-inflammatory effects of THC detected in three SIV-infected RMs (JI45, JT80 and IV90) used for the gingival gene (RNA-seq) and miRNA expression studies is representative of the global anti-inflammatory effects of THC.
Figure 7.
THC treatment decreased IDO1 pathway tryptophan metabolites in plasma of chronically SIV-infected RMs. Plasma levels of THC (a), THC carboxylic acid (b), THC carboxylic acid glucuronide (c), kynurenine (d), kynurenate (kynurenic acid) (e) and quinolinate (quinolinic acid) (f) in VEH/SIV (n = 6) or THC/SIV (n = 5) treated RMs at 5 MPI and uninfected control RMs (n = 16). The open circle represents an outlier because it is beyond the limit of the upper and lower quartile for that particular metabolite. A p-value of <0.05 was considered significant.
Long-term THC administration preserved members of phylum Firmicutes and Bacteroidetes and prevented depletion of Prevotella, Lactobacillus, and Bifidobacteria species in ART naïve chronic SIV-infected RMs
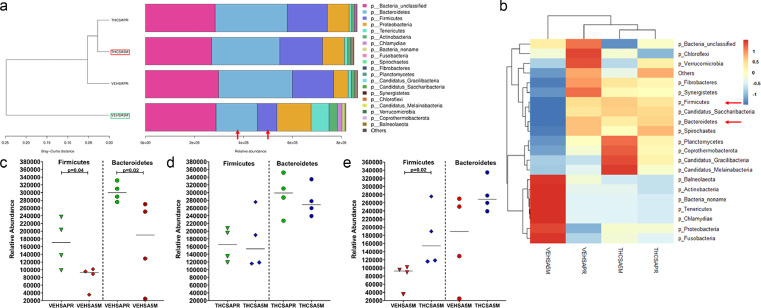
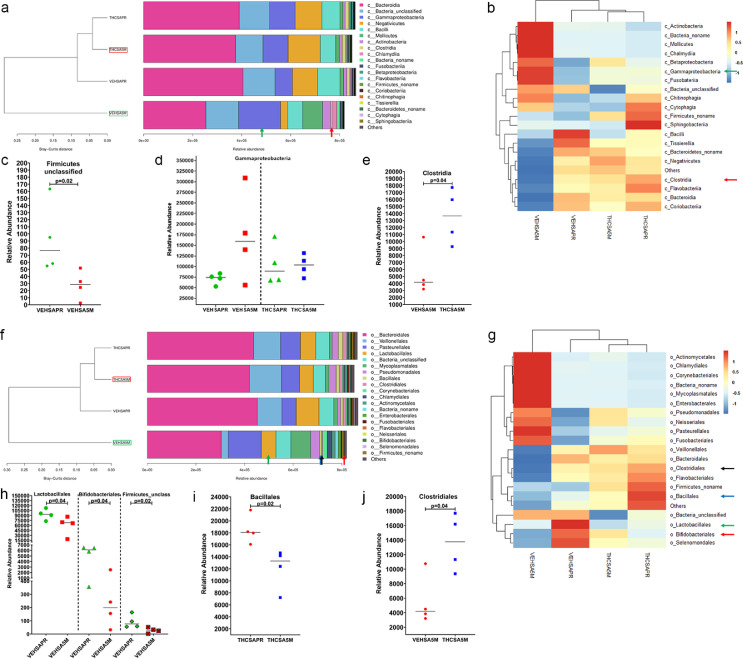
The significantly elevated expression of immune activation genes such as TLR2, GSDMD, LYN and IDO1 and the concurrent downregulation of the anti-microbial DUOX1 in the gingiva of VEH-untreated/SIV RMs indirectly suggested the existence of oral/salivary dysbiosis. In addition, we previously showed reduced protein expression of AGR2, WFDC2 and TSC22D3 in minor salivary glands (MiSGs) of chronic SIV-infected RMs(6), which could impair mucin production (consequence of reduced AGR2 expression) leading to MiSG hypofunction. Since MiSGs produce high concentrations of secretory IgA53,54, we hypothesized that their functional impairment could compromise innate immune defenses in the oral cavity causing an imbalance in the relative proportions of beneficial and harmful bacteria leading to salivary dysbiosis. To test this hypothesis, we performed metagenomic profiling of the salivary microbiome before and again at 5 months post-infection (MPI) in VEH/SIV (n = 4) and THC/SIV (n = 4) RMs (Table 1). We also collected saliva swabs at necropsy (6 MPI), however, the DNA yields in some of the samples were low and hence the 5 MPI samples were used for this study. Based on the Bray-Curtis distance, it is clear that at the 5 MPI time point, THC/SIV RMs (red box in Figure 8a) clustered closer to the preinfection time point than VEH/SIV (green box in Figure 8a) RMs. No differences in alpha or beta diversity were detected.
Figure 8.
Long-term THC treatment maintained the relative abundance of Firmicutes and Bacteroidetes in saliva of chronically SIV-infected RMs. The taxonomy cluster (a) and heat map (b) of top 21 oral bacterial phyla before and at 5 MPI in vehicle (VEH) or THC treated chronically SIV-infected RMs. Dot plots show relative abundance of phylum Firmicutes and Bacteroidetes in saliva of VEH/SIV (c) and THC/SIV (d) at 5 MPI compared to their respective preinfection time points and in THC/SIV (e) relative to VEH/SIV RMs. VEHSAPR- VEH/SIV preinfection; VEHSA5M- VEH/SIV 5 MPI; THCSAPR- THC/SIV preinfection; THCSA5M- THC/SIV 5 MPI. A p-value of <0.05 was considered significant.
The two most abundant phyla, Firmicutes and Bacteroidetes in the oral cavity were significantly depleted (p<0.05) at 5 MPI in saliva of VEH/SIV (red arrows in Figure 8a, b and Figure 8c) but not in THC/SIV RMs (Figure 8d) relative to their respective preinfection time point. At 5 MPI, relative to THC/SIV RMs, Firmicutes were significantly (p<0.05) depleted and Bacteroidetes were markedly reduced in VEH/SIV RMs (Figure 8e). At the level of bacterial class, Firmicutes unclassified showed significant (p<0.05) reduction in the saliva of VEH/SIV RMs compared to their preinfection time point (Figure 9c). Although statistically non-significant, Gammaproteobacteria showed marked expansion in saliva of VEH/SIV at 5 MPI but not in THC/SIV RMs compared to their respective preinfection time points (green arrow in Figure 9a, b and Figure 9d). Interestingly, the relative abundance of Clostridia, an important commensal obligate anerobic bacteria in the oral cavity was significantly higher in THC/SIV compared to VEH/SIV RMs at 5 MPI (red arrow in Figure 9a, b and Figure 9e).
Figure 9.
Clostridia (class) and Bifidobacteria (order) are relatively abundant in saliva of THC-treated chronically SIV-infected RMs. The taxonomy cluster (a, f) and heat map (b, g) of top 21 oral bacterial class (a, b) and order (f, g) before and at 5 MPI in VEH- or THC-treated chronically SIV-infected RMs. Dot plots show relative abundance of Firmicutes unclassified (c), gammaproteobacteria (d) in saliva of VEH/SIV (c, d) and THC/SIV (d) compared to their respective preinfection timepoints and Clostridia (e) in THC/SIV relative to VEH/SIV RMs at 5 MPI. Relative abundance of Lactobacillales, Bifidobacteriales, Firmicutes unclassified (h), Bacillales (i) in saliva of VEH/SIV (h) and THC/SIV (i) compared to their respective preinfection time points and Clostridiales (j) in THC/SIV relative to VEH/SIV RMs at 5 MPI. VEHSAPR- VEH/SIV preinfection; VEHSA5M- VEH/SIV 5 MPI; THCSAPR- THC/SIV preinfection; THCSA5M- THC/SIV 5 MPI. A p-value of <0.05 was considered significant.
When looking at bacterial order, Lactobacillales (green arrow in Figure 9f, g and Figure 9h), Bifidobacteriales (red arrow in Figure 9g, and Figure 9h), and Firmicutes unclassified (Figure 9h) showed significant depletion in VEH/SIV RMs at 5 MPI compared to their preinfection time point. Interestingly, the relative abundance of Bacillales (blue arrow in Figure 9f, g and Figure 9i) was significantly reduced in THC/SIV RMs at 5 MPI relative to their preinfection time point. Similar to that observed at the level of bacterial class, the relative abundance of Clostridiales was significantly higher in THC/SIV compared to VEH/SIV RMs (black arrow in Figure 9f, g and Figure 9j).
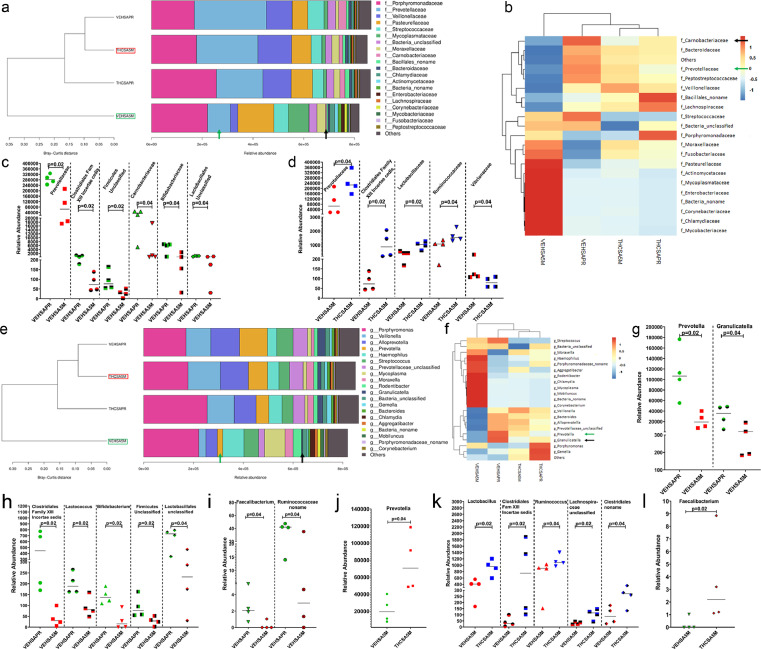
At the family level, the relative abundance of Prevotellaceae (green arrow in Figure 10a, b and Figure 10c), Clostridiales Family XIII incertae cedis (Figure 10c), Firmicutes unclassified (Figure 10c), Carnobacteriaceae (black arrow in Figure 10a, b and Figure 10c), Bifidobacteriaceae and Lactobacillus unclassified (Figure 10c) showed significant (p<0.05) depletion in VEH/SIV RMs at 5 MPI compared to their preinfection time point. Interestingly, while Prevotellaceae, Clostridiales Family XIII incertae cedis, Lactobacillaceae and Ruminococcaceae showed significantly (p<0.05) greater abundance in THC/SIV RMs compared to VEH/SIV RMs, Vibrionaceae showed the opposite trend (Figure 10d).
Figure 10.
Prevotellaceae, Carnobacteriaceae, Bifidobacteriaceae (family) and Prevotella, Granulicatella, Lactococcus, Bifidobacterium and Faecalibacterium (genus) are significantly depleted in saliva of VEH-treated /SIV-infected RMs. The taxonomy cluster (a, e) and heat map (b, f) of top 21 oral bacterial family (a, b) and genus (e, f) before and at 5 MPI in VEH- or THC-treated chronically SIV-infected RMs. Dot plots show relative abundance of Prevotellaceae, Carnobacteriaceae, and Bifidobacteriaceae (c), and Prevotellaceae, Lactobacillaceae and Ruminococcaceae (d) in saliva of VEH/SIV c) compared to their preinfection timepoints or in THC/SIV (d) relative to VEH/SIV RMs at 5 MPI. Dot plots show relative abundance of Prevotella, Granulicutella (g), Lactococcus, Bifidobacterium (h), Faecalibacterium, Ruminococcaceae_noname (i), Prevotella (j), Lactobacillus, Ruminococcus (k) and Faecalibacterium (l) in saliva of VEH/SIV (g, h, i) compared to their preinfection timepoints and in THC/SIV (j, k, l) relative to VEH/SIV RMs at 5 MPI. VEHSAPR- VEH/SIV preinfection; VEHSA5M- VEH/SIV 5 MPI; THCSAPR- THC/SIV preinfection; THCSA5M- THC/SIV 5 MPI. A p-value of <0.05 was considered significant.
At the genus level, Prevotella, Granulicatella (green and black arrows in Figure 10e, f and Figure 10g), Clostridiales Family XIII incertae cedis, Lactococcus, Bifidobacterium, Firmicutes Unclassified, Lactobacillales unclassified (Figure 10h), Faecalibacterium and Ruminococcaceae_noname (Figure 10i) showed significantly (p<0.05) reduced abundance in VEH/SIV RMs at 5 MPI compared to their preinfection time point. When comparing VEH/SIV and THC/SIV RMs, Prevotella (Figure 10j), Lactobacillus, Clostridiales Family XIII incertae cedis, Ruminococcus, Lachnospiraceae unclassified, Clostridiales no name (Figure 10k), and Faecalibacterium (Figure 10l) were significantly (p<0.05) reduced in saliva of VEH/SIV RMs.
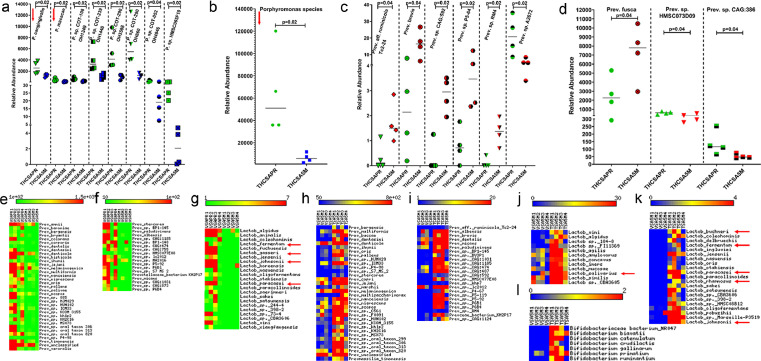
Although several bacterial species showed statistically significant differences in relative abundance at the species level, we focused on Porphyromonas, Prevotella, Lactobacillus and Bifidobacteria species for a couple of reasons. While Porphyromonas species have been implicated as major periodontal pathogens55, 56, 57, 58, Prevotella, Lactobacillus, and Bifidobacteria are part of the normal oral commensal flora59 that play essential roles in maintaining oral homeostasis. As shown in Figure 11a, out of the eight distinct Porphyromonas species, P. cangingivalis and P. macacae, two species strongly associated with periodontitis in companion animals,57 showed significantly reduced abundance in the saliva of THC/SIV RMs at 5 MPI. The relative abundance of Porphyromonas species as a whole was significantly reduced in saliva of THC/SIV RMs (Figure 11b). Unlike Porphyromonas, out of nine Prevotella species (Figure 11c, d) six, including Prevotella fusca showed high relative abundance at 5 MPI (Figure 11c, d). While greater numbers of Prevotella species showed significantly greater abundance, Prevotella. Sp. A2672, Prevotella sp HMSC073D09 and Prevotella sp. CAG:386 showed the opposite trend (Figure 11c, d). Unlike THC/SIV RMs, in VEH/SIV RMs about 56 different Prevotella species were significantly depleted at 5 MPI (Figure 11e, f). Similarly, twenty-three different Lactobacillus species that included the well-characterized probiotic species, L. fermentum, L. gasseri60, L. johnsonii,61,62 and L. paracasei63,64 were significantly depleted in the saliva of VEH/SIV RMs at 5 MPI (red arrows in Figure 11g). Relative to VEH/SIV RMs, the relative abundance of 58 Prevotella species (Figure 11h, i), 33 Lactobacillus species that included the widely used probiotic species L. salivarius,65 L. buchneri,66 L. fermentum, L. paracasei, L. rhamnosus63,64 and L johnsonii61,62 (Figure 11j, k) and seven Bifidobacteria species (Figure 11l) was significantly (p<0.05) higher in saliva of THC/SIV RMs at 5 MPI.
Figure 11.
Long-term THC treatment modulated the relative abundance of pathogenic and commensal bacterial species in saliva of chronically SIV-infected RMs. Dot plots show relative abundance of pathogenic Porphyromonas (a, b) and commensal Prevotella (c, d) species in saliva of THC/SIV RMs at 5 MPI compared to their preinfection time point. Heat maps show relative abundance of commensal Prevotella (e, f, h, i), and Lactobacillus (g, j, k) and Bifidobacteria species (l) in saliva of VEH/SIV (e, f, g) compared to their preinfection time points and THC/SIV (h, i, j, k, l) relative to VEH/SIV RMs at 5 MPI. VEHSAPR- VEH/SIV preinfection; VEHSA5M- VEH/SIV 5 MPI; THCSAPR- THC/SIV preinfection; THCSA5M- THC/SIV 5 MPI. A p-value of <0.05 was considered significant.
Discussion
The advent of combination antiretroviral therapy (cART) and the continued development of new antiretroviral drugs has transformed HIV infection from a fatal infection to a manageable chronic disease for majority of the infected population. Nevertheless, individuals with sustained viral suppression show increased incidence of age-associated comorbidities such as periodontal disease (gingivitis and periodontitis) that negatively impact their long-term survival and overall quality of life.1, 2, 3, 4, 5 Persistent periodontal disease can contribute to the breakdown of the oral epithelial barrier resulting in oral microbial and by-product translocation, systemic immune activation, and markedly increase their risk for developing other comorbidities such as cardiovascular disease, diabetes, arthritis, etc.4,5 Critical unanswered questions include how the gingival tissues respond to this chronic inflammatory environment and what post-transcriptional mechanisms regulate these responses. Further and more importantly, we wanted to determine if the persistent inflammatory responses were amenable to modulation using safe and feasible pharmacological strategies such as cannabinoids. In this study, we report the long-term effects of the naturally occurring phytocannabinoid in the Cannabis sativa plant, namely, THC, and demonstrate its anti-inflammatory and anti-dysbiotic effects in the gingiva and saliva, respectively, of cART naïve chronically SIV-infected RMs.
Given the lack of information on the molecular pathology of HIV/SIV-induced gingival mucosal dysfunction, we first profiled mRNA expression in gingival tissues of chronic SIV-infected RMs. Consistent with inflammatory signaling emanating from increased viral replication, we found gene expression signatures indicative of a host response reflective of viral replication, epithelial barrier disruption, proinflammatory signaling and concurrent bacterial dysbiosis in the gingiva. The latter signature is supported by the bimodal expression of two anti-microbial enzymes DUOX1 and IDO1 detected exclusively in the gingiva of VEH-untreated SIV RMs. While DUOX1 gene and protein expression was significantly downregulated, IDO1 showed significant upregulation. The strong protein expression of DUOX1 in the gingival epithelium of uninfected control and THC/SIV RMs aptly positions it to perform critical mucosal defense and regulate local immune responses. Specifically, DUOX1 derived hydrogen peroxide diffuses into the apical surface, where it is utilized by lactoperoxidase to oxidize its substrate thiocyanate into the antimicrobial hypothiocyanite.46,47 DUOX1 protein expression was also detected in gingival lamina propria macrophages, where the respiratory burst it creates works as an important defense mechanism against bacteria.46 Accordingly, the significant downregulation of DUOX1 in VEH-untreated/SIV RMs may contribute to gingival/periodontal disease, potentially resulting in impaired DUOX1-mediated innate immune and anti-microbial responses involved in gingival epithelial homeostasis.
Under reduced DUOX1 protein levels, the upregulation of the anti-microbial enzyme IDO1 may represent a compensatory anti-viral and anti-bacterial effector mechanism in the gingiva. Specifically, IDO1 is an intracellular non-secretory enzyme that catalyzes the production of kynurenine metabolites from tryptophan.67,68 Since many microbes require the essential amino acid tryptophan, its conversion by IDO1 expressing host cells to the bactericidal kynurenine is considered an innate immune mechanism to suppress microbial growth and proliferation. Despite its antimicrobial function, the induction and maintenance of IDO1 protein expression has also been reported to initiate and maintain a heightened inflammatory state in progressive HIV/SIV infection18 and that early ART initiation normalized kynurenine/tryptophan ratios to levels observed in healthy individuals.69 In this context, the absence of SIV-mediated DUOX1 downregulation and IDO1 upregulation in the gingiva of THC/SIV macaques are novel and important findings as it indirectly suggests subdued proinflammatory signaling and reduced abundance or proliferation of oral pathogenic bacteria. Although THC treatment reduced the expression of IDO1 in the gingiva, we predicted THC to exert similar effects systemically and not be limited to the gingiva. To test this possibility, we quantified tryptophan metabolite levels in archived plasma samples collected from a separate group of VEH/SIV, THC/SIV and uninfected control RMs. Consistent with the ability of THC to suppress IDO1 expression, we detected reduced levels of kynurenine, kynurenate and the neurotoxic quinolinate in plasma of THC/SIV relative to VEH/SIV RMs. These data suggest that THC may either inhibit the activity of enzymes downstream of the kynurenine step or reduce the levels of kynurenine available for downstream metabolite formation. Moreover, unlike chemical IDO1 inhibitors,70 naturally occurring plant-derived cannabinoids with pleiotropic (additional beneficial) effects that did not produce any detectable adverse effects despite twice-daily dosing for more than a year offers a safe approach to dampen HIV/SIV driven chronic immune activation by reducing IDO1 activity.
Similarly, CXCL10 (3.2 fold high) and MMP12 (8.7 fold high) mRNAs that were significantly upregulated in the gingiva of VEH-untreated/SIV RMs have been shown to promote bone resorption and extracellular matrix degradation in periodontitis.28, 29, 30, 31 In addition, elevated expression of VCAM1 and TLR233,34 have been associated with the severity of periodontal disease. Interestingly, the reduction of CXCL10 mRNA expression in the gingiva of THC/SIV RMs, suggests that cannabinoids may protect against periodontal bone loss. Overall, these findings provide new knowledge on the pathogenesis of gingival mucosal dysfunction in HIV/SIV infection and have translational relevance for mitigating periodontal (gingivitis/periodontitis) inflammation in PLWH who do not receive cART or completely suppress the virus under cART and other chronic inflammatory diseases of the oral mucosa.
To identify a potential mechanism regulating differential gene expression by THC, we profiled miRNA expression in the gingiva of all three treatment groups. Similar to gene expression data, several notable miRNAs previously linked to periodontal disease, T cell activation and oral squamous cell carcinoma; miR-21, miR-miR-142–3p, miR-125a-5p, miR-223, and miR-199a-3p were significantly upregulated in the gingiva of VEH-untreated/SIV RMs.48, 49, 50 Interestingly, SIV-mediated upregulation of inflammation-associated miR-21, miR-146a and miR-223 were decreased in the gingiva of THC/SIV RMs compared to VEH /SIV RMs clearly demonstrating the anti-inflammatory effects of THC at both gene and miRNA expression. The bioinformatic identification of predicted miR-125a-5p binding sites on the 3′ mRNA UTR of DUOX1 suggested a possible post-transcriptional gene silencing mechanism under inflammatory conditions. Luciferase reporter assays and miRNA overexpression studies confirmed the ability of miR-125a-5p to regulate DUOX1 post-transcriptionally. Therefore, these data collectively suggest that gingival mucosal dysfunction is also characterized by significant dysregulation of inflammation-associated miRNA expression and miR-125a-5p can potentially bind and post-transcriptionally downregulate DUOX1 protein expression. Although miR-125a-5p-mediated DUOX1 downregulation may weaken the gingival epithelial anti-microbial defense, this may be a protective host response to prevent mutagenic effects as prolonged DUOX1-dependent H2O2 production has been shown to induce DNA double-strand breaks, which can cause genomic instability and promote neoplastic transformation.71
Unlike DUOX1, none of the downregulated miRNAs were predicted to target IDO1. Therefore, we investigated the endocannabinoid receptor mechanisms associated with the inhibitory effects of THC on IDO1 protein expression in primary gingival epithelial cells. Although chronic THC administration effectively suppressed IDO1 mRNA and protein expression in gingival tissues of SIV-infected RMs, under in vitro conditions, blockade of the CB1R was required for THC to suppress rIFN-γ induced IDO1 protein expression in HGEPp cells. The significant increase in IDO1 protein expression following CB2R blockade amply suggests that CB1R is not involved in relaying the inhibitory effects of THC but may enhance rIFN-γ-induced IDO1 protein expression in HGEPp cells. Further, the finding that THC displayed greater efficacy at the CB1R than CB2R under in vitro conditions72, may partly explain why CB1R blockade was needed to suppress rIFN-γ-induced IDO1 protein expression. Combined, these findings suggest that THC inhibits rIFN-γ-induced IDO1 protein expression by primarily signaling through the CB2R in HGEPp cells. Interestingly, these findings agree with those of a recent study that demonstrated the role of CB2R in promoting periodontal cell adhesion and migration in an in vitro wound-healing assay.73 However, under in vivo conditions, THC may likely inhibit IDO1 expression by blocking IFN-γ production in the gingival lamina propria resident T cells and macrophages signaling via the CB2R, the predominant cannabinoid receptor expressed ubiquitously on immune cells.74 Accordingly, reduced IFN-γ production is expected to result in decreased IDO1 protein expression in the gingival epithelium of THC/SIV RMs. These translational findings may have broader implications for the clinical management of comorbidities like cardiometabolic disease in PLWH on cART as IDO1 inhibition improved insulin sensitivity, reduced endotoxemia, and chronic inflammation, and positively regulated lipid metabolism in the liver, and adipose tissue in the context of metabolic syndrome and cardiovascular disease.75,76
Given that chronic inflammation promotes dysbiosis,77 we next hypothesized that inhibition of MiSG6 and gingival inflammatory signaling and preservation of AGR2, WFDC2, TSC22D3 in MiSGs6 and suppression of IDO1 protein expression by THC may prevent or reduce salivary dysbiosis. Consistent with this postulation, members of the phylum Firmicutes and Bacteroidetes, two major phyla that comprise >80% of the oral bacteria, were significantly depleted in VEH/SIV but not in THC/SIV RMs at 5 MPI. Although statistically non-significant, Gammaproteobacteria, a class under phylum proteobacteria reported to translocate into the systemic circulation and induce systemic immune activation during chronic HIV/SIV infection was markedly expanded in VEH/SIV but not in THC/SIV RMs at 5 MPI relative to their respective preinfection time points. More importantly, VEH/SIV RMs showed significantly reduced abundance of bacteria belonging to the family Prevotellaceae, Carnobacteriaceae and Bifidobactericeae at 5 MPI compared to their preinfection timepoint. In vitro studies have demonstrated the ability of Carnobacteriaceae, a group of bacteriocin producing lactic acid bacteria to reduce the virulence of Listeria monocytogenes on colonic epithelial cells.78 Further, members of Carnobacteriaceae were proven to be effective as a probiotic in food-animal production79, 80, 81 suggesting that they may exert a similar effect on the oral epithelial barrier. Furthermore, members of the family Bifidobacteriaceae are part of the oral commensal bacteria known to exert beneficial effects in the oral cavity and are currently being used as probiotics.
Among the several genera that showed significantly reduced abundance in VEH/SIV RMs were Prevotella, Granulicatella, Bifidobacteria and Lactobacillus. Interestingly, Granulicatella biofilms have been shown to strengthen the human gingival epithelial barrier function82. About 56 different Prevotella species were significantly depleted in the saliva of VEH/SIV RMs but not in THC/SIV RMs. This is significant because Prevotella is the predominant genera found in the saliva that falls under phylum Bacteriodetes59. Similarly, 23 different Lactobacillus species that included the well-characterized L. gasseri, L. johnsonii, and L. paracasei were significantly depleted in the saliva of VEH/SIV RMs. Within the THC/SIV group, long-term THC administration significantly reduced the relative abundance of Porphyromonas cangingivalis and Porphyromonas macacae, two species associated with periodontitis in dogs and macaques, respectively. Porphyromonas is a major periodontal pathogen83,84 that has also been linked to reactivation of the HIV reservoir in the oral cavity85. Moreover, Lactobacilli can directly inhibit the growth and expansion of the periodontal pathogen Porphyromonas gingivalis86. Therefore, the inflammation-induced depletion of oral commensal bacteria (Lactobacilli, Bifidobacteria, Prevotella, Clostridia) may help pathogenic bacteria to expand and colonize the gingiva and periodontium, resulting in gingivitis and, if untreated, progression to periodontitis. Such drastic alterations in oral microbiota composition may exacerbate gingival inflammation, eventually leading to breakdown of the oral epithelial barrier integrity resulting in microbial translocation, systemic immune activation, and disease progression.
When comparing VEH/SIV to THC/SIV RMs, long-term THC administration preserved the abundance of bacteria belonging to the phylum Firmicutes and Bacteroidetes, suggesting that THC can preserve the normal bacterial microflora in chronic SIV infection potentially by reducing inflammatory signaling in the oral cavity. Importantly, THC/SIV RMs had significantly higher abundance of Clostridia (class and order), Lactobacillaceae, Ruminococcaceae (both family) and Faecalibacterium (genera) compared to VEH/SIV RMs at 5 MPI. Another key finding is that chronic THC administration significantly prevented the depletion of about 58 different Prevotella, 33 Lactobacilli and 7 Bifidobacteria species compared to VEH/SIV RMs at 5MPI. Among the 33 Lactobacillus species, L. rhamnosus, L. paracasei and L. salivarius have been extensively used in several clinical trials as probiotic therapy to reduce the number/expansion of cariogenic and periodontitis associated bacteria59. The ability of THC to preserve Lactobacilli and Bifidobacteria suggests that it exerts a probiotic-like effect in the oral cavity. While numerous studies have described oral and intestinal dysbiosis in HIV and SIV infection, we are not aware of any study that successfully reversed/reduced or prevented dysbiosis similar to that reported in our study.
Further, reduced availability of nutrients in a chronic inflammatory environment can promote the expansion of pathogenic bacteria equipped with the enzymatic machinery to utilize inflammatory byproducts such as nitrates (nitrate respiration) as respiratory electron acceptors77. Therefore, the decrease in the relative abundance of Porphyromonas coupled with the preservation of Lactobacilli and Bifidobacteria in the oral cavity of THC/SIV RMs may be attributed to the reduced inflammatory state as revealed by the RNA-seq data. The oral microflora, without a doubt, plays a critical role in maintaining homeostasis of the ecosystem in the oral cavity. Our findings suggest that chronic low dose THC can potentially maintain this ecosystem by preventing significant shifts in the relative proportions of beneficial and harmful bacteria. As reported previously in the intestine of SIV-infected RMs, in the present study, the relatively high abundance of Lactobacilli was associated with reduced IDO124 and stable DUOX1 mRNA and protein expression in the gingiva of THC/SIV RMs. A schematic summarizing the anti-inflammatory and anti-dysbiotic effects of THC in the gingiva and saliva, respectively, is shown in Figure 12.
Figure 12.
Proposed anti-inflammatory mechanisms of action of long-term low dose THC in the gingiva during HIV/SIV infection. During chronic HIV/SIV infection, high viral replication in the gingival lamina propria CD4+ T-cells and macrophages leads to increased production of proinflammatory mediators (CXCL10, MMP12), activation of IDO1 and concomitant downregulation of the anti-microbial enzyme DUOX1 and subsequent salivary dysbiosis. Elevated CXCL10 and MMP12 production can exacerbate disease severity by promoting periodontal bone loss and degrading the extracellular matrix. These changes together with reduced expression of gingival epithelial junction proteins like DSC3, can facilitate translocation of microbes and their products into the systemic circulation, thereby, driving chronic systemic immune activation. Concurrent administration of cannabinoids (Δ9-THC) can preserve DUOX1 by blocking miR-125a-5p upregulation and reduce IDO1 protein expression in the apical epithelium via a CB2R-mediated mechanism. In this way, low dose cannabinoids can strengthen oral mucosal defenses and concurrently halt the progression of periodontal disease by blocking the upregulation of CXCL10 and MMP12. By inhibiting inflammation, THC can reduce salivary dysbiosis by preserving the relative abundance of Firmicutes and Bacteroidetes and reducing the relative abundance of pathogenic bacteria (Porphyromonas cangingivalis, Porphyromonas macacae). Furthermore, THC can produce a probiotic-like effect by preserving the relative abundance of oral commensal bacteria like Lactobacilli and Bifidobacteria that can directly compete and inhibit the growth of oral pathogenic bacteria by producing bacteriocins.
While these findings are novel and clinically important, the study has a few limitations that future studies need to address. Group 1 rhesus macaques used for gene and miRNA expression did not receive vehicle treatment, which could be a confounder. Nevertheless, the absence of vehicle treatment is unlikely to impact the studies as we recently demonstrated that vehicle treatment alone did not make any differences to the outcome16. In that study, SIV-infected RMs that did (IH96, HV48, IN24 and JC81) and did not receive (FT11, GH25, HB31, GA19 and HD08) vehicle showed significantly increased DEFA4 and DEFA6 gene expression in the intestine compared to uninfected controls16. Therefore, all nine RMs in that study were categorized as VEH/SIV RMs. In contrast, DEFA4 and DEFA6 expression in the colon of THC treated SIV-infected RMs did not differ from uninfected control macaques. Although the absence of cART treatment in these animals may limit the clinical relevance of the findings, it is important to note that ∼33% of the HIV-infected individuals do not have access to cART and a sizable percentage of individuals receiving cART do not suppress the virus well and therefore, these findings are directly applicable both groups. Based on the conclusions from the present study, we predict similar outcomes in the cART setting, where chronic inflammation persists despite undetectable viremia.
To the best of our knowledge, this is the first study describing genome-wide analysis of differential gene expression in the gingiva during chronic HIV/SIV and its modulation by long-term low dose (0.32 mg/kg) THC administration. Specifically, our findings demonstrate a significant impact of HIV/SIV infection on the gingiva and the potential for miR-125a-5p to downregulate DUOX1 protein expression in the gingival epithelium, post-transcriptionally. These changes may explain the high incidence of periodontal disease experienced by HIV-infected patients2,3. By inhibiting HIV/SIV driven gingival and MiSG inflammation6, low dose THC may concurrently strengthen oral innate immune defenses and prevent significant shifts in salivary microbiota composition without producing any major adverse psychotropic effects or xerostomia. These findings are scientifically and clinically important as they for the first time, demonstrate the long-term anti-inflammatory and anti-dysbiotic effects of cannabinoids in the gingiva/oral cavity in chronic HIV/SIV infection. From a clinical standpoint, these findings inform a feasible pharmacological cannabinoid based strategy (capsules or oil-based medication) to modulate the oral microbiome that is often altered in a wide variety of oral inflammatory diseases including a large number (33%) of PLWH who neither receive cART nor completely suppress the virus under cART87,88. Alternatively, cannabinoids may be incorporated into dentifrices to prevent or control the progression of periodontal disease through inhibition of damaging inflammation mediated by CXCL10, MMP12, interferon-stimulated genes, etc. in both HIV and uninfected individuals. Most importantly, the low dose THC administration should not be equated with cannabis/marijuana smoking as cannabis smoke, and smoke combustion products can increase the risk of dental caries, periodontitis and even development of oral cancers. Finally, these findings will complement ongoing phase 2 human clinical trials investigating the impact of low-dose orally administered cannabinoids in combination with antiretroviral drugs89 on systemic immune activation/inflammation (NCT03550352) and cART pharmacodynamics and neurotoxicity (NCT04800159).
Contributors
MM carried out the overall planning, direction, and design of the experiments. MMW, EL, and MM carried out the day-day sampling scheduling (animal experiments) and performed the microRNA profiling, RT-qPCR validation of miRNAs and genes of interest, immunofluorescence, in vitro experiments and data analysis. XA assisted with confocal image capture and HALO image analysis. SNB assisted with plasma and tissue viral load assays. KS, BJL and CMO assisted with the cell culture, data analysis, microbiome data interpretation and conclusions. MM wrote the manuscript with input from all authors. MMW, KS, BJL, SNB, and CMO provided helpful suggestions and review of the manuscript. All authors read and approved the final version of the manuscript.
Data sharing
RNA-seq and microRNA profiling data has been submitted to GEO. Metagenomics data has been submitted to the SRA. The links with accession numbers are provided in the methods section.
Declaration of interests
All authors declare no conflict of interests.
Acknowledgments
The authors would like to thank, Ronald S. Veazey, Maurice Duplantis, Faith R. Schiro, Cecily C. Midkiff, and Coty Tatum for their technical assistance in the study.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103769.
Appendix. Supplementary materials
References
- 1.Pólvora T.L.S., Nobre Á V.V., Tirapelli C., Taba M., Jr., Macedo L.D., Santana R.C., et al. Relationship between human immunodeficiency virus (HIV-1) infection and chronic periodontitis. Expert Rev Clin Immunol. 2018;14(4):315–327. doi: 10.1080/1744666X.2018.1459571. [DOI] [PubMed] [Google Scholar]
- 2.Ryder M.I., Shiboski C., Yao T.J., Moscicki A.B. Current trends and new developments in HIV research and periodontal diseases. Periodontol 2000. 2020;82(1):65–77. doi: 10.1111/prd.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blignaut E., Rossouw T.M., Becker P.J., Mavuso D.S., Feucht U.D. Gingival recession and localized aggressive periodontitis among HIV-infected children and adolescents receiving antiretroviral therapy. Pediatr Infect Dis J. 2019;38(6):e112–e1e5. doi: 10.1097/INF.0000000000002166. [DOI] [PubMed] [Google Scholar]
- 4.Khumaedi A.I., Purnamasari D., Wijaya I.P., Soeroso Y. The relationship of diabetes, periodontitis and cardiovascular disease. Diabetes Metab Syndr. 2019;13(2):1675–1678. doi: 10.1016/j.dsx.2019.03.023. [DOI] [PubMed] [Google Scholar]
- 5.Bui F.Q., Almeida-da-Silva C.L.C., Huynh B., Trinh A., Liu J., Woodward J., et al. Association between periodontal pathogens and systemic disease. Biomed J. 2019;42(1):27–35. doi: 10.1016/j.bj.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alvarez X., Sestak K., Byrareddy S.N., Mohan M. Long term delta-9-tetrahydrocannabinol administration inhibits proinflammatory responses in minor salivary glands of chronically simian immunodeficieny virus infected rhesus macaques. Viruses. 2020;12(7) doi: 10.3390/v12070713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giavedoni L.D., Chen H.L., Hodara V.L., Chu L., Parodi L.M., Smith L.M., et al. Impact of mucosal inflammation on oral simian immunodeficiency virus transmission. J Virol. 2013;87(3):1750–1758. doi: 10.1128/JVI.02079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katchan V., David P., Shoenfeld Y. Cannabinoids and autoimmune diseases: a systematic review. Autoimmun Rev. 2016;15(6):513–528. doi: 10.1016/j.autrev.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Riggs P.K., Vaida F., Rossi S.S., Sorkin L.S., Gouaux B., Grant I., et al. A pilot study of the effects of cannabis on appetite hormones in HIV-infected adult men. Brain Res. 2012;1431:46–52. doi: 10.1016/j.brainres.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vidot D.C., Lerner B., Gonzalez R. Cannabis use, medication management and adherence among persons living with HIV. AIDS Behav. 2017;21(7):2005–2013. doi: 10.1007/s10461-017-1782-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambrose T., Simmons A. Cannabis, cannabinoids, and the endocannabinoid system-is there therapeutic potential for inflammatory bowel disease? J Crohns Colitis. 2019;13(4):525–535. doi: 10.1093/ecco-jcc/jjy185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naftali T. An overview of cannabis based treatment in Crohn's disease. Expert Rev Gastroenterol Hepatol. 2020;14(4):253–257. doi: 10.1080/17474124.2020.1740590. [DOI] [PubMed] [Google Scholar]
- 13.Perisetti A., Rimu A.H., Khan S.A., Bansal P., Goyal H. Role of cannabis in inflammatory bowel diseases. Ann Gastroenterol. 2020;33(2):134–144. doi: 10.20524/aog.2020.0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Picardo S., Kaplan G.G., Sharkey K.A., Seow C.H. Insights into the role of cannabis in the management of inflammatory bowel disease. Therap Adv Gastroenterol. 2019;12 doi: 10.1177/1756284819870977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandra L.C., Kumar V., Torben W., Vande Stouwe C., Winsauer P., Amedee A., et al. Chronic administration of Delta9-tetrahydrocannabinol induces intestinal anti-inflammatory microRNA expression during acute simian immunodeficiency virus infection of rhesus macaques. J Virol. 2015;89(2):1168–1181. doi: 10.1128/JVI.01754-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar V., Torben W., Mansfield J., Alvarez X., Vande Stouwe C., Li J., et al. Cannabinoid attenuation of intestinal inflammation in chronic SIV-infected rhesus macaques involves T cell modulation and differential expression of micro-RNAs and pro-inflammatory. Genes. Front Immunol. 2019;10:914. doi: 10.3389/fimmu.2019.00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antoniou S. Cannabinoids - high expectations? Br Dent J. 2021;231(2):70. doi: 10.1038/s41415-021-3282-7. [DOI] [PubMed] [Google Scholar]
- 18.Favre D., Mold J., Hunt P.W., Kanwar B., Loke P., Seu L., et al. Tryptophan catabolism by indoleamine 2,3-dioxygenase 1 alters the balance of TH17 to regulatory T cells in HIV disease. Sci Transl Med. 2010;2(32):32ra6. doi: 10.1126/scitranslmed.3000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molina P.E., Winsauer P., Zhang P., Walker E., Birke L., Amedee A., et al. Cannabinoid administration attenuates the progression of simian immunodeficiency virus. AIDS Res Hum Retroviruses. 2011;27(6):585–592. doi: 10.1089/aid.2010.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winsauer P.J., Molina P.E., Amedee A.M., Filipeanu C.M., McGoey R.R., Troxclair D.A., et al. Tolerance to chronic delta-9-tetrahydrocannabinol (Delta(9)-THC) in rhesus macaques infected with simian immunodeficiency virus. Exp Clin Psychopharmacol. 2011;19(2):154–172. doi: 10.1037/a0023000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anders S., Pyl P.T., Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S., McCarthy D.J., Chen Y., Okoniewski M., Smyth G.K., Huber W., et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8(9):1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 24.Vujkovic-Cvijin I., Swainson L.A., Chu S.N., Ortiz A.M., Santee C.A., Petriello A., et al. Gut-resident lactobacillus abundance associates with IDO1 inhibition and Th17 dynamics in SIV-infected macaques. Cell Rep. 2015;13(8):1589–1597. doi: 10.1016/j.celrep.2015.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nisapakultorn K., Makrudthong J., Sa-Ard-Iam N., Rerkyen P., Mahanonda R., Takikawa O. Indoleamine 2,3-dioxygenase expression and regulation in chronic periodontitis. J Periodontol. 2009;80(1):114–121. doi: 10.1902/jop.2009.080315. [DOI] [PubMed] [Google Scholar]
- 26.Pallikkuth S., Parmigiani A., Pahwa S. Role of IL-21 and IL-21 receptor on B cells in HIV infection. Crit Rev Immunol. 2012;32(2):173–195. doi: 10.1615/critrevimmunol.v32.i2.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin X., Wang Z., Wu T., Ma M., Zhang Z., Chu Z., et al. The combination of CXCL9, CXCL10 and CXCL11 levels during primary HIV infection predicts HIV disease progression. J Transl Med. 2019;17(1):417. doi: 10.1186/s12967-019-02172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aldahlawi S., Youssef A.R., Shahabuddin S. Evaluation of chemokine CXCL10 in human gingival crevicular fluid, saliva, and serum as periodontitis biomarker. J Inflamm Res. 2018;11:389–396. doi: 10.2147/JIR.S177188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rath-Deschner B., Memmert S., Damanaki A., de Molon R.S., Nokhbehsaim M., Eick S., et al. CXCL5, CXCL8, and CXCL10 regulation by bacteria and mechanical forces in periodontium. Ann Anat. 2021;234 doi: 10.1016/j.aanat.2020.151648. [DOI] [PubMed] [Google Scholar]
- 30.Björnfot Holmström S., Clark R., Zwicker S., Bureik D., Kvedaraite E., Bernasconi E., et al. Gingival tissue inflammation promotes increased matrix metalloproteinase-12 production by CD200R(low) monocyte-derived cells in periodontitis. J Immunol. 2017;199(12):4023–4035. doi: 10.4049/jimmunol.1700672. [DOI] [PubMed] [Google Scholar]
- 31.Holmström S.B., Lira-Junior R., Zwicker S., Majster M., Gustafsson A., Åkerman S., et al. MMP-12 and S100s in saliva reflect different aspects of periodontal inflammation. Cytokine. 2019;113:155–161. doi: 10.1016/j.cyto.2018.06.036. [DOI] [PubMed] [Google Scholar]
- 32.Almubarak A., Tanagala K.K.K., Papapanou P.N., Lalla E., Momen-Heravi F. Disruption of monocyte and macrophage homeostasis in periodontitis. Front Immunol. 2020;11:330. doi: 10.3389/fimmu.2020.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S., Liu X., Li H., Pan H., Acharya A., Deng Y., et al. Integrated analysis of long noncoding RNA-associated competing endogenous RNA network in periodontitis. J Periodontal Res. 2018;53(4):495–505. doi: 10.1111/jre.12539. [DOI] [PubMed] [Google Scholar]
- 34.Sumedha S., Kotrashetti V.S., Nayak R.S., Nayak A., Raikar A. Immunohistochemical localization of TLR2 and CD14 in gingival tissue of healthy individuals and patients with chronic periodontitis. Biotech Histochem. 2017;92(7):487–497. doi: 10.1080/10520295.2017.1357192. [DOI] [PubMed] [Google Scholar]
- 35.Chen Q., Liu X., Wang D., Zheng J., Chen L., Xie Q., et al. Periodontal inflammation-triggered by periodontal ligament stem cell pyroptosis exacerbates periodontitis. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.663037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhuang J., Wang Y., Qu F., Wu Y., Zhao D., Xu C. Gasdermin-d played a critical role in the cyclic stretch-induced inflammatory reaction in human periodontal ligament cells. Inflammation. 2019;42(2):548–558. doi: 10.1007/s10753-018-0912-6. [DOI] [PubMed] [Google Scholar]
- 37.Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L., et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020 doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han Q., Bradley T., Williams W.B., Cain D.W., Montefiori D.C., Saunders K.O., et al. Neonatal rhesus macaques have distinct immune cell transcriptional profiles following HIV envelope immunization. Cell Rep. 2020;30(5):1553–1569. doi: 10.1016/j.celrep.2019.12.091. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu P., Qi F., Xu Y., Li F., Liu P., Liu J., et al. Age-related rhesus macaque models of COVID-19. Animal Model Exp Med. 2020;3(1):93–97. doi: 10.1002/ame2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lynn T.M., Molloy E.L., Masterson J.C., Glynn S.F., Costello R.W., Avdalovic M.V., et al. SMAD signaling in the airways of healthy rhesus macaques versus rhesus macaques with asthma highlights a relationship between inflammation and bone morphogenetic proteins. Am J Respir Cell Mol Biol. 2016;54(4):562–573. doi: 10.1165/rcmb.2015-0210OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamperschroer C., Goldstein R., Schneider P.A., Kuang B., Eisenbraun M.D. Utilization of lipopolysaccharide challenge in cynomolgus macaques to assess IL-10 receptor antagonism. J Immunotoxicol. 2019;16(1):164–172. doi: 10.1080/1547691X.2019.1656683. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J., Guo M., Rao Y., Wang Y., Xian Q., Yu Q., et al. Mycobacterium tuberculosis Erdman infection of cynomolgus macaques of Chinese origin. J Thorac Dis. 2018;10(6):3609–3621. doi: 10.21037/jtd.2018.05.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakajima T., Amanuma R., Ueki-Maruyama K., Oda T., Honda T., Ito H., et al. CXCL13 expression and follicular dendritic cells in relation to B-cell infiltration in periodontal disease tissues. J Periodontal Res. 2008;43(6):635–641. doi: 10.1111/j.1600-0765.2008.01042.x. [DOI] [PubMed] [Google Scholar]
- 44.Yan K., Lin Q., Tang K., Liu S., Du Y., Yu X., et al. Substance P participates in periodontitis by upregulating HIF-1α and RANKL/OPG ratio. BMC Oral Health. 2020;20(1):27. doi: 10.1186/s12903-020-1017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carvalho-Filho P.C., Moura-Costa L.F., Pimentel A.C.M., Lopes M.P.P., Freitas S.A., Miranda P.M., et al. Apoptosis transcriptional profile induced by porphyromonas gingivalis HmuY. Mediators Inflamm. 2019;2019 doi: 10.1155/2019/6758159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarr D., Tóth E., Gingerich A., Rada B. Antimicrobial actions of dual oxidases and lactoperoxidase. J Microbiol. 2018;56(6):373–386. doi: 10.1007/s12275-018-7545-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Vliet A., Danyal K., Heppner D.E. Dual oxidase: a novel therapeutic target in allergic disease. Br J Pharmacol. 2018;175(9):1401–1418. doi: 10.1111/bph.14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bourbour S., Beheshti M., Kazemian H., Bahador A. Effects of micro RNAs and their targets in periodontal diseases. Infect Disord Drug Targets. 2018;18(3):183–191. doi: 10.2174/1871526518666180405155327. [DOI] [PubMed] [Google Scholar]
- 49.Zhou W., Su L., Duan X., Chen X., Hays A., Upadhyayula S., et al. MicroRNA-21 down-regulates inflammation and inhibits periodontitis. Mol Immunol. 2018;101:608–614. doi: 10.1016/j.molimm.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luan X., Zhou X., Naqvi A., Francis M., Foyle D., Nares S., et al. MicroRNAs and immunity in periodontal health and disease. Int J Oral Sci. 2018;10(3):24. doi: 10.1038/s41368-018-0025-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin S.H., Zhou J.G., Guan X.Y., Bai G.H., Liu J.G., Chen L.W. Development of an miRNA-array-based diagnostic signature for periodontitis. Front Genet. 2020;11 doi: 10.3389/fgene.2020.577585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eliasson L., Carlen A. An update on minor salivary gland secretions. Eur J Oral Sci. 2010;118(5):435–442. doi: 10.1111/j.1600-0722.2010.00766.x. [DOI] [PubMed] [Google Scholar]
- 54.Aframian D.J., Keshet N., Nadler C., Zadik Y., Vered M. Minor salivary glands: clinical, histological and immunohistochemical features of common and less common pathologies. Acta Histochem. 2019;121(8) doi: 10.1016/j.acthis.2019.151451. [DOI] [PubMed] [Google Scholar]
- 55.Jia L., Han N., Du J., Guo L., Luo Z., Liu Y. Pathogenesis of important virulence factors of porphyromonas gingivalis via toll-like receptors. Front Cell Infect Microbiol. 2019;9:262. doi: 10.3389/fcimb.2019.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu W., Zhou W., Wang H., Liang S. Roles of Porphyromonas gingivalis and its virulence factors in periodontitis. Adv Protein Chem Struct Biol. 2020;120:45–84. doi: 10.1016/bs.apcsb.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.do Nascimento Silva A., de Avila E.D., Nakano V., Avila-Campos M.J. Pathogenicity and genetic profile of oral Porphyromonas species from canine periodontitis. Arch Oral Biol. 2017;83:20–24. doi: 10.1016/j.archoralbio.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 58.Lee J.H., Moon J.H., Ryu J.I., Kang S.W., Kwack K.H., Lee J.Y. Antibacterial effects of sodium tripolyphosphate against Porphyromonas species associated with periodontitis of companion animals. J Vet Sci. 2019;20(4):e33. doi: 10.4142/jvs.2019.20.e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y., Wang X., Li H., Ni C., Du Z., Yan F. Human oral microbiota and its modulation for oral health. Biomed Pharmacother. 2018;99:883–893. doi: 10.1016/j.biopha.2018.01.146. [DOI] [PubMed] [Google Scholar]
- 60.Hirasawa M., Kurita-Ochia T. Probiotic potential of lactobacilli isolated from saliva of periodontally healthy individuals. Oral Health Prev Dent. 2020;18(1):563–570. doi: 10.3290/j.ohpd.a44693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jeong D., Kim D.H., Song K.Y., Seo K.H. Antimicrobial and anti-biofilm activities of Lactobacillus kefiranofaciens DD2 against oral pathogens. J Oral Microbiol. 2018;10(1) doi: 10.1080/20002297.2018.1472985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xia B., Yu J., He T., Liu X., Su J., Wang M., et al. Lactobacillus johnsonii L531 ameliorates enteritis via elimination of damaged mitochondria and suppression of SQSTM1-dependent mitophagy in a Salmonella infantis model of piglet diarrhea. Faseb j. 2020;34(2):2821–2839. doi: 10.1096/fj.201901445RRR. [DOI] [PubMed] [Google Scholar]
- 63.Lin X., Chen X., Chen Y., Jiang W., Chen H. The effect of five probiotic lactobacilli strains on the growth and biofilm formation of Streptococcus mutans. Oral Dis. 2015;21(1):e128–e134. doi: 10.1111/odi.12257. [DOI] [PubMed] [Google Scholar]
- 64.Teanpaisan R., Piwat S., Dahlén G. Inhibitory effect of oral Lactobacillus against oral pathogens. Lett Appl Microbiol. 2011;53(4):452–459. doi: 10.1111/j.1472-765X.2011.03132.x. [DOI] [PubMed] [Google Scholar]
- 65.Soares L.G., Carvalho E.B., Tinoco E.M.B. Clinical effect of Lactobacillus on the treatment of severe periodontitis and halitosis: a double-blinded, placebo-controlled, randomized clinical trial. Am J Dent. 2019;32(1):9–13. [PubMed] [Google Scholar]
- 66.Cheon M.J., Lim S.M., Lee N.K., Paik H.D. Probiotic properties and neuroprotective effects of lactobacillus Buchneri KU200793 isolated from Korean fermented foods. Int J Mol Sci. 2020;21(4) doi: 10.3390/ijms21041227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baumgartner R., Forteza M.J. Ketelhuth DFJ. The interplay between cytokines and the Kynurenine pathway in inflammation and atherosclerosis. Cytokine. 2019;122 doi: 10.1016/j.cyto.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 68.Murakami Y., Hoshi M., Imamura Y., Arioka Y., Yamamoto Y., Saito K. Remarkable role of indoleamine 2,3-dioxygenase and tryptophan metabolites in infectious diseases: potential role in macrophage-mediated inflammatory diseases. Mediators Inflamm. 2013;2013 doi: 10.1155/2013/391984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jenabian M.A., El-Far M., Vyboh K., Kema I., Costiniuk C.T., Thomas R., et al. Immunosuppressive tryptophan catabolism and gut mucosal dysfunction following early HIV infection. J Infect Dis. 2015;212(3):355–366. doi: 10.1093/infdis/jiv037. [DOI] [PubMed] [Google Scholar]
- 70.Günther J., Däbritz J., Wirthgen E. Limitations and off-target effects of tryptophan-related IDO inhibitors in cancer treatment. Front Immunol. 2019;10(1801) doi: 10.3389/fimmu.2019.01801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ameziane-El-Hassani R., Talbot M., de Souza Dos Santos M.C., Al Ghuzlan A., Hartl D., Bidart J.M., et al. NADPH oxidase DUOX1 promotes long-term persistence of oxidative stress after an exposure to irradiation. Proc Natl Acad Sci U S A. 2015;112(16):5051–5056. doi: 10.1073/pnas.1420707112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pertwee R.G. The diverse CB1 and CB2 receptor pharmacology of three plant cannabinoids: delta9-tetrahydrocannabinol, cannabidiol and delta9-tetrahydrocannabivarin. Br J Pharmacol. 2008;153(2):199–215. doi: 10.1038/sj.bjp.0707442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kozono S., Matsuyama T., Biwasa K.K., Kawahara K., Nakajima Y., Yoshimoto T., et al. Involvement of the endocannabinoid system in periodontal healing. Biochem Biophys Res Commun. 2010;394(4):928–933. doi: 10.1016/j.bbrc.2010.03.080. [DOI] [PubMed] [Google Scholar]
- 74.Costiniuk C.T., Jenabian M.A. Cannabinoids and inflammation: implications for people living with HIV. Aids. 2019;33(15):2273–2288. doi: 10.1097/QAD.0000000000002345. [DOI] [PubMed] [Google Scholar]
- 75.Taleb S. Tryptophan dietary impacts gut barrier and metabolic diseases. Front Immunol. 2019;10:2113. doi: 10.3389/fimmu.2019.02113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Laurans L., Venteclef N., Haddad Y., Chajadine M., Alzaid F., Metghalchi S., et al. Genetic deficiency of indoleamine 2,3-dioxygenase promotes gut microbiota-mediated metabolic health. Nat Med. 2018;24(8):1113–1120. doi: 10.1038/s41591-018-0060-4. [DOI] [PubMed] [Google Scholar]
- 77.Baumler A.J., Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535(7610):85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pilchova T., Pilet M.F., Cappelier J.M., Pazlarova J., Tresse O. Protective effect of Carnobacterium spp. against listeria monocytogenes during host cell invasion using in vitro HT29 model. Front Cell Infect Microbiol. 2016;6:88. doi: 10.3389/fcimb.2016.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Danielski G.M., Imazaki P.H., Andrade Cavalari C.M., Daube G., Clinquart A., Macedo R.E.F. Carnobacterium maltaromaticum as bioprotective culture in vitro and in cooked ham. Meat Sci. 2020;162 doi: 10.1016/j.meatsci.2019.108035. [DOI] [PubMed] [Google Scholar]
- 80.Pedrozo H.A., Dallagnol A.M., Vignolo G.M., Pucciarelli A.B., Schvezov C.E. Mechanistically inspired kinetic approach to describe interactions during co-culture growth of carnobacterium maltaromaticum and listeria monocytogenes. J Food Sci. 2019;84(9):2592–2602. doi: 10.1111/1750-3841.14754. [DOI] [PubMed] [Google Scholar]
- 81.Wiernasz N., Leroi F., Chevalier F., Cornet J., Cardinal M., Rohloff J., et al. Salmon gravlax biopreservation with lactic acid bacteria: a polyphasic approach to assessing the impact on organoleptic properties, microbial ecosystem and volatilome composition. Front Microbiol. 2019;10:3103. doi: 10.3389/fmicb.2019.03103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shang L., Deng D., Buskermolen J.K., Janus M.M., Krom B.P., Roffel S., et al. Multi-species oral biofilm promotes reconstructed human gingiva epithelial barrier function. Sci Rep. 2018;8(1):16061. doi: 10.1038/s41598-018-34390-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Griffen A.L., Thompson Z.A., Beall C.J., Lilly E.A., Granada C., Treas K.D., et al. Significant effect of HIV/HAART on oral microbiota using multivariate analysis. Sci Rep. 2019;9(1):19946. doi: 10.1038/s41598-019-55703-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang F., He S., Jin J., Dong G., Wu H. Exploring salivary microbiota in AIDS patients with different periodontal statuses using 454 GS-FLX Titanium pyrosequencing. Front Cell Infect Microbiol. 2015;5:55. doi: 10.3389/fcimb.2015.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gonzalez O.A., Li M., Ebersole J.L., Huang C.B. HIV-1 reactivation induced by the periodontal pathogens Fusobacterium nucleatum and Porphyromonas gingivalis involves Toll-like receptor 2 [corrected] and 9 activation in monocytes/macrophages. Clin Vaccine Immunol. 2010;17(9):1417–1427. doi: 10.1128/CVI.00009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khalaf H., Nakka S.S., Sanden C., Svard A., Hultenby K., Scherbak N., et al. Antibacterial effects of Lactobacillus and bacteriocin PLNC8 alphabeta on the periodontal pathogen Porphyromonas gingivalis. BMC Microbiol. 2016;16(1):188. doi: 10.1186/s12866-016-0810-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hanna D.B., Felsen U.R., Ginsberg M.S., Zingman B.S., Beil R.S., Futterman D.C., et al. Increased antiretroviral therapy use and virologic suppression in the bronx in the context of multiple HIV prevention strategies. AIDS Res Hum Retroviruses. 2016;32(10–11):955–963. doi: 10.1089/aid.2015.0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mujugira A., Celum C., Tappero J.W., Ronald A., Mugo N., Baeten J.M. Younger age predicts failure to achieve viral suppression and virologic rebound among HIV-1-infected persons in serodiscordant partnerships. AIDS Res Hum Retroviruses. 2016;32(2):148–154. doi: 10.1089/aid.2015.0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Costiniuk C.T., Saneei Z., Routy J.P., Margolese S., Mandarino E., Singer J., et al. Oral cannabinoids in people living with HIV on effective antiretroviral therapy: CTN PT028-study protocol for a pilot randomised trial to assess safety, tolerability and effect on immune activation. BMJ Open. 2019;9(1) doi: 10.1136/bmjopen-2018-024793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.