Abstract
Whole slide imaging (WSI), an important technique in the field of digital pathology, has recently been the subject of increased interest and avenues for utilization; and with more widespread WSI utilization, there will also be increased interest in and implementation of image analysis techniques. Image analysis includes artificial intelligence (AI) and targeted or hypothesis-driven algorithms. In the overall pathology field, citations related to these topics have increased in recent years. Renal pathology is one anatomic pathology subspecialty that has utilized WSIs and image analysis algorithms; and it can be argued that renal transplant pathology could be particularly suited for WSI and image analysis, since renal transplant pathology is frequently classified using the semiquantitative Banff Classification of Renal Allograft Pathology. Hypothesis-driven/targeted algorithms have been used in the past for the assessment of a variety of features in the kidney (e.g., interstitial fibrosis and tubular atrophy and inflammation); and in recent years, research has particularly increased in the area of AI/machine learning for the identification of glomeruli, for histologic segmentation, and other applications. Deep learning is the form of machine learning most often used for such AI approaches to the “big data” of pathology WSIs, and deep learning methods such as artificial neural networks (ANNs)/convolutional neural networks (CNNs) are utilized. Unsupervised and supervised AI algorithms can be employed to accomplish image or semantic classification. In this review, AI and other image analysis algorithms applied to WSIs are discussed; and examples from renal pathology are covered, with an emphasis on renal transplant pathology.
Keywords: Digital Pathology, Artificial Intelligence, Machine Learning, Image Analysis, Renal Transplant Pathology
Introduction
Background
Computational techniques for the analysis of pathology material have expanded over the past decades, and this is evidenced by a general trend toward an increase in publications per year in PubMed using a variety of search terms (Figures 1 and 2 and Table 1). The Digital Pathology Association has defined “digital pathology” as “tools and systems to digitize pathology slides and associated meta-data, their storage, review, analysis, and enabling infrastructure”1; and “digital pathology” is sometimes considered a topic in the larger field of “computational pathology” 1, 2. However, broader definitions of “digital pathology” are sometimes used to include any number of computational techniques applied to pathology, particularly anatomic pathology, including whole slide imaging (WSI), algorithms for dedicated morphometric analysis, algorithms employing artificial intelligence (AI)/machine learning, natural language processing (NLP), and computerized processing of data from novel microscopic techniques (e.g., Fourier–transform infrared [FTIR] and other IR, multispectral imaging, and second harmonic generation microscopy) 3–7. Using this broader definition of “digital pathology” does bring about some overlap with the term “computational pathology”; however, it can be posited that an inclusive definition of “digital pathology” does have some advantages. This and other key definitions are shown in Table 2. Definitions in the table and throughout this paper are based on our own experience, expert group publications 1, 8, and useful reviews 9–16; however, we recognize that variable definitions are provided and used in other publications and are likely in flux.
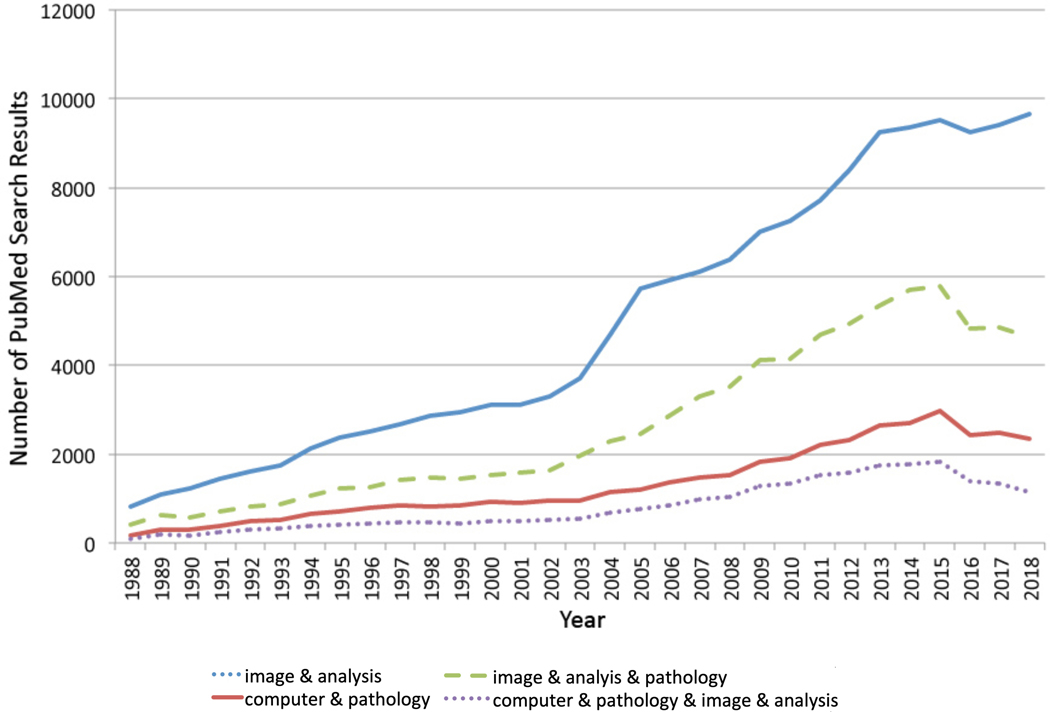
Figure 1:
Publications per year based on the PubMed search terms specified below are shown. The “&” in the figure key designates that the “AND” Boolean operator used to combine the specified terms in the PubMed Advanced Search Builder.
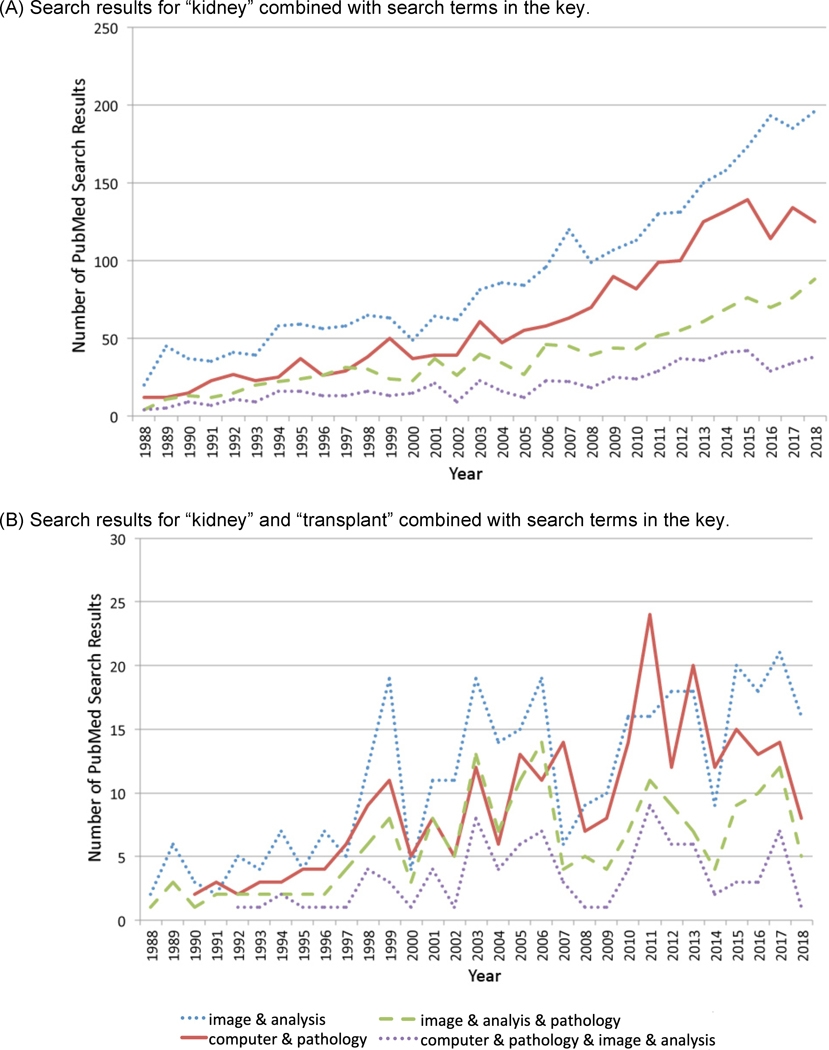
Figure 2:
Publications per year pertaining to the kidney and kidney transplantation based on the PubMed search terms specified below are shown. The “&” in the figure key designates that the “AND” Boolean operator used to combine the specified terms in the PubMed Advanced Search Builder.
Table 1:
PubMed publications for combinations of the specified search terms are available as specified below for the years 1988–2018. The number of publications are graphed in Figure 1 (italics) and Figure 2 (bold).
| PubMed search terms Base | Number of Search Results | ||
|---|---|---|---|
| Only base terms on left | Terms on left & “kidney” | & “kidney & “transplant” | |
| “image” & “analysis” | 167,828 | 3,108 | 369 |
| “image” & “analysis” & “pathology” | 44,005 | 1,254 | 188 |
| “computer” & “pathology” | 88,603 | 2,108 | 290 |
| “computer” & “pathology” & “image” & “analysis” | 26,389 | 653 | 94 |
| “algorithm” & “pathology” | 40,775 | 1,331 | 258 |
| “algorithm” & “image” & “pathology” | 12,945 | 252 | 23 |
| “artificial” & “intelligence” | 31,663 | 51 | 8 |
| “digital” & “pathology” | 21,784 | 572 | 117 |
| “digital” & “pathology” & “image” & “analysis” | 3,625 | 96 | 27 |
| “whole” & “slide” & “image” | 876 | 33 | 12 |
The numbers are based on PubMed searches on April 8, 2020.
“&” above designates the “AND” Boolean operator used in the PubMed Advanced Search Builder.
Table 2:
Definitions for common image analysis topics are shown.
| Term | Definition |
|---|---|
| Algorithm | Set of actions to solve a specific problem defined in a mathematical formula and/or conducted by computer program commands or humans (e.g., in care pathways) |
| Artificial intelligence (AI) | Computer science field dealing with programming techniques imparting “intelligent” human behavior and abilities to computers (e.g., data analysis, prediction, decision, and the simulation of other activities) |
| Artificial neural networks (ANNs) | Computer-based analysis relying on the determination of the optimal combination of multiple forms of interconnected data manipulation similar to the structural connections of human brain neurons |
| Computational pathology | Computer utilization for multiparametric biologic data analysis and prediction, frequently with the goal of improving healthcare |
| Computer vision | Computer science area investigating how high-level understanding of digital images can be obtained by computers |
| Convolutional neural networks (CNNs) | Artificial neural network type frequently applied to image analysis (e.g., medical image recognition) and natural language processing |
| Deep learning | Machine learning form in which a computer “learns” the best combination of data manipulation procedures defined by artificial neural networks applied to representative data |
| Digital Pathology* | Computational techniques applied to pathology, particularly anatomic pathology, including WSI and novel microscopy, algorithms with dedicated morphometric analysis, artificial intelligence (AI)/machine learning, or natural language processing (NLP) |
| Machine learning | AI branch in which data analysis can be performed and computer programs “learn” to perform tasks or prediction through pattern identification via exposure to representative data (input features and output data labels) |
| Pixel | Two-dimensional (2D) unit, which when assembled into a matrix, composes a digital image (short for “picture element”), as opposed to a voxel, which is the basic 3D unit composing a digital volume |
| Recurrent neural networks (RNNs) | ANN alternative that inter-relates all inputs, as opposed to ANNs, which typically processes inputs independently. Often used when dealing with temporal dynamic data. |
| Supervised (versus Unsupervised) Learning | Supervised learning by an algorithm involves exposure to representative data and labeled/tagged/classified responses as opposed to unsupervised learning, which involves learning by an algorithm from examples without any associated label |
| Whole slide imaging (WSI) | Image storage method that allows the acquisition and storage of the entire tissue sample on a histology slide (as opposed to “static” images of single fields of view) |
This definition of “digital pathology” is somewhat inclusive and overlaps with “computational pathology”, as discussed in the text.
2D: Two-dimensional, 3D: Three-dimensional, AI: artificial intelligence, ANN: Artificial neural network, CNN: Convolutional neural network, RNN: Recurrent neural network, WSI: whole slide image/imaging
The aim of this publication is to provide an introduction to these topics, particularly with regard to image analysis; and examples will be provided primarily from the area of kidney (renal) transplant pathology, which is a field that has been the focus of much of our work and is an area that we believe presents unique opportunities for the application of computerized image analysis. With an apology to groups working in this field that may have been missed, we have mainly provided examples from our group and groups we have encountered in our work in this domain.
Algorithm Types
For the purposes of this discussion, algorithms are categorized into “hypothesis-driven” or “targeted” algorithms and artificial intelligence (AI) algorithms, which can be considered more data-driven (Figure 3); and a brief discussion is included below, followed by examples of each.
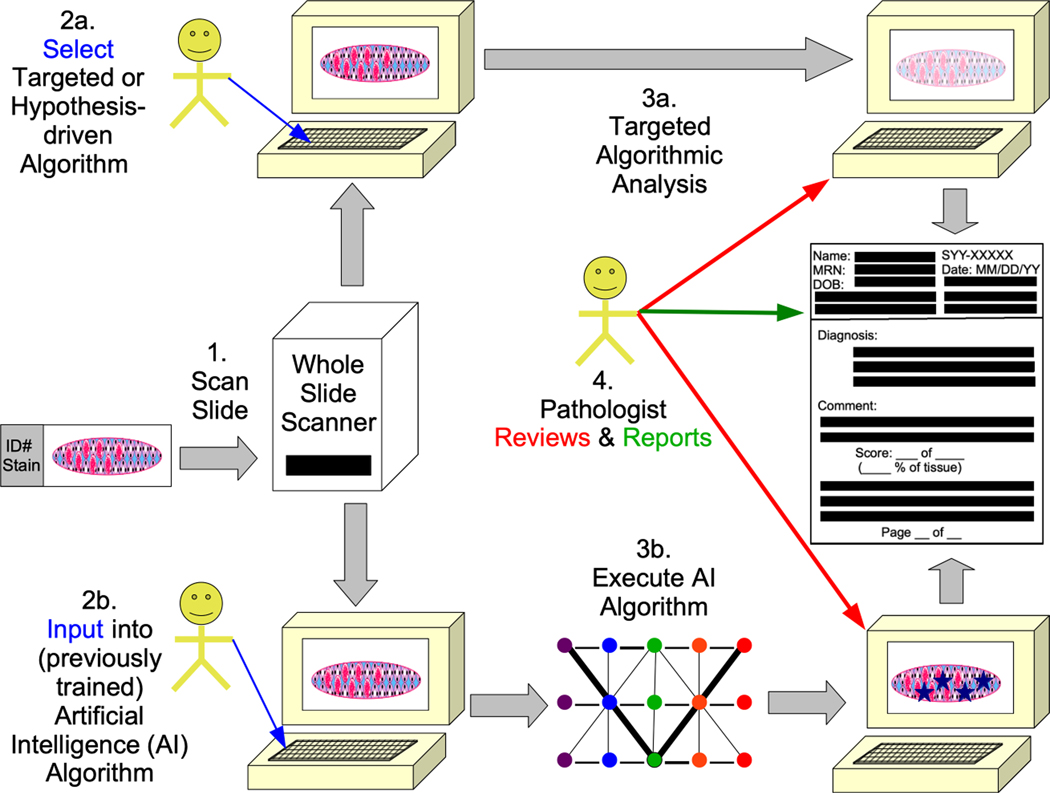
Figure 3:
A summary of the application digital pathology is shown in different forms of whole slide image (WSI) analysis (based in part on a prior publication from our group 21 and others 9, 10. Slides are scanned into WSIs (1). In a targeted or hypothesis-driven algorithmic approach (2a), specific algorithms are run (3a). When artificial intelligence (AI) algorithms are used (3b), the previously trained AI algorithms (e.g., neural networks with differentially weighted nodes and connections) are executed on the image (3b). For both targeted and AI, pathologists (or other trained individuals) review the results in some manner and eventually report the results for patient care or research.
Hypothesis-driven/Targeted Algorithms
“Hypothesis-driven” or “targeted” algorithms rely on relatively simple computer instructions (that is, simple compared to AI algorithms) programmed to perform set tasks or mathematical calculations; and in the field of digital pathology, these algorithms can be used to analyze WSIs. “Targeted” algorithms are also referred to as “handcrafted” algorithms or “real” intelligence algorithms because they are intuitively devised using a hypothesis-driven approach based on prior knowledge of the target morphology, disease mechanisms, and/or pathogenesis.
The positive pixel count algorithm (PPC) is probably the simplest type of “targeted” algorithm that essentially “counts” image pixels (the smallest divisible unit of a digital image) considered “positive” with regard to certain human-defined color/hue parameters; and “negative” pixels are also tabulated, allowing the determination of the percent of positive pixels in a given image. In this manner, the PPC algorithm can be used to assess features such as renal interstitial fibrosis (Figure 4). PPC can be applied to any image, but typically works best when applied to special histologic stains such as trichrome, Sirius red, or collagen immunohistochemistry for interstitial fibrosis 17–19. PPC can find uses outside of renal images, examples include its application to measure parameters such as steatosis in the liver on routine histologic stains 20 and extent and intensity of immunohistochemistry staining 21.
Figure 4:
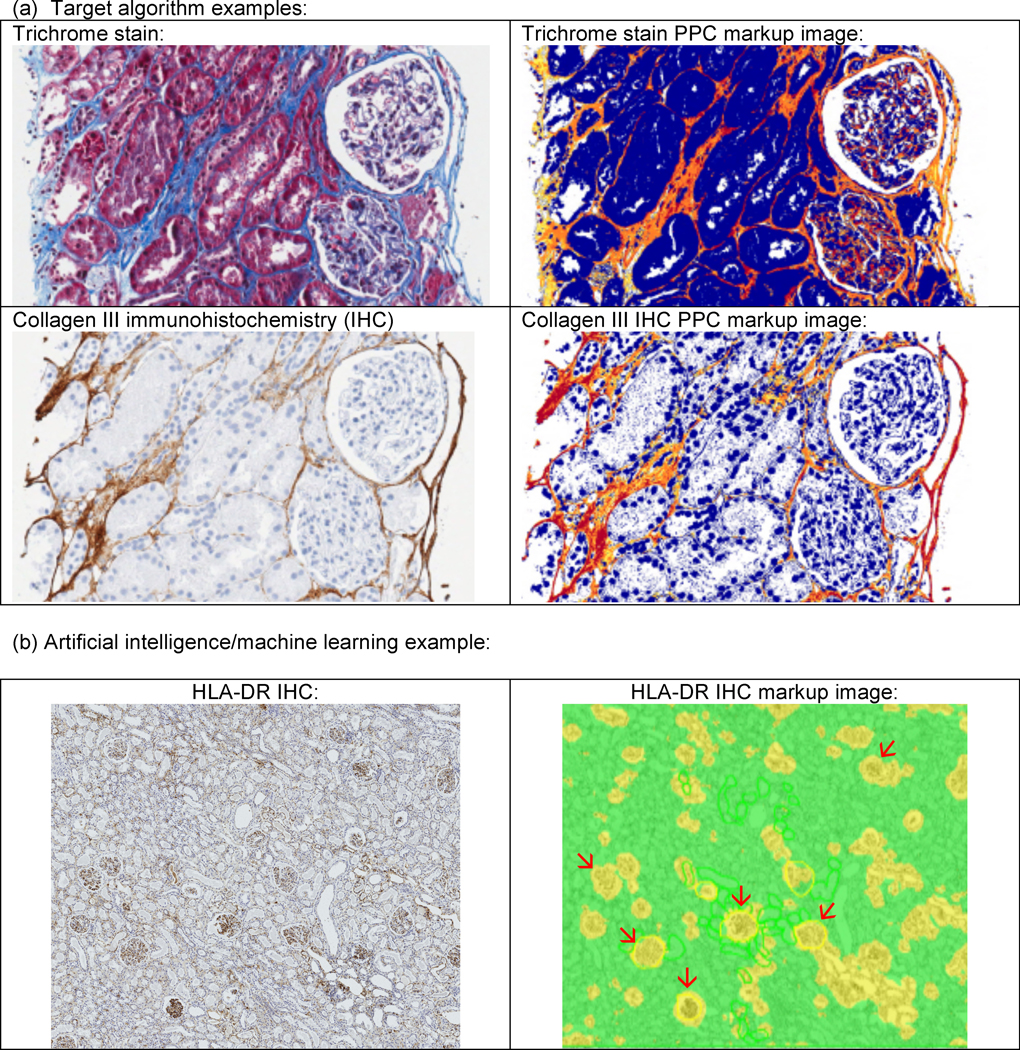
Examples of image analysis of the kidney are shown. In the upper panels (a), examples of a positive pixel count (PPC) algorithm to detected fibrous areas on trichrome and collagen III immunohistochemistry (IHC) are depicted. In the markup images showing the algorithm analysis depicted on the right, tissue considered “positive” is marked up as yellow, orange, or red, in that order with increasing positivity of match to the algorithm parameters. In the lower panel (b), an example of glomerular detection conducted on Human Leukocyte Antigen (HLA)-DR IHC using the Leica/Aperio GENIE algorithm is shown. In the markup images showing the algorithm analysis depicted on the right, areas classified as glomeruli by the algorithm are depicted in yellow; and selected glomeruli in the field are pointed out with red arrows. It can be appreciated that some smaller yellow areas amidst the remaining renal parenchyma (in green) do not represent glomeruli, showing that additional algorithm training is needed.
More complex targeted algorithms have been developed for a variety of histologic features composed of multiple pixels that in aggregate form a histologic object / feature considered important or of interest to an anatomist, pathologist, or other researcher or clinician. These include algorithms for cell counting; and these can be utilized to assess a variety of cell types, such as interstitial inflammation 22. Algorithms have also been developed to detect other histologic parameters such as the microvasculature, allowing assessment of microvessel size, density, and other parameters 19.
Data-Driven/Artificial Intelligence Algorithms
AI algorithms are effectively data-driven, since they don’t necessarily require pathologists or other users to choose particular hypothesis-driven steps for analysis. AI can be used for a variety of specimens to achieve a number of goals. For example, AI can be used for automated tumor detection and grading; immunohistochemistry scoring; predicting mutation status; and other diagnostic, prognostic, and theranostic support 23–25. Deep learning is a major AI method used for pathology images. Deep learning is a form of machine learning; and in turn, machine learning is a branch of AI 26–31. In the learning process and in subsequent application, machine learning can process large quantities of data, thus exhibiting applicability to “big data”; and in contrast to targeted or hypothesis-driven algorithms, the need for “big data” more acutely applies to data-driven algorithms such as machine learning 29, 31, 32.
When applying deep learning methods in pathology, artificial neural networks (ANNs) can be used to tackle a wide variety of problems. The concept of ANNs has been around for several decades 26–30. ANNs allow “learning” by computers in a process that loosely recapitulates the structure of neurons in the human brain. Multiple forms of data manipulation are applied to the input data (digital images for the purposes of most of this article), and the best possible combination of data manipulation steps (essentially the neuronal connections) is determined through the process of “back propagation” in which the neuronal connections are given preferential weight based on their ability to produce optimal performance output. Convolutional neural networks (CNNs) are an ANN type frequently applied to image analysis such as medical image recognition and natural language processing 26–30.
AI algorithms, in the realm of images, can be roughly broken down into unsupervised and supervised learning algorithms. Unsupervised learning only requires the image to be provided, and the model’s goal is to find relationships between the images based on the image content alone. More common in pathology research is the use of supervised learning. In this approach it is critical to have expertly labeled images, with the labels being defined by the question being asked. In renal pathology these labels could be a binary / categorical label, like whether or not a glomerulus is present in an image (e.g., a field of view in a WSI). These categorical type labels are associated with classification AI models. In contrast, the label could be the delineation of the glomerular boundary, and the model would be aimed at predicting which pixels lie within or outside glomeruli (also known as, semantic segmentation or pixel-level classification). Supervised learning algorithms are more commonly used in pathology today, and a big part of this process is the generation of expertly labeled datasets to train the models. Various tools are available to annotate WSIs, and some tools are also available to manage large annotation projects with multiple annotators. One example is the Digital Slide Archive (DSA) with the HistomicsTK web interface, a resource developed with contributions from members of our group and used in our current projects. Some of these tools allow the utilization of complex AI algorithms that combine different type of data and metadata sources (such as images, structured data [demographics, laboratory values, genetics, etc.], textual data [sometimes through natural language processing (NLP), etc.] in order to predict a diagnosis or outcome 32–36.
AI with regard to digital pathology can also be categorized as either image classification or semantic classification. Image classification is useful when high-level classification is needed without the absolute requirement for interpretability. In semantic segmentation, each pixel is assigned a class. This is most suitable for problems where differentiation between various objects of the same class is not necessary. Some methods combine object detection, classification, and segmentation and assign each pixel a class and object identification. Recent object detection +/− classification +/− segmentation deep learning models include Faster R-CNN and Mask R-CNN. These are useful when individual objects tend to be close together 32, 37, 38.
Kidney Transplant Pathology Examples
It can be postulated that the kidney is uniquely positioned as a fertile area for the application of image analysis and artificial intelligence because quantitative data is often included in kidney biopsy reports (e.g., the number of total glomeruli, the number/proportion of sclerotic glomeruli, the extent of tubulointerstitial and vascular scarring, etc.) 9. This is particularly true in the area of renal transplant pathology, where the Banff classification of allograft pathology is often applied. The Banff classification includes semiquantitative scores for various histologic features (e.g., tubulitis [t], endarteritis [v], interstitial inflammation [i, ti, i-IFTA], etc.) that are assembled to reach a diagnosis, most notably the absence/presence of allograft rejection 39, 40. As in the overall fields of medicine and pathology, there has been a general trend toward an increasing number of publications per year (Figure 2 and Table 1) when the PubMed search terms “kidney” and “transplant” are combined with the terms shown in Figure 1. Thus algorithms can potentially quite useful in aiding the interpretation of renal allograft specimens. Even without algorithms, morphometric assessment of Banff histologic features can be directly performed on WSIs, as has been done by a group at the Mayo Clinic in a method they term “computer-assisted morphometics” (CAM) 41. A Banff Digital Pathology Working Group (DPWG) was recently formed to explore the use of digital pathology techniques in transplant pathology; therefore, interest in AI and other algorithms in renal transplant pathology will likely increase in the future 42.
Hypothesis-driven/Targeted Algorithms
Hypothesis-driven/targeted image analysis algorithms have been used to assess a number of parameters in the kidney. For example, renal interstitial fibrosis has been quantitated using computational algorithms devised by a number of groups 17–19, 43–62 (Table 2 and Figure 4). In general, these prior studies have used PPC or thresholding-type algorithms to obtain the area of tissue involved by fibrosis. Individuals at Stanford University and colleagues have investigated the utility of image analysis in the assessment of fibrosis and have correlated these findings with the influence of immunosuppression and have primarily utilized Sirius red staining in these studies 52, 53, 63, 64. Computerized quantitation has been conducted on other stains, including trichrome stains by groups from Barcelona 57, 65 and France 45–47, 66, 67.
Our group has examined the utility of PPC algorithms in the quantitation of fibrosis on trichrome and collagen III immunohistochemistry 17–19 and Sirius red 18. A Banff working group on renal interstitial fibrosis assessment showed a great deal of intraobserver variability amongst an international group of pathologists with regard to standard practices and the quantitation of interstitial fibrosis; however, this working group did show some promise in the use of the collagen III immunohistochemistry image analysis used by our group for the assessment of fibrosis 17. In addition to interstitial fibrosis, targeted algorithms have also been used by our group and others to assess microvessel density 19, inflammation 22, and other features.
In a targeted image analysis pipeline, more complex structures can also be assessed. In particular, glomeruli have been detected by using computer vision techniques. For example, a group primarily at the University of Buffalo detected glomeruli boundaries with a computational pipeline consisting of Gabor filtering, Gaussian blurring, F-testing, and other algorithmic steps 68.
Targeted algorithms can be used for digital 3-dimensional (3D) reconstruction of anatomic features. For example, arteries and other anatomic features can be reconstructed in 3D using histological slides containing sequential serial sections. In such an approach, one group examined chronic allograft vasculopathy in a heart transplantation model using “virtual coronary arteriography”, and other disorders can also likely be structurally examined using similar methods 69, 70.
Multiplex immunostaining can be used in conjunction with image analysis to characterize various cells, particularly immune cells 14, 71–75 and use automated image analysis to perform “-omics”-type assessment of tissue 73, 74. This multiplex immunostaining is similar to methods being used for cancer research 76, 77.
Artificial Intelligence Algorithms
Glomerular detection has been the focus of much of the initial AI work directed toward kidney histology specimens. Commercial algorithms are available that can be trained to do this work. For example, we conducted preliminary studies on glomerular detection in our group using the Leica Aperio GENIE algorithm (Figure 4, previously unpublished) 30. However, many of the published methods for glomerular detection have utilized data analysis pipelines and algorithms refined by their own groups, as discussed below.
A group in Japan proposed a novel image descriptor – rectangular histogram of oriented gradients (Segmental HOG) – and used it to train a support vector machine (SVM) model to classify desmin-stained images with and without glomeruli 78. The University of Buffalo group used local binary pattern (LBP) features to train a SVM model 79. Convolutional neural networks (CNNs) have also been used to detect glomerular versus non-glomerular tissue in PAS-stained slides by a group primarily in Spain 80 and trichrome-stained slides by a group at Boston University 81 and a group of collaborators from the Medical College of Wisconsin, Wake Forest University, and the University of Michigan 82. Another CNN model was used by a group at Washington University for the identification of sclerotic and nonsclerotic glomeruli on frozen section slides, suggesting that it could have utility in the determination of suitability of donor kidneys for transplantation 83.
The University of Buffalo group and their collaborators have established what they term a “human-in-the-loop” or “Human AI Loop (H-AI-L)” approach. This method focuses on an iterative process of annotating ground truth for CNN segmentation problems, training a model, predicting on new images, correcting prediction, and adding newer predictions to training dataset. Using this approach they reduce the annotation burden required of pathologists, and they used this approach to establish an AI pipeline for the segmentation of the kidney 84. Using these methods, this group also recapitulated the classification of diabetic glomerulosclerosis using a scalable AI pipeline85. This is similar to other ways to interactively improve algorithm performance being developed by our group in which uncertainty in the algorithm analysis provides the user with images the algorithm is most unsure about for correction. Such an active learning method provides a ways for humans can correct algorithmic errors 31.
Deep learning has been utilized for the assessment of kidney specimens, employing multi-class semantic segmentation of periodic acid-Schiff (PAS)-stained kidney tissue sections by collaborators from Radboud University Medical Center (UMC) in The Netherlands, Amsterdam UMC, Massachusetts General Hospital/Massachusetts Institute of Technology/Harvard University, the Mayo Clinic, and Linköping University in Sweden; and this provided quantitative data that could potentially be useful in research on kidney disease. In this study, CNN-based lesion quantification correlated well with semi-quantitative scoring by renal pathologists with minimal interobserver variability observed 86. Segmentation has also been explored by another group, who used 20 deep learning-based methods to identify glomeruli, tubules, arteries, arterioles, and peritubular capillaries using hematoxylin and eosin, PAS, trichrome, and silver stains in the multi-institutional NEPTUNE study. They found that PAS-stained whole slide images yielded the best concordance between pathologists and deep learning segmentation across all structures 87.
Interstitial fibrosis has been specifically quantitated using AI-based approaches as well. For example, Kolachalama et al at Boston University have established associations with AI detection of pathological fibrosis with renal survival using the GoogLeNet Inception deep learning model, deployed with the TensorFlow program 88.
Discussion
AI can be a powerful tool in pathology, particularly renal transplantation pathology, as highlighted in this publication. In light of applications similar to those highlighted here, “augmented intelligence” has been suggested as a better term for the “AI” acronym than “artificial intelligence” 89, 90 since it is hoped that image analysis and other digital pathology techniques can help “augment” the skills and work of pathologists and other medical professionals. Rather than replacing pathologists, the hope is that these techniques will be complementary to human work rather than replacing it; and it is conceivable that these techniques will even enhance current approaches toward medical problems.
In the realm of transplantation, as mentioned previously, the Banff Digital Pathology Working Group has laid forth goals for more widespread implementation of digital pathology as well as standardization of digital pathology efforts. One goal of this effort is the establishment of a WSI bank (optimally including clinical and ground truth annotations) so that different groups working on AI and other digital image analysis algorithms can compare the performance of their algorithms to other groups 42 similar to the CAMELYON challenges that tested the ability of multiple groups to detect lymph node metastasis in breast cancer 91, 92.
Investment in digital pathology technologies can be costly; however, with the benefits afforded by digital pathology workflow improvements, it has been shown that such investment can be cost effective 93, 94. Some WSI experts have advocated for joint radiology/pathology efforts to host the large images that WSI scanners produce because both pathology and radiology are subspecialties that frequently deal with image-intensive data 95–98. Ultimately, WSI image analysis using targeted and AI algorithms can be incorporated into data-rich, quantitative reports (Figure 4) that provide decision support to improve patient care 21.
Computational techniques are likely to gain more widespread use in the future, as evidenced by our search of the literature, which showed an increase in citations in recent years (Table 1 and Figures 1–2). The utility of digital techniques has been highlighted in unique ways during the recent COVID-19 pandemic, since remote work and other adaptations have been permitted; and it is likely that digital pathology, AI, and other techniques will help address future threats to clinical services, research, and education 99–103. We highlight transplant renal pathology as an area of particular interest for these techniques; however, it is likely that they will extend to essentially pathology subspecialties to at least some degree. Regulatory hurdles may need to be overcome for widespread application of AI algorithms, addressing consensus recommendations and legal concerns (e.g., with the European Union [EU], the College of American Pathologists [CAP], and the Food and Drug Administration [FDA]); and at the current time AI algorithms are mostly for “research use only”, particularly in nephropathology 16. As these issues are addressed and as the field moves forward, we foresee exciting possibilities for pathologists, clinicians, and ultimately patients, who are likely to receive enhanced care through precision medicine-based approaches.
Table 3:
Examples of image analysis studies primarily from the field of renal transplantation employing either hypothesis-driven/targeted or artificial intelligence (AI)/machine learning algorithms are shown.
| Parameter(s) Assessed | Material/Stain(s) Assessed | Description | Ref.(s) |
|---|---|---|---|
| Hypothesis-Driven or Targeted Algorithms: | |||
| IF | TC (Masson), SR, and SMA IHC | IF IA on allograft biopsies correlated with GFR and urine total protein | 43 |
| IF | TC (Masson) | IF IA correlates with serum Cr in IgA nephropathy and membranoproliferative glomerulonephritis (MPGN) | 44 |
| IF | TC (Masson) | IF IA in renal allograft patients receiving cyclosporine correlated with worsened Cr | 45 |
| IF | TC (Light Green) | IF IA in renal allograft patients randomized to cyclosporine or conversion to sirolimus | 46 |
| IF | TC (Light green) | Quantitative IF in sequential renal allograft renal biopsies correlated with eGFR | 47 |
| IF | SR and collagen | Renal IF correlates with presence of TGF-β, decorin, SMA, and interstitial collagens | 48–51 |
| IF | SR | SR IA predicted long-term renal allograft function and time to graft failure | 52 |
| IF | SR | SR IA predicted long-term renal allograft function (decreased GFR) | 53 |
| IF | SR | IF was not significantly different between non-heart-beating and conventional heart-beating donor kidneys | 54 |
| IF | SR | IF scoring predicts survival and Cr in lupus nephritis | 55 |
| IF | SR | IA-based application (Fibrosis HR) for IF and glomerular morphometry | 56 |
| IF | SR | IF measurements using digital imaging coupled with point counting correlated with GFR | 57 |
| IF | SR | SR IF measurement combined with ultrasound measurements of renal artery resistance index helped predict “chronic allograft nephropathy” correlated with decreased GFR | 58 |
| IF | CIII IHC | IF by a semiautomatic system correlate with GFR in protocol renal transplant biopsies | 59 |
| IF | CIII IHC | IF measurements by a semiautomatic system correlate with GFR in protocol renal transplant biopsies | 60 |
| IF | TC (Masson) | IF IA and VA of cyclosporine (CsA) therapy effects | 61, 62 |
| IF | CIII IHC, TC, and SR | CIII IHC, TC, SR IA, and GFR correlated with each other and with VA | 18 |
| IF | TC, PAS, & IHC for CIII & CD34 | Renal cortical and medullary IF, epithelial area, & microvessel density were correlated using IA and VA | 19 |
| Gloms | H&E, TC, PAS, Congo red, & Jones silver | Gabor filtering, Gaussian blurring, and statistical-based and other algorithmic steps were used to segment gloms in various stains | 68 |
| Artificial intelligence (AI)/machine learning algorithms | |||
| Gloms | H&E | Local binary pattern (LBP) support vector machine (SVM)-based glom detection | 79 |
| Gloms | Desmin IHC | Rectangular histogram of oriented gradients (Rectangular HOG) for glom detection | 78 |
| Gloms | Frozen H&E | Automated identification of sclerotic and nonsclerotic glomeruli using deep learning | 83 |
| Gloms | PAS | Diabetic glomerulosclerosis could be classified with CNNs | 85 |
| Gloms | PAS | CNN distinguished between Gloms & Non-Gloms | 80 |
| Gloms | TC | CNN segmentation of gloms | 81 |
| Gloms | TC | CNN localization of injured and noninjured gloms | 82 |
| Gloms, Tub, Int, Banff scoring | PAS | DL-based segmentation of Gloms, Tub, Int, other features, & Banff scores correlated with pathologist assessment | 86 |
| IF | TC | AI IF detection associates with renal survival using CNNs | 88 |
| Segmentation | H&E & PAS | “Human AI Loop (H-AI-L)” method decreased the annotation burden required of pathologists while still allowing for the AI-based segmentation of the kidney, prostate, & radiology data | 84 |
| Segmentation | H&E, PAS, TC, & Silver | DL segmentation of Gloms, Tub, arteries, arterioles, and peritubular capillaries | 87 |
AI: artificial Intelligence, CIII: collagen III, CNN: convolutional neural networks, Cr: creatinine, DL: deep learning, eGFR: estimated GFR, GFR: glomerular filtration rate, Glom/Gloms: glomerulus (glomerular)/glomeruli, H&E: hematoxylin and eosin, IHC: immunohistochemistry, IF: interstitial fibrosis, IA: image analysis, Int: interstitium, MPGN: membranoproliferative glomerulonephritis, PAS: periodic acid–Schiff, Ref(s): references, SMA: smooth muscle actin, SR: Sirius red, TC: trichrome, TGF-β: transforming growth factor, Tub: tubules, VA: visual analysis.
Acknowledgements
ABF, JV, MAM, LADC, DG, and JH (Principal Investigator) are supported in part by the National Institutes of Health (NIH) National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) 1R21DK122229–01 and an Emory University Synergy Grant. LADC and DG are supported in part by the NIH National Cancer Institute (NCI) grants U01CA220401 and U24CA19436201.
Abbreviations
- 3D
3-Dimensional
- AI
Artificial Intelligence
- ANN
Artificial Neural Network
- CNN
Convolutional Neural Network
- IA
Image Analysis
- IHC
Immunohistochemistry
- IF
Interstitial Fibrosis
- NLP
Natural Language Processing
- PAS
Periodic Acid-Schiff
- PPC
Positive Pixel Count
- SVM
Support Vector Machine
- WSI
Whole Slide Image/Imaging
Footnotes
Disclosure/Statement of Competing Financial Interests
The authors of this manuscript have no conflicts of interest to disclose as described by Histopathology. Commercial programs are mentioned in this publication only because they are common and/or our group has access to them, and their mention does not imply a specific endorsement of their use.
Conflict of interest: The authors declare no conflicts of interest related to this manuscript.
References
- 1.Abels E, Pantanowitz L, Aeffner F et al. Computational pathology definitions, best practices, and recommendations for regulatory guidance: A white paper from the digital pathology association. J Pathol 2019;249;286–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Feldman M, Carter AB et al. Computational pathology. Arch Pathol Lab Med 2015;140;41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fereidouni F, Griffin C, Todd A, Levenson R. Multispectral analysis tools can increase utility of rgb color images in histology. J Opt 2018;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torres R, Velazquez H, Chang JJ et al. Three-dimensional morphology by multiphoton microscopy with clearing in a model of cisplatin-induced ckd. J Am Soc Nephrol 2016;27;1102–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qian HS, Weldon SM, Matera D et al. Quantification and comparison of anti-fibrotic therapies by polarized srm and shg-based morphometry in rat uuo model. PLoS One 2016;11;e0156734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vuiblet V, Fere M, Gobinet C, Birembaut P, Piot O, Rieu P. Renal graft fibrosis and inflammation quantification by an automated fourier-transform infrared imaging technique. J Am Soc Nephrol 2016;27;2382–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varma VK, Kajdacsy-Balla A, Akkina SK, Setty S, Walsh MJ. A label-free approach by infrared spectroscopic imaging for interrogating the biochemistry of diabetic nephropathy progression. Kidney Int 2016;89;1153–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blue Ridge Academic Health Group. Separating fact from fiction: Recommendations for academic health centers on artificial and augmented intelligence Atlanta: Emory University, 2019. [Google Scholar]
- 9.Santo BA, Rosenberg AZ, Sarder P. Artificial intelligence driven next-generation renal histomorphometry. Curr Opin Nephrol Hypertens 2020;29;265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boor P. Artificial intelligence in nephropathology. Nat Rev Nephrol 2020;16;4–6. [DOI] [PubMed] [Google Scholar]
- 11.Roeder AH, Cunha A, Burl MC, Meyerowitz EM. A computational image analysis glossary for biologists. Development 2012;139;3071–3080. [DOI] [PubMed] [Google Scholar]
- 12.Deo RC. Machine learning in medicine. Circulation 2015;132;1920–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mueller JP, Massaron L. Artificial intelligence for dummies. Sebastopol, California: For Dummies/ O’Reilly Media, Inc., 2018. [Google Scholar]
- 14.Wood-Trageser MA, Lesniak AJ, Demetris AJ. Enhancing the value of histopathological assessment of allograft biopsy monitoring. Transplantation 2019;103;1306–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barisoni L, Lafata KJ, Hewitt SM, Madabhushi A, Balis UGJ. Digital pathology and computational image analysis in nephropathology. Nat Rev Nephrol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becker JU, Mayerich D, Padmanabhan M et al. Artificial intelligence and machine learning in nephropathology. Kidney Int 2020;98;65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farris AB, Chan S, Climenhaga J et al. Banff fibrosis study: Multicenter visual assessment and computerized analysis of interstitial fibrosis in kidney biopsies. Am J Transplant 2014;14;897–907. [DOI] [PubMed] [Google Scholar]
- 18.Farris AB, Adams CD, Brousaides N et al. Morphometric and visual evaluation of fibrosis in renal biopsies. J Am Soc Nephrol 2010;22;176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farris AB, Ellis CL, Rogers TE, Lawson D, Cohen C, Rosen S. Renal medullary and cortical correlates in fibrosis, epithelial mass, microvascularity, and microanatomy using whole slide image analysis morphometry. PLoS One 2016;11;e0161019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee MJ, Bagci P, Kong J et al. Liver steatosis assessment: Correlations among pathology, radiology, clinical data and automated image analysis software. Pathol Res Pract 2013;209;371–379. [DOI] [PubMed] [Google Scholar]
- 21.Farris AB, Cohen C, Rogers TE, Smith GH. Whole slide imaging for analytical anatomic pathology and telepathology: Practical applications today, promises, and perils. Arch Pathol Lab Med 2017;141;542–550. [DOI] [PubMed] [Google Scholar]
- 22.Moon A, Smith GH, Kong J, Rogers TE, Ellis CL, Farris ABB 3rd. Development of cd3 cell quantitation algorithms for renal allograft biopsy rejection assessment utilizing open source image analysis software. Virchows Arch 2017;472;259–269. [DOI] [PubMed] [Google Scholar]
- 23.Moxley-Wyles B, Colling R, Verrill C. Artificial intelligence in pathology: An overview. . Diagnostic Histopathology 2020;513–520. [Google Scholar]
- 24.Niazi MKK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence. Lancet Oncol 2019;20;e253–e261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serag A, Ion-Margineanu A, Qureshi H et al. Translational ai and deep learning in diagnostic pathology. Front Med (Lausanne) 2019;6;185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Behnke S. Hierarchical neural networks for image interpretation. Berlin: Springer-Verlag, 2003. [Google Scholar]
- 27.MacKay DJC. Information theory, inference, and learning algorithms. Cambridge, United Kingdom: Cambridge University Press, 2003. [Google Scholar]
- 28.Crick F. The recent excitement about neural networks. Nature 1989;337;129–132. [DOI] [PubMed] [Google Scholar]
- 29.Obermeyer Z, Emanuel EJ. Predicting the future - big data, machine learning, and clinical medicine. N Engl J Med 2016;375;1216–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loupart M-L. Leica biosystems - tissue classifications | tissuepathology.Com. In Kaplan KJ ed. Tissue Classification. tissuepathology.com, 2019. [Google Scholar]
- 31.Nalisnik M, Amgad M, Lee S et al. Interactive phenotyping of large-scale histology imaging data with histomicsml. Sci Rep 2017;7;14588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amgad M, Elfandy H, Hussein H et al. Structured crowdsourcing enables convolutional segmentation of histology images. Bioinformatics 2019;35;3461–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gutman DA, Cobb J, Somanna D et al. Cancer digital slide archive: An informatics resource to support integrated in silico analysis of tcga pathology data. J Am Med Inform Assoc 2013;20;1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gutman DA, Khalilia M, Lee S et al. The digital slide archive: A software platform for management, integration, and analysis of histology for cancer research. Cancer Res 2017;77;e75–e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dimitriou N, Arandjelovic O, Caie PD. Deep learning for whole slide image analysis: An overview. Front Med (Lausanne) 2019;6;264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mobadersany P, Yousefi S, Amgad M et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc Natl Acad Sci U S A 2018;115;E2970–E2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ueda D, Shimazaki A, Miki Y. Technical and clinical overview of deep learning in radiology. Jpn J Radiol 2019;37;15–33. [DOI] [PubMed] [Google Scholar]
- 38.Amgad M, Stovgaard ES, Balslev E et al. Report on computational assessment of tumor infiltrating lymphocytes from the international immuno-oncology biomarker working group. NPJ Breast Cancer 2020;6;16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haas M, Loupy A, Lefaucheur C et al. The banff 2017 kidney meeting report: Revised diagnostic criteria for chronic active t cell-mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am J Transplant 2018;18;293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roufosse C, Simmonds N, Clahsen-van Groningen M et al. A 2018 reference guide to the banff classification of renal allograft pathology. Transplantation 2018;102;1795–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Denic A, Morales MC, Park WD et al. Using computer-assisted morphometrics of 5-year biopsies to identify biomarkers of late renal allograft loss. Am J Transplant 2019;19;2846–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farris AB, Moghe I, Wu S et al. Banff digital pathology working group: Going digital in transplant pathology. Am J Transplant 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diaz Encarnacion MM, Griffin MD, Slezak JM et al. Correlation of quantitative digital image analysis with the glomerular filtration rate in chronic allograft nephropathy. Am J Transplant 2004;4;248–256. [DOI] [PubMed] [Google Scholar]
- 44.Kostadinova-Kunovska S, Petrusevska G, Jovanovic R et al. Histomorphometric analysis of fibrosis in the renal interstitial compartment. Prilozi 2005;26;51–59. [PubMed] [Google Scholar]
- 45.Servais A, Meas-Yedid V, Buchler M et al. Quantification of interstitial fibrosis by image analysis on routine renal biopsy in patients receiving cyclosporine. Transplantation 2007;84;1595–1601. [DOI] [PubMed] [Google Scholar]
- 46.Servais A, Meas-Yedid V, Toupance O et al. Interstitial fibrosis quantification in renal transplant recipients randomized to continue cyclosporine or convert to sirolimus. Am J Transplant 2009;9;2552–2560. [DOI] [PubMed] [Google Scholar]
- 47.Servais A, Meas-Yedid V, Noel LH et al. Interstitial fibrosis evolution on early sequential screening renal allograft biopsies using quantitative image analysis. Am J Transplant 2011;11;1456–1463. [DOI] [PubMed] [Google Scholar]
- 48.De Heer E, Sijpkens YW, Verkade M et al. Morphometry of interstitial fibrosis. Nephrol Dial Transplant 2000;15 Suppl 6;72–73. [DOI] [PubMed] [Google Scholar]
- 49.Vleming LJ, Baelde JJ, Westendorp RG, Daha MR, van Es LA, Bruijn JA. Progression of chronic renal disease in humans is associated with the deposition of basement membrane components and decorin in the interstitial extracellular matrix. Clin Nephrol 1995;44;211–219. [PubMed] [Google Scholar]
- 50.Abrass CK, Berfield AK, Stehman-Breen C, Alpers CE, Davis CL. Unique changes in interstitial extracellular matrix composition are associated with rejection and cyclosporine toxicity in human renal allograft biopsies. Am J Kidney Dis 1999;33;11–20. [DOI] [PubMed] [Google Scholar]
- 51.Verkade MA, Bajema IM, De Heer E, Bruijn JA. Decorin and tgf-beta-1 protein expression in renal disease: A morphometric analysis. J Am Soc Nephrol 1999;10;584 (abstract). [Google Scholar]
- 52.Grimm PC, Nickerson P, Gough J et al. Computerized image analysis of sirius red-stained renal allograft biopsies as a surrogate marker to predict long-term allograft function. J Am Soc Nephrol 2003;14;1662–1668. [DOI] [PubMed] [Google Scholar]
- 53.Pape L, Henne T, Offner G et al. Computer-assisted quantification of fibrosis in chronic allograft nephropaty by picosirius red-staining: A new tool for predicting long-term graft function. Transplantation 2003;76;955–958. [DOI] [PubMed] [Google Scholar]
- 54.Bains JC, Sandford RM, Brook NR, Hosgood SA, Lewis GR, Nicholson ML. Comparison of renal allograft fibrosis after transplantation from heart-beating and non-heart-beating donors. Br J Surg 2005;92;113–118. [DOI] [PubMed] [Google Scholar]
- 55.Hunter MG, Hurwitz S, Bellamy CO, Duffield JS. Quantitative morphometry of lupus nephritis: The significance of collagen, tubular space, and inflammatory infiltrate. Kidney Int 2005;67;94–102. [DOI] [PubMed] [Google Scholar]
- 56.Masseroli M, O’Valle F, Andujar M et al. Design and validation of a new image analysis method for automatic quantification of interstitial fibrosis and glomerular morphometry. Lab Invest 1998;78;511–522. [PubMed] [Google Scholar]
- 57.Moreso F, Seron D, Vitria J et al. Quantification of interstitial chronic renal damage by means of texture analysis. Kidney Int 1994;46;1721–1727. [DOI] [PubMed] [Google Scholar]
- 58.Pape L, Mengel M, Offner G, Melter M, Ehrich JH, Strehlau J. Renal arterial resistance index and computerized quantification of fibrosis as a combined predictive tool in chronic allograft nephropathy. Pediatr Transplant 2004;8;565–570. [DOI] [PubMed] [Google Scholar]
- 59.Nicholson ML, Bailey E, Williams S, Harris KP, Furness PN. Computerized histomorphometric assessment of protocol renal transplant biopsy specimens for surrogate markers of chronic rejection. Transplantation 1999;68;236–241. [DOI] [PubMed] [Google Scholar]
- 60.Nicholson ML, McCulloch TA, Harper SJ et al. Early measurement of interstitial fibrosis predicts long-term renal function and graft survival in renal transplantation. Br J Surg 1996;83;1082–1085. [DOI] [PubMed] [Google Scholar]
- 61.Ruiz P, Kolbeck PC, Scroggs MW, Sanfilippo F. Associations between cyclosporine therapy and interstitial fibrosis in renal allograft biopsies. Transplantation 1988;45;91–95. [DOI] [PubMed] [Google Scholar]
- 62.Ruiz P, Kolbeck PC, Scroggs MW, Bollinger RR, Sanfilippo F. Cyclosporine therapy and the development of interstitial fibrosis in renal allografts. Transplant Proc 1988;20;807–811. [PubMed] [Google Scholar]
- 63.Sund S, Grimm P, Reisaeter AV, Hovig T. Computerized image analysis vs semiquantitative scoring in evaluation of kidney allograft fibrosis and prognosis. Nephrol Dial Transplant 2004;19;2838–2845. [DOI] [PubMed] [Google Scholar]
- 64.Grimm PC, Nickerson P, Gough J et al. Quantitation of allograft fibrosis and chronic allograft nephropathy. Pediatr Transplant 1999;3;257–270. [DOI] [PubMed] [Google Scholar]
- 65.Seron D, Moreso F. Protocol biopsies in renal transplantation: Prognostic value of structural monitoring. Kidney Int 2007;72;690–697. [DOI] [PubMed] [Google Scholar]
- 66.Meas-Yedid V, Servais A, Noel LH et al. New computerized color image analysis for the quantification of interstitial fibrosis in renal transplantation. Transplantation 2011;92;890–899. [DOI] [PubMed] [Google Scholar]
- 67.Servais A, Noel LH, Roumenina LT et al. Acquired and genetic complement abnormalities play a critical role in dense deposit disease and other c3 glomerulopathies. Kidney Int 2012;82;454–464. [DOI] [PubMed] [Google Scholar]
- 68.Ginley B, Tomaszewski JE, Yacoub R, Chen F, Sarder P. Unsupervised labeling of glomerular boundaries using gabor filters and statistical testing in renal histology. J Med Imaging (Bellingham) 2017;4;021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fonyad L, Shinoda K, Farkash EA et al. 3-dimensional digital reconstruction of the murine coronary system for the evaluation of chronic allograft vasculopathy. Diagn Pathol 2015;10;16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Farahani N, Braun A, Jutt D et al. Three-dimensional imaging and scanning: Current and future applications for pathology. J Pathol Inform 2017;8;36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feng S, Bucuvalas JC, Demetris AJ et al. Evidence of chronic allograft injury in liver biopsies from long-term pediatric recipients of liver transplants. Gastroenterology 2018;155;1838–1851 e1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pantanowitz L, Wiley CA, Demetris A et al. Experience with multimodality telepathology at the university of pittsburgh medical center. J Pathol Inform 2012;3;45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Isse K, Lesniak A, Grama K, Roysam B, Minervini MI, Demetris AJ. Digital transplantation pathology: Combining whole slide imaging, multiplex staining and automated image analysis. Am J Transplant 2012;12;27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Isse K, Grama K, Abbott IM et al. Adding value to liver (and allograft) biopsy evaluation using a combination of multiplex quantum dot immunostaining, high-resolution whole-slide digital imaging, and automated image analysis. Clin Liver Dis 2010;14;669–685. [DOI] [PubMed] [Google Scholar]
- 75.Calvani J, Terada M, Lesaffre C et al. In situ multiplex immunofluorescence analysis of the inflammatory burden in kidney allograft rejection: A new tool to characterize the alloimmune response. Am J Transplant 2020;20;942–953. [DOI] [PubMed] [Google Scholar]
- 76.Yagi Y, Aly RG, Tabata K et al. Three-dimensional histologic, immunohistochemical and multiplex immunofluorescence analysis of dynamic vessel co-option of spread through air spaces (stas) in lung adenocarcinoma. J Thorac Oncol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hossain MS, Hanna MG, Uraoka N et al. Automatic quantification of her2 gene amplification in invasive breast cancer from chromogenic in situ hybridization whole slide images. J Med Imaging (Bellingham) 2019;6;047501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kato T, Relator R, Ngouv H et al. Segmental hog: New descriptor for glomerulus detection in kidney microscopy image. BMC Bioinformatics 2015;16;316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Simon O, Yacoub R, Jain S, Tomaszewski JE, Sarder P. Multi-radial lbp features as a tool for rapid glomerular detection and assessment in whole slide histopathology images. Sci Rep 2018;8;2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gallego J, Pedraza A, Lopez S et al. Glomerulus classification and detection based on convolutional neural networks. J. Imaging 2018;4;20. [Google Scholar]
- 81.Kannan S, Morgan LA, Liang B et al. Segmentation of glomeruli within trichrome images using deep learning. Kidney Int Rep 2019;4;955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bukowy JD, Dayton A, Cloutier D et al. Region-based convolutional neural nets for localization of glomeruli in trichrome-stained whole kidney sections. J Am Soc Nephrol 2018;29;2081–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marsh JN, Matlock MK, Kudose S et al. Deep learning global glomerulosclerosis in transplant kidney frozen sections. IEEE Trans Med Imaging 2018;37;2718–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lutnick B, Ginley B, Govind D et al. An integrated iterative annotation technique for easing neural network training in medical image analysis. Nat Mach Intell 2019;1;112–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ginley B, Lutnick B, Jen KY et al. Computational segmentation and classification of diabetic glomerulosclerosis. J Am Soc Nephrol 2019;30;1953–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hermsen M, de Bel T, den Boer M et al. Deep learning-based histopathologic assessment of kidney tissue. J Am Soc Nephrol 2019;30;1968–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jayapandian CP, Chen Y, Janowczyk AR et al. Development and evaluation of deep learning-based segmentation of histologic structures in the kidney cortex with multiple histologic stains. Kidney Int 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kolachalama VB, Singh P, Lin CQ et al. Association of pathological fibrosis with renal survival using deep neural networks. Kidney Int Rep 2018;3;464–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.American Medical Association. Augmented intelligence (ai) - american medical association. 2019. [Google Scholar]
- 90.Carter S, Nielsen M. Using artificial intelligence to augment human intelligence. Distill: Google Brain Team and YC Research, 2017. [Google Scholar]
- 91.Litjens G, Bandi P, Ehteshami Bejnordi B et al. 1399 h&e-stained sentinel lymph node sections of breast cancer patients: The camelyon dataset. Gigascience 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ehteshami Bejnordi B, Veta M, Johannes van Diest P et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017;318;2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hanna MG, Reuter VE, Samboy J et al. Implementation of digital pathology offers clinical and operational increase in efficiency and cost savings. Arch Pathol Lab Med 2019;143;1545–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hanna MG, Reuter VE, Hameed MR et al. Whole slide imaging equivalency and efficiency study: Experience at a large academic center. Mod Pathol 2019;32;916–928. [DOI] [PubMed] [Google Scholar]
- 95.Aeffner F, Zarella MD, Buchbinder N et al. Introduction to digital image analysis in whole-slide imaging: A white paper from the digital pathology association. J Pathol Inform 2019;10;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zarella MD, Bowman D, Aeffner F et al. A practical guide to whole slide imaging: A white paper from the digital pathology association. Arch Pathol Lab Med 2019;143;222–234. [DOI] [PubMed] [Google Scholar]
- 97.Friedman BA. Orchestrating a unified approach to information management. Radiol Manage 1997;19;30–36. [PubMed] [Google Scholar]
- 98.Sorace J, Aberle DR, Elimam D, Lawvere S, Tawfik O, Wallace WD. Integrating pathology and radiology disciplines: An emerging opportunity? BMC Med 2012;10;100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Madrigal E. Going remote: Maintaining normalcy in our pathology laboratories during the covid-19 pandemic. Cancer Cytopathol 2020;128;321–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hanna MG, Reuter VE, Ardon O et al. Validation of a digital pathology system including remote review during the covid-19 pandemic. Mod Pathol 2020;33;2115–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Browning L, Fryer E, Roskell D et al. Role of digital pathology in diagnostic histopathology in the response to covid-19: Results from a survey of experience in a uk tertiary referral hospital. J Clin Pathol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Browning L, Colling R, Rakha E et al. Digital pathology and artificial intelligence will be key to supporting clinical and academic cellular pathology through covid-19 and future crises: The pathlake consortium perspective. J Clin Pathol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stathonikos N, van Varsseveld NC, Vink A et al. Digital pathology in the time of corona. J Clin Pathol 2020;73;706–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Karn U. An intuitive explanation of convolutional neural networks. 2016. [Google Scholar]