Abstract
Aims/hypothesis
Normal cellular prion protein (PrPC) is a conserved mammalian glycoprotein found on the outer plasma membrane leaflet through a glycophosphatidylinositol anchor. Although PrPC is expressed by a wide range of tissues throughout the body, the complete repertoire of its functions has not been fully determined. The misfolded pathogenic isoform PrPSc (the scrapie form of PrP) is a causative agent of neurodegenerative prion diseases. The aim of this study is to evaluate PrPC localisation, expression and trafficking in pancreases from organ donors with and without type 1 diabetes and to infer PrPC function through studies on interacting protein partners.
Methods
In order to evaluate localisation and trafficking of PrPC in the human pancreas, 12 non-diabetic, 12 type 1 diabetic and 12 autoantibody-positive organ donor tissue samples were analysed using immunofluorescence analysis. Furthermore, total RNA was isolated from 29 non-diabetic, 29 type 1 diabetic and 24 autoantibody-positive donors to estimate PrPC expression in the human pancreas. Additionally, we performed PrPC-specific immunoblot analysis on total pancreatic protein from non-diabetic and type 1 diabetic organ donors to test whether changes in PrPC mRNA levels leads to a concomitant increase in PrPC protein levels in human pancreases.
Results
In non-diabetic and type 1 diabetic pancreases (the latter displaying both insulin-positive [INS(+)] and -negative [INS(−)] islets), we found PrPC in islets co-registering with beta cells in all INS(+) islets and, strikingly, unexpected activation of PrPC in alpha cells within diabetic INS(−) islets. We found PrPC localised to the plasma membrane and endoplasmic reticulum (ER) but not the Golgi, defining two cellular pools and an unconventional protein trafficking mechanism bypassing the Golgi. We demonstrate PrPC co-registration with established protein partners, neural cell adhesion molecule 1 (NCAM1) and stress-inducible phosphoprotein 1 (STI1; encoded by STIP1) on the plasma membrane and ER, respectively, linking PrPC function with cyto-protection, signalling, differentiation and morphogenesis. We demonstrate that both PRNP (encoding PrPC) and STIP1 gene expression are dramatically altered in type 1 diabetic and autoantibody-positive pancreases.
Conclusions/interpretation
As the first study to address PrPC expression in non-diabetic and type 1 diabetic human pancreas, we provide new insights for PrPC in the pathogenesis of type 1 diabetes. We evaluated the cell-type specific expression of PrPC in the human pancreas and discovered possible connections with potential interacting proteins that we speculate might address mechanisms relevant to the role of PrPC in the human pancreas.
Keywords: Cellular prion protein, Protein trafficking, Stress-induced phosphoprotein 1, Type 1 diabetes, Type 1 diabetes-dependent endocrine cell expression
Introduction
The cellular prion protein (PrPC), encoded by the gene PRNP, is a glycophosphatidylinositol-anchored cell-surface glycoprotein expressed in numerous cell types [1–4]. PrPC has been the subject of intense study since identification of the scrapie form of the prion protein, PrPSc, as an infectious agent [1, 5–7] transmissible through pathogenic conversion of the host-expressed PrPC. As a pathogenic, alternatively folded aggregate, PrPSc is linked to a host of neurodegenerative disorders affecting animals and humans, including: scrapie; transmissible mink encephalopathy (TME); chronic wasting disease (CWD); bovine spongiform encephalopathy (BSE); feline spongiform encephalopathy (FSE); exotic ungulate encephalopathy (EUE); camel spongiform encephalopathy (CSE) and in humans disorders classified as Kuru, Creutzfeldt-Jakob disease (CJD), variant CJD (vCJD), Gerstmann–Sträussler–Scheinker syndrome (GSS) and fatal familial insomnia (FFI) [8–11]. Recently, Corbett et al [12] have demonstrated a PrPc requirement in the toxicity of α-synuclein, amyloid-β and tau in induced pluripotent stem cell (iPSC)-derived neurons from patients with Alzheimer’s, Lewy body and Pick’s disease.
Although numerous studies have attempted to clarify PrPC cellular function, including 3D protein structure and generation of several gene ablation models, a definitive biochemical function for PrPC has not been elucidated [3, 8, 13]. Potential roles for PrPC include modulation of signalling pathways, immune and hippocampal function, neuroprotection, T lymphocyte proliferation and anti-apoptotic activity, together with metal ion, mitochondrial and glucose homeostasis [3, 8].
Historically, Atouf et al [14] reported that Prnp gene expression was hormonally controlled in rodent pancreatic endocrine cell lines and primary islets. More recently, implementing a PrPC overexpressing mouse model, Ashok and Singh [15] demonstrated PrPC expression in mouse pancreatic beta cells, promoting iron uptake through divalent metal transporters, thus modulating glucose homeostasis. Their results showed PrPC silencing in 1.1B4 cells resulted in depletion of intracellular iron with GLUT2 and insulin upregulation. Using a rat model and mice lacking the prion protein, Strom et al [16, 17] reported that PrPC is ubiquitously expressed in all four major endocrine cell types within rat islets and identified ‘cytosolic inclusions’ containing PrPC exclusively in a subpopulation of insulin-producing beta cells; the study also implicated involvement of PrPC in glucose homeostasis. Similarly, Amselgruber et al [18] demonstrated that PrPC is strongly expressed in fetal and adult bovine pancreatic islets. PrPC-null and -overexpressing mice fed a high-fat diet presented with insulin resistance including hyperglycaemia, hyperinsulinaemia and obesity [19, 20]. The majority of studies identify PrPC expression in endocrine cells of rodents and ungulates, with some suggesting homeostatic functions for PrPC. Furthermore, given the wealth of PrPC data in the brain, and since neurons and pancreatic beta cells share numerous similarities [21], the question arises whether PrPC has a role in the pathogenesis of type 1 diabetes. Therefore, given the resemblance between neurons and beta cells, we investigated the cell-type specific expression of PrPC in human pancreas and tested possible connections with potential interacting proteins to address mechanisms relevant to the role of PrPC in the human pancreas. To our knowledge, this study is the first attempt to evaluate such connections, expression and localisation of cellular prion protein in unaffected and type 1 diabetic human pancreases.
Methods
Human organ donors
Pancreas samples were obtained from nPOD (www.jdrfnpod.org). Procedures were approved by the University of Florida Institutional Review Board and the United Network for Organ Sharing (UNOS) according to federal guidelines, and informed consent obtained from each donor’s legal representative. For each donor, a medical chart review was performed and C-peptide measured with type 1 diabetes diagnosis confirmed according to ADA guidelines. Demographics, length of hospitalisation, and organ transport duration information was obtained from hospital records or UNOS (ESM Tables 1 and 2). Cases were selected to represent type 1 diabetic donors with residual insulin-positive [INS(+)] islets, and type 2 diabetic, autoantibody-positive (AAb+) and non-diabetic control donors. Randomisation was not performed whereas blinding was only utilised in the immunoblot analysis. No data were excluded.
RNA extraction and qRT-PCR
Human pancreas tissue was cryo-preserved by flash freezing or in RNAlater (Qiagen, USA), ~16 h from cross clamp. Total RNA was isolated as described [22] including treatment with DNase 1 and analysed by quantitative RT-PCR (qRT-PCR). RNA concentrations were determined using a Nanodrop 2000C (Thermo Scientific, USA) and, when necessary, integrity verified by visualisation of ribosomal RNA by gel electrophoresis and ethidium bromide staining. Samples were confirmed to be free of DNA contamination in no-reverse-transcriptase controls with an exon/intron primer pair. cDNA was produced with Superscript II (Invitrogen, USA) using oligo dT priming and subsequently utilised for qRT-PCR using Thermo Luminaris qPCR Master Mix (Thermo Scientific, USA). One microgram of total RNA was used for each 20 μl cDNA reaction, which was then diluted to 200 μl and 2 μl of diluted cDNA was employed for each 25 μl qRT-PCR reaction containing 600 nmol/l of each primer pair. Individual qRT-PCR reactions were carried out in duplicate in a Bio-Rad MyiQ system. Normalisation factors, based on the geometric mean of three reference genes, were applied to the expression of all genes in respective pancreases, with the fold difference reported as the ratio of the means (type 1 diabetic/non-diabetic). All qRT-PCR parameters were standardised in accordance with the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines [23].
Immunohistochemistry and immunofluorescence
Formalin-fixed paraffin-embedded sections (4 μm) were evaluated by immunolocalisation, after deparaffinisation and rehydration, blocking, and incubation with primary antibodies to cellular prion (Sigma-Aldrich, USA, #HPA042754, 1:100), insulin (Agilent, USA, #IR002, undiluted), glucagon (Abcam, USA, #ab10988, 1:1600), somatostatin (Agilent, USA, #A0566, 1:1000), stress-inducible phosphoprotein 1 (STI1, Novus, USA, #H00010963-M11, 1:50) and neural cell adhesion molecule 1 (NCAM1, Bio-Rad, USA, #MCA2693GA, 1:50). Detection of primary antibody was performed by multiplex staining using polymer horseradish peroxidase (Akoya Biosciences, USA, HRP, #ARH1001EA) followed by tyramine amplification (TSA detection system, Akoya Biosciences, USA, NEL741001KT/744001KT/745001K). To assess amyloid-like plaques, slides were stained with Congo Red (Newcomer Supply Copper, USA, #9103) according to the manufacturer’s protocol. Consecutive slides were stained for PrPC primary antibody and then stained with 0.05% thioflavin T (in 50% ethanol) for 8 min at room temperature. To assess immunolocalisation, slides were deparaffinised, rehydrated and subjected to heat-induced antigen retrieval. Slides were blocked using 5% normal goat serum and incubated in primary antibodies to Golgin A2 (GM130; Abcam, USA, #ab169276, 1:50), Wolfram syndrome 1 (WFS1; LSBio, USA, #LS-B14378, 1:200) and CD99 (BioLegend, USA, #318002, 1:2000) in Dako diluent (Dako, USA, #S080983-2) overnight. Primary antibodies were visualised with H+L Alexa Fluor secondary antibodies (Thermo Scientific, USA), followed by nuclear staining with DAPI counterstain. Slides were analysed using a BZ-X710 Keyence florescence microscope, USA.
Tissue homogenisation
Human pancreases were homogenised in modified RIPA buffer (50 mmol/l Tris-HCl pH 7.4, 150 mmol/l NaCl, 5 mmol/l EDTA, 1 mmol/l EGTA, 1% NP-40, 0.5% sodium deoxycholate and 0.1% SDS) with protease and phosphatase inhibitor cocktails (Thermo Scientific, USA, #78430 and #78428, respectively) using a Tissue Tearor (BioSpec, USA), further disrupted with a Vibra-Cell sonicator (Sonics & Materials, USA) and placed on ice for 15 min after each sonication step. Tissue homogenates were centrifuged at 14,000 g for 20 min at 4°C and supernatants collected for protein concentration assay using a Pierce BCA assay kit (Thermo Scientific, USA, #23227).
Immunoblotting
Forty micrograms of protein lysates in Laemmli’s buffer containing 2.5% β-mercaptoethanol was boiled for 5 min and loaded on 4–20% gradient Mini-PROTEAN TGX Stain-Free gels (Bio-Rad, USA, #456-8095). Gels were activated using a Gel DOC EZ imager (Bio-Rad, USA) and transferred onto nitrocellulose membranes (Li-Cor, USA, #926-31092). Membranes were scanned with the Gel DOC™ EZ imager and total protein staining visualised and quantified using Image Lab software version 5.2.1 (Bio-Rad, USA). Membranes were washed and blocked for 1 h at room temperature with Intercept Blocking Buffer (Li-Cor, USA, #927–60001) and incubated at 4°C overnight with mouse anti-PrPC monoclonal antibody (BioLegend, USA, #SIG-39620), in Intercept Antibody Diluent (Li-Cor, USA, #927–60001). Membranes were washed with Tris-buffered saline containing 0.1% Tween 20 (TBST) and incubated with goat anti-mouse IRDye 800CW (Li-Cor, USA, #926–32210;) secondary antibody for 1 h at room temperature. Membranes were washed 3 times with TBST at 5 min intervals. Immunoreactive bands were densitometrically analysed using Odyssey infrared scanner software version 3.1 (Li-Cor, USA). PrPc monoclonal antibody specificity was validated in human brain tissue lysate (Fig. 4b), consistent with published reports [24].
Fig. 4.
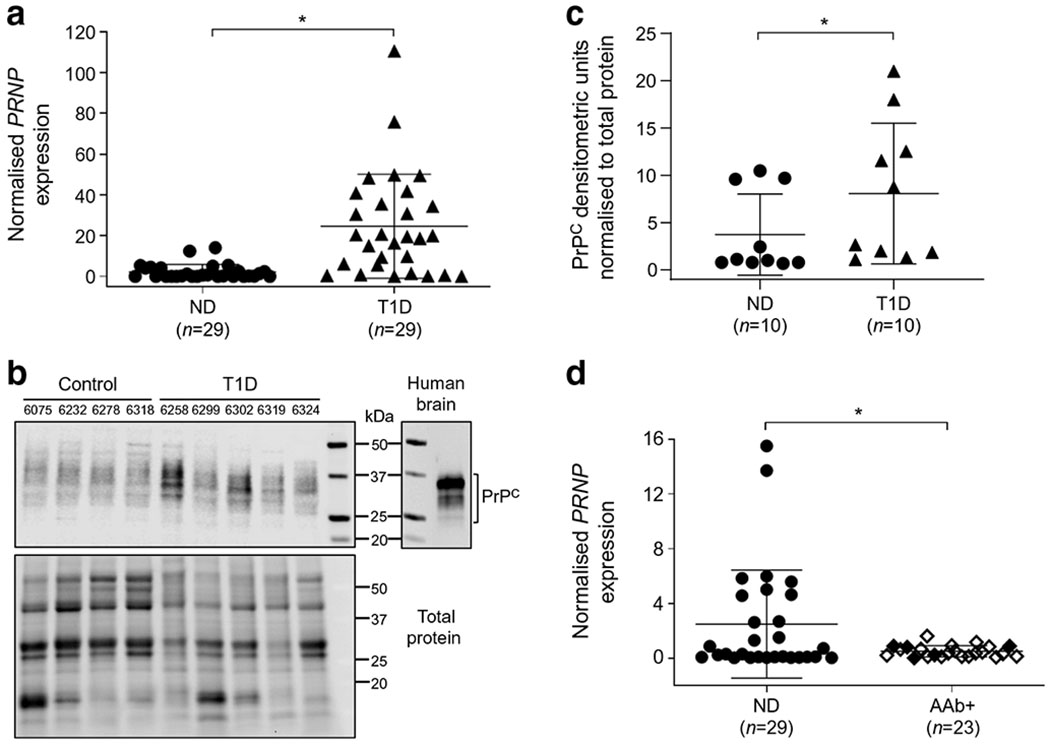
PRNP gene and PrPC protein expression in non-diabetic, type 1 diabetic and AAb+ organ donor human pancreases. PRNP mRNA expression levels were measured by qRT-PCR from isolated total pancreatic RNA from 29 non-diabetic, 29 type 1 diabetic and 23 AAb+ donors. (a) PRNP gene expression in type 1 diabetes revealed a highly significant increase of 10.85-fold (p ≤ 0.0001) compared with non-diabetic donors. (b) Representative PrPC-specific immunoblot analysis on total pancreatic protein isolated from non-diabetic and type 1 diabetic organ donors was performed to test changes in PrPC mRNA levels. Right panel: the migration pattern of PrPC in human brain extracts; left panel: immunoblot analysis of purified human pancreas extracts of four representative non-diabetic and five representative type 1 diabetic donors. Full immunoblot for PrPC and total protein can be found in ESM Fig. 3. (c) Densitometric analysis data from immunoblot shown in (b) combined with additional data (6 more non-diabetic; 5 more type 1 diabetic donors) normalised to total protein, illustrating a concomitant increase in PrPC levels of 2.16-fold (p = 0.0433). (d) In contrast, AAb+ donors exhibited significantly lower PRNP mRNA levels when compared with non-diabetic donors (0.579-fold) (p = 0.012; white diamond, single AAb+; black diamond, multiple AAb+). *p < 0.05. ND, non-diabetic; T1D, type 1 diabetic
Quantification and statistical analysis
Statistical variables, including exact n, mean and SD are reported in the figures and figure legends. Data were judged statistically significant when p < 0.05 by a two-sample t test with Welch’s correction for qRT-PCR and immunoblot analyses. Data were analysed and graphed using GraphPad Prism software version 8 (GraphPad, USA).
Results
Cellular localisation of PrPC in human pancreases
To investigate the cellular localisation of PrPC in human pancreas, PrPC, insulin-producing beta cells, glucagon-producing alpha cells and somatostatin-producing delta cells were studied by immunofluorescence on paraffin-embedded tissue sections. We studied 12 non-diabetic and 12 type 1 diabetic pancreases (the latter all containing both residual INS-positive [INS(+)] and INS-negative [INS(−)] islets), together with 12 AAb+ human organ donor pancreases. Pancreases were obtained from the Network for Pancreatic Organ Donors with Diabetes (nPOD) biobank, with organ donor metadata and de-identified patient numbering (ESM Tables 1 and 2). We analysed PrPC expression in islets of non-diabetic pancreases (Fig. 1a) compared with the expression in INS(+) (Fig. 1e) and INS(−) islets (Fig. 1i) from type 1 diabetic donors. High PrPC immunoreactivity was observed in islets of non-diabetic and type 1 diabetic donors with a low level of expression in the exocrine pancreas (Fig. 1a, e, i). We deduced that in nondiabetic and INS(+) islets of type 1 diabetic donors, PrPC was expressed mainly in beta cells, based on co-expression with insulin (Fig. 1c, g for non-diabetic and type 1 diabetic islets, respectively). We confirmed co-registration of PrPC with insulin in beta cells by 3D image reconstruction analysis (Fig. 1d, h). Of particular note, even in the absence of insulin, PrPC continues to be expressed in a large number of cells in type 1 diabetic INS(−) islets (Fig. 1i, k, l), implicating its co-expression in an alternative endocrine cell type. These immunofluorescence results were consistently observed in all 12 non-diabetic and type 1 diabetic donor pancreases.
Fig. 1.
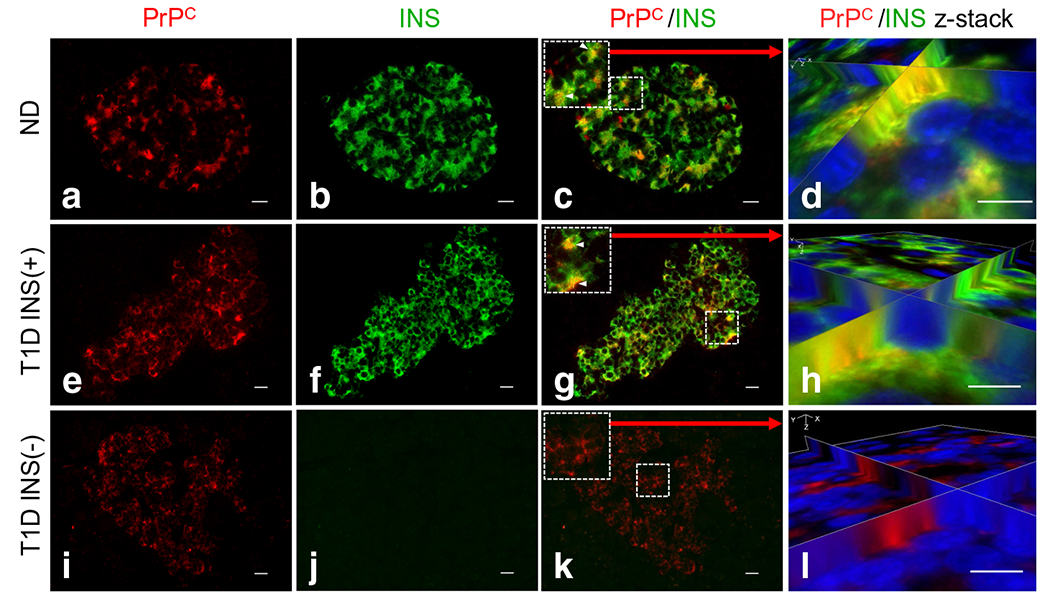
Cellular localisation of PrPC in beta cells of human pancreases based on immunofluorescence analysis, (a, e, i) PrPC was observed to be highly expressed in pancreatic islets of all three islet types [non-diabetic; type 1 diabetic INS(+); type 1 diabetic INS(−)] with low level expression in the exocrine pancreas, (a–d) PrPC expression in non-diabetic donors localises to pancreatic islets and co-localises with beta cells (c). Co-registration of PrPC/INS cells was confirmed using z-stack 3D image reconstruction analysis (d). (e, i) In our analysis of PrPC in type 1 diabetic pancreas, we separated images from donors that contain both INS(+) (e) and INS(−) islets (i). (e)–h) As with non-diabetic donors, type 1 diabetic INS(+) islets show co-registration between PrPC and INS cells (g, h). (i–l) In the absence of insulin [INS(−)], PrPC continues to be expressed in a large number of cells (i, k). Scale bars, 20 αm. INS, insulin; ND, non-diabetic, T1D, type 1 diabetic
We found no co-registration of PrPC with glucagon in both non-diabetic and type 1 diabetic INS(+) islets (Fig. 2a–j). However, by contrast, PrPC was found expressed in alpha cells in type 1 diabetic INS(−) islets (Fig. 2k–o), suggesting that, in the absence of beta cells, PrPC expression is activated in alpha cells. We also evaluated PrPC expression in conjunction with somatostatin and found no evidence of PrPC expression in somatostatin-positive delta cells in either non-diabetic or type 1 diabetic pancreases (ESM Fig. 1).
Fig. 2.
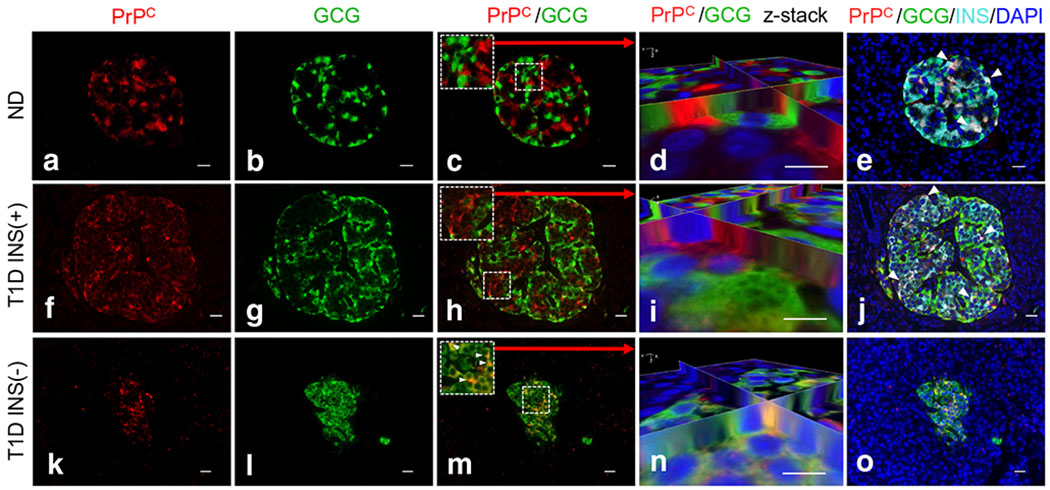
Cellular localisation of PrPC in alpha cells of human pancreases based on immunofluorescence analysis. (a)–j) We found no co-registration of PrPC with glucagon in either non-diabetic or type 1 diabetic INS(+) islets (k)–o) but did find co-localisation of PrPC with insulin (white arrowheads e and j). PrPC was found to be co-localised with alpha cells in type 1 diabetic INS(−) islets, suggesting that PrPC expression is activated in alpha cells in the absence of beta cells in these islets. Co-registration was confirmed using z-stack 3D image reconstruction analysis shown in (n). Scale bars, 20 μm. GCG, glucagon; INS, insulin; ND, non-diabetic, T1D, type 1 diabetic
Intracellular localisation of PrPC in the human pancreas
The majority of data on PrPC trafficking is derived from results in the brain and spinal cord. In neurons, PrPC is synthesised and processed following a pathway similar to other membrane or secreted proteins [8]. Synthesis originates on endoplasmic reticulum (ER)-bound ribosomes where a C-terminal, glycosyl-phosphatidylinositol (GPI) lipid anchor is added, followed by transit through the Golgi apparatus leading to GPI-dependent anchoring on the extracellular leaflet of lipid rafts [25]. To evaluate PrPC intracellular localisation within human pancreatic endocrine cells, we performed co-staining of PrPC with compartment-specific markers for ER (WFS1; [26]), Golgi (GM130; [27]) and plasma membrane (CD99; [28]). PrPC was found to co-register with WFS1 (Fig. 3a, d, g) and CD99 (Fig. 3c, f, i) but not GM130 (Fig. 3b, e, h), based on 3D image reconstruction analysis, indicating that PrPC is associated mainly with the ER and plasma membrane of beta cells in non-diabetic and type 1 diabetic INS(+) islets and in alpha cells in type 1 diabetic INS(−) islets. These results clearly demonstrate that PrPC is not associated with the Golgi apparatus, as shown in Fig. 3b–h, together with additional patient data in ESM Fig. 2, suggesting that PrPC in pancreatic endocrine cells is synthesised in the ER utilising an unconventional secretory pathway that bypasses the Golgi to reach the plasma membrane [29, 30].
Fig. 3.
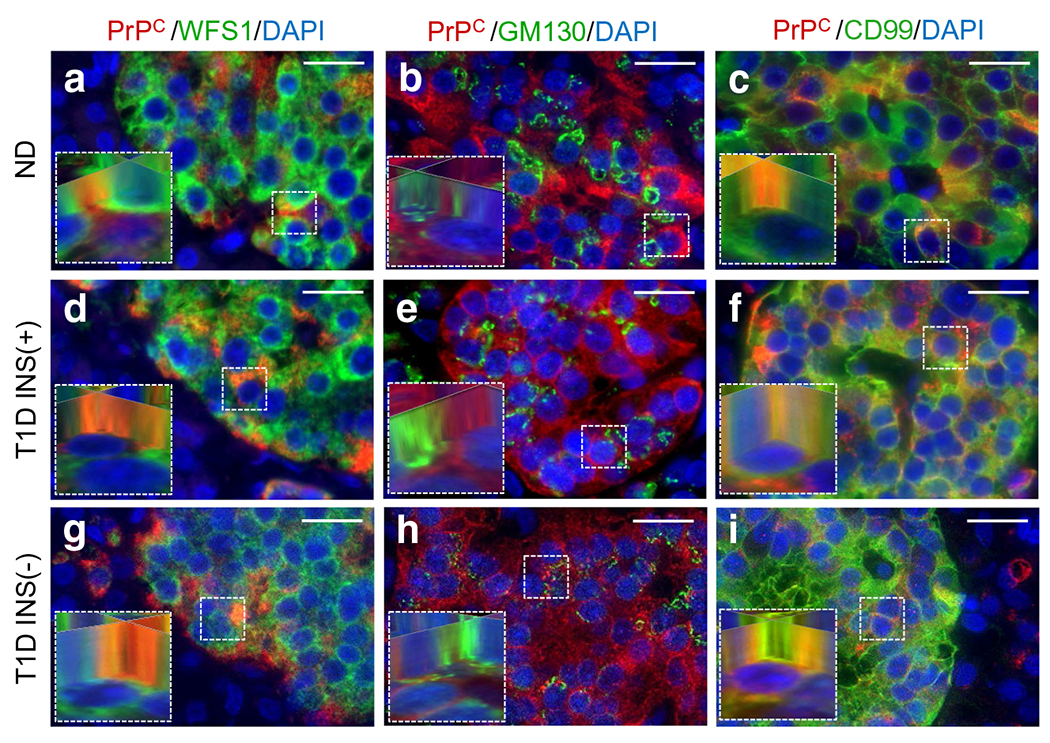
Intracellular localisation of PrPC in the human pancreas. Specific co-staining of PrPC and cellular markers for ER (WFS1), Golgi (GM130) and cell membrane (CD99) staining was implemented to assess PrPC intracellular localisation within endocrine cells of the human pancreas. PrPC was found to co-register with WFS1 (a, d, g) and CD99 (c, f, i) but not GM130 (b, e, h; ESM Fig. 2) based on 3D image reconstruction analysis (insets), indicating that PrPC is mainly associated with the ER and plasma membrane of pancreatic endocrine cells. Scale bars, 20 μm. ND, non-diabetic, T1D, type 1 diabetic
PRNP gene and PrPC protein expression in non-diabetic, type 1 diabetic and AAb+ organ donor human pancreases
To evaluate PRNP gene expression, mRNA expression levels were measured using qRT-PCR from total RNA isolated from freshly frozen pancreases from 29 non-diabetic and 29 type 1 diabetic organ donors (Fig. 4a). qRT-PCR was performed following strict MIQE guidelines by normalisation with the geometric mean of three reference genes [23]. The quantitative fold difference in gene expression between these two groups is reported as the ratio of the means. PRNP gene expression in type 1 diabetic donors revealed a highly significant increase of 10.85-fold; p ≤ 0.0001 compared with non-diabetic donors (Fig. 4a). To test whether changes in PRNP mRNA levels leads to a concomitant increase in PrPC protein levels, we performed PrPC-specific immunoblot analysis on total pancreatic protein from non-diabetic and type 1 diabetic organ donors (Fig. 4b) with images for the intact blot and total protein stain included in ESM Fig. 3. The right panel illustrates the migration pattern for PrPC in a human brain extract, for comparison (Fig. 4b, right). Consistent with the mRNA results (Fig. 4a), Fig. 4c provides a quantitative assessment of PrPC protein levels from ten non-diabetic and ten type 1 diabetic donors, normalised to total protein, illustrating a significant increase of 2.16-fold between the type 1 diabetic and the non-diabetic groups.
We also evaluated PRNP gene expression in 23 AAb+ donor pancreases, from individuals at risk for type 1 diabetes (as the propensity for disease onset increases in the presence of multiple autoantibodies), and found a significantly decreased level of mRNA expression compared with the non-diabetic cohort (0.579-fold; p ≤ 0.012; Fig. 4d). Given that PrPC may function as an anti-apoptotic or cytoprotective protein [3], we also evaluated the expression of PrPC in AAb+ donors by immunofluorescence and, as with the non-diabetic and type 1 diabetic donors, PrPC was expressed primarily in islets and specifically in beta cells, based on co-registration with insulin and not glucagon (ESM Fig. 4). We also observed that in the majority of the 12 studied AAb + donors, PrPC levels in pancreatic islets were consistently lower compared with non-diabetic islets, as observed for patient 6450 using identical imaging parameters (ESM Fig. 4).
PrPC does not co-register with protein aggregates in type 1 diabetic pancreases
The hallmarks of prion-related diseases include protein aggregates and amyloid plaques, which in most cases positively stain for PrPSc [4, 8–11]. Similarly, islet amyloid has been most strongly associated with type 2 diabetes pathogenesis along with type 1 diabetes and cystic fibrosis-related diabetes (CFRD) [31–36]. To evaluate whether PrPC co-registers with islet-associated protein aggregates previously documented in type 1 diabetic pancreases [31, 33], we identified three type 1 diabetic pancreases displaying protein aggregates from among our 12 type 1 diabetic donors, based on detection of amyloid using Congo Red stain [37] (Fig. 5a–c). A pancreatic section from a type 2 diabetic donor with known presence of amyloid was used as a positive control (Fig. 5d). To investigate whether these protein aggregates co-registered with PrPC, we took consecutive sections from each of these donors and co-stained with a PrPC antibody and thioflavin T (an additional amyloid marker) [38], with co-detection by fluorescence microscopy (Fig. 5e–h). Thioflavin T-positive regions of each islet correlated with the Congo Red staining; however, we found no colocalisation of PrPC with the thioflavin T-positive regions in either the type 1 or type 2 diabetic pancreases.
Fig. 5.
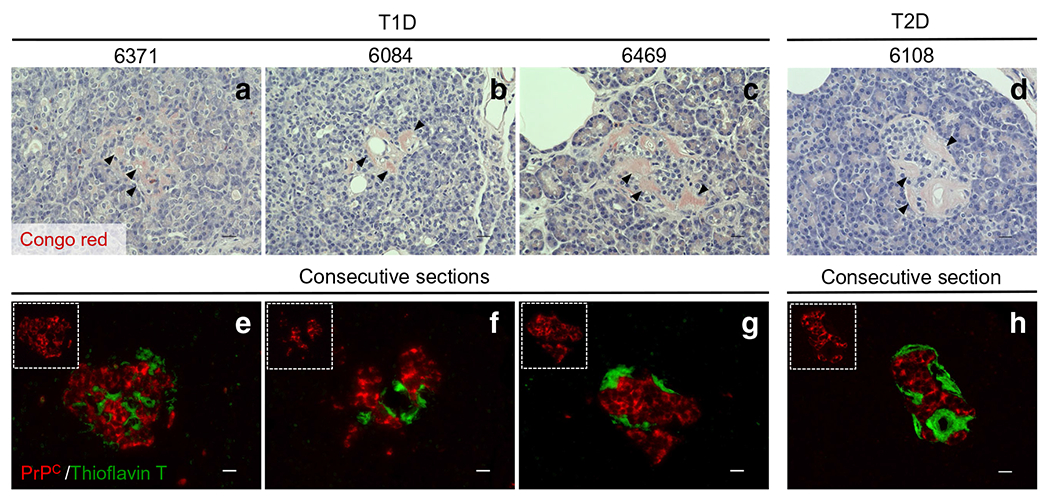
Assessment of possible interactions between PrPC and protein aggregates in type 1 and type 2 diabetic donors. Among our 12 type 1 diabetic organ donors, three cases displayed amyloid-like plaques. In these three cases, we evaluated whether PrPC co-localised with these islet-associated protein aggregates. (a)–d) The pathology stain, Congo Red, was used to identify amyloid/protein aggregates in the three type 1 diabetic donors as well as one type 2 diabetic donor (positive control). Black arrows show positivity for amyloid-like plaques (pink colour) in pancreatic islets. (e)–h) To visualise amyloid-like plaques and PrPC expression together, consecutive slides were stained with thioflavin T (green) and PrPC (red). We found no colocalisation of PrPC-positive islet cells with the thioflavin T regions in either the type 1 or the type 2 diabetic pancreas samples. Scale bars, 20 μm. T1D, type 1 diabetic; T2D, type 2 diabetic
STI1 and NCAM1 co-register with PrPc in islets: implications for stress protection and cell morphogenesis
With no consensus on the physiological function of PrPC [8], a powerful approach to infer function is by studying potential PrPC binding proteins with known biological roles [13]. We utilised the Protein–Protein Interaction Networks Functional Enrichment Analysis software, STRING 11.0 (https://string-db.org), to generate a PrPC-predicted human protein–protein interaction model (ESM Fig. 5). Based on this model and our own corroborating literature review, STI1 (encoded by STIP1) and NCAM1 (also known as CD56) were especially promising PrPC binding partners [39, 40].
As a co-chaperone protein, STI1 coordinates the functions of heat-shock protein 70 (HSP70) and 90 (HSP90) in protein folding [41], whereas NCAM1 is a cell adhesion protein and member of the immunoglobulin superfamily directing cell–cell and cell–matrix interactions [42]. Zanata et al [43] demonstrated that interaction of PrPC with STI1 activated anti-apoptotic pathways providing significant neuroprotection, as well as effects on neuritogenesis and survival of hippocampal neurons [44]. Mehrabian et al [45] have demonstrated that, during epithelial-to-mesenchymal transitions (EMT), PRNP transcription is increased >10-fold and correlates with the polysialylation of NCAM1, controlled by PrPC.
To investigate the expression of these prion interacting proteins in human pancreas, we stained pancreatic tissue sections from non-diabetic and type 1 diabetic donors for STI1, and found that STI1 is highly expressed in nondiabetic and type 1 diabetic INS(+) and INS(−) islets, together with lower levels in the exocrine pancreas (Fig. 6b, f, j). Moreover, we observed that STI1 highly co-registers with PrPC primarily in the cytosol in islets, regardless of their insulin positivity, as shown in the 3D reconstruction images (Fig. 6d, h, l). As expected, co-registration of STI1 with insulin- and glucagon-positive cells was also observed in all three islet types (ESM Fig. 6). Interestingly, we determined that STIP1 mRNA levels in type 1 diabetic donors were notably increased compared with the non-diabetic cohort (Fig. 6m: 13.42-fold; p ≤ 0.0001). We also compared STIP1 mRNA levels in non-diabetic and AAb+ donors, which showed a modest but significant increase (Fig. 6n: 2.09-fold; p = 0.00641. We similarly investigated NCAM1 and found that, in contrast to STI1, PrPC co-registers with NCAM1 primarily at the cell membrane (Fig. 7a–l), thus establishing separate interacting partners for the two cellular pools of PrPC in the human pancreas, with potentially independent cellular roles.
Fig. 6.
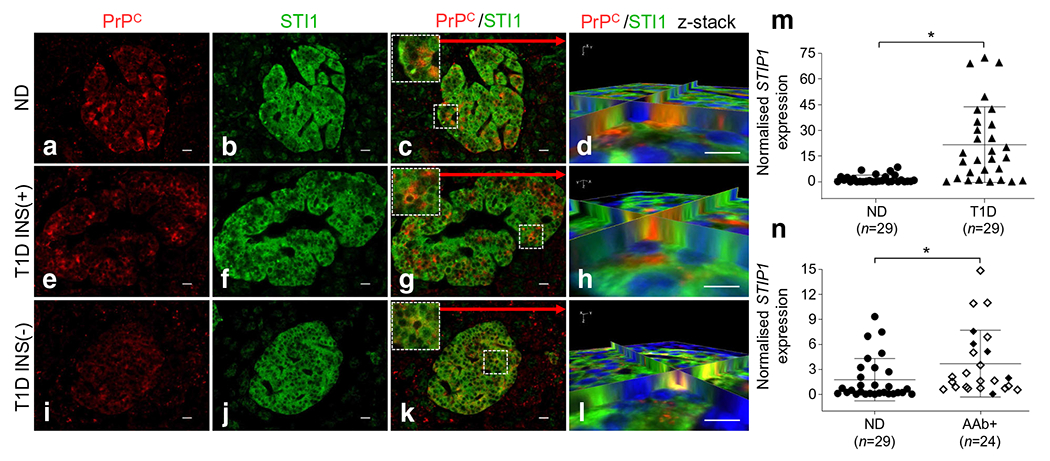
STI1 co-registers with PrPC in islets. (a, e, i) PrPC is expressed in non-diabetic, type 1 diabetic INS(+) and type 1 diabetic INS(−) islets. (b, f, j) STI1 is highly expressed in the pancreatic islets of non-diabetic, type 1 diabetic INS(+) and type 1 diabetic INS(−) islets. (d, h, l) We found that STI1 highly co-registers with PrPC in all three groups, as shown in z-stack 3D reconstruction images. (j, k, l) Even with no insulin expression in INS(−) islets, STI1 expression is still evident, with co-registration with glucagon-positive cells (ESM Fig. 6). Scale bars, 20 μm. (m) STIP1 total mRNA levels in type 1 diabetic donors is significantly increased compared with non-diabetic donors 13.42-fold (p ≤ 0.0001). (n) We also compared mRNA levels in AAb+ donors to non-diabetic donors and observed increased STIP1 expression of 2.09-fold (p = 0.0064) compared with non-diabetic donors (white diamond, single AAb+; black diamond, multiple AAb+). *p < 0.05. ND, non-diabetic; T1D, type 1 diabetic
Fig. 7.
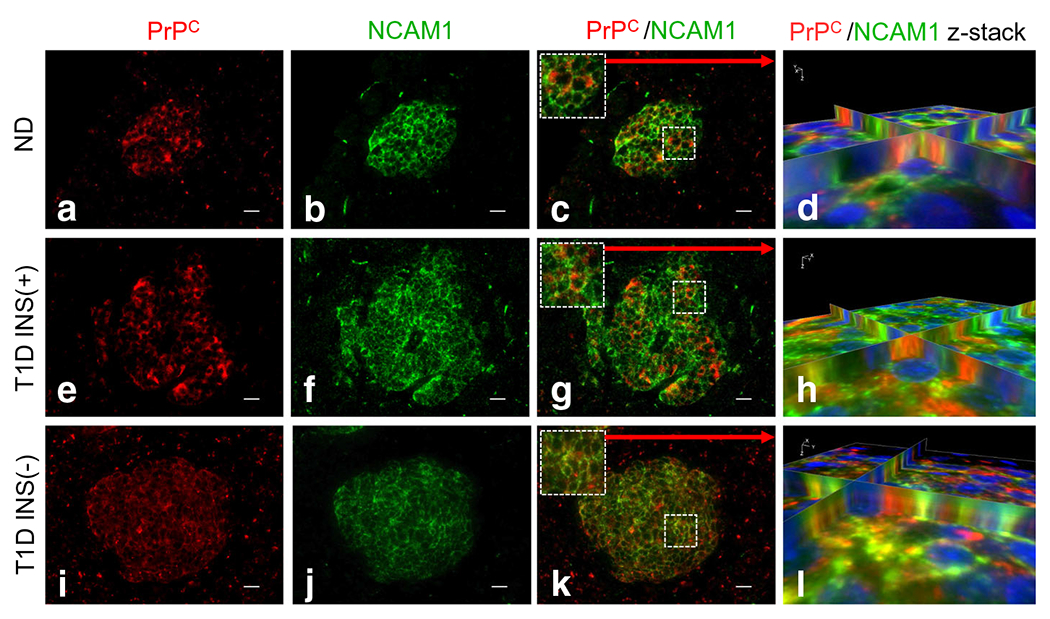
NCAM1 co-registers with PrPC primarily in the cell membrane. (a, e, i) PrPC is expressed in non-diabetic, type 1 diabetic INS(+) and type 1 diabetic INS(−) islets. (b, f, j) NCAM1 is highly expressed in the pancreatic islets of non-diabetic, type 1 diabetic INS(+) and type 1 diabetic INS(−) islets. (c, g, k) We found that NCAM1 highly co-registers with PrPC in all three groups as shown in z-stack 3D reconstruction images (d, h, l). Scale bars, 20 μm. ND, non-diabetic; T1D, type 1 diabetic
Discussion
The yin and yang prion proteins, PrPC and PrPSc, have established themselves as singularly famous and infamous, in that the normal protein, after years of intense investigation lacks a consensus function, whilst the concept of an infectious protein particle remains a difficult concept to grasp. Previous studies implicated a unique expression of PrPC in rodent and ungulate pancreatic islets, with a potential functional role for PrPC in glucose homeostasis [15–18]. For example, with regard to normal or pathophysiological states, several studies in PrPC knockout mice have demonstrated a role for PrPC in gluco-regulation [17], together with a PrPC-mediated modulation of glucose homeostasis through regulation of intracellular iron stores in pancreatic beta cells [15]. We have corroborated these findings by showing similar in situ expression of PrPC in non-diabetic human islets in organ donor pancreases, specifically in beta cells, and extended these results to demonstrate that PrPC is also expressed in type 1 diabetic islets that still harbour INS(+) beta cells. Strikingly, islets lacking beta cells [INS(−)] are still positive for PrPC, but have switched expression of PrPC to alpha cells. These results may implicate a beta cell regeneration mechanism relevant to the observation by Chakravarthy et al [46] that in type 1 diabetes patients, loss of DNA methyltransferase 1 (DNMT1) and aristaless related homeobox (ARX) in subsets of glucagon-expressing alpha cells leads to a population of cells expressing insulin and other beta cell-specific factors. The INS(−) islets may therefore be initiating a regeneration or survival mechanism involving PrPC-positive alpha cells as beta cell progenitors. This is consistent with a purported role for PrPC in pluripotency and stem cell differentiation [47, 48] and consistent with PrPC interaction with NCAM1, discussed below.
As one might expect, PrPC is expressed in beta cells in islets from AAb+ pancreases, identically to our non-diabetic cohort. However, close visual examination of numerous islets from each of our 12 AAb+ organ donor pancreases indicate that a vast majority, although not quantified, appear to have diminished PrPC expression compared with non-diabetic islets. This observation is in complete alignment with significantly diminished expression of PRNP mRNA in AAb+ pancreases observed from these donors. Of note, equally diminished expression is observed between AAb+ donors with single or multiple autoantibodies, with, in fact, ~75% of our cohort derived from single AAb+ donors. As a component of the cellular differentiation machinery, the reduction in PrPC expression in AAb+ cohort could signal a prelude to a reduced capacity for beta cell regeneration, prior to extensive beta cell loss associated with insulin therapy accompanying type 1 diabetes diagnosis.
Another unique observation in the human pancreas from our studies indicates two cellular pools of PrPC, localised to either the ER or plasma membrane, with the lack of expression in the Golgi apparatus in both beta and alpha cells. Conventional protein trafficking for secreted, integral membrane and GPI-anchored proteins involves synthesis in the ER, transit through the Golgi and vesicular transport of each of these protein classes to the plasma membrane [49, 50]. On the basis of our results, we propose that PrPC in the human pancreas may use an unconventional protein trafficking mechanism that bypasses the Golgi, employing either a Golgi reassembly stacking protein 2 (GRASP55) or HSP70–DnaJ heat shock protein family (Hsp40) member C14 (DNAJC14) transport pathway [51].
In addition to the type 1 diabetes-dependent transition of PrPC expression from beta to alpha cells in INS(−) islets, we have demonstrated a significant increase in PRNP transcription and protein synthesis in type 1 diabetic pancreases compared with our non-diabetic cohort. With no biological function for PrPC to rationalise the increased levels in pancreases from the type 1 diabetic cohort, we studied the expression of PrPC-interacting partners with known biological function to help provide a link to type 1 diabetes pathophysiology. Foremost, we found that PrPC within its two distinct cellular pools co-registers with the co-chaperone STI1 in the ER/cytosol, whilst the PrPC membrane fraction interacts with the cell adhesion protein, NCAM1. The existence of these two protein-protein complexes provides the opportunity to hypothesise on potential roles for PrPC in type 1 diabetes pathogenesis. Based on its interaction with STI1 numerous studies have implicated the stress-protective properties of PrPC [8, 43, 44, 52, 53]. In our results, not only did we observe type 1 diabetes-dependent increases in PrPC expression, but we also demonstrate a parallel increase in gene expression for its binding partner STIP1 in the type 1 diabetic pancreas. Therefore, increased levels of the PrPC–STI1 complex may be an attempt by type 1 diabetic islets to induce cytoprotective measures circumventing further endocrine cell loss, potentially offering a therapeutic opportunity to stimulate alpha to beta cell replenishment. Furthermore, it has been shown that PrPC can protect against Bax-mediated cell death in human primary neurons and cancer cells by inhibiting Bax activation through a pro-apoptotic conformational change and subsequent cytochrome C release [8, 54, 55]. This cytoprotective role may also be important in type 1 diabetes, based on recent evidence that loss of Bax and Bak protect against beta cell death under conditions of glucolipotoxicity and chronic ER stress [56].
Separately, co-registration of PrPC at the membrane with NCAM1 may support a putative function in cell signalling, differentiation and morphogenesis in the human pancreas [39, 40, 45]. For example, Santuccione et al [57] have shown that PrPC mediates the recruitment of NCAM1 to lipid rafts leading to cellular signalling through activation of the proto-oncogene, tyrosine kinase, FYN. In relation to the comparative evolution of neurons and beta cells, PrPC also regulates the neuronal polysialylation of NCAM1 thus controlling cell migration, axonal and dendritic projection, and synaptic targeting along with morphogenetic reprogramming specific to the epithelial-to-mesenchymal transition [58]. Linden [59] has, thus, consolidated results detailing a variety of potential PrPC functions and proposed that PrPC may propagate in lipid rafts a flexible cell-surface scaffold for the recruitment of a variety of signalling modules, thus providing a dynamic and adaptable platform for roles in physiology, behaviour and pathophysiology.
Our results on PrPC expression and tissue and intracellular localisation, as well as PrPC-interacting protein partners in non-diabetic, type 1 diabetic and AAb + pancreases provide potential new insights into the role PrPC may play in the pathogenesis of type 1 diabetes. A schematic summary of our results is depicted in Fig. 8. We recognise that future efforts using alternative methods (e.g. isolated islets, surrogate beta cell lines, pancreatic slice cultures) may afford advances into an improved mechanistic understanding. It is, however, exciting to speculate that intervention through PrPC protein–ligand interactions could be therapeutic in type 1 diabetes. However, without a universally accepted function, further mechanistic studies in models of type 1 diabetes remain highly speculative.
Fig. 8.
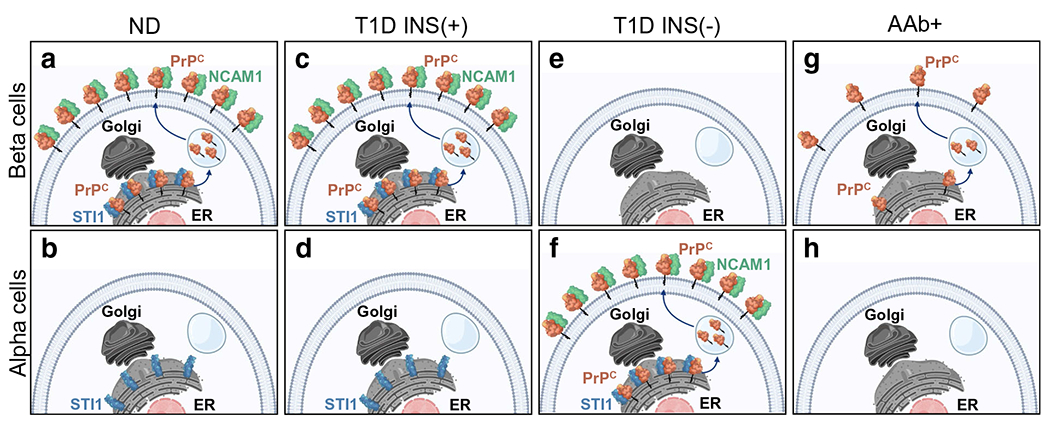
Schematic summary of human PrPC cellular localisation, trafficking and interacting partners in beta and alpha cells in non-diabetic, type 1 diabetic and AAb+ human pancreas. (a) A summary of all the results in non-diabetic islets, including: PrPC co-registration with beta cells; PrPC existence in two cellular pools with interaction of PrPC with NCAM1 on the plasma membrane and STI1 in the ER, along with the proposal that PrPC must traffic from the ER to the plasma membrane bypassing transit through the Golgi. For illustration, the unconventional protein trafficking mechanism is depicted using vesicle-mediated transit from the ER to the plasma membrane. (a, c, g) PrPC co-registers with beta cells in non-diabetic, type 1 diabetic INS(+) islets and AAb+ islets, with no co-registration with alpha cells (b, d, h). (e, f) PrPC co-registers with alpha cells in type 1 diabetic INS(−) islets. (g) PrPC is expressed at significantly lower levels in AAb+ beta cells compared with non-diabetic and INS(+) islets (a, c). Note: STI1 and NCAM1 were not measured in AAb+ (g, h). Graphic design created in biorender.com (https://biorender.com)
Supplementary Material
Research in context.
What is already known about this subject?
The normal cellular prion protein (PrPC) is a conserved mammalian glycoprotein found on the outer plasma membrane and it is expressed by a wide range of tissues throughout the body
The complete repertoire of its functions has not been fully determined, but a folded aggregate version (PrPSc) is linked to neurodegenerative disorders affecting both animals and humans
PrPC has been linked to modulation of signalling pathways, neuroprotection, anti-apoptotic activity and glucose homeostasis, among other things
What is the key question?
Does PrPC localise in human pancreases from organ donors with and without type 1 diabetes, and does it interact with partner proteins to affect cell function?
What are the new findings?
PrPC is highly expressed in the pancreatic islets of non-diabetic and autoantibody-positive donors and of donors with type 1 diabetes and with insulin-positive islets, and co-registers almost exclusively with beta cells
In pancreatic endocrine cells PrPC is synthesised in the endoplasmic reticulum, but utilises an unconventional secretory pathway that bypasses the Golgi to reach the plasma membrane
Cellular prion protein co-registers with STI1 partner protein suggesting a PrPC–STI1 complex that might induce cytoprotective measures to circumvent further endocrine cell loss
How might this impact on clinical practice in the foreseeable future?
Our results provide potential new insights into the role that PrPC may play in the pathogenesis of type 1 diabetes
Acknowledgements
The authors would like to thank the organ donors and their families for their precious contributions to nPOD for research, without which this work would not be possible. Organ Procurement Organizations (OPO) partnering with nPOD to provide research resources are listed at http://www.jdrfnpod.org//for-partners/npod-partners/.
Funding
Research reported in this publication was supported by the network for Pancreatic Organ donors with Diabetes (nPOD; RRID-SCR_014541), a collaborative type 1 diabetes research project sponsored by JDRF (nPOD:5-SRA-2018-557-Q-R) and The Leona M. & Harry B. Helmsley Charitable Trust (grant no. 2018PG-T1D053), as well as NIH grant 1UC4DK108132-01, principal investigator MAA. In addition, the Organ Procurement Organizations (OPO) partnering with nPOD to provide research resources are listed at http://www.jdrfnpod.org//for-partners/npod-partners/.
Abbreviations
- AAb+
Autoantibody-positive
- ER
Endoplasmic reticulum
- GM130
Golgin A2
- GPI
Glycosyl-phosphatidylinositol
- HSP
Heat-shock protein
- INS(+)
Insulin-positive
- INS(−)
Insulin-negative
- MIQE
Minimum Information for Publication of Quantitative Real-Time PCR Experiments
- NCAM1
Neural cell adhesion molecule 1
- PrPC
Cellular prion protein
- PrPSc
Scrapie form of prion protein (pathogenic, alternatively folded aggregate)
- qRT-PCR
Quantitative RT-PCR
- STI1
Stress-inducible phosphoprotein 1
- UNOS
United Network for Organ Sharing
- WFS1
Wolfram syndrome 1
Footnotes
Supplementary Information The online version contains peer-reviewed but unedited Supplementary material available at https://doi.org/10.1007/s00125-021-05501-8.
Authors’ relationships and activities The authors declare that there are no relationships or activities that might bias, or be perceived to bias, their work.
Data availability
Application for datasets generated during and/or analysed during the current study may be considered by the corresponding author on reasonable request.
References
- 1.Prusiner SB (1998) The prion diseases. Brain Pathol 8(3):499–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wulf MA, Senatore A, Aguzzi A (2017) The biological function of the cellular prion protein: an update. BMC Biol 15(1):34. 10.1186/s12915-017-0375-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkinson CJ, Zhang K, Munn AL, Wiegmans A, Wei MQ (2016) Prion protein scrapie and the normal cellular prion protein. Prion 10(1):63–82. 10.1080/19336896.2015.1110293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kovacs GG, Budka H (2008) Prion diseases: from protein to cell pathology. Am J Pathol 172(3):555–565. 10.2353/ajpath.2008.070442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prusiner SB (1982) Novel proteinaceous infectious particles cause scrapie. Science 216(4542):136–144. 10.1126/science.6801762 [DOI] [PubMed] [Google Scholar]
- 6.Otero A, Duque Velasquez C, Johnson C et al. (2019) Prion protein polymorphisms associated with reduced CWD susceptibility limit peripheral PrP(CWD) deposition in orally infected white-tailed deer. BMC Vet Res 15(1):50. 10.1186/s12917-019-1794-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryskalin L, Busceti CL, Biagioni F et al. (2019) Prion protein in glioblastoma multiforme. Int J Mol Sci 20(20). 10.3390/ijms20205107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castle AR, Gill AC (2017) Physiological functions of the cellular prion protein. Front Mol Biosci 4:19. 10.3389/fmolb.2017.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tee BL, Longoria Ibarrola EM, Geschwind MD (2018) Prion diseases. Neurol Clin 36(4):865–897. 10.1016/j.ncl.2018.07.005 [DOI] [PubMed] [Google Scholar]
- 10.Asher DM, Gregori L (2018) Human transmissible spongiform encephalopathies: historic view. Handb Clin Neurol 153:1–17. 10.1016/B978-0-444-63945-5.00001-5 [DOI] [PubMed] [Google Scholar]
- 11.Kovacs GG, Budka H (2009) Molecular pathology of human prion diseases. Int J Mol Sci 10(3):976–999. 10.3390/ijrns10030976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corbett GT, Wang Z, Hong W et al. (2020) PrP is a central player in toxicity mediated by soluble aggregates of neurodegeneration-causing proteins. Acta Neuropathol 139(3):503–526. 10.1007/s00401-019-02114-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westergard L, Christensen HM, Harris DA (2007) The cellular prion protein (PrP(C)): its physiological function and role in disease. Biochim Biophys Acta 1772(6):629–644. 10.1016/j.bbadis.2007.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atouf F, Scharfmann R, Lasmezas C, Czemichow P (1994) Tight hormonal control of PrP gene expression in endocrine pancreatic cells. Biochem Biophys Res Commun 201 (3):1220–1226. 10.1006/bbrc.1994.1835 [DOI] [PubMed] [Google Scholar]
- 15.Ashok A, Singh N (2018) Prion protein modulates glucose homeostasis by altering intracellular iron. Sci Rep 8(1):6556. 10.1038/s4f598-018-24786-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strom A, Wang GS, Reimer R, Finegood DT, Scott FW (2007) Pronounced cytosolic aggregation of cellular prion protein in pancreatic beta-cells in response to hyperglycemia. Lab Investig 87(2):139–149. 10.1038/labinvest.3700500 [DOI] [PubMed] [Google Scholar]
- 17.Strom A, Wang GS, Scott FW (2011) Impaired glucose tolerance in mice lacking cellular prion protein. Pancreas 40(2):229–232. 10.1097/mpa.0b013e3181f7e547 [DOI] [PubMed] [Google Scholar]
- 18.Amselgruber WM, Buttner M, Schlegel T, Schweiger M, Pfaff E (2006) The normal cellular prion protein (PrPc) is strongly expressed in bovine endocrine pancreas. Histochem Cell Biol 125(4):441–448. 10.1007/s00418-005-0089-6 [DOI] [PubMed] [Google Scholar]
- 19.de Brito G, Lupinacci FC, Beraldo FH et al. (2017) Loss of prion protein is associated with the development of insulin resistance and obesity. Biochem J 474(17):2981–2991. 10.1042/BCJ20170137 [DOI] [PubMed] [Google Scholar]
- 20.Zhu C, Schwarz P, Abakumova I, Aguzzi A (2015) Unaltered prion pathogenesis in a mouse model of high-fat diet-induced insulin resistance. PLoS One 10(12):e0144983. 10.1371/journal.pone.0144983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eberhard D (2013) Neuron and beta-cell evolution: learning about neurons is learning about beta-cells. Bioessays 35(7):584. 10.1002/bies.201300035 [DOI] [PubMed] [Google Scholar]
- 22.Visner GA, Dougall WC, Wilson JM, Burr IA, Nick HS (1990) Regulation of manganese superoxide dismutase by lipopolysaccharide, interleukin-1, and tumor necrosis factor. Role in the acute inflammatory response. J Biol Chem 265(5):2856–2864. 10.1016/S0021-9258(19)39880-1 [DOI] [PubMed] [Google Scholar]
- 23.Bustin SA, Benes V, Garson JA et al. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55(4):611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 24.Torres M, Cartier L, Matamala JM, Hernandez N, Woehlbier U, Hetz C (2012) Altered prion protein expression pattern in CSF as a biomarker for Creutzfeldt-Jakob disease. PLoS One 7(4):e36159. 10.1371/journal.pone.0036159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis V, Hooper NM (2011) The role of lipid rafts in prion protein biology. Front Biosci (Landmark Ed) 16:151–168. 10.2741/3681 [DOI] [PubMed] [Google Scholar]
- 26.Takeda K, Inoue H, Tanizawa Y et al. (2001) WFS1 (Wolfram syndrome 1) gene product: predominant subcellular localization to endoplasmic reticulum in cultured cells and neuronal expression in rat brain. Hum Mol Genet 10(5):477–484. 10.1093/hmg/10.5.477 [DOI] [PubMed] [Google Scholar]
- 27.Puthenveedu MA, Bachert C, Puri S, Lanni F, Linstedt AD (2006) GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat Cell Biol 8(3):238–248. 10.1038/ncb1366 [DOI] [PubMed] [Google Scholar]
- 28.Dworzak MN, Fritsch G, Buchinger P et al. (1994) Flow cytometric assessment of human MIC2 expression in bone marrow, thymus, and peripheral blood. Blood 83(2):415–425. 10.1182/blood.V83.2.415.415 [DOI] [PubMed] [Google Scholar]
- 29.Grieve AG, Rabouille C (2011) Golgi bypass: skirting around the heart of classical secretion. Cold Spring Harb Perspect Biol 3(4):a005298. 10.1101/cshperspect.a005298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gee HY, Noh SH, Tang BL, Kim KH, Lee MG (2011) Rescue of DeltaF508-CFTR trafficking via a GRASP-dependent unconventional secretion pathway. Cell 146(5):746–760. 10.1016/j.cell.2011.07.021 [DOI] [PubMed] [Google Scholar]
- 31.Beery ML, Jacobsen LM, Atkinson MA, Butler AE, Campbell-Thompson M (2019) Islet amyloidosis in a child with type 1 diabetes. Islets 11(2):44–49. 10.1080/19382014.2019.1599707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Couce M, O’Brien TD, Moran A, Roche PC, Butler PC (1996) Diabetes mellitus in cystic fibrosis is characterized by islet amyloidosis. J Clin Endocrinol Metab 81 (3):1267–1272. 10.1210/jcem.81.3.8772610 [DOI] [PubMed] [Google Scholar]
- 33.Maloy AL, Longnecker DS, Greenberg ER (1981) The relation of islet amyloid to the clinical type of diabetes. Hum Pathol 12(10):917–922. 10.1016/s0046-8177(81)80197-9 [DOI] [PubMed] [Google Scholar]
- 34.Mukherjee A, Morales-Scheihing D, Butler PC, Soto C (2015) Type 2 diabetes as a protein misfolding disease. Trends Mol Med 21 (7):439–449. 10.1016/j.molmed.2015.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukherjee A, Soto C (2017) Prion-like protein aggregates and type 2 diabetes. Cold Spring Harb Perspect Med 7(5):a024315. 10.1101/cshperspect.a024315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta D, Leahy JL (2014) Islet amyloid and type 2 diabetes: over-production or inadequate clearance and detoxification? J Clin Invest 124(8):3292–3294. 10.1172/JCI77506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yakupova EI, Bobyleva LG, Vikhlyantsev IM, Bobylev AG (2019) Congo Red and amyloids: history and relationship. Biosci Rep 39(1):BSR20181415. 10.1042/BSR20181415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xue C, Lin TY, Chang D, Guo Z (2017) Thioflavin T as an amyloid dye: fibril quantification, optimal concentration and effect on aggregation. R Soc Open Sci 4(1): 160696. 10.1098/rsos.160696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.da Fonseca ACC, Matias D, Geraldo LHM et al. (2020) The multiple functions of the co-chaperone stress inducible protein 1. Cytokine Growth Factor Rev. 10.1016/j.cytogfr.2020.06.003 [DOI] [PubMed] [Google Scholar]
- 40.Schmitt-Ulms G, Legname G, Baldwin MA et al. (2001) Binding of neural cell adhesion molecules (N-CAMs) to the cellular prion protein. J Mol Biol 314(5):1209–1225. 10.1006/jmbi.2000.5183 [DOI] [PubMed] [Google Scholar]
- 41.Hernandez MP, Sullivan WP, Toft DO (2002) The assembly and intermolecular properties of the hsp70-Hop-hsp90 molecular chaperone complex. J Biol Chem 277(41):38294–38304. 10.1074/jbc.M206566200 [DOI] [PubMed] [Google Scholar]
- 42.Sytnyk V, Leshchyns’ka I, Schachner M (2017) Neural cell adhesion molecules of the immunoglobulin superfamily regulate synapse formation, maintenance, and function. Trends Neurosci 40(5):295–308. 10.1016/j.tins.2017.03.003 [DOI] [PubMed] [Google Scholar]
- 43.Zanata SM, Lopes MH, Mercadante AF et al. (2002) Stress-inducible protein 1 is a cell surface ligand for cellular prion that triggers neuroprotection. EMBO J 21(13):3307–3316. 10.1093/emboj/cdf325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopes MH, Hajj GN, Muras AG et al. (2005) Interaction of cellular prion and stress-inducible protein 1 promotes neuritogenesis and neuroprotection by distinct signaling pathways. J Neurosci 25(49):11330–11339. 10.1523/JNEUROSCI.2313-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mehrabian M, Brethour D, Wang H, Xi Z, Rogaeva E, Schmitt-Ulms G (2015) The prion protein controls polysialylation of neural cell adhesion molecule 1 during cellular morphogenesis. PLoS One 10(8):e0133741. 10.1371/journal.pone.0133741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chakravarthy H, Gu X, Enge M et al. (2017) Converting adult pancreatic islet alpha cells into beta cells by targeting both Dnmt1 and Arx. Cell Metab 25(3):622–634. 10.1016/j.cmet.2017.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miranda A, Ramos-Ibeas P, Pericuesta E, Ramirez MA, Gutierrez-Adan A (2013) The role of prion protein in stem cell regulation. Reproduction 146(3):R91–R99. 10.1530/REP-13-0100 [DOI] [PubMed] [Google Scholar]
- 48.Steele AD, Emsley JG, Ozdinler PH, Lindquist S, Macklis JD (2006) Prion protein (PrPc) positively regulates neural precursor proliferation during developmental and adult mammalian neurogenesis. Proc Natl Acad Sci U S A 103(9):3416–3421. 10.1073/pnas.0511290103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viotti C (2016) ER to Golgi-dependent protein secretion: the conventional pathway. Methods Mol Biol 1459:3–29. 10.1007/978-1-4939-3804-9_1 [DOI] [PubMed] [Google Scholar]
- 50.Maeda Y, Kinoshita T (2011) Structural remodeling, trafficking and functions of glycosylphosphatidylinositol-anchored proteins. Prog Lipid Res 50(4):411–424. 10.1016/j.plipres.2011.05.002 [DOI] [PubMed] [Google Scholar]
- 51.Rabouille C (2017) Pathways of unconventional protein secretion. Trends Cell Biol 27(3):230–240. 10.1016/j.tcb.2016.11.007 [DOI] [PubMed] [Google Scholar]
- 52.Ostapchenko VG, Beraldo FH, Mohammad AH et al. (2013) The prion protein ligand, stress-inducible phosphoprotein 1, regulates amyloid-beta oligomer toxicity. J Neurosci 33(42):16552–16564. 10.1523/JNEUROSCI.3214-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beraldo FH, Ostapchenko VG, Xu JZ et al. (2018) Mechanisms of neuroprotection against ischemic insult by stress-inducible phosphoprotein-1/prion protein complex. J Neurochem 145(1):68–79. 10.1111/jnc.14281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roucou X, Giannopoulos PN, Zhang Y, Jodoin J, Goodyer CG, LeBlanc A (2005) Cellular prion protein inhibits proapoptotic Bax conformational change in human neurons and in breast carcinoma MCF-7 cells. Cell Death Differ 12(7):783–795. 10.1038/sj.cdd.4401629 [DOI] [PubMed] [Google Scholar]
- 55.Gill AC, Castle AR (2018) The cellular and pathologic prion protein. Handb Clin Neurol 153:21–44. 10.1016/B978-0-444-63945-5.00002-7 [DOI] [PubMed] [Google Scholar]
- 56.White SA, Zhang LS, Pasula DJ, Yang YHC, Luciani DS (2020) Bax and Bak jointly control survival and dampen the early unfolded protein response in pancreatic beta-cells under glucolipotoxic stress. Sci Rep 10(1):10986. 10.1038/s41598-020-67755-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santuccione A, Sytnyk V, Leshchyns’ka I, Schachner M (2005) Prion protein recruits its neuronal receptor NCAM to lipid rafts to activate p59fyn and to enhance neurite outgrowth. J Cell Biol 169(2):341–354. 10.1083/jcb.200409127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mehrabian M, Hildebrandt a Schmitt-Ulms G (2016) NCAM1 polysialylation: the prion protein’s elusive reason for being? ASN Neuro 8(6): 1759091416679074. 10.1177/1759091416679074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Linden R (2017) The biological function of the prion protein: a cell surface scaffold of signaling modules. Front Mol Neurosci 10:77. 10.3389/fnmol.2017.00077 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Application for datasets generated during and/or analysed during the current study may be considered by the corresponding author on reasonable request.


