Abstract
Hierarchical genotyping approaches can provide insights into the source, geography and temporal distribution of bacterial pathogens. Multiple hierarchical SNP genotyping schemes have previously been developed so that new isolates can rapidly be placed within pre-computed population structures, without the need to rebuild phylogenetic trees for the entire dataset. This classification approach has, however, seen limited uptake in routine public health settings due to analytical complexity and the lack of standardized tools that provide clear and easy ways to interpret results. The BioHansel tool was developed to provide an organism-agnostic tool for hierarchical SNP-based genotyping. The tool identifies split k-mers that distinguish predefined lineages in whole genome sequencing (WGS) data using SNP-based genotyping schemes. BioHansel uses the Aho-Corasick algorithm to type isolates from assembled genomes or raw read sequence data in a matter of seconds, with limited computational resources. This makes BioHansel ideal for use by public health agencies that rely on WGS methods for surveillance of bacterial pathogens. Genotyping results are evaluated using a quality assurance module which identifies problematic samples, such as low-quality or contaminated datasets. Using existing hierarchical SNP schemes for Mycobacterium tuberculosis and Salmonella Typhi, we compare the genotyping results obtained with the k-mer-based tools BioHansel and SKA, with those of the organism-specific tools TBProfiler and genotyphi, which use gold-standard reference-mapping approaches. We show that the genotyping results are fully concordant across these different methods, and that the k-mer-based tools are significantly faster. We also test the ability of the BioHansel quality assurance module to detect intra-lineage contamination and demonstrate that it is effective, even in populations with low genetic diversity. We demonstrate the scalability of the tool using a dataset of ~8100 S. Typhi public genomes and provide the aggregated results of geographical distributions as part of the tool’s output. BioHansel is an open source Python 3 application available on PyPI and Conda repositories and as a Galaxy tool from the public Galaxy Toolshed. In a public health context, BioHansel enables rapid and high-resolution classification of bacterial pathogens with low genetic diversity.
Keywords: bacterial typing, contamination detection, genotyping, k-mer, software, SNP
Data Summary
BioHansel is a Python 3 application available as PyPI, Conda and Galaxy Tool Shed packages. It is an open source application distributed under the Apache License, Version 2.0. The source code is available at https://github.com/phac-nml/biohansel. The BioHansel user guide is available at https://bio-hansel.readthedocs.io/en/readthedocs/.
Impact Statement.
Hierarchical genotyping approaches provide multi-resolution nomenclatures that assist in surveillance and outbreak detection, and that contribute contextual information on pathogen lineages to provide insights into their source, geography and temporal distribution. Predefined hierarchical SNP genotyping schemes have been developed for numerous organisms, but uptake of these approaches has been limited due to the lack of tools to readily incorporate new schemes and update them as new lineages are identified. To successfully apply these techniques in a public health context, the typing results must be readily interpretable, reliable and easy to communicate. BioHansel addresses these needs by requiring minimal parameters from the user and providing clear genotyping results that have been verified by the built-in quality assurance module. BioHansel is an organism-agnostic tool for fast, flexible and readily interpretable hierarchical genotyping of haploid genomes, requiring minimal computational resources.
Introduction
Public health, animal health, food safety and environmental authorities around the world are actively working to operationalize whole genome sequencing (WGS) technologies for their infectious disease diagnostics, surveillance, and outbreak detection and response programmes. Data analysis is one of the biggest challenges facing the adoption of WGS for these applications due to its complexity and the need for bioinformatics support for analysis and interpretation of the data [1, 2]. For example, in a foodborne pathogen outbreak situation, there are extreme pressures upon public health investigators to rapidly and accurately identify the outbreak source in order to avoid potentially severe health, financial and legal repercussions of incorrect source attribution. Genomic epidemiology of outbreak-related pathogens using WGS involves contextualizing the bacterial isolates in a broader population [3–5], which requires a plethora of software tools. The results of the WGS analysis then must be distilled into a readily communicable format for epidemiologists, clinicians and food safety officials [6]. Genomic data analysis pipelines that serve these mission-critical programmes must implement robust, reproducible and computationally tractable analytical methods that generate accurate and informative results for end-users without extensive bioinformatics expertise [6].
Numerous analytical approaches exist to quantify the relatedness between bacterial isolates using WGS data, with gene-by-gene and SNP-based approaches being the two most commonly used in public health [2, 7–9]. The most appropriate approach is determined based on the biology of the organism and the specific purpose of the resulting analysis [7, 10]. An SNP-based approach provides maximal resolution between organisms with low levels of genetic diversity because the analysis includes intergenic regions and does not compress multiple genetic changes into a single allele. To perform SNP-based analyses of genomic data, the raw sequence reads typically must be mapped against a common reference sequence using one of the numerous tools available for short-read alignment [9, 11, 12]. Variant calling is then performed on the quality-filtered reference-mapped reads, considering both the individual base quality and mapping scores [9, 11, 13]. An SNP-based approach can comprehensively identify all variants with respect to the reference sequence, but requires significant computational resources and the results obtained can differ depending on the software and parameter settings used [14]. There are numerous SNP identification pipelines available including CFSAN [15], Lyve-SET [11], snippy [16] and SNVPhyl [9]. Each pipeline has its own complex set of parameters and working hypotheses. Additionally, in a recent review of 41 different SNP identification pipelines, it was demonstrated that the selection of the appropriate reference genome for SNP calling was shown to have a dramatic effect on the variants identified, in addition to the variability introduced by the pipeline [12]. The need to map to a common reference sequence limits portability of the results because the optimal reference could be different depending on the scope of the analysis (whole species, serotype or sub-lineage). Furthermore, isolate relatedness is examined using a phylogenetic tree that must be recomputed every time a new isolate is added: this is very computationally expensive and can limit scalability.
Hierarchal SNP genotyping approaches divide populations into lineages and sub-lineages based on pre-defined mutations which are used to place new isolates phylogenetically without the need to construct a new phylogeny each time. Additionally, they provide a nested hierarchical nomenclature that is easily incorporated into line lists and spreadsheet databases that are common tools utilized by epidemiologists during outbreak investigations. Hierarchical nomenclatures have been developed for multiple bacterial pathogens with low genetic diversity such as Mycobacterium tuberculosis (MTB) [4], Salmonella enterica subsp. enterica serovar Typhi (S. Typhi) [3], Salmonella enterica subsp. enterica serovar Heidelberg [17] and Bordetella pertussis [18]. However, there is currently no generic and flexible tool available to perform hierarchical SNP genotyping. Consequently, the usability of novel SNP genotyping schemes is severely limited unless the authors also create a tool that can specifically support their scheme. Of the tools that are currently available, SnapperDB implements a hierarchical SNP genotyping nomenclature termed an ‘SNP address’, which is dynamically updated as new isolates are incorporated into the central database [8]. SnapperDB does not incorporate a model of evolution and its analytical results cannot be replicated without using an identical central database of isolates [8]. This is in contrast to gene-by-gene approaches for which there are many tools, including ARIBA [19], chewBBACA [20], MentaLiST [21] and SRST2 [22], that can readily incorporate new schemes. SNP genotyping pipelines need to become similarly portable in order to be adopted by cross-jurisdictional programmes.
The massive volume of genomic data being produced also necessitates the development of faster and more scalable approaches for comparing genomes [5, 23]. Many of these faster methods for genome comparisons are based on k-mers, which are small subsequences of a known, defined length. Mash was amongst the first k-mer-based tools able to rapidly estimate the genetic relatedness between two genomes, doing so in a fraction of the time required by alignment-based methods [23]. Numerous other k-mer tools have been developed over the last few years to identify sequence variations between genomes. One such toolset is the Split K-mer Analysis (SKA) suite, which is designed as a comprehensive set of k-mer-based tools for routine genomic epidemiology analysis of highly similar genomes [5]. The k-mer-based approach to genome comparison in SKA is highly congruent with mapping-based approaches and requires substantially fewer computational resources [5].
Contamination is another important and frequently encountered concern when using WGS for microbial genomic applications. Contamination can confound subtype assignment and be troublesome to detect [24, 25]. Mixed infections, which present similarly to contamination, can also be difficult to detect [26, 27]. The most common sources of contamination include improperly isolated genomic material, environmental contamination, the libraries used to prepare the genomic material for sequencing and DNA barcode ‘cross-talk’ generated during the sequencing step [3–5]. Contamination detection pipelines are therefore commonly applied to newly generated WGS data prior to analysis. These pipelines normally assign reads to taxa using fast phylotyping tools such as Kraken [28], Centrifuge [29] or Kaiju [30]. However, these tools typically cannot reliably assign taxa beyond the species level and thus are unsuitable for identifying genomic contamination from the same lineage [31, 32]. In contrast to the many tools available to detect contamination between species, there are fewer tools available for identifying intra-lineage contamination in microbial populations with low sequence diversity [27]. To the best of our knowledge, ConFindr is the only organism-agnostic tool which identifies both inter- and intra-species contamination by searching for the presence of multiple variant bases per position in a set of conserved single-copy ribosomal protein-coding genes, or in a user-defined gene schema [24]. Illumina sequence data contain noise in addition to low levels of contamination [33] that can occur within a flow cell, so ConFindr uses a requirement of three polymorphic sites to reduce false positive results [24]. However, ConFindr is unable to detect contamination between very closely related isolates which do not have genetic diversity in their ribosomal multilocus sequebcing type (rMLST) loci [24]. Due to the hierarchical nature of SNP genotyping schemes and the fact that they are a curated set of markers for delineating populations with low genetic diversity, the presence of incompatible sets of SNPs can readily indicate issues with a given sample such as contamination, mixed infection or recombination events. Incompatible SNPs can represent base heterozygosity or the presence of SNP states that should only occur in known combinations in specific lineages. Thus, a hierarchical SNP-based genotyping scheme can provide the means to detect contamination and reliably identify mixed infections even between the most closely related genotypes in a pathogen population that has very low genetic diversity.
Here we present BioHansel, an organism-agnostic, k-mer-based genotyping tool that can readily incorporate new and updated hierarchical SNP genotyping schemes. BioHansel was designed with a focus on producing easily interpretable results, so as to eliminate the need for end users to have extensive bioinformatics expertise. BioHansel reports provide the user with the hierarchical genotype call, along with clearly interpretable quality control (QC) messages as to the presence of intra-strain contamination or low confidence in the genotyping result. In this study we benchmark the genotyping and contamination detection performance of two k-mer based tools, BioHansel and SKA [5], and two reference mapping tools, Genotyphi [3] and TBProfiler [34], using two published SNP schemes used for genotyping: S. Typhi and MTB. Furthermore, we demonstrate the scalable nature of BioHansel by genotyping ~8100 public S. Typhi read sets and highlighting temporal and geographical trends in the data. The results of the global S. Typhi analysis are provided with BioHansel genotyping reports to contextualize each genotype.
Methods
BioHansel design and implementation
BioHansel is a fast and flexible k-mer-based genotyping tool designed to support a wide variety of hierarchical genotyping schemes. The software is implemented in Python 3 with open-source Python library dependencies. The tool uses the Aho-Corasick algorithm [35], implemented in the pyahocorasick Python library (https://github.com/WojciechMula/pyahocorasick), to search assemblies or raw Illumina fastq reads in linear time for a predefined set of variable-length k-mers. The software provides three levels of result reporting: a simplified report containing a genotype call along with the reliability of that result, a results summary providing information about specific k-mers identified in relation to those expected for the targeted pathogen, and a report with additional details such as the target k-mer frequency, for troubleshooting purposes.
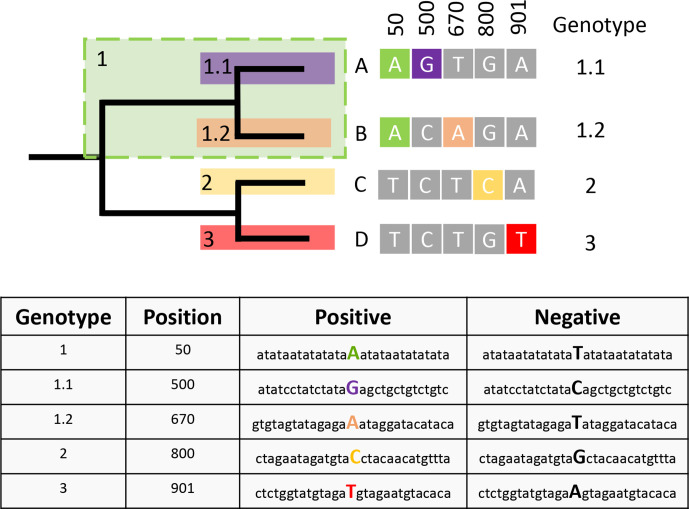
BioHansel takes a query DNA sequence or sequences, and a genotyping scheme as input. A small BioHansel-compatible genotyping scheme, based on variant positions against a reference genome, is presented in Fig. 1. In a typical BioHansel genotyping scheme, lineage-defining SNP positions are encoded as pairs of short DNA sequences each containing one of two possible nucleotides of the variant base surrounded by a genome sequence that is conserved across the whole pathogen population targeted by the genotyping scheme. SKA utilizes a similar concept, which is defined as a split k-mer pair with a variable base [6]. BioHansel also requires that lineage designations adhere to a strict hierarchical structure, in which each clade (or lineage) must be defined by at least one exclusive variant shared by all clade members. Each nested sub-clade must possess the variant(s) defining the parent clade, as well as additional exclusive variant(s) shared amongst all members of the sub-clade. In the example shown in Fig. 1, samples A and B belong to the parent clade 1, as they both contain the variant base A in position 50, which is not present in any other clade, and therefore serves as the distinguishing variant defining clade 1 (e.g. genotype 1). Samples A and B further represent sub-clades 1.1 and 1.2 respectively, defined by additional variants at genome positions 500 and 670 (Fig. 1). In BioHansel, the k-mer that is present in the genotype/lineage being defined is called the positive k-mer, and the paired k-mer that is present at that genome position in the rest of the pathogen population (outside of the defined genotype/lineage) is called the negative k-mer. In the given example, the positive k-mer for genotype 1 contains the variant base A in the middle of the conserved flanking sequences, while the middle base is a T in the paired negative k-mer (Fig. 1). BioHansel uses the presence of both positive and negative k-mers in a scheme to support the quality assurance module.
Fig. 1.
Phylogenetic representation of a BioHansel-compatible hierarchical SNP genotyping scheme based on genome-wide variant positions. Samples A and B belong to the same parent genotype 1 so they contain the same defining SNP at position 50. The other genotyping SNPs are exclusive to their corresponding type. Genotyping split k-mers for BioHansel are derived by extracting the sequence from the reference sequence around the variant position. The positive k-mer is used to define the presence of a genotype level and should only be found in members of a genotype. The negative k-mer would be present in members which are not part of the genotype. A BioHansel scheme uses the genome position of the variant as the unique ID for the split k-mer pair combined with the pair’s corresponding genotype.
Developing a new scheme is highly complex and requires extensive expertise in phylogenomics. However, adapting existing hierarchical SNP schemes to work with BioHansel only requires a phylogenetic tree showing the evolutionary relationships between the defined genotypes, and knowing the position of each genotype-defining SNP in the genome along with its conserved flanking sequences (Fig. 1). We adapted two previously published hierarchical genotyping schemes for MTB [4] and S. Typhi [3] using split k-mers centred on the SNPs defining each genotype. We selected a k-mer length of 33 for the BioHansel schemes, which is short enough to fit within a single Illumina read while still providing sufficient specificity. However, BioHansel does not impose a fixed k-mer length, and although not required in these instances, BioHansel also supports the use of indels in the genotype-defining k-mers. The nomenclature of the MTB and Typhi schemes required some modifications so that the schemes followed a strict hierarchy, which is required for BioHansel. The published scheme genotypes are maintained for reporting through the inclusion of a metadata table which maps the adapted genotypes to the genotypes present in the original schemes [4, 5]. When adapting an existing SNP scheme for use in BioHansel, the selected k-mers should have highly conserved sequences flanking the variant base. However, BioHansel does support the inclusion of degenerate bases in k-mers, which can be necessary if multiple polymorphisms are present across the population in the genome sequence surrounding a genotype-defining SNP base.
A key feature of BioHansel is that it allows genotypes to be defined by multiple redundant k-mer targets in each scheme, contrary to MLST schemes where each locus is independent and missing data can prevent the call of an MLST type. This redundancy allows for a certain proportion of missing data in a successful genotype call. Unlike MLST, split k-mer hierarchal typing is not suitable for high genetic diversity or recombinant organisms, as the quality of the data is assessed by the presence of both positive and negative k-mers. For example, when a high proportion of the scheme k-mers are missing in some genotypes in the population, BioHansel’s ability to assess the quality of these genotype calls is limited. As a default, BioHansel allows up to 5% of scheme k-mers to be missing from a dataset for a successful genotype call, but this is configurable by the user. Recombinant organisms similarly are not suited to this analytical approach since BioHansel assumes strict vertical inheritance.
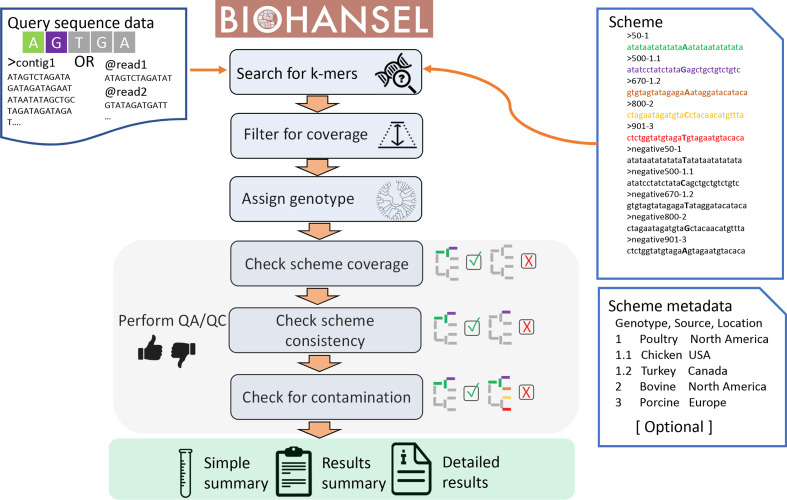
The overall BioHansel workflow is presented in Fig. 2, using an example based on the small scheme described in Fig. 1. BioHansel takes as input fasta- or fastq-formatted sequence data, along with a fasta-formatted scheme file listing the genotyping split k-mers, as well as an optional metadata table. If raw Illumina data (in fastq format) are provided, a filtering step is performed to remove low-frequency k-mers which are probably the result of sequencing noise. Genotype assignment uses only the identified positive k-mers, and the overall genotype is determined by the most resolved genotyping k-mer(s) identified; i.e. the k-mer(s) associated with the genotype(s) that have the highest number of hierarchical levels. In the example shown in Fig. 2, positive k-mers are identified for both genotype 1 and 1.1. BioHansel reports the highest resolution genotype found for a given query. Since positive k-mers were found for both genotype 1 and genotype 1.1 in the example given in Fig. 2, that query sample was determined to be genotype 1.1. The negative k-mers are only used for QC purposes: assessing the completeness of the dataset for the pathogen targeted by the scheme, the reliability of the final genotype call and the presence of contamination.
Fig. 2.
BioHansel genotyping workflow. Query sequence data in fasta or fastq format are provided to the tool with a corresponding scheme and optional scheme metadata table. The scheme provides a set of k-mers and places them in a hierarchy. BioHansel searches for the specified k-mers in the query data. Fastq data are filtered based on coverage to remove low-abundance k-mers. BioHansel examines all the identified positive k-mers to find the most resolved genotype, the deepest in the hierarchy, which will be the overall genotyping call. The genotyping results are then evaluated through the QA/QC module to determine if a sample has an adequate number of scheme k-mers to consider the result as reliable. The identified k-mers are examined for consistency with the scheme hierarchy. In the current example, where the dataset possesses the DNA bases A, G, T, G and A in the five target positions defined in the genotyping scheme (see also Fig. 1), a genotype designation of 1.1 is consistent with the hierarchy, since the positive k-mers for both genotype 1 and genotype 1.1 would be present. A sample would be inconsistent if any of the parent genotype k-mers were missing or if the negative version of the k-mer was present. The QA/QC module in BioHansel also can identify intra-strain contamination by looking for the presence of both positive and negative versions of the same k-mer, or the presence of positive k-mers from other genotypes. In the current example, if the positive k-mer for genotype 2 was also identified, it would indicate a contaminated sample.
Our focus in designing BioHansel was to provide readily interpretable results to users with limited bioinformatics experience. Therefore, we developed a QC module to give information on the reliability of user-supplied sequence data along with BioHansel’s genotyping results. BioHansel’s reports provide information about scheme coverage, genotype consistency and contamination (Fig. 2). Using both positive and negative k-mers, BioHansel determines how many of the split k-mer pairs were identified in the sample as a measure of scheme coverage. One of the strengths of using hierarchical schemes is the ability to identify incompatible combinations of SNPs which may be indicative of contamination or recombination events. The QC module leverages the hierarchical nature of the schemes to look for inconsistencies in the genotyping call, which could indicate a problem with the sample. If the query sample was missing the positive k-mer for genotype 1, but contained the positive k-mer for 1.1, then the sample would be listed as inconsistent since there is not support for the parent genotype in the lineage. The presence of incompatible genotyping targets in the same sample is reported as intra-strain contamination, for example when positive k-mers that are outside the hierarchy for the identified genotype are detected in the dataset. Similarly, if both the positive and the negative k-mers for the same SNP position are identified, this is flagged as potential contamination. Following the example shown in Fig. 2, the presence of positive k-mers for positions 50, 500 and 901 in the same query sample would generate a contamination flag warning that both genotype 1.1 and genotype 3 may be present in the sample.
Benchmarking strain selection
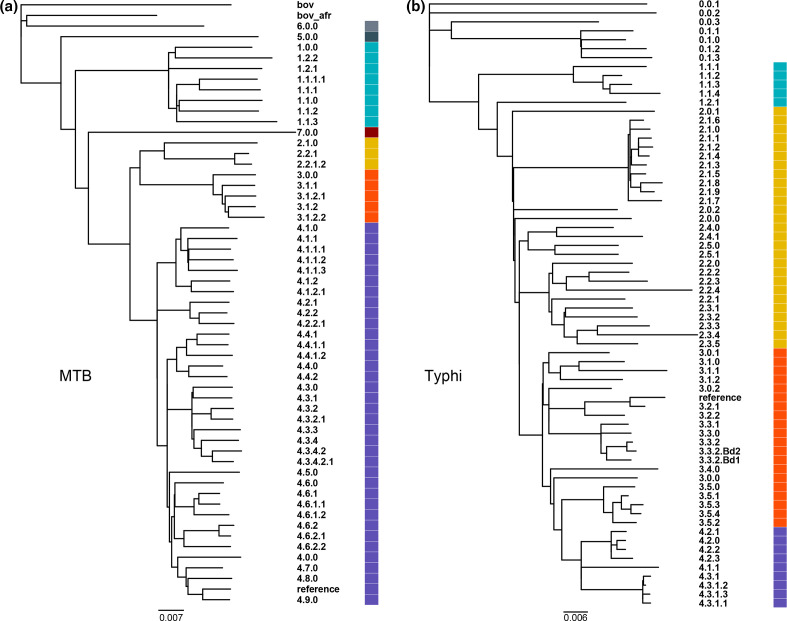
A testing panel of isolates was selected for both the Typhi and MTB schemes with one representative sample selected per available genotype from NCBI. Candidates for each genotype were identified from https://tbdr.lshtm.ac.uk/ for MTB and https://pathogen.watch/ for S. Typhi. The panel includes 58 MTB genotypes and 67 S. Typhi genotypes (Table S1, available in the online version of this article). Where there were multiple representatives per genotype, a random isolate was selected which was over 40× coverage (Table S1). Phylogenetic analyses were done using the SNVPhyl pipeline (Workflow SNVPhyl v1.1 Paired-End, released in Galaxy) [9]. Variants from the datasets listed in Table S1 were identified against the reference genomes used for each scheme; from strain CT18 (GenBank Accession NC_003198) for S. Typhi, and from strain H37Rv (GenBank Accession NC_000962.3) for MTB, using the following parameters: minimum coverage=8; minimum mean mapping quality=30 (default); SNV abundance ratio=0.75 (default); SNV density filtering search window size=20, and using default settings for all other parameters . The maximum-likelihood phylogeny (Fig. 3) was generated using 17092 sites (MTB) or 6288 sites (S. Typhi) over the core genome representing 87.55% (MTB) or 88.91% (S. Typhi) of the genome sequence of the respective reference strains. Using the raw Illumina reads, the genotype for each isolate was determined using either TBProfiler v. 2.8.12 [34] or genotyphi [3] (Table S1). Coverage levels, quality and read length varied considerably across the benchmarking strains. To enable more consistent benchmarking, we developed synthetic read sets using assemblies from the original samples. We assembled each sample using unicycler v. 4.6.0 [36] and then generated synthetic Illumina reads using ART Illumina v. 2.5.8 [37] with 250 bp MiSeq v. 1 reads, with 350 bp inserts, at the desired coverage levels (1× to 50×, as described below).
Fig. 3.
Maximum-likelihood phylogenetic trees of benchmarking isolates representing the diversity of genotypes defined in the MTB (a) and Typhi (b) schemes. Each tree is labelled such that each isolate is labelled with its genotype and a colour representing the first level of the corresponding scheme. Bars, approximatively 120 SNPs (a) and 38 SNPs (b), indicating that the genetic diversity of the S. Typhi population represented by these genotypes is approx. 3× lower than that of MTB.
Genotyping benchmarking
We compared the computational resource requirements of the read mapping-based tools genotyphi and TBProfiler against the k-mer-based tools BioHansel v. 2.5.0 and SKA v. 1.0.0 [5]. We measured the accuracy of the k-mer-based tools by comparing their genotyping results to those of genotyphi and TBProfiler. Note that while we designed BioHansel to be a genotyping tool based on predefined sets of k-mers, SKA is a set of tools designed to perform generic k-mer-based analysis of genomic data for organisms with small haploid genomes [5]. The feature of SKA that is most comparable to BioHansel is the typing module, which will report MLST types based on a profile and set of allele sequences [5]. In order to compare BioHansel genotyping with the SKA typing module, we converted the BioHansel-compatible Typhi and MTB schemes into an MLST format where each variant position was treated as a locus with its positive k-mer labelled as allele 1 and its negative k-mer as allele 0. The k-mers identified by the SKA typing module were manually inspected for concordance with the ground truth genotype. A Nextflow workflow has been developed for performing the computational resource benchmarking and is available online at: https://github.com/peterk87/nf-bh-typing-comparison.
Intra-strain contamination benchmarking
We examined the ability of BioHansel v. 2.5.0 and ConFindr v. 0.7.1 [24] to detect intra-strain contamination using the same synthetic MTB and S. Typhi datasets used for the genotyping benchmarking. We constructed seven levels of contamination in read sets with 50× genome coverage depth, yielding the following levels of contamination: 1× (2%), 2× (4%), 5× (10%), 10× (20%), 15× (30%), 20× (40%) and 25× (50%), using reads generated by ART Illumina v. 2.5.8. BioHansel genotyping was performed using the default parameters. ConFindr v. 0.7.1 was run using the default parameters with the exception of an explicit flag of –rmlst to maintain consistency between the two schemes, since there is a core gene MLST scheme for Salmonella but not for Mycobacterium . ConFindr was run on the forward and reverse (R1 and R2) synthetic read sets separately, and the detection of contamination was considered valid (ContamStatus=TRUE) for each read set when a minimum number of three contaminating single nucleotide variants (SNVs) were detected in both R1 and R2.
Global S. Typhi data analyses
All 8139 public isolates identified as S. Typhi by SISTR [38] in Enterobase [39–41] as of May 2020 were genotyped using BioHansel and assessed for contamination using BioHansel. The biosamples’ geographical metadata were downloaded directly from NCBI and standardized through manual curation. The genotyping results were summarized to generate a BioHansel-compatible metadata table listing the earliest and latest collection dates and primary geographical location associated with a given genotype. This metadata table is included with BioHansel and genotype-specific information is provided to the user when they run the built-in Typhi scheme in BioHansel.
Results and discussion
Genotyping benchmarking
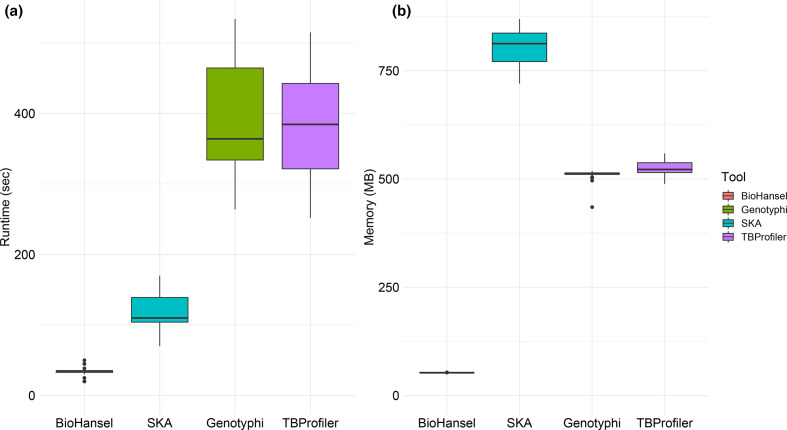
The genotyping results of SKA and BioHansel were completely concordant with the results obtained from TBProfiler and genotyphi for the panel of public isolates from MTB and S. Typhi (Table S1). Thus, both k-mer methods using the selected 33 bp split k-mers provided the same genotyping results as traditional reference mapping-based SNP calling procedures (Table S1). We then examined the runtimes and peak memory usage of the four tools against genotype synthetic WGS datasets (Fig. 4). Both k-mer tools were considerably faster than the reference mapping-based SNP calling tools, with BioHansel taking on average 16 s to process a sample and SKA requiring 109 s, while the read mapping-based approaches, genotyphi and TBProfiler, took an average of 285 and 297 s, respectively. BioHansel was 19 times faster than both reference mapping approaches at processing WGS datasets, while SKA was 2.6 times faster. BioHansel used the least memory with an average of 52 MB, but SKA used the most at 805 MB. Genotyphi had similar memory usage to TBProfiler, with an average of 510 and 525 MB, respectively. Since SKA does not have multi-threading support, we only examined single thread performance for all the tools. SKA also requires a sketching stage before the typing module can be used, so the timing results for this tool represent both sketching and typing. Even when in a single-threaded mode, BioHansel was found to genotype WGS datasets seven times faster than SKA while using 95% less memory.
Fig. 4.
Boxplots of the runtime (a) and peak memory usage (b) comparison of four tools, BioHansel, Genotyphi, SKA and TBProfiler, on synthetic Illumina fastq data with a fixed coverage of 50×. BioHansel and SKA results are based on datasets representing both MTB and Typhi schemes (N=129) while genotyphi and TBProfiler results are only based on datasets representing either the Typhi (N=67) or the MTB (N=62) scheme, respectively.
The k-mer-based tools SKA and BioHansel are designed for different purposes: SKA is designed as a set of multiple tools for analysis of bacterial genomes, while BioHansel is designed as a specialized hierarchical genotyping tool that can use redundant k-mers to identify a genotype. SKA and BioHansel differ particularly in their ability to handle missing data. The ability of a genotyping tool to handle missing data allows for increased scheme resolution, since k-mers that are highly prevalent in a target population can be used for genotyping even if they are not present in all the samples. For example, when adapting the MTB scheme to work with SKA, the absence of some targets from specific lineages rendered the tool unable to issue a genotype call for isolates from these lineages, unless the scheme was constricted to include only the targets found in those individual lineages. For instance, all members of MTB genotypes 1.X.X were missing the positive and negative k-mer for MTB genotype 4.5 and SKA could not type any of these samples without that target being removed from the scheme before testing. No such changes were needed for BioHansel to type the samples. Since Illumina sequence data often do not cover the complete genome of an organism, and regions may be missing due to differences in library preparation or quality of DNA template [42, 43], inclusion of redundant genetic features for genotyping can increase the robustness of a genotyping scheme. SKA and BioHansel are highly complementary tools for analysis of low genetic diversity populations, since SKA is suited to performing in-depth genomic epidemiological analyses including variant calling and clustering, while BioHansel provides clearly interpretable genotyping and quality metrics.
Contamination benchmarking
We examined the ability of BioHansel and ConFindr to detect intra-lineage contamination for the genotypes represented in the MTB and S. Typhi datasets. The tools were tested on artificial WGS datasets created from a pairwise mixing of the different genotypes. ConFindr is able to detect inter-species contamination as well as intra-species contamination [24], but BioHansel can only detect contamination within the population targeted by its genotyping scheme, so we restricted our analyses to the intra-lineage contamination detection. Both the MTB and S. Typhi populations have limited genetic diversity, and a tool’s ability to detect mixed samples decreases with the level of genetic divergence between the genotypes present in the mixed sample. ConFindr requires three or more SNPs in the rMLST genes [24] in order to detect contamination, so we first performed all pairwise combinations of each genotype at a 1:1 ratio to determine which genotype combinations are sufficiently divergent genetically to be detected by both ConFindr and BioHansel. The S. Typhi dataset included 67 genotypes for a total of 2211 pair-wise comparisons. Of these pairs, 168 had at least three contaminating rMLST SNPs in both directions (R1 and R2) in the artificial read sets and were retained for the tool comparison experiment. The MTB dataset contained 58 genotypes and had a total of 1711 pair-wise comparisons. Of these, 852 had a minimum of three contaminating rMLST SNPs detected in both read sets and were retained for the tool comparison experiment. The increased number of viable comparisons in the MTB scheme is probably because a greater genetic distance separates each of the genotypes defined by the scheme, which spans two highly variant clades [44]. In contrast, the Typhi scheme only subtypes within a single Salmonella serotype and dissects several recent clonal expansions into closely related sub-lineages (Fig. 3).
Here we want to point out that for this very low genetic diversity dataset, BioHansel has an advantage over ConFindr since a single target conflict is enough for BioHansel to consider a sample contaminated, whereas ConFindr requires three conflicts. BioHansel uses the genotyping schemes for quality assurance and requires a single incompatible k-mer to detect contamination. Consequently, BioHansel detected contamination in all the genotype pairs examined in this study. In practice, however, if the contaminants have a high genetic diversity and are not covered by the BioHansel genotyping scheme, as in the case of inter-species contamination, BioHansel would probably fail to detect the contamination, as the genotyping k-mers are unlikely to be conserved between species. In contrast, ConFindr uses a mapping approach which is more tolerant of diversity and thus can readily detect inter-species contamination. BioHansel and ConFindr are therefore highly complementary tools since each tool has strengths at different levels of genetic divergence.
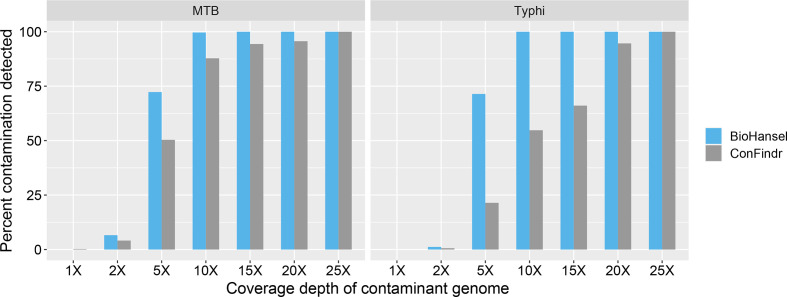
Impact of sequencing coverage on contamination detection
The ability to detect a contaminant in a WGS dataset depends on the level of contamination, so we determined the limits of detection of contamination for ConFindr and BioHansel at varying levels of contaminant genome coverage depth. We confirmed that neither BioHansel nor ConFindr reported any contamination with the simulated read sets which contained a single genotype. Using the genotype combinations retained above, which were detectable by both ConFindr and BioHansel, we used a fixed level of 50× coverage and examined six levels of contamination (Fig. 5). ConFindr is designed to detect levels of intra-species contamination of at least 5% (~2.5× in our artificial datasets), whereas a k-mer frequency of 8 (equivalent to 16% contamination for a 50× dataset) is required for BioHansel to produce a valid k-mer identification as a default setting. Both tools should therefore fail to detect contamination below 5% (2.5×). Interestingly, both tools were able to detect contamination events in 4–7% of the 852 artificial datasets of MTB and approx. 1 % of the 168 artificial datasets of S. Typhi at only 2× (4%) contaminant coverage (Fig. 5). Variation of coverage depth across the genome was expected due to random sampling by ART Illumina and seqtk, which explains why the detection thresholds were met in a small number of the artificial datasets. At 20× coverage depth, both tools detected contamination in >94% of the artificial datasets (Fig. 5). As expected, BioHansel identifies contamination in >99.5% of samples with at least 10× (20%) coverage of the contaminant, but surprisingly, there are several cases where ConFindr fails to identify contamination below 20× (40%) contaminant coverage, with a sharper drop in detection at lower contamination levels in the (less genetically diverse) S. Typhi datasets (Fig. 5). Intra-species contamination has been shown to cause errors in estimation of genetic distances for SNP calling and MLST workflows, especially at levels of 20% and greater of the total number of reads [45], and both tools reliably detect contamination at this level or higher. We recommend that ConFindr and BioHansel be used in combination to detect different types of contamination, with ConFindr capable of identification between species and within genetically diverse species [24, 45], and with BioHansel providing finer scale intra-lineage contamination detection.
Fig. 5.
Bar plot of contamination detection of BioHansel and ConFindr using datasets with different levels of contamination (1–25× coverage depth of contaminant genotype) in a fixed level of 50× Illumina genome coverage depth. Results are aggregated for 852 MTB and 168 Typhi pair-wise combinations where both ConFindr and BioHansel could detect contamination at the 25× coverage depth (50% contamination).
Global Salmonella Typhi analysis
A total of 8139 WGS datasets identified as Salmonella Typhi by Enterobase were downloaded from the NCBI Sequence Read Archive and analysed with both genotyphi and BioHansel. We found that the genotyping results of a large public genotyphi dataset by BioHansel were >99.8% concordant with those from genotyphi. A total of 153 samples failed BioHansel QC, 43 had warnings and 7943 passed QC (Table S2). Of the samples which failed BioHansel QC, 61 were identified as contaminated with multiple genotypes, indicating that intra-lineage contamination is present at a relatively low level in public data for S. Typhi. Enterobase does filter poor quality data and inter-species contaminated read-sets as part of its workflow [40], so a higher rate of contamination may be observed if an unfiltered public dataset was examined. We then compared the genotyping results obtained by both tools for the 7943 samples that passed BioHansel QC, and identified only 13 instances (0.16%) where the genotyping results from BioHansel did not agree with those of genotyphi. A manual inspection of the bam files for these 13 samples showed support for the BioHansel genotype calls. For example, we uncovered a problem in genotyphi with the identification of lineage 3.5.3, which is actually a sub-lineage of 3.5.4 (Fig. 4). We specifically adapted the nomenclature of this genotype in BioHansel by nesting it under 3.5.4, as the isolates from lineage 3.5.3 all possess both the variant bases for 3.5.3 and for its parent lineage 3.5.4. However, genotyphi calls all isolates from genotype 3.5.3 as 3.5.4 because they occur at the same rank according to their genotyping nomenclature. These results show that BioHansel’s genotyping calls are accurate and compare favourably with those of the current gold-standard method.
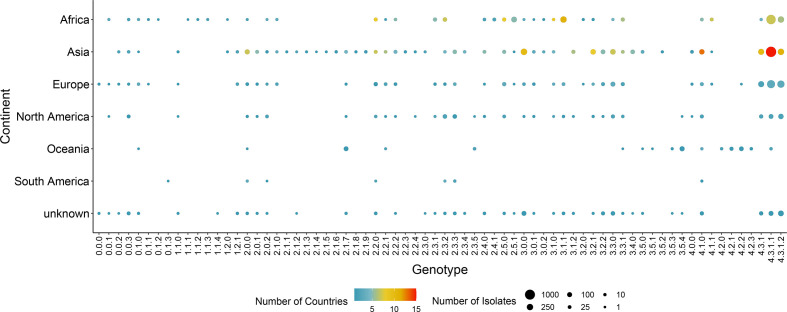
We then examined the genotype composition and geographical distribution of all these publicly available S. Typhi datasets (Fig. 6, Table S2). We found that the genotypes 4.3.1, 4.3.1.1 and 4.3.1.2 account for 53 % of all the public S. Typhi datasets and are distributed primarily in Asia and Africa (Fig. 6). These results are consistent with the original analyses of S. Typhi distribution, which found that clade 4.3.1 (H58) was highly prevalent and showed a ubiquitous geographical distribution [3]. Geographical associations can be found for many lineages; for example, in this dataset, lineage 4.2.X is found exclusively in Oceania and has a very strong association with Fiji, perhaps reflecting a local evolution of the lineage (Fig. 6, Table S2). Typhoid fever is not endemic in North America and Europe [3], so none of the S. Typhi genotypes is specific to these geographical locations. It is therefore likely that S. Typhi isolates from North America and Europe represent travel-associated cases. Using this global S. Typhi analysis, we generated a contextual metadata file indicating the earliest isolation date associated with the public datasets of each genotype, along with the geographical location that predominated in each genotype (Table S2). This information is provided along with the BioHansel genotyping results of S. Typhi datasets and can be used to inform potential geographical and temporal associations for an isolate assigned to a given genotype.
Fig. 6.
Balloon plot of 7943 global S. Typhi isolates showing the associations between genotypes and geography at the level of continents. The size of a point indicates the number of samples and the colour of a point indicates the number of discrete countries contained within it.
Conclusions
BioHansel is a rapid and accurate genotyping tool producing results that are fully concordant with those of reference mapping approaches. BioHansel can be used with a variety of hierarchical SNP typing schemes and provides a clear quality assessment of the typing results for public health professionals. Through the quality assurance module, BioHansel addresses the need for detection of intra-lineage contamination in low genetic diversity organisms. BioHansel increases the accessibility of published SNP genotyping schemes by removing both the requirement for development of a separate tool for each scheme and the necessity for high-end computer equipment. Contextual information can also be derived from large-scale population analyses and can be included with BioHansel reports to provide insights for hypothesis generation in genomic epidemiology.
Supplementary Data
Funding information
This work was supported by the Genomics Research and Development Initiative (GRDI) of the Government of Canada (Grants ID 2256344 and 2267905) and the Public Health Agency of Canada.
Acknowledgements
We thank Jonathan Looi (BioHansel Read-the-Docs); Gary Tong (BioHansel Read-the-Docs); Adrian Zetner (bioinformatics support); Aaron Petkau (Galaxy and IRIDA workflow development).
Author contributions
G.L., Conceptualization, Funding acquisition, Methodology, Data curation, Formal analysis, Investigation, Supervision, Project administration, Writing – original draft, Visualization. P.K., Conceptualization, Software, Methodology, Supervision, Formal analysis, Writing – review and editing. J.R., Conceptualization, Software, Methodology, Data curation, Formal analysis, Investigation, Funding acquisition, Writing – original draft, Visualization. P.M., Supervision, Software, Methodology. J.S., Software, Methodology, Writing – original draft. D.K., Data curation, Formal analysis, Investigation. M.A.R., Data curation, Formal analysis, Investigation. M.G., Software, Methodology, Formal analysis. D.H., Software, Investigation. D.S., Software, Methodology, Data curation, Investigation. N.K., Writing – review and editing. C.R.L., Software, Methodology, Resources. K.B., Software, Methodology. E.T., Funding acquisition, Supervision, Writing – review and editing. C.Y., Supervision, Project administration, Writing – review and editing. K.Z., Funding acquisition, Investigation, Formal analysis. A.N., Funding acquisition, Writing – review and editing. R.P.J., Funding acquisition, Project administration, Supervision, Writing – review and editing. G.V.D., Funding acquisition, Project administration, Resources, Supervision, Writing – review and editing. J.H.E.N., Funding acquisition, Project administration, Supervision, Writing – review and editing.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: MLST, multilocus sequence typing; MTB, Mycobacterium tuberculosis; QC, quality control; SNP, Single Nucleotide Polymorphism; SNV, single nucleotide variant; WGS, whole genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Two supplementary tables are available with the online version of this article.
References
- 1.Deurenberg RH, Bathoorn E, Chlebowicz MA, Couto N, Ferdous M, et al. Application of next generation sequencing in clinical microbiology and infection prevention. J Biotechnol. 2017;243:16–24. doi: 10.1016/j.jbiotec.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 2.Nadon C, Walle V, Gerner-Smidt P, Campos J, Chinen I, et al. PulseNet International: Vision for the implementation of whole genome sequencing (WGS) for global food-borne disease surveillance. Euro Surveill Bull Eur Sur Mal Transm Eur Commun Dis Bull. 2017;22:30544. doi: 10.2807/1560-7917.ES.2017.22.23.30544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong VK, Baker S, Connor TR, Pickard D, Page AJ, et al. An extended genotyping framework for Salmonella enterica serovar Typhi, the cause of human typhoid. Nat Commun. 2016;7:12827. doi: 10.1038/ncomms12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coll F, McNerney R, Guerra-Assunção JA, Glynn JR, Perdigão J, et al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun. 2014;5:4812. doi: 10.1038/ncomms5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris SR. SKA: Split Kmer Analysis Toolkit for Bacterial Genomic Epidemiology. bioRxiv. 2018:453142. [Google Scholar]
- 6.PHG Foundation Pathogen genomics into practice. 2020. https://www.phgfoundation.org/report/pathogen-genomics-into-practice
- 7.Jagadeesan B, Baert L, Wiedmann M, Orsi RH. Comparative analysis of tools and approaches for source tracking Listeria monocytogenes in a food facility using whole-genome sequence data. Front Microbiol. 2019;10:947. doi: 10.3389/fmicb.2019.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dallman T, Ashton P, Schafer U, Jironkin A, Painset A, et al. SnapperDB: a database solution for routine sequencing analysis of bacterial isolates. Bioinformatics. 2018;34:3028–3029. doi: 10.1093/bioinformatics/bty212. [DOI] [PubMed] [Google Scholar]
- 9.Petkau A, Mabon P, Sieffert C, Knox NC, Cabral J, et al. SNVPhyl: a single nucleotide variant phylogenomics pipeline for microbial genomic epidemiology. Microb Genom. 2017;3:e000116. doi: 10.1099/mgen.0.000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearce ME, Alikhan N-F, Dallman TJ, Zhou Z, Grant K, et al. Comparative analysis of core genome MLST and SNP typing within a European Salmonella serovar Enteritidis outbreak. Int J Food Microbiol. 2018;274:1–11. doi: 10.1016/j.ijfoodmicro.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katz LS, Griswold T, Williams-Newkirk AJ, Wagner D, Petkau A, et al. A comparative analysis of the Lyve-SET phylogenomics pipeline for genomic epidemiology of foodborne pathogens. Front Microbiol. 2017;8:375. doi: 10.3389/fmicb.2017.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bush SJ, Foster D, Eyre DW, Clark EL, De Maio N, et al. Genomic diversity affects the accuracy of bacterial single-nucleotide polymorphism-calling pipelines. GigaScience. 2020;9:giaa007. doi: 10.1093/gigascience/giaa007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshimura D, Kajitani R, Gotoh Y, Katahira K, Okuno M, et al. Evaluation of SNP calling methods for closely related bacterial isolates and a novel high-accuracy pipeline: BactSNP. Microb Genomics. 2019;5:e000261. doi: 10.1099/mgen.0.000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cameron DL, Di Stefano L, Papenfuss AT. Comprehensive evaluation and characterisation of short read general-purpose structural variant calling software. Nat Commun. 2019;10:3240. doi: 10.1038/s41467-019-11146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis S, Pettengill JB, Luo Y, Payne J, Shpuntoff A, et al. CFSAN SNP Pipeline: an automated method for constructing SNP matrices from next-generation sequence data. PeerJ Comput Sci. 2015;1:e20. doi: 10.7717/peerj-cs.20. [DOI] [Google Scholar]
- 16.Seeman, Torsten snippy: fast bacterial variant calling from NGS reads. 2020. https://github.com/tseemann/snippy
- 17.Labbé G, Rankin MA, Robertson J, Moffat J, Giang E, et al. Targeting discriminatory SNPs in Salmonella enterica serovar Heidelberg genomes using RNase H2-dependent PCR. J Microbiol Methods. 2019;157:81–87. doi: 10.1016/j.mimet.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 18.van Gent M, Bart MJ, van der Heide HGJ, Heuvelman KJ, Kallonen T, et al. SNP-based typing: a useful tool to study Bordetella pertussis populations. PLoS One. 2011;6:e20340. doi: 10.1371/journal.pone.0020340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt M, Mather AE, Sánchez-Busó L, Page AJ, Parkhill J, et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genomics. 2017;3:e000131–e000131. doi: 10.1099/mgen.0.000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silva M, Machado MP, Silva DN, Rossi M, Moran-Gilad J, et al. chewBBACA: A complete suite for gene-by-gene schema creation and strain identification. Microb Genomics. 2018;4:e000166. doi: 10.1099/mgen.0.000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feijao P, Yao H-T, Fornika D, Gardy J, Hsiao W, et al. MentaLiST - a fast MLST caller for large MLST schemes. Microb Genom. 2018;4 doi: 10.1099/mgen.0.000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ, et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Low AJ, Koziol AG, Manninger PA, Blais B, Carrillo CD. ConFindr: rapid detection of intraspecies and cross-species contamination in bacterial whole-genome sequence data. PeerJ. 2019;7:e6995. doi: 10.7717/peerj.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wyllie DH, Robinson E, Peto T, Crook DW, Ajileye A, et al. Identifying mixed Mycobacterium tuberculosis infection and laboratory cross-contamination during mycobacterial sequencing programs. J Clin Microbiol. 2018;56:e00923-18. doi: 10.1128/JCM.00923-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohl TA, Utpatel C, Schleusener V, Filippo MRD, Beckert P, et al. MTBseq: a comprehensive pipeline for whole genome sequence analysis of Mycobacterium tuberculosis complex isolates. PeerJ. 2018;6:e5895. doi: 10.7717/peerj.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anyansi C, Keo A, Walker BJ, Straub TJ, Manson AL, et al. QuantTB – a method to classify mixed Mycobacterium tuberculosis infections within whole genome sequencing data. BMC Genomics. 2020;21:80. doi: 10.1186/s12864-020-6486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D, Song L, Breitwieser FP, Salzberg SL. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 2016;26:1721–1729. doi: 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menzel P, Ng KL, Krogh A. Fast and sensitive taxonomic classification for Metagenomics with kaiju. Nat Commun. 2016;7:11257. doi: 10.1038/ncomms11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sczyrba A, Hofmann P, Belmann P, Koslicki D, Janssen S, et al. Critical Assessment of Metagenome Interpretation—a benchmark of metagenomics software. Nat Methods. 2017;14:1063–1071. doi: 10.1038/nmeth.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye SH, Siddle KJ, Park DJ, Sabeti PC. Benchmarking metagenomics tools for taxonomic classification. Cell. 2019;178:779–794. doi: 10.1016/j.cell.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wright ES, Vetsigian KH. Quality filtering of Illumina index reads mitigates sample cross-talk. BMC Genomics. 2016;17:876. doi: 10.1186/s12864-016-3217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phelan JE, O’Sullivan DM, Machado D, Ramos J, Oppong YEA, et al. Integrating informatics tools and portable sequencing technology for rapid detection of resistance to anti-tuberculous drugs. Genome Med. 2019;11:41. doi: 10.1186/s13073-019-0650-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aho AV, Corasick MJ. Efficient string matching: an aid to bibliographic search. Commun ACM. 1975;18:333–340. [Google Scholar]
- 36.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang W, Li L, Myers JR, Marth GT. ART: a next-generation sequencing read simulator. Bioinformatics. 2012;28:593–594. doi: 10.1093/bioinformatics/btr708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VPJ, et al. The Salmonella In Silico Typing Resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLOS ONE. 2016;11:e0147101. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alikhan N-F, Zhou Z, Sergeant MJ, Achtman M. A genomic overview of the population structure of Salmonella . PLOS Genetics. 2018;14:e1007261. doi: 10.1371/journal.pgen.1007261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Z, Alikhan N-F, Mohamed K, Fan Y, Agama Study Group. et al. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020;30:138–152. doi: 10.1101/gr.251678.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Achtman M, Zhou Z, Alikhan N-F, Tyne W, Parkhill J, et al. Genomic diversity of Salmonella enterica -The UoWUCC 10K genomes project. Wellcome Open Res. 2020;5:223. doi: 10.12688/wellcomeopenres.16291.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uelze L, Borowiak M, Deneke C, Szabó I, Fischer J, et al. Performance and accuracy of four open-source tools for in silico serotyping of Salmonella spp. based on whole-genome short-read sequencing data. Appl Environ Microbiol. 2020;86:e02265-19. doi: 10.1128/AEM.02265-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S, Zhang S, Deng X. GC content-associated sequencing bias caused by library preparation method may infrequently affect Salmonella serotype prediction using SeqSero2. Appl Environ Microbiol. 2020;86:e00614-20. doi: 10.1128/AEM.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riojas MA, McGough KJ, Rider-Riojas CJ, Rastogi N, Hazbón MH. Phylogenomic analysis of the species of the Mycobacterium tuberculosis complex demonstrates that Mycobacterium africanum, Mycobacterium bovis, Mycobacterium caprae, Mycobacterium microti and Mycobacterium pinnipedii are later heterotypic synonyms of Mycobacterium tuberculosis . Int J Syst Evol Microbiol. 2018;68:324–332. doi: 10.1099/ijsem.0.002507. [DOI] [PubMed] [Google Scholar]
- 45.Pightling AW, Pettengill JB, Wang Y, Rand H, Strain E. Within-species contamination of bacterial whole-genome sequence data has a greater influence on clustering analyses than between-species contamination. Genome Biol. 2019;20:286. doi: 10.1186/s13059-019-1914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.