Abstract
Analysing the flanking sequences surrounding genes of interest is often highly relevant to understanding the role of mobile genetic elements (MGEs) in horizontal gene transfer, particular for antimicrobial-resistance genes. Here, we present Flanker, a Python package that performs alignment-free clustering of gene flanking sequences in a consistent format, allowing investigation of MGEs without prior knowledge of their structure. These clusters, known as ‘flank patterns’ (FPs), are based on Mash distances, allowing for easy comparison of similarity across sequences. Additionally, Flanker can be flexibly parameterized to fine-tune outputs by characterizing upstream and downstream regions separately, and investigating variable lengths of flanking sequence. We apply Flanker to two recent datasets describing plasmid-associated carriage of important carbapenemase genes (bla OXA-48 and bla KPC-2/3) and show that it successfully identifies distinct clusters of FPs, including both known and previously uncharacterized structural variants. For example, Flanker identified four Tn4401 profiles that could not be sufficiently characterized using TETyper or MobileElementFinder, demonstrating the utility of Flanker for flanking-gene characterization. Similarly, using a large (n=226) European isolate dataset, we confirm findings from a previous smaller study demonstrating association between Tn1999.2 and bla OXA-48 upregulation and demonstrate 17 FPs (compared to the 5 previously identified). More generally, the demonstration in this study that FPs are associated with geographical regions and antibiotic-susceptibility phenotypes suggests that they may be useful as epidemiological markers. Flanker is freely available under an MIT license at https://github.com/wtmatlock/flanker.
Keywords: antimicrobial resistance (AMR), bioinformatics, mobile genetic element (MGE), plasmid, whole-genome sequencing
Data Summary
National Center for Biotechnology Information (NCBI) accession numbers for all sequencing data used in this study are provided in Table S1 (available with the online version of this article). The analysis performed in this article can be reproduced in a binder environment provided on the Flanker GitHub page (https://github.com/wtmatlock/flanker). Accession numbers for the MEFinder and TETyper outputs are provided in Table S1.
Impact Statement.
The global dissemination of antimicrobial-resistance genes (ARGs) has in part been driven by carriage on mobile genetic elements (MGEs) such as transposons and plasmids. However, our understanding of these MGEs remains poor, partly due to their high diversity. This means current referenced-based approaches are often inappropriate. Flanker is a fast software tool that overcomes this barrier by de novo clustering of ARG flank diversity by sequence similarity. We demonstrate the utility of Flanker by associating bla OXA-48 and bla KPC-2/3 flanking sequences with geographical regions and resistance phenotypes.
Introduction
The increasing incidence of antimicrobial resistance (AMR) in clinical isolates poses a threat to all areas of medicine [1–3]. AMR genes (ARGs) are found in a diverse range of genetic contexts, bacterial species, and in both clinical and non-clinical environments (e.g. agricultural, refuse and natural ecosystems) [4–7]. However, the mechanisms underpinning the dissemination of many ARGs between these reservoirs remain poorly understood, limiting the efficacy of surveillance and the ability to design effective interventions. Usually, ARGs are spread vertically, either via chromosomal integration or stable association of a plasmid within a clonal lineage, or by horizontal gene transfer (HGT) through mobile genetic elements (MGEs), e.g. transposons or plasmids [8]. HGT can accelerate the rate of ARG acquisition, both within and across species [9–11].
The epidemiology of ARGs can, therefore, involve multiple levels, from clonal spread to MGEs. There are many existing software tools to facilitate epidemiological study of bacterial strains [12–16], whole plasmids [17, 18] and smaller MGEs [19, 20]. Several tools and databases exist for the annotation of non-plasmid MGEs such as insertion sequences (ISs) and transposons [19, 20], but all rely on comparisons to reference sequences, so are limited to known diversity. Reference-free tools for analysing MGE diversity would, therefore, be a useful addition. Here, we describe Flanker, a simple, reference-free tool to investigate MGEs by analysing the flanking sequences of ARGs.
The flanking sequences (hereafter, flanks) around an ARG that has been mobilized horizontally may act as signatures of relevant MGEs and support epidemiological analyses. However, these flanks can contain a great deal of structural variation due to their evolutionary history. Where a single known MGE is under investigation, it is possible to specifically type this element (for example, using TETyper [19]) or align flanks against a known ancestral form after the removal of later structural variation [21]. However, often multiple structures may be involved. This is particularly true for ARGs that move frequently on a variety of MGEs. Studies of different ARGs often choose different ad hoc approaches to extract flanks and cluster genetic structures. Examples include hierarchical clustering of isolates carrying an ARG based on short-read coverage of known ARG-carrying contigs [22], assigning assembled contigs into 'clustering groups' based on gene presence and synteny [23] or iterative ‘splitting’ of flanks based on pairwise nucleotide blast identity [24]. A consistent and simple approach for this task would not only avoid repeated method development, but also aid comparison between methods developed for specific ARGs.
To address this problem, we developed Flanker, a pipeline to analyse the regions around a given ARG in a consistent manner. Flanker flexibly extracts the flanks of a specified gene from a dataset of contigs, then clusters these sequences using Mash distances to identify consistent structures [25]. Flanker is available as a documented Python and Bioconda package released under the MIT open-source license. Source code has been deposited at https://github.com/wtmatlock/flanker and documentation at https://flanker.readthedocs.io/en/latest/.
Methods
Flanker
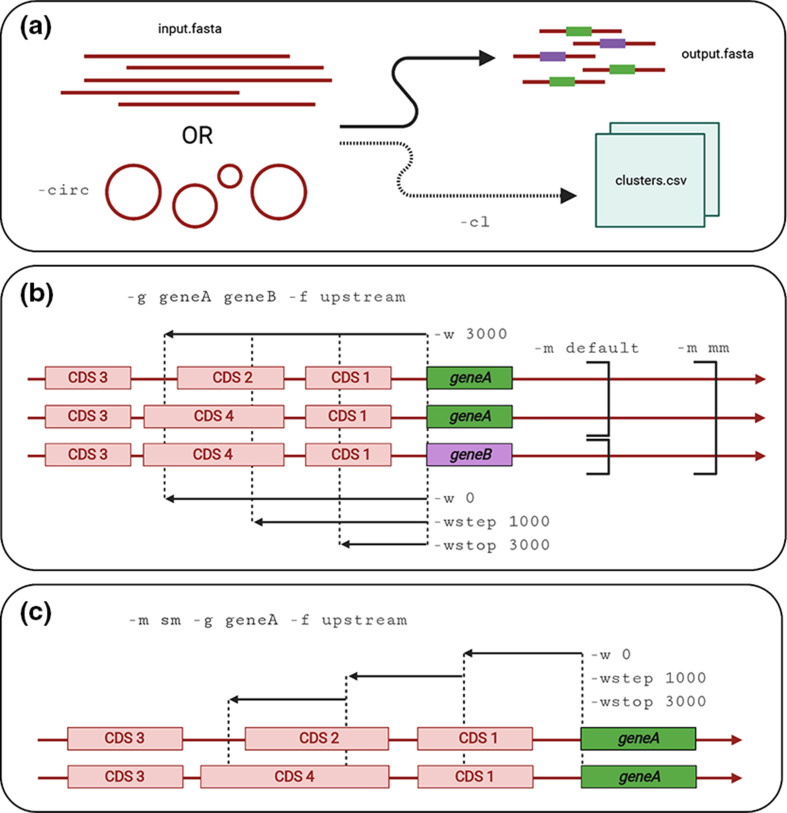
The Flanker package contains two basic modules: the first extracts a region of length w around an annotated gene of interest, and the second clusters such regions based on a user-defined Mash distance threshold (default --threshold 0.001; Fig. 1a). Within each FASTA/multi-FASTA format input file, the location of the gene of interest is first determined using the Abricate annotation tool [26]. Flanks around the gene (optionally including the gene itself to enable complete alignments with --include_gene) are then extracted and written to a FASTA format file using Biopython [27]. Flanker gives users the option to either extract flanks using a single window (defined by length in bp) or multiple windows from a start position (--window) to an end position (--wstop) in fixed increments (--wstep). Flanks may be extracted from upstream, downstream or on both sides of the gene of interest (--flank). Corrections are also made for circularized genomes where the gene occurs close to the beginning or end of the sequence (--circ mode) and for genes found on both positive and negative strands. The clustering module groups flanks of user-defined sequence lengths together based on a user-defined Mash [25] distance threshold (--threshold) of user-defined sequence lengths.
Fig. 1.
Schematic of Flanker’s modes and parameters. (a) Flanker uses Abricate to annotate the gene of interest in input sequences and outputs associated flanking sequences, optionally clustering (-cl) these on a user-defined Mash distance threshold. It can take linear or circularized sequences. (b) In this example, genes geneA and geneB have been queried (-g geneA geneB), and only the upstream flank is desired (-f upstream). The top single black arrow represents choosing a single window of length 3000 bp (-w 3000), whereas the bottom three black arrows represent stepping in 1000 bp windows from 0 to 3000 bp (-w 0 -wstep 1000 -wstop 3000). The default mode (-m default) extracts flanks for all annotated alleles separately, but the multi-allelic mode (-m mm) extracts flanks for all alleles in parallel. (c) Flanker has a supplementary salami mode (-m sm), which outputs non-contiguous blocks of sequence with a start point, step size and end point (-w 0 -wstep 1000 -wstop 3000), represented by the three black arrows.
In default mode (--mode default), Flanker considers multiple gene queries in turn. In multi-allelic mode (--mode mm), Flanker considers all genes in the list for each window (Fig. 1b). Multiple genes can be queried by either a space-delimited list in the command line (--gene geneA geneB), or a newline-delimited file with the list of genes option (--list_of_genes). A supplementary module ‘salami mode’ (--mode sm) is provided to allow comparison of non-contiguous blocks from a start point (--window), step size (--wstep) and end point (--wstop) (Fig. 1c).
Datasets
To validate Flanker, demonstrate its application and provide a comparison with existing tools, we used two recent datasets of complete plasmids (derived from hybrid long-/short-read assemblies) containing carbapenemase genes of clinical importance [23, 28]. The first dataset comprised 51 complete bla OXA-48-harbouring plasmids; 42/51 came from carbapenem-resistant Escherichia coli and Klebsiella pneumoniae isolates from patients in the Netherlands [28] and 9/51 from EuSCAPE (a European surveillance programme investigating carbapenem resistance in Enterobacterales ) [23]. The second dataset comprised 50 bla KPC-2 or bla KPC-3-( K. pneumoniae carbapenemase)-harbouring plasmids in carbapenem-resistant K. pneumoniae isolated from the Netherlands [28] (8/50) and as part of the EuSCAPE study (42/50) [23]. The EuSCAPE dataset [23, 29] additionally contains a large collection of short-read sequencing data for Klebsiella spp. isolates alongside meropenem-susceptibility data. This was used to demonstrate additional possible epidemiological applications of the Flanker tool by evaluating whether specific flank patterns (FPs) were more likely to be associated with phenotypic meropenem resistance.
Mash distances
Pairwise distances between flanks were calculated using Mash (version 2.2.2) [25]. Mash reduces sequences to a fixed-length MinHash sketch, which is used to estimate the Jaccard distance between k-mer content. It also gives the Mash distance, which ranges from 0 (~identical sequences) to 1 (~completely dissimilar sequences). We used the default Mash parameters in all analyses. The Mash distance was developed to approximate the rate of sequence mutation between genomes under a simple evolutionary model, and explicitly does not model more complex processes. We use it here for fast alignment-free clustering of sequences and do not draw any direct conclusions about evolution from pairwise comparisons.
Clustering
To cluster the flanks, Flanker generates an adjacency matrix weighted by Mash distances. It then thresholds this matrix to retain edges weighted less than or equal to the defined threshold. This is then used to construct a graph using the Python NetworkX library [30] and clusters are defined using the nx.connected_components function, which is analogous to single linkage. This is a similar methodology to that used by the Assembly Dereplicator tool [31] (from which Flanker re-uses several functions). However, Flanker aims to assign all flanks to a cluster rather than to deduplicate by cluster.
Cluster validation
We validated the output of flanking sequence-based clustering using a PERMANOVA (permutational analysis of variance) test, implemented with the Adonis function from the Vegan package (version 2.7.5) [32] in R. Only flanks in clusters of at least two members were considered; 42/51 and 48/50 of bla OXA-48 and bla KPC-2/3 flanks, respectively. The formula used was Mash dist ~cluster, with the ‘Euclidean’ method and 999 permutations.
Comparison to existing methods/application
We compared the classifications of TETyper (v1.1) [19] and MEFinder (v1.0.3) [20] to those produced by Flanker for 500 and 5000 bp flanks around bla KPC-2/3 genes. TETyper was run using the –threads 8 and --assemblies options with the Tn4401 reference and SNP/structural profiles provided in the package and MEFinder was run in Abricate [26] using the –mincov 10 option. For comparisons of the proportions of resistant isolates per FP, isolates were classified as resistant or sensitive using the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoint for meropenem (>8 mg l−1) [33].
Data visualization
All figures were made using BioRender (https://biorender.com) and the R packages ggplot2 (v3.3.0) [34], gggenes (v0.4.0) [35] and ggtree (v2.4.1) [36]. Prokka (v1.14.6) [37] was used to annotate Flanker output. Mashtree (v1.2.0) [38] was used to construct a visual representation of Mash distances between whole plasmid genomes. Plasmidfinder was used to detect the presence/absence of plasmid types using Abricate (version 1.01) with --mincov 80 and --minid 80 [39]. Galileo AMR (https://galileoamr.arcbio.com/mara/) was used to visualize the transposon variants. Figures can be reproduced using the code in the GitHub repository (https://github.com/wtmatlock/flanker).
Results
Clustering validation and comparison with TETyper/MEFinder
The clustering mode was validated numerically with a PERMANOVA test (Mash dist ~cluster: bla OXA-48 P value <0.001, bla KPC2/3 P value <0.001; see Methods). Figs 2 and 3 also provide a visual comparison of an alignment of genes (Gene Graphical Representation panel) to the FP.
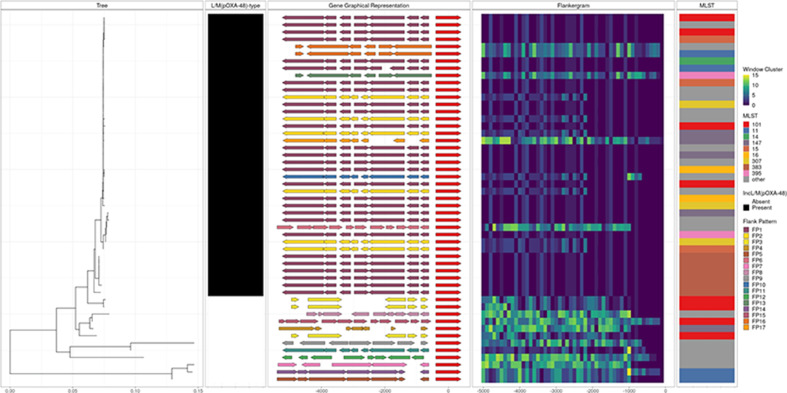
Fig. 2.
Flanking regions 5000 bp upstream of bla OXA-48 in plasmids from K . pneumoniae isolates. The Tree panel is a neighbour-joining tree reconstructed from Mash distances between complete sequences of plasmids carrying the bla OXA-48 gene. The second panel indicates the presence/absence of a L/M(pOXA-48)-type plasmid. The Gene Graphical Representation panel schematically represents coding regions in the 5000 bp sequence upstream of the bla OXA-48 gene, which is shown in red. Other genes are coloured according to the FP, which considers the overall pattern of all 100 bp window clusters up to 2200 bp (the approximate upstream limit of Tn1999). The Flankergram panel shows window clusters of all groups over each 100 bp window between 0 and 5000 bp. The dotted line at 2200 bp indicates the approximate point of upstream divergence between several FPs. The MLST panel shows K. pneumoniae multilocus sequence types, with those occurring once labelled ‘other’. FPs are numbered in ascending order according to abundance in the hybrid assemblies. Data used to make this figure came from the Dutch CPE surveillance and EuSCAPE hybrid assembly datasets.
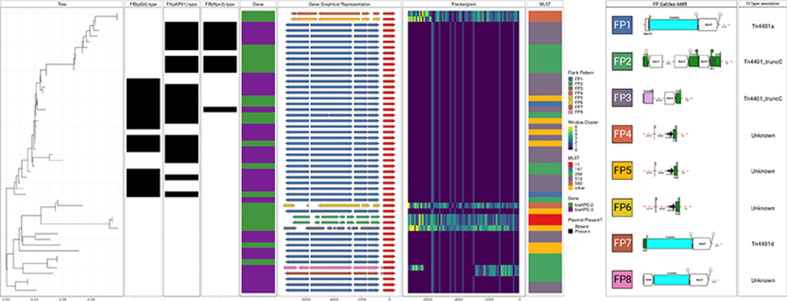
Fig. 3.
Flanking regions 7200 bp upstream of bla KPC-2/3 in plasmids from K. pneumoniae isolates. The Tree panel is a neighbour-joining tree reconstructed from Mash distances between complete sequences of plasmids carrying the bla KPC-2/3 gene. The next three panels indicate the presence/absence of FIB(pQ1I)-, FII(pKP91)- and FIB(Kpn3)-type plasmids. The Gene column indicates which bla KPC allele (2 or 3) is present. The Gene Graphical Representation panel schematically represents coding regions in the 7200 bp sequence region upstream of the bla KPC-2/3 gene, which is shown in red. Other genes are coloured according to the FP, which here takes into account the overall pattern of all 100 bp window groups (shown in the Flankergram panel) over the full 7200 bp region upstream of blaKPC-2/3 . The Flankergram shows window clusters over each 100 bp window between 0 and 7200 bp. The MLST panel shows K. pneumoniae multilocus sequence types, with those occurring once labelled ‘other’. The final two panels show the Galileo AMR and the TETyper outputs for the eight FPs, respectively. The FPs are numbered in ascending order according to abundance in the hybrid assemblies.
Of the two existing tools we compared in evaluating the flanks around bla KPC2/3, TETyper was by far the slowest (1172 s), whereas MEFinder, run in Abricate, and Flanker took 7 and 11 s, respectively (benchmarked on 5000 bp upstream flanks on a cluster with Intel Skylake 2.6 GHz chips). MEFinder was able to detect Tn4401, but could not provide any further structural resolution and was unable to classify 6/50 (12 %) 500 bp and 1/50 (2 %) 5000 bp flanks. TETyper structural profiles were consistent with Flanker when analysing 500 and 5000 bp upstream regions (Fig. 3), though Flanker split a group of six isolates with the TETyper structural profile 1–7127|7202–10006 into four groups (Table S2). To map our FPs to the established nomenclature, we additionally compared the output of Flanker to that of TETyper when the latter was given the entire Tn4401 sequence (i.e. by evaluating the typical 7200 bp Tn4401-associated flank upstream of blaKPC ). Flanker and TETyper classifications of Tn4401 regions were broadly consistent (Table S2), though this analysis demonstrated the potential benefit of the reference-free approach of Flanker, which showed that four non-Tn4401 structural profiles ('unknown' in TETyper) were distinct from each other. In addition, TETyper classified three flanks as Tn4401_truncC-1, whereas Flanker resolved this cluster into two distinct groups (Table S2).
Application to plasmids carrying bla OXA-48
The carbapenemase gene bla OXA-48 has been shown to be disseminated by Tn1999-associated structures (~5 kb, see detailed review by Pitout et al. [40]) nested in L/M-type plasmids, and as part of an IS1R-associated composite transposon containing bla OXA-48 and part of Tn1999, namely Tn6237 (~21.9 kb), which has been implicated in the chromosomal integration of bla OXA-48 [29, 41]. It has been recently demonstrated that most bla OXA-48-like genes in clinical isolates in Europe are carried on highly similar L/M(pOXA-48)-type plasmids, with evidence of both horizontal and vertical transmission across a diverse set of sequence types [23]. Whilst Tn1999-like flanking regions are relatively well characterized [40], in this example we chose an initial arbitrary upstream window of 5000 bp to simulate a scenario in which there is no prior knowledge. Inspection of a plot of window clusters (i.e. as shown in the Flankergram in Fig. 2) demonstrates that Flanker output allows the empirical identification of the position ~2200 bp upstream of bla OXA-48 as an important point of structural divergence without requiring annotation (as shown at ~2200 along the x-axis, where the window cluster colour schemes diverge), corresponding to the edge of Tn1999 at its expected position.
Using complete plasmids from the Netherlands [28]/EuSCAPE [23] hybrid assembly datasets, Flanker identified 17 distinct FPs in the 2200 bp upstream sequence of bla OXA-48 of which 7 occurred in L/M(pOXA-48)-type plasmids (Fig. 2, Table S3). To investigate the association of phenotypic carbapenem resistance with bla OXA-48 FPs, we created a Mash sketch using one randomly chosen representative per group and screened an Illumina-sequenced collection of European carbapenemase-resistant Klebsiella isolates [29] (n=425) [Mash screen, assigning FP based on the top hit (median identity=1.00; range 0.97–1.00)]. Two FPs (FP6 and FP16) accounted for 338/425 (80 %) of isolates; both were widely distributed across Europe. Of the 226 isolates with meropenem-susceptibility data available, those belonging to FP6 were proportionally more meropenem resistant compared to FP16 [70/135 (52 %) vs 6/44 (14 %), exact P value <0.001; Fig. S1]. Annotation (using Galileo AMR; see Methods) of these revealed that whereas FP16 contains Tn1999, FP6 contains Tn1999.2, which has been previously described as creating a strong promoter that produces twofold higher enzymatic activity [42].
Application to plasmids carrying bla KPC-2/3
David et al. showed that bla KPC-2/3 genes have been disseminated in European K. pneumoniae clinical isolates via a diverse collection of plasmids in association with a dominant clonal lineage, ST258/512, which accounted for 230/312 (74 %) of bla KPC-associated isolates in the EuSCAPE collection [23]. bla KPC has largely been associated with variants of a ~10 kb transposon, Tn4401 [43, 44]. From the combined EuSCAPE [23] and Dutch CPE collection [28] of 50 hybrid assembled KPC-containing plasmids, Flanker identified eight distinct FPs over a 7200 bp window upstream of bla KPC-2/3 (Fig. 3, Table S2). This window length was chosen to capture the entire Tn4401 sequence upstream of blaKPC .
Considering Mash containment of the eight representative FPs within the EuSCAPE short-read assemblies dataset, 346/442 (78 %) belonged to FP1 (corresponding to isoform Tn4401a). Whilst FP1 was widely distributed across Europe, FP2 (corresponding to Tn4401_truncC) and FP7 (corresponding to Tn4401d) appeared more geographically restricted: FP2 to Spain (5/5, 100 %) and FP7 to Israel (19/59, 32 %) and Portugal (34/59, 58 %) with isolates also found in Poland and Germany (n=2 each) and Italy and Austria (n=1) (Table S4). Of the 442 short-read assemblies, 274 had meropenem MIC data available for analysis. There was no evidence of a difference in the proportion of isolates resistant to meropenem between FP1 and FP7 [202/238 (85 %) vs 23/25 (92 %), exact P value=0.5; Table S5], though there was incomplete susceptibility data for isolates from both groups [108/346 (31 %) for FP1 and 38/63 (60 %) for FP7].
Discussion
We present Flanker, a fast and flexible Python package for analysing gene flanking sequences. We anticipate that this kind of analysis will become more common as the number of complete reference-grade, bacterial assemblies increase. Our analysis of data from the EuSCAPE project suggests that FPs might be useful epidemiological markers when evaluating geographical associations of sequences. Additionally, we validated findings of a small (n=7) PCR-based study on a large (n=226) European dataset, confirming an association between Tn1999.2 and increased meropenem resistance. A key advantage compared to existing tools is that there is no reliance on reference sequences or prior knowledge. Despite analysing only a relatively small number (n=50) of complete bla KPC-containing plasmids, there were four distinct FPs that TETyper classified as 'unknown' because their profiles had not been previously characterized. Similarly, we identified 17 FPs associated with bla OXA-48 in contrast to the five structural variants of Tn1999 currently described in the literature.
TETyper works well when alleles/structural variants are known but can only classify a single transposon type at a time and requires manual curation when this is not the case. The observed diversity of flanking sequences is likely to continue to increase and manual curation of naming schemes will be arduous to maintain. MEFinder, however, is a quick screening tool that can search a large library of known mobile elements but lacks sequence-level resolution. Whilst Flanker overcomes these challenges, users may need to perform downstream analysis to interpret its output. We hope that Flanker will be complementary to these and other similar existing tools by reducing the dimensionality of large datasets and identifying smaller groups of sequences to focus on in detail. Though we have developed Flanker for ARGs, Abricate allows use of custom databases meaning any desired genes of interest could be analysed. Accurate outputs from Flanker will be dependent on the quality of input assemblies, and on the correct annotation of the gene of interest.
In summary, we present Flanker, a tool for comparative genomics of gene flanking regions that integrates several existing tools (Abricate, Biopython, NetworkX) in a convenient package with a simple command-line interface.
Supplementary Data
Funding information
W.M. is supported by a scholarship from the Medical Research Foundation National PhD Training Programme in Antimicrobial Resistance Research (MRF-145-0004-TPG-AVISO). S.L. is a Medical Research Council Clinical Research Training Fellow (MR/T001151/1). L.P.S. is a Sir Henry Wellcome Postdoctoral Fellow (220422/Z/20/Z). A.S.W. and T.E.A.P. are National Institute for Health Research (NIHR) Senior Investigators. The computational aspects of this research were funded by the NIHR Oxford Biomedical Research Centre with additional support from the Wellcome Trust Core Award grant number 203141/Z/16/Z. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR nor the Department of Health. The research was supported by the NIHR Health Protection Research Unit in Healthcare Associated Infections and Antimicrobial Resistance (NIHR200915) at the University of Oxford in partnership with Public Health England (PHE) and by the Oxford NIHR Biomedical Research Centre.
Acknowledgements
The authors thank the EuSCAPE and Dutch CPE surveillance groups for making their data publicly available.
Author contributions
Contributions have been attributed by the CRediT system as follows. Conceptualization: W.M., S.L., L.P.S., N.S. Methodology: W.M., S.L. Software: W.M., S.L., B.C. Validation: W.M., S.L. Formal Analysis: W.M., S.L. Investigation: W.M., S.L. Resources: D.C., T.E.A.P., A.S.W., N.S., S.H. Data Curation: S.L., W.M. Writing – Original Draft Preparation: S.L., W.M., L.P.S., N.S. Writing – Review and Editing: S.L., W.M., L.P.S., B.C., N.S., D.C., T.E.A.P., A.S.W., S.H. Visualization: S.L., W.M. Supervision: L.P.S., N.S., T.E.A.P., A.S.W., D.C. Project Administration: S.L., W.M., N.S., L.P.S. Funding: T.E.A.P., D.C., A.S.W., N.S.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: ARG, antimicrobial-resistance gene; FP, flank pattern; MGE, mobile genetic element; NIHR, National Institute for Health Research.
All supporting data, code and protocols have been provided within the article or through supplementary data files. One supplementary figure and five supplementary tables are available with the online version of this article.
References
- 1.Lipworth S, Vihta K-D, Chau K, Barker L, George S, et al. Molecular epidemiology of Escherichia coli and Klebsiella species bloodstream infections in Oxfordshire (UK) 2008-2018. medRxiv. 2021 doi: 10.1101/2021.01.05.20232553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vihta K-D, Stoesser N, Llewelyn MJ, Quan TP, Davies T, et al. Trends over time in Escherichia coli bloodstream infections, urinary tract infections, and antibiotic susceptibilities in Oxfordshire, UK, 1998–2016: a study of electronic health records. Lancet Infect Dis. 2018;18:1138–1149. doi: 10.1016/S1473-3099(18)30353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buetti N, Atkinson A, Marschall J, Kronenberg A, Swiss Centre for Antibiotic Resistance (ANRESIS) Incidence of bloodstream infections: a nationwide surveillance of acute care hospitals in Switzerland 2008–2014. BMJ Open. 2017;7:e013665. doi: 10.1136/bmjopen-2016-013665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thanner S, Drissner D, Walsh F. Antimicrobial resistance in agriculture. mBio. 2016;7:e02227–15. doi: 10.1128/mBio.02227-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wyres KL, Holt KE. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr Opin Microbiol. 2018;45:131–139. doi: 10.1016/j.mib.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Collis RM, Burgess SA, Biggs PJ, Midwinter AC, French NP, et al. Extended-spectrum beta-lactamase-producing enterobacteriaceae in dairy farm environments: a New Zealand perspective. Foodborne Pathog Dis. 2019;16:5–22. doi: 10.1089/fpd.2018.2524. [DOI] [PubMed] [Google Scholar]
- 7.Velasova M, Smith RP, Lemma F, Horton RA, Duggett NA, et al. Detection of extended-spectrum β-lactam, AmpC and carbapenem resistance in Enterobacteriaceae in beef cattle in Great Britain in 2015. J Appl Microbiol. 2019;126:1081–1095. doi: 10.1111/jam.14211. [DOI] [PubMed] [Google Scholar]
- 8.von Wintersdorff CJH, Penders J, van Niekerk JM, Mills ND, Majumder S, et al. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front Microbiol. 2016;7:173. doi: 10.3389/fmicb.2016.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Passarelli-Araujo H, Palmeiro JK, Moharana KC, Pedrosa-Silva F, Dalla-Costa LM, et al. Genomic analysis unveils important aspects of population structure, virulence, and antimicrobial resistance in Klebsiella aerogenes . FEBS J. 2019;286:3797–3810. doi: 10.1111/febs.15005. [DOI] [PubMed] [Google Scholar]
- 10.Nakamura K, Murase K, Sato MP, Toyoda A, Itoh T, et al. Differential dynamics and impacts of prophages and plasmids on the pangenome and virulence factor repertoires of Shiga toxin-producing Escherichia coli O145:H28. Microb Genom. 2020;6:000323. doi: 10.1099/mgen.0.000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decano AG, Downing T. An Escherichia coli ST131 pangenome atlas reveals population structure and evolution across 4,071 isolates. Sci Rep. 2019;9:17394. doi: 10.1038/s41598-019-54004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ, et al. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seemann T. Mlst. 2019. [ Jul 12; 2019 ]. https://github.com/tseemann/mlst accessed.
- 14.Lam MMC, Wick RR, Wyres KL, Holt KE. Genomic surveillance framework and global population structure for Klebsiella pneumoniae . bioRxiv. 2020 doi: 10.1101/2020.12.14.422303. [DOI] [Google Scholar]
- 15.Beghain J, Bridier-Nahmias A, Le Nagard H, Denamur E, Clermont O. ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb Genom. 2018;4:000192. doi: 10.1099/mgen.0.000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lees JA, Harris SR, Tonkin-Hill G, Gladstone RA, Lo SW, et al. Fast and flexible bacterial genomic epidemiology with PopPUNK. Genome Res. 2019;29:304–316. doi: 10.1101/gr.241455.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robertson J, Nash JHE. MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb Genom. 2018;4:000206. doi: 10.1099/mgen.0.000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acman M, van Dorp L, Santini JM, Balloux F. Large-scale network analysis captures biological features of bacterial plasmids. Nat Commun. 2020;11:2452. doi: 10.1038/s41467-020-16282-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheppard AE, Stoesser N, German-Mesner I, Vegesana K, Walker AS, et al. TETyper: a bioinformatic pipeline for classifying variation and genetic contexts of transposable elements from short-read whole-genome sequencing data. Microb Genom. 2018;4:000232. doi: 10.1099/mgen.0.000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johansson MHK, Bortolaia V, Tansirichaiya S, Aarestrup FM, Roberts AP, et al. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J Antimicrob Chemother. 2021;76:101–109. doi: 10.1093/jac/dkaa390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang R, van Dorp L, Shaw LP, Bradley P, Wang Q, et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1 . Nat Commun. 2018;9:1179. doi: 10.1038/s41467-018-03205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ludden C, Raven KE, Jamrozy D, Gouliouris T, Blane B, et al. One health genomic surveillance of Escherichia coli demonstrates distinct lineages and mobile genetic elements in isolates from humans versus livestock. mBio. 2019;10:e02693-18. doi: 10.1128/mBio.02693-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.David S, Cohen V, Reuter S, Sheppard AE, Giani T, et al. Integrated chromosomal and plasmid sequence analyses reveal diverse modes of carbapenemase gene spread among Klebsiella pneumoniae . Proc Natl Acad Sci USA. 2020;117:25043–25054. doi: 10.1073/pnas.2003407117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Acman M, Wang R, van Dorp L, Shaw LP, Wang Q, et al. Role of the mobilome in the global dissemination of the carbapenem resistance gene blaNDM. bioRxiv. 2021 doi: 10.1101/2021.01.14.426698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seemann T. Abricate. 2019. [ Jul 05; 2019 ]. https://github.com/tseemann/abricate accessed.
- 27.Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendrickx APA, Landman F, de Haan A, Witteveen S, van Santen-Verheuvel MG. blaOXA-48-like genome architecture among carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in the Netherlands. Microb Genom. 2021;7 doi: 10.1099/mgen.0.000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.David S, Reuter S, Harris SR, Glasner C, Feltwell T. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol. 2019;4:1919–1929. doi: 10.1038/s41564-019-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hagberg A, Swart P S, Chult D. Exploring network structure, dynamics, and function using NetworkX. Los Alamos National Lab.(LANL), Los Alamos, NM (United States. 2008. https://www.osti.gov/biblio/960616
- 31.Wick R. Assembly-dereplicator. Github. 2021. [ Feb 02; 2021 ]. https://github.com/rrwick/Assembly-Dereplicator accessed.
- 32.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, et al. vegan: Community Ecology Package. 2019. https://CRAN.R-project.org/package=vegan
- 33.EUCAST European committee on antimicrobial susceptibility testing. 2021. https://www.eucast.org/clinical_breakpoints/
- 34.Wickham H. ggplot2: Elegant Graphics for Data Analysis. 2016. https://ggplot2.tidyverse.org
- 35.Wilkins D. gggenes: Draw Gene Arrow Maps in “ggplot2.”. 2019. https://CRAN.R-project.org/package=gggenes
- 36.Yu G, Smith DK, Zhu H, Guan Y, Lam TT. Ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 37.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 38.Katz L, Griswold T, Morrison S, Caravas J, Zhang S. Mashtree: a rapid comparison of whole genome sequence files. JOSS. 2019;4:1762. doi: 10.21105/joss.01762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pitout JDD, Peirano G, Kock MM, Strydom K-. A, Matsumura Y. The global ascendency of OXA-48-type carbapenemases. Clin Microbiol Rev. 2019;33:e00102-19. doi: 10.1128/CMR.00102-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beyrouthy R, Robin F, Delmas J, Gibold L, Dalmasso G, et al. IS1R-mediated plasticity of IncL/M plasmids leads to the insertion of blaOXA-48 into the Escherichia coli chromosome. Antimicrob Agents Chemother. 2014;58:3785–3790. doi: 10.1128/AAC.02669-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carrër A, Poirel L, Eraksoy H, Cagatay AA, Badur S, et al. Spread of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in Istanbul, Turkey. Antimicrob Agents Chemother. 2008;52:2950–2954. doi: 10.1128/AAC.01672-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, et al. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22:686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cuzon G, Naas T, Nordmann P. Functional characterization of Tn4401, a Tn3-based transposon involved in blaKPC gene mobilization. Antimicrob Agents Chemother. 2011;55:5370–5373. doi: 10.1128/AAC.05202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.