Abstract
Our understanding of the host component of sepsis has made significant progress. However, detailed study of the microorganisms causing sepsis, either as single pathogens or microbial assemblages, has received far less attention. Metagenomic data offer opportunities to characterize the microbial communities found in septic and healthy individuals. In this study we apply gradient-boosted tree classifiers and a novel computational decontamination technique built upon SHapley Additive exPlanations (SHAP) to identify microbial hallmarks which discriminate blood metagenomic samples of septic patients from that of healthy individuals. Classifiers had high performance when using the read assignments to microbial genera [area under the receiver operating characteristic (AUROC=0.995)], including after removal of species ‘culture-confirmed’ as the cause of sepsis through clinical testing (AUROC=0.915). Models trained on single genera were inferior to those employing a polymicrobial model and we identified multiple co-occurring bacterial genera absent from healthy controls. While prevailing diagnostic paradigms seek to identify single pathogens, our results point to the involvement of a polymicrobial community in sepsis. We demonstrate the importance of the microbial component in characterising sepsis, which may offer new biological insights into the aetiology of sepsis, and ultimately support the development of clinical diagnostic or even prognostic tools.
Keywords: bacteraemia, machine learning, metagenomics, sepsis, contamination, kitome, blood metagenomics, SHAP
Data Summary
All relevant source code and parsed datasets used can be found on GitHub (https://github.com/cednotsed/Polymicrobial-Signature-of-Sepsis). The raw sequence data for each study can be found from NCBI SRA and the European Nucleotide Archive repository with the accessions listed in Table 1. The authors confirm all supporting data, code and protocols have been provided within the article or through supplementary data files.
Table 1.
Summary of metagenomic datasets.
Sample sizes indicated here are those after all quality control steps have been applied. Grumaz-16/19 is a combined dataset comprising Grumaz-16 and Grumaz-19.
|
Study |
Dataset alias |
Accession |
Sepsis definition |
Sequencing technique |
Sample size |
|
|---|---|---|---|---|---|---|
|
Septic |
Healthy |
|||||
|
Single datasets | ||||||
|
Grumaz et al. [25] |
Grumaz-19 |
PRJEB21872 PRJEB30958 |
Sepsis-2 |
Shotgun |
50 |
– |
|
Grumaz et al. [23] |
Grumaz-16 |
PRJEB13247 |
Sepsis-2 |
Shotgun |
7 |
15 |
|
Gosiewski et al. [24] |
Gosiewski-17 |
Requested from authors |
Sepsis-1 |
16S (paired-end) |
56 |
23 |
|
Blauwkamp et al. [22] |
Karius |
PRJNA507824 |
Sepsis-1 |
Shotgun |
117 |
170 |
|
Combined datasets | ||||||
|
All single datasets |
Pooled |
All accessions |
Sepsis-1 and Sepsis-2 |
Shotgun and 16S (paired-end) |
230 |
208 |
|
Grumaz et al. [23] and Grumaz et al. [25] |
Grumaz-16/19 |
PRJEB13247 PRJEB21872 PRJEB30958 |
Sepsis-2 |
Shotgun |
57 |
15 |
Impact Statement.
In this work, we analysed publicly available metagenomics datasets, comparing the patterns of microbial DNA in the blood plasma of septic patients relative to that of healthy individuals. As a technical contribution to (meta)genomic medicine, we demonstrate the application of a state-of-the-art machine learning technique to computationally identify putative contaminant taxa, which confound metagenomic investigations of blood infections. Additionally, the main contribution of our work is to show that septic infections tend to be polymicrobial rather than unimicrobial in nature. Polymicrobial interactions are known to alter infectious disease progression, severity and the host’s response to treatment. As such, our conclusions justify further work into characterising the microbial component of sepsis, and how it may be leveraged for management of sepsis in a clinical setting.
Introduction
Sepsis poses a significant challenge to public health and was listed as a global health priority by the World Health Organisation (WHO) in 2017. In the same year, 48.9 million cases of sepsis and 11 million deaths were recorded worldwide [1], having a particular impact in low- and low-to-middle income countries [2].
Current research efforts have predominately focused on understanding the host’s response to sepsis. Indeed, all contemporary definitions of sepsis focus on the host’s response and resulting systemic complications. The 1991 Sepsis-1 definition described sepsis as a systemic inflammatory response syndrome (SIRS) caused by infection, with patients being diagnosed with sepsis if they fulfil at least two SIRS criteria and have a culture-confirmed infection [3]. The 2001 Sepsis-2 definition then expanded the scope of SIRS to include more symptoms [4]. More recently, the 2016 Sepsis-3 definition sought to differentiate between mild and severe cases of dysregulated host responses, describing sepsis as a life-threatening organ dysfunction as a result of infection [5]. Significant progress has been made in understanding how dysregulation occurs [6] and the long-term impacts of sepsis [7, 8]. Additionally, early-warning tools have been developed based on patient healthcare records [9–11] and clinical checklists [12, 13]. However, the focus on the host component of sepsis may overlook the important role of microbial composition in the pathogenesis of the disease.
Due to the severity of sepsis, current practice considers identification of a single pathogen sufficient to warrant a diagnosis, without consideration of other, potentially relevant, species in the bloodstream. Upon diagnosis, infections are rapidly treated with broad-spectrum antibiotics. However, blood cultures, the current recommended method of diagnosis before antimicrobial treatment [14], are known to yield false negatives due to certain microorganisms failing to grow in culture [15], particularly in samples with low microbial loads [16]. Culture-based methods, while useful in a clinical context, may therefore under-estimate the true number of causative pathogens infecting septic patients.
Sepsis is a highly heterogeneous disease which consists of both a host component and a microbial component. While the former has been widely studied, the latter appears to represent a largely untapped source of information that could further advance our understanding of sepsis. Several diseases manifest as a result of interactions in a polymicrobial community. For example, microbial interactions in lung, urinary tract and wound infections are all known to contribute to differing disease outcomes (reviewed by Tay et al. [17]). These findings suggest that the microbial component of sepsis may also be crucial to understanding its pathogenesis.
Current technologies to investigate the presence of polymicrobial communities have some major limitations. As noted previously, culture-based methods have a high false negative rate. Furthermore, without knowledge of the range of microorganisms that infect blood, co-culture experiments to study microbial interactions prove difficult. For PCR-based technologies, the use of species-specific primers (e.g. SeptiFast [18]) necessitates a priori knowledge of microbial sequences endogenous to septic blood. Lastly, metagenomic sequencing is ubiquitously prone to environmental contamination. This can include DNA from viable cells introduced during sample collection, sample processing or DNA present in laboratory reagents [19–21]: the so-called ‘kitome’. As such, it can be difficult to determine which microorganisms are truly endogenous to the sample, and at what abundance.
In this study, we sought to expand our understanding of the full microbial component of sepsis. Multiple statistical and state-of-the-art machine learning techniques were applied to metagenomic sequencing data published by Blauwkamp et al. [22] (henceforth Karius study) from 117 sepsis patients and 170 healthy individuals. To circumvent the problem of potential contamination in metagenomic data, we developed and applied a novel computational contamination reduction technique. We also externally validated our findings using external hold-out datasets comprising three other independent sepsis cohorts. Taken together, our results provide strong evidence for a polymicrobial signature of sepsis and the utility of metagenomic sequencing for the investigation of blood-borne infections.
Methods
Datasets
Our primary analysis involved published shotgun metagenomic sequence data from the Karius study [22]. As detailed in this study, patients were diagnosed with sepsis if they presented with a temperature > 38 °C or < 36 °C, at least one other SIRS criterion and evidence of bacteraemia. Bacteraemia was confirmed via clinical microbiological testing performed within 7 days after collection of the blood samples. The list of pathogens identified by such tests (which we refer to as ‘culture-confirmed’ pathogens) can be found on GitHub (github.com/cednotsed/Polymicrobial-Signature-of-Sepsis/blob/master/datasets/karius_parsed_metadata.csv) and corresponds to Table S5 in the Karius study. This included tissue, fluid and blood cultures, serology, and nucleic acid testing. The clinical outcome of each patient was not reported in the original study. Seven of the 117 septic patients were found to have more than one ‘culture-confirmed’ pathogen identified by microbiological testing (Table S5 in the Karius study). According to the Karius study, healthy individuals were ‘screened for common health conditions including infectious diseases through a questionnaire and standard blood donor screening assays’. We believe this to be reasonable grounds for ruling out bloodstream infections in healthy patients (i.e. of non-septic origin).
To determine if the findings of our primary analysis were applicable beyond the Karius dataset, we also used metagenomic sequencing data from three other independent sepsis cohorts [23–25], where participants were recruited under different sepsis definitions, and samples were sequenced using different sequencing strategies (single datasets; Table 1). All four datasets were combined to yield the Pooled dataset (combined datasets; Table 1), which was used to determine if models could perform well given data from diverse sources. To further test the generalizability of our models, we held out one dataset and used it to evaluate models trained on the remaining datasets (see section ‘Holdout cross-validation’). Since Grumaz-16 did not contain samples from healthy individuals, it had to be combined with Grumaz-19 to form a single holdout dataset named Grumaz-16/19 (combined datasets; Table 1). In this case, Karius and Gosiewski-17 were used for model training and optimization while Grumaz-16/19 was used for evaluation. We will henceforth refer to each dataset by its dataset alias as shown in Table 1.
Data pre-processing
As described in the Karius study, input circulating free DNA was sequenced using a NextSeq500 (75-cycle PCR, 1×75 nt). Raw Illumina sequencing reads were demultiplexed by bcl2fastq (v2.17.1.14; default parameters) and quality trimmed using Trimmomatic (v0.32) [26] retaining reads with a quality (Q-score) above 20. Mapping and alignment were performed using Bowtie (v2.2.4) [27]. Human reads were identified by mapping to the human reference genome and removed prior to deposition in NCBI’s Sequence Read Archive (PRJNA507824).
For Grumaz-16 and Grumaz-19, BBMap (v38.79) [28] was used to trim adapter sequences, remove reads with a Q-score below 20 and remove reads mapping to a masked human hg19 reference (https://tinyurl.com/yya4xmrg). For the Gosiewski-17 dataset, we performed the same pre-processing steps as reported in the associated study [24]. Briefly, primers and adapters were removed using Cutadapt (v1.18) [29], paired reads were merged using ea-utils (v1.1.2.537) [30], merged reads and forward unmerged fastq files were concatenated, and reads with a Q-score below 20 were removed using BBMap.
Taxonomic classification of all shotgun sequencing data was performed using Kraken 2 (v2.0.9-beta; default parameters) [31] with the maxikraken2_1903_140 GB database (https://tinyurl.com/y7zfg9kr). To mitigate potential misclassification of closely related species (e.g. Escherichia coli and Shigella species) during taxonomic assignment, we considered only microbial abundance at the genus rank for downstream analyses. For the Gosiewski-17 dataset, Kraken 2 with a Kraken 2-built Silva database was used instead of conventional 16S amplicon metagenomic classification methods [32]. Read assignments for all ‘culture-confirmed’ bacterial pathogens using the maxikraken2_1903_140 GB and Kraken 2-built Silva databases are shown in Fig. S1. While the relative number of reads assigned to each bacterial genus showed some inconsistencies, this hardly affected the classifier performance of septic and healthy patients (Fig. S2). This suggests that our model is fairly robust to heterogeneity which may be introduced by the classification step. For downstream analyses, we use the genera assignments based on the Kraken 2-built Silva database for the 16S Gosiewski-17 samples. Additionally, all unclassified reads were excluded from the analyses.
Unexpectedly, for the Karius dataset, a small number of reads were assigned to the genus Homo, which was possibly due to misclassification. Mapping of all reads in the Karius sequencing data found just 873 bases with 96 % identity to a masked human hg19 reference (https://tinyurl.com/yya4xmrg), with an average of 0.3 reads per sample (range: 0–7 reads). Since human reads were already removed in the bioinformatics workflow of the Karius study, we did not perform an additional human read removal step to avoid introducing biases in the data.
The ouput of taxonomic assignment is a data matrix with samples represented in rows and taxa in columns (i.e. features). Each element in the matrices represented the total number of reads assigned to each taxon, which we loosely refer to as ‘abundance’. The set of taxa used in each analysis will henceforth be referred to as the ‘feature space’. Where a single dataset was used to train a single model, the feature space comprised all microbial taxa identified during taxonomic assignment. Where multiple datasets were used in tandem to train a single model, the feature space comprised the microbial taxa common to all datasets. Feature spaces that have not undergone any statistical removal of microbial taxa are denoted by Neat.
Model training, optimization and nested cross-validation
To assess the suitability of taxonomic assignments for discriminating between septic and healthy blood metagenomic samples, gradient-boosted tree classifiers were trained and evaluated using the data matrices parsed from the Kraken 2 taxonomic assignments. The task of all classifiers was to predict if a sample belonged to a septic or healthy individual given the read counts assigned to microbial taxa. Classifiers were trained with a binary-logistic loss function and implemented using XGBoost API (v0.90) [33]. Model optimization was performed using a randomised hyperparameter optimization protocol [34] with 1000 samples, implemented using RandomizedSearchCV in the Scikit-learn API (v0.23.1) [35]. The test error of each model was estimated using a nested, stratified, 10×10-fold cross-validation procedure. Nested cross-validation was necessary to obtain an unbiased estimate for test error since hyperparameter optimisation was required [36]. Briefly, in each iteration of the outer cross-validation loop, a tenth of the data is held-out. The remaining data are used in an inner cross-validation loop where a search for the best set of hyperparameters is performed. The held-out data in the outer loop are then used to evaluate the model with the best set of hyperparameters identified in the inner loop. Separately, a hyperparameter optimisation protocol was performed using the entire dataset, yielding the hyperparameter set that maximises the receiver operating characteristic curve (AUROC) metric. This hyperparameter set was then used for downstream analyses.
Holdout cross-validation
To determine if our models were generalisable across the different sepsis cohorts, the data from three of four sepsis cohorts were combined for model training and hyperparameter optimisation. The test error of each optimized model was then estimated using the holdout dataset. We refer to this protocol as ‘holdout cross-validation’. For holdout cross-validation, precision, recall and the area under the precision-recall curve (AUPRC) were used as performance metrics since they are more informative when used on imbalanced test sets [37]. Any statistical filtering of features (see sections ‘SHAP decontamination’ and ‘Simple decontamination’) was performed before model evaluation.
Model interpretation
To interpret models, each feature in a single sample was assigned a SHAP (SHapley Additive exPlanations) value, which corresponds to the change in a sample’s predicted probability score (i.e. probability of sepsis) when the feature is either present or absent. Using SHAP values therefore allows the decomposition of predicted probability scores for each sample into the sum of contributions from individual genera. The relative importance of each feature was inferred via its mean absolute SHAP value across all samples. A higher mean absolute SHAP value implies that the feature has a larger impact on the model predictions. SHAP values were computed using TreeExplainer, part of the shap library (v0.34.0) [38]. For every model, SHAP values were computed for the whole dataset by setting the feature_pertubation parameter to ‘interventional’.
SHAP Decontamination
SHAP Decontamination was performed in two main steps. First, genera that are not currently identified as known human pathogens were removed. This selection was based on a study by Shaw et al. [39], who considered a ‘human pathogen’ to be any microbial species for which there is evidence in the literature that it can cause infection in humans, sometimes in a single patient. The list of known human pathogens used can be found on GitHub (https://github.com/cednotsed/Polymicrobial-Signature-of-Sepsis/blob/master/datasets/pathogen_list.csv) and was downloaded from FigShare [40]. Second, a classifier was optimised and trained on genera abundance (Neat feature spaces). SHAP values for model predictions on the dataset were then calculated. Genera with a negative Spearman’s correlation between their corresponding SHAP values and abundances were removed. Spearman’s correlations were calculated using spearmanr as part of the SciPy library (v1.4.1) [41]. A new classifier was then retrained using the previously optimized set of parameters but with this new reduced feature space. This process was repeated iteratively until the number of genera retained remained constant. The resultant feature space is denoted by CR.
To test the hypothesis that genera containing true pathogens are positively associated with sepsis, we inspected the SHAP values and read counts assigned to the genera corresponding to cases of each type of ‘culture-confirmed’ infection (e.g. SHAP value/read count assigned to Escherichia for only Escherichia -positive samples) using the Karius-Neat feature space. The SHAP values were all greater or equal to zero apart from a single sample which had a negative SHAP value for Mycobacterium (Fig. S3). The assigned read counts were non-zero except for one sample with a ‘culture-confirmed’ fungal Candida glabrata infection reported (SRR8288759). These findings suggest that SHAP values can successfully recover experimentally identified pathogens.
Simple Decontamination
We also employed a more direct, model-free contaminant removal technique (Simple Decontamination) that follows the same underlying premise of SHAP Decontamination. In this procedure, genera in the Neat feature space that were significantly (P < 0.05) more abundant in healthy controls than septic samples were considered contaminants and removed. The resultant feature space is denoted by SD.
Microbial networks
Microbial co-occurrence networks were constructed using the SparCC algorithm [42], implemented in the SpiecEasi package (v1.1.0) [43] and visualized using Igraph (v1.2.5) [44]. SparCC was used to account for compositionality that could lead to spurious correlations. Separate networks were constructed for the genera assignments of septic and healthy metagenomes. To determine the microbial associations present exclusive to septic samples, a corrected sepsis network was produced. This network was constructed by subtracting all edges of the healthy network from the sepsis network. Only co-occurrence relationships where the SparCC correlations exceed 0.2 were retained. The Karius-SD feature space was used as input.
Results
Metagenomic sequencing can be used to discriminate septic from healthy samples
The performance of all classifiers is summarized in Table 2. Models were first trained and evaluated using 117 septic patients and 170 healthy individuals in the Karius study (Table 1). Classifiers could discriminate between sepsis from healthy samples using the read counts assigned to each microbial taxon (Karius-Neat model; AUROC=0.995). Classifiers performed similarly well when using a more diverse dataset comprising data pooled from all four sepsis cohorts (Pooled-Neat model; AUROC=0.982). We also tested the effect of normalising assigned read counts by the total per-sample count. Such normalisation resulted in reduced classification performance (Karius-normalised model; AUROC=0.943) and so was not performed for the rest of the models tested.
Table 2.
Summary of models trained
Models were optimized and evaluated via a nested cross-validation protocol. The prefix and suffix of each model name corresponds to the dataset and contamination reduction technique applied, respectively. Neat, SD and CR refer to the feature spaces with no decontamination, Simple Decontamination, and SHAP Decontamination applied, respectively (see Methods). Karius-Without corresponds to the SHAP-decontaminated feature space after claimed ‘culture-confirmed’ pathogens are excluded. Karius-Only refers to the feature space containing only genera with ‘culture-confirmed’ pathogens as features.
|
No. of features |
Feature space |
Model performance |
||
|---|---|---|---|---|
|
Precision |
Recall |
AUROC |
||
|
1564 |
Karius-Neat |
0.976 |
0.983 |
0.995 |
|
1564 |
Karius-normalised |
0.956 |
0.932 |
0.943 |
|
111 |
Karius-SD |
0.896 |
0.787 |
0.942 |
|
25 |
Karius-CR |
0.883 |
0.810 |
0.942 |
|
22 |
Karius-Without |
0.803 |
0.727 |
0.915 |
|
22 |
Karius-Only |
0.929 |
0.862 |
0.950 |
|
685 |
Pooled-Neat |
0.950 |
0.939 |
0.982 |
|
21 |
Pooled-CR |
0.870 |
0.796 |
0.904 |
SHAP can be used to remove putative sequencing contaminants
Accurate characterization of the microbial component of sepsis requires discrimination between a true biological signal and that arising from putative environmental contamination in metagenomes. We developed and applied a procedure to remove biologically irrelevant genera from the feature space, which we refer to as SHAP Decontamination (CR; see Methods). Briefly, we leveraged SHAP – a state-of-the-art machine learning technique for interpreting ‘black-box’ classifiers [38] – to determine how the read counts assigned to a genus (i.e. feature) influence model predictions for each sample. In doing so, we selectively removed putative contaminants from the feature spaces obtained from taxonomic classification. We illustrate this for a single ‘culture-confirmed’ E. coli -positive sample in the Karius dataset (Fig. S4).
To evaluate the effectiveness of this approach, we compared SHAP Decontamination to a simpler statistical method for the removal of putative pathogens, which we call Simple Decontamination (SD; see Methods). For the Karius dataset, application of SHAP Decontamination resulted in a pruned feature space of 25 genera while Simple Decontamination resulted in 111 genera. The resultant Karius-CR and Karius-SD feature spaces, respectively, shared 21 genera in common. Classifiers trained on either of the Karius-CR or Karius-SD feature space had similarly high performance (Table 2, Karius-CR/SD; AUROC=0.942), despite the large reduction in the number of features. This suggests that computational decontamination efficiently removes redundancy in the metagenomic feature space. Furthermore, SHAP Decontamination appears to be more efficient, as demonstrated by the equivalent classification performance, but higher number of removed putative contaminant genera than Simple Decontamination.
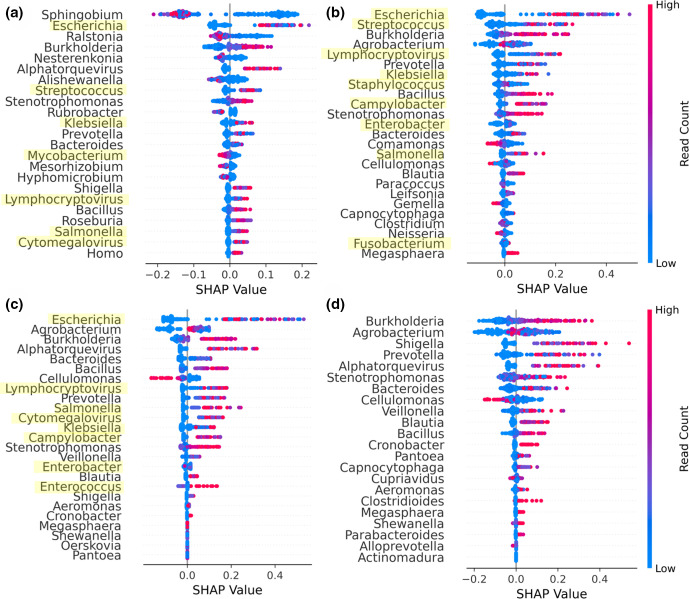
Separately, we observed that the Karius-CR model comprised almost all genera associated with sepsis at higher abundance. Additionally, genera such as Sphingobium , Mesorhizobium and Ralstonia were highly important features in the Karius-Neat feature space (Fig. 1a), though not present in either the Karius-SD or the Karius-CR feature space (Fig. 1b, c). These genera are likely to be contaminants since they contribute negatively to the predicted probability of sepsis at high abundance, and have been previously ascribed as common sequencing contaminants [19]. Of the 25 genera in the Karius-CR feature space, eight corresponded to genera containing clinically ‘culture-confirmed’ pathogens (see Methods). Notably, Escherichia and Enterobacter , which are both ‘culture-confirmed’ pathogens but also common contaminants [19], were retained in both decontaminated feature spaces. These findings collectively suggest that computational decontamination procedures were removing putative contaminants while selectively retaining biologically important genera.
Fig. 1.
Model interpretation and performance. (a) Plot summarizing the SHAP values across all samples for the most important features ranked by the mean absolute SHAP value (highest at the top) for Karius-Neat, (b) Karius-SD, (c) Karius-CR and (d) Karius-Without models. Each point represents a single sample. Points with similar SHAP values were stacked vertically for visualization of point density and were coloured according to the magnitude of the feature values (i.e. read counts). Genera that contained ‘culture-confirmed’ pathogens are highlighted in yellow.
Evidence for a polymicrobial community
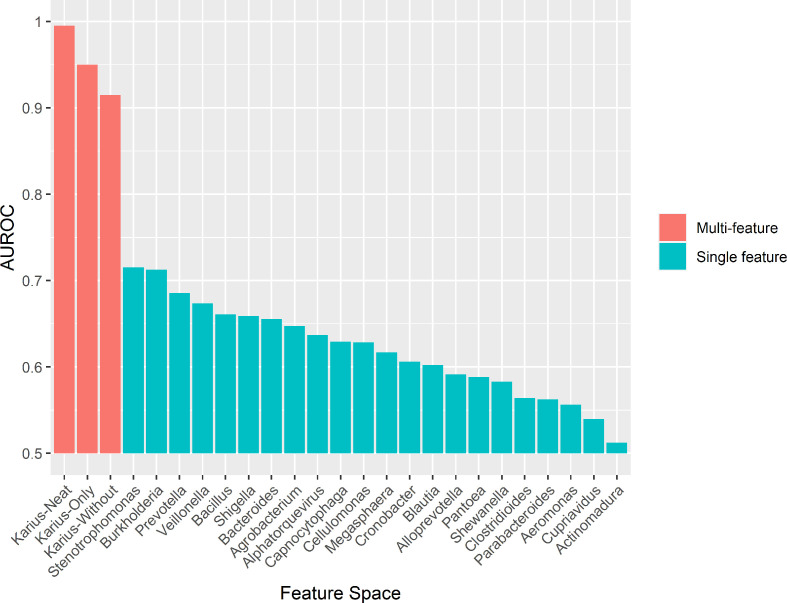
Having assessed the biological relevance of microbial predictors of sepsis, we provide several pieces of evidence supporting a polymicrobial model of sepsis; that is, that there are sets of microbial genera that delineate septic from healthy blood metagenomes, rather than just individual pathogens. Most notably, a classifier trained on the Karius dataset using the SHAP-decontaminated feature space but with all genera containing clinically identified pathogens (henceforth ‘culture-confirmed’ pathogens; see Methods) removed performed well (Karius-Without model; AUROC=0.915), suggesting the presence of these species alone does not capture the full microbial signal of sepsis. Visualization of the SHAP values for this model (Fig. 1d) confirmed that most genera had positive associations with sepsis at higher abundances. To test if any single features in the Karius-Without model were driving the high classification performance, we trained and evaluated multiple single-feature classifiers with each genus in the Karius-Without feature space. Additionally, we trained a classifier on genera containing ‘culture-confirmed’ pathogens as features only (Karius-Only). Fig. 2 shows the performance of the multi-feature Karius-Neat, Karius-Without and Karius-Only models compared to single-feature models. All multi-feature models performed better than those relying on single-feature models.
Fig. 2.
Comparison of performance (AUROC) for the multi-feature models (Karius-Neat, Karius-Only, Karius-Without feature space) and single-feature models (x-axis). Models were optimised and evaluated using the nested cross-validation protocol.
We then trained classifiers on the pooled dataset to determine if our results were unique to the Karius dataset or whether they were portable to other sepsis cohorts. Current metagenomics datasets are limited in their suitability for external validation due to the use of different sequencing technologies, differing sepsis definitions and small sample sizes. However, despite the pooled dataset comprising multiple data sources from different studies, the classifier still performed well (Pooled-Neat model, AUROC=0.982; Pooled-CR model, AUROC=0.904). This suggests strongly that there is a generalisable microbial signature which can be leveraged across metagenomic datasets.
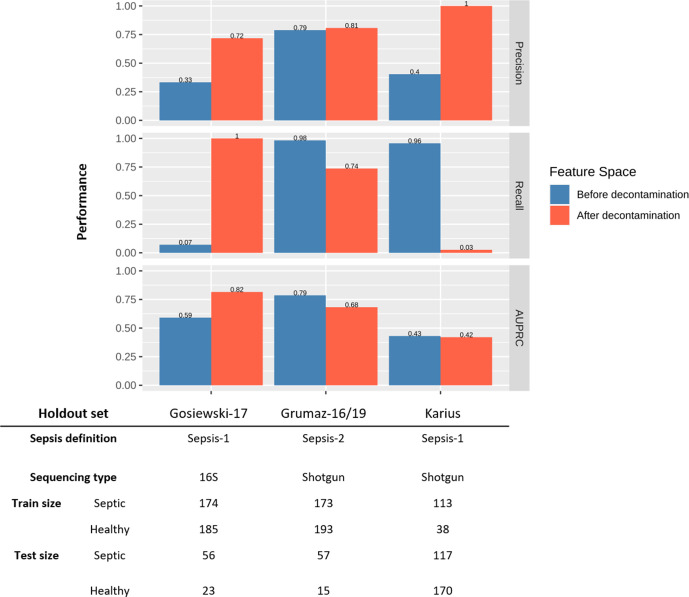
To more formally test the generalisability of the observed polymicrobial signature, we used holdout cross-validation (see Methods). Most notably, the classifier trained on shotgun metagenomic data and tested on 16S data as the holdout set (Gosiewski-17) did not perform well. However, after SHAP Decontamination, classification performance improved markedly. Interestingly, this performance increase was not observed when using the other datasets as holdout sets (Fig. 3). Indeed, the classifier trained with Grumaz-16/19 as the holdout set performed well before SHAP Decontamination, but relatively worse after. Additionally, holding out the Karius dataset resulted in poor classification performance both before and after SHAP Decontamination. A possible explanation for SHAP Decontamination lowering classification performance when Grumaz-16/19 is used as the test set is that septic cases recruited in these studies were based on different sepsis definitions, which may involve a different set of pathogens and reflect different aetiologies. Separately, the poor performance observed when the Karius dataset is used as the test set can be attributed to the highly imbalanced training dataset (Fig. 3).
Fig. 3.
Generalisability of models across sepsis cohorts. Model performance before and after SHAP Decontamination determined via holdout cross-validation (see Methods). The table appended describes the sepsis definition used, sequencing type and test size for each holdout dataset, and the corresponding size of the training data.
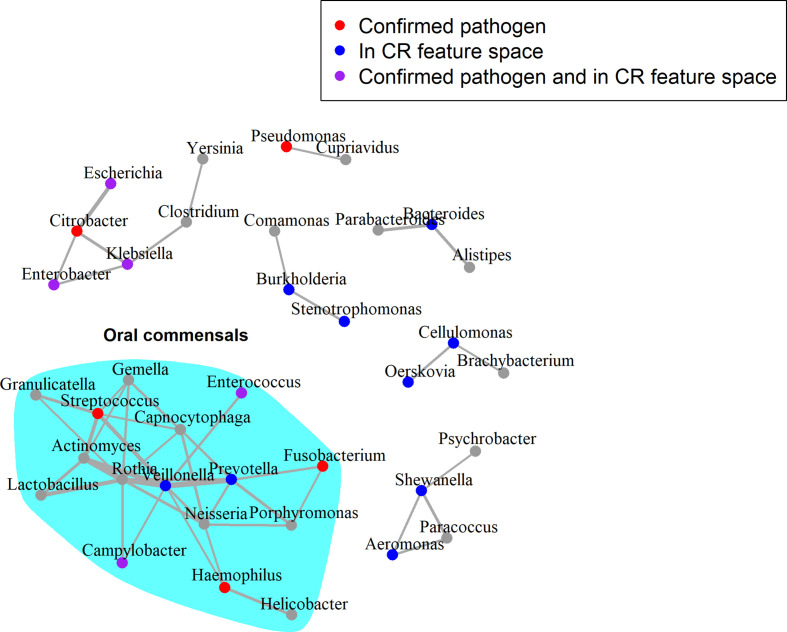
Lastly, microbial co-occurrence networks were used to identify relationships between genera that were exclusive to samples from septic patients. Two genera are said to co-occur if an increase in the abundance of one is associated with an increase in the abundance of the other. The presence of such relationships would lend weight to the polymicrobial nature of sepsis infections. The Karius-SD feature space was used in this analysis to corroborate previous analyses using the Karius-CR feature space. Multiple co-occurrence relationships between genera were present in the corrected network including those containing 10 of the 22 ‘culture-confirmed’ pathogens and 14 of the 25 genera in the Karius-CR feature space (Fig. 4). Interestingly, we detected a group of co-occurring genera associated with the oral cavity (Fig. 4), as suggested by the Human Oral Microbiome Database [45] (accessed 15 July 2020) and the current literature [46–49]. This was also present in the corrected network when the Pooled-SD feature space was used as input (Fig. S5).
Fig. 4.
Corrected microbial co-occurrence network for genera assigned in sepsis metagenomes. Input data correspond to the Karius-SD feature space. The edges in this network represent those in the septic network that were not present in the healthy network. The widths of edges are weighted by the strength of the SparCC correlations. Nodes are coloured as per the legend at the top, with ‘culture-confirmed’ pathogens those experimentally shown to be implicated in sepsis. The layout of the graph was generated using the Fruchterman–Reingold algorithm.
Discussion
The polymicrobial signature of sepsis
Our work demonstrates a clear polymicrobial signal in sepsis, where multiple, co-occuring, genera can be used to discriminate blood metagenomes of septic patients from that of healthy controls. The high performance of the Karius-Only model highlights that genera containing ‘culture-confirmed’ pathogens were very useful in delineating septic from healthy samples. More importantly, the Karius-Without model, which had these genera removed, also performed well, suggesting that the abundance of microbial genera that were not amongst the ‘culture-confirmed’ pathogens are also highly relevant to delineate septic from healthy samples. Furthermore, the single-feature models performed poorly, highlighting that no genus is solely responsible for the high classification performance of the Karius-Without model, further supporting the polymicrobial nature of sepsis infections.
We also demonstrate that the polymicrobial signal we detected is generalisable across datasets, first by nested cross-validation with all datasets pooled (Pooled-CR model) and then with holdout cross-validation using the Gosiewski-17 or Grumaz-16/19 datasets as test sets. The increased performance after SHAP Decontamination when holding out 16S data (Gosiewski-17) suggests that the retained set of genera allow a markedly more generalizable decision boundary to be learnt, even across sequencing techniques.
Additionally, the multiple co-occurrence relationships between genera detected suggest that there may be a distinct microbial community that tends to be present during sepsis infection. Although our networks were inferred computationally, published evidence supports possible synergies between some of the co-occurring genera we detected. For example, Stenotrophomonas and Burkholderia are known to play a collective role in the pathogenesis of cystic fibrosis [50]. Additionally, Klebsiella pneumoniae was found to be able to transmit extended-spectrum beta-lactamase genes to Citrobacter freundii and E. coli [51], potentiating synergism during polymicrobial infections. Furthermore, using fluorescence in-situ hybridisation, interspecies spatial associations were found between Prevotella , Veillonella , Streptococcus , Gemella , Rothia and Actinomyces in dental biofilms [52]. The tendency of bacteria of these genera to aggregate in biofilms agrees with the their strong correlations in the corrected sepsis network (Fig. 3). Moreover, bacterial cells from the genera Prevotella and Actinomyces were found to be in contact with the most number of bacterial species, suggesting that they were key players in maintaining intercellular adhesion and hence biofilm maturation in the oral cavities [52]. This was recapitulated in the corrected sepsis network, where the two genera are central nodes in the oral commensal cluster (Fig. 3). These examples suggest that the co-occurrence relationships we computationally detected may reflect genuine biological relationships.
Notably, the presence of a densely connected cluster of oral colonisers — some of which were identified to have interspecies spatial associations [52] – may point to a potential reservoir of sepsis pathogens. This also suggests the possibility of opportunistic infections from the human microbiota and dysbioses that could affect disease severity, given that oral infections are a known risk factor for systemic disease [53, 54]. This hypothesis is in line with the reported changes in nasal microbiomes in septic individuals [55] and the associations of intestinal dysbiosis with increased susceptibility to sepsis [56]. If these hypotheses were validated, the microbiome profiles of patients might offer opportunities to assess a patient’s risk of developing sepsis prior to onset. Further investigation of the interactions between different clusters of genera in the corrected sepsis network, together with expanding our analyses to future datasets, may yield valuable insights into the underlying biology of sepsis infections and ultimately inform treatment.
The need to account for environmental contamination
Contamination from environmental sources poses one of the greatest challenges for metagenomic investigations of microbial communities, particularly in low-biomass and clinical samples [20, 57]. It is therefore crucial to discriminate between contaminants and biologically relevant taxa and to remove putative contaminants to protect against spurious signals.
The main premise behind SHAP Decontamination is that pathogens should occur at higher abundance in septic patients relative to healthy controls. This is because we expect most infections to be characterised by the proliferation of microorganisms [58, 59] and, as such, true pathogenic genera should contribute to a higher predicted probability of sepsis at higher abundances. Consequently, the abundance of contaminant taxa would demonstrate a negative Spearman’s correlation with their corresponding SHAP values. This allows putative contaminant genera to be computationally detected and removed. Our results demonstrate the efficacy of our post-hoc contamination reduction technique called SHAP Decontamination in removing redundancy in the feature space while selectively retaining taxa involved in sepsis. It is likely that the taxa removed in this procedure would in principle include commensals and environmental contaminants introduced during sample collection or preparation. As such, application of this technique provides greater confidence that the polymicrobial signals we observed were not largely driven by contaminants.
We appreciate that a more rigorous evaluation of this technique, particularly with mock communities, will be required. As an alternative to our contamination reduction technique, statistical decontamination techniques identifying inverse relationships between the assigned abundance of taxa and sample DNA concentration [60, 61] could be used. However, this method was not applicable for our study since the sample DNA concentrations in the datasets used were not reported.
Potential for metagenomics-based diagnostics
Although we do not claim to have developed a model sufficiently robust for immediate diagnostic purposes, our results highlight the clear promise of metagenomics-informed diagnostic models, which have also been suggested by previous studies [22, 62, 63]. To put the high performance of our models in context, Mao et al. [9] reported that InSight, a model trained on vital signs of patients, had a diagnostic AUROC of 0.92 using Sepsis-2 as the ground truth. They also reported that the Modified Early Warning Score (MEWS), Sequential Organ Failure Assessment (SOFA) and SIRS had an AUROC of 0.76, 0.63 and 0.75 respectively. Additionally, a classifier trained on nasal metagenomes of septic and healthy samples had an AUROC of 0.89 with Sepsis-3 as the ground truth [55]. Notably, it is difficult to compare the performance of models trained with labels generated by different definitions of sepsis, which is also inherently a highly heterogeneous disease. Further, the discrepancies in model performance could be due to differences in the size of training and testing datasets. At the very least, our results suggest that the microbial component of sepsis alone contains sufficient information for the diagnosis of sepsis. A crucial next step will be to generate larger datasets, from more diverse sources, to allow the training of more robust and generalisable models for diagnostic or prognostic use.
Limitations
We identified several limitations in our study. First, metagenomic sequencing involves measurements of circulating free DNA and not of viable microorganisms in blood. As such, the detection of DNA from multiple taxa does not necessarily represent the true number or abundance of active taxa present. However, multiple studies have demonstrated high concordance of targeted [64] or shotgun metagenomic sequencing with culture [22, 62, 65]. This suggests some level of agreement between the presence of microbial cells and their DNA in blood. Additionally, given its higher sensitivity and throughput, metagenomic sequencing appears to be the best tool currently available for gaining insights into polymicrobial infections.
Though our results suggest the importance of multiple genera in delineating metagenomes of septic patients from that of healthy controls, the aetiological contributions of these genera and their ecological relationships cannot be inferred. Such hypotheses must be confirmed experimentally. It is also important to keep in mind that the models presented in this study are not prognostic in nature, in that they were not trained to predict the onset or progression of sepsis. However, furthering our understanding of the microbial component of sepsis may prove useful in the development of better prognostic tools.
Some genera such as Escherichia and Enterobacter contain both biologically relevant genera and common sequencing contaminants. As such it is expected that a proportion of DNA molecules, and hence sequencing reads, may have come from contamination rather than microorganisms endogenous to blood. The abundance of these microorganisms, as detected by metagenomic approaches, may differ from the true abundance.
Additionally, k-mer-based approaches may be less accurate for taxonomic classification compared to, for example, Bayesian sequence read-assignment methods [66]. As such, we used taxonomic assignments at the genus level which were shown to be, in general, more reliable than that at the species level [67]. We also appreciate that k-mer-based classification approaches are significantly faster [68], which may provide clinically relevant turnaround times that are important in sepsis diagnostics.
Finally, we acknowledge the relatively small size of the datasets used in our analyses. As a result, the models presented in this study are not yet robust enough to be used in a clinical context. A larger and more diverse dataset is required to develop such models. This is to ensure that models can learn a more generalisable decision boundary for accurate sepsis diagnosis.
Irrespective of these limitations, our results nonetheless demonstrate the importance of considering the full polymicrobial component of sepsis and suggest that a metagenomics-based approach may provide biological and clinical insights supporting the future development of rapid diagnostic tools.
Future directions and implications of polymicrobial sepsis
A major next step forward would be to elucidate the functional role of the polymicrobial communities we identify in sepsis. One key hypothesis is whether there are different clusters of microbial communities in different sepsis aetiologies. Evidence for discriminatory microbial signals in different manifestations of disease would facilitate sepsis to be redefined to also include a microbial component. However, to robustly test such hypotheses, a much larger sepsis cohort must be recruited to provide adequate statistical power, particularly considering the true number of sepsis subtypes is unknown. The associations between detected polymicrobial communities and disease severity could also be investigated. To do so, anonymised healthcare records with detailed curation of the clinical outcomes and treatment history of each patient would be required. In addition, pre-infection data from animal models holds promise to identify taxa relevant for the early detection of sepsis, which can be an important bottleneck to good patient outcomes.
It would also be valuable to investigate how identified polymicrobial communities may change during the course of infection. This can be done via analysis of microbial community dynamics [69] using longitudinal metagenomic sampling. By monitoring the change in microbial composition along the course of infection, ecological relationships between pathogens can be inferred. Additionally, this would allow for a better identification of key taxa involved in sepsis at the level of the microbial species together with the presence of particular antimicrobial resistance genes. Lastly, co-culture experiments [70] could be performed to elucidate interactions between pathogens. These could also be paired with metabolomic approaches, which may be useful in identifying possible synergies or antagonisms between microbial species [69, 71, 72].
The advent of large-scale metagenomic sequencing of clinical samples offers new opportunities to better characterize the pathogens contributing to systemic infections, and unlike culture-based methods are not limited to organisms that are fast-growing or culturable. In this study, we demonstrate the promise of a metagenomics-based approach to sepsis. Our results provide evidence that septic infections should be considered as polymicrobial in nature, comprising multiple co-occurring pathogens indicative of disease. Our findings thus pave the way for more microbial-focused models of sepsis, with long run potential to inform early detection, clinical interventions and improve patient outcomes.
Supplementary Data
Funding information
L.v.D. and F.B. were funded by the UCL and Partner Hospitals: AI in Healthcare Funding Call 2019.
Author contributions
Development of the methods, data curation, analysis and visualization were performed by C.C.S.T. All authors were involved in conceptualization, drafting and revision of the manuscript. L.V.D. and F.B. contributed equally to this work as co-senior authors.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
No human participants or samples were involved in this study.
Footnotes
Abbreviations: AUPRC, area under the precision-recall curve; AUROC, area under the receiver operating characteristic; MEWS, modified early warning score; SHAP, SHapley Additive exPlanations; SIRS, systemic inflammatory response syndrome; SOFA, sequential organ failure assessment; WHO, World Health Organisation.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Five supplementary figures and five supplementary tables are available with the online version of this article.
References
- 1.Rudd KE, Johnson SC, Agesa KM, Shackelford KA, Tsoi D, et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: analysis for the Global Burden of Disease Study. Lancet. 2020;395:200–211. doi: 10.1016/S0140-6736(19)32989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwizera A, Baelani I, Mer M, Kissoon N, Schultz MJ, et al. The long sepsis journey in low-and middle-income countries begins with a first step… but on which road? Springer. 2018 doi: 10.1186/s13054-018-1987-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Chest. 1992;101:1644–1655. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 4.Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, et al. 2001 sccm/esicm/accp/ats/sis international sepsis definitions conference. Intensive Care Med. 2003;29:530–538. doi: 10.1007/s00134-003-1662-x. [DOI] [PubMed] [Google Scholar]
- 5.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3. Jama. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Poll T, van de Veerdonk FL, Scicluna BP, Netea MG. The immunopathology of sepsis and potential therapeutic targets. Nat Rev Immunol. 2017;17:407–420. doi: 10.1038/nri.2017.36. [DOI] [PubMed] [Google Scholar]
- 7.Venet F, Monneret G. Advances in the understanding and treatment of sepsis-induced immunosuppression. Nat Rev Nephrol. 2018;14:121–137. doi: 10.1038/nrneph.2017.165. [DOI] [PubMed] [Google Scholar]
- 8.Ammer-Herrmenau C, Kulkarni U, Andreas N, Ungelenk M, Ravens S, et al. Sepsis induces long-lasting impairments in CD4+ T-cell responses despite rapid numerical recovery of T-lymphocyte populations. PLoS One. 2019;14:e0211716. doi: 10.1371/journal.pone.0211716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mao Q, Jay M, Hoffman JL, Calvert J, Barton C, et al. Multicentre validation of a sepsis prediction algorithm using only vital sign data in the emergency department, general ward and ICU. BMJ Open. 2018;8:e017833. doi: 10.1136/bmjopen-2017-017833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor RA, Pare JR, Venkatesh AK, Mowafi H, Melnick ER, et al. Prediction of in‐hospital mortality in emergency department patients with sepsis: a local big data–driven, machine learning approach. Acad Emerg Med. 2016;23:269–278. doi: 10.1111/acem.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nemati S, Holder A, Razmi F, Stanley MD, Clifford GD, et al. An interpretable machine learning model for accurate prediction of sepsis in the ICU. Crit Care Med. 2018;46:547–553. doi: 10.1097/CCM.0000000000002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henry KE, Hager DN, Pronovost PJ, Saria S. A targeted real-time early warning score (TREWScore) for septic shock. Sci Transl Med. 2015;7:299ra122. doi: 10.1126/scitranslmed.aab3719. [DOI] [PubMed] [Google Scholar]
- 13.Smith GB, Prytherch DR, Meredith P, Schmidt PE, Featherstone PI. The ability of the National Early Warning Score (NEWS) to discriminate patients at risk of early cardiac arrest, unanticipated intensive care unit admission, and death. Resuscitation. 2013;84:465–470. doi: 10.1016/j.resuscitation.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 14.Rhodes A, Evans LE, Alhazzani W, Levy MM, Antonelli M, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Intensive Care Med. 2017;43:304–377. doi: 10.1007/s00134-017-4683-6. [DOI] [PubMed] [Google Scholar]
- 15.Klaerner HG, Eschenbach U, Kamereck K, Lehn N, Wagner H, et al. Failure of an automated blood culture system to detect nonfermentative gram-negative bacteria. J Clin Microbiol. 2000;38:1036–1041. doi: 10.1128/JCM.38.3.1036-1041.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benjamin RJ, Wagner SJ. The residual risk of sepsis: modeling the effect of concentration on bacterial detection in two‐bottle culture systems and an estimation of false‐negative culture rates. Transfusion. 2007;47:1381–1389. doi: 10.1111/j.1537-2995.2007.01326.x. [DOI] [PubMed] [Google Scholar]
- 17.Tay WH, Chong KKL, Kline KA. Polymicrobial–host interactions during infection. J Mol Biol. 2016;428:3355–3371. doi: 10.1016/j.jmb.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Westh H, Lisby G, Breysse F, Böddinghaus B, Chomarat M, et al. Multiplex real-time PCR and blood culture for identification of bloodstream pathogens in patients with suspected sepsis. Clin Microbiol Infect. 2009;15:544–551. doi: 10.1111/j.1469-0691.2009.02736.x. [DOI] [PubMed] [Google Scholar]
- 19.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glassing A, Dowd SE, Galandiuk S, Davis B, Chiodini RJ. Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathog. 2016;8:24. doi: 10.1186/s13099-016-0103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiss S, Amir A, Hyde ER, Metcalf JL, Song SJ, et al. Tracking down the sources of experimental contamination in microbiome studies. Genome Biol. 2014;15:564. doi: 10.1186/s13059-014-0564-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blauwkamp TA, Thair S, Rosen MJ, Blair L, Lindner MS, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol. 2019;4:663–674. doi: 10.1038/s41564-018-0349-6. [DOI] [PubMed] [Google Scholar]
- 23.Grumaz S, Stevens P, Grumaz C, Decker SO, Weigand MA, et al. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 2016;8:73. doi: 10.1186/s13073-016-0326-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gosiewski T, Ludwig-Galezowska AH, Huminska K, Sroka-Oleksiak A, Radkowski P, et al. Comprehensive detection and identification of bacterial DNA in the blood of patients with sepsis and healthy volunteers using next-generation sequencing method-the observation of DNAemia. Eur J Clin Microbiol Infect Dis. 2017;36:329–336. doi: 10.1007/s10096-016-2805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grumaz S, Grumaz C, Vainshtein Y, Stevens P, Glanz K, et al. Enhanced performance of next-generation sequencing diagnostics compared with standard of care microbiological diagnostics in patients suffering from septic shock. Crit Care Med. 2019;47:e394–e402. doi: 10.1097/CCM.0000000000003658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bushnell B. BBMap: a Fast, Accurate, Splice-Aware Aligner. Berkeley, CA (United States: Lawrence Berkeley National Lab (LBNL; 2014. [Google Scholar]
- 29.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 30.Aronesty E. Comparison of sequencing utility programs. Open Bioinforma J. 2013;7 doi: 10.2174/1875036201307010001. [DOI] [Google Scholar]
- 31.Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu J, Salzberg S. Ultrafast and accurate 16S microbial community analysis using Kraken 2. bioRxiv. 2020 doi: 10.1186/s40168-020-00900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen T, Guestrin C. Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining. 2016. Xgboost: A scalable tree boosting system; pp. 785–794. [Google Scholar]
- 34.Bergstra J, Bengio Y. Random search for hyper-parameter optimization. J Mach Learn Res. 2012;13:281–305. [Google Scholar]
- 35.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, et al. Scikit-learn: Machine learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 36.Cawley GC, Talbot NLC. On over-fitting in model selection and subsequent selection bias in performance evaluation. J Mach Learn Res. 2010;11:2079–2107. [Google Scholar]
- 37.Saito T, Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One. 2015;10:e0118432. doi: 10.1371/journal.pone.0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lundberg SM, Erion G, Chen H, DeGrave A, Prutkin JM, et al. From local explanations to global understanding with explainable AI for trees. Nat Mach Intell. 2020;2:56–67. doi: 10.1038/s42256-019-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaw LP, Wang AD, Dylus D, Meier M, Pogacnik G, et al. The phylogenetic range of bacterial and viral pathogens of vertebrates. Mol Ecol. 2020:n/a(n/a. doi: 10.1111/mec.15463. [DOI] [PubMed] [Google Scholar]
- 40.Shaw L. The phylogenetic range of bacterial and viral pathogens of vertebrates: dataset and supplementary material [Internet] 2020. https://figshare.com/articles/dataset/The_phylogenetic_range_of_bacterial_and_viral_pathogens_of_vertebrates_dataset_and_supplementary_material/8262779 [DOI] [PubMed]
- 41.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8:e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurtz ZD, Müller CL, Miraldi ER, Littman DR, Blaser MJ, et al. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput Biol. 2015;11:e1004226. doi: 10.1371/journal.pcbi.1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal, complex Syst. 2006;1695:1–9. [Google Scholar]
- 45.Chen T, Yu W-H, Izard J, Baranova O, Lakshmanan A, et al. The Human Oral microbiome database: A web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010;2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pytko-Polonczyk J, Konturek SJ, Karczewska E, Bielański W, Kaczmarczyk-Stachowska A. Oral cavity as permanent reservoir of Helicobacter pylori and potential source of reinfection. J Physiol Pharmacol an Off J Polish Physiol Soc. 1996;47:121–129. [PubMed] [Google Scholar]
- 47.Periasamy S, Kolenbrander PE. Mutualistic biofilm communities develop with Porphyromonas gingivalis and initial, early, and late colonizers of enamel. J Bacteriol. 2009;191:6804–6811. doi: 10.1128/JB.01006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cephas KD, Kim J, Mathai RA, Barry KA, Dowd SE, et al. Comparative analysis of salivary bacterial microbiome diversity in edentulous infants and their mothers or primary care givers using pyrosequencing. PLoS One. 2011;6:e23503. doi: 10.1371/journal.pone.0023503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen H, Jiang W. Application of high-throughput sequencing in understanding human oral microbiome related with health and disease. Front Microbiol. 2014;5:508. doi: 10.3389/fmicb.2014.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saiman L, Chen Y, San Gabriel P, Knirsch C. Synergistic activities of macrolide antibiotics against Pseudomonas aeruginosa, Burkholderia cepacia, Stenotrophomonas maltophilia, and Alcaligenes xylosoxidans isolated from patients with cystic fibrosis. Antimicrob Agents Chemother. 2002;46:1105–1107. doi: 10.1128/AAC.46.4.1105-1107.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sánchez MU, Bello HT, Domínguez MY, Mella SM, Zemelman RZ, et al. Transference of extended-spectrum beta-lactamases from nosocomial strains of Klebsiella pneumoniae to other species of Enterobacteriaceae . Rev Med Chil. 2006;134:415–420. doi: 10.4067/s0034-98872006000400002. [DOI] [PubMed] [Google Scholar]
- 52.Valm AM, Welch JLM, Rieken CW, Hasegawa Y, Sogin ML, et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc Natl Acad Sci U S A. 2011;108:4152–4157. doi: 10.1073/pnas.1101134108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar PS. From focal sepsis to periodontal medicine: a century of exploring the role of the oral microbiome in systemic disease. J Physiol. 2017;595:465–476. doi: 10.1113/JP272427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar PS. Oral microbiota and systemic disease. Anaerobe. 2013;24:90–93. doi: 10.1016/j.anaerobe.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 55.Tan X, Liu H, Long J, Jiang Z, Luo Y, et al. Septic patients in the intensive care unit present different nasal microbiotas. Future Microbiol. 2019;14:383–395. doi: 10.2217/fmb-2018-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haak BW, Prescott HC, Wiersinga WJ. Therapeutic potential of the gut microbiota in the prevention and treatment of sepsis. Front Immunol. 2018;9:2042. doi: 10.3389/fimmu.2018.02042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bharucha T, Oeser C, Balloux F, Brown JR, Carbo EC, et al. STROBE-metagenomics: a STROBE extension statement to guide the reporting of metagenomics studies. Lancet Infect Dis. 2020;20:e251–e260. doi: 10.1016/S1473-3099(20)30199-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Casadevall A, Pirofski L. Host-pathogen interactions: basic concepts of microbial commensalism, colonization, infection, and disease. Infect Immun. 2000;68:6511–6518. doi: 10.1128/IAI.68.12.6511-6518.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balloux F, van Dorp L. Q&A: What are pathogens, and what have they done to and for us? BMC Biol. 2017;15:1–6. doi: 10.1186/s12915-017-0433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jervis-Bardy J, Leong LEX, Marri S, Smith RJ, Choo JM, et al. Deriving accurate microbiota profiles from human samples with low bacterial content through post-sequencing processing of Illumina MiSeq data. Microbiome. 2015;3:19. doi: 10.1186/s40168-015-0083-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grumaz S, Hoffmann A, Vainshtein Y, Kopp M, Grumaz S, et al. Rapid next-generation sequencing-based diagnostics of bacteremia in septic patients. J Mol Diagn. 2020;22:405–418. doi: 10.1016/j.jmoldx.2019.12.006. [DOI] [PubMed] [Google Scholar]
- 63.Sanderson ND, Street TL, Foster D, Swann J, Atkins BL, et al. Real-time analysis of nanopore-based metagenomic sequencing from infected orthopaedic devices. BMC Genomics. 2018;19:714. doi: 10.1186/s12864-018-5094-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salipante SJ, Sengupta DJ, Rosenthal C, Costa G, Spangler J, et al. Rapid 16S rRNA next-generation sequencing of polymicrobial clinical samples for diagnosis of complex bacterial infections. PLoS ONE. 2013;8:e65226. doi: 10.1371/journal.pone.0065226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brenner T, Decker SO, Grumaz S, Stevens P, Bruckner T, et al. Next-generation sequencing diagnostics of bacteremia in sepsis (Next GeneSiS-Trial): study protocol of a prospective, observational, noninterventional, multicenter, clinical trial. Medicine (Baltimore) 2018;97:e9868. doi: 10.1097/MD.0000000000009868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morfopoulou S, Plagnol V. Bayesian mixture analysis for metagenomic community profiling. Bioinformatics. 2015;31:2930–2938. doi: 10.1093/bioinformatics/btv317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McIntyre ABR, Ounit R, Afshinnekoo E, Prill RJ, Hénaff E, et al. Comprehensive benchmarking and ensemble approaches for metagenomic classifiers. Genome Biol. 2017;18:182. doi: 10.1186/s13059-017-1299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simon HY, Siddle KJ, Park DJ, Sabeti PC. Benchmarking metagenomics tools for taxonomic classification. Cell. 2019;178:779–794. doi: 10.1016/j.cell.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Faust K, Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol. 2012;10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 70.Ponomarova O, Patil KR. Metabolic interactions in microbial communities: untangling the Gordian knot. Curr Opin Microbiol. 2015;27:37–44. doi: 10.1016/j.mib.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 71.Freilich S, Zarecki R, Eilam O, Segal ES, Henry CS, et al. Competitive and cooperative metabolic interactions in bacterial communities. Nat Commun. 2011;2:1–7. doi: 10.1038/ncomms1597. [DOI] [PubMed] [Google Scholar]
- 72.Kumar M, Ji B, Zengler K, Nielsen J. Modelling approaches for studying the microbiome. Nat Microbiol. 2019;4:1253–1267. doi: 10.1038/s41564-019-0491-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.