Abstract
Purpose of review:
High density lipoproteins (HDL) are a heterogeneous family of particles that contain distinct complements of proteins that define their function. Thus, it is important to accurately and sensitively identify proteins associated with HDL. Here we highlight the HDL Proteome Watch Database which tracks proteomics studies from different laboratories across the world.
Recent findings:
In 45 published reports, almost 1000 individual proteins have been detected in preparations of HDL. Of these, 251 have been identified in at least three different laboratories. The known functions of these consensus HDL proteins go well beyond traditionally recognized roles in lipid transport with many proteins pointing to HDL functions in innate immunity, inflammation, cell adhesion, hemostasis and protease regulation, and even vitamin and metal binding.
Summary:
The HDL proteome derived across multiple studies using various methodologies provides confidence in protein identifications that can offer interesting new insights into HDL function. We also point out significant issues that will require additional study going forward.
Keywords: High density lipoproteins, Subspecies, Proteome, Cardiovascular disease, Mass spectrometry, Immunity, Hemostasis, Lipid transport
1. Introduction
The concentration of high-density lipoprotein cholesterol (HDL-C) in plasma has long been associated with protection from cardiovascular disease (CVD), particularly in large scale human studies [1]. The most widely accepted basis for this was HDL’s ability to mediate the process of reverse cholesterol transport, i.e. the removal of excess peripheral cell cholesterol and return to the liver for catabolism [2]. However, HDL has also been strongly associated with other cardioprotective effects such as prevention of lipoprotein oxidation and anti-inflammatory functions (recently reviewed here [3,4]). Despite this, across-the-board elevation of HDL-C levels by pharmacologic or genetic means does not appear to reduce CVD risk [5]. At the same time, awareness is rising that HDL is not a single entity [6]. The particles in this family are heterogeneous, varying dramatically in size, charge, density and composition. Beyond the common dominant proteins apolipoprotein A-I (APOA1) and A-II (APOA2), proteomic studies have identified hundreds of individual proteins and upwards of 200 lipid species [7] in preparations of human HDL. While many HDL proteins are known to be highly exchangeable, careful fractionation studies show that some of these proteins are disproportionally distributed across different HDL subspecies that remain stably distinct in the circulation (for a more in-depth discussion of HDL subspeciation see [6]). With this recognition, efforts are being made to track specific HDL subspecies, identified by their unique protein constituents, and their relationships to disease [8]. These studies highlight the importance of accurately and sensitively identifying HDL particle composition. In this review, we will focus on advancement in understanding the HDL proteome since our last review on this topic in 2013 [9]. Other recent reviews have done an excellent job of highlighting instrumentation, quantitation and statistical issues with understanding the HDL proteome [10]. Thus, we will primarily focus on insights into HDL function that can be gleaned from assessing multiple proteomic studies from a variety of different laboratories and techniques.
2. The HDL Proteome Watch Database
As of this writing, we have tracked 45 published reports of shotgun proteomics studies of HDL [11–52]. The identifications vary widely with one laboratory identifying only 13 proteins while another found 487. The reasons for this variation include the resolution of the pre-MS peptide separation techniques, sensitivity of the MS equipment as technology advances as well as choices made during data analysis. Typically, these studies are subject to at least two major sources of error in HDL protein identification. First, each laboratory tends to find distinct contaminants that show up in their samples. These likely come from sample carry over between MS runs in which small amounts of peptides stay associated with tubing or columns during chromatographic separations. For example, our instrumentation at the University of Cincinnati tends to report trace amounts of human titin, one of the largest translated proteins known, in some of our samples. This typically does not affect high scoring, abundant proteins but does increase the likelihood that low abundance contaminants can be assigned to HDL erroneously within a given analysis. These contaminants tend to be characteristic of individual labs depending on the types of samples they analyze routinely.
Second, the quality of the HDL starting material contributes significantly to the final results. HDL is a good example of the observer effect problem in science. One can liken the problem of accurately characterizing the circulating HDL proteome to counting different varieties of butterflies contained in an opaque box. An accurate accounting requires opening the box. But how this is done can profoundly affect the results depending on how many butterflies escape before they are counted. Similarly, one has to isolate HDL from plasma before characterizing the proteome. The choice of isolation method can have significant effects on the results. The most common method of isolating HDL is by salt density ultracentrifugation between 1.063 and 1.210 g/ml [53,54]. This method provides the namesake for the lipoprotein class, and in a technical sense is the only method whose product can truly be called ‘HDL’. However, repeated centrifugation can systematically reduce the APOA1 content of HDL particles [55]. Furthermore, Munroe and colleagues demonstrated that the rotor speed (i.e. centrifugal force) can have significant impacts on the protein composition and final density of isolated HDL particles [56]. In a head to head comparison, KBr-based methods for HDL isolation resulted in fewer stainable proteins by 2-D gel electrophoresis compared to non-ionic density gradient methods using D2O/sucrose [57], suggesting that high ionic strength may affect final HDL composition. In addition, in our experience, the degree of separation of the HDL density band from the >1.21 g/ml “bottom” fraction - which contains all non-lipoprotein blood components - can vary between laboratories. Variables such length of the centrifugation, geometry of the tubes and rotors used, method for isolating HDL density bands, and a host of other factors can contribute significantly to the ultimate proteome measured in the final preparations. In addition, recent studies have cautioned that KBr-based density ultracentrifugation approaches can co-isolate extracellular vesicles (EVs) along with HDL [58].
Other laboratories have taken more gentle approaches such as gel filtration chromatography of plasma [59,60]. These separations are usually treated to a second purification method to eliminate similarly sized plasma species that are not lipid associated, i.e. the use of calcium silica hydrate (CSH) resins to isolate phospholipid containing lipoproteins from each gel filtration fraction [17]. This method can isolate phospholipid-rich lipoprotein particles that elute in the same size range as UC-isolated HDL. But, as the method is insensitive to particle density, these isolates are technically not considered ‘HDL’. Nevertheless, size-based definitions of HDL-like lipoproteins have been widely used clinically with the advent of nuclear magnetic resonance [61] and ion mobility analysis [62]. While the gel filtration method is less likely to lose proteins due to high ionic strength or g-forces, it is susceptible to non-lipoprotein contaminants, particularly those that non-specifically bind the CSH resins [17]. The method also dilutes the lipoprotein samples substantially which could be envisioned to affect the distribution of proteins that may be in equilibrium between particle-bound and soluble states.
Immunoaffinity methods of HDL isolation suffer from two drawbacks. First, by their very nature, they introduce a bias inherent to the antibody that is being used to pull down the lipoproteins. Anti-APOA1 antibodies are most commonly employed [63,64,22], which in theory should capture >95% of lipidated particles in the HDL size and density range, but non-APOA1 containing species have been reported [65]. Indeed, mice [66] and humans [67] lacking APOA1 still contain some phospholipid or cholesterol that isolates at HDL densities. Additionally, antibody specificity for certain conformations of the target protein may fail to capture other conformations, a particular worry with highly conformationally dynamic proteins like APOA1. Immunoaffinity methods also tend to be susceptible to contaminants that non-specifically bind either the antibody or its immobilization matrix. Multidimensional electrophoresis separations of plasma may suffer from stripping of proteins from the lipid during the separation and contaminations from comigrating non-lipidated contaminants. We note the benefits and limitations of the various methods used to isolate HDL (or HDL-like particles) above not to favor or disparage any given technique, but to point out that there is no one perfect method for isolating phospholipid-rich lipoprotein particles in plasma. It follows that it is not wise to rely on any one study or any one method of particle isolation with respect to a complete understanding of the HDL proteome. Recognizing this, we started the HDL Proteome Watch Database in 2011. It can be found at (https://homepages.uc.edu/~davidswm/HDLproteome.html).
Our objective in creating the database was simple. We sought to accumulate proteomics data from individual laboratories and present them for comparison with standardized protein identification nomenclature. We also carefully tracked major factors that impact the results such as method of HDL isolation, instrumentation used, any pre-MS treatments or separations, as well as the peptide identification algorithms used. We surveyed the literature and included proteomic studies that met the following criteria: 1) The study was designed with a non-biased approach to identifying the totality of HDL proteins (sometimes referred to as a shotgun approach). Targeted studies that reported only specific proteins (usually done for quantitation purposes), though scientifically valid for other purposes, were excluded from the database to avoid biasing the protein hit counts across the studies. 2) Some attempt must have been made to isolate the lipoproteins from non-lipidated plasma proteins. By far, the most common method for HDL isolation was density ultracentrifugation (30 of the 45 studies). But we also included those that isolated HDL sized species by gel filtration chromatography followed by pull down of lipid containing particles by lipid affinity resins (7 studies). Other included studies performed 1-D or 2-D gel electrophoresis (2 studies) or immunoprecipitation with antibodies to APOA1 (6 studies). The idea was to cast as wide of a net as possible to factor out biases inherent to any given isolation method. 3) The protein identification data needed to be publicly available either in the paper itself, a supplement or via response to an inquiry. Several published studies were not included in the database due to our inability to obtain the original protein identification data from the authors.
As of this writing, the database covers 45 qualifying proteomics studies from 2005 to June 2021. The database is formatted as an Excel spreadsheet with consensus protein names listed vertically and the published studies listed horizontally in chronological order (Fig. 1). Each laboratory is assigned a letter code. If a protein was identified in a given study, the laboratory code is entered in the appropriate cell. The UniProtKB consensus protein names are cross-referenced with as many alternate names as could be found along with the protein ID (ex. APOA1_HUMAN) and accession number (ex. P02647) and the molecular weight of the intact protein. The accession numbers are hyperlinked to the appropriate UniProtKB entry page so that proteins of interest can be quickly researched. Each published report is also hyperlinked to the relevant PubMed entry for facile access to the study and its details. The database also tracks the location of the laboratory where the mass spectrometry was performed, the patient population and any disease states used to obtain the HDL samples, the type of HDL analyzed (Total HDL, vs. HDL3 for example), the method used for HDL isolation, any pre-MS separation strategies such as electrophoresis, the type of MS instrumentation used, and finally the database used for protein identifications.
Fig. 1.
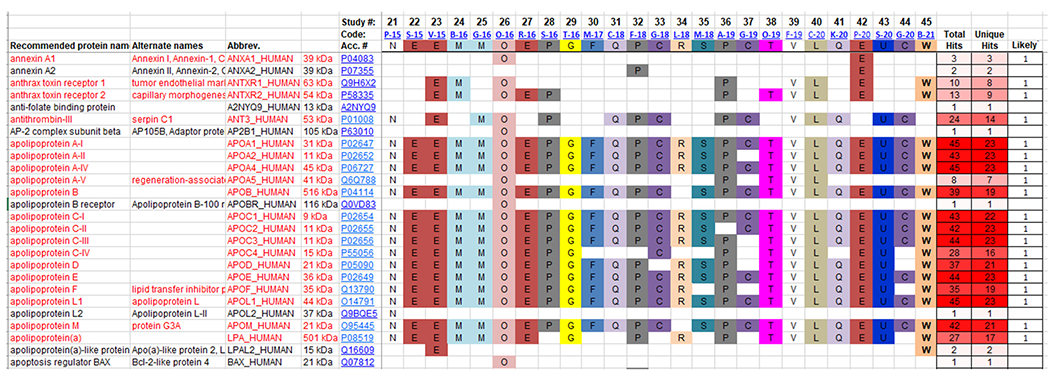
Screenshot of a portion of the HDL Proteome Watch Database. The database is in the form of an Excel spreadsheet with all proteins identified in any covered study listed on the left with alternate names, standard accession identifiers and UniProtKB codes. All cataloged studies are listed across the top (latest 24 studies are shown here) with a study code (ex. P-15) that is hyperlinked to the PubMed entry for the relevant published paper. The study code is comprised of the first letter of the first author’s last name and the year in which the study was published. Each laboratory is assigned a colored letter code. If a given protein appeared in a given study, the laboratory letter code appears in the appropriate column and row. For example, in the above shown portion of the database, annexin A1 was identified in 2 laboratories (Code O; Greifswald, Germany and Code E; University of Washington, US). The number of times a given protein was identified is shown on the right with “unique hits” (the number of times that protein was seen in different laboratories). The final righthand column contains a 1 if the protein met our criterion of appearing in at least three independent studies from three different laboratories. Currently “likely” HDL proteins are listed in red text.
3. The power of combining studies across labs
Across all tracked studies, 936 proteins have been reported to be found in HDL preparations from 35 laboratories. We adopted the convention that a “likely” HDL protein had to have been identified in at least three independent studies from three different laboratories; 251 proteins currently satisfy this criterion. They are listed in Fig. 2 along with the number of studies in which they were identified. This cutoff is admittedly arbitrary, but this low bar was chosen as a way to factor out laboratory-specific contaminants to the extent possible, while still capturing low-detection proteins that may have true functional roles in minor HDL subspecies. Our philosophy was to err on the side of inclusion rather than risk factoring out proteins that may truly be present and have potentially important functions. As such, the database includes proteins that some investigators dismiss as contaminants. For example, numerous human skin keratins have appeared in multiple studies. These probably result from manipulation of the samples and are most likely not true HDL proteins. But we report them nevertheless. The list of “likely” HDL constituents contains entries that one might find surprising. For example, anthrax toxin receptors 1 and 2, L-selectin and talin-1 are transmembrane proteins that one would expect to be associated with cells. Profilin-1 is an intracellular protein that interacts with the cytoskeleton [68]. Each of these proteins was identified in more studies than apolipoprotein A-V, an extremely low abundance protein that was clearly shown to be HDL associated by immunoblot techniques [69]. Why would these proteins (or parts of them) be found in HDL? The answers are not yet clear, but we feel it is important to report the dataset without bias or preconceptions as to whether the protein “should” be in HDL. This ensures that individual users have the most information available for any future analyses. We also implore anyone considering publishing a proteomic study to resist the temptation to arbitrarily factor out valid hits duly identified by a pre-designed data analysis workflow.
Fig. 2.
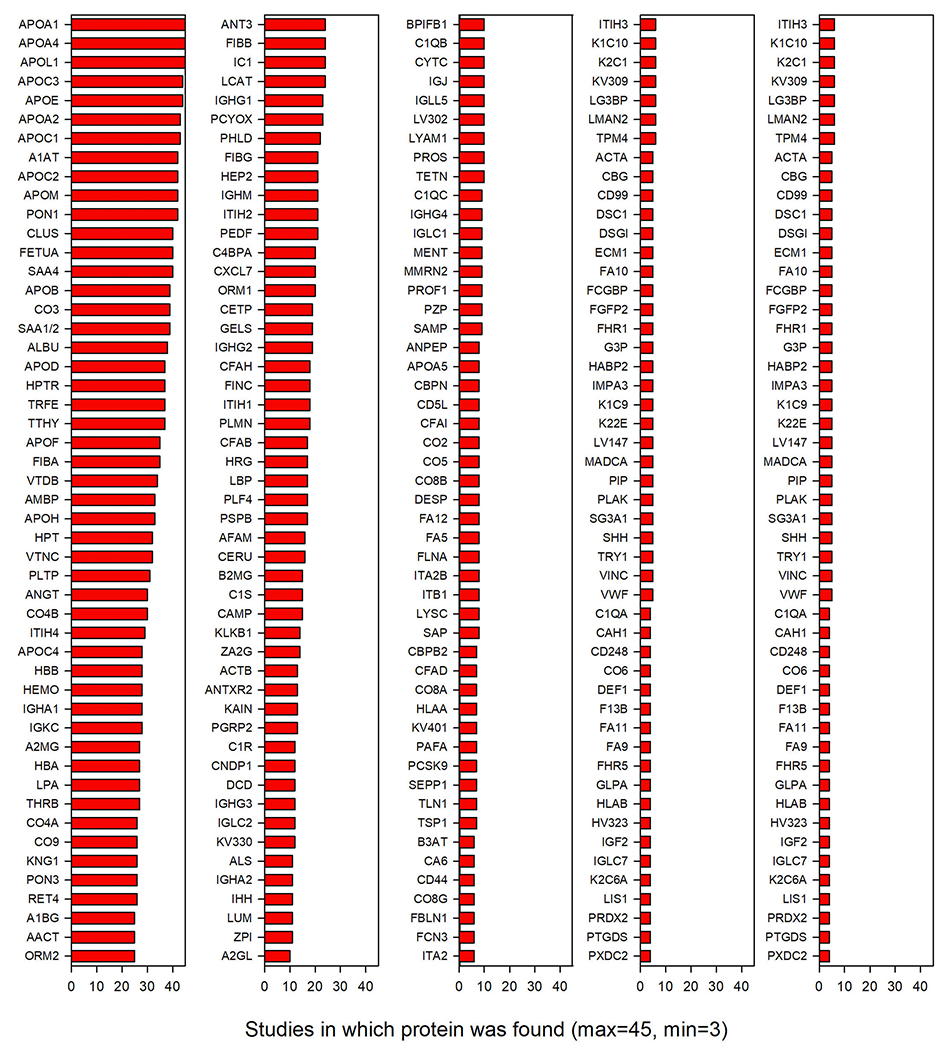
Frequency of observation of 251 “likely” HDL-associated proteins. The abbreviated protein name is shown on the left with the bar indicating the number of studies that identified it in HDL. 3 proteins were identified in all 45 of the publications that met our criterial for inclusion. Proteins with less than three identifications from three different labs are likely contaminants.
4. Effect of isolation method on proteins identified
To determine how isolation method impacts which proteins are detected on HDL, we analyzed protein detection across the four common particle isolation methods. Of the 251 consensus HDL-associated proteins, 91 have been detected by all HDL isolation methods evaluated (Fig. 3A). Most were detectable in UC isolated HDL (230), while fewer were detected by other techniques (Table 1). In fact, 48 proteins (about 20% of the consensus HDL proteome) are only detected on HDL isolated by UC. This may indicate that, while UC is often considered the most physically harsh isolation technique, many protein interactions are maintained during this process. High salt conditions have been suggested to promote protein-lipid interaction for some proteins. Therefore, it cannot be ruled out that these conditions may drive protein-HDL interactions that are not present under physiological salt concentrations. Notably, all proteins detected by gel filtration have also been validated by detection on HDL isolated by different methods. However, a couple of key proteins involved in lipid transport on HDL (CETP and LCAT) have not been detected on HDL isolated by gel filtration, yet have been detected on HDL isolated by all other methods (Supplemental Table 1; underlined). One limitation of our analysis is the overrepresentation of UC studies compared to the other isolation methods (Table 1). The greater number of UC studies likely contributes to the observation that more proteins are detected by this approach. However, examination of the number of proteins reported per study revealed that, although UC had more total protein identifications and a large number of unique proteins, the average number of proteins detected in any single study was similar to that from the other isolation techniques (Fig. 3B). This would suggest that the dramatically greater detection and perhaps some of the unique ID’s could be due to a cumulative effect and not a systematic methodological effect.
Fig. 3.
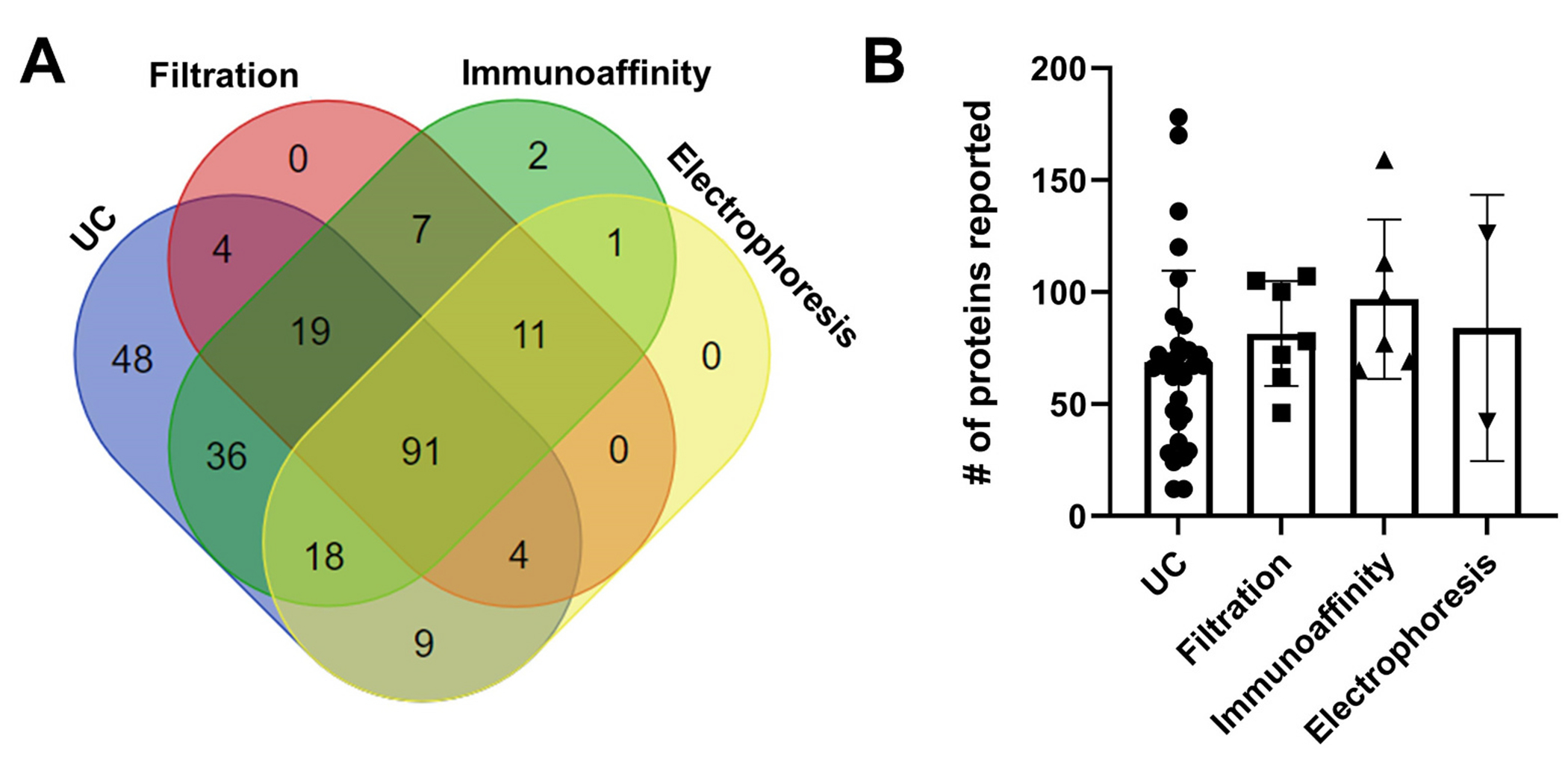
Detection of HDL-associated proteins by different isolation methods. (A) The 251 “likely” HDL-associated proteins from 45 studies were analyzed to determine if HDL isolation method has an impact on which proteins are detected. This Venn diagram displays the counts of proteins detected by each method and the overlap in detection across methods. Only 91 proteins were detected by all isolation methods and 48 were detected only in studies where HDL was isolated by ultracentrifugation (UC). The protein identities within each cluster are listed in Supplementary Table 1. (B) Examination of the total number of identified proteins reported by each of the 45 studies, grouped by HDL isolation method, reveals that roughly the same number of proteins is detected on average regardless of isolation method. Statistical comparisons were not attempted due to large differences in sample size across groups.
Table 1.
Count of HDL proteins detected by different isolation methods.
| Isolation method | Number of studies | Total proteins | Unique proteins |
|---|---|---|---|
| UC | 30 | 230 | 48 |
| Filtration | 7 | 137 | 0 |
| Immunoaffinity | 6 | 186 | 2 |
| Electrophoresis | 2 | 134 | 0 |
| Combined total | 45 | 251 | - |
5. Functional analysis of the HDL proteome
We performed a gene ontology keyword enrichment analysis on the 251 consensus HDL proteins. Five of the most populated clusters are shown in Fig. 4. Clearly, many of the consensus proteins are associated with lipid transport including many of the classic “apo” proteins known to be components of HDL for half a century. Similarly, many proteins have clear roles in inflammation and acute phase response including the serum amyloid A isoforms 1 and 2 and APOA4. Continuing trends noted previously, increasing numbers of HDL proteins are involved in immunity and anti-bacterial functions. This includes the growing list of immunoglobulins consistently detected in HDL as well as proteins in the complement pathway (see insets in Fig. 4). Many recent additions to the list appear to be involved in hemostasis and protease inhibition as well as cell and heparin binding. Not shown in Fig. 4 are clusters with smaller numbers of proteins. This includes proteins such as afamin, vitamin D binding protein and retinol binding protein that appear to be involved in vitamin binding and transport. There are also several proteins like ceruloplasmin, hemopexin, serotransferrin and His-rich glycoprotein that are associated with metal ion binding. There are fewer numbers of proteins associating with these functional groups, but judging from their consistent appearance in many independent studies, they are probably present in HDL in relatively high abundance.
Fig. 4.
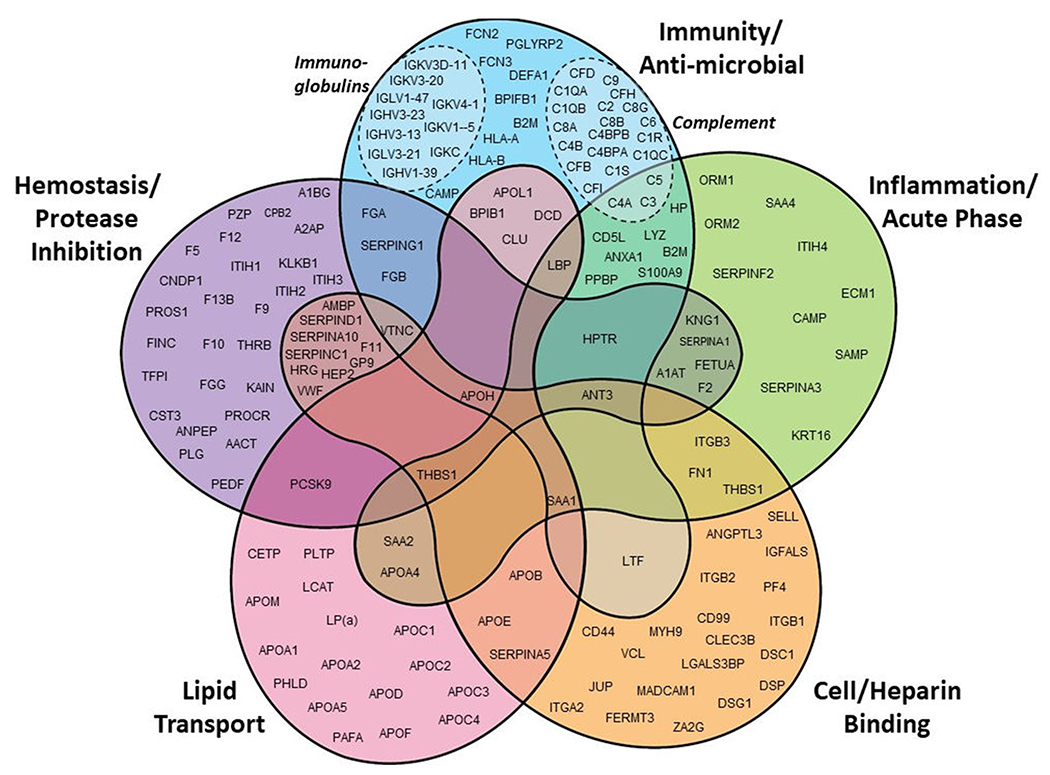
Gene Ontology (GO) keyword functional analysis of “likely” HDL proteins. Each protein was assigned to a rough functional classification based on keyword associations using the DAVID Bioinformatics Resource (DAVID.ncifcrf.gov). This is not a complete listing of all likely proteins as some were associated with functions that fell outside these major groupings.
The inescapable conclusion from Fig. 4 is that HDL is designed for much more than lipid transport. The evidence indicates that HDL serves as an extracellular platform that hosts proteins that mediate a variety of functions in biology and pathology. One might liken this situation as a lower complexity analog of the plasma membrane (PM) of a cell. In addition to its role as a barrier, the PM also serves as a platform that limits diffusion of transmembrane proteins to two dimensions, dramatically increasing the chances for complex formation and synergistic activity. HDL may serve this role for secreted plasma proteins by acting as a nexus for amphipathic proteins that need each other to execute their biological functions. Indeed, a remarkable example of this HDL protein synergy is seen in the HDL subfraction/subspecies called trypanosome lytic factor (TLF, see below). Similar arguments can be made for the various proteases and protease inhibitors involved in hemostasis that have been localized to HDL [70]. The proximity of these factors may facilitate both the activation and resolution of the various proteolytic events that occur during the clotting cascade.
Given the large number of proteins identified in HDL, it is impractical to review the known roles of each protein. Information is sparse as to how some of these lower abundance proteins affect HDL function. However, the field is transitioning from what we would call a lipoprotein-centric, or holo-particle view of HDL, to a more granular protein-specific way of looking at HDL function. HDL used to be thought of in terms of a singular component – its cholesterol content – as a marker for the totality of its beneficial functions. However, interventional clinical trials have demonstrated that small molecule therapies which raise HDL cholesterol across the board failed to deliver the health benefits suggested by epidemiological studies in large populations (recently reviewed here [5]). Since most HDL particles contain some amount of cholesterol, this metric is insufficient for tracking the functionality of minor compositional subspecies that may perform varied roles in different physiological processes. Fortunately, laboratories are beginning to explore compositionally defined subspecies, i.e. HDL sub-particles that are distinguished by the presence of a minor protein components identified in preparations of total HDL. Below, we highlight some examples of advances that have focused on the roles of proteomic distinguishers of HDL subspecies that appeared in the last 3–5 years for each functional category listed in Fig. 4.
6. Examples of roles of specific minor HDL proteins
6.1. Lipid transport
Most of the classic “apo”lipoproteins listed in Fig. 4 are known to play important roles in the structure, remodeling or targeting of HDL particles in the circulation. APOA1 and APOA2, are the key structural scaffold proteins present on almost all HDL particles (APOA1) or about 60% of them (APOA2). APOA1 seems to be the major factor that solubilizes phospholipid into HDL size particles. It is critical for interactions with important lipid remodeling factors like lecithin:cholesterol acyl transferase (LCAT), which esterifies cholesterol to cholesteryl ester, or cholesteryl ester transfer protein (CETP), which mediates the exchange of neutral lipid esters between HDL and the apoB-containing lipoproteins. However, recent studies have shown that certain minor apolipoprotein-defined subspecies of HDL may play important roles in lipid transport, specifically the process of reverse cholesterol transport (RCT) whereby cholesterol accumulated in the periphery is returned to the liver for catabolism. Morton et al. [71] tracked the metabolic fate of endogenously labeled proteins on HDL isolated by immunoaffinity chromatography. They found that APOE-containing HDL could expand in size in circulation and be quickly cleared, consistent with a role in RCT. Non-APOE-HDL did not expand and was cleared more slowly. However, when APOC3 was present on APOE-HDL, their expansion was curtailed and particle clearance was inhibited. The authors speculated that APOE-HDL plays important roles in the transport of cholesterol out of the vessel wall and is thus atheroprotective, but the presence of APOC3 on these particles negates that protection. When this was investigated in a large prospective population-based study, they found that APOE-HDL was inversely related to CVD risk, but only the fraction that did not contain APOC3.
6.2. Hemostasis/protease inhibition
HDL has been shown in epidemiological studies to be inversely associated with the risk of thrombosis [72,73]. It can modulate cholesterol levels and signaling pathways in platelets to regulate their association (recently reviewed here [74,75]). It also acts on endothelial cells to downregulate platelet aggregation. Until recently, much of our understanding has been derived from whole-particle HDL studies in which the role of individual components has not been clear. Recently, Xu and colleagues elucidated a direct role of the minor HDL protein APOA4 in the inhibition of thrombosis [76]. They showed that APOA4 bound to the platelet integrin αIIbβ3 competitively inhibits the early steps of the prothrombotic cascade. Interestingly, since APOA4 levels are sensitive to lipid absorption in the gut, it may act to inhibit the transient platelet hyperactivity that occurs postprandially. The authors also identified specific residues in the N-terminal domain of APOA4 responsible for the effect.
6.3. Inflammation/acute phase response
Fig. 4 shows that the HDL proteome is highly enriched in proteases and protease inhibitors. Gordon and Remaley analyzed these and determined that the majority of HDL proteases were serine proteases and most of the inhibitors identified at the time target serine proteases (i.e. SERPINS [70]). SERPINS regulate a variety of proteases involved in inflammation, hemostasis and complement activation [77]. The colocalization of proteases and their inhibitors on HDL strongly suggests co-ordination of activity on these circulating platforms. HDL may also act as a shuttle to get SERPINS into areas of the vasculature they might not reach on their own. For example, the SERPIN alpha-1-antitrypsin (A1AT), an acute phase protein, is a potent inhibitor of proteases secreted by neutrophils and macrophages in the inflamed vessel wall. These proteases, including elastase, can destroy extracellular matrix in the intima and media leading to intraplaque neovascularization and plaque rupture [78]. Since HDL has been shown to transcytose across the endothelium [79], it may act as a conduit for getting A1AT to the site of inflammation to prevent runaway destruction of the underlying extracellular matrix [70]. Indeed, HDL bound A1AT, can inhibit the degradation of smooth muscle cell extracellular matrix to prevent their apoptosis [80]. Gordon et al. also showed that statin treatment in humans raised the levels of A1AT in HDL and that this enrichment reduced inflammatory cytokine production in inflamed macrophages [59].
The Proteome Watch List contains inhibitors of other classes of proteases as well. For example, Cystatin-C (found in 10 studies) is an inhibitor of cysteine proteases. Cystatin-C deficient mice on an APOE deficient background exhibited increased extracellular matrix fragmentation and dilated thoracic and abdominal aortae compared to controls [81]. The cathepsin cysteine protease family members are found in human atherosclerotic lesions [82], suggesting another role by which HDL shuttled protease inhibitors could contribute to vascular protection.
Another interesting complex of HDL proteins that may play a role in inflammation is centered around the phospholipid-transfer protein (PLTP). Cheung and colleagues used immune-affinity chromatography to isolate PLTP complexes from human plasma. They found that PLTP was associated with a lipid-poor complex that contained some 24 proteins (all present on the Watchlist) of which 14 were strongly associated with acute and chronic inflammation [83]. Being able to transfer bacterial lipopolysaccharide (LPS) between lipoproteins, the authors speculated that PLTP plays a protective role in endotoxemia by shuttling LPS to HDL for neutralization. Interestingly, the activity of PLTP seemed to depend on the size of the complex in which it is associated with smaller complexes of about 160 kDa containing active PLTP while larger complexes of ~520 kDa contained an inactivated form. Importantly, these complexes also contained proteins in the complement system as well as those known to play roles in coagulation and thrombosis.
6.4. Immunity/anti-microbial
The complement system plays crucial roles in innate immunity by attacking foreign substances and organisms in the body [84]. Almost half of the known complement proteins appear on the HDL Watchlist, strongly suggesting that HDL acts as a coordinating platform for at least part of the complement cascade. Levels of certain complement factors have been associated with carotid intima-media thickening in subjects with systemic lupus erythematosus [85]. These investigators also found an intriguing relationship between certain complement factors and small HDL particles. Using orthogonal separations of human plasma, we previously identified a co-migration cluster of human complement factors within HDL composed of C1s which is important in the formation of the C1 serine protease complex responsible for activation of downstream targets in the classical complement cascade [19]. This includes complement C4, another member of this cluster. The presence of both these factors on HDL subspecies may facilitate complement activation because the factors do not have to diffuse in three dimensions to find each other.
In addition to complement, some proteins in HDL have been shown to play remarkably specific roles in innate immunity. The classic example is the trypanosome lytic factor (TLF) subspecies of HDL. These particles contain APOA1, haptoglobin-related protein (Hrp) and APOL1. Current understanding [86,87] holds that Hrp mediates TLF binding to a heme receptor in the flagellar pocket of an invading trypanosome. Once internalized, APOL1 finds its way to the plasma membrane and forms an unregulated channel, causing a cascading osmotic pressure imbalance that lyses the cell [88]. HDL plays a crucial role in this pathway by packaging Hrp and APOL1 into a lethal, mobile package that can access the parasite in the circulation.
More recently, Han et al. showed that the tissue origin of HDL production as well as the presence of particular protein constituents may play a critical role in preventing liver injury from toxic lipopolysaccharide derived from the gut [89]. They showed that enterically derived HDL is primarily shuttled to the liver via the portal vein prior to joining the bulk of circulating HDL. Certain small HDL3 species were found to be enriched in LPS binding protein (LBP), a commonly identified protein on the HDL Watchlist. These particles were capable of binding free LPS, possibly derived from dietary intake, and masking it from initiating inflammatory reactions in liver Kupfer cells. The authors suggested that these particles may neutralize other microbial antagonists such as lipoteichoic acid among others. Thus, particular proteomically defined HDL subspecies produced by the gut play a direct role in liver injury from dietary derived microbes. This is further evidence of the diverse immune and anti-inflammatory roles played by various HDL subspecies.
6.5. Cell/heparin binding
HDL contains many protein components that are capable of directly binding cells, extracellular matrix, cell receptors/transporters, or cell surface proteoglycans. A well-studied example is APOE which contains both a heparin binding site and a receptor binding site for the LDL receptor [90]. Numerous other proteins in the HDL Watchlist have documented abilities to bind heparin. HDL can also participate in cell signaling pathways that result in the down regulation of cell adhesion molecules to attenuate the association of macrophages and neutrophils [91]. With regard to specific proteome components, recent work has shown that the HDL component vitronectin plays an important role in stabilization of neutrophils to damaged areas of the endothelium and eventually cross to the perivascular space [92]. Vitronectin interacts with plasminogen activator inhibitor-1 and low-density lipoprotein receptor-related protein-1 on the neutrophil surface. This complex is proposed to trigger a signaling cascade that localizes integrins on the cell surface that facilitate binding to the endothelium. The role of HDL-bound vitronectin in this process has not been fully elucidated, but one possibility is that it acts as a sink for vitronectin to prevent inappropriate neutrophil localization.
Shingosine-1-phosphate (S1P) is a bioactive sphingolipid that is a ligand for a family of receptors that mediate a variety of cellular processes. In endothelial cells, one S1P signaling pathway controls cellular nitric oxide production to control vasodilation [93]. This lipid is sequestered in HDL subspecies that contain apolipoprotein M (APOM), a minor apolipoprotein known to bind S1P [94]. ApoM is a member of an interesting set of HDL associated proteins (including paraoxonase 1 and haptoglobin-related protein) that can interact with lipoproteins through a retained signal peptide [95]. ApoM-containing HDL particles were shown to produce an anti-inflammatory response in human primary endothelial cells including limiting the expression of certain cell adhesion molecules. Importantly, S1P delivered in other ways, such as complexed to albumin, was much less efficient in modulatiing cell adhesion molecule expression [93]. There is also a developed literature indicating that S1P/apoM subspecies play a key role in liver inflammation/regeneration (reviewed here [96]).
7. Progress in relating HDL proteome members to cardiovascular disease
While accurately characterizing the HDL lipoproteome is interesting and important for understanding the biology of HDL, one would ultimately like to exploit a subset of proteome members as biomarkers or even causative agents in HDL’s protective role in disease. Given the established inverse associations between HDL-C and HDL cholesterol efflux capacity and CVD, several groups have attempted to relate levels of HDL proteome members to the incidence of CVD. The Proteome Watchlist contains 9 studies where the HDL proteome was evaluated across a spectrum of CVD conditions ranging from hypercholesterolemia to CHF. Six of these studies included a healthy control group and were considered below with the goal to link specific HDL proteins to CVD.
A total of 927 HDL associated proteins were identified in the six CVD studies. Twenty-six proteins were found in all six studies. While all protein abundance differences were based on MS data, there were wide differences with respect to the protein quantification methods used (peptide counting vs. other label-free methods) and statistical rigor with respect to multiple comparisons. There were also differences in HDL isolation method across these studies which, as discussed above, can impact HDL composition or protein detection by MS. Accepting the data as is, there were 109 proteins found to differ in abundance between healthy controls and CVD subjects across all studies. Fig. 5 shows these proteins with an arrow to indicate if their levels went up or down in CVD patients vs. healthy controls. Most of the 109 proteins were observed only in a single study. 13 proteins were found to move in the same direction in 2 of the studies. Two (APOA4 and APOC4) were found to change in the same direction in 3 studies. In 4/6 studies, complement C3 [52,97,98] and serum amyloid A [97,52,46] were found to be enriched in HDL in individuals with CVD compared to controls. Interestingly, APOA4 was statistically different in four studies, but two reported higher [98,52] and three reported lower concentrations [97,46]. Therefore, while each of these 6 studies alone found multiple proteins that exhibited different abundances between control and diseased individuals, only a handful reproduced between studies.
Fig. 5.
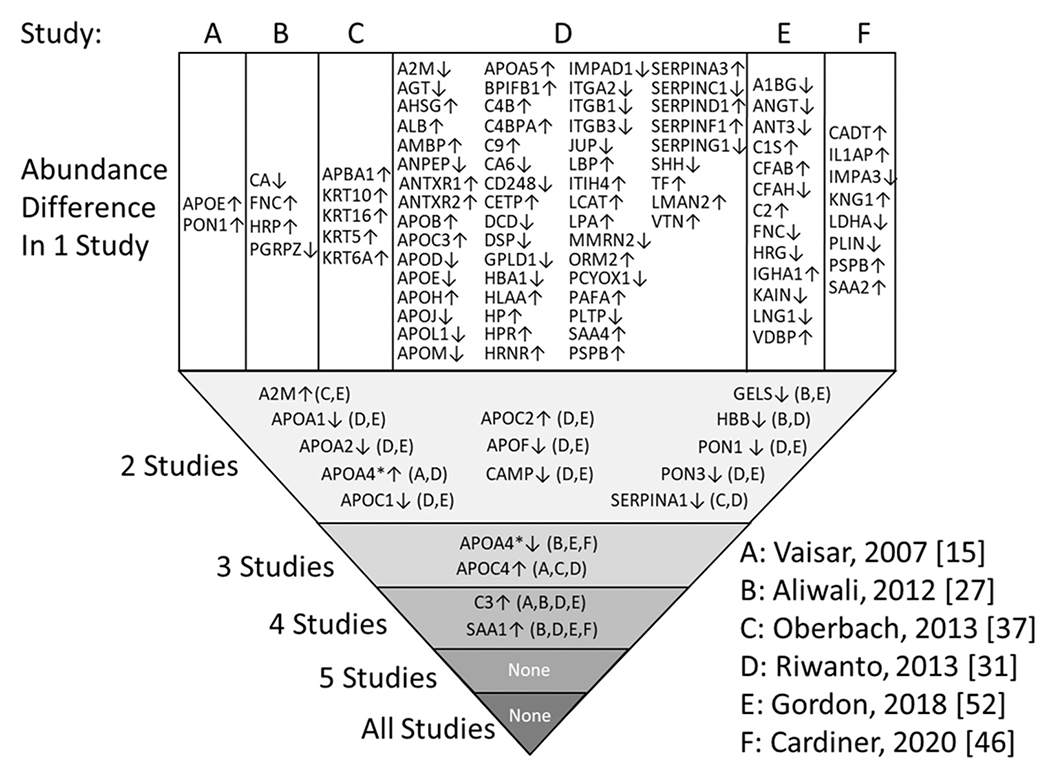
Protein abundance differences between normal and CVD patients across 6 studies. For each of the six studies, A-F, proteins found to have changed abundance by MS quantitation are shown. The arrow indicates the direction protein abundance differed between controls and CVD subjects. Protein differences identified once are at the top, listed for each study. Protein changes that reproduced between studies are shown progressing downward. APOA4 is starred because it reproducibly went down in CVD in some studies, but up in others. Reported proteins that were not considered “likely” HDL proteins as defined here are not listed. Note: No consideration was given for differences in methodology or criteria for indicating a difference in protein abundance between the studies. In some cases, controls were compared to stable CAD and acute coronary syndrome (ACS). In such case, we used the ACS data.
The inability to reproducibly relate levels of specific HDL proteome members to vascular disease states is disappointing. One conclusion, of course, is that HDL and its proteome does not play a major role in development/protection from CVD. However, given the overlapping pathways between HDL and the etiology of CVD in Fig. 4, it seems unlikely that there would be no consistent HDL cargo changes in this disease. Several technical issues previously raised by Vaisar et al. [99] could be responsible. Heterogeneity in the studied diseases could certainly contribute to variability between studies. The nature of the HDL source material and method of isolation likely also contribute as some evaluated total HDL while others used the density subfraction HDL3. Finally, differences in quantitation procedures and lack of universal standards also undoubtedly contribute to inter-laboratory variability in these types of studies. This outlines the clear need for setting universal standards and protocols for lipoprotein proteomics studies. We would also caution that, given the complexity of HDL subspeciation, it may not be enough to simply track the ups and downs of individual proteins with respect to disease states. It is likely that specific subspecies, defined by specific combinations of HDL bound proteins, could be operational factors behind HDL’s varied anti-atherogenic properties. In such case, differential pairings of proteins in normal and diseased physiology may be the key, without major changes in any one component protein level across all of plasma or total HDL.
8. Fragments of proteins vs. intact proteins in HDL
Identifications reported by shotgun MS often require only the detection of two tryptic peptides from a protein. Assuming an average tryptic peptide fragment size of about 8 amino acids, this could represent a small percentage of the whole protein. While this is suitable for high-confidence determination of protein identity, it is possible that reported protein identifications result from peptide fragments of partially degraded proteins. This limitation of shotgun proteomics studiesis often not considered. The reporting of a protein identification in the HDL proteome based on shotgun MS is not evidence that the intact, functionally active protein exists on HDL. This point was made explicitly by Hortin et al. [13] who showed that that HDL can contain dozens of small peptide components ranging from 1000 to 5000 Da representing many of the proteins found in the Watchlist. As the relationships between individual proteins of interest on HDL are examined, analyses of protein integrity should be included. This may be particularly relevant given the abundance of proteolytic enzymes on HDL and the presence of components of the coagulation and complement cascades, which often involve sequential cleavage of precursor proteins to generate active peptide fragments.
9. Conclusions and ways forward
Tremendous strides are being made in sensitively identifying the totality of the HDL proteome. As more studies are added to the Watchlist, and as MS technologies continue to improve, more and more proteins will join the HDL club. Many of these will undoubtedly fall into the major functional categories shown in Fig. 4 while others may surprise and begin to identify even more diverse functions for HDL. At some point, though, we may reach a level of diminishing returns. We suggest that it may be wise to turn our attention to understanding what a particular protein brings to the table when it comes to defining HDL subspecies function. Although we have good ideas about what many of these HDL proteins do in isolation, there is precious little data about what they do in the context of HDL and other proteome members. Many of the proteins in the Watchlist are abundant plasma proteins that undoubtedly exist to a significant extent as soluble proteins in addition to the fraction traveling with HDL. What drives their physical interaction with HDL? Are there common domains or motifs among HDL-interacting plasma proteins that facilitate interaction? Do these proteins have different functional profiles when lipid-free or lipid-bound? Are their functions altered when they share a lipid platform with another interacting protein? These questions will require new and clever ways to isolate and functionally study protein-defined HDL subspecies. This may take the shape of targeted immunoaffinity approaches or chemical cross-linking techniques that capture specific protein:protein interactions and allow for the detailed examination of HDL protein interactome networks. It may be possible to derive antibodies that identify discontinuous epitopes across interacting HDL proteins, or those that can bind specific conformational features characteristic of specific HDL subspecies. Once this hard work is done, it may become possible to more completely relate specific HDL subspecies to specific functions that are relevant to disease progression or protection.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health National Heart, Lung, and Blood Institute, R01HL67093, R01HL155601, P01HL128203, and R01HL153118 to W.S.D., R01HL157260 to A.S.S., and K22HL141299 to S.M.G. This work was also supported by a Harold S. Geneen Charitable Trust Award for Coronary Heart Disease Research to S.M.G. The authors wish to thank Lance Morton for invaluable advice and formatting the database and Diksha Bedi for assistance in tracking down the published proteomics studies.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: W. Sean Davidson reports financial support was provided by National Institutes of Health. Amy Shah reports financial support was provided by National Institutes of Health. Scott Gordon reports financial support was provided by the National Instituted of Health and the Harold S Geneen Charitable Trust.
Abbreviations:
- ABCA1
ATP binding cassette transporter A1
- ALT
advanced lipid testing
- CE
cholesteryl ester
- CETP
cholesteryl ester transfer protein
- CHD
coronary heart disease
- CVD
cardiovascular disease
- HDL
high density lipoprotein
- LDL
low density lipoprotein
- NMR
nuclear magnetic resonance
- RCT
reverse cholesterol transport
- TG
triglyceride
- T2D
Type 2 Diabetes
- TLF
trypanosome lytic factor
- VLDL
very low-density lipoprotein
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbalip.2021.159072.
References
- [1].Kannel WB, Castelli WP, McNamara PM, Serum lipid fractions and risk of coronary heart disease. The Framingham study, Minn. Med 52 (8) (1969) 1225–1230. [PubMed] [Google Scholar]
- [2].Adorni MP, Ronda N, Bernini F, Zimetti F, High density lipoprotein cholesterol efflux capacity and atherosclerosis in cardiovascular disease: pathophysiological aspects and pharmacological perspectives, Cells 10 (3) (2021), 10.3390/cells10030574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Brites F, Martin M, Guillas I, Kontush A, Antioxidative activity of high-density lipoprotein (HDL): mechanistic insights into potential clinical benefit, BBA Clin. 8 (2017) 66–77, 10.1016/j.bbacli.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lemmers RFH, van Hoek M, Lieverse AG, Verhoeven AJM, Sijbrands EJG, Mulder MT, The anti-inflammatory function of high-density lipoprotein in type II diabetes: a systematic review, J. Clin. Lipidol 11 (3) (2017) 712–724, 10.1016/j.jacl.2017.03.013, e5. [DOI] [PubMed] [Google Scholar]
- [5].Barter P, Genest J, HDL cholesterol and ASCVD risk stratification: a debate, Atherosclerosis 283 (2019) 7–12, 10.1016/j.atherosclerosis.2019.01.001. [DOI] [PubMed] [Google Scholar]
- [6].Davidson WS, Cooke AL, Swertfeger DK, Shah AS, The difference between high density lipoprotein subfractions and subspecies: an evolving model in cardiovascular disease and diabetes, Curr. Atheroscler. Rep 23 (6) (2021) 23, 10.1007/s11883-021-00925-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kontush A, Lhomme M, Chapman MJ, Unraveling the complexities of the HDL lipidome, J. Lipid Res 54 (11) (2013) 2950–2963, 10.1194/jlr.R036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sacks FM, Liang L, Furtado JD, Cai T, Davidson WS, He Z, et al. , Protein-defined subspecies of HDLs (High-density Lipoproteins) and differential risk of coronary heart disease in 4 prospective studies, Arterioscler. Thromb. Vasc. Biol 40 (11) (2020) 2714–2727, 10.1161/ATVBAHA.120.314609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shah AS, Tan L, Long JL, Davidson WS, Proteomic diversity of high density lipoproteins: our emerging understanding of its importance in lipid transport and beyond, J. Lipid Res 54 (10) (2013) 2575–2585, 10.1194/jlr.R035725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ronsein GE, Vaisar T, Deepening our understanding of HDL proteome, Expert Rev. Proteomics 16 (9) (2019) 749–760, 10.1080/14789450.2019.1650645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Karlsson H, Leanderson P, Tagesson C, Lindahl M, Lipoproteomics II: mapping of proteins in high-density lipoprotein using two-dimensional gel electrophoresis and mass spectrometry, HDL Compos. 5 (5) (2005) 1431–1445. [DOI] [PubMed] [Google Scholar]
- [12].Heller M, Stalder D, Schlappritzi E, Hayn G, Matter U, Haeberli A, Mass spectrometry-based analytical tools for the molecular protein characterization of human plasma lipoproteins, Proteomics 5 (10) (2005) 2619–2630. [DOI] [PubMed] [Google Scholar]
- [13].Hortin GL, Shen RF, Martin BM, Remaley AT, Diverse range of small peptides associated with high-density lipoprotein 340 (3) (2006) 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Rezaee F, Casetta B, Levels JH, Speijer D, Meijers JC, Proteomic analysis of high-density lipoprotein, Proteomics 6 (2) (2006) 721–730. [DOI] [PubMed] [Google Scholar]
- [15].Vaisar T, Tang C, Babenko I, Hutchins P, Wimberger J, Suffredini AF, et al. Inflammatory remodeling of the HDL proteome impairs cholesterol efflux capacity, J. Lipid Res 56 (8) (2015) 1519–1530, 10.1194/jlr.M059089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Vaisar T, Pennathur S, Green PS, Gharib SA, Hoofnagle AN, Cheung MC, et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL 117 (3) (2007) 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gordon SM, Deng J, Lu LJ, Davidson WS, Proteomic characterization of human plasma high density lipoprotein fractionated by gel filtration chromatography 9 (2010) 5239–5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Davidson WS, Silva RA, Chantepie S, Lagor WR, Chapman MJ, Kontush A, Proteomic analysis of defined HDL subpopulations reveals particle-specific protein clusters: relevance to antioxidative function, Arterioscler. Thromb. Vase. Biol 29 (6) (2009) 870–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gordon SM, Deng J, Tomann AB, Shah AS, Lu LJ, Davidson WS, Multidimensional co-separation analysis reveals protein-protein interactions defining plasma lipoprotein subspecies, Mol. Cell. Proteomics 12 (11) (2013) 3123–3134, 10.1074/mcp.M113.028134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gordon SM, Davidson WS, Urbina EM, Dolan LM, Heink A, Zang H, et al. The effects of type 2 diabetes on lipoprotein composition and arterial stiffness in male youth, Diabetes 62 (8) (2013) 2958–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Melchior JT, Street SE, Andraski AB, Furtado JD, Sacks FM, Shute RL, et al. Apolipoprotein A-II alters the proteome of human lipoproteins and enhances cholesterol efflux from ABCA1, J. Lipid Res 58 (7) (2017) 1374–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Furtado JD, Yamamoto R, Melchior JT, Andraski AB, Gamez-Guerrero M, Mulcahy P, et al. Distinct proteomic signatures in 16 HDL (High-density Lipoprotein) subspecies, Arterioscler. Thromb. Vasc. Biol 38 (12) (2018) 2827–2842, 10.1161/ATVBAHA.118.311607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Holzer M, Birner-Gruenberger R, Stojakovic T, El-Gamal D, Binder V, Wadsack C, et al. Uremia alters HDL composition and function 22 (9) (2011) 1631–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Trieb M, Horvath A, Birner-Gruenberger R, Spindelboeck W, Stadlbauer V, Taschler U, et al. Liver disease alters high-density lipoprotein composition, metabolism and function, Biochim. Biophys. Acta 1861 (7) (2016) 630–638, 10.1016/j.bbalip.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kopecky C, Haidinger M, Birner-Grunberger R, Darnhofer B, Kaltenecker CC, Marsche G, et al. Restoration of renal function does not correct impairment of uremic HDL properties, J. Am. Soc. Nephrol 26 (3) (2015) 565–575, 10.1681/ASN.2013111219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sreckovic I, Birner-Gruenberger R, Obrist B, Stojakovic T, Scharnagl H, Holzer M, et al. Distinct composition of human fetal HDL attenuates its anti-oxidative capacity, Biochim. Biophys. Acta 1831 (4) (2013) 737–746, 10.1016/j.bbalip.2012.12.015. [DOI] [PubMed] [Google Scholar]
- [27].Alwaili K, Bailey D, Awan Z, Bailey SD, Ruel I, Hafiane A, et al. The HDL proteome in acute coronary syndromes shifts to an inflammatory profile 1821 (3) (2012) 405–415. [DOI] [PubMed] [Google Scholar]
- [28].Mange A, Goux A, Badiou S, Patrier L, Canaud B, Maudelonde T, et al. HDL proteome in hemodialysis patients: a quantitative nanoflow liquid chromatography-tandem mass spectrometry approach, PLoSOne. 7 (3) (2012), e34107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Watanabe J, Charles-Schoeman C, Miao Y, Elashoff D, Lee YY, Katselis G, et al. Proteomic profiling following immunoaffinity capture of high-density lipoprotein: association of acute-phase proteins and complement factors with proinflammatory high-density lipoprotein in rheumatoid arthritis, Arthritis Rheum. 64 (6) (2012) 1828–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Weichhart T, Kopecky C, Kubicek M, Haidinger M, Doller D, Katholnig K, Serum amyloid A in uremic HDL promotes inflammation, J. Am. Soc. Nephrol 23 (5) (2012) 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Riwanto M, Rohrer L, Roschitzki B, Besler C, Mocharla P, Mueller M, et al. Altered activation of endothelial anti- and proapoptotic pathways by high-density lipoprotein from patients with coronary artery disease: role of high-density lipoprotein-proteome remodeling, Circulation 127 (8) (2013) 891–904, 10.1161/CIRCULATIONAHA.112.108753. [DOI] [PubMed] [Google Scholar]
- [32].Godzien J, Ciborowski M, Armitage EG, Jorge I, Camafeita E, Burillo E, et al. A single in-vial dual extraction strategy for the simultaneous lipidomics and proteomics analysis of HDL and LDL fractions, J. Proteome Res 15 (6) (2016) 1762–1775, 10.1021/acs.jproteome.5b00898. [DOI] [PubMed] [Google Scholar]
- [33].Burillo E, Jorge I, Martinez-Lopez D, Camafeita E, Blanco-Colio LM, Trevisan-Herraz M, et al. Quantitative HDL proteomics identifies Peroxiredoxin-6 as a biomarker of human abdominal aortic aneurysm, Sci. Rep 6 (2016) 38477, 10.1038/srep38477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Jorge I, Burillo E, Mesa R, Baila-Rueda L, Moreno M, Trevisan-Herraz M, et al. The human HDL proteome displays high inter-individual variability and is altered dynamically in response to angioplasty-induced atheroma plaque rupture, J. Proteome 106 (2014) 61–73, 10.1016/j.jprot.2014.04.010. [DOI] [PubMed] [Google Scholar]
- [35].Pedret A, Catalan U, Fernandez-Castillejo S, Farras M, Valls RM, Rubio L, et al. Impact of virgin olive oil and phenol-enriched virgin olive oils on the HDL proteome in hypercholesterolemic subjects: a double blind, randomized, controlled, cross-over clinical trial (VOHF Study), PLoS One 10 (6) (2015), e0129160, 10.1371/journal.pone.0129160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Shao B, de Boer I, Tang C, Mayer PS, Zelnick L, Afkarian M, et al. A cluster of proteins implicated in kidney disease is increased in high-density lipoprotein isolated from hemodialysis subjects, J. Proteome Res 14 (7) (2015) 2792–2806, 10.1021/acs.jproteome.5b00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Oberbach A, Adams V, Schlichting N, Heinrich M, Kullnick Y, Lehmann S, et al. Proteome profiles of HDL particles of patients with chronic heart failure are associated with immune response and also include bacteria proteins, Clin. Chim. Acta 453 (2016) 114–122, 10.1016/j.cca.2015.12.005. [DOI] [PubMed] [Google Scholar]
- [38].Andraski AB, Singh SA, Lee LH, Higashi H, Smith N, Zhang B, et al. Effects of replacing dietary monounsaturated fat with carbohydrate on HDL (High-density Lipoprotein) protein metabolism and proteome composition in humans, Arterioscler. Thromb. Vase. Biol 39 (11) (2019) 2411–2430, 10.1161/ATVBAHA.119.312889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Singh SA, Andraski AB, Pieper B, Goh W, Mendivil CO, Sacks FM, et al. Multiple apolipoprotein kinetics measured in human HDL by high-resolution/accurate mass parallel reaction monitoring, J. Lipid Res 57 (4) (2016) 714–728, 10.1194/jlr.D061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Collier TS, Jin Z, Topbas C, Bystrom C, Rapid affinity enrichment of human apolipoprotein A-I associated lipoproteins for proteome analysis, J. Proteome Res 17 (3) (2018) 1183–1193, 10.1021/acs.jproteome.7b00816. [DOI] [PubMed] [Google Scholar]
- [41].Lv P, Zhao M, Liu Y, Jin H, Cui W, Fan C, et al. Apolipoprotein C-III in the high-density lipoprotein proteome of cerebral lacunar infarction patients impairs its anti-inflammatory function, Int. J. Mol. Med 41 (1) (2018) 61–68, 10.3892/ijmm.2017.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mathew AV, Li L, Byun J, Guo Y, Michailidis G, Jaiswal M, et al. Therapeutic lifestyle changes improve HDL function by inhibiting myeloperoxidase-mediated oxidation in patients with metabolic syndrome, Diabetes Care 41 (11) (2018) 2431–2437, 10.2337/dc18-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gourgari E, Ma J, Playford MP, Mehta NN, Goldman R, Remaley AT, et al. Proteomic alterations of HDL in youth with type 1 diabetes and their associations with glycemic control: a case-control study, Cardiovasc. Diabetol 18 (1) (2019) 43, 10.1186/s12933-019-0846-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Okada T, Ohama T, Takafuji K, Kanno K, Matsuda H, Sairyo M, et al. Shotgun proteomic analysis reveals proteome alterations in HDL of patients with cholesteryl ester transfer protein deficiency, J Clin Lipidol. 13 (2) (2019) 317–325, 10.1016/j.jacl.2019.01.002. [DOI] [PubMed] [Google Scholar]
- [45].Florens N, Calzada C, Delolme F, Page A, Guebre Egziabher F, Juillard L, et al. Proteomic characterization of high-density lipoprotein particles from non-diabetic hemodialysis patients, Toxins (Basel) 11 (11) (2019), 10.3390/toxins11110671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cardner M, Yalcinkaya M, Goetze S, Luca E, Balaz M, Hunjadi M, Structure-function relationships of HDL in diabetes and coronary heart disease 5 (1) (2020), 10.1172/jci.insight.131491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kheniser KG, Osme A, Kim C, Ilchenko S, Kasumov T, Kashyap SR, Temporal dynamics of high-density lipoprotein proteome in diet-controlled subjects with type 2 diabetes, Biomolecules 10 (4) (2020), 10.3390/biom10040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Plubell DL, Fenton AM, Rosario S, Bergstrom P, Wilmarth PA, Clark WM, et al. High-density lipoprotein carries markers that track with recovery from stroke, Circ. Res 127 (10) (2020) 1274–1287, 10.1161/CIRCRESAHA.120.316526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Holzer M, Wolf P, Curcic S, Birner-Gruenberger R, Weger W, Inzinger M, et al. Psoriasis alters HDL composition and cholesterol efflux capacity, J. Lipid Res 53 (8) (2012) 1618–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ronsein GE, Reyes-Soffer G, He Y, Oda M, Ginsberg H, Heinecke JW, Targeted proteomics identifies Paraoxonase/Arylesterase 1 (PON1) and apolipoprotein cs as potential risk factors for hypoalphalipoproteinemia in diabetic subjects treated with fenofibrate and rosiglitazone, Mol. Cell. Proteomics 15 (3) (2016) 1083–1093, 10.1074/mcp.M115.054528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Begue F, Tanaka S, Mouktadi Z, Rondeau P, Veeren B, Diotel N, et al. Altered high-density lipoprotein composition and functions during severe COVID-19, Sci. Rep 11 (1) (2021) 2291, 10.1038/s41598-021-81638-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gordon SM, Chung JH, Playford MP, Dey AK, Sviridov D, Seifuddin F, et al. High density lipoprotein proteome is associated with cardiovascular risk factors and atherosclerosis burden as evaluated by coronary CT angiography, Atherosclerosis 278 (2018) 278–285, 10.1016/j.atherosclerosis.2018.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].De Lalla OF, Gofman JW, Ultracentrifugal analysis of serum lipoproteins, Methods Biochem. Anal 1 (1954) 459–478, 10.1002/9780470110171.ch16. [DOI] [PubMed] [Google Scholar]
- [54].Havel RJ, Eder HA, Bragdon JH, The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum, J. Clin. Invest 34 (9) (1955) 1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kunitake ST, Kane JP, Factors affecting the integrity of high density lipoproteins in the ultracentrifuge, J. Lipid Res 23 (6) (1982) 936–940. [PubMed] [Google Scholar]
- [56].Munroe WH, Phillips ML, Schumaker VN, Excessive centrifugal fields damage high density lipoprotein, J. Lipid Res 56 (6) (2015) 1172–1181, 10.1194/jlr.M058735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Stahlman M, Davidsson P, Kanmert I, Rosengren B, Boren J, Fagerberg B, et al. Proteomics and lipids of lipoproteins isolated at low salt concentrations in D2O/sucrose or in KBr, J. Lipid Res 49 (2) (2008) 481–490. [DOI] [PubMed] [Google Scholar]
- [58].Yuana Y, Levels J, Grootemaat A, Sturk A, Nieuwland R, Co-isolation of extracellular vesicles and high-density lipoproteins using density gradient ultracentrifugation, J. Extracell Vesicles 3 (2014), 10.3402/jev.v3.23262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Gordon SM, McKenzie B, Kemeh G, Sampson M, Perl S, Young NS, et al. Rosuvastatin alters the proteome of high density lipoproteins: generation of alpha-1-antitrypsin enriched particles with anti-inflammatory properties, Mol. Cell. Proteomics 14 (12) (2015) 3247–3257, 10.1074/mcp.M115.054031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gordon SM, Deng J, Lu LJ, Davidson WS, Proteomic characterization of human plasma high density lipoprotein fractionated by gel filtration chromatography, J. Proteome Res 9 (10) (2010) 5239–5249, 10.1021/pr100520x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Jeyarajah EJ, Cromwell WC, Otvos JD, Lipoprotein particle analysis by nuclear magnetic resonance spectroscopy, Clin. Lab. Med 26 (4) (2006) 847–870. [DOI] [PubMed] [Google Scholar]
- [62].Caulfield MP, Li S, Lee G, Blanche PJ, Salameh WA, Benner WH, et al. Direct determination of lipoprotein particle sizes and concentrations by ion mobility analysis, Clin.Chem 54 (8) (2008) 1307–1316. [DOI] [PubMed] [Google Scholar]
- [63].McVicar JP, Kunitake ST, Hamilton RL, Kane JP, Characteristics of human lipoproteins isolated by selected-affinity immunosorption of apolipoprotein A-I 81 (5) (1984) 1356–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Cheung MC, Albers JJ, Characterization of lipoprotein particles isolated by immunoaffinity chromatography. Particles containing A-I and A-II and particles containing A-I but no A-II, J. Biol. Chem 259 (19) (1984) 12201–12209. [PubMed] [Google Scholar]
- [65].Duka A, Fotakis P, Georgiadou D, Kateifides A, Tzavlaki K, von Eckardstein L, et al. ApoA-IV promotes the biogenesis of apoA-IV-containing HDL particles with the participation of ABCA1 and LCAT, J. Lipid Res 54 (1) (2013) 107–115, 10.1194/jlr.M030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Moore RE, Navab M, Millar JS, Zimetti F, Hama S, Rothblat GH, et al. Increased atherosclerosis in mice lacking apolipoprotein A-I attributable to both impaired reverse cholesterol transport and increased inflammation, Circ.Res 97 (8) (2005) 763–771. [DOI] [PubMed] [Google Scholar]
- [67].Matsunaga T, Hiasa Y, Yanagi H, Maeda T, Hattori N, Yamakawa K, et al. Apolipoprotein A-I deficiency due to a codon 84 nonsense mutation of the apolipoprotein A-I gene 88 (7) (1991) 2793–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Romeo GR, Moulton KS, Kazlauskas A, Attenuated expression of profilin-1 confers protection from atherosclerosis in the LDL receptor null mouse, Circ. Res 101 (4) (2007) 357–367, 10.1161/CIRCRESAHA.107.151399. [DOI] [PubMed] [Google Scholar]
- [69].van der Vliet HN, Sammels MG, Leegwater AC, Levels JH, Reitsma PH, Boers W, et al. Apolipoprotein A-V: a novel apolipoprotein associated with an early phase of liver regeneration, J. Biol. Chem 276 (48) (2001) 44512–44520, 10.1074/jbc.M106888200. [DOI] [PubMed] [Google Scholar]
- [70].Gordon SM, Remaley AT, High density lipoproteins are modulators of protease activity: implications in inflammation, complement activation, and atherothrombosis, Atherosclerosis 259 (2017) 104–113, 10.1016/j.atherosclerosis.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Morton AM, Furtado JD, Mendivil CO, Sacks FM, Dietary unsaturated fat increases HDL metabolic pathways involving apoE favorable to reverse cholesterol transport, JCI Insight 4 (7) (2019), 10.1172/jci.insight.124620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Deguchi H, Pecheniuk NM, Elias DJ, Averell PM, Griffin JH, High-density lipoprotein deficiency and dyslipoproteinemia associated with venous thrombosis in men, Circulation 112 (6) (2005) 893–899, 10.1161/CIRCULATIONAHA.104.521344. [DOI] [PubMed] [Google Scholar]
- [73].Morelli VM, Lijfering WM, Bos MHA, Rosendaal FR, Cannegieter SC, Lipid levels and risk of venous thrombosis: results from the MEGA-study, Eur. J. Epidemiol 32 (8) (2017) 669–681, 10.1007/s10654-017-0251-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].van der Stoep M, Korporaal SJ, Van Eck M, High-density lipoprotein as a modulator of platelet and coagulation responses, Cardiovasc. Res 103 (3) (2014) 362–371, 10.1093/cvr/cvu137. [DOI] [PubMed] [Google Scholar]
- [75].Cuchel M, Rader DJ, The role of high density lipoproteins in thrombosis, ScientificWorldJournal 2 (2002) 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Xu XR, Wang Y, Adili R, Ju L, Spring CM, Jin JW, et al. Apolipoprotein A-IV binds alphaIIbbeta3 integrin and inhibits thrombosis, Nat. Commun 9 (1) (2018) 3608, 10.1038/s41467-018-05806-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, et al. An overview of the serpin superfamily, Genome Biol. 7 (5) (2006) 216, 10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Dollery CM, Owen CA, Sukhova GK, Krettek A, Shapiro SD, Libby P, Neutrophil elastase in human atherosclerotic plaques: production by macrophages, Circulation 107 (22) (2003) 2829–2836, 10.1161/01.CIR.0000072792.65250.4A. [DOI] [PubMed] [Google Scholar]
- [79].Rohrer L, Cavelier C, Fuchs S, Schluter MA, Volker W, von Eckardstein A, Binding, internalization and transport of apolipoprotein A-I by vascular endothelial cells, Biochim.Biophys.Acta 1761 (2) (2006) 186–194. [DOI] [PubMed] [Google Scholar]
- [80].Ortiz-Munoz G, Houard X, Martin-Ventura JL, Ishida BY, Loyau S, Rossignol P, et al. HDL antielastase activity prevents smooth muscle cell anoikis, a potential new antiatherogenic property, FASEB J. 23 (9) (2009) 3129–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Sukhova GK, Wang B, Libby P, Pan JH, Zhang Y, Grubb A, et al. Cystatin C deficiency increases elastic lamina degradation and aortic dilatation in apolipoprotein E-null mice, Circ. Res 96 (3) (2005) 368–375, 10.1161/01.RES.0000155964.34150.F7. [DOI] [PubMed] [Google Scholar]
- [82].Wu H, Du Q, Dai Q, Ge J, Cheng X, Cysteine protease cathepsins in atherosclerotic cardiovascular diseases, J. Atheroscler. Thromb 25 (2) (2018) 111–123, 10.5551/jat.RV17016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Cheung MC, Vaisar T, Han X, Heinecke JW, Albers JJ, Phospholipid transfer protein in human plasma associates with proteins linked to immunity and inflammation, Biochemistry 49 (34) (2010) 7314–7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Reis ES, Mastellos DC, Hajishengallis G, Lambris JD, New insights into the immune functions of complement, Nat. Rev. Immunol 19 (8) (2019) 503–516, 10.1038/s41577-019-0168-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Parra S, Vives G, Ferre R, Gonzalez M, Guardiola M, Ribalta J, et al. Complement system and small HDL particles are associated with subclinical atherosclerosis in SLE patients, Atherosclerosis 225 (1) (2012) 224–230, 10.1016/j.atherosclerosis.2012.08.029. [DOI] [PubMed] [Google Scholar]
- [86].Wheeler RJ, The trypanolytic factor-mechanism, impacts and applications, Trends Parasitol. 26 (9) (2010) 457–464. [DOI] [PubMed] [Google Scholar]
- [87].Vanhollebeke B, Pays E, The trypanolytic factor of human serum: many ways to enter the parasite, a single way to kill, Mol.Microbiol 76 (4) (2010) 806–814. [DOI] [PubMed] [Google Scholar]
- [88].Greene AS, Hajduk SL, Trypanosome lytic Factor-1 initiates oxidation-stimulated osmotic lysis of trypanosoma brucei brucei, J. Biol. Chem 291 (6) (2016) 3063–3075, 10.1074/jbc.M115.680371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Han YH, Onufer EJ, Huang LH, Sprung RW, Davidson WS, Czepielewski RS, et al. Enterically derived high-density lipoprotein restrains liver injury through the portal vein, Science 373 (6553) (2021), 10.1126/science.abe6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Mahley RW, Ji ZS, Remnant lipoprotein metabolism: key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E, J. Lipid Res 40 (1) (1999) 1–16. [PubMed] [Google Scholar]
- [91].Cockerill GW, Rye KA, Gamble JR, Vadas MA, Barter PJ, High-density lipoproteins inhibit cytokine-induced expression of endothelial cell adhesion molecules, Arteriosder. Thromb. Vase. Biol 15 (11) (1995) 1987–1994. [DOI] [PubMed] [Google Scholar]
- [92].Zuchtriegel G, Uhl B, Pick R, Ramsauer M, Dominik J, Mittmann LA, et al. Vitronectin stabilizes intravascular adhesion of neutrophils by coordinating beta2 integrin clustering, Haematologica (2020), 10.3324/haematol.2019.226241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Persegol L, Verges B, Foissac M, Gambert P, Duvillard L, Inability of HDL from type 2 diabetic patients to counteract the inhibitory effect of oxidised LDL on endothelium-dependent vasorelaxation, Diabetologia 49 (6) (2006) 1380–1386. [DOI] [PubMed] [Google Scholar]
- [94].Christoffersen C, Obinata H, Kumaraswamy SB, Galvani S, Ahnstrom J, Sevvana M, et al. Endothelium-protective sphingosine-1-phosphate provided by HDL-associated apolipoprotein M, Proc. Natl. Acad. Sci. U. S. A 108 (23) (2011) 9613–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Christoffersen C, Ahnstrom J, Axler O, Christensen EI, Dahlback B, Nielsen LB, The signal peptide anchors apolipoprotein M in plasma lipoproteins and prevents rapid clearance of apolipoprotein M from plasma, J. Biol. Chem 283 (27) (2008) 18765–18772, 10.1074/jbc.M800695200. [DOI] [PubMed] [Google Scholar]
- [96].Chen Z, Hu M, The apoM-S1P axis in hepatic diseases, Clin. Chim. Acta 511 (2020) 235–242, 10.1016/j.cca.2020.10.023. [DOI] [PubMed] [Google Scholar]
- [97].Alwaili K, Bailey D, Awan Z, Bailey SD, Ruel I, Hafiane A, et al. The HDL proteome in acute coronary syndromes shifts to an inflammatory profile, Biochim. Biophys. Acta 1821 (3) (2012) 405–415, 10.1016/j.bbalip.2011.07.013. [DOI] [PubMed] [Google Scholar]
- [98].Vaisar T, Pennathur S, Green PS, Gharib SA, Hoofnagle AN, Cheung MC, et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL, J. Clin. Invest 117 (3) (2007) 746–756, 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Vaisar T, Proteomics investigations of HDL: challenges and promise, Curr.Vasc. Pharmacol 10 (4) (2012) 410–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


