Abstract
Background
Chest computed tomography (CT) is crucial in the diagnosis of coronavirus disease 2019 (COVID-19). However, the persistent pandemic and similar CT manifestations between COVID-19 and community-acquired pneumonia (CAP) raise methodological requirements.
Methods
A fully automatic pipeline of deep learning is proposed for distinguishing COVID-19 from CAP using CT images. Inspired by the diagnostic process of radiologists, the pipeline comprises four connected modules for lung segmentation, selection of slices with lesions, slice-level prediction, and patient-level prediction. The roles of the first and second modules and the effectiveness of the capsule network for slice-level prediction were investigated. A dataset of 326 CT scans was collected to train and test the pipeline. Another public dataset of 110 patients was used to evaluate the generalization capability.
Results
LinkNet exhibited the largest intersection over union (0.967) and Dice coefficient (0.983) for lung segmentation. For the selection of slices with lesions, the capsule network with the ResNet50 block achieved an accuracy of 92.5% and an area under the curve (AUC) of 0.933. The capsule network using the DenseNet121 block demonstrated better performance for slice-level prediction, with an accuracy of 97.1% and AUC of 0.992. For both datasets, the prediction accuracy of our pipeline was 100% at the patient level.
Conclusions
The proposed fully automatic deep learning pipeline of deep learning can distinguish COVID-19 from CAP via CT images rapidly and accurately, thereby accelerating diagnosis and augmenting the performance of radiologists. This pipeline is convenient for use by radiologists and provides explainable predictions.
Keywords: Coronavirus disease 2019, Deep learning, Community-acquired pneumonia, Lung computed tomography image, Capsule network
1. Introduction
Coronavirus disease 2019 (COVID-19) was discovered in late 2019, and spread rapidly worldwide in only a few months [1,2]. The novel coronavirus is characterized by a high infectivity, mild symptoms, and a long incubation period. Currently, the number of COVID-19 patients abroad is increasing at a rate of 560,000 per day [3]. Early diagnosis of COVID-19 plays a crucial role in the isolation and treatment of patients [4,5]. The gold standard for the diagnosis of COVID-19 is the real-time polymerase chain reaction (RT-PCR) test for the detection of the novel coronavirus nucleic acid [[6], [7], [8], [9]], which is time-consuming. Patients affected by detoxification concentration must undergo multiple nucleic-acid tests to confirm the diagnosis, during which the virus can spread [10,11]. Lesions can be clearly observed in the chests of patients via computed tomography (CT) or radiography, mainly with ground-glass opacities (GGO) and crazy-paving [[12], [13], [14]]. However, the CT images of community-acquired pneumonia (CAP) and COVID-19 are similar. Thus, it is challenging even for experienced radiologists to distinguish between them. Moreover, radiologists perform long readings, leading to the degradation of the reading quality. Furthermore, misdiagnosis and missed diagnosis are not conducive to the analysis of symptoms [15,16].
Deep learning—particularly convolutional neural networks (CNNs)—demonstrates a significant potential for feature extraction and representation, and has attracted considerable attention for application in classification tasks for COVID-19 and CAP. Some researchers have focused on chest X-rays owing to their high speed and low radiation dose. Oh et al. proposed a patch-based CNN method, which has the advantage of relatively few parameters [17]. Nwosu et al. created a two-channel and half-cultured model based on residual neural networks for the classification of chest X-ray images, with supervised and unsupervised paths [18]. Gulati proposed a new convolutional network model architecture based on a combination of DarkNet and AlexNet for the automatic diagnosis of COVID-19 using patient X-ray images [19]. Haritha et al. created a deep learning model (CheXNet) using a pretrained model (DenseNet121) for the diagnosis of COVID-19 patients [20]. Waheed et al. developed CovidGAN, which adopts an auxiliary classifier generative adversarial network to generate synthetic X-ray images [21]. Rahaman et al. compared the performance of different transfer learning approaches for the identification of COVID-19 using chest X-ray images [22].
In contrast to chest X-rays, CT images have no overlapping tissues; thus, more details can be obtained and reconstructed in different planes. Moreover, the high resolution of CT examination allows dissection-level analysis. Thus, researchers have used chest CT images for COVID-19 analysis. Soares et al. constructed an open dataset of CT scans and applied an available deep learning model to the dataset for classifying pneumonia [23]. Alshazly et al. used CT images from two datasets—related to severe acute respiratory syndrome coronavirus 2 (SARS-COV-2) and COVID-19—to propose a transfer learning strategy based on a custom input of different depth architectures [24]. Gozes et al. proposed a three-dimensional (3D) deep learning framework that can extract two-dimensional (2D) and 3D global features to distinguish COVID-19 from CAP [25]. Ouyang et al. combined online attention with 3D ResNet34 and developed a dual-sampling attention network [26]. Qi et al. proposed a deep represented multiple-instance learning method [27]. However, more advanced deep learning models are required to improve the performance of the differential diagnosis of COVID-19 and CAP.
In this study, inspired by the diagnostic process of radiologists, we propose a fully automated pipeline of CNNs and capsule networks for distinguishing COVID-19 from CAP using CT images. The pipeline comprises LinkNet [28] for lung segmentation, a capsule network for automatically selecting critical slices with infected lesions, a capsule network for distinguishing COVID-19 from CAP at the slice level, and a majority voting module for patient-level prediction.
Our study provides the following novelties and contributions. First, an automatic pipeline was constructed, and excellent performance was achieved for multiple datasets. The pipeline comprises four connected modules, namely lung segmentation, selection of slices with lesions, slice-level prediction, and patient-level prediction. Thus, the results of each module can be inspected. Second, LinkNet was used to accurately segment the lung field with infected lesions. Third, a capsule network was implemented to automatically select critical slices with infected lesions. Fourth, a capsule network was designed for the slice-level prediction of COVID-19. To the best of our knowledge, this type of pipeline mimicking the diagnostic process of radiologists has not been previously reported.
2. Materials and methods
2.1. Dataset
COVID-19 and CAP data were collected from the General Hospital of the Yangtze River Shipping and Affiliated Hospital of Guizhou Medical University. The patients were enrolled between December 2019 and March 2020. After the elimination of abnormal data, the final data included 161 CT scans from 57 patients with COVID-19 and 165 scans from 100 patients with CAP. Chest CT scans were performed using GE LightSpeed16 CT, Toshiba Aquilion ONE CT, Toshiba Aquilion CT, Siemens Somatom Scope CT, Siemens Somatom CT, and Definition AS + CT. The patient information and scanning parameters are presented in Table 1 .
Table 1.
Demographic information of the participants and acquisition parameters for the CT images.
| Information | COVID-19 | CAP | p value |
|---|---|---|---|
| Gender (male/female) | 27/30 | 53/47 | 0.497a |
| Age (years) | 56.1 ± 18.4 | 40.5 ± 20.7 | 6.526 × 10−6 |
| kVp (kV) | 120 | – | |
| Slice thickness (mm) | 5 | – | |
| Pixel size (mm) | 0.763 ± 0.067 | 0.697 ± 0.105 | 5.738 × 10−5 |
| X-ray tube current (mA) | 216.4 ± 23.5 | 207.1 ± 89.2 | 0.456b |
| Exposure (mA*s) | 60.4 ± 45.0 | 103.7 ± 37.2 | 1.995 × 10−9 |
p was calculated via a chi-square test.
p was calculated via a two-sample t-test.
All the COVID-19 subjects were diagnosed via RT-PCR tests. In the CAP group, 69 patients obtained etiological confirmation from a specialized laboratory (57 bacterial, 9 viral pneumonia, and 3 mycoplasma); 31 patients did not have etiological confirmation, but the possibility of false negatives was eliminated through strict epidemiological investigations, numerous RT-PCR tests, and clinical outcomes.
The second dataset, which included 110 patients, was obtained from the China Consortium of Chest CT Image Investigation (CC-CCII) dataset [29]. The third dataset, which included 538 COVID-19 patients (9997 slices), was acquired from The Cancer Imaging Archive (TCIA) Collections [30].
2.2. Overview of study procedure
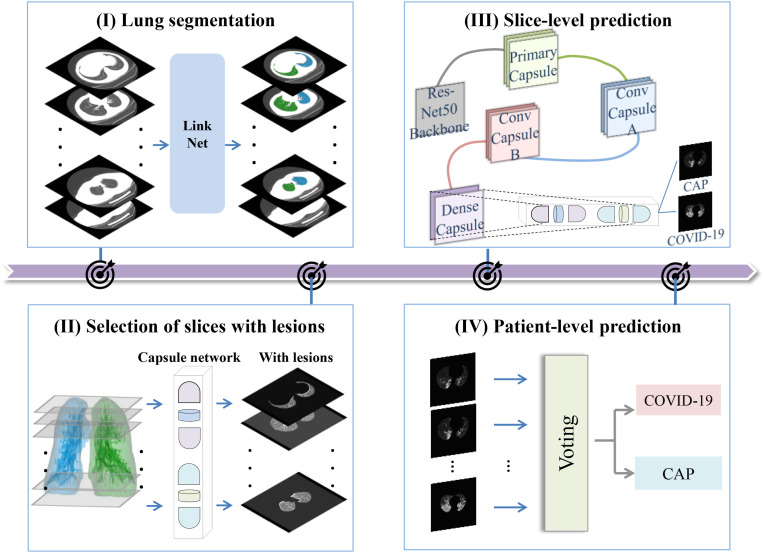
Inspired by the diagnostic process of radiologists, a fully automated pipeline is proposed for distinguishing COVID-19 from CAP via CT images (Fig. 1 ). The pipeline consists of four modules: (1) lung segmentation (radiologists initially devote attention to the lung field during the diagnosis of lung diseases), (2) selection of slices with lesions (radiologists shift their attention from the lung field to the lesions), (3) slice-level prediction (radiologists typically perform a preliminary diagnosis based on the observation results for a specific slice), and (4) patient-level prediction (radiologists provide the final diagnosis after integration of information from all suspected slices). The details of each module of the pipeline are presented in the following sections.
Fig. 1.
Overview of the proposed pipeline including four modules: (I) Lung segmentation, (II) selection of slices with lesions, (III) slice-level prediction, and (IV) patient-level prediction.
2.3. Lung segmentation
It was important to preprocess the data, as they originated from different hospitals. First, the CT scans were unified in a fixed window (window level = −300 HU, window width = 1400 HU). Second, the pixel intensity in the CT scan was normalized and adjusted between 0 and 1.
Using Pulmonary Toolkit (https://www.tomdoel.com/software/), the mask of the lung field in 161 CT scans of COVID-19 was drawn semiautomatically. During the preparation of masks, all structures following the secondary bronchial were included in the lung field. The preprocessed data and obtained mask of the lung field were used to train and evaluate the lung segmentation module.
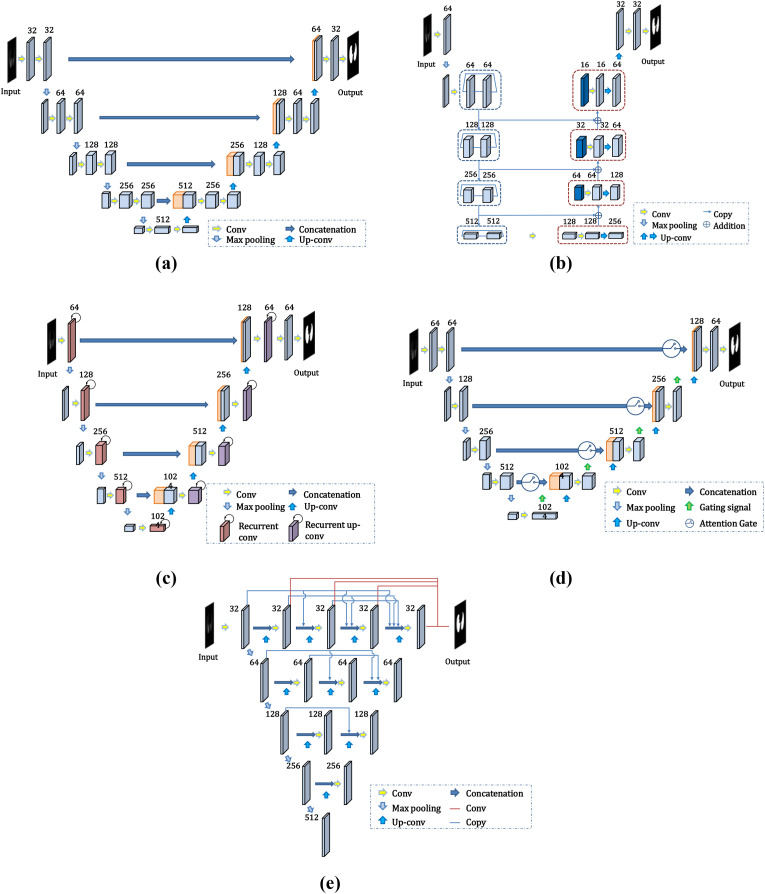
Considering their good performance in medical image segmentation, five deep CNNs—U-Net [31], LinkNet, Recurrent Residual CNN-based U-Net (R2U-Net) [32], Attention U-Net [33], and U-Net++ [34,35]—were applied to our lung segmentation module. The performances of the different networks were compared to identify the most suitable structure. The network architecture is depicted in Fig. 2 .
Fig. 2.
Architecture of the deep CNN for lung segmentation: (a) U-Net; (b) LinkNet; (c) R2U-Net; (d) Attention U-Net; (e) U-Net++.
U-Net [31] was presented at the Medical Image Computing and Computer Assisted Intervention (MICCAI) Society in 2015. It has been widely applied in medical image segmentation. In this study, preprocessed slices with sizes of 512 × 512 pixels were fed into the network. As shown in Fig. 2(a), the encoder of the network contains four pairs of convolutional layers and pooling layers with 32, 64, 128, and 256 channels, and the decoder contains four upsampling layers. The most significant advantage of U-Net is that the skip connection between the layers in the encoder and decoder helps each upsampling result to be combined with low-level features, such that features of different scales are fused and the edge segmentation is more accurate.
Instead of recovering lost location information through the maximum index of the pooling layer, LinkNet [28] directly connects the encoder to the decoder by replacing the “concatenation” operation in U-Net with the “addition” operation (Fig. 2(b)). This operation bypasses the spatial information and improves the segmentation accuracy while reducing the processing time. The architecture of LinkNet is similar to that of U-Net. The encoder of the U-Net network is replaced by a residual module in LinkNet. To improve the accuracy of the network and minimize the training parameters, the residual module of ResNet34 was used as the network encoder in this study.
R2U-Net [32] integrates the structure of the recurrent neural network (RNN) and ResNet into U-Net. Its performance in the segmentation of retinal blood vessels is better than that of U-Net [31]. The convolutional layers in both the encoder and decoder are replaced by an RNN with an embedded ResNet50 module (Fig. 2(c)).
Attention U-Net [33] is also based on the U-Net model and introduces the attention mechanism. An attention gate is inserted into each skip connection between the layers in the encoder and decoder (Fig. 2(d)). The information from the convolutional layer of the encoder and the bottom layer of the decoder is input into the attention gate. The output of the attention gate is concatenated to the upsampling layer to emphasize the salient region of this layer and improve the model performance.
U-Net++ [34] is a deeply supervised encoder–decoder network that consists of a few U-Net sub-networks with different depths (Fig. 2(e)). This architecture takes advantage of redesigned skip pathways and deep supervision. The skip pathways are used to reduce the semantic gap between the feature maps of the encoder and decoder sub-networks, which makes it easier for the optimizer to solve simple optimization problems.
We selected binary cross entropy (BCE) as a loss function for the four segmentation networks. This loss function is used to solve the dichotomy problem, and can be expressed as
| (1) |
where , , and represent the number of slices, output of the network obtained using the sigmoid function, and label, respectively.
The CT images of both COVID-19 and CAP patients may contain lesions, and the presence of holes in the segmentation results of these networks cannot be avoided. In this study, the morphological filling method was used. The “FindContours” function in the OpenCV library was used. The maximum edge information of the segmented lung field was used to improve the results for avoiding the loss of key lesion information in the lung region in the subsequent classification network.
2.4. Selection of slices with lesions
For COVID-19 and CAP, infected lesions seldom occupy the entire lung field. The attention of radiologists is devoted to CT slices with lesions during diagnosis. A capsule network [36] was designed and trained to classify all the slices into two categories—with and without lesions—after lung segmentation.
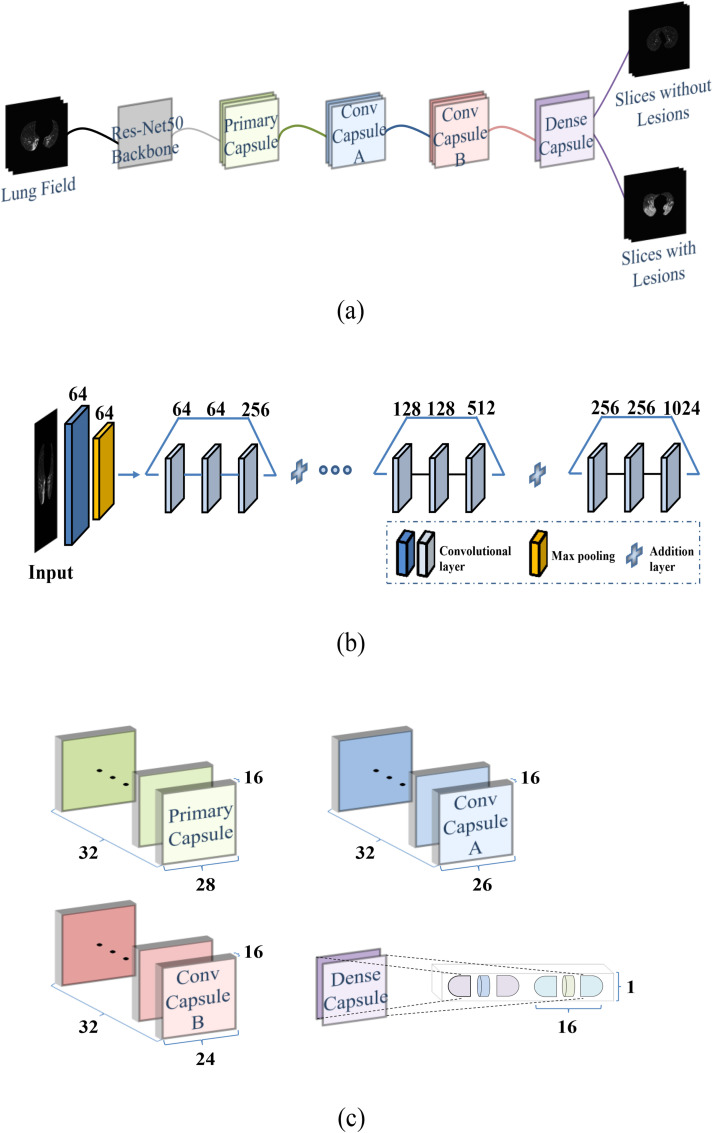
As shown in Fig. 3 , the capsule network for slice selection consisted of a convolutional block, a primary capsule layer, two convolutional capsule layers (A and B), and a dense capsule layer. The size of the input image was 512 × 512 pixels. The first three stages of the pretrained ResNet50 [37] were used as convolutional blocks (Fig. 3(b)). In total, 1024 features were output from the convolutional block and transmitted to the primary capsule layer. Each capsule of the primary capsule layer and two convolution capsule layers was a 1 × 16 vector (Fig. 3(c)). The output vector of the bottom capsule was multiplied by the weight matrix to calculate the prediction vector (i.e., high-level information), and was then transmitted to the upper capsule. Finally, a dense capsule layer was connected. Each of the two output categories had a 1 × 16 capsule. The prediction category of the image was determined according to its Frobenius norm.
Fig. 3.
Structure of the capsule network for slice selection: (a) Overall structure; (b) Pretrained ResNet50 without fully connected layers; (c) Capsule architecture.
In this study, the dynamic routing mechanism of three iterations was used to update the parameters of the weight matrix. However, the dynamic routing mechanism could not completely replace the back propagation. Thus, we used the spread loss function to train the back propagation.
| (2) |
here, and represent the activation values of the target and the ith position from the target, respectively.
2.5. Slice-level prediction of COVID-19 and CAP
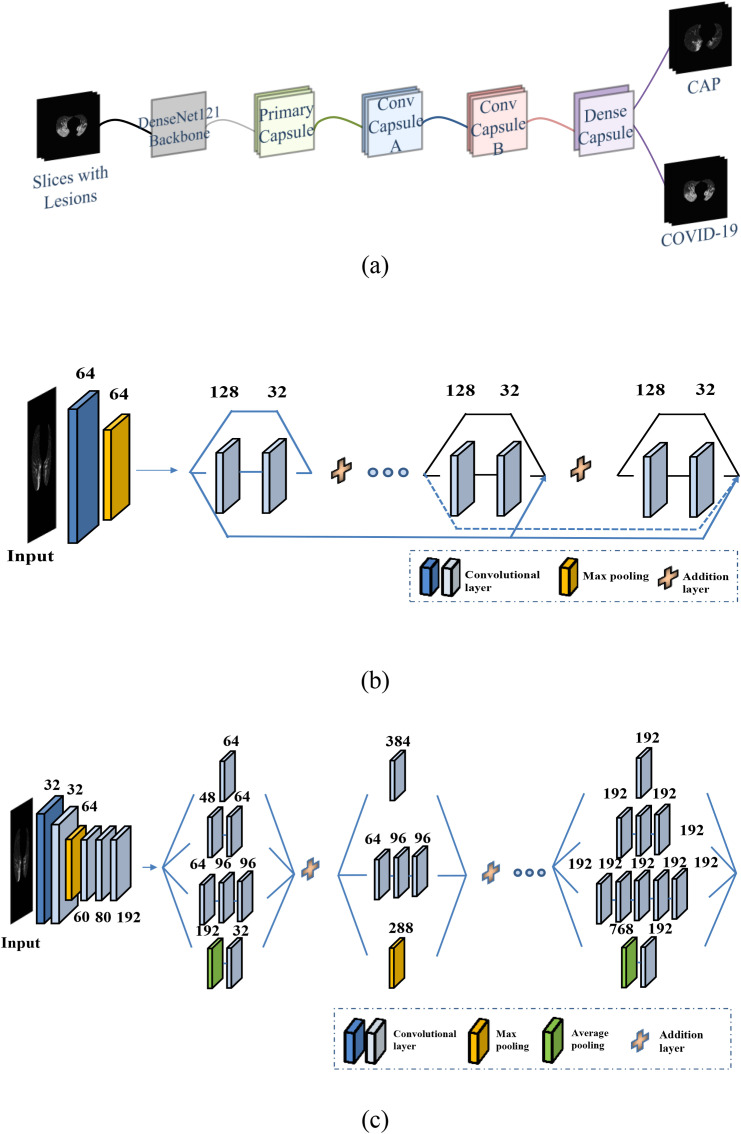
Using the selected slices with lesions as inputs, another capsule network was trained for the slice-level prediction of COVID-19 and CAP. Except for the convolutional block, which was a pretrained DenseNet121 block [38], the architectures of this capsule network were identical to those of the network for slice selection (Fig. 4 (a)). The previous ResNet50 and pretrained Inception-V3 were also used as the convolutional blocks in the capsule network for slice-level prediction. The detailed architectures of DenseNet121 and Inception-V3 [39] are depicted in Fig. 4(b) and (c), respectively. The same spread loss function (Equation (2)) was used to train the three models. The performances of the models using the three different convolutional blocks were compared.
Fig. 4.
Structure of the capsule network for slice-level prediction of COVID-19 and CAP: (a) Overall structure; (b) Pretrained DenseNet121 without fully connected layers; (c) Pretrained Inception-V3 without fully connected layers.
2.6. Patient-level prediction
After the slice-level predictions, two majority voting methods were used to obtain the final patient-level prediction of COVID-19 or CAP. The first method is hard majority voting, wherein each slice with lesions from one patient provides a vote. The final prediction is determined according to the minority obeying the majority. The second method is soft majority voting, wherein for each slice with lesions, the probability of two categories is obtained by inputting the norm of the two capsules (i.e., the output of the capsule network) into a SoftMax operation. The final prediction is determined by comparing the sums of the probabilities of the two categories for all slices with lesions from one patient.
2.7. Comparative experiments
Three categories of comparative experiments were conducted. The objective of the first experiment was to determine whether lung segmentation improves the performance at the slice and patient levels. The input of the capsule network for slice-level prediction was changed to the original CT images without lung segmentation, and all other modules in the pipeline were maintained unchanged.
The objective of the second experiment was to determine whether the selection of slices with lesions is useful. After lung segmentation, all slices were directly used to train and evaluate the capsule network for slice-level prediction.
The objective of the third experiment was to determine whether the introduction of the capsule network concept affects the accuracy of COVID-19 and CAP classification. In this experiment, in the last step of the process, we used the traditional DenseNet121 block instead of the capsule network for classification.
2.8. Training and evaluation of models
During the training of the lung segmentation network, we marked the lung fields on 161 CT scans of COVID-19. A total of 10,280 image slices were obtained and divided into training, validation, and test datasets in the ratio of 7:1:2.
For the capsule network for the selection of slices with lesions, 19,781 slices (6712 slices with COVID-19 or CAP lesions; 13,069 slices without lesions) were collected. Among these slices, 17,356 were used for training and validation, and the remaining slices (approximately 1/10 of all slices) were used for testing.
For the capsule network for slice-level prediction, 6712 slices with lesions were divided into training, validation, and test sets in the ratio of 8:1:1. All the slices in the test set were obtained from 34 scans (9 COVID-19 and 25 CAP); no slice from these scans appeared in the training and validation sets. Moreover, the data from the CC-CCII dataset were divided into training, validation, and test sets in the ratio of 7:1:2.
Seven performance metrics were used to evaluate the different models: the intersection over union (IoU), Dice coefficient, area under the curve (AUC), accuracy, precision, sensitivity, and specificity.
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
here, TP, TN, FP, and FN denote a true positive, true negative, false positive, and false negative, respectively.
After training and validation, the final optimized parameter was obtained. The learning rate, batch size, and number of epochs were 0.0001, 1, and 21, respectively. Data augmentation and early stopping were conducted in the experiment to alleviate the overfitting. When the accuracy of the validation dataset did not increase within seven epochs, the early stopping operation occurred.
The experiments were implemented using the PyTorch library. The Pulmonary Toolkit for the preparation of lung field masks in MATLAB 2016b was used, with a Windows 10 system. The workstation used for the implementation had an Intel Core I7-9700 3.00 GHz central processing unit (CPU) and an NVIDIA GeForce RTX 2080 Ti graphics processing unit (GPU).
3. Results
3.1. Lung segmentation
Table 2 presents the performances of the different lung segmentation models with regard to the IoU and Dice coefficient. LinkNet exhibited the largest IoU (0.967) and Dice coefficient (0.983) among the five segmentation models (U-Net, LinkNet, R2U-Net, Attention U-Net, and U-Net++), whereas R2U-Net exhibited the lowest IoU and Dice coefficient.
Table 2.
Performances of the five lung segmentation networks.
| Model | IoU | Dice coefficient |
|---|---|---|
| U-Net [40] | 0.962 | 0.980 |
| LinkNet | 0.967 | 0.983 |
| R2U-Net [32] | 0.928 | 0.962 |
| Attention U-Net | 0.951 | 0.974 |
| U-Net++ [34,35] | 0.936 | 0.966 |
*Bold font indicates the network with the best performance.
Fig. 5 depicts examples of lung segmentation using different networks for the same slice with infected lesions. In example slice 1, there was an under-segmented region indicated by an arrow for U-Net, R2U-Net, Attention U-Net, and U-Net++. Additionally, there were several holes in the results obtained using R2U-Net and Attention U-Net. In example slice 2, under-segmented regions were observed for U-Net, R2U-Net, and Attention U-Net. Moreover, there were several holes in the results obtained using R2U-Net, and parts of the bed of the CT scanner were incorrectly segmented as the lung field.
Fig. 5.
Examples of lung segmentation using different networks.
3.2. Selection of slices with lesions
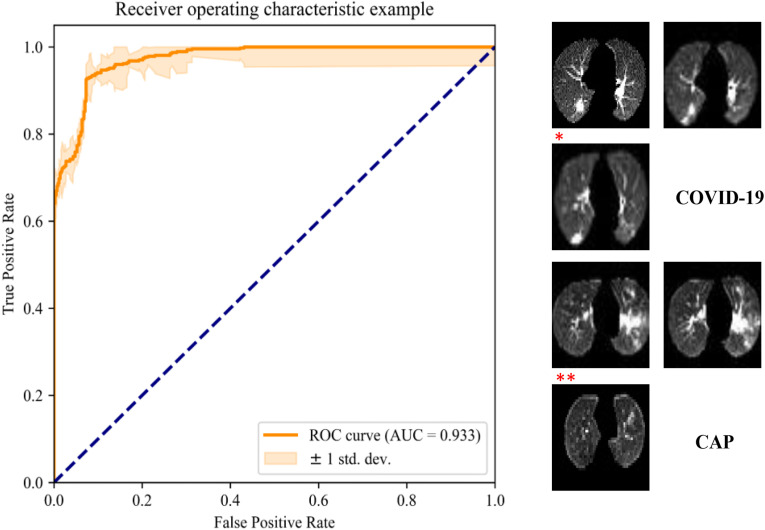
Fig. 6 depicts the receiver operating characteristic (ROC) curve and examples of the automatic selection of slices with lesions. The capsule network with the ResNet50 block achieved an accuracy of 92.5% and AUC of 0.933. Slices with obvious COVID-19 or CAP lesions were correctly selected. The slice marked with an asterisk is an example of a case of COVID-19 with small lesions, which was not selected. The slice marked with two asterisk symbols represents a case of CAP without significant lesions, which was incorrectly selected.
Fig. 6.
ROC curve of the deep capsule network for automatic selection of slices with lesions and classification results (* indicates an example of a slice with a small COVID-19 lesion that was incorrectly classified as a slice without lesions, and ** indicates an example of a slice without apparent CAP lesions that was incorrectly classified as a slice with lesions).
3.3. Prediction for our laboratory dataset
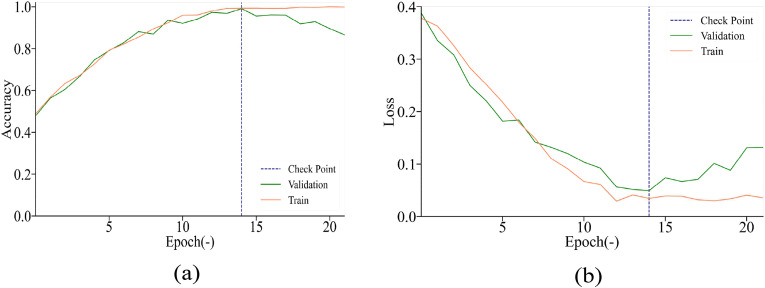
Fig. 7 depicts the accuracy and loss for our laboratory dataset. The accuracy in the training and validation process was high, and the check point indicated that the accuracy in the validation process did not increase after seven epochs. The loss in the training and validation process decreased rapidly, indicating that the parameters selected in our model were sufficient. Table 3 presents the performances of the three capsule networks for slice-level prediction with the pretrained DenseNet121, Inception-V3, and ResNet50 network blocks. The capsule network using the DenseNet121 block exhibited the best performance. It had the fewest training parameters and achieved an accuracy of 97.1% and an AUC of 0.992.
Fig. 7.
Accuracy and loss for our laboratory dataset.
Table 3.
Performance comparison of slice-level prediction models with different pretraining blocks.
| Model | Params. (M) | Precision | Accuracy | Sensitivity | Specificity | AUC |
|---|---|---|---|---|---|---|
| ResNet50 | 9.63 | 0.965 | 0.981 | 0.997 | 0.966 | 0.983 |
| Inception | 9.92 | 0.939 | 0.923 | 0.900 | 0.945 | 0.973 |
| DenseNet121 | 8.04 | 0.979 | 0.971 | 0.959 | 0.981 | 0.992 |
* Bold font indicates the highest value among the three models.
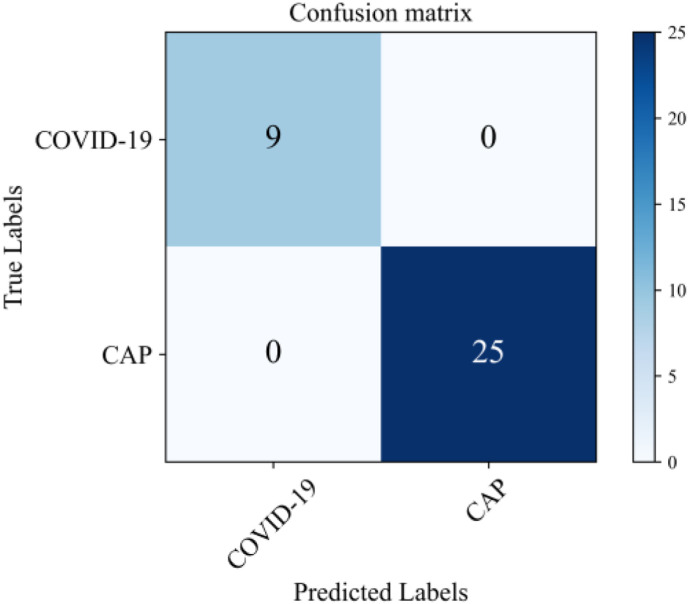
The confusion matrix for the final patient-level prediction is shown in Fig. 8 . The prediction accuracy of the pipeline can be 100%, whereas the diagnosis accuracies of two radiologists (A and B) were 65.1% and 66.8%, respectively. Thus, the performance of our pipeline was remarkable.
Fig. 8.
Confusion matrix of patient-level prediction of COVID-19 and CAP for our laboratory dataset.
3.4. Prediction for two other public datasets
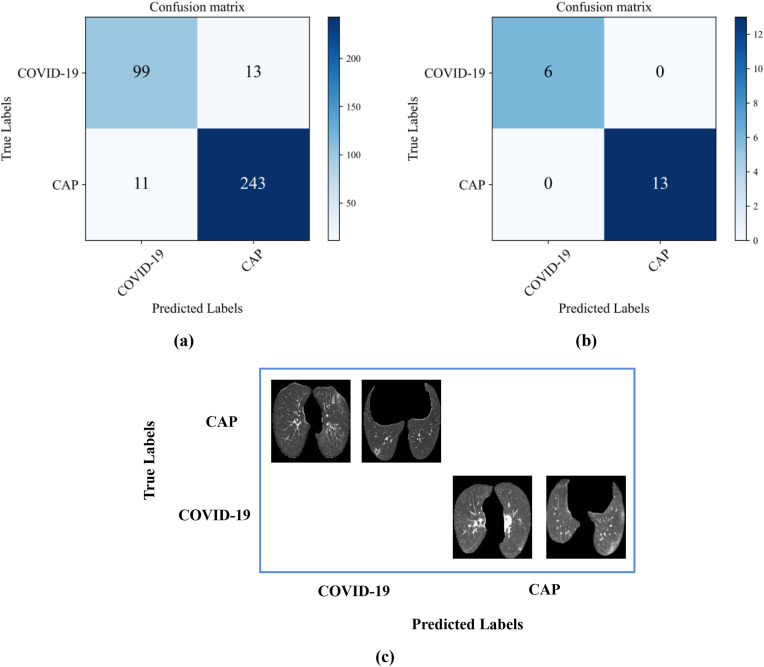
For the CC-CCII dataset, the confusion matrix of the pipeline is shown in Fig. 9 . The accuracy can be 93.4%, while the AUC can be 0.876 for the slice-level prediction. The accuracy of the proposed method for patient-level prediction reached 100%, which was higher than those of the radiologists (A: 73.7%; B: 78.9%). Thus, the proposed strategy is robust and applicable to multiple datasets.
Fig. 9.
Results for the prediction of COVID-19 and CAP with the CC-CCII dataset: (a) Confusion matrix at the slice level; (b) Confusion matrix at the patient level; (c) Two examples from the CC-CCII dataset that were incorrectly diagnosed.
For the dataset from TCIA Collections, the accuracy was 94.8% and 96.7% for slice- and patient-level prediction, respectively.
3.5. Results of comparative experiment
Without lung segmentation before the slice-level prediction of COVID-19 and CAP, the accuracy was reduced to 86.32% and the AUC was reduced to 0.880. At the patient level, the accuracy was reduced to 78.57%.
Without the selection of slices with lesions, the accuracy of slice-level prediction was merely 78.5%, which was approximately 20% lower than that of the model with slice selection. The AUC was reduced to 0.654. At the patient level, the accuracy was reduced to 85.71%.
When the traditional DenseNet121 block was used for slice-level prediction, the accuracy was 94.7% and the AUC was 0.960 (inferior to those of the capsule network). Correspondingly, the accuracy at the patient level was reduced from 97.1% to 100%.
3.6. Comparison of our method and current state-of-the-art studies
Table 4 presents a performance comparison of our pipeline and other state-of-the-art methods for distinguishing COVID-19 from CAP. The state-of-the-art methods include [27], 2D CNN methods [41], BigBiGAN framework [42], Pretrained EfficientNet-b7 [43], and 3D ResNet34 with attention modules [26].
Table 4.
Performance of our method and state-of-the-art methods.
| Study | Key aspects | Performance |
|---|---|---|
| Our method |
|
Accuracy = 0.971 |
|
Sensitivity = 0.959 | |
|
Specificity = 0.981 AUC = 0.992 | |
|
||
|
||
|
||
| Qi et al., 2021 [27] |
|
Accuracy = 0.959 Sensitivity = 0.972 |
|
Specificity = 0.941 | |
|
AUC = 0.955 | |
| Javaheri et al., 2021 [41] |
|
Accuracy = 0.933 |
|
Sensitivity = 0.909 | |
|
Specificity = 1.00 | |
|
AUC = 0.94 | |
| Song et al., 2020 [42] |
|
Sensitivity = 0.92 |
|
Specificity = 0.91 | |
|
AUC = 0.972 | |
| Basset et al., 2021 [43] |
|
Accuracy = 0.968 |
|
AUC = 0.988 | |
|
||
|
||
| Ouyang et al., 2020 [26] |
|
Accuracy = 0.875 |
|
Sensitivity = 0.869 | |
|
Specificity = 0.901 | |
|
AUC = 0.944 | |
|
Our method achieved an accuracy of 0.971 at the patient level, outperforming the aforementioned state-of-the-art methods. Because the steps of the proposed pipeline mimic the workflow of radiologists, the pipeline is more applicable in medicine than the other methods.
4. Discussion
4.1. COVID-19 pandemic and challenges (distinguishing COVID-19 from CAP)
COVID-19 was detected in December 2019 and then rapidly spread worldwide [44]. In the following year, the delta variant emerged, which is more contagious and pathogenic than the original strain [45]. Although vaccines have been widely used and distributed among large populations [46], it is critical to diagnose and treat affected patients at an early stage [47]. The gold standard for COVID-19 diagnosis is the RT-PCR test [48,49]. However, this test is time-consuming and has limitations in underdeveloped areas [50]. Thus, a quick and simple approach to distinguish COVID-19 from CAP is required.
Song et al. proposed an end-to-end classification method using a dataset acquired from two different hospitals that included 201 CT images (COVID-19: 98; non-COVID-19 pneumonia: 103) [42]. The state-of-the-art BigBiGAN framework was used for feature extraction, and a support vector machine was employed as the classifier, resulting in an AUC of 0.972. In our previous work, we proposed a method based on multiple-instance learning for distinguishing COVID-19 from CAP [27]. In that study, the pretrained ResNet50 block with finetuning was employed for deep feature representation, and the k-nearest neighbor method was used to generate the final result. An accuracy of 95% and an AUC of 0.943 were achieved. The proposed pipeline can achieve a performance comparable to those of the state-of-the-art methods. Thus, we developed an automatic pipeline of CNNs and capsule networks to distinguish COVID-19 from CAP using CT images. This pipeline can help radiologists to classify COVID-19 and CAP. CT is one of the most widely used imaging methods in clinical practice [[51], [52], [53], [54]] and plays an important role in the diagnosis of CAP and epidemiological studies [55]. GGO, consolidation, and peripheral and bilateral involvement have been observed in CT images of COVID-19 [56]. Radiologists have a high specificity but moderate sensitivity for distinguishing COVID-19 from CAP, which implies missed diagnoses of COVID-19 [57]. Thus, the deep learning is effective for screening [41].
4.2. Pipeline mimicking radiological diagnosis
To mimic the diagnostic process of radiologists, our pipeline has four modules: (1) lung segmentation, (2) selection of slices with lesions, (3) slice-level prediction, and (4) patient-level prediction. Therefore, the pipeline is convenient for use by radiologists. End-to-end deep learning models have been proposed for distinguishing COVID-19 from CAP [41,42,58]. Compared with that of these models, the proposed pipeline has better explanatory power, as radiologists can conveniently check the output of each module and confirm the final results. Thus, the pipeline with four modules provides explainable predictions.
4.3. Lung segmentation
Deep learning networks have been used to segment the lung field in a preprocessing procedure before the prediction, and high accuracies have been achieved [59,60]. In our study, the lung segmentation module improved the classification performance for COVID-19 and CAP. This is because all lesions in the lung field and tissues outside the lung field can interfere with the feature representation in the capsule network.
We compared the performances of U-Net, LinkNet, R2U-Net, Attention U-Net, and U-Net++ for lung segmentation. LinkNet outperformed the other four networks. It achieved a Dice coefficient of 0.983 and an IoU of 0.967, which were larger than or comparable to previous results obtained using DenseNet161 U-Net [61], LungSeg-Net [62], and three-stage segmentation [63]. In LinkNet, the residual module was employed in the encoder of the network to represent the high-level semantic information of the CT images, which contributed to the outstanding segmentation results. Although R2U-Net, which includes the recurrent convolution module, and Attention U-Net, which includes the attention mechanism, were derived from U-Net, their lung segmentation performances were inferior to that of U-Net. This may be because the R2U-Net and Attention U-Net models were trained from scratch with our small dataset and cannot be fully trained.
4.4. Selection of slices with lesions
The selection of slices with lesions is useful for distinguishing COVID-19 from CAP with capsule networks via CT images. CNN models have been trained to classify COVID and CAP without selecting lesion slices [23,64]. However, in this study, a specific module was introduced to identify the slices with lesions so that the value could be fed into the network in our pipeline. This significantly increased the accuracy of slice-level prediction (by approximately 20%). The patient-level prediction accuracy increased by 14% with the selection of slices with lesions, mainly because lesions do not necessarily spread throughout the lung field. For patients, labeling slices without lesions reduces the workload of radiologists and improves the classification accuracy.
4.5. Advantages of capsule network
The capsule network reflects the spatial information of images better than CNNs [65,66]. According to the comparative experiment, the accuracy of the capsule network can reach 97.1%, which is higher than that without a capsule block. This is mainly because compared with the scalar value produced by the CNN models, the vector output by the capsule network can better represent the features. The vector formula used by the capsule network can offset the deficiency of the CNN and help the network represent the features in a strong and lightweight manner [67].
4.6. Limitations and future studies
Our study has several limitations. First, the number of datasets considered is small. Although the capsule network is a lightweight network, overfitting may occur because of the small number of datasets, which may limit the generalization and robustness of the pipeline. Second, COVID-19 is distinguished from CAP, but other clinical phenotypes of COVID-19 and CAP are not considered, as the numbers of cases of different clinical phenotypes of COVID-19 are unequally distributed in the dataset and the COVID-19 phenotypes of some patients are unclear. Third, healthy individuals may also have opacities in the lung field. The influence of this condition is unknown, because a healthy control group was not included in our study.
With the increase in the number of patients in the dataset, the clinical type of CAP and severity of COVID-19 is expected to be balanced, and the pipeline is expected to exhibit a better generalization ability. More advanced methods, such as Deep Bayes-SqueezeNet [68], deep convolutional generative adversarial networks [69], and ensemble learning [70], are required to improve the detection performance for COVID-19. Additionally, these methods demonstrate potential for use in the segmentation of lung infections and prognosis for COVID-19 patients in the future [71,72].
5. Conclusion
In this study, a fully automatic deep learning pipeline that can rapidly and accurately distinguish COVID-19 from CAP using CT images was developed. The performance of the pipeline was improved by adding modules for lung segmentation and the selection of slices with lesions. The capsule network in the pipeline effectively represented the deep features of COVID-19 lesions in CT images. Because the four modules mimic the diagnostic process of radiologists, the pipeline is convenient for use by radiologists and provides explainable predictions. The proposed pipeline can accelerate diagnosis and augment the performance of radiologists.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was waived because this was a prospective study.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Acknowledgements
Funding: This work was supported by the National Natural Science Foundation of China [grant numbers 82072008, 81671773, and 61672146]; the Fundamental Research Funds for the Central Universities [grant numbers N2119010, N2124006-3]; the Key R&D Program Guidance Projects in Liaoning Province [grant number 2019JH8/10300051]; and Foshan Special Project of Emergency Science and Technology Response to COVID-19 [grant number 2020001000376].
References
- 1.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. 2020;296:E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pm A., Mae F., Mb C., Es D., Rstg H., Jjm B., Hlh U.K. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395 doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johns Hopkins Coronavirus Resource Center https://gisanddata.maps.arcgis.com/apps/dashboards/
- 4.Mobiny A., Cicalese P.A., Zare S., Yuan P., Abavisani M., Wu C.C., Ahuja J., Groot P.D., Nguyen H.V. 2020. Radiologist-Level COVID-19 Detection Using CT Scans with Detail-Oriented Capsule Networks.https://ui.adsabs.harvard.edu/abs/2020arXiv200407407M [Google Scholar]
- 5.Yang X., Yu Y., Xu J., Shu H., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296:E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiang F., Wang X., He X., Peng Z., Yang B., Zhang J., Zhou Q., Ye H., Ma Y., Li H. Antibody detection and dynamic characteristics in patients with coronavirus disease 2019. Clin. Infect. Dis. 2020;71:1930–1934. doi: 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan Y., Li X., Yang G., Fan J., Tang Y., Zhao J., Long X., Guo S., Zhao Z., Liu Y. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J. Infect. 2020;81:e28–e32. doi: 10.1016/j.jinf.2020.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majidi H., Niksolat F. Chest CT in patients suspected of COVID-19 infection: a reliable alternative for RT-PCR. Am. J. Emerg. Med. 2020;38:2730–2732. doi: 10.1016/j.ajem.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu S., Gao Y., Niu Z., Jiang Y., Li L., Xiao X., Wang M., Fang E.F., Menpes-Smith W., Xia J. Weakly supervised deep learning for covid-19 infection detection and classification from ct images. IEEE Access. 2020;8:118869–118883. doi: 10.1109/ACCESS.2020.3005510. [DOI] [Google Scholar]
- 11.Farooq M., Hafeez A. Covid-resnet: a deep learning framework for screening of covid19 from radiographs. 2020. https://ui.adsabs.harvard.edu/abs/2020arXiv200314395F arXiv preprint arXiv:2003.14395.
- 12.Li J., Zhao G., Tao Y., Zhai P., Chen H., He H., Cai T. Multi-task contrastive learning for automatic CT and X-ray diagnosis of COVID-19. Pattern Recogn. 2021;114:107848. doi: 10.1016/j.patcog.2021.107848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meng H., Xiong R., He R., Lin W., Hao B., Zhang L., Lu Z., Shen X., Fan T., Jiang W. CT imaging and clinical course of asymptomatic cases with COVID-19 pneumonia at admission in Wuhan, China. J. Infect. 2020;81:e33–e39. doi: 10.1016/j.jinf.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ardakani A.A., Kwee R.M., Mirza-Aghazadeh-Attari M., Castro H.M., Kuzan T.Y., Altintoprak K.M., Besutti G., Monelli F., Faeghi F., Acharya U.R., Mohammadi A. A practical artificial intelligence system to diagnose COVID-19 using computed tomography: a multinational external validation study. Pattern Recogn. Lett. 2021;152:42–49. doi: 10.1016/j.patrec.2021.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rangarajan K., Muku S., Garg A.K., Gabra P., Shankar S.H., Nischal N., Soni K.D., Bhalla A.S., Mohan A., Tiwari P. European radiology; 2021. Artificial Intelligence–Assisted Chest X-Ray Assessment Scheme for COVID-19; pp. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai H.X., Hsieh B., Xiong Z., Halsey K., Choi J.W., Tran T.M.L., Pan I., Shi L.-B., Wang D.-C., Mei J. Performance of radiologists in differentiating COVID-19 from non-COVID-19 viral pneumonia at chest CT. Radiology. 2020;296:E46–E54. doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh Y., Park S., Ye J.C. Deep learning covid-19 features on cxr using limited training data sets. IEEE Trans. Med. Imag. 2020;39:2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 18.Nwosu L., Li X., Qian L., Kim S., Dong X. Semi-supervised learning for COVID-19 image classification via ResNet. 2021. https://ui.adsabs.harvard.edu/abs/2021arXiv210306140N arXiv preprint arXiv:2103.06140.
- 19.Gulati A. medRxiv; 2020. LungAI: A Deep Learning Convolutional Neural Network for Automated Detection of COVID-19 from Posteroanterior Chest X-Rays. [DOI] [Google Scholar]
- 20.Haritha D., Pranathi M.K., Reethika M. ICCCS); 2020. COVID Detection from Chest X-Rays with DeepLearning: CheXNet, 2020 5th International Conference on Computing, Communication and Security. [DOI] [Google Scholar]
- 21.Waheed A., Goyal M., Gupta D., Khanna A., Al-Turjman F., Pinheiro P.R. 2021. CovidGAN: Data Augmentation Using Auxiliary Classifier GAN for Improved Covid-19 Detection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rahaman M.M., Li C., Yao Y., Kulwa F., Zhao X. Identification of COVID-19 samples from chest X-Ray images using deep learning: a comparison of transfer learning approaches. J. X Ray Sci. Technol. 2020;28 doi: 10.3233/XST-200715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angelov P., Almeida Soares E. MedRxiv; 2020. SARS-CoV-2 CT-scan Dataset: A Large Dataset of Real Patients CT Scans for SARS-CoV-2 Identification. [DOI] [Google Scholar]
- 24.Alshazly H., Linse C., Barth E., Martinetz T. Explainable covid-19 detection using chest ct scans and deep learning. Sensors. 2021;21:455. doi: 10.3390/s21020455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gozes O., Frid-Adar M., Greenspan H., Browning P.D., Zhang H., Ji W., Bernheim A., Siegel E. Rapid ai development cycle for the coronavirus (covid-19) pandemic: initial results for automated detection & patient monitoring using deep learning ct image analysis. 2020. https://ui.adsabs.harvard.edu/abs/2020arXiv200305037G arXiv preprint arXiv:2003.05037.
- 26.Ouyang X., Huo J., Xia L., Shan F., Liu J., Mo Z., Yan F., Ding Z., Yang Q., Song B. Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans. Med. Imag. 2020;39:2595–2605. doi: 10.1109/tmi.2020.2995508. [DOI] [PubMed] [Google Scholar]
- 27.Qi S., Xu C., Li C., Tian B., Xia S., Ren J., Yang L., Wang H., Yu H. DR-MIL: deep represented multiple instance learning distinguishes COVID-19 from community-acquired pneumonia in CT images. Comput. Methods Progr. Biomed. 2021:106406. doi: 10.1016/j.cmpb.2021.106406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaurasia A., Culurciello E. 2017 IEEE Visual Communications and Image Processing (VCIP) IEEE; 2017. Linknet: exploiting encoder representations for efficient semantic segmentation; pp. 1–4. [DOI] [Google Scholar]
- 29.Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., Zha Y., Liang W., Wang C., Wang K. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181:1423–1433. doi: 10.1016/j.cell.2020.04.045. e1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark K., Vendt B., Smith K., Freymann J., Kirby J., Koppel P., Moore S., Phillips S., Maffitt D., Pringle M. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J. Digit. Imag. 2013;26:1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ronneberger O., Fischer P., Brox T. Springer; 2015. U-net: Convolutional Networks for Biomedical Image Segmentation, International Conference on Medical Image Computing and Computer-Assisted Intervention; pp. 234–241. [DOI] [Google Scholar]
- 32.Alom M.Z., Hasan M., Yakopcic C., Taha T.M., Asari V.K. Recurrent residual convolutional neural network based on U-net (R2U-net) for medical image segmentation. 2018. [DOI]
- 33.Oktay O., Schlemper J., Folgoc L.L., Lee M., Heinrich M., Misawa K., Mori K., McDonagh S., Hammerla N.Y., Kainz B. Attention u-net: learning where to look for the pancreas. 2018. https://ui.adsabs.harvard.edu/abs/2018arXiv180403999O arXiv preprint arXiv:1804.03999.
- 34.Zhou Z., Siddiquee M.M.R., Tajbakhsh N., Liang J. In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. DLMIA 2018, ML-CDS. Stoyanov D., et al., editors. vol. 11045. Springer; Cham: 2018. Unet++: a nested u-net architecture for medical image segmentation. Lecture Notes in Computer Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Z., Siddiquee M.M.R., Tajbakhsh N., Liang J. Unet++: redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans. Med. Imag. 2019;39:1856–1867. doi: 10.1109/TMI.2019.2959609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sabour S., Frosst N., Hinton G.E. NIPS; 2017. Dynamic Routing between Capsules. [Google Scholar]
- 37.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [DOI] [Google Scholar]
- 38.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Densely connected convolutional networks; pp. 4700–4708. [Google Scholar]
- 39.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Rethinking the inception architecture for computer vision; pp. 2818–2826. [Google Scholar]
- 40.Shaziya H., Shyamala K., Zaheer R. IEEE; 2018. Automatic Lung Segmentation on Thoracic CT Scans Using U-Net Convolutional Network, 2018 International Conference on Communication and Signal Processing (ICCSP) pp. 643–647. [Google Scholar]
- 41.Javaheri T., Homayounfar M., Amoozgar Z., Reiazi R., Homayounieh F., Abbas E., Laali A., Radmard A.R., Gharib M.H., Mousavi S.A.J. CovidCTNet: an open-source deep learning approach to diagnose covid-19 using small cohort of CT images. NPJ Digit. Med. 2021;4:1–10. doi: 10.1038/s41746-021-00399-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song J., Wang H., Liu Y., Wu W., Dai G., Wu Z., Zhu P., Zhang W., Yeom K.W., Deng K. End-to-end automatic differentiation of the coronavirus disease 2019 (COVID-19) from viral pneumonia based on chest CT. Eur. J. Nucl. Med. Mol. Imag. 2020;47:2516–2524. doi: 10.1007/s00259-020-04929-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abdel-Basset M., Hawash H., Moustafa N., Elkomy O.M. Pattern Recognition Letters; 2021. nnTwo-Stage Deep Learning Framework for Discrimination between COVID-19 and Community-Acquired Pneumonia from Chest CT Scans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S., Lau E.H., Wong J.Y. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J.M. Should the world collaborate imminently to develop neglected live attenuated vaccines for COVID‐19? J. Med. Virol. 2021 doi: 10.1002/jmv.27335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pandey K., Thurman M., Johnson S.D., Acharya A., Johnston M., Klug E.A., Olwenyi O.A., Rajaiah R., Byrareddy S.N. Mental health issues during and after COVID-19 vaccine era. Brain Res. Bull. 2021 doi: 10.1016/j.brainresbull.2021.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.López C.E.B. Detection of COVID-19 and other pneumonia cases using convolutional neural networks and X-ray images. Ing. Invest. 2022;42:e90289. doi: 10.15446/ing.investig.v42n1.90289. e90289. [DOI] [Google Scholar]
- 48.Hasan M.K., Alam M.A., Dahal L., Elahi M.T.E., Roy S., Wahid S.R., Marti R., Khanal B. medRxiv; 2020. Challenges of Deep Learning Methods for Covid-19 Detection Using Public Datasets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001017. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie H., Shan H., Cong W., Liu C., Zhang X., Liu S., Ning R., Wang G. Deep efficient end-to-end reconstruction (DEER) network for few-view breast CT image reconstruction. IEEE Access. 2020;8:196633–196646. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brenner D.J., Hall E.J. Computed tomography—an increasing source of radiation exposure. N. Engl. J. Med. 2007;357:2277–2284. doi: 10.1056/NEJMra072149. [DOI] [PubMed] [Google Scholar]
- 53.Subhalakshmi R., Balamurugan S.A.a., Sasikala S. Deep learning based fusion model for COVID-19 diagnosis and classification using computed tomography images. Concurr. Eng. 2021 doi: 10.1177/1063293X211021435. 1063293X211021435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhuyan H.K., Chakraborty C., Shelke Y., Pani S.K. COVID‐19 diagnosis system by deep learning approaches. Expet Syst. 2021 doi: 10.1111/exsy.12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim Y., Lee M.-J., Kim M.-J. Comparison of radiation dose from X-Ray, CT, and PET/CT in paediatric patients with neuroblastoma using a dose monitoring program. Eur. Congr. Radiol.-ECR 2015. 2015 doi: 10.5152/dir.2015.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., Cui J., Xu W., Yang Y., Fayad Z.A. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020;295:202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Z., Xiao Y., Li Y., Zhang J., Lu F., Hou M., Liu X. Automatically discriminating and localizing COVID-19 from community-acquired pneumonia on chest X-rays. Pattern Recogn. 2021;110:107613. doi: 10.1016/j.patcog.2020.107613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oulefki A., Agaian S., Trongtirakul T., Laouar A.K. Automatic COVID-19 lung infected region segmentation and measurement using CT-scans images. Pattern Recogn. 2021;114 doi: 10.1016/j.patcog.2020.107747. 107747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao C., Xu Y., He Z., Tang J., Zhang Y., Han J., Shi Y., Zhou W. Lung segmentation and automatic detection of COVID-19 using radiomic features from chest CT images. Pattern Recogn. 2021:108071. doi: 10.1016/j.patcog.2021.108071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bougourzi F., Contino R., Distante C., Taleb-Ahmed A. Recognition of COVID-19 from CT scans using two-stage deep-learning-based approach: CNR-IEMN. Sensors. 2021;21:5878. doi: 10.3390/s21175878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qiblawey Y., Tahir A., Chowdhury M.E., Khandakar A., Kiranyaz S., Rahman T., Ibtehaz N., Mahmud S., Maadeed S.A., Musharavati F. Detection and severity classification of COVID-19 in CT images using deep learning. Diagnostics. 2021;11:893. doi: 10.3390/diagnostics11050893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pawar S.P., Talbar S.N. LungSeg-Net: lung field segmentation using generative adversarial network. Biomed. Signal Process Control. 2021;64:102296. doi: 10.1016/j.bspc.2020.102296. [DOI] [Google Scholar]
- 63.Osadebey M., Andersen H.K., Waaler D., Fossaa K., Martinsen A.C., Pedersen M. Three-stage segmentation of lung region from CT images using deep neural networks. BMC Med. Imag. 2021;21:1–19. doi: 10.1186/s12880-021-00640-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu X., Lu S., Guo L., Wang S.-H., Zhang Y.-D. ResGNet-C: A graph convolutional neural network for detection of COVID-19. Neurocomputing. 2021;452:592–605. doi: 10.1016/j.neucom.2020.07.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang W., Tang P., Zhao L. Remote sensing image scene classification using CNN-CapsNet. Rem. Sens. 2019;11:494. doi: 10.3390/rs11050494. [DOI] [Google Scholar]
- 66.Paoletti M.E., Haut J.M., Fernandez-Beltran R., Plaza J., Plaza A., Li J., Pla F. Capsule networks for hyperspectral image classification. IEEE Trans. Geosci. Rem. Sens. 2018;57:2145–2160. doi: 10.1109/TGRS.2018.2871782. [DOI] [Google Scholar]
- 67.Wang C., Wu Y., Wang Y., Chen Y. Scene recognition using deep softpool capsule network based on residual diverse branch block. Sensors. 2021;21:5575. doi: 10.3390/s21165575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ucar F., Korkmaz D. COVIDiagnosis-Net: deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med. Hypotheses. 2020;140:109761. doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Monkam P., Qi S., Ma H., Gao W., Yao Y., Qian W. Detection and classification of pulmonary nodules using convolutional neural networks: a survey. IEEE Access. 2019;7:78075–78091. doi: 10.1109/ACCESS.2019.2920980. [DOI] [Google Scholar]
- 70.Zhang B., Qi S., Monkam P., Li C., Yang F., Yao Y.-D., Qian W. Ensemble learners of multiple deep CNNs for pulmonary nodules classification using CT images. IEEE Access. 2019;7:110358–110371. doi: 10.1109/ACCESS.2019.2933670. [DOI] [Google Scholar]
- 71.Wu Q., Wang S., Li L., Wu Q., Qian W., Hu Y., Li L., Zhou X., Ma H., Li H. Radiomics analysis of computed tomography helps predict poor prognostic outcome in COVID-19. Theranostics. 2020;10:7231. doi: 10.7150/thno.46428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mu N., Wang H., Zhang Y., Jiang J., Tang J. Progressive global perception and local polishing network for lung infection segmentation of COVID-19 CT images. Pattern Recogn. 2021;120:108168. doi: 10.1016/j.patcog.2021.108168. [DOI] [PMC free article] [PubMed] [Google Scholar]