Abstract
Examining volumetric differences of the amygdala and anterior-posterior regions of the hippocampus is important for understanding cognition and clinical disorders. However, the gold standard manual segmentation of these structures is time and labor-intensive. Automated, accurate, and reproducible techniques to segment the hippocampus and amygdala are desirable. Here, we present a hierarchical approach to multi-atlas segmentation of the hippocampus head, body and tail and the amygdala based on atlases from 195 individuals. The Open Vanderbilt Archive of the temporal Lobe (OVAL) segmentation technique outperforms the commonly used FreeSurfer, FSL FIRST, and whole-brain multi-atlas segmentation approaches for the full hippocampus and amygdala and nears or exceeds inter-rater reproducibility for segmentation of the hippocampus head, body and tail. OVAL has been released in open-source and is freely available.
Keywords: hippocampus, amygdala, temporal lobe, multi-atlas segmentation
1. Introduction
The hippocampus and amygdala are critically important structures for learning, memory and emotion. Volumetric changes in the hippocampus have been linked to neurological and psychiatric disorders including Alzheimer’s disease [1], epilepsy [2], schizophrenia [3], and depression [4]. Although most studies to date have focused on identifying changes in overall volume, the hippocampus is not a unitary structure. It can be divided along the transverse axis into subfields (CA1-4, dentate gyrus and subiculum) and along the longitudinal axis into anterior (head) and posterior (body and tail) subregions. The anterior and posterior hippocampal subregions differ in structural connectivity, function, and gene expression [5–7]. A growing body of work has begun to demonstrate the significance of structural variation along the longitudinal axis of the hippocampus in basic cognitive functions and across development [8–10]. Current models suggest that the posterior regions of the hippocampus are specialized for “fine-grained” information and spatial processing, whereas the anterior hippocampus is involved in emotional or global information processing and scene construction [5, 7]. The amygdala is an adjacent medial temporal structure that is preferentially connected to the anterior hippocampus [11], consistent with a role for both structures in emotional and motivational processing [5, 12]. Structural changes in the amygdala have been identified in schizophrenia [3] and depression [13]. Despite the importance of understanding the nature and implications of structural variation in these medial temporal lobe regions, there are methodological barriers to accurate volumetric measurement of both structures.
The gold standard for quantification of temporal lobe volumes has traditionally depended upon labor-intensive manual segmentation, requiring approximately 3 hours per subject for the hippocampus and amygdala. However, the advancement of large-scale and longitudinal imaging studies such as the Human Connectome Project, Alzheimer’s Disease Neuroimaging Initiative, and UK BioBank necessitates the development of low-cost, reliable automated segmentation methods. Several open-source, automatic techniques have been developed for segmentation of the hippocampus and amygdala using standard, T1-weighted MR images. One of the most common techniques, FreeSurfer, reconstructs the cortical surface and subcortical brain structures with an energy model [14]. Another common technique is FSL FIRST, which uses a Bayesian shape and appearance model to segment subcortical structures [15]. Approaches like FSL FIRST and Freesurfer tend to result in less accurate segmentations since they only incorporate one atlas and thus are more susceptible to biases and errors in registration. A third approach is a multi-atlas segmentation [16–18]. Other approaches, such as Automated Segmentation of Hippocampal Subfields (ASHS) delineate the hippocampus and its subfields, but require collection of an additional MR sequence outside of a standard T1-weighted MR protocol [19].
Multi-atlas segmentation (MAS) can provide a robust and accurate segmentation of a target image [16]. Previous works have used MAS to segment a wide variety of anatomical targets, from the whole brain / cortex and optic nerve to the abdomen and other structures [17, 20–22]. A typical multi-atlas segmentation procedure involves non-rigidly registering ten or more atlases, image volumes paired with expertly labeled structures of interest, to a target image to be segmented. These registered target images are then joined together to create a representation that is more accurate than any individually registered atlas. One typical assumption of many studies is that 30 atlases are sufficient to produce a maximally accurate segmentation [16].
In this work, we present the Open Vanderbilt Archive of the temporal Lobe (OVAL). OVAL is a fully automated segmentation approach using 195 atlases to produce an accurate segmentation of the hippocampus head, body, and tail and the amygdala. Briefly, OVAL uses a whole-brain multi-atlas segmentation method to localize the hippocampus and amygdala. OVAL then registers the 195 atlases to the localized target images and fuses them following a standard MAS protocol. Secondarily, OVAL allows us to test the assumption that 30 atlases are enough for optimal multi-atlas segmentation, and we show that 30 atlases produce inferior results to using the entire population of 195 atlases. These data contributed in de-identified form to the MICCAI Medical Segmentation Decathlon Challenge (http://medicaldecathlon.com).
2. Methods
2.1. Overview
The OVAL algorithm produces segmentations of target images using 195 atlases of the hippocampus and amygdala. The atlases are generated from 195 manually delineated hippocampi (dataset 1) and automatically segmented amygdalae defined from training data in a second population of 35 subjects with manually delineated amygdalae (dataset 2). Briefly, the 195 subjects with manual hippocampus segmentations were segmented with the 35 amygdala atlases following the protocol outlined below. These atlases are then cropped to a bounding box around each temporal lobe, resulting in 195 left and 195 right hippocampus and amygdala atlases. For a given target image, the atlases are used in a MAS framework to segment the amygdala, hippocampus head, and posterior. Finally, an anatomical landmark defined from whole-brain segmentation is used to split the hippocampus posterior into the body and tail. All processing steps in this algorithm are done automatically and presented in an open source implementation.
2.2. Subjects
Dataset 1 consisted of MR images acquired in 90 healthy adults and 105 adults with a non-affective psychotic disorder (56 schizophrenia; 32 schizoaffective disorder; 17 schizophreniform disorder) taken from the Psychiatric Genotype/Phenotype Project data repository at Vanderbilt University Medical Center (Table 1). Patients were recruited from the Vanderbilt Psychotic Disorders Program and controls were recruited from the surrounding community. All participants were assessed with the Structured Clinical Interview for DSM-IV [23] New York: Biometrics Research, New York State Psychiatric Institute (2002). Dataset 2 included 35 subjects recruited as part of a study on temperament (Clauss et al., 2014). All subjects were free from significant medical or neurological illness, head injury and active substance use or dependence. The Vanderbilt University Institutional Review Board approved both studies.
Table 1.
Subject demographics.
| Dataset 1: Hippocampus | Dataset 2: Amygdala | |||
|---|---|---|---|---|
| Psychosis | Control | |||
| N | 105 | 90 | 18 | 17 |
| Age, years (Mean ± SD) | 34.62 ± 12.38 | 33 ± 11.33 | 24.6 ± 5.1 | 23.6 ± 4.8 |
| Gender (Female/Male) | 37/68 | 41/49 | 12/6 | 10/7 |
| Race (White/Black/Other) | 63/37/5 | 60/26/4 | 13/1/3 | 15/2/0 |
2.3. Image Acquisition and Manual Tracing
Structural images were acquired with a 3D T1-weighted MPRAGE sequence (TI/TR/TE = 860/8.0/3.7 ms; 170 sagittal slices; voxel size = 1.0mm3). All images were collected on a Philips Achieva scanner (Philips Healthcare, Inc., Best, The Netherlands). Manual tracing of the head, body, and tail of the hippocampus on images from dataset 1 was completed following a previously published protocol [8, 24]. For the purposes of this study, the term hippocampus includes the hippocampus proper (CA1-4 and dentate gyrus) and parts of the subiculum, together more often termed the hippocampal formation [25]. The last slice of the head of the hippocampus was defined as the coronal slice containing the uncal apex. The resulting 195 labeled images are hereafter referred to as hippocampus atlases. Note that the term hippocampus posterior refers to the union of the body and the tail. Manual delineation of the amygdala on images from dataset 2 was completed as described in [26]. The 35 labeled images from dataset 2 are hereafter referred to as amygdala atlases.
2.3. Whole-Brain Segmentation
Whole-brain segmentation (WBS) was carried out on target images from dataset 1. First, 45 atlases labeled with the BrainCOLOR protocol (www.neuromorphometrics.com) were affinely registered to each target image with Niftyreg [27]. The 15 atlases geodesically closest to the target were selected and these atlases were non-rigidly registered to the target image using the Advanced Normalization Tools (ANTs) Symmetric Normalization (SyN) algorithm [28]. The 15 registered atlases were fused with the hierarchical Non-Local Spatial STAPLE algorithm [17, 29, 30]. Finally, the segmentation was refined with corrected learning following [31]. The resulting segmentation contained 132 labels including the hippocampus and amygdala in both hemispheres, along with 98 other cortical structures. This segmentation acts as a guiding mechanism for segmentation of the hippocampus and amygdala.
2.4. Atlas Creation
To create a set of atlases labeled with both the amygdala and hippocampus, the 35 amygdala atlases were non-rigidly registered to the 195 hippocampus atlases and the registered atlases were fused with joint label fusion (JLF). The resulting amygdala labels were added to the hippocampus atlases where they did not conflict with manual hippocampus labels.
The hippocampus atlases were then segmented with the WBS described in §2.3. For each atlas, the WBS was used to determine a bounding box around the hippocampus and amygdala for each hemisphere; the bounding box was dilated 5mm in each direction to assure the full true hippocampus and amygdala was included. The bounding box was then used to extract the atlas image and label volume localized to the region around the hippocampus and amygdala. This resulted in 195 hippocampus and amygdala (HA) atlases for each hemisphere.
2.5. OVAL Segmentation
The OVAL method results in lateralized segmentations of the amygdala, hippocampus head, body, and tail. The algorithm requires an input T1-weighted MRI volume and a WBS. First, the input T1-weighted volume is cropped to the left hippocampus and amygdala by its WBS, herein the left target image. The 195 left HA atlases are non-rigidly registered to the left target image with NiftyReg and the ANTs SyN algorithm [27, 28]. The atlases are fused with JLF and the posterior probability volumes for amygdala, hippocampus head, and posterior are considered [32]. At voxels where the sum of the probability of these labels exceeds 0.5, the label of these three with the highest probability is chosen
where Li is the label decision at voxel i, is the sum of the probability of amygdala, hippocampus head, and posterior at I, is the probability of label X at i, and S is the set of labels of interest, amygdala, hippocampus head, and hippocampus posterior. This correction primarily applies to voxels near the boundary of two structures, for instance the hippocampus head and posterior, where JLF shows a posterior probability less than 0.5 for the background, but the probability of the head or posterior does not exceed the probability of background. For instance, a case where the probability of hippocampus head is 0.35, hippocampus posterior is 0.25, and background is 0.4. This procedure is then repeated for the right hippocampus.
After the segmentation of the amygdala, hippocampus head, and hippocampus posterior is complete, the final step in the segmentation is splitting the posterior into the body and tail. For the left hippocampus, the most posterior point on the left thalamus is identified from the WBS by finding the point on the thalamus nearest to the mean location of the left occipital lobe. Next, a line is fit through the coordinates of the voxels of the full hippocampus, defined by the OVAL segmentation. Lastly, a plane is fit through the posterior point of the thalamus, orthogonal to the line through the hippocampus. The points of the posterior hippocampus posterior to the plane are then defined as the tail and the points anterior to the plane defined as the body.
2.6. Experimental Design
Three experiments were considered to test the accuracy of OVAL compared with other segmentation approaches. First, we compared OVAL’s accuracy for whole hippocampal segmentation to manual segmentation by two expert human raters. A set of 10 atlases, distinct from the training population, was labeled with the hippocampus segmentation protocol (§2.2) by two expert raters, creating a human rater reproducibility dataset, herein the hippocampus testing atlases. A separate set of 35 atlases, distinct from the training population and the hippocampus reproducibility population, was labeled with the amygdala protocol (§2.2), herein the amygdala testing atlases. These two datasets were segmented with the WBS described in (§2.3), FreeSurfer (v5.1.0), FSL (v5.0) FIRST, OVAL with 30 atlases (OVAL-30), and OVAL with 195 atlases (OVAL) to test OVAL’s accuracy. Second, we evaluated the accuracy of OVAL in identifying the hippocampus head, body and tail on the 10 hippocampus testing atlases with OVAL-30 and OVAL to test OVAL’s accuracy on the hippocampus head, body, and tail. Finally, the BrainCOLOR, FreeSurfer, and FSL FIRST segmentation approaches do not use exactly the same labeling protocol as the manual segmentations used to train OVAL [33]. Thus, we cannot necessarily conclude that OVAL is a better approach than the other techniques. The Kirby21 multi-modal reproducibility dataset is a set of 21 subjects scanned twice in immediate succession. We used the Kirby21 dataset to compare OVAL’s intra-subject reproducibility of the amygdala and whole hippocampus segmentation to BrainCOLOR, FreeSurfer, and FSL FIRST.
3. Results
3.1. Whole Hippocampus and Amygdala Segmentation
The hippocampus testing atlases and amygdala testing atlases were first segmented with the WBS, identified as BrainCOLOR in figures and results. These atlases were segmented with FreeSurfer using their standard reconstruction and FSL FIRST with standard parameters. Finally, the atlases were segmented with OVAL and OVAL-30. The OVAL hippocampus segmentations were reduced to whole hippocampus by collapsing hippocampus head, body, and tail into one label. Dice similarity coefficient (DSC) and mean surface distance (MSD) were calculated between each segmentation and the hippocampus and amygdala testing atlases.
For DSC of the hippocampus testing atlases, OVAL outperformed FSL FIRST, FreeSurfer, BrainCOLOR, OVAL-30, and human reproducibility for left hippocampus for rater 1, left hippocampus rater 2, and right hippocampus rater 2 (p<0.05 Wilcoxon sign-rank test; Figure 1). For right hippocampus rater 1, OVAL and human reproducibility performed comparably and outperformed other techniques (p<0.05 Wilcoxon sign-rank test). For MSD of the hippocampus testing atlases, human reproducibility outperformed other techniques for the left and right hippocampus for rater 1, OVAL-30 and OVAL outperformed all other automated techniques (p<0.05 Wilcoxon sign-rank test; Figure 1). For the left and right hippocampus for rater 2, OVAL-30, OVAL, and human reproducibility all performed statistically similarly and outperformed all other techniques (p<0.05 Wilcoxon sign-rank test; Figure 1).
Figure 1:
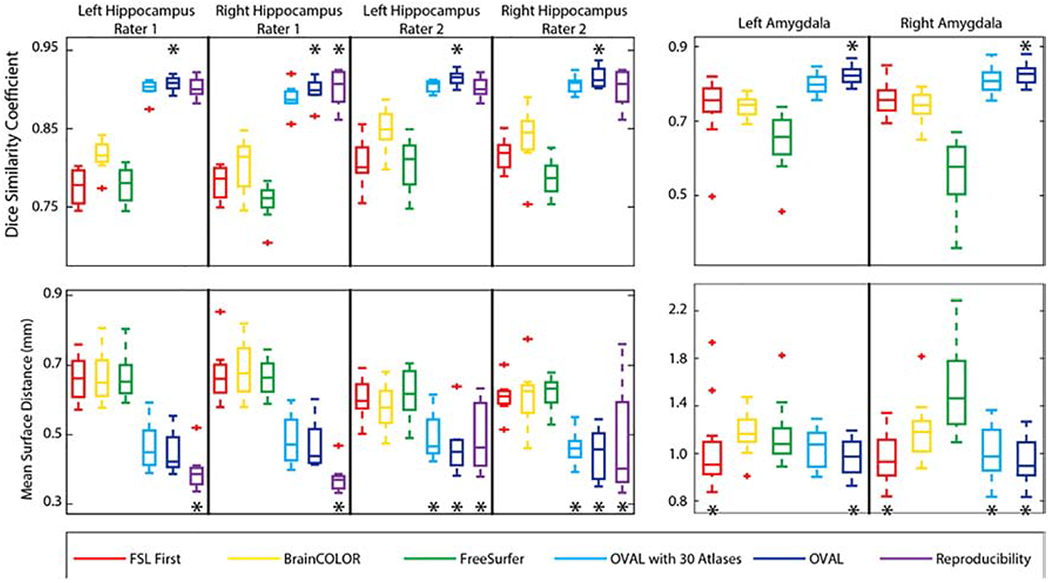
Quantitative segmentation results for the whole hippocampus and amygdala. OVAL outperforms all other segmentation techniques in terms of DSC for the left hippocampus in both raters, the right hippocampus in rater 2, and the left and right amygdala (p<0.05, *). OVAL outperforms all other techniques for the right hippocampus of rater 1 except human reproducibility, which performs comparably statistically. Human reproducibility outperforms all other techniques in MSD for the left and right hippocampus for rater 1 (p<0.05, *). OVAL and OVAL-30 outperform all other automated techniques for those structures. OVAL, OVAL-30, and human reproducibility perform statistically comparable for the left and right hippocampus for rater 2 and outperform all other techniques (p<0.05, *). OVAL and FSL FIRST perform statistically similarly for the left amygdala and outperform all other segmentation approaches (p<0.05, *). OVAL, OVAL-30, and FSL FIRST perform statistically similarly for the right amygdala and outperform all other segmentation approaches (p<0.05, *).
For DSC of the amygdala testing atlases, OVAL outperformed FSL FIRST, FreeSurfer, BrainCOLOR, and OVAL-30 for both left and right amygdala (p<0.05 Wilcoxon sign-rank test). For MSD of the amygdala testing atlases, OVAL and FSL FIRST outperformed BrainCOLOR, FreeSurfer, and OVAL-30 for the left amygdala (p<0.05 Wilcoxon sign-rank test; Figure 1). For the right amygdala, OVAL, OVAL-30, and FSL FIRST all performed statistically similarly and outperformed BrainCOLOR and FreeSurfer (p<0.05 Wilcoxon sign-rank test; Figure 1). For MSD of the amygdala, FSL FIRST and OVAL resulted in a significantly lower MSD for the left amygdala compared with FreeSurfer, BrainCOLOR, and OVAL-30; for the right amygdala, FSL FIRST, OVAL-30, and OVAL resulted in a significantly lower MSD than BrainCOLOR and FreeSurfer (p<0.05; Figure 1).
3.2. Hippocampus Head, Body, Tail Segmentation
The hippocampus testing atlases were segmented with OVAL, following §2.5, and OVAL-30. Since no other approach provides a segmentation of the hippocampus head, body, and tail, only these two approaches were compared against reproducibility. Since the hippocampus testing atlases only segmented the head and posterior, the tail was split from the body following the protocol in §2.6. For simplicity, only the results with respect to rater 1 are presented (Figure 2).
Figure 2:
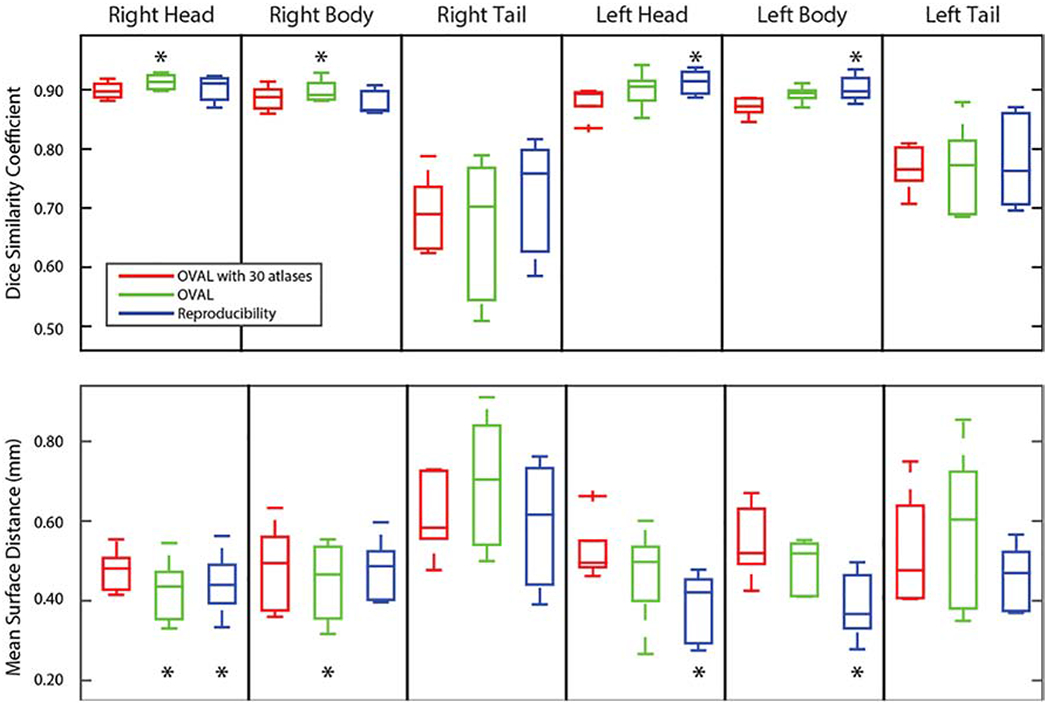
Quantitative segmentation results for the whole hippocampus head, body, and tail. OVAL outperformed OVAL-30 and human reproducibility on the right head and body in Dice Similarity Coefficient (p<0.05; *). No technique showed significant improvement on the right or left tail. Human reproducibility outperformed OVAL and OVAL-30 (p<0.05, *), though OVAL outperformed OVAL-30. In mean surface distance, OVAL and human reproducibility outperformed OVAL-30 for the right head, OVAL outperformed all other techniques for the right body, no technique outperformed any other for the right and left tail, and human reproducibility outperformed the other techniques for the left and right head (p<0.05, *).
For the right head and posterior, OVAL significantly outperformed human reproducibility and OVAL-30 in DSC (p<0.05 Wilcoxon sign-rank test). OVAL significantly outperformed human reproducibility and OVAL-30 in MSD of the right head and OVAL and human reproducibility outperformed OVAL-30 in MSD of the right tail (p<0.05 Wilcoxon sign-rank test). For the left head and posterior, human reproducibility outperformed OVAL and OVAL-30 in DSC and MSD, though OVAL outperformed OVAL-30 on both of these structures (p<0.05 Wilcoxon sign-rank test). For the right and left tail, no technique performed significantly better in DSC and MSD.
3.3. Whole Hippocampus and Amygdala Reproducibility
We examined the intra-subject reproducibility of OVAL compared to BrainCOLOR, FreeSurfer, and FSL FIRST segmentation approaches using the Kirby21 multi-modal reproducibility dataset. The two T1-weighted MPRAGEs for each subject were segmented with the BrainCOLOR multi-atlas segmentation (§2.3), FreeSurfer, FSL FIRST, and OVAL. To assess the reproducibility of each technique, the volume of the amygdala and whole hippocampus were calculated. The average volume () and absolute percent volume difference () between each scanning session was calculated for each subject. The percent volume difference of OVAL was significantly lower than all other techniques for all structures (p<0.05 Wilcoxon sign-rank test; Figure 3). In the hippocampus, OVAL had an average percent volume difference of 0.75 and 0.66 for the left and right, respectively. For the amygdala, OVAL had an average percent volume similarity of 2.67 and 3.10 for the left and right, respectively.
Figure 3:
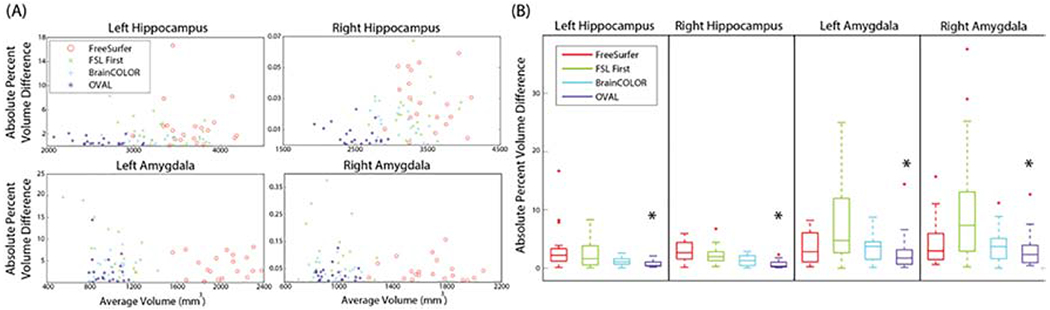
Reproducibility results for segmentation of the full hippocampus and amygdala on the Kirby 21 multi-modal reproducibility dataset. There was no effect shown of volume on percent volume difference (A). OVAL produced the lowest percent volume difference between reproducibility segmentations for all structures (B, p<0.05, *).
3.4. Full Brain Hippocampus and Amygdala Segmentation
Finally, we examined the impact of using a full brain registration and segmentation as opposed to the cropped segmentation proposed in §2.5. For the full brain segmentation, the same registration and segmentation algorithms were used as in §2.5 with the complete set of 195 atlases. For the head and body of the hippocampus, the full brain segmentation did not result in a significant change in DSC (p>0.05 Wilcoxon sign-rank test; Figure 4). For the amygdala, the full brain segmentation produced a significant improvement in DSC (p<0.05 Wilcoxon sign-rank test) likely due to difference in amygdala protocol between the BrainCOLOR protocol used in §2.3 and §2.5 for the cropping region identification. The full brain segmentation with 195 atlases took over 200 hours when run in serial whereas the OVAL segmentation took under 90 minutes.
Figure 4:
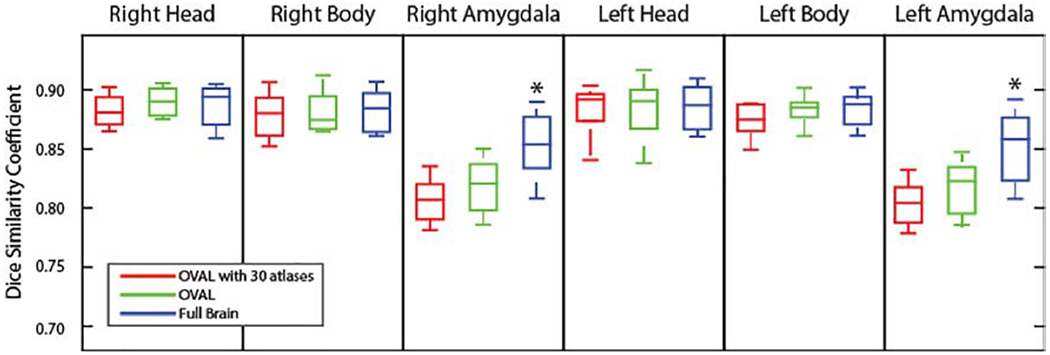
Quantitative segmentation results for the hippocampus head and body and the amygdala comparing OVAL and OVAL with 30 atlases to a full brain segmentation. OVAL and the full brain segmentation did not show significant differences in DSC (p>0.05 Wilcoxon sign-rank test) for either the head or body of the hippocampus. On the other hand, the full brain segmentation resulted in a significant increase in DSC for both side of the amygdala (p<0.05 Wilcoxon sign-rank test; *).
4. Discussion
In this work, we presented the OVAL algorithm for segmentation of the hippocampus and amygdala. First, we presented labeling protocols for the hippocampus head and body and the amygdala. Second, we created an atlas population of 195 subjects with manually traced hippocampi and automatically segmented amygdalae. Third, we presented the OVAL segmentation algorithm which, for a given target image, uses an initialization of the temporal lobe from a whole-brain segmentation to efficiently perform the segmentation.
Qualitatively, the OVAL segmentation tends to produce a segmentation of the amygdala with smoother boundaries than the atlas definitions since several of the atlas boundaries are defined by global landmarks instead of boundaries visible in contrast (Figure 5). In general, OVAL performed comparably with human reproducibility and outperformed OVAL-30. Qualitatively, OVAL segmentations are typically within 1mm of the manual segmentation at all voxels including the boundary between the body and head (Figure 6). Third, since FreeSurfer, FSL FIRST, BrainCOLOR, and OVAL use different segmentation protocols, these segmentation techniques were evaluated for reproducibility with the Kirby21 multi-modal reproducibility dataset. OVAL showed the lowest average percent volume difference of any technique, indicating that it is the most reproducible of any algorithm tested.
Figure 5:
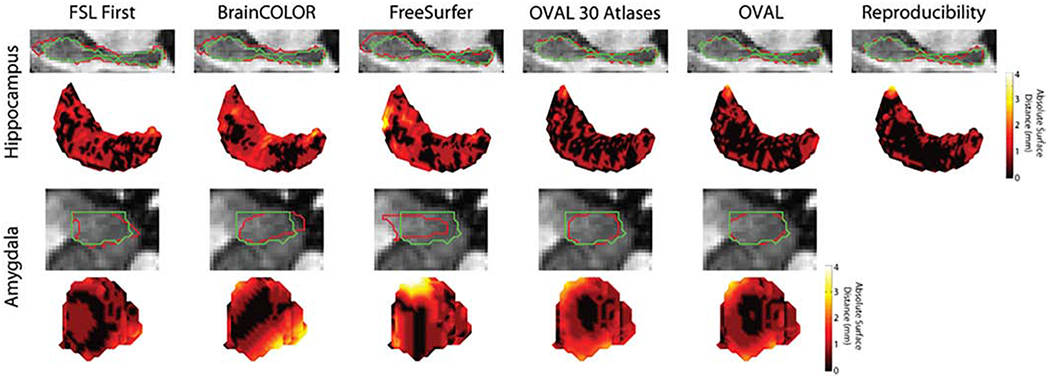
Median qualitative segmentation results for the whole hippocampus and amygdala; red represents the estimated segmentation and green is the manual label. FSL FIRST, BrainCOLOR, and FreeSurfer all showed large surface distances up to 4mm for both the hippocampus and amygdala. OVAL and OVAL-30 were typically within 1mm distance on the hippocampus, though OVAL produced more consistent results than OVAL with 30 atlases. On the amygdala, OVAL and OVAL-30 captured the overall contour of the amygdala.
Figure 6:
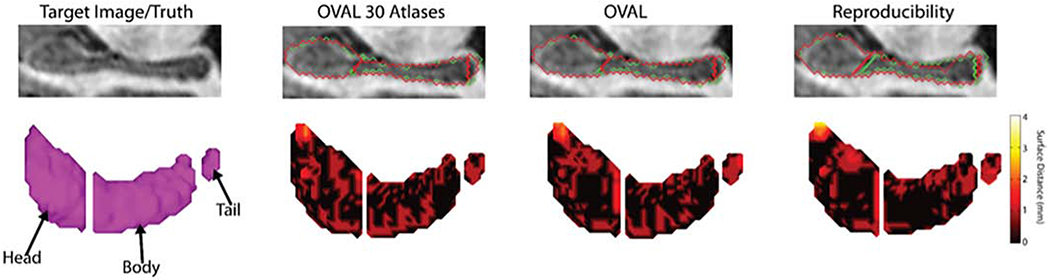
Median qualitative segmentation results for the hippocampus head, body, and tail. Green represents the true segmentation and red represents the estimate. Human reproducibility defined a different point for the head/body split and rater 2 under-segmented the tail of the hippocampus and the tip of the head compared with rater 1. OVAL-30 produced more local errors than OVAL. Images were rotated along the axis of the hippocampus, gaps between the head, body, and tail are exaggerated for visualization.
Limitations of this work include the variance in human understanding and definition of temporal lobe labels and the generalizability of the subject populations. OVAL was trained on data from the indicated manual labeling protocols. When compared against prior methods, OVAL is superior due to the size and quality of the dataset and the slight improvements in foreground/background segmentation. However, the prior methods were trained on datasets that were not labeled by the same manual raters, hence, OVAL has an intrinsic advantage. To help enable future fair comparisons, we have publicly released the temporal lobe training and testing data from these studies. The age range of OVAL atlases spans the relatively young adult range. Future studies would be needed to examine the applicability of the OVAL approach in children and elderly populations and populations with significant noise and signal artifacts. While we consider both healthy and patient groups in OVAL, the robustness is yet to prove. OVAL can be employed to numerous works that compare psychotic disorder patients and healthy control participants to study the early Stage of Psychosis for early diagnosis and understand the prognostic of the clinical outcome on the Hippocampus and Amygdala [34–39]. Another future step is to expand OVAL with a new imaging modality [40] to segment hippocampal subfields, as various research groups have focused on the disease-related changes of hippocampal subfields morphology in psychotic disorders [41–45]. The next promising future direction is integrating OVAL and cortical reconstruction [46] to study shape differences of the hippocampus in schizophrenia [47–49].
With the advent of large-scale cross-sectional and longitudinal neuroimaging studies [50, 51], there is a strong need for highly reproducible, low-cost structural image analysis methods. Detailed, single-subject level characterization of the hippocampus and amygdala is critical for understanding the pathophysiology of many psychiatric and neurological diseases. As shown in the current work, these structures have proven challenging for previously available automated segmentation approaches. OVAL presents an accurate and reproducible segmentation of the hippocampus and amygdala, two of the most studied structures in the human brain. Furthermore, this work demonstrates that the use of 30 atlases is insufficient to produce optimal segmentations of these structures. The OVAL algorithm and atlases are available in open source at https://www.nitrc.org/projects/oval/.
Goals:
Present labeling protocols for the hippocampus head and body and amygdala.
created an atlas population of 195 subjects with manually traced hippocampi and automatically segmented amygdalae
presents presented the OVAL segmentation algorithm which, for a given target image, uses an initialization of the temporal lobe from a whole-brain segmentation to efficiently perform the segmentation
6. Acknowledgements
This research was supported by NSF CAREER 1452485 (Landman), R01NS095291 (Dawant), R01MH70560 (Heckers), K01MH083052 (Blackford), T32LM012412 (Malin), and the Charlotte and Donald Test Fund. This research was conducted with the support from Intramural Research Program, National Institute on Aging, NIH. This study was also supported by NIH 5R01NS056307, 5R21NS082891 and in part using the resources of the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University, Nashville, TN. This project was supported in part by the National Center for Research Resources, Grant UL1 RR024975-01, and is now at the National Center for Advancing Translational Sciences, Grant 2 UL1 TR000445-06. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
5 Disclosures
No conflicts of interest, financial or otherwise, are declared by the authors.
7 References
- [1].Barnes J et al. , “A meta-analysis of hippocampal atrophy rates in Alzheimer’s disease,” Neurobiol Aging, vol. 30, no. 11, pp. 1711–1723, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Keller SS and Roberts N, “Voxel - based morphometry of temporal lobe epilepsy: An introduction and review of the literature,” Epilepsia, vol. 49, no. 5, pp. 741–757, 2008. [DOI] [PubMed] [Google Scholar]
- [3].van Erp TG et al. , “Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium,” Molecular psychiatry, vol. 21, no. 4, pp. 547–553, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Arnone D, McIntosh A, Ebmeier K, Munafò M, and Anderson I, “Magnetic resonance imaging studies in unipolar depression: systematic review and meta-regression analyses,” European Nearopsychopharmacology, vol. 22, no. 1, pp. 1–16, 2012. [DOI] [PubMed] [Google Scholar]
- [5].Poppenk J, Evensmoen HR, Moscovitch M, and Nadel L, “Long-axis specialization of the human hippocampus,” Trends in cognitive sciences, vol. 17, no. 5, pp. 230–240, 2013. [DOI] [PubMed] [Google Scholar]
- [6].Strange BA, Witter MP, Lein ES, and Moser EI, “Functional organization of the hippocampal longitudinal axis,” Nature Reviews Neuroscience, vol. 15, no. 10, pp. 655–669, 2014. [DOI] [PubMed] [Google Scholar]
- [7].Zeidman P and Maguire EA, “Anterior hippocampus: the anatomy of perception, imagination and episodic memory,” Nature Reviews Neuroscience, vol. 17, no. 3, pp. 173–182, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Woolard AA and Heckers S, “Anatomical and functional correlates of human hippocampal volume asymmetry,” Psychiatry Research: Neuroimaging, vol. 201, no. 1, pp. 48–53, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].DeMaster D, Pathman T, Lee JK, and Ghetti S, “Structural development of the hippocampus and episodic memory: developmental differences along the anterior/posterior axis,” Cerebral cortex, vol. 24, no. 11, pp. 3036–3045, 2013. [DOI] [PubMed] [Google Scholar]
- [10].Lee JK, Johnson EG, and Ghetti S, “Hippocampal Development: Structure, Function and Implications,” in The Hippocampus from Cells to Systems: Springer, 2017, pp. 141–166. [Google Scholar]
- [11].McDonald AJ and Mott DD, “Functional neuroanatomy of amygdalohippocampal interconnections and their role in learning and memory,” Journal of neuroscience research, vol. 95, no. 3, pp. 797–820, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Janak PH and Tye KM, “From circuits to behaviour in the amygdala,” Nature, vol. 517, no. 7534, pp. 284–292, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hamilton JP, Siemer M, and Gotlib IH, “Amygdala volume in major depressive disorder: a meta-analysis of magnetic resonance imaging studies,” ed: Nature Publishing Group, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fischl B, “FreeSurfer,” Neuroimage, vol. 62, no. 2, pp. 774–781, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Patenaude B, Smith SM, Kennedy DN, and Jenkinson M, “A Bayesian model of shape and appearance for subcortical brain segmentation,” Neuroimage, vol. 56, no. 3, pp. 907–922, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Iglesias JE and Sabuncu MR, “Multi-Atlas Segmentation of Biomedical Images: A Survey,” arXiv preprint arXiv:1412.3421, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Asman AJ and Landman BA, “Hierarchical performance estimation in the statistical label fusion framework,” Med Image Anal, vol. 18, no. 7, pp. 1070–81, October 2014, doi: 10.1016/j.media.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Collins DL and Pruessner JC, “Towards accurate, automatic segmentation of the hippocampus and amygdala from MRI by augmenting ANIMAL with a template library and label fusion,” Neuroimage, vol. 52, no. 4, pp. 1355–1366, 2010. [DOI] [PubMed] [Google Scholar]
- [19].Yushkevich PA et al. , “Nearly automatic segmentation of hippocampal subfields in in vivo focal T2-weighted MRI,” Neuroimage, vol. 53, no. 4, pp. 1208–1224, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Harrigan RL et al. , “Robust optic nerve segmentation on clinically acquired computed tomography,” Journal of Medical Imaging, vol. 1, no. 3, pp. 034006–034006, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Panda S et al. , “Evaluation of multiatlas label fusion for in vivo magnetic resonance imaging orbital segmentation,” Journal of Medical Imaging, vol. 1, no. 2, pp. 024002–024002, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Xu Z et al. , “Efficient multi-atlas abdominal segmentation on clinically acquired CT with SIMPLE context learning,” Medical image analysis, vol. 24, no. 1, pp. 18–27, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].First MB, Spitzer RL, Gibbon M, and Williams JB, “Structured clinical interview for DSM-IV-TR axis I disorders, research version, patient edition,” SCID-I/P, 2002. [Google Scholar]
- [24].Pruessner J et al. , “Volumetry of hippocampus and amygdala with high-resolution MRI and three-dimensional analysis software: minimizing the discrepancies between laboratories,” Cerebral cortex, vol. 10, no. 4, pp. 433–442, 2000. [DOI] [PubMed] [Google Scholar]
- [25].Amaral DG and Witter M, “The three-dimensional organization of the hippocampal formation: a review of anatomical data,” Neuroscience, vol. 31, no. 3, pp. 571–591, 1989. [DOI] [PubMed] [Google Scholar]
- [26].Blackford JU, Clauss JA, Avery SN, Cowan RL, Benningfield MM, and VanDerKlok RM, “Amygdala–cingulate intrinsic connectivity is associated with degree of social inhibition,” Biological psychology, vol. 99, pp. 15–25, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ourselin S, Roche A, Subsol G, Pennec X, and Ayache N, “Reconstructing a 3D structure from serial histological sections,” Image and vision computing, vol. 19, no. 1, pp. 25–31,2001. [Google Scholar]
- [28].Avants BB, Tustison NJ, Song G, Cook PA, Klein A, and Gee JC, “A reproducible evaluation of ANTs similarity metric performance in brain image registration,” (in eng), NeuroImage, Evaluation Studies Research Support, N.I.H., Extramural vol. 54, no. 3, pp. 2033–44, February 1 2011, doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Asman AJ and Landman BA, “Formulating spatially varying performance in the statistical fusion framework,” IEEE transactions on medical imaging, vol. 31, no. 6, pp. 1326–36, June 2012, doi: 10.1109/TMI.2012.2190992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Asman AJ and Landman BA, “Non-local statistical label fusion for multi-atlas segmentation,” Med Image Anal, vol. 17, no. 2, pp. 194–208, February 2013, doi: 10.1016/j.media.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang H et al. , “A learning-based wrapper method to correct systematic errors in automatic image segmentation: consistently improved performance in hippocampus, cortex and brain segmentation,” NeuroImage, vol. 55, no. 3, pp. 968–985, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang H, Suh JW, Das SR, Pluta JB, Craige C, and Yushkevich PA, “Multi-atlas segmentation with joint label fusion,” Pattern Analysis and Machine Intelligence, IEEE Transactions on, vol. 35, no. 3, pp. 611–623, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Boccardi M et al. , “Survey of protocols for the manual segmentation of the hippocampus: preparatory steps towards a joint EADC-ADNI harmonized protocol,” Journal of Alzheimer’s Disease, vol. 26, no. s3, pp. 61–75, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Roeske MJ, McHugo M, Vandekar S, Blackford JU, Woodward ND, and Heckers S, “Incomplete hippocampal inversion in schizophrenia: prevalence, severity, and impact on hippocampal structure,” Molecular Psychiatry, pp. 1–10, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Avery SN, McHugo M, Armstrong K, Blackford JU, Woodward ND, and Heckers S, “Stable habituation deficits in the early stage of psychosis: a 2-year follow-up study,” Translational psychiatry, vol. 11, no. 1, pp. 1–10, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Avery SN et al. , “Relational memory in the early stage of psychosis: a 2-year follow-up study,” Schizophrenia Bulletin, vol. 47, no. 1, pp. 75–86, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].McHugo M, Armstrong K, Roeske MJ, Woodward ND, Blackford JU, and Heckers S, “Hippocampal volume in early psychosis: a 2-year longitudinal study,” Translational psychiatry, vol. 10, no. 1, pp. 1–10, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Avery SN, McHugo M, Armstrong K, Blackford JU, Woodward ND, and Heckers S, “Disrupted habituation in the early stage of psychosis,” Biological Psychiatry: Cognitive Neuroscience and Neuroimaging, vol. 4, no. 11, pp. 1004–1012, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].McHugo M et al. , “Hyperactivity and reduced activation of anterior hippocampus in early psychosis,” American Journal of Psychiatry, vol. 176, no. 12, pp. 1030–1038, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bao S et al. , “Registration-based image enhancement improves multi-atlas segmentation of the thalamic nuclei and hippocampal subfields,” Magnetic resonance imaging, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sahakyan L et al. , “Anterior vs posterior hippocampal subfields in an extended psychosis phenotype of multidimensional schizotypy in a nonclinical sample,” Schizophrenia Bulletin, vol. 47, no. 1, pp. 207–218, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Prestia A et al. , “Hippocampal and amygdalar local structural differences in elderly patients with schizophrenia,” The American Journal of Geriatric Psychiatry, vol. 23, no. 1, pp. 47–58, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kawano M et al. , “Hippocampal subfield volumes in first episode and chronic schizophrenia,” PloS one, vol. 10, no. 2, p. e0117785, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ota M et al. , “Structural differences in hippocampal subfields among schizophrenia patients, major depressive disorder patients, and healthy subjects,” Psychiatry Research: Neuroimaging, vol. 259, pp. 54–59, 2017. [DOI] [PubMed] [Google Scholar]
- [45].Provenzano FA et al. , “Hippocampal pathology in clinical high-risk patients and the onset of schizophrenia,” Biological psychiatry, vol. 87, no. 3, pp. 234–242, 2020. [DOI] [PubMed] [Google Scholar]
- [46].Huo Y et al. , “Consistent cortical reconstruction and multi-atlas brain segmentation,” NeuroImage, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kalmady SV et al. , “Clinical correlates of hippocampus volume and shape in antipsychotic-naïve schizophrenia,” Psychiatry Research: Neuroimaging, vol. 263, pp. 93–102, 2017. [DOI] [PubMed] [Google Scholar]
- [48].Mamah D, Harms MP, Barch DM, Styner MA, Lieberman J, and Wang L, “Hippocampal shape and volume changes with antipsychotics in early stage psychotic illness,” Frontiers in psychiatry, vol. 3, p. 96, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ho B-C and Magnotta V, “Hippocampal volume deficits and shape deformities in young biological relatives of schizophrenia probands,” Neuroimage, vol. 49, no. 4, pp. 3385–3393, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Addington J et al. , “North American prodrome longitudinal study (NAPLS 2): overview and recruitment,” Schizophrenia research, vol. 142, no. 1, pp. 77–82, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Jack CR et al. , “The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods,” Journal of Magnetic Resonance Imaging, vol. 27, no. 4, pp. 685–691, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]


