Abstract
One SARS-CoV-2-positive sample demonstrated impaired detection of the N1 target by RT-PCR using US CDC primer/probe sets. A 3 nucleotide deletion was discovered that overlaps the forward primer binding site. This finding underscores the importance of continued SARS-CoV-2 mutation surveillance and assessment of the impact on diagnostic test performance.
Keywords: SARS-CoV-2 NAAT, SARS-CoV-2 mutation
1. Research note
Nucleic acid amplification tests (NAAT) remain the mainstay for the laboratory diagnosis of the SARS-CoV-2 infection. The high sensitivity and specificity of these tests rely on carefully designed primer/probe sets targeting the conserved regions of the viral genome. One month after the first report of COVID-19, the viral genome was made available to the public (Zhu et al., 2020). Many primer/probe sets were designed based on the original sequences (Wuhan-Hu-1, NC_045512.2) and soon adopted by clinical laboratories and in vitro diagnostics manufacturers. As the pandemic continues, the virus continues to evolve. Changes in the viral genome, in particular the spike gene, have been associated with increased transmissibility and virulence, reduced susceptibility to convalescent or vaccinated sera, while changes in S gene and other regions of the genome, such as the N or E gene, have been reported to adversely impact the performance of some NAATs resulting in reduced test sensitivity (Amato et al., 2021; Artesi et al., 2020; Bal et al., 2021; Hasan et al., 2021; Hasan et al., 2021; Lee et al., 2021; Leelawong et al., 2021; Vanaerschot et al., 2020; Wollschlager et al., 2021; Zannoli et al., 2021; Ziegler et al., 2020).
The US CDC 2019-Novel Coronavirus Real-time RT-PCR Diagnostic Panel, distributed and released in February 2020, was the first test to achieve US Food & Drug Administration (FDA) - Emergency Use Authorization (EUA) for detection of SARS-CoV-2 in the United States. This panel originally targeted 3 regions of the N gene and consisted of 3 assays: N1, N2, and N3; N1 and N2 assays were designed to specifically detect SARS-CoV-2 while N3 assay can recognize viruses within the subgenus Sarbecovirus (Lu et al., 2020). The N3 target was later dropped from the panel following reports of low specificity. Subsequently, many commercial and lab-developed tests used the N1, N2 primer/probe sets to detect SARS-CoV-2. The clinical microbiology laboratories at Nationwide Children's Hospital adopted the US CDC N1 and N2 primer/probe sets and fully validated and implemented the testing for clinical use with FDA EUA granted in April 2020 (NCH EUA LDT). Here we report one incidence that demonstrates poor N1 gene assay performance relative to the N2 assay.
In October 2021, a nasopharyngeal sample from a 2 year-old hospitalized female patient presenting to the emergency room with pneumonia was submitted for routine diagnostic SARS-CoV-2 testing. The sample tested positive with the NCH EUA LDT, but test results showed discrepant Ct profiles with a Ct value of 37.1 for the N1 assay and 17.1 for the N2 assay. Repeat testing confirmed this finding. This sample was reported as positive for SARS-CoV-2. Additional testing results using the Xpert Xpress SARS-CoV-2 assay (Cepheid, Sunnyvale, CA), BioFire FilmArray Respiratory Panel v2.1 (Salt Lake City, UT) and BD BioGX SARS-CoV-2 assay (Franklin Lakes, NJ) are presented in Table 1 . This sample presented similar discrepant Ct profiles in the BD BioGx SARS-CoV-2 assay which also utilizes US CDC primer/probe sets. The large Ct value difference between the N1 and N2 assays with the NCH EUA LDT prompted further investigation as mutation(s) in the primer and/or probe binding sites in the N gene were suspected.
Table 1.
SARS-CoV-2 NAAT Test results for the sample suspected of N gene mutation.
| Test Platform | Test Results |
|||
|---|---|---|---|---|
| Gene Target | Ct value or positivity | Gene Target | Ct value or positivity | |
| NCH EUA LDT | N1 | 37.1 | N2 | 17.1 |
| BD BioGX | N1 | 32.4 | N2 | 16.8 |
| Cepheid | E | 19.1 | N | 19.3 |
| BioFire | S | Positive | M | Positive |
Whole genome sequencing was performed on the residual eluate. Briefly, NEBNext ARTIC SARS-CoV-2 FS Library Prep Kit was used for target enrichment and library preparation following the VarSkip Short Express Protocol (New England Biolabs, Ipswich MA). The library was sequenced (76 × 8 × 8 × 76) using the Illumina MiniSeq, and binary base call (BCL) sequence files were converted to FASTQ for analysis using the SARS-CoV-2 Illumina workflow in the CLC Genomics Workbench (Qiagen, Hilden, Germany). The genetic lineages of the sequences was determined using Phylogenetic Assignment of Named Global Outbreak Lineages (Pangolin, pangolin.cog-uk.io) and Nextclade (clades.nextstrain.org).
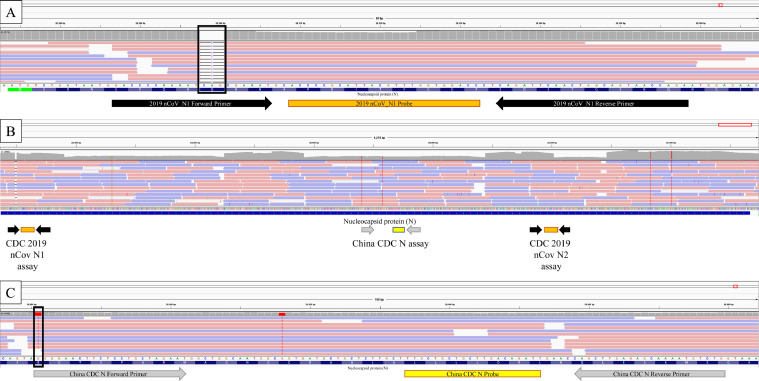
WGS revealed a deletion of 3 nucleotides located within the forward primer binding site of the N1 assay (28298-28300 delCAG), resulting in a deletion of the amino acid Glutamine at position 9 of Nucleocapsid protein (Fig. 1 A). Lineage assignment for this sample is consistent with the Delta variant belonging to AY.44 sublineage (Pango v.3.1 16, 2021-10-18). The sequence was uploaded to GISAID (EPI_ISL_5750423). Investigating publicly available databases (GISAID, www.gisaid.org and NCBI www.ncbi.nlm.nih.gov; accessed 11/2/2021), we found 2 reports of this deletion; both identified in Germany collected on 2/21/2021 (EPI_ISL_1147393) and 3/4/2021 (EPI_ISL_1570530). Unlike the sample we identified, these 2 samples are highly similar and belong to lineage B.1.160 (Pango v.3.1 16, 2021-10-18).
Fig. 1.
Sequence reads aligned to the SARS-CoV-2 genome (MN908947.3) are visualized in the integrated genome viewer (IGV). The snapshot view illustrates forward (red arrows) and reverse (blue arrows) read alignments between genome coordinates NC_045512.2:28,272-29,500, representing sequence coverage depth at each base position (gray bars above read alignments). A. The US CDC NAAT N1 assay illustrating the nucleotide sequence, N gene protein translation, primer (black arrows), and probe binding (orange box) sites are the IGV tracks below the sequence read track. A three-nucleotide deletion (r.28,898-28,300) results in an in-frame 9Q deletion localized to the N1-F primer binding site (black box -3-) with a concomitant loss of sequence coverage (missing gray columns). B. An IGV snapshot of the US CDC NAAT N1 and N2 assays and the China CDC NAAT N assay. The N1 and N2 primers (black arrows) and probes (oranges boxes) flank the China CDC NAAT N assay (primers are gray arrows, and the probe is the yellow box). C. The China CDC NAAT N assay illustrating the nucleotide sequence, N gene protein translation, primer (gray arrows), and probe binding (yellow box) sites are the IGV tracks below the sequence read track. The reported location of the GGG to AAC mutation at positions r.28,882-28,884 (adjacent to the black box and the 28,880 G>T substitution) impacts the forward primer binding site 5’ GGGGAACTTCTCCTGCTAGAAT and results in poor assay performance. (Color version of figure is available online).
The N gene of SARS-CoV-2 encodes the nucleocapsid protein, a major structural component of the virus, and is relatively well-conserved; it is the target of many diagnostic assays for COVID-19, including NAATs, antigen, and antibody-based tests. Mutations in the N-gene that affect NAAT performance have been previously reported. Notably, a GGG-AAC mutation in the N gene (28881-28883) overlaps the forward primer binding site of the Chinese CDC N assay (Fig. 1C). This mutation was found in a significant portion of SARS-CoV-2 sequences in GISAID and renders the inclusivity of this assay as low as 63.89% in a recent analysis (Gand et al., 2021). Single nucleotide polymorphisms C29200T, C29200A, C29197T and G29227T overlapping the US CDC N2 probe or reverse primer have also been reported. They may be associated with the failure of amplification of the N gene by the Xpert Xpress SARS-CoV-2 assay (Hasan et al., 2021; Leelawong et al., 2021; Ziegler et al., 2020). The impact of these SNPs on the performance of the US CDC N2 assay has yet to be determined.
In testing over 50,000 samples since implementation, we have only detected this in one sample. To our knowledge, this is the first report of a mutation in the N gene that affects US CDC N1 assay. This mutation will similarly impact diagnostic test performance that utilizes the US CDC N1 primer/probe, exemplified by results from testing of our sample with the BD BioGX SARS-CoV-2 assay. Although the 3-nucleotide deletion reported here has been identified twice previously from Germany, it is unlikely to represent many of the circulating strains. Using a multi-target approach, mutations that affect 2 set of primer/probes will not cause a false-negative result. Nevertheless, laboratories should be aware of the assay targets and their assays’ primer/probe binding sites and regularly monitor inclusivity against currently circulating strains. Our finding underscores the importance of continuous SARS-CoV-2 mutation surveillance, regular assessment of diagnostic test performance as the virus continues evolving, and including multiple targets in the assay to maintain maximal inclusivity.
Acknowledgements
We would like to thank the staff at the NCH Institution of Genomic Medicine for sequencing the specimen and Clinical microbiology laboratories for their dedication during the COVID19 pandemic.
Funding
This research was partially funded by the Nationwide Foundation Pediatric Innovation Fund (No. 40315-0004).
Declaration of competing interest
The authors report no conflicts of interest relevant to this article.
Author Contributions
Huanyu Wang: Study design, data curation and analysis, writing, reviewing and editing
Sophonie Jean: data analysis, reviewing and editing
Sarah A. Wilson: Genome library preparation, Illumina sequencing, data processing, data reporting
Jocelyn M. Lucyshyn: Genome library preparation, Illumina sequencing, data processing, data reporting
Sean McGrath: Genome profiling assay development, genome analysis development, data review, writing, editing
Richard K. Wilson: Genome profiling assay development, genome analysis development, data review, writing, editing
Vincent Magrini: Genome profiling assay development, genome analysis development, data review, writing, editing
Amy L Leber: Study design, writing, reviewing and editing
References
- Amato L., Jurisic L, Puglia I, Di Lollo V, Curini V, Torzi G, et al. Multiple detection and spread of novel strains of the SARS-CoV-2 B.1.177 (B.1.177.75) lineage that test negative by a commercially available nucleocapsid gene real-time RT-PCR. Emerg Microbes Infect. 2021;10(1):1148–1155. doi: 10.1080/22221751.2021.1933609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artesi M., Bontems S, Gobbels P, Franckh M, Maes P, Boreux R, et al. A Recurrent mutation at position 26340 of SARS-CoV-2 is associated with failure of the E gene quantitative reverse transcription-PCR utilized in a commercial dual-target diagnostic assay. J Clin Microbiol. 2020;58(10):e01598–20. doi: 10.1128/JCM.01598-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal A., Destras G, Gaymard A, Stefic K, Marlet J, Eymieux S, et al. Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69-V70, France, August to December 2020. Euro Surveill. 2021;26(3) doi: 10.2807/1560-7917.ES.2021.26.3.2100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gand M., Vanneste K, Thomas I, Van Gucht S, Capron A, Herman P, et al. Deepening of in silico evaluation of SARS-CoV-2 detection RT-qPCR assays in the context of new variants. Genes (Basel) 2021;12(4) doi: 10.3390/genes12040565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan M.R., Sundararaju S, Manickam C, Mirza F, Al-Hail H, Lorenz S, et al. A novel point mutation in the N gene of SARS-CoV-2 may affect the detection of the virus by reverse transcription-quantitative PCR. J Clin Microbiol. 2021;59(4) doi: 10.1128/JCM.03278-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan R., Hossain ME, Miah M, Hasan MM, Rahman M, Rahman MZ. Identification of novel mutations in the N gene of SARS-CoV-2 that adversely affect the detection of the virus by reverse transcription-quantitative PCR. Microbiol Spectr. 2021;9(1) doi: 10.1128/spectrum.00545-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Won D, Kim CK, Ahn J, Lee Y, Na H, et al. Novel indel mutation in the N gene of SARS-CoV-2 clinical samples that were diagnosed positive in a commercial RT-PCR assay. Virus Res. 2021;297 doi: 10.1016/j.virusres.2021.198398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leelawong M., Mitchell SL, Fowler RC, Gonzalez E, Hughes S, Griffith MP, et al. SARS-CoV-2 N gene mutations impact detection by clinical molecular diagnostics: reports in two cities in the United States. Diagn Microbiol Infect Dis. 2021;101(3) doi: 10.1016/j.diagmicrobio.2021.115468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Wang L, Sakthivel SK, Whitaker B, Murray J, Kamili S, et al. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26(8):1654–1665. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanaerschot M., Mann SA, Webber JT, Kamm J, Bell SM, Bell J, et al. Identification of a polymorphism in the N gene of SARS-CoV-2 that adversely impacts detection by reverse transcription-PCR. J Clin Microbiol. 2020;59(1) doi: 10.1128/JCM.02369-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollschlager P., Todt D, Gerlitz N, Pfaender S, Bollinger T, Sing A, et al. SARS-CoV-2 N gene dropout and N gene Ct value shift as indicator for the presence of B.1.1.7 lineage in a commercial multiplex PCR assay. Clin Microbiol Infect. 2021;27(9) doi: 10.1016/j.cmi.2021.05.025. :1353e1−1353e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannoli S., Dirani G, Taddei F, Gatti G, Poggianti I, Denicolo A, et al. A deletion in the N gene may cause diagnostic escape in SARS-CoV-2 samples. Diagn Microbiol Infect Dis. 2021;102(1) doi: 10.1016/j.diagmicrobio.2021.115540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler K., Steininger P, Ziegler R, Steinmann J, Korn K, Ensser A. SARS-CoV-2 samples may escape detection because of a single point mutation in the N gene. Euro Surveill. 2020;25(39) doi: 10.2807/1560-7917.ES.2020.25.39.2001650. [DOI] [PMC free article] [PubMed] [Google Scholar]