Abstract
Thyroid hormone receptors are encoded by the TRα (NR1A1) and TRβ (NR1A2) loci. These genes are transcribed into multiple variants whose functions are unclear. Analysis by gene inactivation in mice has provided new insights into the functional complexity of these products. Different strategies designed to modify the TRα locus have led to strikingly different phenotypes. In order to analyze the molecular basis for these alterations, we generated mice devoid of all known isoforms produced from the TRα locus (TRα0/0). These mice are viable and exhibit reduced linear growth, bone maturation delay, moderate hypothermia, and reduced thickness of the intestinal mucosa. Compounding TRα0 and TRβ− mutations produces viable TRα0/0β−/− mice, which display a more severe linear growth reduction and a more profound hypothermia as well as impaired hearing. A striking phenotypic difference is observed between TRα0/0 and the previously described TRα−/− mice, which retain truncated TRΔα isoforms arising from a newly described promoter in intron 7. The lethality and severe impairment of the intestinal maturation in TRα−/− mice are rescued in TRα0/0 animals. We demonstrate that the TRΔα protein isoforms, which are natural products of the TRα locus, are the key determinants of these phenotypical differences. These data reveal the functional importance of the non-T3-binding variants encoded by the TRα locus in vertebrate postnatal development and homeostasis.
Thyroid hormone receptors (TRs), encoded by the TRα and TRβ loci, mediate the action of triiodothyronine (T3) in all vertebrates. These nuclear proteins activate or repress the transcription of target genes by recruiting several protein complexes which modify the structure of chromatin (15). In mammals, T3 exerts homeostatic and developmental functions. Thyroid hormone (TH) deprivation leads to developmental (growth retardation, impaired neurogenesis) (8, 23, 36) or metabolic (reduced oxygen consumption, bradycardia, weakness, fatigue) disorders (13). Point mutations in the TRβ gene result in syndromes known as resistance to thyroid hormone (30). Recently, the introduction of mutations in the genes encoding these receptors in mice has allowed further understanding of the mechanisms mediating the physiological actions of T3 (17, 19). The TRβ locus encodes the TRβ1 and TRβ2 receptors. A new receptor, TRβ3, and its non-DNA-binding TRΔβ3 variant have recently been described. The latter isoforms are proposed to arise from a novel promoter located upstream of the exons encoding the DNA binding domain (41). Genetic invalidation of the TRβ2 gene leads to pituitary resistance to TH with elevated thyrotropin (TSH) and thyroxine (T4) levels in serum (1). Knocking out all TRβ1, TRβ2, and TRβ3/Δβ3 genes reproduces this phenotype (10, 14) but also induces the loss of auditory function, due to delayed expression of a potassium channel in the cochlea (11), and results in impaired transcriptional response to T3 in liver (38). The TRα locus encodes the ubiquitous TRα1 receptor and a splice variant, TRα2, which cannot bind T3 and behaves as a weak inhibitor of TRs in transfection assays (21, 22). We have previously shown that, in a limited number of tissues, the TRα locus also encodes two transcripts, previously characterized by Northern blotting, RNase protection, and reverse transcription (RT)-PCR analysis in embryonic stem (ES) cells, driven by a promoter located in intron 7, which generate TRΔα1 and TRΔα2 proteins. These proteins do not bind DNA or T3; hence, they are inhibitors of the activities of TRs and retinoic acid receptors in transfection assays (6). In vivo genetic modifications of the TRα locus result in different phenotypes, according to the precise location of the mutation. In TRα−/− mice, the expression of TRα1 and TRα2 transcripts is abolished, but the TRΔα transcripts are still expressed (12). These mice suffer from hypothyroidism and display several developmental defects, including growth retardation, altered intestinal development (28), impaired T- and B-lymphocyte maturation (3), expansion of cartilage regions in long bones, and finally growth arrest and death shortly after weaning (12). Mice which lack the TRα1 and TRΔα1 products (TRα1−/−) have been reported to exhibit mild hypothyroxinemia, bradycardia, and lower body temperature, but they display normal growth and life span (39). The lethal phenotype of TRα−/− mice compared to the viability of TRα1−/− mice might be attributed to the deeper hypothyroxinemia of the former. The involvement of TH as the cause of this phenotypic discrepancy can be ruled out. Indeed, although TRα−/−β−/− and TRα1−/−β−/− mice both lack the known TRs and have very high serum levels of TH, TRα−/−β−/− mice die upon weaning (14), whereas TRα1−/−β−/− mice are viable (16). Hence, the phenotypic differences must be ascribed to structural differences between the TRα mutant alleles. They could be attributed either to the TRα2 isoform, which is expressed in TRα1−/− but not in TRα−/− mice, or to the balance between the TRα and TRΔα isoforms. In an attempt to determine the role of the non-T3-binding products, we have generated a new mouse strain harboring a targeted deletion designed to prevent the expression of all transcripts from the TRα locus. In this paper we describe the phenotype of these TRα0/0 mutant mice and point out similarities and striking differences with previously described TRα1−/− and TRα−/− mice. This comparative analysis brings a new piece in the genetic analysis of the complex TRα locus and sheds new light on the respective functions of the TRα products in body growth, bone maturation, thermogenesis, audition, and intestinal maturation.
MATERIALS AND METHODS
Targeted disruption of TRα gene.
The TRα0-targeting vector was constructed using pGNAβ as starting plasmid (gift from P. Brulet, Institut Pasteur, Paris, France). The 3′ arm homologous to TRα was amplified by PCR from ENS ES cell DNA (14), using the Expand High Fi system (Roche). The 5′ arm was cloned from a λEMBL4 library (gift from J. P Magaud). The thymidine kinase gene was inserted downstream of the 3′ arm.
ENS ES cells were electroporated with 40 μg of linearized targeting vector and then selected with 250 μg of G418 (Gibco-BRL) per ml and 0.2 μM ganciclovir. Targeted clones were confirmed by Southern blot analysis using PvuII enzyme and a 900-bp PCR fragment extending from the 3′ end of exon 3 to the 5′ end of exon 4 of TRα as a probe. Two positive clones were injected into blastocysts of C57BL/6 recipient mice. TRα0/+ animals were derived in an inbred 129sv background. TRα0/0 mice were obtained by intercrossing heterozygous animals. Mouse breeding and handling were carried out in a certified animal facility, according to procedures approved by the local animal care and use committee.
Purification of RNA, Northern blotting RNase protection assay, and RT-PCR analysis.
Total RNA from distal small intestine or pituitary was isolated by the improved acid-guanidine-phenol-chloroform method. For Northern blot analysis, single-stranded DNA probes were generated by a 30-cycle reaction using Taq DNA polymerase from Eurobio using 10 ng of TRα (exon 8), growth hormone (GH), or hypoxanthine phosphoribosyltransferase (HPRT) PCR fragments as a template, and 20 ng of a specific antisense primer: oligonucleotides α15A (CAGCCTGCAGCAGAGCCACTTCCG), GHa (GTCAAACTTGTCATAGGTTTG), and HPRTa (CACAGGACTAGAACACCTGC) were used as primers to generate the TRα, GH, and HPRT probes, respectively.
RNase protection was performed using reagents from Ambion (Austin, Tex.) according to the manufacturer's instructions. HPRT and TRα cDNA fragments were generated by RT-PCR and inserted into a pGEMt easy vector (Promega). Oligonucleotides HPRTs (GCTGGTGAAAAGGACCTCT) and HPRTa (see above) were used to generate the HPRT fragment, and TRαCs (CTCTGTGATCCTGCTGTTCCACAG) and TRα1a (CGACTTTCATGTGGAGGAAG) were used to generate the TRα cDNA.
RT was performed as described in reference 12. cDNA (0.05 μg) was used for each PCR with Eurobio Taq. To allow the detection of TRα gene products (TRα1, TRα2, TRΔα1, and TRΔα2) by RT-PCR, we used a sense primer within exon 8 (α2′, CTGCCTTGCGAAGACCAGATC) and an antisense primer within exon 9 (α3, GCGGTGGTTGACGTAGTGCTC). All the oligonucleotides were purchased from MWG (Ebersberg, Germany).
Morphological analysis of long bones and growth plate cartilage.
Skin and muscle were dissected from forelimbs and hindlimbs of 3-week-old wild-type and TRα0/0 null mice. For anatomical analysis of long bones, one upper and one lower limb from each animal were fixed in 80% ethanol for 1 to 4 days and transferred to 95% ethanol for a further 1 to 8 days, followed by acetone for 1 to 2 days at 4°C. Fixed limbs were stained with alizarin red and alcian blue 8GX (0.3% alcian blue in 70% ethanol [1 ml]–0.1% alizarin red in 95% ethanol [1 ml]–glacial acetic acid [1 ml]–70% ethanol [17 ml]) for 2 days at 37°C and transferred sequentially for 2- to 4-day periods through solutions of 1% KOH, 1% KOH–20% glycerol, 1% KOH–40% glycerol, 1% KOH–60% glycerol, and 1% KOH–80% glycerol at 4°C until, finally, destained limbs were stored in 100% glycerol at 4°C prior to analysis and photography under a dissecting microscope. In these preparations, mineralized bone is stained by alizarin red and cartilage is stained by alcian blue 8GX. For mineralization analysis, tibias were dissected out, fixed in 70% alcohol, embedded in paraffin, and cut (7 μm), and undecalcified sections were stained according to the standard protocol (26). For histological analyses, the paired limbs were fixed in 10% neutral buffered formalin for 4 to 6 days and subsequently decalcified for 2 days in 10% formic acid–10% neutral buffered formalin at 20°C. Limbs were embedded in paraffin and 3-μm sections were cut, deparaffinized in xylene, and rehydrated. Sections were stained with hematoxylin and eosin or alcian blue 8GX and van Gieson to visualize cartilage matrix mucopolysaccharides in blue and collagen-containing bone osteoid in red, according to standard histological protocols.
Body temperature.
The system we used, ELAMS (Electronic Laboratory Animal Monitoring System by BioMedic Data Systems, Inc., Seaford, Del.), consisted of implantable programmable temperature transponders (IPTT-100) and a portable data scanner (DAS-5007). The transponders were implanted subcutaneously in the backs of three 8- to 12-week-old males from each genotype. Animals were maintained under anesthesia with midazolam. All animals were kept in a climate-controlled room with 12-h alternating light and dark cycles. Temperature measurements were initiated 3 days after the implantation and were recorded in duplicate at 10 a.m., while the animals were moving freely in their cage with the lid removed, and the scanner was placed within 5 cm of the transponder. Body temperature monitoring continued for up to 7 days.
Temperature was measured rectally at the same time in TRα0/0β−/− by means of a digital recording with a thermistor system. Values were not significantly different from those recorded by the transponder (data not shown).
Growth measurements.
Male mice anesthetized with methoxyflurane were weighed and measured (nose to tail base) weekly from 2 to 9 weeks of age. Mice were housed five per cage, and food and water were provided ad libitum. The numbers of mice used from each genotype were as follows: wild type, 19 to 29; TRβ−/−, 6 to 17; and TRα0/0, 10 to 22.
Hormone measurements.
Serum T4 and TSH were measured by radioimmunoassays as previously described (29).
Hearing tests.
The auditory brainstem response (ABR) was used to assess the auditory sensitivity by a procedure that has been described in detail elsewhere (35). Tucker-Davis Technologies (Gainesville, Fla.) hardware and software (AeP and Siggen packages) were employed. Briefly, mice were anesthetized with sodium pentobarbital (Nembutal, 1.2 μl per g of body weight intraperitoneally [i.p.]) and chlorprothixene (Taractan, 0.5 μl/g i.p.) and kept warm with a heating pad. The average response (waveform) per 100 presentations of tone bursts (1 ms rise and fall; 3 ms duration; rate, 21 Hz; frequencies of 4, 8, 12, 16, and 24 kHz) was recorded using a subdermal needle electrode placed in the scalp. Each tone frequency was presented in descending intensity steps beginning at 80-dB sound pressure level and ending when a visually discernible ABR waveform could no longer be detected. Thresholds were then determined to within 5 dB for each frequency (35). The numbers of mice (60 to 90 days old) tested from each genotype were as follows: wild type, 7; TRβ−/−, 9; TRα0/0, 7; and combined TRβ−/−TRα0/0, 4.
Morphological staining and immunohistochemistry.
Three-week-old mice were killed by cervical dislocation and the small intestines were immediately removed. Proximal jejunums and distal ilea were collected separately and fixed in Carson's solution overnight at 4°C. They were then embedded in paraffin and 5-μm sections were applied on polylysine-coated slides. For morphological observations, after dewaxing and rehydration, the slides were stained with hematoxylin and eosin.
Immunochemistry experiments were performed with a monoclonal antibody (Novocastra Laboratories) for Ki-67 detection. Polyclonal antibody PAI-211 (Affinity Bioreagents, Inc.) for TRα1 and TRΔα1 protein localization and another recognizing the N-terminal part of both TRα1 and TRα2 proteins (generous gift of D. Baas [4]) were used in combination with secondary biotinylated antibody and streptavidin-peroxidase detection system (Histomouse; Zymed). The tissue was counterstained lightly with hematoxylin. Before incubation with the primary antibody, the slides were immersed in 0.01 M citrate buffer, pH 6, and microwaved for 15 min.
Cells, plasmids, transfections, and Western blotting.
HeLa cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. The plasmids containing the cDNAs of human TRα1 (pSG5hTRα1), rat TRβ1 (pSG5rTRβ1), mouse TRΔα1 (pSG5mTRΔα1), FlagTRΔα1 (CMV-FmTRΔα1), and mouse TRΔα2 (pSG5mTRΔα2) have been described previously (6). HeLa cells were transfected using a calcium phosphate procedure as previously described (7) and harvested after 48 h. Lactacystin was a gift from Satoshi Omura (Tokyo, Japan). Western blotting was performed as previously described (6). We used the no. 21 polyclonal antibody (gift of J. Ghysdael) as described in reference 5 to detect TRα1 and TRΔα1 proteins and the M2 anti-Flag antibody (IBI) to detect the Flag-TRΔα1 protein.
RESULTS
The TRα0/0 mutation abolishes the production of all products from the TRα locus.
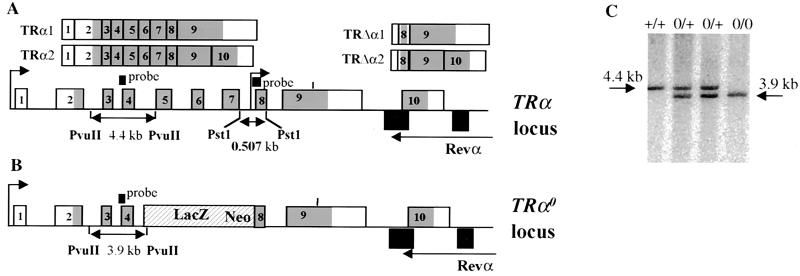
To generate mice deprived of any TRα gene product (TRα1 and TRα2 and their truncated counterparts TRΔα1 and TRΔα2), a lacZ neor cassette (12) was introduced by homologous recombination between exon 4 and exon 8 of the TRα gene (Fig. 1A) to generate the TRα0 allele (Fig. 1B). This removed the DNA binding region of TRα1 and TRα2, preventing the production of a chimeric protein that could act in a dominant-negative manner upon TRβ. It also removed the internal promoter in intron 7 which has been shown to drive the transcription of TRΔα1 and TRΔα2 (6). The TRα0 mutated allele was introduced into ES cells by homologous recombination. The structure of the modified allele was checked by Southern blotting (Fig. 1C). Two independent positive clones were injected into host blastocysts. Heterozygous mice were derived from an inbred 129Sv genetic background. They were then intercrossed to obtain TRα0/0 mice.
FIG. 1.
Targeted disruption of the TRα gene by homologous recombination. (A) Structure of the TRα gene and of the various isoforms encoded by this gene. The two transcription start sites are indicated by upper arrows. The differential splice site in exon 9 is indicated by a vertical bar. Exons are numbered starting downstream to the distal promoter. Grey, coding regions. The structure of the transcripts is shown. (B) The targeted TRα0 allele contains a deletion extending from the middle part of intron 4 to the beginning of exon 8. The probe used for Southern blot analysis and the size of the fragment detected after digestion with PvuII are indicated. (C) Detection of wild-type (+/+), heterozygous (0/+), and homozygous (0/0) littermates by Southern blot analysis.
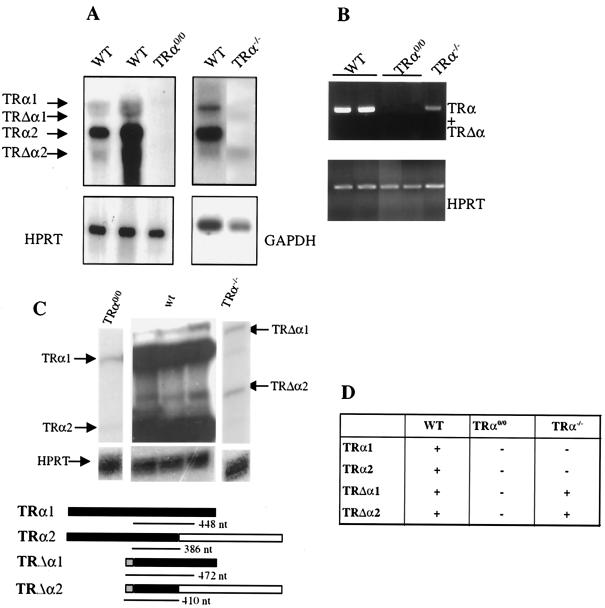
We checked for the expression of TRα transcripts in wild-type, TRα0/0, and previously described TRα−/− mice (12). This analysis was performed on intestine RNA where the four TRα isoforms have been shown to be expressed. Northern blot analysis (Fig. 2A) showed that all four transcripts could be identified in the distal ileum of wild-type mice. In TRα−/− mice, TRα1 and TRα2 RNAs were undetectable, whereas the TRΔα1 and TRΔα2 transcripts were still detected. In TRα0/0 mice, neither TRα nor TRΔα transcripts were detected. In agreement with these data, RT-PCR did not allow the detection of transcripts containing exons 8 and 9 in the intestines of TRα0/0 mice (Fig. 2B). RNase protection analysis (Fig. 2C) revealed that the TRα1 and TRα2 isoforms, which are highly expressed in wild-type samples, are almost undetectable in TRα−/− and TRα0/0 samples. Occasionally, the 448- and 386-nucleotide fragments were detected in the mutant mice, corresponding to transcription reading through the stop sites within the lacZ neor cassette. It is noteworthy that RNase protection showed that wild-type ileum contained similar amounts of TRα1 and TRα2 transcripts. Underrepresentation of the large TRα1 transcript in Northern blot experiments may have been caused by poor transfer onto nylon membranes. Two protected fragments, whose sizes correspond to the TRΔα1 and TRΔα2 isoforms, were detected in wild-type and TRα−/− but not in TRα0/0 samples. It is thus concluded that in the distal ileum of wild-type mice, four TRα isoforms are expressed, including TRΔα1 and TRΔα2, which are expressed at much lower levels than TRα1 and TRα2. The TRα0 mutation completely abolishes the transcription from the TRα locus, in contrast with the TRα mutation which still allows the production of TRΔα transcripts, as previously described (Fig. 2D).
FIG. 2.
Expression of the different transcripts encoded by the TRα locus in the distal ilea of the different mutants. (A) A Northern blot assay was performed using 20 μg of total RNA isolated from the distal ilea of animals carrying the indicated genotypes. The radiolabeled probe hybridized to both TRα and TRΔα transcripts. A glyceraldehyde-3-phosphate dehydrogenase or a hypoxanthine phosphorybosyl transferase probe was used as an internal control. (B) Representative semiquantitative RT-PCR analysis of mRNAs encoded by the TRα locus in wild-type (WT), TRα−/−, and TRα0/0 mice. The primers used allowed the amplification of both TRα and TRΔα transcripts. HPRT was used as an internal control. (C) RNase protection assay. Twenty micrograms of total distal ileum RNA from either wild-type, TRα0/0, or TRα−/− mice was mixed with 75 fmol of HPRT and 75 fmol of TRα RNA probes. The sizes of the protected TRα fragments and the positions of the TRα transcript isoforms are indicated. (D) Summary of the TRα and TRΔα mRNA expression in the distal ileum of each genotype.
TRα0/0 and TRα0/0β−/− mice display normal embryonic development and are viable.
It has previously been shown that TRα−/− and TRα−/−β−/− mice were born in Mendelian proportions but died after weaning (12, 14). Lethality was found in 129SV, BALB/c, and C57BL/6 genetic backgrounds after eight backcrosses. When TRα0/+ mice were intercrossed, among 346 born pups, 98 (28.3%), 170 (49%), and 78 (22.5%) were, respectively, wild-type, heterozygous, and homozygous mutant progenies. Unlike the TRα−/− mice, all TRα0/0 mice survived over the weaning period and the mortality in the adult population was not different from that observed in the wild type. Male and female TRα0/0 mice were fertile. Viability and fertility were observed in C57BL/6 and 129SV backgrounds. TRα0/0 mice were crossed with TRβ−/− mice to obtain TRα0/+β−/− progenies. These mice were then intercrossed and their progenies contained TRα0/0β−/+ and TRα0/+β−/− animals that were all viable. Because of the low fertility displayed by TRα0/+β−/− animals, only TRα0/0β−/+ mice were used to further generate double-homozygous TR mutants (TRα0/0β−/−). The proportion of TRα0/0β−/− progenies was 19.5%. This slight deviation from Mendelian ratios may reflect marginal embryonic lethality or occasional improper maternal care for TRα0/0β−/− newborn mice. However, this result indicates that TRs are not absolutely required for embryonic viability. TRα0/0β−/− mice survived beyond weaning time and were viable for at least six months. Only occasionally were progenies obtained from TRα0/0β−/− intercrossings or from TRα0/0β−/− males or females mated with wild-type partners, suggesting that these animals have a very low fertility.
Deregulation of the pituitary-thyroid axis in TRα0/0β−/− mice.
The serum levels of T3 and T4 are strictly controlled by the direct hormonal feedback on TSH-releasing hormone (TRH) and TSH secretion, in the hypothalamus and the pituitary, respectively (20, 24, 29). We and others have previously demonstrated that TRβ plays a key role in this feedback loop since TRβ−/− mice display high T3, T4, and TSH levels (11, 14, 1). The role of TRα in this regulation has not been clearly established. TRα1−/− mice demonstrated only a slight decrease of T4 and TSH concentrations in males (39), whereas TRα−/− mice presented a progressive but severe reduction of circulating T4 and T3 between the third and the fourth week after birth (12). In TRα0/0 adult males, basal concentration of T4, but not T3 or TSH, is slightly but significantly reduced compared to wild-type mice. Furthermore, TSH and T4 suppression by increasing doses of T3 is more profound in TRα0/0 than in wild-type mice (25). In adult TRα0/0β−/− mice, the concentrations of serum T4, T3, and TSH were increased 14-fold, 13-fold, and more than 200-fold respectively, compared to those of wild-type mice. These levels are comparable with those previously reported for TRα1−/−β−/− animals (11-, 30-, and 60- to 160-fold, respectively) (16) or 3-week-old TRα−/−β−/− mice (7-, 20-, and more than 100-fold, respectively) (14).
Reduced growth rate and delayed bone maturation in TRα0/0 and TRα0/0β−/− mice.
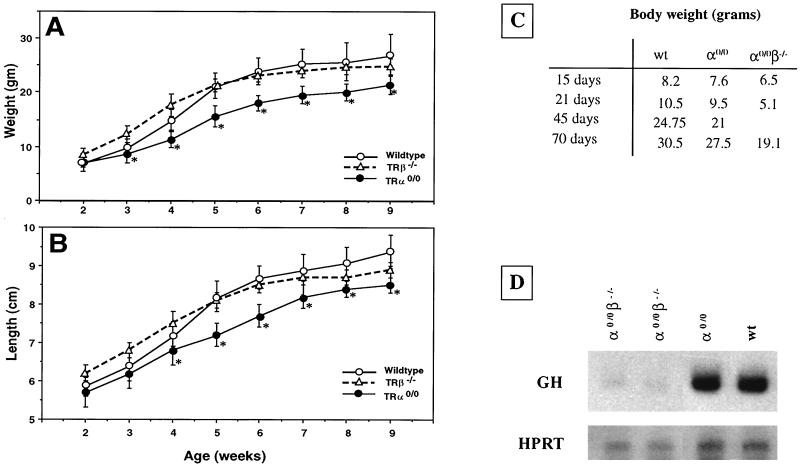
The growth of TRα0/0 males and their wild-type littermates was monitored over a two-month period. TRα0/0 mice showed a reduced increase of body weight compared to wild-type animals, at all ages analyzed, in both 129Sv (not shown) and C57BL/6 genetic backgrounds (Fig. 3A). The time course of linear growth displayed similar differences (Fig. 3B), showing that alteration of body growth fully accounts for the weight differences and that the TRα0 mutation does not result in an abnormal weight-to-length ratio. TRβ−/− mice displayed no difference in weight or length growth compared to that of the wild type (Fig. 3A and B)
FIG. 3.
Comparison of growth in wild-type and TRα- and TRβ-deficient mice. Weight (A) and length (B) were measured weekly from 2 to 9 weeks and plotted as means ± standard deviations. The numbers of animals are given in Materials and Methods. ∗, P < 0.01 for wild-type and TRα0/0 mice. (C) Body weights of wild-type, TRα0/0, and TRα0/0β−/− mice (in grams). (D) Northern blot analysis of pituitary GH mRNA from wild-type, TRα0/0, and TRα0/0β−/− mice. One to two micrograms of total RNA from individual pituitaries was loaded. Single-stranded DNA probes for HPRT and GH were used.
The growth of TRα0/0β−/− mice was further reduced compared to the TRα0/0 mutants, though no difference could be observed at birth. Weight reductions of 50 and 30% were observed in mice at weaning time and in adult mice, respectively, compared to wild-type mice (Fig. 3C).
The synthesis of GH has been shown to be regulated by TH and to be decreased in TRα1−/−β−/− mice (16), which display marked growth retardation. We compared the expressions of the GH mRNA in the pituitary glands of wild-type, TRα0/0, and TRα0/0β−/− mice. As shown in Fig. 3D, the amount of GH RNA was not altered in TRα0/0 mice but was markedly reduced in double-knockout animals. These data suggest that the reduced growth observed in TRα0/0 mice may not be due to a reduced production of GH.
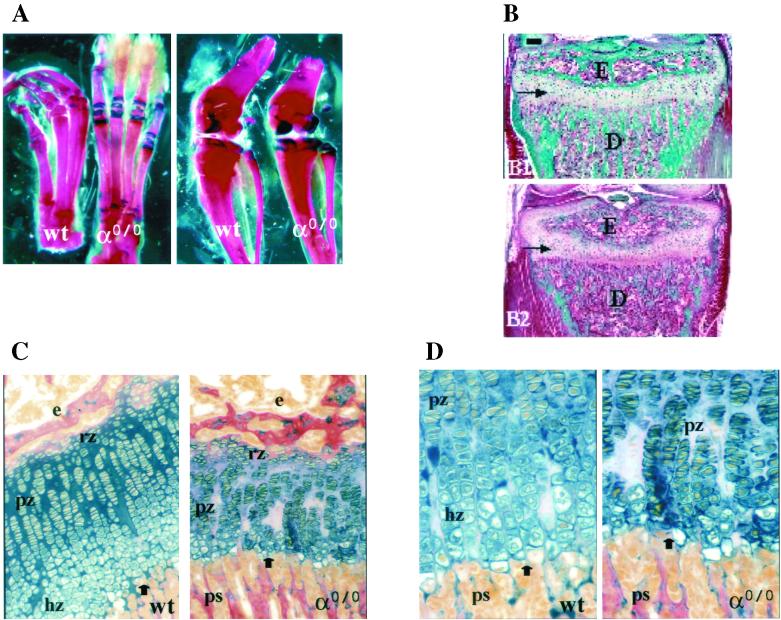
Since linear growth was affected in TRα0/0 mice, we studied endochondral ossification in their long bones. For this purpose, anatomical and histological analyses were performed. In 3-week-old TRα0/0 animals at weaning, the ends of metatarsals and phalanges of the foot, the condyles and growth plate of the femur, and the growth plates of the tibia and fibula stained positively for cartilage with alcian blue (Fig. 4A). In wild-type mice, alcian blue staining was markedly diminished and replaced by alizarin red staining in these areas, indicating that bone formation and mineralization were more advanced than in TRα0/0 animals. Furthermore, ossification of the patella was delayed in TRα0/0 mice relative to that in wild-type mice (Fig. 4A). Similar evidence of delayed bone formation and mineralization was seen in the upper limbs of 3-week-old TRα0/0 mice (data not shown). Goldner staining of undecalcified longitudinal sections of tibias revealed that the ossification of the epiphyses was delayed in TRα0/0 mice, and the density of trabecular bone in the metaphyses was markedly lower than in control mice (Fig. 4B). Histological analyses demonstrated that tibial growth plates from TRα0/0 mice were grossly disorganized compared to those of wild-type animals (Fig. 4B, C, and D), and similar findings were observed in other growth plates throughout the upper and lower limbs of TRα0/0 mice (data not shown). In particular, proliferating growth plate chondrocytes failed to form discrete columns and the hypertrophic zone was markedly diminished and morphologically indistinct in TRα0/0 mice compared to wild-type mice, suggesting that hypertrophic chondrocyte differentiation failed to progress normally in TRα0/0 mice. Furthermore, the degree of alcian blue staining of the growth plate cartilage matrix in TRα0/0 mice was diminished and patchy compared to that in wild-type animals, supporting the view that epiphyseal growth plate architecture is disorganized and endochondral ossification is disrupted in TRα0/0 mice (Fig. 4C and D). In TRα0/0β−/− mice, a similar phenotype was observed (data not shown). These alterations were similar to those described previously for TRα−/− and TRα−/−β−/− mice (12, 14) and are reminiscent of abnormalities in epiphyseal growth plates of hypothyroid rats (34).
FIG. 4.
Morphological appearance of lower limbs and tibial growth plates in 3-week-old wild-type (wt) and TRα gene knockout (α0/0) animals. (A) Lower-limb skeletal mounts stained with alizarin red and alcian blue 8GX showing feet (left) and knee joint including lower femur and upper tibia with fibula (right); magnification, ×8. Red staining shows mineralized bone and blue shows cartilage growth plates; the patella is shown to the right of each knee joint and is stained red in the wt, indicating that mineralization is complete, but blue in TRα0/0, demonstrating the persistence of cartilage. (B) Longitudinal medial bone sections stained with modified Goldner of TRα+/+ (wt) and TRα0/0 mice. Mineralized matrix is stained in green. E, epiphysis; D, diaphysis; horizontal arrows, growth plate. (C and D) alcian blue 8GX and van Gieson staining of proximal tibial growth plates from wt and TRα0/0 mice; a section through the whole growth plate (magnification, ×200) is shown in panel C and a section containing the distal half of the growth plate (magnification, ×400) is shown in panel D. Black arrows, resorption front which separates the growth plate from underlying primary spongiosum; e, epiphysis; ps, primary spongiosum; rz, reserve zone chondrocytes; pz, proliferative zone; hz, hypertrophic zone.
TRα0/0β−/− mice display severe hypothermia.
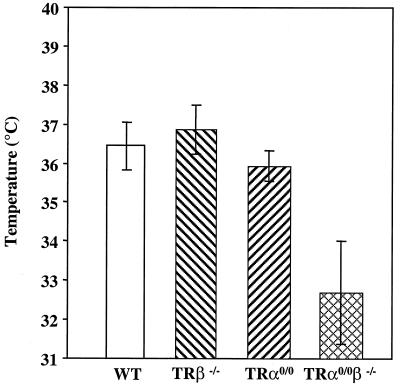
TH plays a crucial role in the control of thermogenesis, and hypothyroidism results in decreased oxygen consumption and heat production (13). We examined the temperatures of adult wild-type and mutant mice, using both a rectal probe and a temperature-sensitive probe implanted under the skin. The two techniques provided similar data. The mean body temperature of TRβ−/− mice was similar to that of wild-type mice, but it was 0.4°C lower in TRα0/0 mice (Fig. 5). Interestingly, the mean body temperature of TRα0/0β−/− animals was dramatically decreased (−4°C) relative to that of the wild type (Fig. 5). These data suggest functional redundancy between the TRβ and the TRα loci in the control of basal body temperature.
FIG. 5.
Body temperature was measured at 10 a.m. with implantable transponders for 7 days in at least 3 male mice aged 8 to 12 weeks for each of the wild-type (WT) TRβ−/−, TRα0/0, and TRα0/0β−/− genotypes. Each bar represents a mean value (± standard deviation). Body temperatures of TRα0/0 and TRα0/0β−/− animals were statistically different from those of wild-type control animals (P < 0.001 and P < 0.001, respectively, by the Student t test).
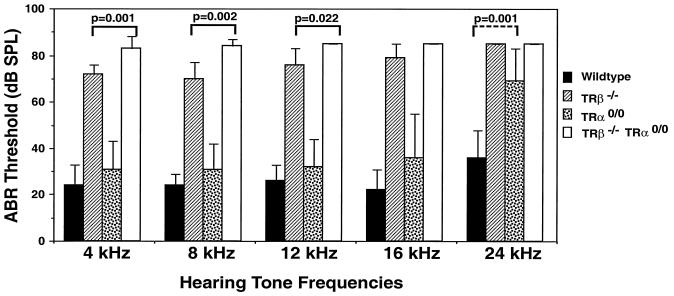
Decrease of auditory sensitivity in TRα0/0 mice and deafness in TRα0/0β−/− mice.
Several authors have described deafness in mice lacking both TRβ1 and TRβ2, but not in mice lacking TRβ2 only or TRα1 (1, 11), suggesting that TRβ1 is the only mediator of this T3-dependent function. However, the role of TRα2 and the effect of the double knockout on the integrity of hearing have not been documented. We therefore measured the ABR threshold in TRα0/0 and TRα0/0β−/− mice. Whereas at low or middle frequencies TRα0/0 mice had a normal hearing threshold, they exhibited a marked loss of sensitivity at high tone frequencies (Fig. 6). In TRα0/0β−/− mice, the loss of hearing threshold was significantly more severe than that observed in TRβ−/− animals. These data suggest that, although the TRβ1 receptor is the major mediator of T3 action, in addition the integrity of the TRα gene is required to achieve full function of the auditory apparatus.
FIG. 6.
ABR threshold in wild-type and TR gene knockout mice. The average response per 100 presentations of tone bursts (1 ms rise and fall; 3 ms duration; rate, 21 Hz; frequencies of 4, 8, 12, 16, and 24 kHz) were recorded. The numbers of animals are given in Materials and Methods. At all frequencies the difference between wild-type and TRβ−/− and TRα0/0β−/− mice was P < 0.001. Solid lines, P values for differences between TRβ−/− and TRα0/0β−/−; dashed line, P values for differences between the wild type and TRα0/0.
The intestinal phenotype is mildly altered in TRα0/0 mice compared to that of TRα−/− mice.
In mammals, the small intestine is lined by a continuously renewable population of epithelial cells. The pluripotent crypt stem cells give rise to immature cells that migrate and differentiate in the villus compartment and die after exfoliation at the villus tip. It was previously demonstrated that TRα−/− mice displayed a strong impairment of postnatal intestinal development (14, 28). It is noteworthy that the intestine is one of the few organs where the TRΔα and the TRα isoforms are coexpressed. In order to differentiate functions of the TRα1/α2 and TRΔα1/Δα2 isoforms in the intestine epithelium, we carried out a comparative investigation of morphofunctional parameters in TRα0/0, TRα−/−, and wild-type mice.
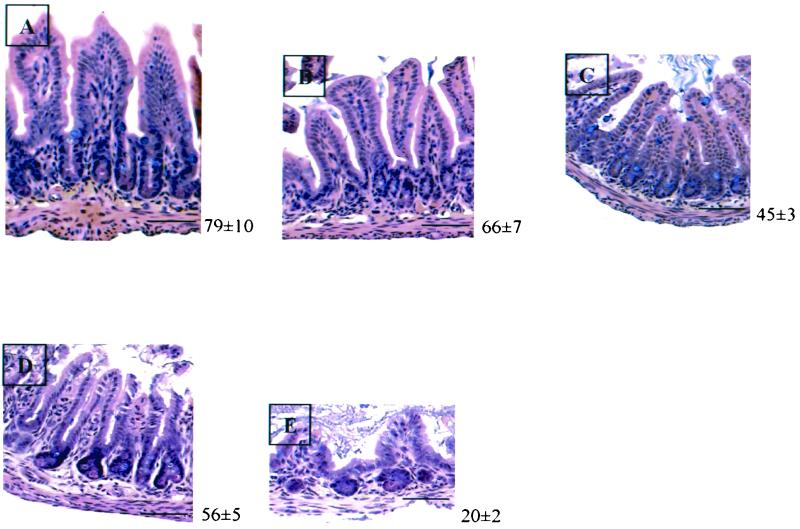
The size of the small intestine mucosa was reduced in TRα0/0 mutants compared to that of wild-type animals. This was mainly due to the reduced lengths of the villi, resulting from a decreased number of epithelial cells per crypt-villus axis (Fig. 7A and B). These features were more strongly affected in the TRα−/− mice (Fig. 7C). The proliferation and differentiation parameters of the epithelial cells also indicated a stronger impairment in TRα−/− than in TRα0/0 mice (27a). In contrast to the TRα−/− mutant animals that displayed a thinning of the smooth muscular layers (12, 28), TRα0/0 mutants appeared normal in this respect. Taken together, these data strongly suggest that the severity of the intestinal phenotype correlates mostly with the persistence of the TRΔα transcripts and not with the absence of TRα1 and TRα2 isoforms.
FIG. 7.
Morphological appearance of distal small intestine in 3-week-old animals. Sections (5 μm) from wild-type (A), TRα0/0 (B), TRα−/− (C), TRα0/0β−/− (D), and TRα−/−β−/− (E) mice were stained with hematoxylin and eosin. The numbers at the right of each picture indicate the number of epithelial cells per crypto-villus axis. Statistical analysis indicated that the number of the epithelial cells in TRα−/−, TRα0/0β−/−, and TRα−/−β−/− animals was significatively decreased compared to that in wild-type mice (P < 0.001, P < 0.05, and P < 0.0001, respectively, by the Student t test; n = 5). In addition, the number of epithelial cells was also significantly decreased in TRα−/−β−/− compared to TRα−/− mice (P < 0.001 by the Student t test). Bar, 30 μm.
The intestinal phenotype is more severe in TRα−/−β−/− than in TRα0/0β−/− mice.
It was previously shown that combining the TRα− and TRβ− mutations in TRα−/−β−/− mice results in a more dramatic alteration of the structure of the distal ileum than that observed in TRα−/− mice (14, 28). When we analyzed the distal ilea of TRα0/0β−/− versus those of TRα0/0 mice, no obvious morphological difference was observed (Fig. 7B and D) and no statistically significant difference was found in the number of epithelial cells per crypt-villus axis. In contrast, in the ilea of TRα−/−β−/− mice, the number of epithelial cells was further decreased compared with that of TRα−/− mice (Fig. 7C and E) and the morphology was dramatically altered (Fig. 7E), in agreement with previous observations. The number of proliferating cells in the crypts of TRα0/0β−/− mice (8 ± 1) was decreased to the same extent as in TRα0/0 animals (7 ± 1). In contrast, this number was more reduced in TRα−/−β−/− (2 ± 1) than in TRα−/− animals (5 ± 1). The analysis of epithelial cell differentiation markers also demonstrated similar features in TRα0/0 and TRα0/0β−/− animals but showed a stronger impairment in TRα−/−β−/− than in TRα−/− mice. In summary, the knockout of the TRβ gene does not worsen the phenotype observed in TRα0/0 mice, but clearly enhances the alterations observed in TRα−/− mice. These data rule out functional redundancy between TRs in the control of postnatal development of the intestine. They suggest instead that the expression of the TRΔα isoforms, which is deleterious in the absence of TRα1 and TRα2, has even more toxic effects in the absence of all TRs.
Identification of the TRΔα proteins in the distal ileum by immunohistochemistry.
The intestinal epithelium is composed of proliferating and undifferentiated cells which form the crypt compartment and of mature cells located in the villi. In order to analyze the presence of the TRΔα proteins in the intestinal mucosa and their pattern of expression along the crypt-villus axis, we performed immunocytochemical analysis. Since TRΔα proteins do not contain any peptide sequence which would differentiate them from their complete TRα counterpart, we had to compare the patterns in distal ilea from wild-type, TRα−/−, and TRα0/0 mice, using the following antisera: anti-Cα1 raised against the common C-terminal part of TRα1 and TRΔα1, anti-Cα2 raised against the common C-terminal part of TRα2 and TRΔα2 but different from TRα1, and anti-Nα raised against the N-terminal part common to TRα1 and TRα2. No staining was obtained with the anti-Cα2 antibody in any genotypes (data not shown), whereas the expression of TRα2 and TRΔα2 mRNAs has been demonstrated by semiquantitative RT-PCR (28), Northern blotting, and RNase protection (this study) in wild-type mice. As this antiserum was used successfully in immunocytochemical detection of TRΔα2 and TRα2 proteins in transfected cells (data not shown), the failure to detect these proteins in the small intestine probably results from their very low concentration in this tissue.
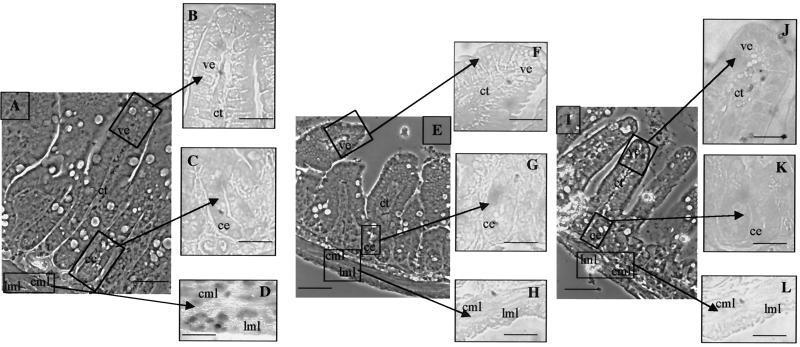
Using the anti-Nα antiserum, we observed a nuclear labeling in muscle layers of wild-type mice (Fig. 8A to D), but no staining was observed in the epithelium or mesenchyme (Fig. 8B and C). No positive cells were detected in TRα0/0 or TRα−/− mice (Fig. 8E to H and I to L). By semiquantitative RT-PCR, it was previously shown that in smooth muscle layers only the TRα1 RNA was detected (28). Consequently, the signal detected with the anti-Nα antibody in this tissue can be attributed to TRα1. In the same experiment, the TRα1 mRNA was found in the epithelium and connective tissues, whereas the TRα2 mRNA was found only in the epithelium (28); therefore, we assume that the corresponding proteins are expressed in these tissue regions but at levels undetectable by this technique.
FIG. 8.
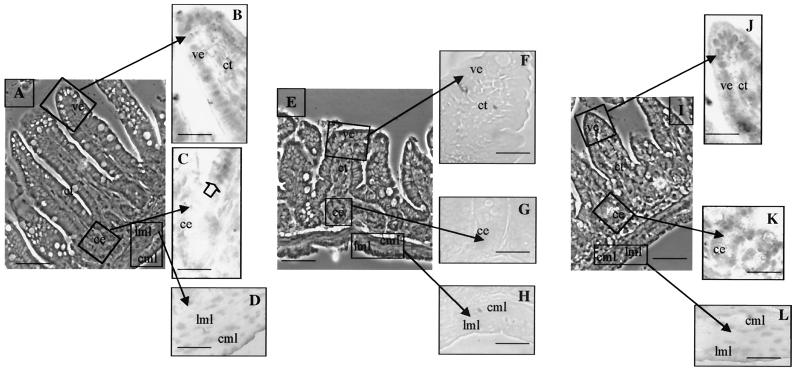
Immunolocalization of TRα1/TRα2 proteins. The pattern of expression of TRα proteins of distal ileum was analyzed in paraffin-embedded sections (5 μm) in wild-type (A to D), TRα0/0 (E to H), and TRα−/− (I to L) 3-week-old mice. The antibody used recognizes the N-terminal part common to TRα1 and TRα2 (anti-Nα) on wild-type sections (B to D), TRα0/0 (F to H), and TRα−/− (J to L). Technical controls included the use of phosphate-buffered saline or nonimmune serum instead of the primary antibody. (A, E, and I) Phase contrast low magnification; bar, 30 μm; (B to D, F to H, and J to L) bright field high magnification; bar, 70 μm. ce, crypt epithelium; ve, villus epithelium; ct, connective tissue; cml, circular muscle layer; lml, longitudinal muscle layer.
Using the anti-Cα1 antibody, we detected a strong nuclear signal in the differentiated epithelial cells of the villi in wild-type mice (Fig. 9A and B). Only a faint signal could be detected in the undifferentiated epithelial cells of crypts, in the lamina propria, and in smooth muscle cells (Fig. 9C and D). However, unlike the signal detected with the anti-Nα reagent, this staining persisted in TRα−/− mice (Fig. 9I to L), which retain the expression of TRΔα transcripts, and disappeared in TRα0/0 mice (Fig. 9E to H). These observations demonstrate that the protein labeled by the anti-Cα1 reagent is TRΔα1. They also suggest that, unlike the anti-Nα antibody, the anti-Cα1 antibody is unable to recognize the TRα1 protein, presumably due to the small amount of this protein in the distal ileum, and to the reduced accessibility of the target epitope in the tight conformation of the ligand binding domain. Altogether, these data indicate that in wild-type mice, the TRα1 protein is mainly located in the nuclei of smooth muscle cells, whereas the TRΔα1 protein is mainly present in the nuclei of differentiated epithelial cells. It is worth noting that in TRα−/− undifferentiated and differentiated epithelial cells have similar nuclear labeling.
FIG. 9.
Immunolocalization of TRΔα1 protein. Its pattern of expression in distal ileum was analyzed in paraffin-embedded sections (5 μm) in wild-type (WT) (A to D), TRα0/0 (E to H), and TRα−/− (I to L) 3-week-old mice. The antibody recognized the C-terminal sequence common to TRα1 and TRΔα1 (anti-CαI) on WT (B to D), TRα0/0 (F to H) and TRα−/− (J to L). Technical controls included the use of phosphate-buffered saline or the antibody preincubated with an excess of the recognized peptide. (A, E, and I) Phase contrast low magnification; bar, 30 μm; (B to D, F to H, and J to L) bright field high magnification; bar, 70 μm. ce, crypt epithelium; ve, villus epithelium; ct, connective tissue; cml, circular muscle layer; lml, longitudinal muscle layer. The open arrow in C indicates the stronger nuclear staining of cells outside the crypts.
The activity of TRΔα proteins is down-regulated by TRα1 via the proteasome pathway.
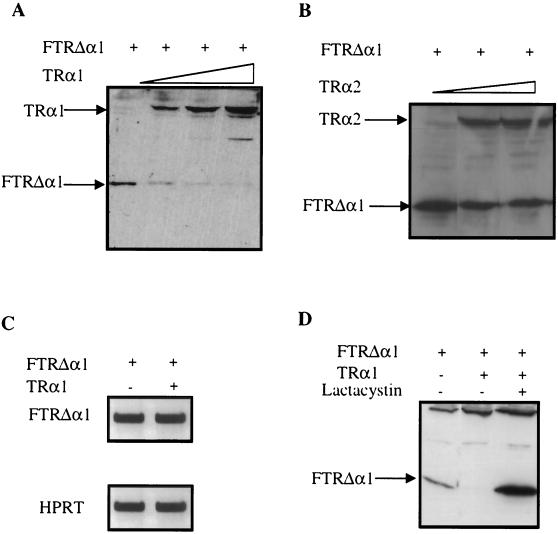
Our data show that the deleterious activity of the TRΔα isoforms is triggered by the inactivation of the TRα1 and TRα2 genes. To clarify the molecular mechanisms which could account for this gene inactivation-dependent activity, we tested the possibility that TRα1 and TRα2 could interfere with the synthesis or degradation of the TRΔα isoforms, thereby modulating their activity. For this purpose, we measured the expression of the TRΔα1 protein in HeLa cells after cotransfection of a TRΔα1 expression vector together with either an empty vector or a vector expressing either TRα1 or TRα2. Figure 10 shows that, while the amount of TRΔα1 transcripts was unaffected by the coexpression of the TRα1 protein (Fig. 10C), the amount of TRΔα1 protein was reduced by increasing amounts of TRα1 (Fig. 10A). This effect was independent of the presence of T3 (data not shown). In contrast to these data, the expression of small amounts of TRα2 did not produce any decrease in the amount of TRΔα1 protein (Fig. 10B). It is noteworthy that expression of TRβ1 did not change the amount of TRΔα1 (data not shown). In order to show that degradation, and not the rate of protein synthesis, accounted for the reduction in the amount of the TRΔα1 protein, we tested the abilities of different permeant protease inhibitors to prevent this effect. When cotransfected cells were treated with lactacystin, a potent inhibitor of the proteasome pathway (9), the amount of TRα1 protein was not changed, but the amount of TRΔα1 protein was considerably increased, reaching levels higher than those observed in the absence of TRα1 (Fig. 10D). These data suggest that TRα1 triggers the degradation of the TRΔα1 protein through a proteasome-dependent pathway.
FIG. 10.
Regulation of the stability of TRΔα1 protein in HeLa cells. (A) Effects of increasing amounts of the expression vector pSG5hTRα1 (0, 1.5, 5, and 15 μg) on the TRΔα1 protein (plasmid CMV-FmTRΔα1, 0.3 μg) as analyzed by Western blotting. HeLa cells were lysed after 48 h of transient transfection. The no. 21 antibody diluted 1:1,000 revealed both TRα1 and FTRΔα1 proteins as indicated by the arrows. (B) Effects of increasing amounts of the expression vector pSG5hTRα2 (0, 1.5, 5, and 15 μg) on the TRΔα1 protein (plasmid CMV-FmTRΔα1, 0.3 μg) as analyzed by Western blotting. (C) Effect of cotransfection in HeLa cells of 15 μg of pSG5hTRα1 and 0.3 μg of CMV-FmTRΔα1 on FTRΔα1 mRNA expression analyzed by RT-PCR. RNA was prepared from HeLa cells after 48 h of transient transfection. HPRT was used as internal control. (D) Effect of lactacystin addition (10−5 M) in culture medium of HeLa cells transiently transfected with 0.3 μg of CMV-FmTRΔα1 and/or 15 μg of pSG5hTRα1 expression vectors. The crude protein lysate was analyzed by Western blotting and the FTRΔα1-specific band was revealed with an anti-Flag antibody diluted 1:1,000. The bands in the middle and upper parts of the picture represent nonspecific staining. In transient transfection experiments a total of 15 μg of DNA/dish has been used; when necessary the pSG5 empty vector has been added.
DISCUSSION
The TRα locus encodes the TRα1 receptor, whose targeted inactivation results in bradycardia and a slight decrease in body temperature (39). This locus also generates the TRα2 variant, which cannot bind T3, poorly heterodimerizes with RXRs, and does not contain any transactivation domain. Transfection studies have shown that the TRα2 protein can inhibit the transcriptional activity of TRs. The TRα2 transcript is present in all mammalian tissues and is generally more abundant than the TRα1 transcript, at least at the level of whole organ analysis (22). Despite these many data, the in vivo function of the TRα2 protein remains unknown. In addition to TRα1 and TRα2, we have identified the TRΔα1 and TRΔα2 transcripts, which are expressed in murine embryonic stem cells and in intestine, lung, and brain tissues of adult mice. The proteins encoded by these transcripts do not bind T3 or DNA but inhibit the transcriptional activity of several nuclear receptors when they are associated with RXR partners (6). Their in vivo function is unknown. Several genetic modifications of the TRα locus which have been generated in a mouse now provide tools with which to obtain insights into the functions of these different products. The goal of this discussion is to draw some conclusions from the comparison of the phenotypes observed in the different TRα gene knockout mice. For the sake of clarity, we have indicated in Fig. 2C the TRα isoforms which are expressed or abolished in the different knockout strains.
TRα1 and TRβs cooperate to regulate TSH production.
Abolishing a TRβ1, TRβ2, TRβ3 and TRΔβ3, or TRβ2 only has been shown to result in altered TH-dependent feedback regulation of TSH production by T3 (1, 37). In contrast, abolishing the TRα1/Δα1 isoforms in TRα1−/− mice (R. E. Weiss, O. Chassande, P. E. Macchia, K. Cua, J. Samarut, and S. E. Refetoff, submitted for publication), TRα1/α2 in TRα−/− mice (12), or TRα1/α2/Δα1/Δα2 in TRα0/0 mice (25) results in mild alterations in the serum concentrations of TH and TSH, indicating that the TRα isoforms may play minor roles in this feedback control. However, TRα1−/− mice display decreased T4 and TSH levels, suggestive of secondary hypothyroidism, while TRα0/0 mice have decreased T4 but normal TSH levels, indicating increased sensitivity of the pituitary to TH. Moreover, careful examination of central responses to varying doses of T3 in TRα0/0 mice reveals increased sensitivity to TH (25). Altogether, these data suggest that the TRα2 isoform weakly antagonizes the activity of TRβs in thyrotroph cells. In the absence of any true TRs, in TRα1−/−β−/− (16), TRα−/−β−/− (14), and TRα0/0β−/− mice, TSH reaches very high levels despite large amounts of TH, independently of the presence of non-T3-binding TR isoforms. This shows that in thyrotroph cells, TRα2 could modulate TSH production only in the presence of a TRβ-mediated T3 response.
TRα1, TRα2, and TRβs cooperate to maintain basal body temperature.
Mice lacking the TRα1/Δα1 isoforms (TRα1−/−) have a body temperature 0.5°C lower than that of wild-type mice (18). We show here that TRα0/0 mice display a similar deficit, suggesting that abolishing TRα2 does not alter thermogenesis, at least under basal conditions. TRβ−/− mice have normal body temperatures suggesting that the contribution of the TRβ isoforms in the T3-dependent basal thermogenesis is minor or compensated for by TRα1. Surprisingly, the comparison between body temperature deficits observed in TRα1−/−β−/− (− 0.5°C) and TRα0/0β−/− (−4°C) mice reveals potentially important thermogenic functions for the TRα2 isoform. Since no difference in body temperature is observed between TRα1−/− and TRα0/0 mice, this function is unraveled only in the absence of functional TR. The above data lead to the conclusion that the effects of TH on basal body temperature are mediated by the TRα and TRβ nuclear receptors, that TRα1, TRβs, and TRα2 isoforms exercise redundant functions in the control of thermogenesis, and that the persistence of only one of these isoforms is sufficient to maintain body temperature within a physiologic range.
TRα2 is essential for normal body growth and bone maturation.
Previous publications and our data have shown that in TRα−/−β−/−, TRα1−/−β−/−, and TRα0/0β−/− mice, the total absence of functional TH receptors results in a reduced growth rate and delayed bone maturation, independently of the presence of non-TH-binding isoforms from the TRα locus. This consensus is in agreement with the bone maturation delay observed in hypothyroid rats (34). This simple observation contrasts with the complexity resulting from the analysis of the functions of individual TRα and TRβ isoforms. Previous reports indicate that TRβ−/− and TRα1−/− mice have normal growth rates and bone maturation (10, 14, 16, 39). In contrast, combining some of the TR mutations results in a profound disruption of these processes, revealing functional redundancies between TR isoforms. One redundancy clearly occurs with TRα1 and TRβ, which are both expressed in bone (2, 5, 31, 34) and which in combined but not individual knockouts result in bone maturation delay. The selective abolishing of TRα1 in TRα1−/− mice does not result in obvious impairment of linear growth or bone maturation. However, in TRα−/− (12) and TRα0/0 (this paper) mice, devoid of both TRα1 and TRα2 isoforms, growth retardation and delayed skeletal development are observed. The skeletal phenotype of TRα0/0 mice includes retarded ossification, failure of progression of hypertrophic chondrocyte differentiation, and disorganization of epiphyseal growth plate architecture, all features similar to those seen in hypothyroidism (34). Importantly, circulating basal TH concentrations in TRα0/0 mice were only mildly impaired and GH concentrations were unchanged compared to those of wild-type mice, supporting the notion that the deletion of the TRα gene fully accounts for this phenotype. On consideration of the TR isoforms that are expressed in the different knockout strains (Fig. 2C), the skeletal phenotypes described can be proposed to result primarily from the abolishing of TRα2, or the concomitant abolishing of both TRα1 and TRα2. Only when data concerning the TRα2-specific mutation are available will it be possible to test this hypothesis further. Nevertheless, our data reveal the important role of TRα2 in body growth and maturation of long bones. An interesting observation is that in bone, abolishing TRα2, an isoform that is known to weakly inhibit T3 responses in vitro, paradoxically results in a phenotype which is similar to the skeletal consequences of profound hypothyroidism. This situation contrasts with the increased T3 response observed in the pituitary of TRα0/0 mice (25). The alteration of growth and bone maturation observed in TRα1−/−β−/− and TRα0/0β−/− mice could be attributed to impaired GH synthesis since these animals exhibit decreased amounts of GH transcripts in pituitary (reference 16 and the present study). In TRα0/0 and TRα−/− mice, however, the GH transcript is expressed at a normal level; therefore, the bone phenotype may be the direct consequence of the TRα mutation within bone cells.
Deafness caused by loss of the TRβ gene is not rescued by deletion of the TRα gene.
In addition to confirming the importance of TRβ in achieving normal hearing function, the present studies demonstrate that TRα may also play a role in the development of normal hearing. In fact, a reduced hearing ability was observed in TRα0/0 mice compared to that of wild-type mice at higher frequencies. Although some elevation of ABR thresholds at 24 kHz is typical of C57BL/6J mice older than 2 to 3 months, the observed hearing loss in the TRα0/0 mice is highly significant. Moreover, the TRα0/0β−/− mouse was more severely deaf than the TRβ−/− mouse. While these observations confirm the important role of TRβ for the development of auditory function (12), they have also identified a contribution of TRα to the full integrity of hearing.
Unbalanced expression of TRΔα isoforms results in improper intestinal development and precocious lethality.
We have previously described the lethal phenotype of TRα−/− mice. The TRα− mutation abolishes the TRα1 and TRα2 transcripts but retains TRΔα transcripts. We show in this study that TRΔα transcripts are expressed in the small intestines of wild-type and TRα−/− but not of TRα0/0 mice. Accordingly, using anti-α1 C-terminal antibody, we detect the TRΔα isoforms in the intestinal epithelial cells of wild-type and TRα−/− but not TRα0/0 mice. This is the first time that the specificity of the staining observed in an immunocytochemical analysis of TRs is assessed by genetic arguments. Therefore, analysis of the products of the TRα locus reveals that the only molecular difference between the TRα−/− and TRα0/0 mutations is the expression of the TRΔα isoforms. Immunocytochemical analysis further shows that the expression of the TRΔα1 protein is expanded in the TRα−/− mice, extending all along the crypt and the villus. In vivo observations show that abolishing TRα1 and TRα2 in TRα−/− mice confers deleterious effects on the TRΔα isoforms, suggesting that, under normal conditions (wild-type mice), the TRα products down-modulate the activity of the TRΔα isoforms. Transfection experiments suggest that one mechanism by which TRs could control the activity of TRΔα proteins is by triggering their rapid removal from the cells through the proteasome-mediated degradation. This mechanism would explain the increased intensity and extended expression domain of the TRΔα1 protein in the absence of TRα1 in the intestines of TRα−/− mice. The mechanism by which TRΔα proteins exert their toxic effects in the developing intestine is unclear. The persistence of a severe intestinal phenotype upon inactivation of the TRβ gene in TRα−/−β−/− mice implies that the molecular targets of the TRΔα proteins are not only the TRs. We have previously suggested that other nuclear receptors, such as retinoic acid receptors, could be antagonized by TRΔα products.
At the level of the whole organism, the lethal phenotype observed in TRα−/− and TRα−/−β−/− mice is indeed due to the persistence of TRΔα isoforms since it can be rescued by their abolition in TRα0/0 and TRα0/0β−/− mice. Thus, the severe intestinal abnormalities and the lethality observed in TRα−/− and TRα−/−β−/− mice are consequences of the persistence of TRΔα isoforms, which are no longer under the control of the TRα1 protein. These genetic and molecular analyses suggest that the TRΔα isoforms have powerful, dose-dependent toxic effects and that their level of expression must be tightly controlled. Altogether, these genetic analyses show that the non-T3-binding isoforms encoded from the TRα locus have important developmental and homeostatic functions.
In addition, our present and previously published data allow the prediction that a potential homozygous mutation in the TRα locus in humans, which would abolish all TRα isoforms, would be expected to result in a mild phenotype mainly characterized by (i) increased sensitivity to TH in the thyrotroph and liver and (ii) reduced sensitivity to TH in heart and bone, manifested as bradycardia, growth retardation, and bone maturation delay. Mutations restricted to the N-terminal moiety of the gene, sparing the TRΔα isoforms, are expected to have severe pathologic consequences.
ACKNOWLEDGMENTS
We thank Christelle Morin, Nadine Aguilera, and Djamel Belgarbi for animal handling, J. P. Magaud, J. Ghysdael, P. Brulet, and S. Omura for kindly providing genomic libraries, antibodies, plasmids, and lactacystin, respectively. We thank F. Flamant and J. M. Vanacker for helpful advice.
This work was supported in part by CNRS and grants from Fondation pour la Recherche Médicale and Human Frontier Scientific Program, and by grants from NIH (DK 15070 [S.R.], AG07554 [J.F.W.], MH61090 [J.F.W.]) and the Seymour J. Abrams Thyroid Research Center. G.R.W. was supported by an MRC Career Establishment grant (G9803002) and a Wellcome Trust Project grant (050570). M. Hara was supported by an FRM fellowship.
REFERENCES
- 1.Abel E D, Boers M-E, Pazos-Moura C, Moura E, Kaulbach H, Zakaria M, Lowell B, Radovick S, Liberman M C, Wondisford F. Divergent roles for thyroid hormone receptor β isoforms in the endocrine axis and auditory system. J Clin Investig. 1999;104:291–300. doi: 10.1172/JCI6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu E O, Bord S, Horner A, Chatterjee V K K, Compston J E. The expression of thyroid hormone receptors in human bone. Bone. 1997;21:137–142. doi: 10.1016/s8756-3282(97)00097-5. [DOI] [PubMed] [Google Scholar]
- 3.Arpin C, Pihlgren M, Fraichard A, Aubert D, Samarut J, Chassande O, Marvel J. Effects of T3Rα1 and T3Rα2 gene deletion on T and B lymphocyte development. J Immunol. 2000;164:152–160. doi: 10.4049/jimmunol.164.1.152. [DOI] [PubMed] [Google Scholar]
- 4.Baas D, Fressinaud C, Ittel M E, Reeber A, Dalençon D, Puymirat J, Sarlièvre I L. Expression of thyroid hormone receptor isoforms in rat oligodendrocyte cultures. Effect of 3,5,3′-triiodo-l-thyronine. Neurosci Lett. 1994;176:47–51. doi: 10.1016/0304-3940(94)90868-0. [DOI] [PubMed] [Google Scholar]
- 5.Ballock R T, Mita B C, Zhou X, Chen D H-C, Mink L M. Expression of thyroid hormone receptors isoforms in rat growth plate cartilage in vivo. J Bone Miner Res. 1999;14:1550–1556. doi: 10.1359/jbmr.1999.14.9.1550. [DOI] [PubMed] [Google Scholar]
- 6.Chassande O, Fraichard A, Gauthier K, Flamand F, Legrand C, Savatier P, Laudet V, Samarut J. Identification of transcripts initiated from an internal promoter in the c-erbAα locus that encode inhibitors of retinoic acid receptor-α and triiodothyronine receptor activities. Mol Endocrinol. 1997;11:1278–1290. doi: 10.1210/mend.11.9.9972. [DOI] [PubMed] [Google Scholar]
- 7.Desbois C, Aubert D, Legrand C, Pain B, Samarut J. A novel mechanism of action for v-erbA: abrogation of the inactivation of transcription factor AP-1 by retinoic acid and thyroid hormone receptors. Cell. 1991;67:731–740. doi: 10.1016/0092-8674(91)90068-a. [DOI] [PubMed] [Google Scholar]
- 8.Dussault J H, Ruel J. Thyroid hormones and brain development. Annu Rev Physiol. 1987;49:321–334. doi: 10.1146/annurev.ph.49.030187.001541. [DOI] [PubMed] [Google Scholar]
- 9.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S L. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 10.Forrest D, Hanebuth E, Smeyne R J, Everds N, Stewart C L, Wehner J M, Curran T. Recessive resistance to thyroid hormone in mice lacking thyroid hormone receptor beta: evidence for tissue-specific modulation of receptor function. EMBO J. 1996;15:3006–3015. [PMC free article] [PubMed] [Google Scholar]
- 11.Forrest D, Erway L C, Ng L, Altschuler R, Curran T. Thyroid hormone receptor beta is essential for development of auditory function. Nat Genet. 1996;13:354–357. doi: 10.1038/ng0796-354. [DOI] [PubMed] [Google Scholar]
- 12.Fraichard A, Chassande O, Plateroti M, Roux J P, Trouillas J, Dehay C, Legrand C, Gauthier K, Kedinger M, Malaval L, Rousset B, Samarut J. The TRα gene encoding a thyroid hormone receptor is essential for post-natal development and thyroid hormone production. EMBO J. 1997;16:4412–4420. doi: 10.1093/emboj/16.14.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franklyn J A. Metabolic changes in hypothyroidism. In: Braverman L E, Utiger R D, editors. The thyroid. Philadelphia, Pa: Lippincott, Williams and Wilkins; 2000. pp. 833–836. [Google Scholar]
- 14.Gauthier K, Chassande O, Plateroti M, Roux J P, Legrand C, Pain B, Rousset B, Weiss R, Trouillas J, Samarut J. Different functions for the thyroid hormone receptors TRα and TRβ in the control of thyroid hormone production and post-natal development. EMBO J. 1999;18:623–631. doi: 10.1093/emboj/18.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glass C K, Rosenfeld M G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 16.Göthe S, Wang Z, Ng L, Kindblom J M, Campos Barros A, Ohlsson C, Vennström B, Forrest D. Mice devoid of all known thyroid hormone receptors are viable but exhibit disorders of pituitary-thyroid axis, growth, and bone maturation. Genes Dev. 1999;13:1329–1341. doi: 10.1101/gad.13.10.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hashimoto K, Curty F H, Borges P P, Lee C E, Abel E D, Elmquist J K, Cohen R N, Wondisford F E. An unliganded thyroid hormone receptor causes severe neurological dysfunction. Proc Natl Acad Sci USA. 2001;98:3998–4003. doi: 10.1073/pnas.051454698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansson C, Gothe S, Forrest D, Vennstrom B, Thoren P. Cardiovascular phenotype and temperature control in mice lacking thyroid hormone receptor-beta or both alpha1 and beta. Am J Physiol. 1999;276:H2006–H2012. doi: 10.1152/ajpheart.1999.276.6.H2006. [DOI] [PubMed] [Google Scholar]
- 19.Kaneshige M, Kaneshige K, Zhu X-G, Dace A, Garrett L, Carter T A, Kazlauskaite R, Pankratz D G, Wynshaw-Boris A, Refetoff S, Weintraub B, Willinham M C, Barlow C, Cheny S-Y. Mice with a targeted mutation in the thyroid hormone β receptor gene exhibit impaired growth and resistance to thyroid hormone. Proc Natl Acad Sci USA. 2000;97:13209–13214. doi: 10.1073/pnas.230285997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langlois M-F, Zanger K, Monden T, Safer J D, Hollenberg A N, Wondisford F E. A unique role of the b2 thyroid hormone receptor isoform in negative regulation by thyroid hormone. J Biol Chem. 1997;272:24927–24933. doi: 10.1074/jbc.272.40.24927. [DOI] [PubMed] [Google Scholar]
- 21.Lazar M A, Hodin R A, Chin W W. Human carboxyl-terminal variant of α-type c-erbA inhibit trans-activation by thyroid hormone receptors without binding thyroid hormone. Proc Natl Acad Sci USA. 1989;86:7771–7774. doi: 10.1073/pnas.86.20.7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazar M A. Thyroid hormone receptors: multiple forms, multiple possibilities. Endocr Rev. 1993;14:184–193. doi: 10.1210/edrv-14-2-184. [DOI] [PubMed] [Google Scholar]
- 23.Legrand J. Thyroid hormone effects on growth and development. In: Hennemann G, editor. Thyroid hormone metabolism. Rotterdam, The Netherlands: Dekker; 1986. pp. 503–534. [Google Scholar]
- 24.Lezoualc'h F, Hassan A H, Giraud P, Loeffler J P, Lee S L, Demeneix B. Assignment of the β-thyroid hormone receptor to 3,5,3′-triiodothyronine-dependent inhibition of transcription from the thyrotropin-releasing hormone promoter in chick hypothalamic neurons. Mol Endocrinol. 1992;6:1797–1804. doi: 10.1210/mend.6.11.1480171. [DOI] [PubMed] [Google Scholar]
- 25.Macchia P E, Takeuchi Y, Kawai T, Cua K, Gauthier K, Chassande O, Seo H, Hayashi Y, Samarut J, Murata Y, Weiss R E, Refetoff S. Increased sensitivity to thyroid hormone in mice with complete deficiency of thyroid hormone receptor α. Proc Natl Acad Sci USA. 2001;98:349–354. doi: 10.1073/pnas.011306998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meunier P J. A yearly survey of developments in the field of bone and mineral metabolism. Amsterdam, The Netherlands: W. A. Peck; 1983. [Google Scholar]
- 27.Nuclear Receptors Committee. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- 27a.Plateroti M, Gauthier K, Domon-Dell C, Freund J-N, Samarut J, Chassande O. Functional interference between thyroid hormone receptor α (TRα) and natural truncated TRΔα isoforms in the control of intestine development. Mol Cell Biol. 2001;21:4761–4772. doi: 10.1128/MCB.21.14.4761-4772.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plateroti M, Chassande O, Fraichard A, Gauthier K, Freund J-N, Samarut J, Kedinger M. Involvement of T3Rα- and β-receptor subtypes in mediation of T3 functions during post-natal murine intestinal development. Gastroenterology. 1999;116:1367–1378. doi: 10.1016/s0016-5085(99)70501-9. [DOI] [PubMed] [Google Scholar]
- 29.Pohlenz J, Maqueem A, Cua K, Weiss R E, Van Sande J, Refetoff S. Improved radioimmunoassay for measurement of mouse TSH in serum: strain differences in TSH concentration and thyrotroph sensitivity to thyroid hormone. Thyroid. 1999;9:1265–1271. doi: 10.1089/thy.1999.9.1265. [DOI] [PubMed] [Google Scholar]
- 30.Refetoff S. Resistance to thyroid hormone. In: Braverman L E, Utiger R D, editors. The thyroid. Philadelphia, Pa: Lippincott, Williams and Wilkins; 2000. pp. 1028–1043. [Google Scholar]
- 31.Robson H, Siebler T, Stevens D A, Shalet S M, Williams G R. Thyroid hormone acts directly on growth plate chondrocytes to promote hypertrophic differentiation and inhibit clonal expansion and cell proliferation. Endocrinology. 2000;141:3887–3897. doi: 10.1210/endo.141.10.7733. [DOI] [PubMed] [Google Scholar]
- 32.Savatier P, Lapillonne H, van Grunsven L, Rudkin B B, Samarut J. Withdrawal of differentiation of inhibitory activity/leukemia inhibitory factor up-regulates D-type cyclins and cyclin-dependent kinase inhibitors in mouse embryonic stem cells. Oncogene. 1996;12:309–322. [PubMed] [Google Scholar]
- 33.Scanlon M F, Toft A D. Regulation of thyrotropin secretion. In: Braverman L E, Utiger R D, editors. Werner and Ingbar's the thyroid: a fundamental and clinical text. 7th ed. New York, N.Y: Lippincott-Raven; 1996. pp. 220–240. [Google Scholar]
- 34.Stevens D A, Hasserjian R P, Robson H, Siebler T, Shalet S M, Williams G R. Thyroid hormones regulate hypertrophic chondrocyte differentiation and expression of parathyroid hormone-related protein and its receptor during endochondral bone formation. J Bone Miner Res. 2000;15:2431–2442. doi: 10.1359/jbmr.2000.15.12.2431. [DOI] [PubMed] [Google Scholar]
- 35.Turner J G, Willott J F. Exposure to an augmented acoustic environment alters progressive hearing loss in DBA/2J mice. Hear Res. 1998;118:101–113. doi: 10.1016/s0378-5955(98)00024-0. [DOI] [PubMed] [Google Scholar]
- 36.Weiss R E, Refetoff S. Effect of thyroid hormone on growth: lessons from the syndrome of resistance of thyroid hormone. Endocrinol Metab Clin N Am. 1996;25:719–730. doi: 10.1016/s0889-8529(05)70349-2. [DOI] [PubMed] [Google Scholar]
- 37.Weiss R E, Forrest D, Pohlenz J, Cua K, Curran T, Refetoff S. Thyrotropin regulation by thyroid hormone in thyroid hormone receptor beta-deficient mice. Endocrinology. 1997;138:3624–3629. doi: 10.1210/endo.138.9.5412. [DOI] [PubMed] [Google Scholar]
- 38.Weiss R E, Murata Y, Cua K, Hayashi Y, Seo H, Refetoff S. Thyroid hormone action on liver, heart and energy expenditure in thyroid hormone receptor β deficient mice. Endocrinology. 1998;139:4945–4952. doi: 10.1210/endo.139.12.6412. [DOI] [PubMed] [Google Scholar]
- 39.Wikström L, Johansson C, Salto C, Barlow C, Campos Barros A, Baas F, Forrest D, Thoren P, Vennström B. Abnormal heart rate and body temperature in mice lacking thyroid hormone receptor α1. EMBO J. 1998;17:455–461. doi: 10.1093/emboj/17.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams G R, Robson H, Shalet S M. Thyroid hormone actions on cartilage and bone: interactions with other hormones at the epiphyseal plate and effects on linear growth. J Endocrinol. 1998;157:391–403. doi: 10.1677/joe.0.1570391. [DOI] [PubMed] [Google Scholar]
- 41.Williams G R. Cloning and characterization of two novel thyroid hormone receptor β isoforms. Mol Cell Biol. 2000;20:8329–8342. doi: 10.1128/mcb.20.22.8329-8342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]