Mini-Abstract
RNA is dynamic and can fold into multiple alternative conformations. A new study shows that the cell can control mRNA translation by regulating the stoichiometry of alternative RNA structures formed from extremely conserved 5’UTR sequences and that this control may contribute to embryonic development.
Extremely conserved elements are long stretches of DNA that match almost identically with corresponding regions in other vertebrate genomes1. While there is evident pressure to conserve protein coding sequences, most of these hyperconserved elements are found in non-coding regions of the genome adding to the mystery of their role and origin. To date, most efforts to characterize hyperconserved elements have focused on intergenic sequences and their potential role as transcriptional enhancers2. Comparatively, their impact on post-transcriptional regulation remains largely unexplored. While a few examples of the functional roles of hyperconserved elements in long non-coding RNAs3 and alternatively spliced cassette exons4,5 exist, how these conserved elements regulate RNA functions is poorly understood. In this issue, Byeon et al.6 demonstrate that hyperconserved 5’ untranslated regions (UTRs) control non-canonical mRNA translation across cell-types and coordinate an unexpected RNA structure-driven regulatory mechanism connecting extreme conservation to function.
Development and non-canonical translation
Non-canonical translation initiation occurs when an mRNA recruits ribosomes independently of the 5’-cap and canonical machinery (i.e., the eIF4F complex)7. This mode of translation initiation is essential for many RNA viruses, which encode internal ribosome entry sites (IRESs) that directly mobilize ribosomes for the translation of uncapped viral RNAs. IRESs are also found in a subset of capped cellular mRNAs8. The better-known role of cellular IRESs is to maintain translation of genes required for the adequate cellular response to various stresses when canonical translation is suppressed9. To date, little is known about the role of cellular IRESs in the absence of stress.
Recently, a conserved IRES in the Hoxa9 5’UTR was shown to regulate the spatiotemporal expression of Hoxa9 and to be detrimental to embryonic development10. These findings suggest that IRESs could act as regulators of physiological gene expression programs that orchestrate vertebrate development, but their prevalence and molecular mechanism remained uncharted territory. To evaluate the extent of translation regulation by cis-regulatory elements, Byeon et al. carried out a large-scale analysis of 5’UTRs and identified >500 hyperconserved 5’UTR (h5UTR) regions across 60 vertebrate genomes. Interestingly, these were enriched in genes critical for embryonic development. CRISPR-mediated endogenous deletion of five h5UTRs led to decreased mRNA translation for each, suggesting that h5UTRs harbor translation enhancers. In a tour de force, Byeon et al. cloned a library of 253 full-length h5UTRs into a bicistronic reporter construct and measured the level of non-canonical translation mediated by these h5UTRs in five different cell types, representing different lineages and differentiation trajectories in the developing embryo. Remarkably, 90 out of the 253 h5UTRs tested (36%) showed high non-canonical translation activity in at least one cell type. Of those, 36 h5UTRs exhibited variable activities across different cell types. These results suggest that h5UTR regions are enriched in enhancers of non-canonical translation with differential activities across cell types and cellular environments (Fig. 1a). These characteristics are reminiscent of transcriptional enhancers that are well established controllers of the spatiotemporal transcription of genes shaping vertebrate development. Therefore, conserved 5’UTR enhancers of non-canonical translation may act as key controllers of the spatiotemporal translation of genes essential for embryonic development. However, while these results begin to address the prevalence of this novel class of regulatory elements, the molecular mechanisms by which they confer regulation remain largely undescribed.
Figure 1. h5UTRs may act as key regulators of translation during vertebrate development.
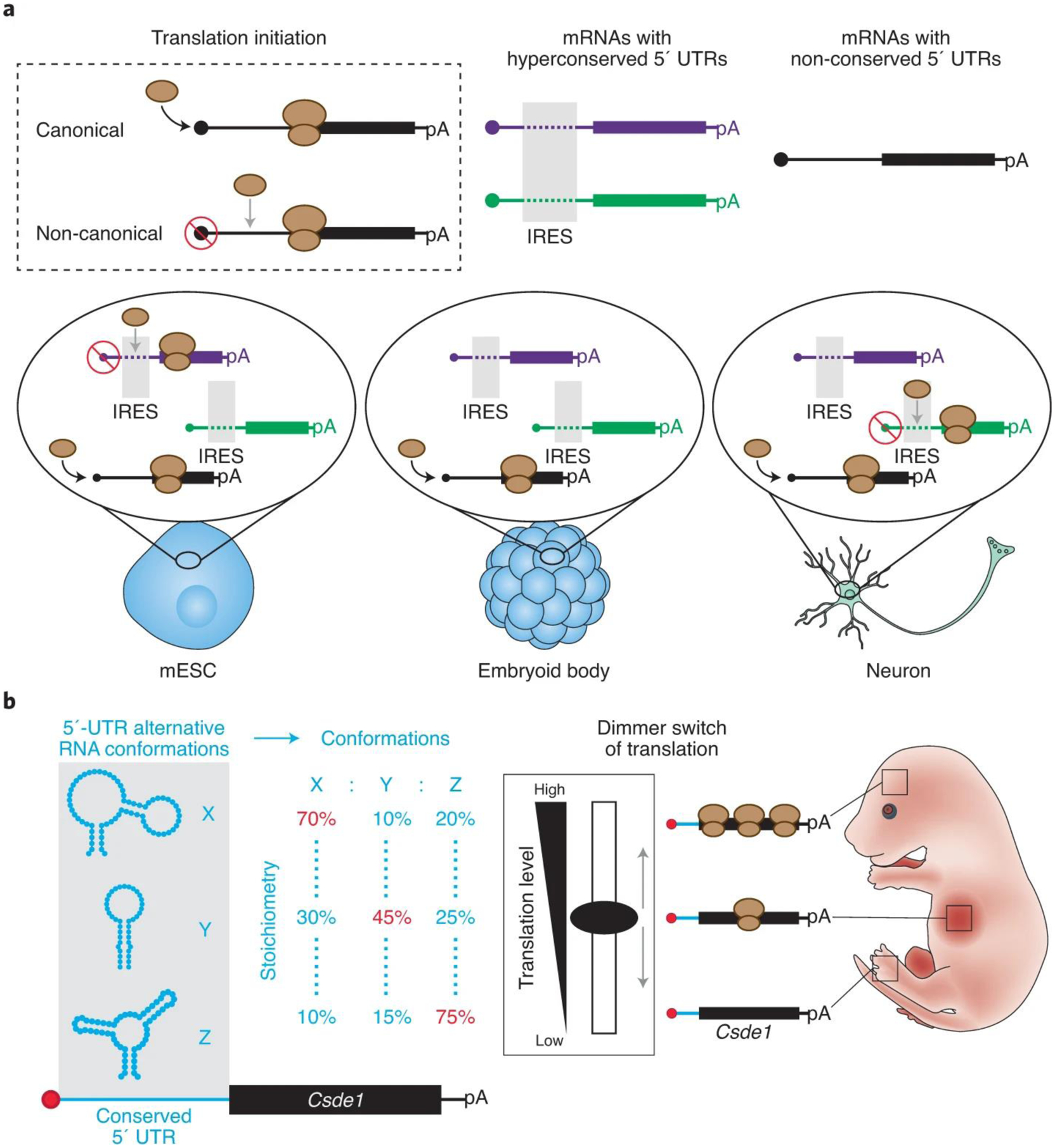
a, Hyperconserved 5’UTRs are enhancers of non-canonical expression with cell type-dependent activities. b, The Csde1 hyperconserved 5’UTR forms an ensemble of RNA conformations. The dynamics of this RNA folding landscape acts as a structure-dependent dimmer switch of non-canonical translation. This RNA dimmer switch may control the spatiotemporal translation of Csde1 during embryonic development.
Structural stoichiometry’s key role
RNA follows a hierarchical folding pathway, where primary sequences form secondary structures that guide tertiary interactions and 3D RNA structures11. Because RNA structure directly dictates RNA activity, it is paramount to understand how RNA regulatory elements fold to elucidate their molecular mechanisms. This is particularly true for IRES activities that have been shown to rely on the formation of specific RNA structures7. RNA molecules are also extremely dynamic and a single sequence can often follow different folding pathways to form different RNA conformations. The choice between one folding pathway and another is directly influenced by the cellular environment (e.g., pH, molecular crowding, RNA helicases and other factors)12. RNA helicases are conserved remodelers of RNA structures and are central to post-transcriptional regulation. To date, which RNA regions are remodeled by helicases in the cell and how they impact RNA regulation remain mostly unknown. To address the extent of h5UTR structural remodeling in the cell, Byeon et al. used dimethyl sulfate probing to analyze the effect of inhibiting RNA helicase activity on the structures of 69 h5UTRs. They found that the structures of 20 out of 69 h5UTRs (29%) were significantly remodeled in the presence of specific cellular helicases. Most of these elements were in mRNAs encoding genes annotated as critical for embryonic development. One of the most noticeable structural reorganizations was observed in the Csde1 5’UTR. Further characterization of Csde1 5’UTR structural dynamics using a new methodology (in-cell mutate-and-map (icM2)) revealed the formation of three alternative conformations (Fig. 1b). Strikingly, the stoichiometry of each conformation was sensitive to RNA helicase activity as well as to single nucleotide mutations in the extremely conserved region. Remarkably, these changes had direct impacts on mRNA translation. While this in-depth analysis of RNA structural dynamics and function was limited to one h5UTR, it suggests that Csde1 mRNA translation is fine-tuned through the relative proportion of multiple conformations in the RNA structural landscape of its 5’UTR (Fig. 1b).
Thus, work from Byeon et al. sheds new light on why mysterious hyperconserved elements in the transcriptome exist in some non-coding regions. First impressions would not suggest that RNA structures could explain such strong sequence conservation pressure. This is because sequence covariation in paired regions can result in the formation of the same structure by different sequences. However, in a context where the regulatory activity of a region relies on the relative proportion of an ensemble of RNA conformations, changing one base pair for another in the stem of one conformation will perturb the folding of the others and alter the overall regulation. In other words, these results support a new paradigm where hyperconserved sequences encode unique RNA folding landscapes that toggle between different alternative conformations in response to cellular environmental changes and act as molecular dimmer switches of RNA activities (Fig. 1b). Importantly, these regulatory RNA structure dimmer switches may be detrimental to embryonic development. The combination of novel approaches to deconvolute the various RNA conformations formed by a single sequence (e.g. icM2, PAIR-MaP13, DREEM14 and DRACO15) coupled with functional assays will reveal the importance of RNA structural dynamics on RNA-mediated gene regulation. Further characterization of these RNA dimmer switches and the cellular trans-factors that control them will uncover novel regulatory pathways shaping the spatiotemporal expression of genes in various biological systems.
References
- 1.Katzman S et al. Human Genome Ultraconserved Elements Are Ultraselected. Science 317, 915–915 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Visel A et al. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet 40, 158–160 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calin GA et al. Ultraconserved Regions Encoding ncRNAs Are Altered in Human Leukemias and Carcinomas. Cancer Cell 12, 215–229 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Lareau LF, Inada M, Green RE, Wengrod JC & Brenner SE Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446, 926–929 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Pirnie SP, Osman A, Zhu Y & Carmichael GG An Ultraconserved Element (UCE) controls homeostatic splicing of ARGLU1 mRNA. Nucleic Acids Res 45, 3473–3486 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byeon GW et al. Functional and structural basis of extreme conservation in vertebrate 5′ untranslated region. Nat Genet 53, 729–741 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leppek K, Das R & Barna M Functional 5’ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biology 19, 158–174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mokrejš M et al. IRESite—a tool for the examination of viral and cellular internal ribosome entry sites. Nucleic Acids Res 38, D131–D136 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lacerda R, Menezes J & Romão L More than just scanning: the importance of cap-independent mRNA translation initiation for cellular stress response and cancer. Cell Mol Life Sci 74, 1659–1680 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue S et al. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature 517, 33 38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reymond C, Beaudoin J-D & Perreault J-P Modulating RNA structure and catalysis: lessons from small cleaving ribozymes. Cell Mol Life Sci 66, 3937 3950 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganser LR, Kelly ML, Herschlag D & Al-Hashimi HM The roles of structural dynamics in the cellular functions of RNAs. Nat Rev Mol Cell Bio 20, 474–489 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mustoe AM, Lama NN, Irving PS, Olson SW & Weeks KM RNA base-pairing complexity in living cells visualized by correlated chemical probing. Proc National Acad Sci 116, 24574–24582 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomezsko PJ et al. Determination of RNA structural diversity and its role in HIV-1 RNA splicing. Nature 1–5 (2020) doi: 10.1038/s41586-020-2253-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morandi E et al. Genome-scale deconvolution of RNA structure ensembles. Nat Methods 18, 249–252 (2021). [DOI] [PubMed] [Google Scholar]


