Abstract
Although most gastrointestinal stromal tumors (GISTs) exhibit activating mutations in either KIT or PDGFRA, rare cases have shown to be driven by gene fusions involving kinases, mainly involving NTRK3, and rarely BRAF or FGFR1. BRAF gene rearrangements have been described in only two patients to date, as separate case reports. In addition, BRAF V600E mutation is an uncommon but established oncogenic pathway in GIST. In this report, we describe two new GIST cases harboring novel BRAF fusion genes, arising in two young-adult women (37 and 40 years of age) in the small bowel and distal esophagus, both with a spindle cell phenotype. The small bowel GIST measured 2.8 cm and showed a high cellularity and a mitotic rate of 20/50 HPFs, while the esophageal lesion measured 7 cm and 1/50 HPFs. Immunohistochemically, both tumors showed diffuse reactivity for DOG1, while KIT/CD117 was weakly positive in the small bowel GIST and completely negative in the esophageal tumor. Based on these findings, the latter case was misinterpreted as a low-grade myxoid leiomyosarcoma, as it showed a myxoid stroma, reactivity for SMA and focal positivity for desmin. Archer FusionPlex revealed a fusion between BRAF with either AGAP3 or MKRN1 gene partners. Moreover, MSK-IMPACT DNA targeted sequencing confirmed both fusions but did not identify additional mutations. In one case with available material, the BRAF gene rearrangement was also validated by FISH. The recognition of BRAF fusion-positive GISTs is critical as it may be associated with a low level of KIT expression and may result in diagnostic challenges with significant impact on therapeutic management. The clinical benefit with KIT inhibitors, such as imatinib, remains to be determined.
Keywords: BRAF, fusion, gastrointestinal stromal tumors, KIT
1 |. INTRODUCTION
Gastrointestinal stromal tumor (GIST) is the most common mesenchymal tumor arising in the gastrointestinal tract. Over the last two decades, the molecular abnormalities underpinning these tumors have been discovered, with up to 85–92% of cases in adults harboring mutually exclusive gain-of function KIT or PDGFRA mutations.1–3 Instead, gastric tumors in the pediatric and young adult patients are often wild-type for these mutations4 and harbor alterations resulting in a deficiency in the succinate dehydrogenase (SDH) complex.2,5,6 Moreover, these SDH-deficient GISTs form the underlying pathogenetic basis of syndromic cases such as Carney triad and Carney– Stratakis syndrome.7,8 Patients with Type I Neurofibromatosis (NF-1) also develop GIST at an increased frequency, often multifocal, typically in the small bowel and associated with interstitial cell of Cajal hyperplasia, driven by NF-1 rather than KIT or PDGFRA mutations.9,10 Additionally, BRAF V600E mutations have been found in KIT-expressing GISTs lacking KIT/PDGFRA/SDH abnormalities, and in rare cases of imatinib-resistant GIST.11,12 In the last few years, as a result of wide application of targeted RNA sequencing in clinical practice, a small subset of GIST driven by gene fusions resulting in oncogenic kinase activation has been identified, including a handful of cases with FGFR1 and NTRK3 fusions.13,14 In this study, we report on two BRAF-fusion positive spindle cell GISTs which posed diagnostic challenges due to low or absent KIT expression. Thus, further investigation unmasking molecular alterations in these tumors can facilitate accurate classification and detect tumors unlikely to respond to current targeted therapy (Table 1).
TABLE 1.
Summary of clinical, pathologic, and molecular findings of four BRAF-fusions positive GIST
| Charo et al23 | Vanden Bempt et al3 | Case 1 | Case 2 | |
|---|---|---|---|---|
| Age | 34 | 64 | 40 | 37 |
| Gender | F | M | F | F |
| Site | Small intestine | Stomach | Small intestine | Esophagus |
| Size (cm) | 14 | 2.5 | 2.8 | 7.3 |
| Morphology | Spindle | Spindle | Spindle | Spindle |
| Mitotic rate | 3/50 HPF | ≤5/50HPF | 20/50 HPF | <1/50 HPF |
| IHC | CD117 positive DOG1 positive |
N/A | CD117 weakly pos DOG1 positive 18 mo NED |
CD117 negative DOG1 positive SMA positive Desmin rare pos ER negative 36 mo/NED |
| Fusion |
PRKAR1B-BRAF PRKAR1B exon 9-BRAF exon 9 |
TRIM4-BRAF TRIM4 exon 6-BRAF exon 10 |
AGAP3-BRAF AGAP3 exon 11-BRAF exon 10 |
MKRN1-BRAF MKRN1 exon 4-BRAF exon 11 |
| Survival/therapy | N/A | N/A | 18 mo NED Surgery |
36 mo NED Surgery |
Abbreviations: mo, months; NA, not available; NED, no evidence of disease; pos, positive.
2 |. MATERIALS AND METHODS
Tissue was analyzed prospectively in the course of management as clinical cases of patients referred to our institution for continued care. The tumors were subjected to morphologic and immunohistochemical analysis, targeted RNA sequencing (Archer FusionPlex), MSK-IMPACT, and fluorescence in situ hybridization (FISH). Significant clinical follow-up is not yet available due to the recent nature of the cases. This study was approved by the institutional review board.
2.1 |. Immunohistochemistry
The immunohistochemical analysis was performed on formalin-fixed, paraffin-embedded (FFPE) tissue sections of 4 μm thickness. Antibody epitope retrieval was performed using standard protocols for the following markers: KIT (CD117), DOG1, S100, SOX10, cytokeratin AE1: AE3, desmin, and smooth muscle actin (SMA). BRAF immunohistochemistry for BRAF V600E was performed in one case with available tissue.
2.2 |. Fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) on interphase nuclei from paraffin-embedded 4-μm sections was performed applying custom probes using bacterial artificial chromosomes (BAC) covering and flanking genes of interest. A BAC clone for BRAF was chosen according to UCSC genome browser (http://genome.ucsc.edu), as previously described.15,16 The BAC clone was obtained from BACPAC sources of Children's Hospital of Oakland Research Institute (CHORI; Oakland, CA; https://bacpacresources.org/). DNA from individual BACs was isolated according to the manufacturer's instructions, labeled with different fluorochromes in a nick translation reaction, denatured, and hybridized to pretreated slides. Slides were then incubated, washed, and mounted with DAPI in an antifade solution, as previously described.16 The genomic location of each BAC set was verified by hybridizing them to normal metaphase chromosomes. Two hundred successive nuclei were examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by the Isis 5 software (Metasystems, Newton, MA). A positive score was interpreted when at least 20% of the nuclei showed a split-apart signal in the break-apart assay. Nuclei with an incomplete set of signals were omitted from the score.
2.3 |. Targeted RNA sequencing
RNA is extracted from formalin-fixed paraffin-embedded tumor material followed by cDNA synthesis. cDNA libraries were made using the ArcherTM FusionPlexTM standard protocol and supplied reagents, including Archer® Universal RNA Reagent Kit for Illumina® (Catalog #AK0040–8), as previously described.17 Fusion unidirectional gene specific primers were designed to target specific exons in 62 genes known to be involved in chromosomal rearrangements based on current literature. At the end of the two-PCR steps the final targeted amplicons were sequenced (2 × 150 bp) on an Illumina MiSeq sequencer. Data analysis was performed using the ArcherTM analysis software settings.17
2.4 |. Next generation sequencing (MSK-IMPACT)
The IMPACT next generation sequencing platform has been described in detail previously.18 It is an FDA-approved hybridization capture-based genomic sequencing assay performed in a Clinical Laboratory Improvement Amendments-certified laboratory that examines all exons and selected introns of 468 cancer-associated genes. Genomic alterations detected on IMPACT are annotated according to the OncoKB database,19 a precision oncology knowledge base denoting the oncogenic effects and predictive significance of molecular alterations. Genomic data and OncoKB annotations were visualized in cBioPortal for Cancer Genomics.20,21
3 |. RESULTS
3.1 |. Case reports
3.1.1 |. Case 1
The patient was a 40 year-old woman who was incidentally found to have a mass involving the small intestine on imaging on a workup for a pyelonephritis. The patient underwent a laparoscopic exploration and an en-bloc resection of the proximal jejunal mass. On gross examination, the tumor measured 2.8 × 2.5 × 2.0 cm and showed a solid, white cut-surface. Microscopically, the tumor was vaguely multi-nodular and composed of intersecting fascicles of spindle cells with pale eosinophilic cytoplasm and uniform fusiform nuclei. Focally, cytoplasmic vacuoles and skeinoid fibers were observed. (Figure 1). The mitotic count was brisk (20/50 HPFs), but necrosis was not identified. Immunohistochemical analysis showed that the tumor cells were positive for DOG1 (Figure 1) and weakly positive for KIT/CD117. Based on the high-risk features, patient was started on adjuvant imatinib therapy for 1 month and then discontinued once the molecular results revealed no mutations in the KIT/PDGRFA genes. The patient is free of recurrence 18 months since diagnosis.
FIGURE 1.
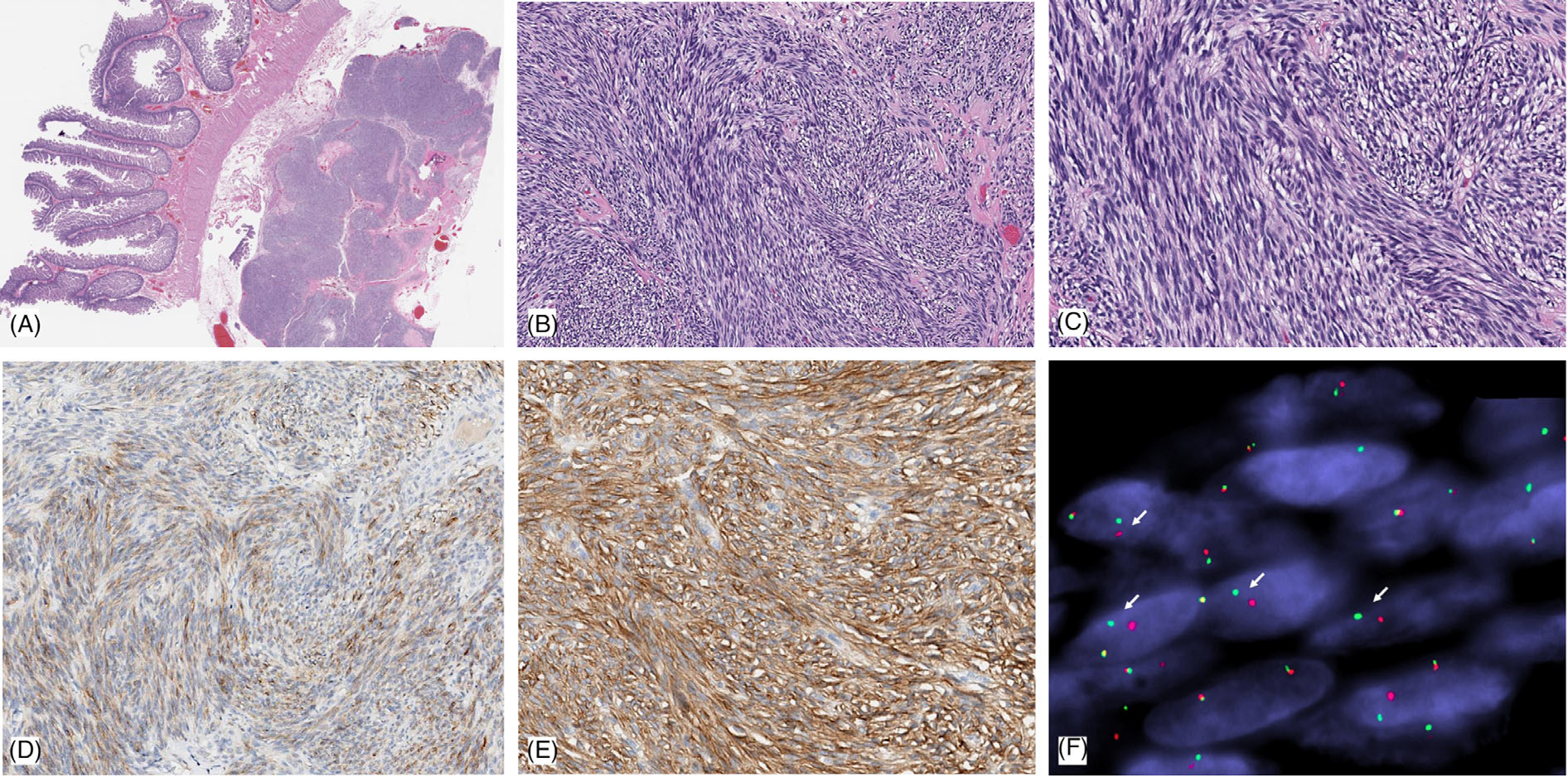
Pathologic features of case 1: small intestinal GIST harboring a novel AGAP3-BRAF fusion. Low power view shows a lobulated tumor within the muscularis propria and subserosal layer of the small bowel (A). Intermediate power showing highly cellular, intersecting fascicles of spindle cells (B); while at high power show eosinophilic cytoplasm, intracytoplasmic vacuoles, and monomorphic nuclei with fine chromatin. (C). Immunohistochemically the tumor showed weak staining for KIT/CD117 (D), while there was diffuse, strong staining for DOG1 (E). (F). FISH shows break-apart red (centromeric) and green (telomeric) in keeping with a BRAF gene rearrangement (the narrow, fixed gaps between the break-apart signals support an intrachromosomal inversion)
3.1.2 |. Case 2
The patient was a 37 year-old woman who presented with dysphagia and was found to have a large mass involving the distal esophagus. The patient underwent a video-assisted thoracoscopic resection of the mass. On gross examination, the tumor appeared well-circumscribed and measured 7.3 × 5.5 × 4.0 cm, with a pale-yellow, glistening cut surface. Microscopic examination revealed a tumor with well-defined borders, compose spindle cells with fibrillary eosinophilic cytoplasm and uniform fusiform nuclei, embedded in a predominantly myxoid stroma (Figure 2). Mitotic figures were rare (1 MF/50 HPFs) and necrosis was not present. Surgical margins were free of involvement. Immunohistochemically the tumor was positive for DOG1, CD34, SMA, and focal reactivity to desmin (Figure 2). Other stains, including KIT/CD117, AE1:AE3, EMA, S100, ALK, and inhibin were negative. The patient is free of disease 36 months since initial diagnosis without further therapy.
FIGURE 2.
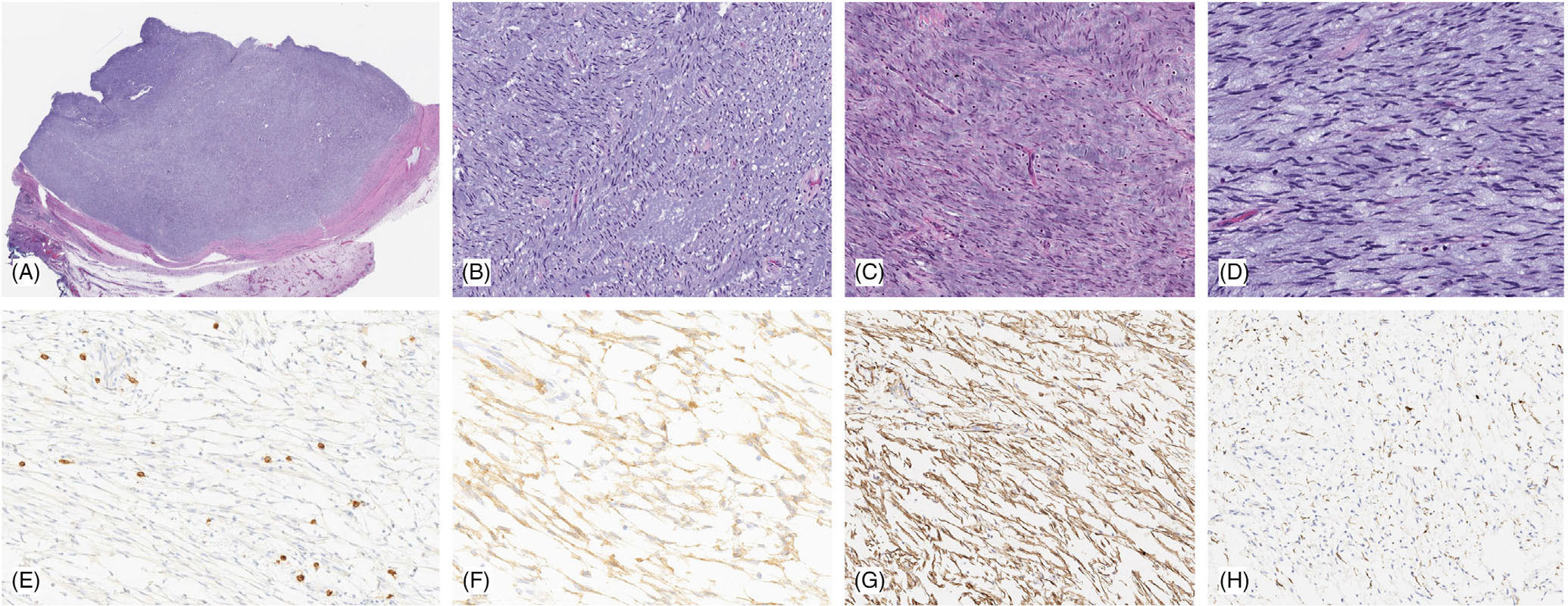
Pathologic features of distal esophageal GIST with MKRN1-BRAF fusion. Low power shows a well-circumscribed lesion surrounded by a fibrous capsule (A), which is composed of loose fascicles of bland spindle cells with scant eosinophilic cytoplasm and ovoid unform nuclei with fine chromatin (B,C). The tumor is associated with extensive myxoid stroma and scattered mast cells (D). Immunohistochemically the tumor cells were negative for KIT/CD117 (E) (which highlights the stromal mast cells, as internal positive control), while diffusely positive for DOG1 (F) and SMA (G), and only rare cells label with desmin (H)
3.2 |. Molecular findings
In case 1, both Archer FusionPlex and IMPACT testing showed the presence of an AGAP3-BRAF fusion gene. The Archer FusionPlex confirmed the fusion transcript involving AGAP3 exon 11 and BRAF exon 10 (Figure 3). The BRAF gene rearrangement was subsequently confirmed by a FISH study.
FIGURE 3.
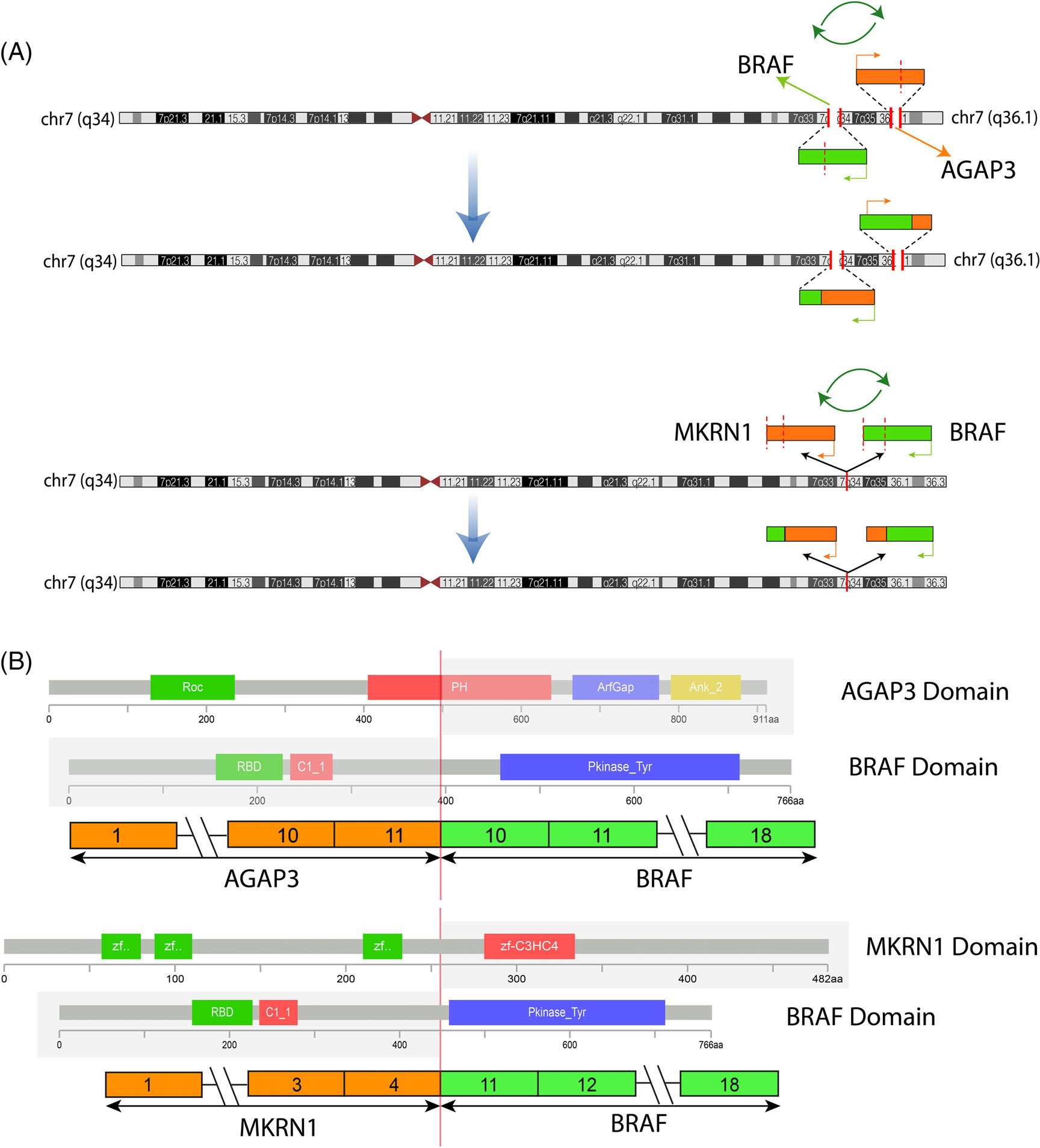
Diagrammatic representation of the two intrachromosomal BRAF fusions. (A). Schematic view of BRAF gene location on 7q34 (green box) and its two fusion partners AGAP3 on 7q36.1 and MKRN1 on 7q34 (orange boxes). The direction of transcription of each gene is shown by an orange or green arrow. Green circular arrows indicate that both fusions result from a complex process of break, inversion, and fusion, resulting in a functional transcript retaining the BRAF kinase domain as the 3' partner in both cases. (B). Upper portion reveals a fusion transcript composed of AGAP3 exon 11 fused to exon 10 of BRAF; while the lower portion shows MKRN1 exon 4 fused to exon 11 of BRAF. In both cases, the projected fusion oncoprotein retains the BRAF kinase domain intact. The protein domains of the participating genes are also displayed
In case 2, both Archer FusionPlex and IMPACT testing showed the presence of an MKRN1-BRAF fusion. The Archer FusionPlex confirmed the fusion transcript involving MKRN1 exon 4 fused to BRAF exon 11 (Figure 3).
Copy number variations were assessed in case 1, where IMPACT identified copy number losses on chromosome arms 1p32, 3p11–13, 3p25, 7q36, and 14q12–31. In both cases, IMPACT did not identify any additional mutations.
4 |. DISCUSSION
Molecular analysis over the past two decades has transformed the diagnosis and management of GIST. The discovery of gain of function mutations in KIT has led to the development of diagnostic immunohistochemistry, provided prognostic indicators, and driven therapeutics.22 In the years following this initial discovery, investigators have further unraveled the molecular drivers in KIT wild-type disease to include PDGFRA/SDH(complex)/NF-1/BRAF mutations.7–11 More recently as result of targeted RNA sequencing rare GIST cases were identified harboring oncogenic kinase gene fusions rather than kinase mutations, specifically involving FGFR1 and NTRK3.13,14 In the study by Shi et al,13 among 24 wild-type GISTs for KIT/PDGFRA/ RAS mutations, two tumors harbored FGFR1 fusions involving (FGFR1-HOOK3 and FGFR1-TACC1; which included most of the FGFR1 kinase fusion domain) and one ETV6-NTRK3. In that series, the patient with small bowel GIST with ETV6-NTRK3 fusion progressed on five lines of therapy, including imatinib, sunitinib, sorafenib, nilotinib, and regorafenib, before the LOXO-101 therapy was instituted based on the genetic findings. The patient showed immediate improvement in his symptoms, with tumor response to LOXO-101 seen at the end of week 8 by PET/CT and an ongoing partial response (44%) according to RECIST 1.1 criteria. One case report of ETV6-NTRK3 fusion positive rectal GIST described by Brenca et al14 showed diffuse and strong reactivity for KIT protein by immunohistochemistry.
Recently, two GIST cases, one each harboring a PRKAR1B-BRAF and TRIM4-BRAF fusion, were reported by two groups of investigators. The PRKAR1B-BRAF fusion occurred in a 14 cm tumor in the small intestine of a 34-year-old woman,23 while the TRIM4-BRAF fusion was identified in a 2.5 cm gastric lesion in a 64 year-old man.3 Detailed pathologic features were not reported.
We report two additional cases of GIST with BRAF gene fusions and present detailed clinical and pathologic features herein. In case 1, the tumor was located in the small bowel and exhibited a tightly packed fascicular growth of spindle cells with pale, eosinophilic cytoplasm. Focal intracytoplasmic vacuoles and skeinoid fibers were present, the latter finding usually observed in small bowel GIST.24,25 The tumor had a high risk of malignancy based on the brisk mitotic rate. In case 2, the tumor was located in the distal esophagus and showed lobulated borders. The tumor exhibited a spindled phenotype with eosinophilic cytoplasm in an abundant myxoid stroma. The mitotic activity was inconspicuous. Despite the distinct clinical presentations, the two cases showed a similar immunophenotype, with DOG1 expression, while KIT/CD117 was either weak/focal (case 1) or completely negative (case 2). In fact, case 2 was initial misdiagnosed as a myxoid leiomyosarcoma at the primary institution, due to the abundant myxoid stroma and the lack of KIT positivity with focal desmin positivity. Positivity of smooth muscle markers is not unusual in GIST and has been previously described in GIST of the esophagus.26 In contrast to SDH-deficient GIST that have a predilection for stomach and NF1-syndromic GIST with predilection for small bowel, it appears that there is no anatomic location preference in the four GIST cases harboring BRAF fusions.
The low or no KIT expression in the two BRAF-fusion positive GIST cases raises important questions regarding the histogenesis of certain molecular variants of GIST (specifically kinase fusion positive GIST) and the utility of KIT immunostaining in this setting. GISTs are believed to arise from interstitial cells of Cajal, which show high levels of KIT immunoexpression.1,27 The majority of GISTs harbor a gain of function mutation in the KIT gene,1 which results in constitutive activation of the KIT tyrosine kinase receptor. In a study investigating 25 KIT immunonegative, morphologically typical GISTs there were 18 tumors harboring PDGFRA mutations, four showing KIT mutations, while the remaining three tumors were considered KIT/PDGFRA wild-type.28 Moreover, two other molecular variants of GIST, including SDH-deficient GIST29 and BRAF V600E-mutant GIST12 also show diffuse positivity for KIT. However, the status of KIT immunoexpression in the context of kinase-fusion positive GISTs is less clear. Intriguingly, in the study by Shi et al,13 the status of KIT/DOG1 expression in the GIST with FGFR1 and NTRK3 fusions was not provided, while the single case report in the study by Brenca et al14 of a rectal GIST with ETV6-NTRK3, both KIT and DOG1 were diffusely and strongly positive by immunohistochemistry. In contrast, a recent report of 8 mesenchymal tumors of the GI tract with NTRK gene rearrangements (including both NTRK1 and NTRK3 fusions) showed none were KIT or DOG1 immunopositive and thus appeared to be unrelated to GIST.30
Moreover, the status of KIT/DOG1 staining was only documented in the small bowel GIST with PRKAR1B-BRAF fusion23 being both positive, while no information was provided in the second case of a gastric GIST.3 Although the weak or absent KIT immunoreactivity in our two cases with BRAF fusions triggered diagnostic challenge, the diffuse DOG1 staining combined with morphologic appearance confirmed the correct diagnosis of GIST. Similarly, DOG1 immunohistochemical stain has proven to be a reliable marker of KIT-negative GIST, particularly in the setting of PDGFRA mutant GISTs or other unusual molecular GIST subtypes.31,32
BRAF is a serine/threonine kinase and a member of the RAF family. Alterations involving the BRAF gene are increasingly recognized in human neoplasia.33 The point mutation resulting in the BRAF V600E mutant is present in 3.9–13% of GIST lacking KIT/PDGFRA mutations.11,12,34 BRAF related fusions have been previously described in other mesenchymal neoplasms, including infantile fibrosarcoma-like tumors35 and myxoinflammatory fibroblastic sarcoma.36 In other cancers, BRAF fusions encode 5' protein partners that contribute coiled-coil or zinc-finger dimerization motifs, which likely produce constitutively activated BRAF dimers capable of driving tumorigenesis and poorly sensitive to RAF inhibitors, but sensitive to inhibition downstream, through MEK (mitogen-activated protein kinase kinase 1 and 2) inhibition.
Finally, the most critical impact of the BRAF fusion alteration in GISTs is the predicted drug resistance to specific tyrosine kinase inhibitor therapy, such as imatinib, the front line targeted therapy in metastatic or locally advanced GIST.22 Equally important are the alternative strategies which can be offered in the setting of this genotype, such as targeted therapies including RAF/pan-kinase inhibitor therapy (sorafenib)37,38 and MEK inhibitor therapy.39
In summary, we describe two cases of GIST with unusual morphologic and immunohistochemical findings and underlying BRAF related gene fusions. Awareness of this molecular variant of GIST may support the use of broader molecular analysis to improve diagnostic accuracy and broaden the scope for targeted therapy.
ACKNOWLEDGMENT
This study was supported by National Institute of Health, Grant Nos. P50 CA217694 (Cristina R Antonescu, Ping Chi), P50 CA 140146-01 (Cristina R Antonescu, Ping Chi), P30 CA008748 (Cristina R Antonescu, Ping Chi), GIST Cancer Research Fund (Cristina R Antonescu, Ping Chi), and Kristin Ann Carr Foundation (Cristina R Antonescu). All authors approve of the submission.
Footnotes
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1.Hirota S, Isozaki K, Moriyama Y, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998; 279(5350):577–580. [DOI] [PubMed] [Google Scholar]
- 2.Corless CL, Fletcher JA, Heinrich MC. Biology of gastrointestinal stromal tumors. J Clin Oncol 2004;22(18):3813–3825. [DOI] [PubMed] [Google Scholar]
- 3.Bempt I Vanden, Borght S Vander, Sciot R, et al. Comprehensive targeted next-generation sequencing approach in the molecular diagnosis of gastrointestinal stromal tumor. Genes Chromosomes Cancer 2021;60(4):239–249. [DOI] [PubMed] [Google Scholar]
- 4.Agaram NP, Laquaglia MP, Ustun B, et al. Molecular characterization of pediatric gastrointestinal stromal tumors. Clin Cancer Res 2008; 14(10):3204–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janeway KA, Kim SY, Lodish M, et al. Defects in succinate dehydrogenase in gastrointestinal stromal tumors lacking KIT and PDGFRA mutations. Proc Natl Acad Sci U S A 2011;108(1):314–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boikos SA, Pappo AS, Killian JK, et al. Molecular subtypes of KIT/PDGFRA wild-type gastrointestinal stromal tumors: a report from the National Institutes of Health gastrointestinal stromal tumor clinic. JAMA Oncol 2016;2(7):922–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasini B, McWhinney SR, Bei T, et al. Clinical and molecular genetics of patients with the Carney-Stratakis syndrome and germline mutations of the genes coding for the succinate dehydrogenase subunits SDHB, SDHC, and SDHD. Eur J Hum Genet 2008;16(1):79–88. [DOI] [PubMed] [Google Scholar]
- 8.Haller F, Moskalev EA, Faucz FR, et al. Aberrant DNA hypermethylation of SDHC: a novel mechanism of tumor development in Carney triad. Endocr Relat Cancer 2014;21(4):567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miettinen M, Fetsch JF, Sobin LH, Lasota J. Gastrointestinal stromal tumors in patients with neurofibromatosis 1: a clinicopathologic and molecular genetic study of 45 cases. Am J Surg Pathol 2006;30(1): 90–96. [DOI] [PubMed] [Google Scholar]
- 10.Maertens O, Prenen H, Debiec-Rychter M, et al. Molecular pathogenesis of multiple gastrointestinal stromal tumors in NF1 patients. Hum Mol Genet 2006;15(6):1015–1023. [DOI] [PubMed] [Google Scholar]
- 11.Huss S, Pasternack H, Ihle MA, et al. Clinicopathological and molecular features of a large cohort of gastrointestinal stromal tumors (GISTs) and review of the literature: BRAF mutations in KIT/PDGFRA wild-type GISTs are rare events. Hum Pathol 2017;62:206–214. [DOI] [PubMed] [Google Scholar]
- 12.Agaram NP, Wong GC, Guo T, et al. Novel V600E BRAF mutations in imatinib-naive and imatinib-resistant gastrointestinal stromal tumors. Genes Chromosomes Cancer 2008;47(10):853–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi E, Chmielecki J, Tang CM, et al. FGFR1 and NTRK3 actionable alterations in "wild-type" gastrointestinal stromal tumors. J Transl Med 2016;14(1):339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brenca M, Rossi S, Polano M, et al. Transcriptome sequencing identifies ETV6-NTRK3 as a gene fusion involved in GIST. J Pathol 2016; 238(4):543–549. [DOI] [PubMed] [Google Scholar]
- 15.Kao YC, Sung YS, Zhang L, et al. BCOR upregulation in a poorly differentiated synovial sarcoma with SS18L1-SSX1 fusion-a pathologic and molecular pitfall. Genes Chromosomes Cancer 2017;56(4):296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antonescu CR, Zhang L, Chang NE, et al. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer 2010;49(12):1114–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu G, Benayed R, Ho C, et al. Diagnosis of known sarcoma fusions and novel fusion partners by targeted RNA sequencing with identification of a recurrent ACTB-FOSB fusion in pseudomyogenic hemangioendothelioma. Mod Pathol 2019;32(5):609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn: JMD 2015;17(3): 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakravarty D, Gao J, Phillips SM, et al. OncoKB: a precision oncology Knowledge Base. JCO Precis Oncol 2017;2017:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2(5):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demetri GD, von Mehren M, Blanke CD, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med 2002;347(7):472–480. [DOI] [PubMed] [Google Scholar]
- 23.Charo LM, Burgoyne AM, Fanta PT, et al. A novel PRKAR1B-BRAF fusion in gastrointestinal stromal tumor guides adjuvant treatment decision-making during pregnancy. J Natl Compr Canc Netw 2018; 16(3):238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Min KW. Small intestinal stromal tumors with skeinoid fibers. Clinicopathological, immunohistochemical, and ultrastructural investigations. Am J Surg Pathol 1992;16(2):145–155. [DOI] [PubMed] [Google Scholar]
- 25.Lopes LF, Ojopi EB, Bacchi CE. Gastrointestinal stromal tumor in Brazil: clinicopathology, immunohistochemistry, and molecular genetics of 513 cases. Pathol Int 2008;58(6):344–352. [DOI] [PubMed] [Google Scholar]
- 26.Lott S, Schmieder M, Mayer B, et al. Gastrointestinal stromal tumors of the esophagus: evaluation of a pooled case series regarding clinicopathological features and clinical outcome. Am J Cancer Res 2015; 5(1):333–343. [PMC free article] [PubMed] [Google Scholar]
- 27.Radenkovic G, Savic V, Mitic D, Grahovac S, Bjelakovic M, Krstic M. Development of c-kit immunopositive interstitial cells of Cajal in the human stomach. J Cell Mol Med 2010;14(5):1125–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medeiros F, Corless CL, Duensing A, et al. KIT-negative gastrointestinal stromal tumors: proof of concept and therapeutic implications. Am J Surg Pathol 2004;28(7):889–894. [DOI] [PubMed] [Google Scholar]
- 29.Italiano A, Chen CL, Sung YS, et al. SDHA loss of function mutations in a subset of young adult wild-type gastrointestinal stromal tumors. BMC Cancer 2012;12:408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atiq MA, Davis JL, Hornick JL, et al. Mesenchymal tumors of the gastrointestinal tract with NTRK rearrangements: a clinicopathological, immunophenotypic, and molecular study of eight cases, emphasizing their distinction from gastrointestinal stromal tumor (GIST). Mod Pathol 2021;34(1):95–103. [DOI] [PubMed] [Google Scholar]
- 31.Liegl B, Hornick JL, Corless CL, Fletcher CD. Monoclonal antibody DOG1.1 shows higher sensitivity than KIT in the diagnosis of gastrointestinal stromal tumors, including unusual subtypes. Am J Surg Pathol 2009;33(3):437–446. [DOI] [PubMed] [Google Scholar]
- 32.West RB, Corless CL, Chen X, et al. The novel marker, DOG1, is expressed ubiquitously in gastrointestinal stromal tumors irrespective of KIT or PDGFRA mutation status. Am J Pathol 2004;165(1):107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417(6892):949–954. [DOI] [PubMed] [Google Scholar]
- 34.Hostein I, Faur N, Primois C, et al. BRAF mutation status in gastrointestinal stromal tumors. Am J Clin Pathol 2010;133(1):141–148. [DOI] [PubMed] [Google Scholar]
- 35.Kao YC, Fletcher CDM, Alaggio R, et al. Recurrent BRAF gene fusions in a subset of pediatric spindle cell sarcomas: expanding the genetic Spectrum of tumors with overlapping features with infantile Fibrosarcoma. Am J Surg Pathol 2018;42(1):28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kao YC, Ranucci V, Zhang L, et al. Recurrent BRAF gene rearrangements in Myxoinflammatory fibroblastic sarcomas, but not Hemosiderotic Fibrolipomatous tumors. Am J Surg Pathol 2017; 41(11):1456–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subbiah V, Westin SN, Wang K, et al. Targeted therapy by combined inhibition of the RAF and mTOR kinases in malignant spindle cell neoplasm harboring the KIAA1549-BRAF fusion protein. J Hematol Oncol 2014;7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross JS, Wang K, Chmielecki J, et al. The distribution of BRAF gene fusions in solid tumors and response to targeted therapy. Int J Cancer 2016;138(4):881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menzies AM, Yeh I, Botton T, Bastian BC, Scolyer RA, Long GV. Clinical activity of the MEK inhibitor trametinib in metastatic melanoma containing BRAF kinase fusion. Pigment Cell Melanoma Res 2015; 28(5):607–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


