Abstract
Pseudoalteromonas (BB2-AT2) is a ubiquitous marine heterotroph, often associated with labile organic carbon sources in the ocean (e.g. phytoplankton blooms and sinking particles). Heterotrophs hydrolyze exported photosynthetic materials, components of the biological carbon pump, with the use of diverse metalloenzymes containing zinc (Zn), manganese (Mn), cobalt (Co), and nickel (Ni). Studies on the metal requirements and cytosolic utilization of metals for marine heterotrophs are scarce, despite their relevance to global carbon cycling. Here, we characterized the Zn, Mn, Co, and Ni metallome of BB2-AT2. We found that the Zn metallome is complex and cytosolic Zn is associated with numerous proteins for transcription (47.2% of the metallome, obtained from singular value decomposition of the metalloproteomic data), translation (33.5%), proteolysis (12.8%), and alkaline phosphatase activity (6.4%). Numerous proteolytic enzymes also appear to be putatively associated with Mn, and to a lesser extent, Co. Putative identification of the Ni-associated proteins, phosphoglucomutase and a protein in the cupin superfamily, provides new insights for Ni utilization in marine heterotrophs. BB2-AT2 relies on numerous transition metals for proteolytic and phosphatase activities, inferring an adaptative potential to metal limitation. Our field observations of increased alkaline phosphatase activity upon addition of Zn in field incubations suggest that such metal limitation operates in sinking particulate material collected from sediment traps. Taken together, this study improves our understanding of the Zn, Mn, Co, and Ni metallome of marine heterotrophic bacteria and provides novel and mechanistic frameworks for understanding the influence of nutrient limitation on biogeochemical cycling.
Graphical Abstract
Graphical Abstract.
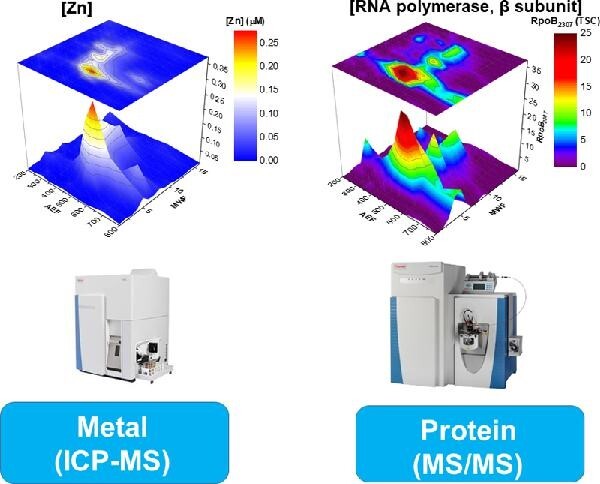
Metalloproteomics was used to understand the Zn, Mn, Co, and Ni metallome of a ubiquitous marine heterotrophic bacterium.
Introduction
The trace elements zinc (Zn), manganese (Mn), cobalt (Co), and nickel (Ni) have important physiological relevance in marine microbes, which utilize sophisticated metalloproteins for metabolic purposes. Concentrations of essential trace metals, such as iron (Fe), can be extremely low in the upper ocean and can influence primary productivity, which removes carbon through the fixation of CO2 by marine phototrophs.1,2 Phototrophic organisms are consumed by zooplanktonic members of the community and decomposed by microbially mediated degradation during their sinking and biogeochemical carbon export. Viral infection contributes to the fate of organic matter, as well, contributing to bacterial heterotrophy and the formation of sinking particles.3–6 Together, the processes of photosynthesis and decomposition of sinking organic matter represent two critical components of the biological pump within the marine environment.7,8 Notably, many of these photosynthetic and carbon export processes are facilitated by metalloenzymes, linking trace metal geochemistry to global biogeochemical changes.9–13
The low concentration of trace metals, such as Fe, has been demonstrated to limit phytoplankton productivity and biomass formation.14–19 However, the scarcity of bioactive metals also has the potential to induce metal substitution, mismetallation, or even repress the key catalytic function. The repression of bacterial proteolytic and β-glucosidase activities due to trace metal limitation has been documented in the marine environment.10,20 The activity of these enzymatic processes dictates the hydrolysis of dissolved organic matter (DOM) in the euphotic zone, and the degradation of particulate organic matter (POM) observed within the meso- and bathypelagic zones.21–27 The cosmopolitan marine heterotroph Pseudoalteromonas, within the order of Alteromonadales, has been isolated from the surface of particles within sediment traps and is often associated with detritus and other marine snow sources (e.g. organic matter falling through the water column from the photic and euphotic zones).28–30Pseudoalteromonas is a model organism for profiling the microbial activity relevant to carbon export and silica ballast dissolution.23,25,31 Moreover, many of the proteases within the genomes of heterotrophic bacteria are metal-associated proteases, including dipeptidases, aminopeptidases, and oligopeptidases. Understanding the cytosolic partitioning and utilization of trace metals within Pseudoalteromonas is instructive in understanding the impacts of trace metal limitation on global, microbially mediated carbon export processes.3,32,33
This study examines the connections between the biogeochemistry of marine heterotrophic bacteria and Zn, Mn, Co, and Ni through investigation of the cellular metalloproteome of Pseudoalteromonas. This method employs two dimensions of chemical separation using anion exchange and size exclusion chromatography on cytosolic metalloproteins after native (nondenaturing) lysis methods.34–40 The resulting subsamples of the purified lysate were characterized by proteomics and inductively coupled plasma mass spectrometry (ICP-MS) to identify protein and element distributions within each sample, which together elucidate statistical correlations of major metal and protein reservoirs, offering putative metalloprotein assignments. This native 2D approach has the advantage of being able to survey the soluble metallome for multiple metals, and providing putative metal assignments that can inspire follow-up studies, as opposed to more focused studies on individual proteins purified to homogeneity for characterization. This article focuses on the Zn, Mn, Co, and Ni metallome of Pseudoalteromonas and builds on our previous findings describing the Fe metallome in this microbe.40
Experimental methods
Cellular metal quota and metalloproteomic analysis
Experimental details for the culturing, cellular metal quota analysis, and metalloproteomic analysis of BB2-AT2 are provided in Mazzotta et al.40 In summary, cultures were grown in marine broth medium in Vineyard Sound seawater at 23°C, and the resulting pellets were stored at −80°C until further processing. For total cellular metal abundances (metallome), pellets were digested in 5% HNO3 and the resulting supernatant solutions were measured by ICP-MS. Metalloproteomic analyses provided native metalloprotein separation involving three steps: (i) native lysis within an anaerobic environment; (ii) separation of the proteins by surface charge via anion exchange chromatography; and (iii) separation of the proteins by molecular weight via size exclusion chromatography. The resulting native extracts were then analyzed by ICP-MS for metal content and high-resolution Orbitrap liquid chromatography mass spectrometry for protein content (after tryptic digestion). Singular value decomposition (SVD), as described in detail in Mazzotta et al., reconstructed the metal abundances and relative protein abundances were compared to identify major uses of metals within the metalloproteome (Equation S1, Fig. S3, and Table S4).40
Mid-log- versus stationary-phase proteome experiments
BB2-AT2 was inoculated into duplicate plastic Erlenmyer flasks with 225 ml of marine broth medium each inoculated with 2.2% volume from a single starter culture. Cultures were grown at 18°C and 70 ml was harvested from each at 10, 22, and 46 h for log phase, late log phase, and stationary phase, respectively. Optical densities at 600 nm (OD600) of the duplicate cultures at 10 h were 0.11 and 0.11, and at 46 h they were 0.86 and 0.94 (Fig. S5). At the 46 h time point, a biofilm was evident on the culture surface. Cells were harvested by centrifugation at 9000 rpm for 15 min at 4°C in an Eppendorf 5810R centrifuge, decanted, and transferred to microcentrifuge tubes and centrifuged at 11 000 rpm for 8 min at 4°C. Supernatants were decanted and pellets were frozen at −80°C until extraction, as previously described.41 From one replicate, 10, 22, and 46 h time points were extracted, while samples from the other replicate were archived for backup use. Briefly, extraction involved resuspending pellets in 100 mM ammonium bicarbonate buffer (pH = 8.0), sonicating on ice, centrifuging, and precipitating supernatants in solvent at −20°C. The precipitated protein was resuspended in a urea buffer, reduced, alkylated, and trypsin digested for mass spectrometry analysis.
For global proteomics analysis of BB2-AT2 cultures, trypsin-digested samples (4 μg protein measured before tryptic digestion via the BCA assay using bovine serum albumin as a standard) were concentrated onto a peptide cap trap and rinsed with 150 μl 0.1% formic acid, 5% acetonitrile (ACN), and 94.9% water before gradient elution through a reverse-phase Magic C18 AQ column (0.2 mm × 150 mm, 3 μm particle size, 200 Å pore size, Michrom Bioresources Inc.) on an Advance HPLC system (Michrom Bioresources Inc.) at a flow rate of 0.75 μl/min. The chromatography consisted of a nonlinear gradient from 5% to 95% of a 0.1% formic acid in acetonitrile for 320 min (starting with 95% 0.1% formic acid in water). A linear ion trap quadrupole mass spectrometer (Thermo Scientific) was used with an Advance CaptiveSpray source (Michrom Bioresources). The linear trap quadrupole (LTQ) was set to perform tandem mass spectrometry on the top five ions using data-dependent settings with a dynamic exclusion window of 30 s, and ions were monitored over a range of 400–2000 mass-to-charge ratio. Each sample was injected three times (technical triplicates), results were averaged, and standard deviation was calculated. All spectral count results are provided in the Supplementary Material, and log-phase and stationary-phase data are presented in the article.
The LTQ mass spectra files were searched using SEQUEST (Bioworks version 3.3, Thermo Inc.). Peptide probability database search results were further processed using Scaffold 2.0 (Proteome Software Inc.) with SEQUEST filters ΔCN > 0.1 and Xcorr vs. charge state of 1.9, 2.4, and 2.9 for +1, +2, and +3 charges, respectively. Spectral counts were normalized across samples in Scaffold.
Sediment trap material incubation experiment
Sediment trap material from METZYME station 5 on the R/V Kilo Moana (KM1128 on 18 October 2011) was collected in the equatorial Central Pacific Ocean at 0°, 158° W using sediment traps.42 The traps were surface tethered and deployed for 3 days at depths of 60, 150, and 500 m. The 500 m sample data are available in the Supplementary Material but, due to low biomass, there was no discernable signal detected and they were not included in our analysis. At each depth, 12 baffled sediment trap tubes (3′′ diameter, 24′′ height) were held in a rack and fitted at the bottom with a funnel and a removable, acid-cleaned 250 ml polyethylene bottle. The bottles were filled with clean brine made by freezing surface seawater collected by a trace metal rosette at sea, all prepared within clean room conditions, and buffered to pH = 8.2 with borate but without additional chemical preservative. Of the 12 tubes, 2 were combined, screened through an acid-washed 350 μm nylon mesh to remove zooplankton, and then gently mixed to homogeneity. Particle material (30 ml) from the trap was combined with 30 ml of filtered seawater (Station 6 at 30 m) into 60 ml polycarbonate bottles used for an experimental treatment replicate. Three treatments were prepared for each depth in quintuplicate: control (no addition), a Zn addition (50 nM, ZnCl2), and the metal chelator disodium ethylenediaminetetraacetic acid (EDTA, 2 × 10–4 M). At time zero, a no-addition treatment was generated and sacrificed immediately. Bottles were incubated in darkness and at either ∼24°C (60 m) or 14°C (150 m) to simulate the mesopelagic conditions of the depths from which the samples originated. After 1.2, 2.2, and 5.0 day time points, subsamples were taken by gently swirling each bottle and removing an aliquot for phosphatase activity. Enz-Chek alkaline phosphatase assay was used in 96-well plate format according to the manufacturer's instructions on a SpectraMax M5 spectrofluorometer (Molecular Probes). The alkaline phosphatase assay consisted of a 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP) solution diluted 50× into 200 mM Tris-HCl, pH 7.8, containing 2 mM sodium azide to produce the 10 mM stock solution. The standard curve for calibration was made using porcine alkaline phosphatase diluted in sterile seawater. Plate assays consisted of 100 μl of sediment trap sample plus 25 μl DiFMUP (40 μM) into a 96-well plate and incubated at 22°C with shaking at 300 rpm with 5 s on and 25 s off. Readings were taken every 30 min over several hours to determine the DiFMUP reaction rate. All sample transfers were conducted in a fabricated clean room space, with acid-rinsed pipette tips, although the 96-well plate and phosphatase reagents were not acid cleaned or trace metal purified, respectively.
Results and discussion
Cellular metal quota and stoichiometries
Whole cell metal quotas of Pseudoalteromonas sp. (BB2-AT2) were compared with the metalloproteomic metal quotas (also see table 3 in Mazzotta et al.).40 While Mazzotta et al. focused on the cellular Fe utilization, this article focuses on the non-Fe cellular constituents, including the following trace metals in differing abundances: Zn, Mn, Ni, Cu, Mo, and Co.
Zn contributed to approximately half of the cellular metal quota, demonstrating a substantial role for Zn, as well as Fe, which was documented in Mazzotta et al., for this marine heterotroph under replete conditions. While there are little metal stoichiometric data beyond Fe in marine heterotrophic bacteria to compare, the metal stoichiometries of marine phytoplankton have been thoroughly characterized.2,8,43–47 Regressions of dissolved metal concentrations in the upper water column against dissolved macronutrient concentrations in the North Pacific, North Atlantic, and Southern Oceans are used to infer ecological stoichiometries of metals in the bulk phytoplankton communities (as they are set by the degradation of biological particulate phases) and can provide additional comparisons to the metal quotas of BB2-AT2.48
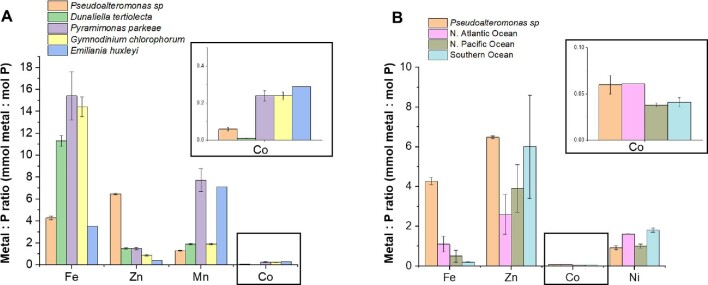
A comparison of Pseudoalteromonas sp. metal stoichiometries with marine phytoplankton cultures characterized by Ho and colleagues indicates as much as 6× the Zn:phosphorus (P) quota relative to other plankton (Fig. 1A).44 Comparison of the Zn:P stoichiometry of Pseudoalteromonas BB2-AT2 with the dissolved-phase ecological stoichiometry (regressions of dissolved metal concentrations in the upper water column against dissolved macronutrient phosphorus) obtained in field studies indicates a particular preference for Zn within this heterotrophic bacterium (Fig. 1B).48 In contrast, some phytoplankton have a greater Mn:P ratio relative to Zn:P; for example, Pyramimonas parkeae has ∼7× the Mn:P ratio to that of BB2-AT2. The Co:P in Pseudoalteromonas is 25–50% of the Co:P content of marine phytoplankton within this comparison. Ni:P ratios of BB2-AT2 were comparable with the North Pacific data (Ni:P = 0.92 for BB2-AT2 and 1 for North Pacific), 1.8× less than that of the North Atlantic and 2× less than that of the Southern Ocean. While Ni has been less studied,49–52 the comparison of these metal quotas implies that heterotrophic communities might possess a multitude of enzymes that require or use Ni. Notably, the Fe:P of BB2-AT2 is comparable with that of the diatom Emiliania huxleyi. The cellular Fe:P concentration ratio is lower than that of green algae, P. parkeae, and ∼3.6× lower than that of Gymnodinium chlorophoroum. Given the replete conditions of BB2-AT2’s growth, the Fe:P is 4× that of the North Atlantic metal:macronutrient and 8-fold that of the North Pacific. The Zn:P in BB2-AT2 was 1.1× that of the Southern Ocean, 2.6× that of the North Atlantic, and 1.5× that of the North Pacific.
Fig. 1.
(A) Comparison of the metal:P stoichiometry of the whole cell metal quota of Pseudoalteromonas sp. (BB2-AT2) with several strains of phytoplankton, characterized by Ho et al. (Ni:P data were unavailable for comparison).44 (B) Comparison of the M:P stoichiometry of BB2-AT2 with the M:P stoichiometries derived from the regression of dissolved metal concentrations in the upper water column against dissolved macronutrient concentrations obtained in the North Atlantic, North Pacific, and Southern Oceans by Twining and Baines (Mn:P data were unavailable for comparison).48
These cellular metal quota data of Pseudoalteromonas BB2-AT2 can be compared with the metalloproteome of this strain.40 The characterization of the metalloproteome consists of the release of the soluble protein pool by native extraction, removal of particulate cell debris by centrifugation, and analysis of the metal content of the resulting supernatant. The sum of the purified metal fractions from the metalloproteome can then be used to estimate the soluble metallome. The decrease in Zn in the soluble metalloproteome compared with the overall cellular Zn quota could be explained by loss of Zn associated with particulate fractions of the cell, including but not limited to exterior membrane proteins. The increased prevalence of Ni within the soluble metalloproteome infers a soluble Ni metalloprotein reservoir within this organism (Table 3 in Mazzotta et al., 2020).
The partitioning of metals, and specifically Zn, within the cytosolic proteins, was probed through metalloproteomics, expanding on the Fe metalloproteome findings for this model heterotroph.40 The native extracts from the BB2-AT2 produced 384 fractions after 2D separation, and each of those subsamples was analyzed by ICP-MS and nano-liquid chromatography-mass spectrometry (LC-MS) for metal and protein content, respectively. Zn showed the most complex metalloproteome, with its large total Zn:P ratio found in the cellular metal quota, while Co, Mn, and Ni were far less complex. The following sections of this article discuss the usage of each metal, as well as categorization with respect to putative cellular function.
Zn metallome: transcription, translation, alkaline phosphatase, and proteolysis
The cytosolic Zn metallome of Pseudoalteromonas was found to be complex (Fig. 2B), featuring numerous metalloproteins in varying abundances. Unlike the Fe metallome of BB2-AT2, where Fe was localized to a small subset of proteins, Zn was distributed throughout the proteome to a variety of proteins. This complexity rendered the metalloproteomic characterization challenging, but major Zn reservoirs were nonetheless apparent and included proteins involved in the following functions: transcription, translation, alkaline phosphatase, and proteolysis. These putative Zn-associated proteins are listed in Table 1 and their details are described subsequently.
Fig. 2.
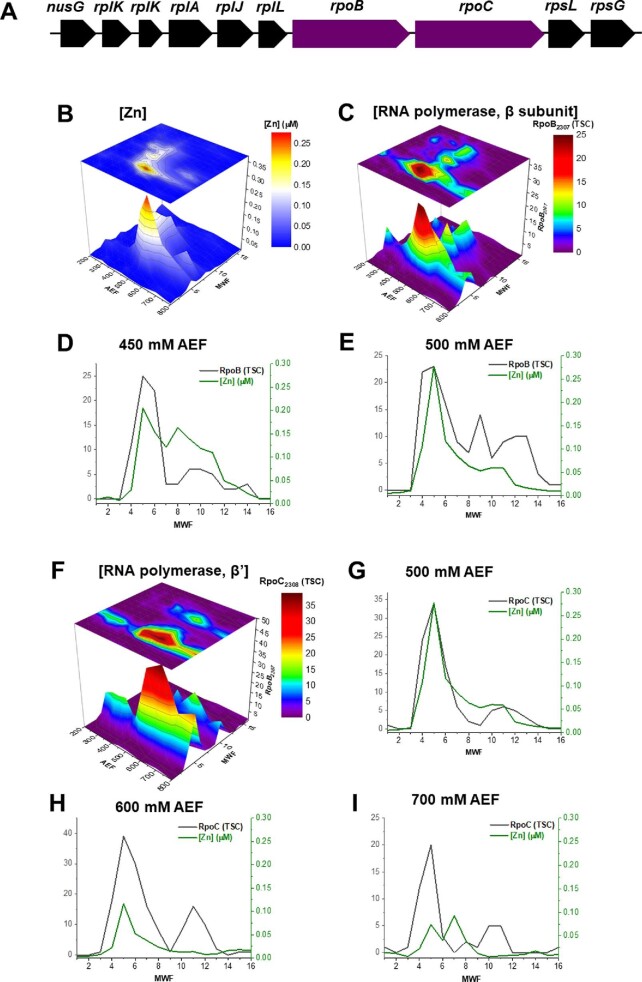
(A) Gene neighborhood diagram showing RpoB and RpoC being flanked by a series of ribosomal proteins. The following panels compare (B) the distribution of cytosolic Zn with (C) that of the distribution of RpoB in metalloproteome space. (D, E) Comparison of the distributions of RpoB and Zn at the 450 and 500 mM NaCl fractions. The distribution of RpoC in metalloproteome space (F) can also be compared with the distributions of the Zn in the cytosolic metalloproteome, within the 500, 600, and 700 mM NaCl fractions (G–I).
Table 1.
Summary of putative metal-associated metalloproteins observed within this analysis of Pseudoalteromonas sp., BB2-AT2, including the fractions where they were observed, their maximum peak maximum, and R2 with various metals, defined at the global maximum of the proteome. AEF, anion exchange fraction; MWF, molecular weight fraction
| R 2 | |||||||||
| Gene no. | Protein | Annotation | AEF (mM NaCl) | MWF (no.) | MWF, [protein]max | Mn | Zn | Co | Ni |
| 2307 | RpoB | β-subunit, DNA-directed RNA polymerase | 200–800 | 4–14 | 5 | 0.41 | 0.82 | 0.52 | 0.51 |
| 2308 | RpoC | β′- subunit, DNA-directed RNA polymerase | 200–700 | 3–14 | 5 | −0.45 | 0.57 | 0.20 | 0.17 |
| 0768 | GltX | Glutamyl tRNA synthetase | 450–500 | 4–11 | 5 | 0.49 | 0.58 | 0.61 | 0.82 |
| 2009 | PhoD | Alkaline phosphatase | 450–600 | 4–10 | 9 | 0.08 | 0.58 | 0.52 | 0.34 |
| 3714 | PhoA | Alkaline phosphatase | 200–700 | 3–14 | 7 | 0.53 | 0.90 | 0.19 | 0.16 |
| 3715 | PhoA | Alkaline phosphatase | 700–800 | 3–14 | 6 | 0.45 | 0.84 | 0.14 | 0.10 |
| 0650 | Pm17 | Peptidase B | 400–500 | 4–12 | 11 | −0.19 | 0.60 | 0.17 | 0.26 |
| 0925 | Pm17 | Peptidase B | 450–800 | 5–10 | 6 | 0.28 | 0.69 | −0.15 | −0.12 |
| 2396 | Pm28 | Glutamate carboxypeptidase | 200–800 | 3–6 | 4 | 0.68 | 0.71 | 0.67 | 0.82 |
| 2101 | Pm3 | Dipeptidase | 300–400 | 8–12 | 8 | −0.11 | 0.67 | 0.29 | −0.04 |
| 1871 | Pm3 | Dipeptidase | 200–800 | 2–7.9 | 9 | 0.14 | 0.15 | −0.28 | −0.28 |
| 2594 | Ps66 | l,d-carboxypeptidase | 400–450 | 8–10 | 8 | −0.11 | 0.64 | 0.26 | −0.07 |
| 2677 | PepN | Aminopeptidase | 350–400 | 8-–11 | 8 | 0.10 | 0.74 | 0.45 | 0.08 |
| 3233 | Pm24 | Prolidase | 350–400 | 7–9 | 8 | −0.09 | 0.69 | 0.19 | −0.08 |
| 0147 | RpsA | Ribosomal protein S1 (30S) | 300–600 | 2–16 | 10 | −0.48 | 0.70 | 0.43 | 0.32 |
| 2512 | RpsB | Ribosomal protein S2 (30S) | 200–800 | 4–7 | 5 | 0.55 | 0.83 | 0.79 | 0.65 |
| 3106 | RpsC | Ribosomal protein S3 (30S) | 200–800 | 4–8 | 5 | 0.52 | 0.45 | 0.43 | 0.72 |
| 2037 | RpsD | Ribosomal protein S4 (30S) | 200–800 | 4–7 | 6 | 0.38 | 0.61 | 0.65 | 0.51 |
| 2305 | RplL | Ribosomal protein L7/L12 (50S) | 250–500 | 7–16 | 8 | −0.25 | 0.28 | 0.28 | 0.23 |
| 3110 | RplD | Ribosomal protein, L4/L1 family (50S) | 200–700 | 4–7 | 5 | 0.33 | 0.32 | 0.30 | 0.67 |
| 3483 | RplM | Ribosomal protein L13 (50S) | 200–500 | 4–6 | 5 | 0.54 | 0.22 | 0.13 | 0.02 |
| 0114 | RplT | Ribosomal protein L20 (50S) | 450–600 | 4–5 | 5 | 0.66 | 0.85 | 0.80 | 0.87 |
| 0116 | MBL | Metallo-β-lactamase | 200–250 | 12–13 | 12 | −0.18 | 0.04 | −0.20 | −0.16 |
| 0291 | Pm24 | Glycyl aminopeptidase | 250–400 | 3–8 | 7 | −0.13 | −0.15 | −0.32 | −0.30 |
| 1846 | Pm20 | Dipeptidase | 300–500 | 4–7 | 7 | −0.10 | −0.46 | −0.23 | −0.18 |
| 2147 | Pm61 | Proline aminopeptidase | 400–450 | 6–8 | 6,7 | 0.64 | 0.55 | 0.06 | −0.05 |
| 2230 | Pm17 | Leucyl aminopeptidase | 400–500 | 6–7 | 4 | 0.25 | 0.41 | 0.74 | 0.49 |
| 2892 | Pm1 | Aminopeptidase | 400–800 | 3–8 | 4 | 0.59 | 0.26 | 0.45 | 0.32 |
| 0632 | Arginase | Arginase | 400–450 | 4–11 | 9 | 0.24 | 0.84 | 0.77 | 0.70 |
| 0549 | ComFB | Annotated: ‘metallopeptidase’ | 300 | 11–12 | 11 | −0.12 | 0.16 | −0.09 | −0.17 |
| 3458 | DUF5117 | ‘Extracellular metal-dependent peptidase’; domain of unknown function | 200–350 | 4–8 | 6 | −0.11 | −0.48 | −0.26 | −0.21 |
| 1566 | Pgm | Phosphoglucomutase | 450–600 | 4–13 | 7,9 | 0.14 | 0.81 | 0.50 | 0.50 |
| 2699 | Cupin_4 | Cupin superfamily protein | 700–800 | 7–12 | 8 | 0.86 | 0.37 | 0.83 | 0.90 |
Two major reservoirs of Zn appear associated with the β and β′ subunits of DNA-directed RNA polymerase subunits, RpoB (BB2AT2_2307) and RpoC (BB2AT2_2308), respectively. RNA polymerase is a well-known metalloenzyme and appears to constitute a major Zn reservoir within Pseudoalteromonas. The rpoB and rpoC genes were located adjacently and flanked on each side by several ribosomal proteins (Fig. 2A). It is possible that these genes are cotranscribed and that one of these ribosomal proteins acts as an internal promoter for the rpoB and rpoC genes, as has been reported in the heterotroph Escherichia coli.53 RpoB and RpoC are comparable in size: ∼150 and 154 kDa, respectively. Proteomic analysis distinguished between these two proteins through identification of tryptic peptides that are unique to each protein (Table S1). Cytosolic Zn and RpoB were well correlated with an R2 value of 0.82 for the 500 mM NaCl anion exchange fraction (Fig. 2C-E). Comparison of cytosolic Zn and RpoC also indicated similar elution profiles, with correlations of R2 value of 0.57 for the 600 mM NaCl anion exchange fraction (Fig. 2F-H). The 700 mM NaCl anion exchange fraction represented a region of poor correlation due to the overlap from other dominant Zn-associated proteins (Fig. 2I.
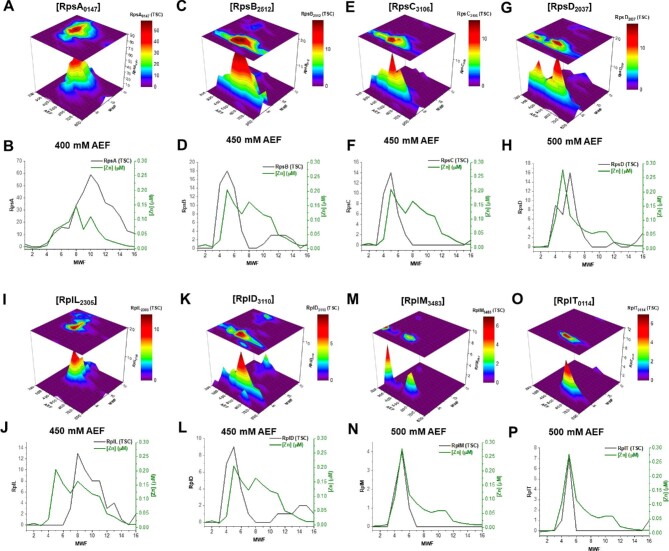
Zn was also associated with several 30S and 50S ribosomal proteins within the 2D metallome of Pseudoalteromonas (Table 1). Ribosomal proteins S1 (RpsA, BB2AT2_0147), S2 (RpsB, BB2AT2_2512), S3 (RpsC, BB2AT2_3106), S4 (RpsD, BB2AT2_2037), L7/L12 (RpIL, BB2AT2_2305), L4/L1 (RpID, BB2AT2_3110), L13 (RpIM, BB2AT2_3483), and L20 (RpIT, BB2AT2_0114) are localized along the 400–500 mM NaCl anion exchange fractions (Fig. 3). These proteins, separated amongst a variety of molecular weight fractions (MWFs), coeluted with major distributions of Zn within the metallome of BB2-AT2. These observations are consistent with reports of Zn-binding domains observed in key translation proteins ensuring proper functioning of bacterial ribosomes.54,55 Linear regressions of these proteins and Zn distributions produced R2 values between 0.28 and 0.85 (Table 1). Noting that these RNA proteins cluster together, these observations may reflect multiprotein ribosomal protein complexes. Comparison of protein size and size exclusion elution profiles did not show a relationship, consistent with this notion of elution of protein complexes (Fig. S2). Moreover, proteins involved in transcription and translation overlapped in their elution profiles within this 2D separation scheme, making their specific contributions to the Zn metallome signal somewhat challenging to disentangle. These putative assignments warrant further investigation, as this Zn-binding component of these fractions may be strictly due to statistical correlations between these protein and metal fractions. Additional chromatographic separation as well as numerical modeling approaches can help refine their relative contributions (discussed subsequently; Equation S1, Fig. S3, and Table S4).
Fig. 3.
Distributions of the (A, B) ribosomal protein S1, RpsA; (C, D) ribosomal protein S2, RpsB; (E, F) ribosomal protein S3, RpsC; (G, H) ribosomal protein S4, RpsD; (I, J) ribosomal protein L4/L1, RpIL; (K, L) ribosomal protein L7/L12, RpID; (M, N) ribosomal protein L13, RpIM; and (O, P) ribosomal protein L20, RpIT are depicted and compared at their anion exchange fraction (AEF) maxima with the distribution of Zn.
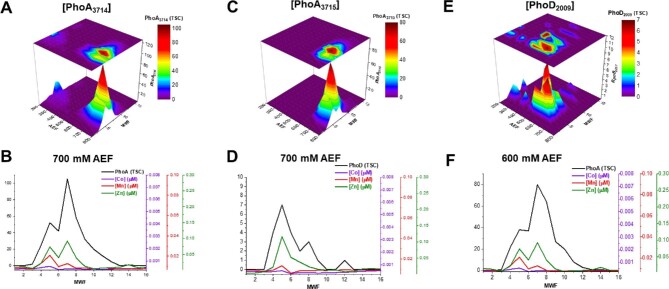
Zn was observed to be associated with three isoforms of the alkaline phosphatase enzyme PhoA (BB2AT2_3714 and BB2AT2_3715) and PhoD (BB2AT2_2009), where two of the three proteins were observed in substantial abundance (with respect to total spectral counts). The majority of PhoA and PhoD eluted along the 700 mM NaCl anion exchange fraction (Fig. 4A–F), offset from the Zn proteins described earlier. Within this anion exchange fraction, Zn and PhoA3714 and PhoA3715 were correlated with R2 values of 0.90 and 0.84, respectively. PhoD was less abundant in total spectral counts relative to the two other alkaline phosphatase enzymes observed within this analysis. The majority of the abundance of PhoD, observed in the 600 mM NaCl fraction (Fig. 4E), correlated with Zn, producing an R2 = 0.58 (Fig. 4F).
Fig. 4.
(A, C, E) Distribution of alkaline phosphatase enzymes in multidimensional chromatographic analysis and (B, D, F) comparison of these metalloproteins (with exclusive selection of the 700 and 600 mM AEFs, since these proteins were observed in highest abundance) compared with their corresponding distributions of Zn, Mn, and Co.
Alkaline phosphatase enzymes have been characterized to bind Zn, as well as Co, so it is plausible to infer cambialism with multiple metal-binding isoforms.56–62 PhoD is a putatively identified cambialistic enzyme, as it features comparable overlap with multiple metals within the chromatographic profiles, as well as comparable R2 values for Zn and Co (R2 = 0.58 and 0.52, respectively). Input from rivers, hydrothermal environments, sedimentary sources, and eolian deposition, where major geochemical sources of Co are present, could present the possibility for alternative metal-binding scenarios in regions of trace metal limitation and requires further investigation.45,63,63–69
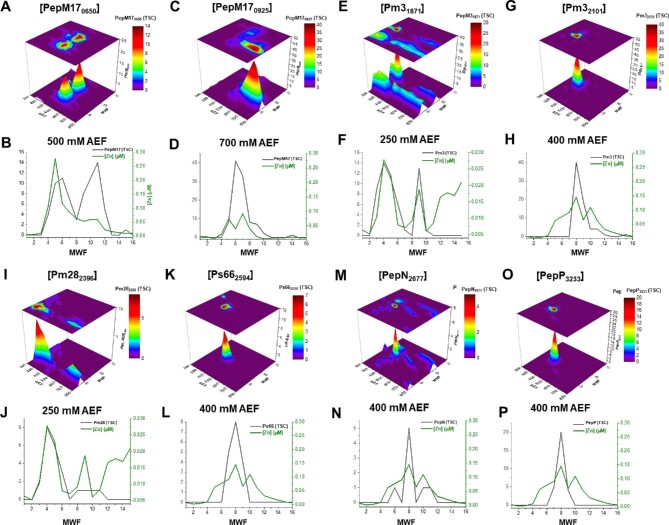
Numerous proteolytic enzymes are associated with Zn in the BB2-AT2 metallome. BB2-AT2 features ∼104 proteolytic enzymes within its genome, which were estimated by summing the proteins annotated as proteases, peptidases, and proteinases (screened via Basic Local Alignment Search Tool (BLAST) for further verification). Among these, 37 were observed within the native metalloproteome (selected metalloproteases shown in Fig. 5). In comparison, 62 proteases were observed in the detergent-based (sodium dodecyl sulfate (SDS)-based) total proteome extraction (calculated from data found in Tables S2 and S3). The difference between the two methodologies may be the result of loss of secreted extracellular proteases upon repeated washing of the cells prior to separation, as well as due to proteases that are associated with the membrane or the periplasm and lost in centrifugation of the particulate biomass fraction. Of the metalloproteases identified in this metalloproteomic analysis, several putatively associated with Zn, Mn, and Co enzymes were determined by comparing the metal and protein distributions (Figs 6 and 7). The most abundant metalloproteases included aminopeptidase-type proteases that function to cleave terminal amino acid linkages, as well as dipeptidases, which function to cleave peptides into dipeptide fragments.
Fig. 5.
Distribution of putatively identified Zn-associated proteolytic enzymes found to be most abundant and comparison (with selection of exclusively the AEF in two dimensions for which the protein is localized in maximum total spectral counts) with that of the distribution of Zn.
Fig. 6.
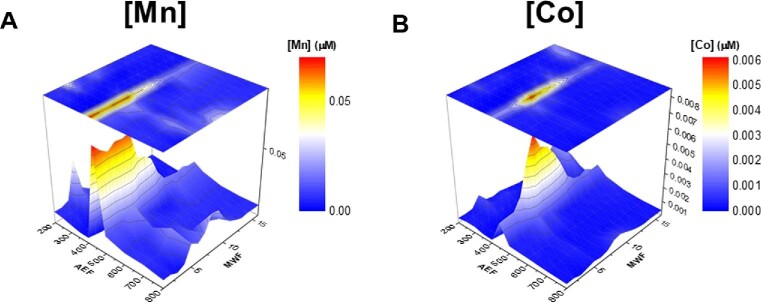
The cytosolic metallome of (A) Mn and (B) Co within Pseudoalteromonas.
Fig. 7.
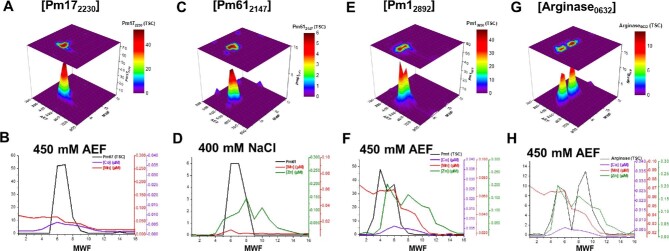
Distributions of cytosolic (A, B) peptidase M17 (Pm17), (C, D) peptidase M6 (Pm610), (E, F) peptidase M1 (Pm1), and (G, H) arginase. Protein distributions are followed with comparisons to the cytosolic Mn, Zn, or Co within the anion exchange fraction of maximum protein abundance.
The Zn metallome described here is the most complex of the metals within the metallome of Pseudoalteromonas. The cytosolic Zn distribution resulted in metal local/global maxima that were composed of multiple metalloproteins as indicated by the elution profiles within the 2D native separation. To account for the contribution of multiple metalloproteins to the metallome signal, we employed singular value decomposition (SVD), as previously reported for the Fe metalloproteome of BB2-AT2 (Equation S1, Fig. S3, and Table S4).40 The SVD enabled an approximation of the percentage contribution of major biochemical functions to the Zn metallome of BB2-AT2, as follows: transcription (47.2%), translation (33.5%), proteolysis (12.8%), and alkaline phosphatase (6.4%).
Mn and Co metallome: proteolysis
The maximum concentrations of Co and Mn within the metallome of Pseudoalteromoas were observed within the 400 and 450 mM NaCl anion exchange fractions (Fig. 6A and B). Distinguishing between Mn- and Co-protein associations in these regions of frequent protein coelution was difficult and putative assignments of these reservoirs are discussed subsequently. Mn within this organism is partitioned among some proteolytic enzymes, as well as the Fe/Mn superoxide dismutase (SOD) protein, which was discussed previously in Mazzotta et al.
Peptidase M17 (BB2AT2_2230, PM17), within the leucyl aminopeptidase family, catalyzes the preferential hydrolysis of leucine-containing peptides at the N-terminus of protein and peptide substrates. The most abundant distribution of PM17, observed along the 450 mM NaCl anion exchange fraction, was correlated to the distribution of metals, producing R2 values of 0.25, 0.41, and 0.74 for Mn, Zn, and Co, respectively (Fig. 7A and B). Peptidase M61 (BB2AT2_2147, PM61), a proline aminopeptidase superfamily protein, hydrolyzes proline-containing dipeptides, and primarily eluted in the 400 mM NaCl anion exchange fraction (Fig. 7C and D). Comparison of PM61 with the distributions of Mn, Zn, and Co produced R2 values of 0.64, 0.55, and 0.06, respectively. PM1 (BB2AT2_2892), within the M1 family of metallopeptidases, features broad-specificity aminopeptidase and glutamyl aminopeptidase activity. Correlation between PM1 and Mn, Zn, and Co revealed R2 values of 0.59, 0.26, and 0.45, respectively (Fig. 7E and 7F). Some of these regression results may be affected by the distribution of more abundant metalloproteins and their shouldering effects. Despite having a high R2 value in PM17, Co was lower in abundance relative to Mn (which itself has a shoulder here), and hence the high R2 should not be interpreted as it being the only potential contributor to this enzyme. SVD methods assist in these putative assignments and complex contributions.
Arginase (BB2AT2_0632) is a putative metal-associated protein. The majority of this protein's abundance is observed along the 450 mM NaCl anion exchange fraction in two distinct distributions between the 4–6 and 8–11 MWFs (Fig. 7G and 7H). The resolution of two distinct protein distributions within the same anion exchange fraction may be indicative of some monomeric or apo form of this arginase. Thus, we sought to correlate metal and protein within these two distributions. Comparison of the correlations (R2) between metal and arginase produced R2 values of 0.24, 0.84, and 0.77 for Mn, Zn, and Co, respectively. Arginase has been reported to form dimers in certain bacteria.70 Given the size exclusion chromatography, it is likely that this distribution within MWF 8–11 is in monomeric form, while MWF 4–6 is the dimeric form. Furthermore, BLAST analysis of the protein sequence indicated an active site composed of Asp and His residues, possibly accommodating a binuclear metal site, as well as the potential for cambialism. From these results, the major Co metalloprotein reservoir does not appear to include a cobalt-containing B12-related protein such as the B12-requiring methionine synthase found within Pseudoalteromonas, which appears to be a B12 auxotroph.71 It is possible that the B12–metalloenzyme coordination was not preserved during separation, or that protein abundance is minor relative to the features described earlier, as was observed in Prochlorococcus.47
Ni metallome: phosphoglucomutase and cupin superfamily proteins
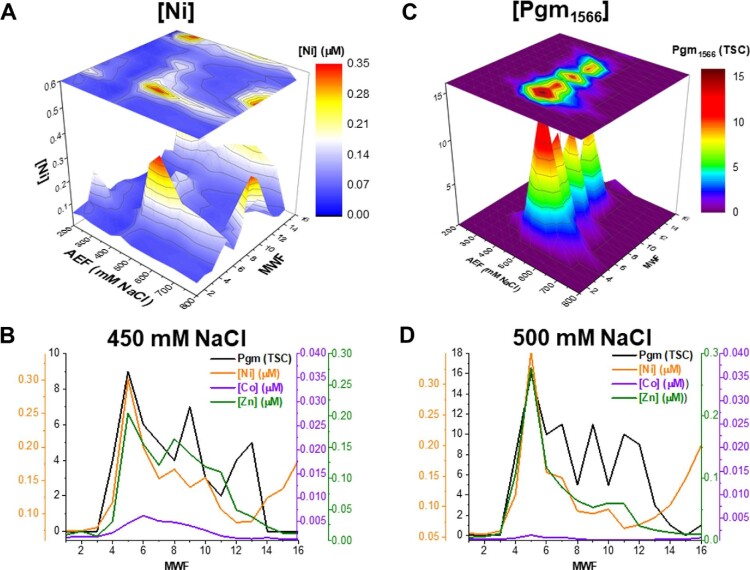
Characterization of the Ni metallome of Pseudoalteromonas revealed two putative assignments (Fig. 8). Phosphoglucomutase (BB2AT2_1566, Pgm) is an enzyme responsible for the interconversion of glucose 1-phosphate and fructose 1-phosphate, which is critical to glucose metabolism. Pgm localized among the 450 and 500 mM NaCl anion exchange fractions (Fig. 8B–D). A comparison of the correlation (R2) between the distribution of Pgm and those of Zn, Co, and Ni produced values of 0.81, 0.50, and 0.50, respectively (Fig. 8C). Pgm and metals Ni and Zn in the 500 mM NaCl anion exchange fraction also showed correlations with coefficient values of 0.50 and 0.80, respectively. The correlation of Pgm and Zn was likely due to the coelution of the dominant ribosomal and transcription-related Zn proteins in this region of the 2D separation (Figs 2–5). Moreover, multiple smaller peaks for Pgm were observed in the size exclusion dimension, which may represent forms of the apo-Pgm or multimers. The abundance of Ni on the Pgm maximum was higher than that of Zn or Co, and reflected the major Ni protein in the metallome. Notably, the relative activity of Pgm observed in various metals follows the trend Ni(II) > Co(II) > Mn(II) > Cd(II) > Zn(II) at pH 7.4, which is also consistent with the observations of different isoforms of this protein.72–74 Additional chromatographic separation and structural biochemistry paired with metalloproteomics could confirm the metal identity of the Pgm active site in marine heterotrophs.
Fig. 8.
(A) Cytosolic Ni within the metallome of Pseudoalteromonas and (C) the protein distribution of a putative Ni-associated metalloenzyme, phosphoglucomutase (Pgm). (B, D) Comparison of the distribution of Ni, Zn, and Co with that of Pgm along the 450 and 500 mM NaCl AEFs reveals a correlation between multiple cytosolic metals, with Ni being the most abundant where Pgm coeluted.
While the Ni-associated SOD, SodN, is observed within the genome of Pseudoalteromonas (sodN, BB2AT2_2930), this protein was not detected within the metalloproteome of this organism under these growth conditions, unlike marine cyanobacteria where it is an important and abundant Ni metalloprotein.49,66 This organism appears reliant upon the Fe-associated SOD under these replete growth conditions.40 Another potential contributor to the Ni metallome of this organism could be urease, which features a binuclear Ni active site. A putative urease 23 kDa accessory protein was observed within the genome of BB2-AT2 (BB2AT2_3099), which does not appear to feature Ni binding in the metalloproteome.
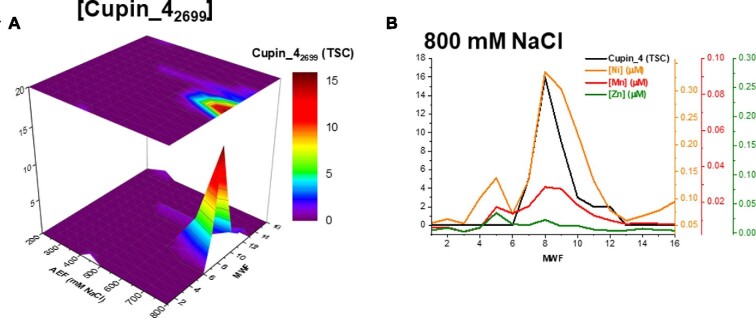
An additional putative Ni metalloenzyme was observed within the cupin superfamily, a series of proteins with diverse functionality. The cupin 4 family protein (BB2AT2_2699) was the only cupin superfamily protein observed within this metalloproteomic analysis, despite two genes of the cupin 2 family of proteins being observed within the genome (BB2AT2_0545, cupin 2 domain-containing protein and BB2AT2_2590, cupin 2 domain-containing protein). This protein was observed to be most abundant along the 800 mM NaCl anion exchange fraction (Fig. 9A). Comparison of this distribution of cupin_4 with those of Ni, Mn, and Zn produced R2 values of 0.91, 0.86, and 0.37, respectively (Fig. 9B). Notably the abundance of Ni at the Ni–cupin maxima (800 mM AE, 8 MWF) had ∼10-fold higher Ni than either Mn or Zn, implying a putative Ni-binding site within this enzyme.
Fig. 9.
(A) Distribution of a cupin superfamily protein, cupin_4, and (B) comparison of this protein with the distribution of Ni, Mn, and Zn within the cytosol.
It is possible that this cupin protein also possesses phosphoglucose isomerase activity, similar to Pgm. Ni-associated cupin-type phosphoglucose isomerases have been characterized within the archaea Archaeoglobus fulgidus, Methanosarcina mazei, and Pyrococcus furiosus, as well as within the bacteria Salmonella enterica, serovar Typhimurium, and Ensifer meliloti.37,75–77 Phylogenetic analysis suggests that these bacterial cupin phosphoglucose isomerases originated from euryarchaeota via lateral gene transfer.77 Sequence analysis via BLAST identified an additional possible function of this cupin superfamily protein as a peptidyl-arginine hydroxylase. However, the function of this protein is currently unknown. Further biochemical analyses are necessary to understand how metal is trafficked to this protein, the catalytic activity of this protein associated with different metals, as well as the metabolic function of this cupin within Pseudoalteromonas.
The results presented here imply underappreciated uses of Ni within marine heterotrophs. Nickel's biochemical functions and its relationship to biogeochemical cycles remain poorly constrained relative to other metals. Its role in the urease and Ni SOD enzymes have been discussed, but here we identify two other putative Ni metalloproteins that appear to be the major reservoirs of Ni within an important marine heterotrophic bacterium.49,78–80,51,66,52 While the Ni-associated SOD and urease are both present within the Pseudoalteromonas genome, they were not observed within this metalloproteome. Pgm and cupin instead appear to form the dominant Ni reservoir under these replete culture conditions.
Global proteome analysis of exponential- and stationary-phase Pseudoalteromonas cells
The global proteome of Pseudoalteromonas BB2-AT2 showed numerous changes during growth progression from exponential phase into stationary phase. Analysis of stationary- and mid-exponential-phase cultures resulted in 675 protein identifications. Notably, the abundances of alkaline phosphatases, proteases, and TonB-dependent receptors were all elevated in the late-exponential-phase cultures, consistent with increased synthesis of degradation enzymes in response to a transition to depleted media in stationary phase (Fig. 10). Similarly, a number of proteases that were observed in the stationary-phase experiment were also found to be present in the metallome. The prominence and dynamic increase in abundance of phosphatases and proteases, compared with the metalloproteome results described earlier, imply that the Zn use in Pseudoalteromonas should increase further in nutrient-depleted conditions.
Fig. 10.
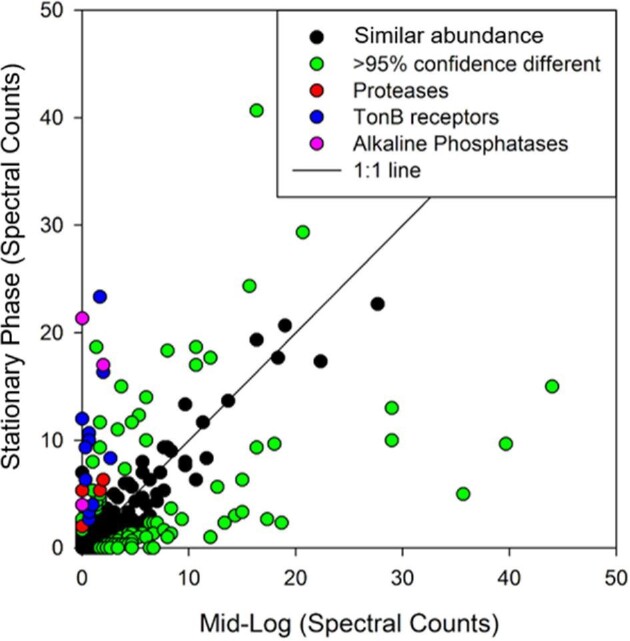
Comparison of protein abundance in Pseudoalteromonas (BB2-AT2) harvested at midlog (exponential) and stationary phases. Spectral counts of technical triplicates were averaged (see the Supplementary Material). Proteins with differing abundances according to Fisher's test 95% confidence interval are shown in green or other colors, while proteins not significantly different between treatments are shown in black. A number of proteases, TonB-dependent transporters, and alkaline phosphatase were more abundant at stationary phase, consistent with an adaptive response to depletion of nutrients.
Sediment trap material incubation experiment
The reliance on multiple transition metals for proteolytic digestion highlights the adaptive potential of Pseudoalteromonas to function in metal-limited environments. Export of sinking particulate material from the upper water column represents an important component of global carbon cycling, yet little is known about the potential for metal limitation for the degradation of this sinking material.10,42,81 We conducted experiments on this material to examine the influence of metals under trace metal conditions in the Central Pacific Ocean. Alkaline phosphatase activity was detected in sinking material collected by sediment traps, with the 60 m sample having ∼10-fold more activity (per milliliter of incubation sample) compared with the 150 m, and activity decreased over 5 days in all treatments (Fig. 11). The reproducibility associated with the quintuplicate analyses was reasonable, and implied some success in producing relatively homogenous treatments despite the challenge of the clean-sediment-trap experimental design.
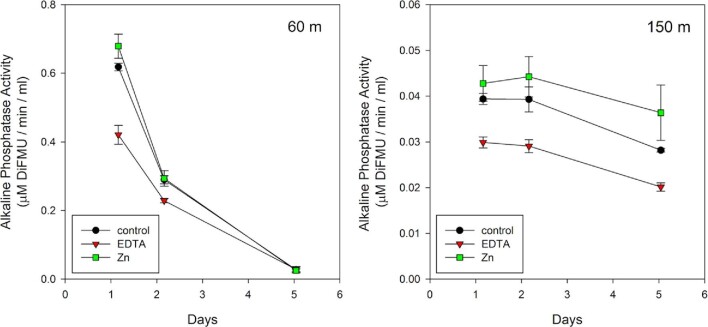
Fig. 11.
Sinking particulate material collected at 60 m (left) and 150 m (right) by the sediment trap was incubated with Zn or EDTA and compared with controls for alkaline phosphatase activity over 5 days. Quintuplicate treatments were averaged and standard deviations shown. Increased and decreased enzyme activity with Zn and EDTA additions, respectively, was observed, consistent with influence on metal demand within the alkaline phosphatase enzymes present.
EDTA and Zn effects were apparent at the early time points in the 60 m experiment, but the phosphatase signal decreased rapidly as biomass rapidly degraded. In contrast, differences in EDTA and Zn treatments were apparent by Day 1 in the 150 m experiment and the trends continued throughout the experiment, implying that these treatments impacted the microbes or their enzymes within 24 h. The addition of EDTA lessened alkaline phosphatase activity in both the 60 and 150 m experiments, likely due to inactivation of the enzyme by removal of the Zn or other divalent cation needed for activity. Activity was 32% lower after 1.2 days and 21% after 2.2 days in the 60 m experiment relative to the control, but could not be differentiated by Day 5. Activity decreased by ∼25% at all time points in EDTA treatments for the 150 m experiment. Zn addition treatments also showed some enhanced phosphatase activity relative to the control in 60 and 150m samples, presumably through increased availability of Zn for enzymatic activity. A 10% increase in phosphatase activity was observed after 1.2 days in the 60 m experiment. Zn additions also resulted in increased phosphatase activity in the 150 m experiment, particularly at the final time point.
Together these laboratory and field experiments imply that the availability of Zn in the upper water column may be an important constraint on remineralization rates and ultimately carbon export flux. Zn availability could be an overlooked parameter controlling the remineralization efficiency and depth of carbon export. In many oceanic waters, Zn is depleted in the euphotic zone, until reaching a ‘zincocline’ that often co-occurs and correlates with the silica distribution.82–84 Moreover, Zn is often complexed by organic ligands in the euphotic zone.85,86 Hence, Zn availability, both in total and by complexation, is likely low in many regions. The remineralization depth is predicted to be important in controlling the air–sea carbon balance, and Zn could be a constraint not only in marine photoautotrophy, but also on the less studied marine heterotrophic processes that contribute to carbon export of the marine biological pump.11,87
Conclusions
Pseudoalteromonas is an abundant marine heterotroph. When grown under replete conditions, it possesses a substantial quota of trace metals, indicating their unique adaptability relative to oligotrophic organisms. Unlike the metallomes of marine phytoplankton that are dominated by Fe, the majority of the metallome of Pseudoalteromonas is devoted to Zn, which facilitates proteolysis, transcription, translation, and phosphorus hydrolysis. SVD analysis enabled an approximation of the percentage contribution of each of the biochemical functions to the Zn metallome. It showed that transcription (47.2%) and translation (33.5%) are major contributions, followed by proteolysis (12.8%) and alkaline phosphatase (6.4%). A variety of proteases were observed to be used by this bacterium, including several putatively identified as Mn-associated, and less that were Co-associated. Future biochemical investigations should aim to clarify the relative importance of heterotrophic bacterial-driven proteolytic transformations, in an effort to better understand the diversity and abundance of these proteolytic systems. This would establish constraints on ectoenzyme activities on dissolved and particulate carbon mineralization within the water column. Modulation of trace metal availability of sinking particulate material showed addition and removal of metals to influence alkaline phosphatase activity, implying the potential for control of remineralization by Zn and other metals. Ni-associated proteins observed within this analysis included Pgm and a cupin superfamily protein, which revealed, for the first time, the Ni utilization of a marine heterotroph. The function of this cupin superfamily protein is currently unknown, but it may play a role in phosphoglucose isomerization. This study improves our understanding of the Zn, Mn, Co, and Ni biogeochemistry of marine heterotrophic bacteria via the characterization of various features of their metalloproteome.
Supplementary Material
Acknowledgements
M.G.M. acknowledges support from the Camille and Henry Dreyfus Foundation Environmental Chemistry Postdoctoral Fellowship. D.T.W. acknowledges support from the Stanley W. Watson Student Fellowship Fund at Woods Hole Oceanographic Institution (WHOI) and the MIT Presidential Graduate Fellowship.
Contributor Information
Michael G Mazzotta, Department of Marine Chemistry and Geochemistry, Woods Hole Oceanographic Institution, Woods Hole, MA 02543, USA.
Matthew R McIlvin, Department of Marine Chemistry and Geochemistry, Woods Hole Oceanographic Institution, Woods Hole, MA 02543, USA.
Dawn M Moran, Department of Marine Chemistry and Geochemistry, Woods Hole Oceanographic Institution, Woods Hole, MA 02543, USA.
David T Wang, Department of Marine Chemistry and Geochemistry, Woods Hole Oceanographic Institution, Woods Hole, MA 02543, USA.
Kay D Bidle, Department of Marine and Coastal Sciences, Rutgers University, New Brunswick, NJ 08901, USA.
Carl H Lamborg, Department of Ocean Sciences, University of California, Santa Cruz, CA 95064, USA.
Mak A Saito, Department of Marine Chemistry and Geochemistry, Woods Hole Oceanographic Institution, Woods Hole, MA 02543, USA.
Funding
Funding for this research was provided by the National Institutes of Health (NIH) (R01-GM135709), the US National Science Foundation (NSF-OCE 1924554, 1850719), and the Gordon and Betty Moore Foundation (3782).
Conflicts of interest
There are no conflicts of interest to declare.
Data availability
Data are available in a repository and can be accessed via https://www.bco-dmo.org/dataset/808610 and https://www.bcodmo.org/dataset/808619.
References
- 1. Morel F. M. M., Hudson R. J. M., Price N. M., Limitation of productivity by trace metals in the sea, Limnol. Oceanogr., 1991, 36 (8), 1742–1755. [Google Scholar]
- 2. Sunda W. G., Trace metal interactions with marine phytoplankton, Biol. Oceanogr., 1989, 6, 411–442. [Google Scholar]
- 3. Smith D. C., Simon M., Alldredge A. L., Azam F., Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution, Nature, 1992, 359 (6391), 139–142. [Google Scholar]
- 4. Suttle C., Ecological, evolutionary, and geochemical consequences of viral infection of cyanobacteria and eukaryotic algae. In: Hurst C. J. (ed.), Viral Ecology, Academic Press, 2000, ch. 6. [Google Scholar]
- 5. Fuhrman J. A., Marine viruses and their biogeochemical and ecological effects, Nature, 1999, 399 (6736), 541–548. [DOI] [PubMed] [Google Scholar]
- 6. Zimmerman A. E., Allison S. D., Martiny A. C., Phylogenetic constraints on elemental stoichiometry and resource allocation in heterotrophic marine bacteria: stoichiometry and resource allocation in marine bacteria, Environ. Microbiol., 2014, 16 (5), 1398–1410. [DOI] [PubMed] [Google Scholar]
- 7. Steinberg D. K., Mooy B., Buesseler K. O., Boyd P. W., Kobari T., Karl D. M., Bacterial vs. zooplankton control of sinking particle flux in the ocean's twilight zone, Limnol. Oceanogr., 2008, 53 (4), 1327–1338. [Google Scholar]
- 8. Sunda W. G., Feedback Interactions between trace metal nutrients and phytoplankton in the ocean, Front. Microbiol., 2012, 3, DOI: 10.3389/fmicb.2012.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tortell P. D., Maldonado M. T., Price N. M., The role of heterotrophic bacteria in iron-limited ocean ecosystems, Nature, 1996, 383 (6598), 330–332. [Google Scholar]
- 10. Fukuda R., Sohrin Y., Saotome N., Fukuda H., Nagata T., Koike I., East–west gradient in ectoenzyme activities in the subarctic Pacific: possible regulation by zinc, Limnol. Oceanogr., 2000, 45 (4), 930–939. [Google Scholar]
- 11. Kwon E. Y., Primeau F., Sarmiento J. L., The impact of remineralization depth on the air–sea carbon balance, Nat. Geosci., 2009, 2 (9), 630–635. [Google Scholar]
- 12. Boyd P. W., Ellwood M. J., Tagliabue A., Twining B.S., Biotic and abiotic retention, recycling and remineralization of metals in the ocean, Nat. Geosci., 2017, 10 (3), 167–173. [Google Scholar]
- 13. Ratnarajah L., Blain S., Boyd P. W., Fourquez M., Obernosterer I., Tagliabue A., Resource colimitation drives competition between phytoplankton and bacteria in the Southern Ocean, Geophys. Res. Lett., 2021, 48 (1), e2020GL088369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martin J. H., Fitzwater S. E., Iron deficiency limits phytoplankton growth in the north-east Pacific subarctic, Nature, 1988, 331 (6154), 341–343. [Google Scholar]
- 15. Martin J. H., Glacial-interglacial CO2 change: the iron hypothesis, Paleoceanography, 1990, 5 (1), 1–13. [Google Scholar]
- 16. Price N. M., Andersen L. F., Morel F. M. M., Iron and nitrogen nutrition of equatorial Pacific plankton, Deep Sea Res., Part I Oceanogr. Res. Pap., 1991, 38, 1361–1378. [Google Scholar]
- 17. Martin J. H., Coale K. H., Johnson K. S., Fitzwater S. E., Gordon R. M., Tanner S. J., Hunter C. N., Elrod V. A., Nowicki J. L., Coley T. L., Barber R. T., Lindley S., Watson A. J., Van Scoy K., Law C. S., Liddicoat M. I., Ling R., Stanton T., Stockel J., Collins C., Anderson A., Bidigare R., Ondrusek M., Latasa M., Millero F. J., Lee K., Yao W., Zhang J. Z., Friederich G., Sakamoto C., Chavez F., Buck K., Kolber Z., Greene R., Falkowski P., Chisholm S. W., Hoge F., Swift R., Yungel J., Turner S., Nightingale P., Hatton A., Liss P., Tindale N. W., Testing the iron hypothesis in ecosystems of the equatorial Pacific Ocean, Nature, 1994, 371 (6493), 123–129. [Google Scholar]
- 18. Coale K. H., Johnson K. S., Fitzwater S. E., Gordon R. M., Tanner S., Chavez F. P., Ferioli L., Sakamoto C., Rogers P., Millero F., Steinberg P., Nightingale P., Cooper D., Cochlan W. P., Landry M. R., Constantinou J., Rollwagen G., Trasvina A., Kudela R., A massive phytoplankton bloom induced by an ecosystem-scale iron fertilization experiment in the equatorial Pacific Ocean, Nature, 1996, 383 (6600), 495–501. [DOI] [PubMed] [Google Scholar]
- 19. Granger J., Price N. M., The importance of siderophores in iron nutrition of heterotrophic marine bacteria, Limnol. Oceanogr., 1999, 44 (3), 541–555. [Google Scholar]
- 20. Hoppe H. G., Use of fluorogenic model substrates for extracellular enzyme activity (EEA) measurement in bacteria, In: Handbook of Methods in Aquatic Microbial Ecology, CRC Press, 1993, [Google Scholar]
- 21. Azam F., Smith D. C., Steward G. F., Hagström A., Bacteria-organic matter coupling and its significance for oceanic carbon cycling, Microb. Ecol., 1994, 28 (2), 167–179. [DOI] [PubMed] [Google Scholar]
- 22. Koike I., Nagata T., High potential activity of extracellular alkaline phosphatase in deep waters of the central Pacific, Deep Sea Res. Part II Top. Stud. Oceanogr., 1997, 44 (9–10), 2283–2294. [Google Scholar]
- 23. Bidle K. D., Azam F., Accelerated dissolution of diatom silica by marine bacterial assemblages, Nature, 1999, 397 (6719), 508–512. [Google Scholar]
- 24. Nagata T., Organic matter–bacteria interactions in seawater, In: Microbial Ecology of the Oceans, John Wiley & Sons, Ltd, 2008, 207–241. [Google Scholar]
- 25. Bidle K. D., Manganelli M., Azam F., Regulation of oceanic silicon and carbon preservation by temperature control on bacteria, Science, 2002, 298 (5600), 1980–1984. [DOI] [PubMed] [Google Scholar]
- 26. McCarren J., Becker J. W., Repeta D. J., Shi Y., Young C. R., Malmstrom R. R., Chisholm S. W., DeLong E. F., Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea, Proc. Natl. Acad. Sci., 2010, 107 (38), 16420–16427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Acinas S. G., Antón J., Rodríguez-Valera F., Diversity of free living and attached bacteria in offshore Western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA, Appl. Environ. Microbiol., 1999, 65 (2), 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ivars-Martinez E., Martin-Cuadrado A.-B., D'Auria G., Mira A., Ferriera S., Johnson J., Friedman R., Rodriguez-Valera F.., Comparative genomics of two ecotypes of the marine planktonic copiotroph Alteromonas macleodii suggests alternative lifestyles associated with different kinds of particulate organic matter, ISME J., 2008, 2 (12), 1194–1212. [DOI] [PubMed] [Google Scholar]
- 29. García-Martínez J., Acinas S. G., Massana R., Rodríguez-Valera F., Prevalence and microdiversity of Alteromonas macleodii-like microorganisms in different oceanic regions, Environ. Microbiol., 2002, 4 (1), 42–50. [DOI] [PubMed] [Google Scholar]
- 30. Hou S., López-Pérez M., Pfreundt U., Belkin N., Stüber K., Huettel B., Reinhardt R., Berman-Frank I., Rodriguez-Valera F., Hess W. R., Benefit from decline: the primary transcriptome of Alteromonas macleodii str. Te101 during Trichodesmium demise, ISME J, 2018, 12 (4), 981–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bidle K. D., Azam F., Bacterial control of silicon regeneration from diatom detritus: significance of bacterial ectohydrolases and species identity, Limnol. Oceanogr., 2001, 46 (7), 1606–1623. [Google Scholar]
- 32. Azam F., Malfatti F., Microbial structuring of marine ecosystems, Nat. Rev. Microbiol., 2007, 5 (10), 782–791. [DOI] [PubMed] [Google Scholar]
- 33. Burd A. B., Frey S., Cabre A., Ito T., Levine N. M., Lønborg C., Long M., Mauritz M., Thomas R. Q., Stephens B. M., Vanwalleghem T., Zeng N., Terrestrial and marine perspectives on modeling organic matter degradation pathways, Global Change Biol., 2016, 22 (1), 121–136. [DOI] [PubMed] [Google Scholar]
- 34. Waldron K. J., A periplasmic iron-binding protein contributes toward inward copper supply, J. Biol. Chem., 2007, 282 (6), 3837. [DOI] [PubMed] [Google Scholar]
- 35. Tottey S., Waldron K. J., Firbank S. J., Reale B., Bessant C., Sato K., Cheek T. R., Gray J., Banfield M. J., Dennison C., Robinson N. J., Protein-folding location can regulate manganese-binding versus copper- or zinc-binding, Nature, 2008, 455 (7216), 1138–1142. [DOI] [PubMed] [Google Scholar]
- 36. Waldron K. J., Robinson N. J., How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol., 2009, 7 (1), 25–35. [DOI] [PubMed] [Google Scholar]
- 37. Cvetkovic A., Menon A. L., Thorgersen M. P., Scott J. W., Poole F. L. 2nd, Jenney F. E. Jr, Lancaster W. A., Praissman J. L., Shanmukh S., Vaccaro B. J., Trauger S. A., Kalisiak E., Apon J. V., Siuzdak G., Yannone S. M., Tainer J. A., Adams M. W., Microbial metalloproteomes are largely uncharacterized, Nature, 2010, 466 (7307), 779–782. [DOI] [PubMed] [Google Scholar]
- 38. Aguirre J. D., Clark H. M., McIlvin M., Vazquez C., Palmere S. L., Grab D. J., Seshu J., Hart P. J., Saito M., Culotta V. C., A manganese-rich environment supports superoxide dismutase activity in a Lyme disease pathogen, Borrelia burgdorferi, J. Biol. Chem., 2013, 288 (12), 8468–8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tarrant E., Riboldi G. P., McIlvin M. R., Stevenson J., Barwinska-Sendra A., Stewart L. J., Saito M. A., Waldron K. J., Copper stress in Staphylococcus aureus leads to adaptive changes in central carbon metabolism, Metallomics, 2019, 11 (1), 183–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mazzotta M. G., McIlvin M. R., Saito M. A., Characterization of the Fe metalloproteome of a ubiquitous marine heterotroph, Pseudoalteromonas (BB2-AT2): multiple bacterioferritin copies enable significant Fe storage, Metallomics, 2020, 12 (5), 654–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Saito M. A., Bulygin V. V., Moran D. M., Taylor C., Scholin C., Examination of microbial proteome preservation techniques applicable to autonomous environmental sample collection, Front. Microbiol., 2011, 2, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Munson K. M., Lamborg C. H., Swarr G. J., Saito M. A., Mercury species concentrations and fluxes in the Central Tropical Pacific Ocean, Glob. Biogeochem. Cycles, 2015, 29 (5), 656–676. [Google Scholar]
- 43. Sunda W. G., Huntsman S. A., Iron uptake and growth limitation in oceanic and coastal phytoplankton, Mar. Chem., 1995, 50 (1–4), 189–206. [Google Scholar]
- 44. Ho T.-Y., Quigg A., Finkel Z., Milligan A. J., Wyman K., Falkowski P., Morel F. M. M., The elemental composition of some phytoplankton, J. Phycol., 2003, 39 (6), 1145–1159. [Google Scholar]
- 45. Saito M. A., Noble A. E., Hawco N., Twining B. S., Ohnemus D. C., John S. G., Lam P., Conway T. M., Johnson R., Moran D., McIlvin M., The acceleration of dissolved cobalt's ecological stoichiometry due to biological uptake, remineralization, and scavenging in the Atlantic Ocean, Biogeosciences, 2017, 14 (20), 4637–4662. [Google Scholar]
- 46. Hawco N. J., Saito M. A., Competitive inhibition of cobalt uptake by zinc and manganese in a pacific Prochlorococcus strain: Insights into metal homeostasis in a streamlined oligotrophic cyanobacterium, Limnol. Oceanogr. 2018, 63 (5), 2229–2249. [Google Scholar]
- 47. Hawco N. J., McIlvin M. M., Bundy R. M., Tagliabue A., Goepfert T. J., Moran D. M., Valentin-Alvarado L., DiTullio G. R., Saito M. A., Minimal cobalt metabolism in the marine cyanobacterium Prochlorococcus, PNAS, 2020, 117 (27), 15740–15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Twining B. S., Baines S. B., The trace metal composition of marine phytoplankton, Annu. Rev. Mar. Sci., 2013, 5 (1), 191–215. [DOI] [PubMed] [Google Scholar]
- 49. Dupont C. L., Barbeau K., Palenik B., Ni uptake and limitation in marine Synechococcus strains, Appl. Environ. Microbiol., 2008, 74 (1), 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dupont C. L., Buck K. N., Palenik B., Barbeau K., Nickel utilization in phytoplankton assemblages from contrasting oceanic regimes, Deep Sea Res. Part I Oceanogr. Res. Pap., 2010, 57 (4), 553–566. [Google Scholar]
- 51. Ho T-Y., Nickel limitation of nitrogen fixation in Trichodesmium, Limnol. Oceanogr., 2013, 58 (1), 112–120. [Google Scholar]
- 52. Glass J. B., Dupont C. L., Oceanic nickel biogeochemistry and the evolution of nickel use. In: Zamble D, Rowinska-Zyrek M, Kozlowski H. (eds.), The Biological Chemistry of Nickel, Royal Society of Chemistry, 2017, ch. 2, 12–26. [Google Scholar]
- 53. Ma J.-C., Newman A. J., Hayward R. S., Internal promoters of the rpoBC operon of Escherichia coli, Mol. Gen. Genet., 1981, 184 (3), 548–550. [DOI] [PubMed] [Google Scholar]
- 54. Kumar M., Kumar S. A. P., Dimkovikj A., Baykal L. N., Banton M. J., Outlaw M. M., Polivka K. E., Hellmann-Whitaker R. A., Zinc is the molecular “switch” that controls the catalytic cycle of bacterial leucyl-tRNA synthetase, J. Inorg. Biochem., 2015, 142, 59–67. [DOI] [PubMed] [Google Scholar]
- 55. Li Y., Sharma M. R., Koripella R. K., Yang Y., Kaushal P. S., Lin Q., Wade J. T., Gray T. A., Derbyshire K. M., Agrawal R. K., Ojha A. K., Zinc depletion induces ribosome hibernation in mycobacteria, PNAS, 2018, 115 (32), 8191–8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Simpson R. T., Vallee B. L., Tait G. H., Alkaline phosphatase of Escherichia coli. Composition, Biochemistry, 1968, 7 (12), 4336–4342. [DOI] [PubMed] [Google Scholar]
- 57. Banci L., Bertini I., Luchinat C., Viezzoli M. S., Wang Y. J., The cobalt(II)-alkaline phosphatase system at alkaline pH, J. Biol. Chem., 1988, 263 (23), 11263–11268. [PubMed] [Google Scholar]
- 58. Wojciechowski C. L., Kantrowitz E. R., Altering of the metal specificity of Escherichia coli alkaline phosphatase, J. Biol. Chem., 2002, 277 (52), 50476–50481. [DOI] [PubMed] [Google Scholar]
- 59. Saito M. A., Moffett J. W., Chisholm S. W., Waterbury J. B., Cobalt limitation and uptake in Prochlorococcus, Limnol. Oceanogr., 2002, 47 (6), 1629–1636. [Google Scholar]
- 60. Shaked Y., Xu Y., Leblanc K., Morel F. M. M, Zinc availability and alkaline phosphatase activity in Emiliania huxleyi: implications for Zn-P co-limitation in the ocean, Limnol. Oceanogr., 2006, 51 (1), 299–309. [Google Scholar]
- 61. Cox A., Saito M., Proteomic responses of oceanic Synechococcus WH8102 to phosphate and zinc scarcity and cadmium additions, Front. Microbiol., 2013, 4, DOI:10.3389/fmicb.2013.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cox A. D., Noble A. E., Saito M. A., Cadmium enriched stable isotope uptake and addition experiments with natural phytoplankton assemblages in the Costa Rica Upwelling Dome, Mar. Chem., 2014, 166, 70–81. [Google Scholar]
- 63. Lane T. W., Morel F. M. M., A biological function for cadmium in marine diatoms, PNAS, 2000, 97 (9), 4627–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Saito M. A., Goepfert T. J., Ritt J. T., Some thoughts on the concept of colimitation: three definitions and the importance of bioavailability, Limnol. Oceanogr., 2008, 53 (1), 276–290. [Google Scholar]
- 65. Saito M. A., Goepfert T. J., Zinc-cobalt colimitation of Phaeocystis antarctica, Limnol. Oceanogr., 2008, 53 (1), 266–275. [Google Scholar]
- 66. Saito M. A., McIlvin M. R., Moran D. M., Goepfert T. J., DiTullio G. R., Post A. F., Lamborg C. H., Multiple nutrient stresses at intersecting Pacific Ocean biomes detected by protein biomarkers, Science, 2014, 345 (6201), 1173–1177. [DOI] [PubMed] [Google Scholar]
- 67. Tagliabue A., Hawco N. J., Bundy R. M., Landing W. M., Milne A., Morton P. L., Saito M. A., The role of external inputs and internal cycling in shaping the global ocean cobalt distribution: insights from the first cobalt biogeochemical model, Glob. Biogeochem. Cycles, 2018, 32 (4), 594–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kellogg M. M., McIlvin M. R., Vedamati J., Twining B. S., Moffett J. W., Marchetti A., Moran D. M., Saito M. A., Efficient zinc/cobalt inter-replacement in northeast Pacific diatoms and relationship to high surface dissolved Co :Zn ratios, Limnol. Oceanogr., 2020, 65 (11), 2557–2582. [Google Scholar]
- 69. Morel F. M. M., Lam P. J., Saito M. A., Trace metal substitution in marine phytoplankton, Annu. Rev. Earth Planet. Sci. 2020, 48 (1), 491–517. [Google Scholar]
- 70. Hwangbo S.-A., Kim J.-W., Jung S.-J., Jin K. S., Lee J.-O., Kim J.-S., Park S.-Y., Characterization of a dimeric arginase From Zymomonas mobilis ZM4, Front. Microbiol., 2019, 10, 2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lee M. D., Walworth N. G., McParland E. L., Fu F.-X., Mincer T. J., Levine N. M., Hutchins D. A., Webb E. A., The Trichodesmium consortium: conserved heterotrophic co-occurrence and genomic signatures of potential interactions, ISME J., 2017, 11 (8), 1813–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ray W. J., Role of bivalent cations in the phosphoglucomutase system. I. Characterization of enzyme-metal complexes, J. Biol. Chem., 1969, 244 (14), 3740–3747. [PubMed] [Google Scholar]
- 73. Ray W. J., Goodin D. S., Ng L., Cobalt(II) and nickel(II) complexes of phosphoglucomutase, Biochemistry, 1972, 11 (15), 2800–2804. [DOI] [PubMed] [Google Scholar]
- 74. Ray W. J., Mildvan A. S., Grutzner J. B., Phosphorus nuclear magnetic resonance studies of phosphoglucomutase and its metal ion complexes, Arch. Biochem. Biophys., 1977, 184 (2), 453–463. [DOI] [PubMed] [Google Scholar]
- 75. Verhees C. H., Tuininga J. E., Kengen S. W. M., Stams A. J., van der Oost J., de Vos W. M., ADP-dependent phosphofructokinases in mesophilic and thermophilic methanogenic Archaea, J. Bacteriol., 2001, 183 (24), 7145–7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hansen T., Schlichting B., Grötzinger J., Swan M. K., Davies C., Schönheit P., Mutagenesis of catalytically important residues of cupin type phosphoglucose isomerase from Archaeoglobus fulgidus, FEBS J., 2005, 272 (24), 6266–6275. [DOI] [PubMed] [Google Scholar]
- 77. Hansen T., Schlichting B., Felgendreher M., Schönheit P., Cupin-type phosphoglucose isomerases (cupin-PGIs) constitute a novel metal-dependent PGI family representing a convergent line of PGI evolution, J. Bacteriol., 2005, 187 (5), 1621–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Price N. M., Morel F. M. M., Colimitation of phytoplankton growth by nickel and nitrogen, Limnol. Oceanogr., 1991, 36 (6), 1071–1077. [Google Scholar]
- 79. Ho T.-Y., Wen L.-S., You C.-F., Lee D.-C., The trace metal composition of size-fractionated plankton in the South China Sea: biotic versus abiotic sources, Limnol. Oceanogr., 2007, 52 (5), 1776–1788. [Google Scholar]
- 80. Allen A. E., Dupont C. L., Oborník M., Horák A., Nunes-Nesi A., McCrow J. P., Zheng H., Johnson D. A., Hu H., Fernie A. R., Bowler C., Evolution and metabolic significance of the urea cycle in photosynthetic diatoms, Nature, 2011, 473 (7346), 203–207. [DOI] [PubMed] [Google Scholar]
- 81. Munson K. M., Lamborg C. H., Boiteau R. M., Saito M. A., Dynamic mercury methylation and demethylation in oligotrophic marine water, Biogeosciences, 2018, 15 (21), 6451–6460. [Google Scholar]
- 82. Bruland K. W., Knauer G. A., Martin J. H., Zinc in north-east Pacific water, Nature, 1978, 271 (5647), 741–743. [Google Scholar]
- 83. Bryan S. P., Marchitto T. M., Testing the utility of paleonutrient proxies Cd/Ca and Zn/Ca in benthic foraminifera from thermocline waters, Geochem. Geophys. Geosyst., 2010, 11 (1), DOI: 10.1029/2009GC002780. [Google Scholar]
- 84. Vance D., Little S. H., de Souza G. F., Khatiwala S., Lohan M. C., Middag R., Silicon and zinc biogeochemical cycles coupled through the Southern Ocean, Nat. Geosci., 2017, 10 (3), 202–206. [Google Scholar]
- 85. Bruland K. W., Complexation of zinc by natural organic ligands in the central North Pacific, Limnol. Oceanogr., 1989, 34 (2), 269–285. [Google Scholar]
- 86. Ellwood M. J., Van den Berg C. M. G., Zinc speciation in the Northeastern Atlantic Ocean, Mar. Chem., 2000, 68 (4), 295–306. [Google Scholar]
- 87. Morel F. M. M., Reinfelder J. R., Roberts S. B., Chamberlain C. P., Lee J. G., Yee D., Zinc and carbon co-limitation of marine phytoplankton, Nature, 1994, 369 (6483), 740–742. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in a repository and can be accessed via https://www.bco-dmo.org/dataset/808610 and https://www.bcodmo.org/dataset/808619.