Abstract
Definitive identification of the species in the Burkholderia cepacia complex by routine clinical microbiology methods is difficult. Phenotypic tests to identify B. multivorans and B. vietnamiensis have been established; more recent work indicates B. stabilis may also be identified by growth characteristics and biochemical tests. However, attempts to identify genomovars I and III have, thus far, proved unsuccessful. Previously, we demonstrated the utility of two primer pairs, directed to the rRNA operon, to specifically identify the B. cepacia complex in a PCR. One of these primer pairs, G1-G2, only amplified a DNA fragment from genomovars I and III and B. stabilis in a PCR with genomic DNA isolated from prototypical strains representing the five genomovars. Sequence analysis of the rRNA operon for all the genomovars indicated that this primer pair targeted a region shared by these isolates. Further analysis revealed a region of heterogeneity between genomovar III and B. stabilis internal to the amplified product of G1-G2. Primers designed to target this region were tested with prototypical strains following an initial amplification with the G1-G2 primer pair. New primers specific for the prototypical genomovar III and B. stabilis were designated SPR3 and SPR4, respectively. Analysis of 93 isolates representing 18 genomovar I, 13 B. multivorans, 36 genomovar III, 11 B. stabilis, and 15 B. vietnamiensis isolates was performed. DNA from all isolates of genomovars I and III and B. stabilis was amplified by G1-G2. Genomovar III isolates yielded a product with SPR3/G1 while B. stabilis amplified with SPR4-G1. Genomovar I isolates were amplified by either SPR3-G1 or SPR4-G1, but not both. B. multivorans yielded a product with SPR3-G1 but not G1-G2, and B. vietnamiensis isolates were negative in all PCRs. Thus using an algorithm with G1-G2, SPR3-G1, and SPR4-G1 primers in a PCR analysis, genomovar III isolates can be separated from B. stabilis and the identity of B. multivorans and B. vietnamiensis can be confirmed.
The gram-negative bacillus Burkholderia cepacia (basonyms Pseudomonas cepacia, P. multivorans, and P. kingii) was originally described in 1950 as the etiological agent responsible for the maceration of onion bulbs (3). In the early 1980s B. cepacia infections were recognized in patients with cystic fibrosis (CF) and chronic granulomatous disease (8, 10). B. cepacia is transmitted from patient to patient (13), and infection with B. cepacia is associated with an increased morbidity and mortality in CF patients (17). Following acquisition, many patients succumb to cepacia syndrome, a necrotizing pneumonia with fever, bacteremia, elevation of erythrocyte sedimentation rate, and leukocytosis, culminating in rapid and fatal clinical deterioration. Other patients begin an incremental decline in pulmonary function that is more rapid than in patients not infected with B. cepacia (8, 18). Clinical intervention is difficult because of the inherent resistance of this organism to a wide range of antimicrobial drugs; thus, the only effective method of infection control is to physically separate infected from noninfected patients. Such an austere approach creates a severe psychosocial stigma for colonized individuals. Recent data indicate that B. cepacia consists of at least five distinct subgroups, originally referred to as genomovars pending definitive biochemical and/or molecular methods to allow discrimination (21). Three of the genomovars can now be identified by a complex phenotypic analysis and have been assigned a binomial designation. Genomovar II is B. multivorans (21), genomovar IV is B. stabilis (20), and genomovar V is the previously described B. vietnamiensis (7). However, isolates of the remaining two genomovars cannot, at present, be distinguished without complex polyphasic analysis of phenotypes, whole-cell protein profiles, and DNA-DNA hybridization (21). Although organisms belonging to each genomovar have been recovered from CF sputum, the majority of clinical isolates are B. multivorans and genomovar III (12). Preliminary studies indicate that isolates associated with increased morbidity and mortality cluster in genomovar III (14). Thus, accurate identification of the genomovars is essential for effective patient management. The utility of PCR for the identification of members of the genus Burkholderia and the B. cepacia complex has been demonstrated (14, 22). In addition, LiPuma et al. also demonstrated identification of B. multivorans, B. vietnamiensis, and a group comprising genomovars I and III and B. stabilis (14). We have developed a PCR-based technique that expands upon this analysis by separating genomovar III and B. stabilis and, in addition, provides a separate species-specific PCR (SS-PCR) for the confirmation of identity of B. multivorans and B. vietnamiensis.
MATERIALS AND METHODS
Bacterial strains.
For 23S rRNA gene sequence determination, B. cepacia complex type strains were obtained from the American Type Culture Collection or the Laboratorium Microbiologie Gent (Ghent, Belgium) (Table 1). All other isolates of B. cepacia complex were obtained from the Burkholderia cepacia Research Laboratory and Repository (University of Michigan, Ann Arbor, Mich.). The identity of each isolate was determined using sodium dodecyl sulfate-polyacrylamide electrophoresis of whole-cell protein extracts, as described by Vandamme et al. (21).
TABLE 1.
B. cepacia strains used for rRNA sequence determination
| Strain | Genomovar or species | Source |
|---|---|---|
| ATCC 25416 | I | Onion |
| LMG 14293 | B. multivorans | CF patient |
| LMG 12614 | III | CF patient |
| LMG 14294 | B. stabilis | CF patient |
| LMG 10929 | B. vietnamiensis | Rice rhizosphere |
Isolation of genomic DNA.
Genomic DNA was purified from cultures of each individual strain using the QIAamp Tissue Kit (QIAGEN, Valencia, Calif.). DNA was resuspended in sterile high-performance liquid chromatography-grade water at a final concentration of 1 μg/ml.
Development of genomovar-specific PCRs.
Primer pair G1-G2 was used in PCR as described by Whitby et al. (23) to amplify an approximately 1.3-kb product from genomovars I and III and B. stabilis. The amplified DNA bands were resolved by electrophoresis in a 0.8% (wt/vol) agarose gel and visualized by UV transillumination after staining with ethidium bromide. The DNA was excised from the gel and purified using Geneclean II (BIO-101, Vista, Calif.), cloned into TOPO-TA vector pCR 2.1 (Invitrogen, Carlsbad, Calif.) and transformed into E. coli Top10 (Invitrogen). This was repeated to give three independent clones of the 1.3-kb spacer region from each isolate. The nucleotide sequence of three independent clones of the amplified 16S-23S spacer region of genomovars I and III and B. stabilis was determined in both directions with multiple internal primers by dideoxy chain termination using an automated sequencer (Applied Biosystems model 373). The consensus sequence of each spacer region was determined using the PILEUP and PRETTY algorithms of the Genetics Computer Group DNA analysis computer package (6). The consensus regions were further analyzed using the BLASTN nucleotide sequence alignment algorithm to determine if putatively genomovar-specific sequences shared identity with other sequenced rRNA operons. For B. stabilis a putatively specific site was determined; genomovars I and III shared homology at this site. Using this putatively specific region, PCR primers SPR3 and SPR4 were designed to encompass the specific nucleotides at the 3′ end (Table 2). Primer pairs SPR3-G1 and SPR4-G1 were optimized in separate PCRs with genomic DNA from prototypic strains of genomovars I and III and B. stabilis. PCRs were performed using the model PTC100 thermocycler (MJ Research Inc., Watertown, Mass.). Each 50-μl reaction mixture contained an 800 pM concentration of each primer, 100 ng of genomic DNA, a 200 μM concentration of each deoxynucleoside triphosphate (dNTP), and 1.25 u of Taq DNA polymerase (Roche Molecular Biochemicals, Indianapolis, Ind.) in a 2 mM MgCl2 PCR buffer. Both PCRs had an initial denaturation at 95°C for 5 min with a subsequent 30-cycle amplification comprising annealing at 66°C for 45 s, extension at 72°C for 2 min, denaturation at 95°C for 45 s, and a final extension of 10 min. Following amplification, 20 μl of each reaction mixture was subjected to electrophoresis in a 0.8% agarose gel in 0.5× Tris-borate-EDTA buffer (pH 8.0). Positive results were assessed by the amplification of a band of approximately 1.2 kb. Negative controls (lacking template DNA) were performed with each PCR assay.
TABLE 2.
Primers used in genomovar determination of the B. cepacia complex
| Primer name | Sequencea | Target siteb |
|---|---|---|
| G1c | GCCATGGATACTCCAAAAGGA | 153–133 (23S) |
| G2c | TCGGAATCCTGCTGAGAGGC | 939–958 (16S) |
| SPR3 | TCGAAAGAGAACCGGCG | 969–985 (16S) |
| SPR4 | TCGAAAGAGAACCGATA | 969–985 (16S) |
| PSL | AGGATTAGATACCCTGGTAGTCCA | 780–803 (16S) |
| PSR | ACTTAACCCAACATCTCACGACAC | 1092–1068 (16S) |
Oligonucleotide sequence, 5′ to 3′. Underlined regions denote variable nucleotides.
Numbers correspond to position in the sequence with GenBank accession numbers X16368 for primer G1; all other numbers correspond to position in the sequence with GenBank accession number X87275.
Primer G1 is also called C-SSR, and primer G2 is also called C-SSF, as described by LiPuma et al. (14).
RESULTS
Sequence determination of the 16S-23S rRNA spacer region.
The nucleotide sequences of each clone were aligned using the Genetics Computer Group PILEUP algorithm to give a consensus sequence for each genomovar. In some cases, the inserts of the three clones from a single genomovar showed differences in size and sequence that corresponded to alternate tRNA operons; such regions were excluded from the consensus sequences. The sequenced region spanned from nucleotide 940 of the 16S rRNA to nucleotide 150 of the 23S rRNA. Examination of the sequences indicated a region of heterogeneity between the genomovar III and the B. stabilis nucleotide sequence, located within the 5′ terminus of the 16S RNA. A span of 3 nucleotides was distinct for each isolate. Genomovar I displayed identity with genomovar III at this region. Using this area of heterogeneity, oligonucleotides were designed to encompass the three variable nucleotides at the 3′ end, for use in PCR with primer G1. These were called SPR3 and SPR4 and were designed to amplify genomovars I and III and B. stabilis, respectively. The primer sequences are given in Table 2.
Development of genomovar-specific PCR.
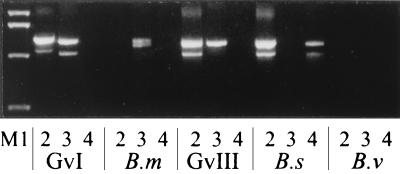
Previously we have shown that PCR with the primer pair G1-G2 specifically amplified the prototypic isolates of genomovars I and III and B. stabilis (23). To further divide this grouping, SPR3 and SPR4 were designed to target a region of heterogeneity inside the 16S rRNA. PCR with the primer pair SPR3-G1 yielded a product for the genomovar III and the genomovar I isolate, while the SPR4-G1 primer pair gave a product using the genomic template of B. stabilis. In each case the amplified DNA fragments were approximately 1.3 kb (Fig. 1).
FIG. 1.
PCR amplification of genomovar I (GvI) B. multivorans (B.m), genomovar III (GvIII), B. stabilis (B.s), and B. vietnamiensis (B.v) using primer pairs G1-G2 (lanes 2), SPR3-G1 (lanes 3), and SPR4-G1 (lanes 4). The molecular marker (lane M1) is a 1-kb ladder; sizes are (from bottom to top) 517, 1,018, 1,636, and 2,036 bp.
To assess the ability of these two primer pairs to correctly identify the genomovars, a panel was examined blindly. The panel consisted of 18 genomovar I isolates, 13 B. multivorans isolates, 36 genomovar III isolates, 11 B. stabilis isolates, and 15 B. vietnamiensis isolates. Genomic DNA template from each was analyzed with G1-G2, SPR3-G1, and SPR4-G1. All isolates of genomovar I and III and B. stabilis were amplified with G1-G2. Additionally, all genomovar III isolates yielded a product with SPR3-G1 while all B. stabilis isolates amplified with SPR4-G1. Each genomovar I isolate was amplified by either SPR3-G1 (11 of 18) or SPR4-G1 (7 of 18), but not both. B. multivorans isolates did not yield a product with G1-G2; however, they all amplified with SPR3-G1. Isolates of B. vietnamiensis did not yield a product in any PCR tested. To determine if this was due to inhibition of the PCR, each B. vietnamiensis genomic template was assessed with the primer pair PSL-PSR, which targets conserved bacterial rRNA regions (4). Products of the correct size were amplified in each case. Due to the primers targeting the 16S-23S rRNA spacer region, the products amplified as a doublet at approximately 1.2 to 1.3 kbp. Thus, using PCR analysis, genomovar III isolates can be separated from B. stabilis and the identity of B. multivorans and B. vietnamiensis is confirmed. Figure 1 shows the results of the individual PCRs for an isolate of each genomovar.
DISCUSSION
CF patients infected with B. cepacia carry a severe psychosocial burden arising from precautions to prevent person-to-person transmission and the poor clinical prognosis associated with this organism. Following DNA hybridization and other taxonomic studies, the original species, B. cepacia, has been divided into five distinct genomovars, of which three, genomovars II, IV, and V, have been given binomial species names. These have become B. multivorans, B. stabilis, and B. vietnamiensis, respectively (20, 21). With the advent of new classifications, numerous questions arise, including whether there is a correlation between genomovar and virulence. Preliminary studies have shown that all the genomovars can cause infection in patients with CF; however, it is becoming increasingly apparent that the majority of isolates from CF patients belong to B. multivorans and genomovar III (14). Unfortunately the genomovars are not easily identified by biochemical reactions, and these studies have been hindered by the complex polyphasic analysis required to determine the genomovar of each isolate (21). Thus, before more complex epidemiologic analysis can be performed a simple and reliable method of genomovar identification is required. Several groups have investigated PCR-based techniques such as SS-PCR and restriction fragment length polymorphism analysis as a tool to identify Burkholderia species and to differentiate the B. cepacia complex genomovars. (1, 2, 4, 5, 9, 11, 14–16, 19, 23, 24). Reliable SS-PCR methods to identify B. multivorans and B. vietnamiensis have been described (14), and recent work indicates that recA-based PCR can identify B. stabilis (20); however, PCR assays to identify genomovars I and III have not been reported. Using terminal regions of the 16S and 23S rRNA genes and the intervening spacer region, we have detected a region of heterogeneity between the genomovar I and III and B. stabilis isolates. This region, which is located near the 5′ end of the 16S rRNA, has three variable consecutive nucleotides that were used to design two oligonucleotide primers. These primers, denoted SPR3 and SPR4, target the variable regions in the prototypical genomovars I and III and B. stabilis, respectively. Since it has been previously demonstrated that the primer pair G1-G2 is specific for genomovars I and III and B. stabilis (14, 23), the G1 primer was used as an anchor in both PCRs. Primer pair SPR3-G1 was specific for genomovars III and I, while primer pair SPR4-G1 amplified a DNA fragment from B. stabilis. The primer pairs were tested against a larger panel of well-characterized B. cepacia complex isolates for which the genomovar had been previously established. The primers accurately identified all the genomovar III and B. stabilis isolates; however, genomovar I isolates showed variable amplification. Some genomovar I isolates amplified with SPR3-G1 while the others amplified with SPR4-G1. In a recent study the majority of genomovar I and B. stabilis strains were found to be closely related, forming a single cluster by whole-cell protein pattern, with a similarity level of 77% (20). All genomovar I isolates gave a product, and no isolate amplified with both sets of primers. The variability of the genomovar I isolates precludes definitive identification of this genomovar by rRNA-directed PCR. However the separation of B. stabilis and certain genomovar I isolates is important since preliminary studies have indicated that the majority of isolates associated with cepacia syndrome belong to genomovar III (14). Thus, amplification of a band from an unknown isolate with SPR4-G1 would indicate that this is not a genomovar III isolate.
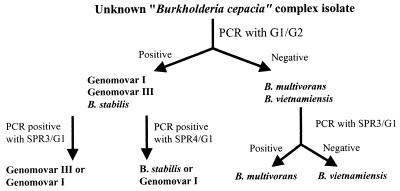
To separate the five genomovars using SS-PCR, an initial PCR with G1-G2 was performed to identify genomovars I and III and B. stabilis. To determine if SPR3-G1 and SPR4-G1 were able to identify genomovar III and B. stabilis directly, we examined a panel of B. multivorans and genomovar V isolates with these primers. All 13 of the genomovar II isolates amplified with SPR3-G1; none amplified with SPR4-G1. None of the 15 genomovar V isolates amplified with either primer pair SPR3-G1 or SPR4-G1. DNA from the latter set of isolates was amplified with the universal primers PSL-PSR, which were used as a control. Thus, the primer pair SPR3-G1 is also capable of identifying B. multivorans and, by a negative result, genomovar V. Figure 1 shows results of the three PCRs with genomic DNA template from the individual prototypic genomovar strains. From this it can be seen that use of the primers G1-G2, SPR3-G1, and SPR4-G1 will allow separation of all the genomovars except genomovar I. Since the primer pairs target the 16S-23S rRNA spacer region, the PCR results in a double band, representing the heterogeneity of this region. Figure 2 details the algorithm of PCRs required to separate the members of the B. cepacia complex. It is interesting that of all the genomovars only isolates currently classified as belonging to genomovar I show heterogeneity of the rRNA gene at this site. Further taxonomic study is required to determine if this represents a distinct division within this genomovar. In summary, we have developed species-specific primers capable of separating genomovar III and B. stabilis isolates, and in conjunction with the PCR primers described by LiPuma et al. (14), these primers can confirm the identify of B. multivorans and B. vietnamiensis isolates. Use of these PCR primers will allow more detailed study of the epidemiology of the B. cepacia complex.
FIG. 2.
PCR algorithm to identify the species and genomovars of the B. cepacia complex.
ACKNOWLEDGMENTS
This work was funded by Cystic Fibrosis Foundation grant STULL97AO awarded to T.L.S. P.W.W. and T.L.S. acknowledge the financial support of the Children's Medical Research Institute.
We thank Jennifer McMenamin, Lauren Pope, and Basharat Muneer for technical help and data analysis.
REFERENCES
- 1.Bauernfeind A, Schneider I, Jungwirth R, Roller C. Discrimination of Burkholderia gladioli from other Burkholderia species detectable in cystic fibrosis patients by PCR. J Clin Microbiol. 1998;36:2748–2751. doi: 10.1128/jcm.36.9.2748-2751.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauernfeind A, Schneider I, Jungwirth R, Roller C. Discrimination of Burkholderia multivorans and Burkholderia vietnamiensis from Burkholderia cepacia genomovars I, III, and IV by PCR. J Clin Microbiol. 1999;37:1335–1339. doi: 10.1128/jcm.37.5.1335-1339.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burkholder W H. Sour skin, a bacterial rot of onion bulbs. Phytopathology. 1950;14:127–130. [Google Scholar]
- 4.Campbell P W I, Phillips J A, III, Heidecker G J, Krishnamani M R S, Zahorchak R, Stull T L. Detection of Pseudomonas (Burkholderia) cepacia using PCR. Pediatr Pulmonol. 1995;20:44–49. doi: 10.1002/ppul.1950200109. [DOI] [PubMed] [Google Scholar]
- 5.Clode F E, Kaufmann M E, Malnick H, Pitt T L. Evaluation of three oligonucleotide primer sets in PCR for the identification of Burkholderia cepacia and their differentiation from Burkholderia gladioli. J Clin Pathol. 1999;52:173–176. doi: 10.1136/jcp.52.3.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillis M, Van T V, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kersters K, Heulin T, Fernandez M P. Polyphasic taxonomy in the genus Burkholderia leading to an amended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Bacteriol. 1995;45:274–289. [Google Scholar]
- 8.Isles A, Maclusky I, Corey M, Gold R, Prober C, Fleming P, Levison H. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J Pediatr. 1984;104:206–210. doi: 10.1016/s0022-3476(84)80993-2. [DOI] [PubMed] [Google Scholar]
- 9.Karpati F, Jonasson J. Polymerase chain reaction for the detection of Pseudomonas aeruginosa, Stenotrophomonas maltophilia and Burkholderia cepacia in sputum of patients with cystic fibrosis. Mol Cell Probes. 1996;10:397–403. doi: 10.1006/mcpr.1996.0055. [DOI] [PubMed] [Google Scholar]
- 10.Lacy D E, Spencer D A, Goldstein A, Weller P H, Darbyshire P. Chronic granulomatous disease presenting in childhood with Pseudomonas cepacia septicaemia. J Infect. 1993;27:301–304. doi: 10.1016/0163-4453(93)92271-w. [DOI] [PubMed] [Google Scholar]
- 11.LiPuma J J. Molecular tools for epidemiologic study of infectious diseases. Pediatr Infect Dis J. 1998;17:667–675. doi: 10.1097/00006454-199808000-00002. [DOI] [PubMed] [Google Scholar]
- 12.LiPuma J J. Burkholderia cepacia: continuing the genomovar story. Pediatr Pulmonol. 1999;28(Suppl. 19):148. [Google Scholar]
- 13.LiPuma J J, Dasen S E, Nielson D W, Stern R C, Stull T L. Person-to-person transmission of Pseudomonas cepacia between patients with cystic fibrosis. Lancet. 1990;336:1094–1096. doi: 10.1016/0140-6736(90)92571-x. [DOI] [PubMed] [Google Scholar]
- 14.LiPuma J J, Dulaney B J, McMenamin J D, Whitby P W, Stull T L, Coenye T, Vandamme P. Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J Clin Microbiol. 1999;37:3167–3170. doi: 10.1128/jcm.37.10.3167-3170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Callaghan E M, Tanner M S, Boulnois G J. Development of a PCR probe test for identifying Pseudomonas aeruginosa and Pseudomonas (Burkholderia) cepacia. J Clin Pathol. 1994;47:222–226. doi: 10.1136/jcp.47.3.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segonds C, Heulin T, Marty N, Chabanon G. Differentiation of Burkholderia species by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene and application to cystic fibrosis isolates. J Clin Microbiol. 1999;37:2201–2208. doi: 10.1128/jcm.37.7.2201-2208.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tablan O C, Carson L A, Cusick L B, Bland L A, Martone W J, Jarvis W R. Laboratory proficiency test results on use of selective media for isolating Pseudomonas cepacia from simulated sputum specimens of patients with cystic fibrosis. J Clin Microbiol. 1987;25:485–487. doi: 10.1128/jcm.25.3.485-487.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor R F, Gaya H, Hodson M E. Pseudomonas cepacia: pulmonary infection in patients with cystic fibrosis. Respir Med. 1993;87:187–192. doi: 10.1016/0954-6111(93)90090-m. [DOI] [PubMed] [Google Scholar]
- 19.Tyler S D, Strathdee C A, Rozee K R, Johnson W M. Oligonucleotide primers designed to differentiate pathogenic pseudomonads on the basis of the sequencing of genes coding for 16S-23S rRNA internal transcribed spacers. Clin Diagn Lab Immunol. 1995;2:448–453. doi: 10.1128/cdli.2.4.448-453.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vandamme P, Mahenthiralingam E, Holmes B, Coenye T, Hoste B, De Vos P, Henry D, Speert D P. Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV) J Clin Microbiol. 2000;38:1042–1047. doi: 10.1128/jcm.38.3.1042-1047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R, Revets H, Lauwers S, Gillis M, Kersters K, Govan J R. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 22.van Pelt C, Verduin C M, Goessens W H F, Vos M C, Tümmler B, Segonds C, Reubsaet F, Verbrugh H, van Belkum A. Identification of Burkholderia spp. in the clinical microbiology laboratory: comparison of conventional and molecular methods. J Clin Microbiol. 1999;37:2158–2164. doi: 10.1128/jcm.37.7.2158-2164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitby P W, Dick H L, Campbell P W, Tullis D E, Matlow A, Stull T L. Comparison of culture and PCR for detection of Burkholderia cepacia in sputum samples of patients with cystic fibrosis. J Clin Microbiol. 1998;36:1642–1645. doi: 10.1128/jcm.36.6.1642-1645.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitby P W, Pope L C, Carter K B, LiPuma J, Stull T L. Species-specific PCR as a tool for the identification of Burkholderia gladioli. J Clin Microbiol. 2000;38:282–285. doi: 10.1128/jcm.38.1.282-285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]