Abstract
Fluro(quinolones) is an important class of antibiotic used widely in both human and veterinary medicine. Resistance to fluro(quinolones) can be acquired by either chromosomal point mutations or plasmid-mediated quinolone resistance (PMQR). There is a lack of studies on the prevalence of PMQR in organisms from environmental sources in Bangladesh. In this study, we investigated the occurrence of PMQR genes in E. coli from various water sources and analysed associations between multi-drug resistance (MDR) and resistance to extended spectrum β-lactam antibiotics. We analysed 300 E. coli isolates from wastewaters of urban live-bird markets (n = 74) and rural households (n = 80), rural ponds (n = 71) and river water samples (n = 75) during 2017–2018. We isolated E. coli by filtering 100 ml of water samples through a 0.2μm cellulose membrane and incubating on mTEC agar media followed by identification of isolated colonies using biochemical tests. We selected one isolate per sample for detection of PMQR genes by multiplex PCR and tested for antibiotic susceptibility by disc diffusion. Clonal relatedness of PMQR-positive isolates was evaluated by enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR). About 66% (n = 199) of E. coli isolates harbored PMQR-genes, predominantly qnrS (82%, n = 164) followed by aac(6’)-lb-cr (9%, n = 17), oqxAB (7%, n = 13), qnrB (6%, n = 11) and qepA (4%, n = 8). Around 68% (n = 135) of PMQR-positive isolates were MDR and 92% (n = 183) were extended spectrum β-lactamase (ESBL)-producing of which the proportion of positive samples was 87% (n = 159) for blaCTX-M-1’ 34% (n = 62) for blaTEM, 9% (n = 16) for blaOXA-1, blaOXA-47 and blaCMY-2, and 2% (n = 4) for blaSHV. Further, 16% (n = 32) of PMQR-positive isolates were resistant to carbapenems of which 20 isolates carried blaNDM-1. Class 1 integron (int1) was found in 36% (n = 72) of PMQR-positive E. coli isolates. PMQR genes were significantly associated with ESBL phenotypes (p≤0.001). The presence of several PMQR genes were positively associated with ESBL and carbapenemase encoding genes such as qnrS with blaCTXM-1 (p<0.001), qnrB with blaTEM (p<0.001) and blaOXA-1 (p = 0.005), oqxAB and aac(6’)-lb-cr with blaSHV and blaOXA-1 (p<0.001), qnrB with blaNDM-1 (p<0.001), aac(6’)-lb-cr with blaOXA-47 (p<0.001) and blaNDM-1 (p = 0.002). Further, int1 was found to correlate with qnrB (p<0.001) and qepA (p = 0.011). ERIC-PCR profiles allowed identification of 84 of 199 isolates with 85% matching profiles which were further grouped into 33 clusters. Only 5 clusters had isolates (n = 11) with identical ERIC-PCR profiles suggesting that PMQR-positive E. coli isolates are genetically heterogeneous. Overall, PMQR-positive MDR E. coli were widely distributed in aquatic environments of Bangladesh indicating poor wastewater treatment and highlighting the risk of transmission to humans and animals.
Introduction
With 700,000 global deaths annually, bacterial infections caused by antimicrobial-resistant organisms are a major public health concern [1]. Antimicrobial-resistant (AMR) infections increase mortality, treatment duration, recovery time, and health care costs. AMR is a One Health problem and addressing the issues related to human and animal health separately or in combination is not enough if the environmental dimensions of the problem are not addressed. The emergence of multidrug-resistant (MDR) organisms in aquatic environments constitutes a major threat for both humans and livestock. Although E. coli is part of the normal flora in humans and animals, pathogenic strains of E. coli cause severe clinical challenges including gastrointestinal tract infection, central nervous system infection, urinary tract and skin and soft tissue infections [2, 3]. These infections become more severe when caused by MDR pathogens [4]. Several recent investigations reported the emergence of MDR bacteria from different host origins including humans, birds, cattle, and fish that increase the need for antimicrobial susceptibility testing to identify the antibiotic of choice as well as screening for emerging MDR strains [5–11].
Quinolones and fluoroquinolones (FQs) are broad-spectrum antibiotics frequently used in human and veterinary health for treatment of both Gram-positive and Gram-negative bacterial infections [12]. Fluro(quinolones) are the third most commonly prescribed antibiotics in Bangladesh for treating outpatients suffering from common cold and fever, infections, diarrhea, and gonorrhea [13, 14]. In livestock production in Bangladesh, flouroquinolone is one of the most commonly used antibiotics, with an estimated consumption of 100 metric ton per year [15]. Ciprofloxacin is widely used as a single drug or in combination with other drugs and is often sold as feed supplements under many different brand names [15]. Farmers use this antibiotic mostly for prophylactic purposes to avert infections or as an alternative to good agricultural practices and as a growth promoter on farm animals [16, 17]. With increasing use of fluoroquinolones, the prevalence of fluoroquinolone resistance has also been increasing. Although quinolone resistance in Enterobacteriaceae is mainly attributed to point mutations in quinolone resistance-determining regions (QRDRs) of the type II topoisomerase genes (gyrase: gyrA, gyrB; and topoisomerase IV: parC, parE), there are an increasing number of reports of plasmid-mediated quinolone resistance (PMQR) determinants associated with low-level resistance to fluoroquinolones [18–21]. Moreover, bacterial pathogens that are positive for PMQR are also more likely to have chromosomal mutations resulting in high level of resistance to the antibiotics [22, 23]. Co-occurrence of PMQR with extended spectrum β-lactamase (ESBL) genes may limit the treatment options for infections caused by ESBL-producing bacteria [24].
Three categories of PMQR genes have been reported based on their mode of action. Such examples include the qnr alleles (qnrA, qnrB, qnrS, qnrC, and qnrD); efflux pump genes (e.g. oqxAB, qepA); and a variant of aminoglycoside acetyl transferase (aac-(6′)-Ib-cr) [25–28]. qnrA, qnrB genes can be carried by large and usually conjugative plasmids, whereas qnrS can be carried by small, mobilizable, and non-conjugative plasmids [18, 29]. However, both types of plasmid can readily disseminate and transmit antibiotic resistance traits among bacterial communities. Another important mechanism of PMQR gene transmission among the bacterial population is via the integrons (int) particularly int1 in Gram-negative bacteria [30, 31]. Close proximity between antibiotic resistance genes and int1 thus enhances mobility by transposition and allows them to become associated with multiple antibiotic-resistant gene (ARG) cassettes and heavy metal and disinfectant resistant genes [18, 32].
The prevalence of PMQR genes has been investigated in different countries across the world. Previous studies in humans, food-producing animals, wild animals, and wastewater samples showed an overall prevalence of PMQR in E. coli of 25% with the highest reported occurrence (49%) in retail turkey from Czech Republic [33]. In China aquatic environmental samples had a 30% prevalence of PMQR-positive E. coli isolates overall; with a prevalence of 28% in hospital-impacted water samples and 37% in aquaculture-impacted river water samples [34]. Limited information is available on the prevalence of PMQR in Bangladesh. PMQR genes were detected in clinical isolates of E. coli and K. pneumoniae largely from wound and urinary tract infections, and in E. coli from cloacal swabs of poultry [35, 36]. Recently, a novel quinolone resistance gene qepA has been detected in E. coli and K. pneumoniae strains isolated from lake and river water samples in Bangladesh [37]. Although previous studies have shown that aquatic environments in Bangladesh particularly drinking water, wastewater, and surface water bodies such as ponds and rivers are heavily contaminated with various faecal pathogens including multi-drug resistant organisms, no studies have estimated the prevalence of PMQR among isolates [38, 39]. In this study, we aimed to investigate water samples from different aquatic environments including wastewater and surface water from both rural and urban areas of Bangladesh to understand the prevalence and distribution of E. coli carrying PMQR along with their resistance patterns against clinically important antibiotics. A further aim was to investigate the association of PMQR genes with ESBL- and carbapenemase-producing genes in environmental E. coli isolates.
Methods and materials
Ethical approval
This research protocol was approved by the Institutional Review Board of the International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b) (protocol number PR-16071).
Study overview
The present study is part of a larger study that simultaneously examined the dynamics of AMR transmission from contaminated outdoor environments such as poultry, soil, surface water, solid waste, and wastewater from urban and rural Bangladesh [40]. Here, we investigated the E. coli isolates collected from wastewater of rural poultry farms and households, and urban live-bird markets, as well as pond water and river water samples from rural areas to determine the presence of PMQR genes and analyse their distribution according to sample types.
Sample collection
A total of 300 water samples including urban wastewater (n = 74), rural wastewater (n = 80) from poultry farms and households, river (n = 75) and pond (n = 71) water samples were collected during 2017–18 following previously described procedures [40]. Briefly, water samples were collected using a sterile plastic bottle filled by plunging downwards about 30 cm below the water surface. Sample bottles were placed in a cool box (4–8°C) and transported to the laboratory within 8 hours of collection for culture.
Isolation and identification of E. coli
About 100 mL of water sample was passed through a 0.2 μm cellulose nitrate filter (Sartorius Stedim Biotech GmbH, Goettingen, Germany) and then the filter was placed in an upright position on modified mTEC agar media (BD Difco, New Jersey, USA). The culture plate was incubated at 37°C for 2 hours and then at 44°C overnight to allow growth of thermotolerant E. coli. mTEC medium contains a chromogen (5-bromo-6-chloro-3-indolyl-β-D-glucuronide), which is catabolized by E. coli producing β-D-glucuronidase to glucuronic acid and produces a red- or magenta-coloured compound. Two E. coli isolates were selected from each water sample and sub-cultured on MacConkey agar (BD Difco, New Jersey, USA) and incubated at 37°C for overnight. The presumptive colonies were identified according to their colony characters, microscopical examination using Gram staining, motility test, and biochemical reactions (oxidase, catalase, indole, lactose fermentation, methyl-red, citrate-utilization, H2S, Voges-Proskauer, and urease tests) as described previously [41]. All E. coli isolates were stored in Tryptone soya broth (Oxoid Limited, Hampshire, England) with 30% glycerol (Sigma-Aldrich, Darmstadt, Germany) and kept at -80°C for future use.
Screening of PMQR-positive isolates by PCR
All E. coli isolates from water samples were investigated for plasmid-mediated quinolone resistance genes (qnrS, qnrB, oqxAB, qepA, aac(6’)-lb-cr, qnrA, qnrC & qnrD) by multiplex-PCR as described previously [42]. The primers and PCR cycling are listed in Table 1.
Table 1. List of primer sequences used in multiplex PCR for the determination of PMQR determinants.
| Target gene | Primer sequences | Amplicon size (bp) | Amplification (30 cycles) | References | ||
|---|---|---|---|---|---|---|
| Denaturation | Annealing | Extension | ||||
| qnrA F | CAGCAAGAGGATTTCTCACG | 630 | 94°C for 30 seconds | 63°C for 90 seconds | 72°C for 90 seconds | Ciesielczuk et al., 2013 [42] |
| qnrA R | AATCCGGCAGCACTATTACTC | |||||
| qnrD F | CGAGATCAATTTACGGGGAATA | 581 | ||||
| qnrD R | AACAAGCTGAAGCGCCTG | |||||
| qnrB F | GGCTGTCAGTTCTATGATCG | 488 | ||||
| qnrB R | GAGCAACGATGCCTGGTAG | |||||
| qnrS F | GCAAGTTCATTGAACAGGGT | 428 | ||||
| qnrS R | TCTAAACCGTCGAGTTCGGCG | |||||
| oqxAB F | CCGCACCGATAAATTAGTCC | 313 | ||||
| oqxAB R | GGCGAGGTTTTGATAGTGGA | |||||
| aac(6’)-lb-cr F | TTGGAAGCGGGGACGGAM | 260 | ||||
| aac(6’)-lb-cr R | ACACGGCTGGACCATA | |||||
| qepA F | GCAGGTCCAGCAGCGGGTAG | 218 | ||||
| qepA R | CTTCCTGCCCGAGTATCGTG | |||||
| qnrC F | GCAGAATTCAGGGGTGTGAT | 118 | ||||
| qnrC R | AACTGCTCCAAAAGCTGCTC | |||||
Confirmation of amplified fragments of PMQR genes by sequencing
The PCR amplified fragment of each PMQR gene found in this study was sequenced using ABI PRISM BigDye Terminator Cycle Sequencing Reaction kit (Applied Biosystems; CA, USA) and ABI PRISM 310 automated sequencer (Applied Biosystems; CA, USA). Before that, PCR products were purified using the PCR Purification kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. BioEdit software was used to analyse the raw sequence reads and for determining the homology with deduced sequence size. All the PMQR gene sequences were searched and confirmed by Basic Local Alignment Search Tool (BLAST). Finally, the sequences were submitted to GeneBank under five accession numbers: qnrS (OL439745), qnrB (OL439744), oqxAB (OK668389), qepA (OK668390) and aac(6’)-lb-cr (OL439743).
Antibiotic susceptibility testing of PMQR-positive E. coli
All PMQR-positive E. coli isolates were tested for antibiotic susceptibility against 16 clinically important antibiotic agents under nine antibiotic classes by disc diffusion method [43]. Commercially available antibiotic discs (Oxoid Limited, Hampshire, England) used for the test were: ampicillin (10 μg), cefotaxime (30 μg), ceftriaxone (30 μg), ceftazidime (30 μg), cefixime (5 μg), cefepime (30 μg),cefoxitin (30 μg), ciprofloxacin (5 μg), nalidixic acid (30 μg), sulfamethoxazole/trimethoprim (25 μg), gentamycin (10 μg), nitrofurantoin(300 μg), imipenem (10 μg), meropenem (10 μg), ertapenem (10 μg) and piperacillin-Tazobactam (10 μg). Results were interpreted as sensitive and resistant, and isolates showing resistance against at least one agent of more than three classes of antibiotics were classified as MDR. Where there was resistance against at least one agent in all classes except two or fewer antibiotic classes were considered as extensively drug-resistant (XDR) [43, 44]. Extended spectrum β-lactamase (ESBL) production was determined by using the combination disk test where a β-lactam inhibitor, clavulanic acid (30/10 μg) with cefotaxime (30 μg) and ceftazidime (30 μg) were used [43].
Detection of ESBL, carbapenemase and integrase encoding genes
All ESBL-producing isolates were screened for blaCTX-M-1, blaCMY-2, blaTEM, blaSHV, blaOXA-1, and blaOXA-47 genes by PCR [45]. All carbapenem resistant isolates were tested for carbapenem resistance genes, blaNDM-1 and blaOXA-48 according to the procedure described earlier [46]. Class 1 integrons were detected by PCR for int1 gene using the primer sequences and PCR conditions as described earlier [47].
Statistical analysis
Data were entered and analysed using Stata (Version 13.0, StataCorp LLC, College Station, TX, USA). Univariate analyses were performed to examine the presence of PMQR genes in different aquatic sources including wastewater, pond water and river water. In bivariate analyses, the prevalence of PMQR genes in different categories of antibiotic resistant E. coli isolates such as MDR, ESBL- and carbapenemase-producers was compared using Chi-square or Fisher’s exact test with Bonferroni correction [48]. Correlations for binary variables were calculated using the ‘cor’ function and using ‘cor.test’ function in R software (version 4.0.2; https://www.r-project.org/). Significant correlations were visualized utilizing the ‘corrplot’ function from the ‘corrplot’ R package [49]. For all analyses, statistical significance was considered as p<0.05.
Phylogenetic analysis using ERIC-PCR
PMQR-positive E. coli isolates were analysed for clonal diversity using enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR). Two primers, ERIC1 (5ʹ-ATGTAAGCTCCTGGGGATTCAC-3ʹ) and ERIC2 (5ʹ-AAGTAAGTGACTGGGGTGAGCG-3ʹ) were used following previously reported procedures [50]. ERIC-PCR amplified products were then separated in 1.5% agarose gel, normalised using the 100 bp DNA ladder as an external reference standard, stained with Midori Green, and visualized by FastGene Blue/Green LED Gel Illuminator (Nippon Genetics, Tokyo, Japan). The TIF formatted image was analyseed with BioNumerics version 4.5 (Applied Maths, Kortrijk, Belgium) to determine phylogenetic similarity among the isolates. Dendrogram clusters based on dice and clustering correlation coefficient showing 85% similarity in banding patterns between E. coli isolates were considered as phylogenetically related [51].
Results
Phenotypic characteristics of E. coli isolated from water samples
Isolates were identified as E. coli based on their morphology and biochemical characteristics. Microscopically, the bacteria appeared as Gram-negative moderate size, motile, and non-sporulated rods. The bacteria grew well on mTEC agar and appeared as red/magenta colour colonies due to β-D-glucuronidase activity which is highly specific for E. coli. On MacConkey agar, bacteria produced characteristic pink colonies due to lactose fermentation. Biochemically, all isolates were positive for catalase, lactose fermentation, indole and methyl-red, tests. Simultaneously, they were negative for cytochrome oxidase, Voges-Proskauer, citrate-utilization, H2S production, and urease tests. All 300 water samples were positive for E. coli.
PMQR genes are highly prevalent in E. coli from water sources
Of 300 E. coli isolates from water samples, 66% (n = 199) were positive for PMQR genes (Table 2). Of these, the majority of isolates were positive for qnrS (82%, n = 164) followed by aac(6’)-lb-cr (9% n = 17), oqxAB (7%, n = 13) qnrB (6%, n = 11) and qepA (4%, n = 8). None of the isolates was positive for qnrA, qnrC & qnrD. qnrS was predominantly detected in rural pond water (90%, n = 45) whereas aac(6’)-lb-cr was found in river water (24%, n = 11). Ten E. coli isolates carried more than one PMQR gene in the following combinations: qnrS+oqxAB (n = 2), qnrS+qnrB+oqxAB (n = 2), qnrB+oqxAB+aac(6)’-lb-cr (n = 1), qnrS+qnrB+aac(6)’-lb-cr (n = 1), qnrS+qepA+oqxAB (n = 1), qnrS+qnrB (n = 1), qnrS+oqxAB (n = 1) and qnrB+oqxAB (n = 1).
Table 2. Prevalence of PMQR genes in E. coli isolates obtained from different aquatic environments.
| Sampling site | Sample Type | Sample number (n) | PMQR positive n (%) | No. (%) of isolates positive for PMQR genes | ||||
|---|---|---|---|---|---|---|---|---|
| qnrS n (%) | qnrB n (%) | oqxAB n (%) | qepA n (%) | aac(6’)-lb-cr n (%) | ||||
| Urban | Wastewater | 74 | 45 (61) | 52 (65) | 3 (4) | 4 (5) | 0 | 2 (3) |
| Rural | Wastewater | 80 | 58 (72) | 37 (50) | 4 (5) | 3 (4) | 3 (4) | 4 (5) |
| pond water | 71 | 50 (70) | 45 (90) | 2 (4) | 3 (6) | 2 (4) | 0 | |
| river water | 75 | 46 (61) | 30 (65) | 2 (4) | 3 (7) | 3 (7) | 11 (24) | |
| Total | 300 | 199 (66) | 164 (82) | 11 (6) | 13 (7) | 8 (4) | 17 (9) | |
*R, resistance; n, number.
PMQR-positive E. coli are predominantly multi-drug resistant
All PMQR-positive isolates (n = 199) were resistant to penicillin followed by 96% resistant to cephamycins and extended spectrum cephalosporins, 48% to fluro(quinolones), 32% to folate pathway inhibitors, 26% to nitrofuran, 23% to β-lactamase inhibitors, 21% to aminoglycoside and 16% resistant to carbapenem (Table 3). Further, the distribution of antibiotic resistance patterns of PMQR positive isolates was analysed according to their sources. Of 199 PMQR positive E. coli isolates, 68% (n = 135) and 14% (n = 27) were MDR and XDR respectively, of which urban wastewater and rural river water showed high abundance. Aminoglycosides, fluoroquinolones, nitrofuran and folate pathway inhibitor drug resistant isolates had significant relationship with their sources (p<0.05) (Table 3).
Table 3. Occurrence of clinically important antibiotic resistance among PMQR-positive E. coli isolates from aquatic environments in Bangladesh.
| Antibiotic classes | Antibiotics tested | No. (%) of E. coli resistant | p value | |||
|---|---|---|---|---|---|---|
| Urban Wastewater (n = 45) | Rural Wastewater (n = 58) | Rural pond water (n = 50) | Rural river water (n = 46) | |||
| Aminoglycoside | Gentamycin | 18 (40) | 7 (12) | 2 (4) | 15 (33) | p<0.05 |
| Antipseudomonal penicillins plus β-lactamase inhibitors | Piperacillin-Tazobactam | 10 (22) | 8 (14) | 6 (12) | 21 (46) | p<0.001 |
| Cephamycins | Cefoxitin | 44 (98) | 56 (97) | 47 (94) | 44 (96) | 0.654 |
| Extended-spectrum cephalosporins | Cefotaxime | 44 (98) | 56 (97) | 45 (90) | 44 (96) | 0.165 |
| Ceftriaxone | 44 (98) | 56 (97) | 45 (90) | 44 (96) | 0.165 | |
| Ceftazidime | 39 (87) | 47 (81) | 38 (76) | 43 (98) | 0.065 | |
| Cefixime | 43 (96) | 56 (97) | 47 (94) | 44 (96) | 0.838 | |
| Cefepime | 44 (98) | 55 (95) | 45 (90) | 44 (96) | 0.277 | |
| Carbapenem | Ertapenem | 9 (20) | 5 (9) | 6 (12) | 10 (22) | 0.343 |
| Meropenem | 10 (22) | 6 (10) | 7 (14) | 9 (19) | 0.363 | |
| Imipenem | 11 (24) | 8 (14) | 6 (12) | 7 (15) | 0.209 | |
| Fluro(quinolone) | Nalidixic acid | 34 (76) | 23 (40) | 19 (38) | 27 (59) | 0.074 |
| Ciprofloxacin | 34 (76) | 20 (34) | 14 (28) | 26 (57) | p<0.05 | |
| Folate pathway inhibitors | Sulfamethoxazole/trimethoprim | 27 (60) | 15 (26) | 11 (22) | 11 (24) | p<0.05 |
| Nitrofuran | Nitrofurantoin | 9 (20) | 8 (14) | 15 (30) | 20 (43) | p<0.05 |
| Penicillin | Ampicillin | 45 (100) | 58 (100) | 50 (100) | 46 (100) | NA |
| MDR (≥3 Ab classes) | 39 (87) | 32 (55) | 29 (58) | 35 (76) | 0.157 | |
| XDR (All antibiotic classes except two or fewer classes) | 8 (18) | 4 (7) | 2 (4) | 13 (28) | p<0.05 | |
A significant proportion of PMQR-positive E. coli were positive for ESBL genes
About 92% (n = 183) of PMQR-positive E. coli were ESBL-producers. Screening of ESBL encoding genes showed that 87% (n = 159) of isolates were positive for blaCTX-M-1, 34% (n = 62) for blaTEM, 9% (n = 17) for blaOXA-1, blaOXA-47 and blaCMY-2 each, and 2% (n = 4) for blaSHV. Among carbapenem resistance, only blaNDM-1 was detected in 10% (n = 20) of the PMQR-positive isolates. None of the isolates were positive for blaOXA-48. Further, the class 1 integron encoding gene, int1, was detected in 37% (n = 73) of the isolates.
Presence of PMQR genes was significantly associated with the ESBL phenotype (p<0.001) of the isolates. At the gene level, blaCTX-M-1 (p<0.001) and blaTEM (p<0.05) were significantly more common in PMQR-positive versus negative isolates (Table 4). A correlation matrix analysis between presence of PMQR genes and presence of ESBL, carbapenemase and class 1 integron encoding genes was conducted (Fig 1). The presence of qnrS was positively associated with blaCTXM-1 (p<0.001) while qnrB was positively associated with blaTEM (p<0.001), blaOXA-1 (p = 0.005) and blaNDM-1 (p<0.001). Detection of PMQR genes oqxAB and aac(6’)-lb-cr in isolates was positively associated with ESBL genes blaSHV and blaOXA-1 (p<0.001 for both). Isolates positive for the PMQR efflux gene, qepA, were predominantly positive for AmpC β-lactamase gene blaCMY (p = 0.016). Isolates carrying aac(6’)-lb-cr gene were more likely to be positive for carbapenem resistance genes blaOXA-47 (p<0.001) and blaNDM-1 (p = 0.002). Unlike antibiotic resistance genes, the presence of Class 1 integron gene Int1 was associated with a diverse group of PMQR genes including qnrB (p<0.001) and qepA (p = 0.011).
Table 4. Association of PMQR genes with ESBL, carbapenemase, integrase genes, and MDR and XDR phenotypes in E. coli isolates.
| Characteristics | No. (%) of E. coli | p value* | |
|---|---|---|---|
| PMQR positive (n = 199) | PMQR negative (n = 101) | ||
| ESBL | 183 (92) | 74 (73) | p<0.001 |
| blaCTXM-1 | 165 (83) | 51 (50) | p<0.001 |
| blaSHV | 5 (3) | 1 (1) | 0.668 |
| blaTEM | 63 (32) | 46 (46) | p<0.05 |
| blaOXA-1 | 17 (9) | 12 (12) | 0.355 |
| blaOXA-47 | 16 (8) | 13 (13) | 0.181 |
| Carbapenemase | 32 (16) | 17 (17) | 0.70 |
| blaNDM-1 | 20 (10) | 16 (16) | 0.145 |
| blaCMY-2 | 16 (8) | 8 (8) | 0.971 |
| blaOXA-48 | 0 | 0 | NA |
| Class 1 integron (int1) | 72 (36) | 40 (40) | 0.562 |
| MDR | 135(68) | 74 (73) | 0.334 |
| XDR | 27 (14) | 20 (20) | 0.160 |
*p values were determined using Chi-square or Fisher’s exact test.
Fig 1. Correlation matrix of the presence of PMQR genes with ESBL and carbapenemase and integron (int1) encoding genes in E. coli.
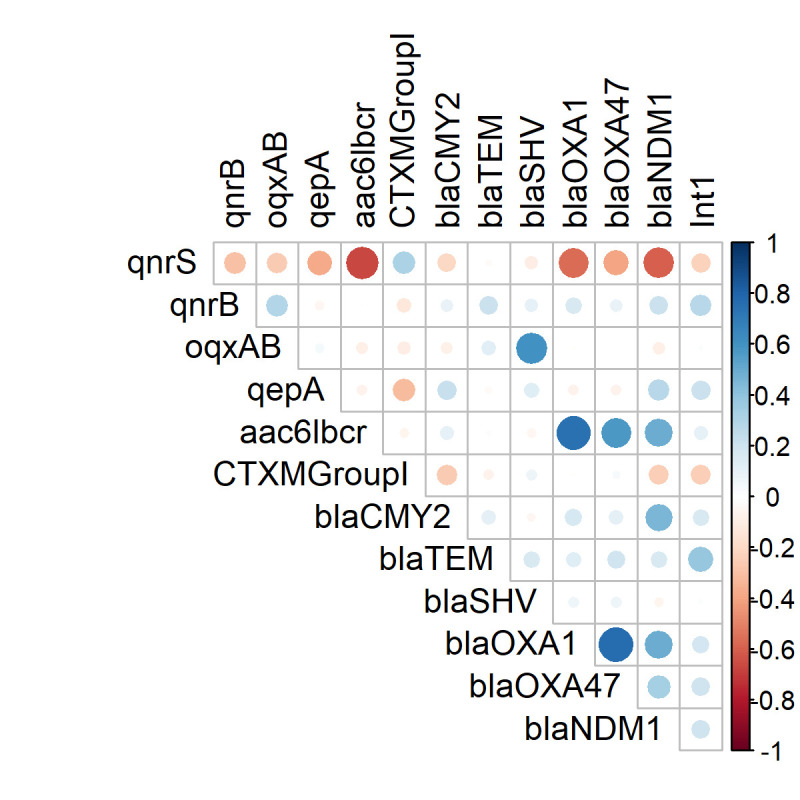
White spaces are not significantly correlated. Blue circles indicated significant positive correlation and red showed significant negative correlation. The size and strength of colour represent the numerical value of the Phi correlation coefficient.
ERIC-PCR analysis
According to the 85% cut-off for similarity in ERIC-PCR banding patterns, 84 of 199 (42%) PMQR-positive E. coli isolates were grouped into 33 clusters (C1-C33) and the isolates in different clusters were randomly distributed irrespective of the sources of isolation (Fig 2). Among these 84 E. coli isolates, only 11 in five clusters, C3, C8, C19, C31 and C33, had identical banding patterns indicating clonal relationships. E. coli isolates from wastewater samples from different locations in urban areas belonged to the same clusters (C8 and C33) whereas isolates from pond and river water samples in rural areas belonged to the same clusters (C3). Further, isolates from pond and river water were grouped in clusters C27 and C46, respectively with isolates from wastewater samples in rural areas.
Fig 2. Dendrogram generated by BioNumerics software, showing distances calculated by the Dice similarity index of ERIC-PCR banding patterns of E. coli strains isolated from various aquatic samples.
The degree of similarity (%) is shown on the scale. The isolates were considered as phylogenetically related based on 85% similarity in ERIC-PCR banding patterns.
Discussion
In Bangladesh, fluro(quinolones) is one of the most frequently used antibiotic classes in both human and veterinary medicine although the major usage is in animal husbandry where it is applied as a feed supplement for prophylaxis and growth promotion [13–16]. A recent report indicates that few farmers use ciprofloxacin for bacterial disease prevention in aquaculture in Bangladesh [52]. Aquatic environments are more likely, therefore, to be contaminated with residual fluro(quinolones) via effluents from both human and animal wastes which may be contributing to the high prevalence of fluoroquinolone resistance in bacterial organisms. In particular, PMQR genes along with ESBL and carbapenemase encoding genes are likely to be carried by the same plasmids that can be shared with other organisms in aquatic environments by horizontal transmission. Except for one investigation of 12 E. coli isolates from water samples reporting 4 positives for qnrS, no other study has characterised environmental isolates for PMQR in Bangladesh [37]. In this study, we found that a significant proportion (66%) of water samples were positive for E. coli carrying PMQR genes, predominantly qnrS. This percentage would have been greater if more than one isolate per sample had been selected. Interestingly, previous studies reported a higher prevalence of qnrS in E. coli isolates from patients with extraintestinal infections, and from poultry cloacal samples in Bangladesh [35, 36]. It is likely that bacterial isolates harboring qnrS from both clinical and poultry sources might be released to the water bodies through human or animal waste due to lack of proper wastewater treatment facilities in Bangladesh [53]. In contrast to our study, studies in other countries such as China, Switzerland, and Poland showed that aac(6’)-lb-cr was the predominant PMQR gene in E. coli isolates from aquatic samples [34, 54–56]. Further investigations of the isolates using a comparative genomic approach will provide more insights into the characteristics of isolates driving this discriminate distribution of PMQR genes.
In the present study, more than 65% of the PMQR positive isolates were resistant to multiple antibiotics including penicillin, cephalosporins, fluro(quinolones), sulfonamides aminoglycosides and carbapenems. A high prevalence of MDR could be associated with the wide range of antibiotics used in the poultry and aquaculture sectors which increase selection pressures for AMR in water bodies. Different mechanisms can be involved in the emergence of MDR E. coli strains such as: 1) shared resistance mechanisms that occur for the antimicrobial agents in the same category, e.g., mutations in penicillin-binding protein and presence of the ß-lactamases. This can also occur for antibiotics in different classes due to the presence of efflux pumps acting on different antibiotics; 2) exposure to multiple antibiotics via routine use of combination therapy and repeated treatment failure and 3) the presence of plasmids that carry resistance genes to multiple antibiotics. We found that the presence of PMQR genes, particularly qnr, in E. coli isolates was associated with the ESBL phenotype (p<0.001) and various β-lactamase encoding genes including blaCMY, blaCTX-M-1, blaCMY, blaTEM and blaOXA-1. This can be explained by carriage of both qnr and ESBL/AmpC genes in the same plasmids as reported by previous studies [57–59]. Apart from qnr, other PMQR genes such as oqxAB were found to associate significantly with blaSHV whereas aac(6’)-lb-cr was associated with blaOXA-1 and blaOXA-47. This finding concurs with earlier reports that indicated the co-occurrence of oqxAB with blaSHV and aac(6’)-lb-cr with blaOXA-1 and blaOXA-47 [59, 60]. The presence of both PMQR and ESBL genes in the same bacterial isolates could be due to the co-selection of isolates in the environment with either of fluoroquinolone or cephalosporins which accentuate further confirmation. Extensive use of quinolones therefore may lead to the emergence of resistance against β-lactams which are important clinically used antibiotics.
Integrons are important mobile genetic elements that carry different antibiotic resistance gene cassettes and play a crucial role in AMR transmission via horizontal gene transfer between different bacterial species [61]. Class 1 integron (int1) is most studied and reported ubiquitously in different enterobacterial species including E. coli [62]. In our study, qnrB and qepA were associated with the presence of int1 (Fig 1) indicating that these genes might be in the gene cassette carried by the integrons. Previous study reported that int1 in E. coli carried gene cassettes encoding resistance to multiple antibiotics including β-lactams (blaOXA-30), trimethoprim (dfrA1, dfrA5, dfrA7, dfrA12, dfrA17), aminoglycosides (aadA1, aadA2, aadA5), chloramphenicol (cmlA) and erythromycin (ereA2) [63]. Findings from our study highlight the need for further investigation to identify whether PMQR genes are located in the class 1 integron of the isolates using whole genome sequencing.
In this work, ERIC-PCR analysis revealed that the PMQR-positive isolates were mostly heterogeneous although a small number of isolates from different sources of water or locations had identical banding patterns indicating their clonal relationship. It could be that certain clonal groups of PMQR-positive ESBL-producing E. coli were predominantly present in the waterbodies, however, further characterization of these clones using next generation sequencing would be useful. Further, comparison of these isolates with clinical isolates could be done to understand the contribution of these widely circulating clones on the burden of antimicrobial resistant infections in the community.
Conclusions
Our study shows a high prevalence of PMQR-positive E. coli in urban and rural waters. These plasmid-mediated isolates were mostly MDR, predominantly ESBL-producing and genetically heterogeneous. The high prevalence of plasmid-mediated quinolone resistance poses a risk for horizontal gene transfer and this, in association with ESBL genes, adds to the threat of AMR transmission via the environment. This study highlights the importance of including surface waters and wastewaters in One Health AMR surveillance programs to understand the emergence and transmission dynamics of AMR and for designing environmental intervention strategies.
Acknowledgments
icddr,b is grateful to the Governments of Bangladesh, Canada, Sweden and the United Kingdom for providing core/unrestricted support.
Data Availability
All relevant data are within the manuscript. In addition, details of isolates, genes and antibiotic susceptibility data are open access and available at the NERC Environmental Data Repository: https://doi.org/10.5285/0239cdaf-deab-4151-8f68-715063eaea45 and https://doi.org/10.5285/dda6dd55-f955-4dd5-bc03-b07cc8548a3d. The nucleotide sequence data of PMQR gene amplified fragments have been deposited in the PubMed GenBank nucleotide sequence database (http://www.ncbi.nlm.nih.gov/) and resulting GenBank under accession numbers were: qnrS (OL439745), qnrB (OL439744), oqxAB (OK668389), qepA (OK668390) and aac(6’)-lb-cr (OL439743).
Funding Statement
This research was funded by the Antimicrobial Resistance Cross Council Initiative supported by the seven research councils in partnership with the Department of Health and Department for Environment Food & Rural Affairs (NERC/ BBSRC/MRC grant number: NE/N019555/1). Dr. Emily K. Rousham received this grant.
References
- 1.O’Neill J. Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations. Available: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf. 2014. [Google Scholar]
- 2.Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. 2010;8(3):207–17. doi: 10.1038/nrmicro2298 [DOI] [PubMed] [Google Scholar]
- 3.Alharbi NS, Khaled JM, Kadaikunnan S, Alobaidi AS, Sharafaddin AH, Alyahya SA, et al. Prevalence of Escherichia coli strains resistance to antibiotics in wound infections and raw milk. Saudi J Biol Sci. 2019;26(7):1557–62. doi: 10.1016/j.sjbs.2018.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med. 2004;10(12):S122–S9. doi: 10.1038/nm1145 [DOI] [PubMed] [Google Scholar]
- 5.Makharita RR, El-Kholy I, Hetta HF, Abdelaziz MH, Hagagy FI, Ahmed AA, et al. Antibiogram and genetic characterization of carbapenem-resistant Gram-negative pathogens incriminated in healthcare-associated infections. Infect Drug Resist. 2020;13:3991–4002. Epub 2020/11/13. doi: 10.2147/IDR.S276975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abolghait SK, Fathi AG, Youssef FM, Algammal AM. Methicillin-resistant Staphylococcus aureus (MRSA) isolated from chicken meat and giblets often produces staphylococcal enterotoxin B (SEB) in non-refrigerated raw chicken livers. Int J Food Microbiol. 2020;328:108669. Epub 2020/06/05. doi: 10.1016/j.ijfoodmicro.2020.108669 [DOI] [PubMed] [Google Scholar]
- 7.Algammal AM, Hetta HF, Elkelish A, Alkhalifah DHH, Hozzein WN, Batiha GE, et al. Methicillin-resistant Staphylococcus aureus (MRSA): One Health perspective approach to the bacterium epidemiology, virulence factors, antibiotic-resistance, and zoonotic impact. Infect Drug Resist. 2020;13:3255–65. Epub 2020/10/17. doi: 10.2147/IDR.S272733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Algammal AM, Hashem HR, Alfifi KJ, Hetta HF, Sheraba NS, Ramadan H, et al. atpD gene sequencing, multidrug resistance traits, virulence-determinants, and antimicrobial resistance genes of emerging XDR and MDR-Proteus mirabilis. Sci Rep. 2021;11(1):9476. Epub 2021/05/06. doi: 10.1038/s41598-021-88861-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Algammal AM, El-Sayed ME, Youssef FM, Saad SA, Elhaig MM, Batiha GE, et al. Prevalence, the antibiogram and the frequency of virulence genes of the most predominant bacterial pathogens incriminated in calf pneumonia. AMB Express. 2020;10(1):99. Epub 2020/05/31. doi: 10.1186/s13568-020-01037-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Algammal AM, Hetta HF, Batiha GE, Hozzein WN, El Kazzaz WM, Hashem HR, et al. Virulence-determinants and antibiotic-resistance genes of MDR-E. coli isolated from secondary infections following FMD-outbreak in cattle. Sci Rep. 2020;10(1):19779. Epub 2020/11/15. doi: 10.1038/s41598-020-75914-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Algammal AM, Hashem HR, Al-Otaibi AS, Alfifi KJ, El-Dawody EM, Mahrous E, et al. Emerging MDR-Mycobacterium avium subsp. avium in house-reared domestic birds as the first report in Egypt. BMC Microbiol. 2021;21(1):237. Epub 2021/08/28. doi: 10.1186/s12866-021-02287-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pham TDM, Ziora ZM, Blaskovich MAT. Quinolone antibiotics. Medchemcomm. 2019;10(10):1719–39. doi: 10.1039/c9md00120d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laizu J, Parvin R, Sultana N, Ahmed M, Sharmin R, Sharmin Z. Prescribing practice of antibiotics for outpatients in Bangladesh: rationality analysis. Am J Pharmacol. 2018;1(1):1008. [Google Scholar]
- 14.Biswas M, Roy DN, Tajmim A, Rajib SS, Hossain M, Farzana F, et al. Prescription antibiotics for outpatients in Bangladesh: a cross-sectional health survey conducted in three cities. Ann Clin Microbiol Antimicrob. 2014;13:15. Epub 2014/04/24. doi: 10.1186/1476-0711-13-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chowdhury R, Haque M, Islam K, Khaleduzzaman A. A review on antibiotics in an animal feed. Bangladesh J Anim Sci. 2009;38(1–2):22–32. [Google Scholar]
- 16.Masud AA, Rousham EK, Islam MA, Alam M-U, Rahman M, Mamun AA, et al. Drivers of antibiotic use in poultry production in Bangladesh: dependencies and dynamics of a patron-client relationship. Front Vet Sci. 2020;7(78). doi: 10.3389/fvets.2020.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rousham EK, Asaduzzaman M, Mozmader T, Amin MB, Rahman M, Hossain MI, et al. Human colonization with extended-spectrum β-lactamase-producing E. coli in relation to animal and environmental exposures in Bangladesh: An observational One Health study. Environ Health Perspect. 2021;129(3):37001. Epub 2021/03/04. doi: 10.1289/EHP7670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev. 2009;22(4):664–89. Epub 2009/10/14. doi: 10.1128/CMR.00016-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacoby GA, Walsh KE, Mills DM, Walker VJ, Oh H, Robicsek A, et al. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob Agents Chemother. 2006;50(4):1178–82. Epub 2006/03/30. doi: 10.1128/AAC.50.4.1178-1182.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacoby GA, Chow N, Waites KB. Prevalence of plasmid-mediated quinolone resistance. Antimicrob Agents Chemother. 2003;47(2):559–62. Epub 2003/01/25. doi: 10.1128/AAC.47.2.559-562.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez-Martínez L, Pascual A, Jacoby GA. Quinolone resistance from a transferable plasmid. Lancet (London, England). 1998;351(9105):797–9. Epub 1998/03/31. doi: 10.1016/S0140-6736(97)07322-4 [DOI] [PubMed] [Google Scholar]
- 22.Wong MH, Chan EW, Liu LZ, Chen S. PMQR genes oqxAB and aac(6’)Ib-cr accelerate the development of fluoroquinolone resistance in Salmonella typhimurium. Front Microbiol. 2014;5:521. Epub 2014/10/18. doi: 10.3389/fmicb.2014.00521 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacoby GA, Strahilevitz J, Hooper DC. Plasmid-mediated quinolone resistance. Microbiol Spectr. 2014;2(5). Epub 2015/01/15. doi: 10.1128/microbiolspec.PLAS-0006-2013 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.WHO. Critically important antimicrobials for human medicine. 2019. [Google Scholar]
- 25.Xiong X, Bromley EH, Oelschlaeger P, Woolfson DN, Spencer J. Structural insights into quinolone antibiotic resistance mediated by pentapeptide repeat proteins: conserved surface loops direct the activity of a Qnr protein from a Gram-negative bacterium. Nucleic Acids Res. 2011;39(9):3917–27. Epub 2011/01/14. doi: 10.1093/nar/gkq1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong MH, Chan EW, Chen S. Evolution and dissemination of OqxAB-like efflux pumps, an emerging quinolone resistance determinant among members of Enterobacteriaceae. Antimicrob Agents Chemother. 2015;59(6):3290–7. Epub 2015/03/25. doi: 10.1128/AAC.00310-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamane K, Wachino J, Suzuki S, Kimura K, Shibata N, Kato H, et al. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob Agents Chemother. 2007;51(9):3354–60. Epub 2007/06/06. doi: 10.1128/AAC.00339-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robicsek A, Strahilevitz J, Jacoby GA, Macielag M, Abbanat D, Park CH, et al. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med. 2006;12(1):83–8. Epub 2005/12/22. doi: 10.1038/nm1347 [DOI] [PubMed] [Google Scholar]
- 29.Cattoir V, Nordmann P. Plasmid-mediated quinolone resistance in Gram-negative bacterial species: an update. Curr Med Chem. 2009;16(8):1028–46. Epub 2009/03/12. doi: 10.2174/092986709787581879 [DOI] [PubMed] [Google Scholar]
- 30.Hansen LH, Jensen LB, Sørensen HI, Sørensen SJ. Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J Antimicrob Chemother. 2007;60(1):145–7. Epub 2007/05/29. doi: 10.1093/jac/dkm167 [DOI] [PubMed] [Google Scholar]
- 31.Gillings MR. Integrons: past, present, and future. Microbiol Mol Biol Rev. 2014;78(2):257–77. Epub 2014/05/23. doi: 10.1128/MMBR.00056-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng Y, Bao X, Ji L, Chen L, Liu J, Miao J, et al. Resistance integrons: class 1, 2 and 3 integrons. Ann Clin Microbiol Antimicrob. 2015;14:45. Epub 2015/10/22. doi: 10.1186/s12941-015-0100-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stokes HW, Gillings MR. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol Rev. 2011;35(5):790–819. doi: 10.1111/j.1574-6976.2011.00273.x [DOI] [PubMed] [Google Scholar]
- 34.Röderova M, Halova D, Papousek I, Dolejska M, Masarikova M, Hanulik V, et al. Characteristics of quinolone resistance in Escherichia coli isolates from humans, animals, and the environment in the Czech Republic. Front Microbiol. 2016;7:2147. Epub 2017/01/26. doi: 10.3389/fmicb.2016.02147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wen Y, Pu X, Zheng W, Hu G. High prevalence of plasmid-mediated quinolone resistance and IncQ Plasmids Carrying qnrS2 gene in bacteria from rivers near hospitals and aquaculture in China. PLoS One. 2016;11(7):e0159418. Epub 2016/07/20. doi: 10.1371/journal.pone.0159418 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan ER, Aung MS, Paul SK, Ahmed S, Haque N, Ahamed F, et al. Prevalence and molecular epidemiology of clinical isolates of Escherichia coli and Klebsiella pneumoniae harboring extended-spectrum β-lactamase and carbapenemase genes in Bangladesh. Microbial Drug Resist (Larchmont, NY). 2018;24(10):1568–79. Epub 2018/06/30. doi: 10.1089/mdr.2018.0063 [DOI] [PubMed] [Google Scholar]
- 37.Mahmud S, Nazir K, Rahman MT. Prevalence and molecular detection of fluoroquinolone-resistant genes (qnrA and qnrS) in Escherichia coli isolated from healthy broiler chickens. Veterinary world. 2018;11(12):1720–4. Epub 2019/02/19. doi: 10.14202/vetworld.2018.1720-1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rahman Z, Islam A, Rashid MU, Johura FT, Monira S, Watanabe H, et al. Existence of a novel qepA variant in quinolone resistant Escherichia coli from aquatic habitats of Bangladesh. Gut Pathog. 2017;9:58. Epub 2017/10/28. doi: 10.1186/s13099-017-0207-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Islam MA, Islam M, Hasan R, Hossain MI, Nabi A, Rahman M, et al. Environmental spread of New Delhi Metallo-β-lactamase-1-producing multidrug-resistant bacteria in Dhaka, Bangladesh. Appl Environ Microbiol. 2017;83(15). doi: 10.1128/aem.00793-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hasan MK, Shahriar A, Jim KU. Water pollution in Bangladesh and its impact on public health. Heliyon. 2019;5(8):e02145–e. doi: 10.1016/j.heliyon.2019.e02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arefin MA, Mallik A. Sources and causes of water pollution in Bangladesh: A technical overview. Bibechana. 2018;15:97–112. [Google Scholar]
- 42.Rousham E, Unicomb L, Wood P, Smith M, Asaduzzaman M, Islam MA. Spatial and temporal variation in the community prevalence of antibiotic resistance in Bangladesh: an integrated surveillance study protocol. BMJ Open. 2018;8(4):e023158. Epub 2018/05/01. doi: 10.1136/bmjopen-2018-023158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.James C, Natalie S. Microbiology. A laboratory manual: Pearson Education; 2014. [Google Scholar]
- 44.Ciesielczuk H, Hornsey M, Choi V, Woodford N, Wareham DW. Development and evaluation of a multiplex PCR for eight plasmid-mediated quinolone-resistance determinants. J Medical Microbiol. 2013;62(Pt 12):1823–7. Epub 2013/09/04. doi: 10.1099/jmm.0.064428-0 [DOI] [PubMed] [Google Scholar]
- 45.Patel J. Performance standards for antimicrobial susceptibility testing: Clinical and Laboratory Standards Institute. 2017. [Google Scholar]
- 46.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81. Epub 2011/07/29. doi: 10.1111/j.1469-0691.2011.03570.x . [DOI] [PubMed] [Google Scholar]
- 47.Amin MB, Sraboni AS, Hossain MI, Roy S, Mozmader TAU, Unicomb L, et al. Occurrence and genetic characteristics of mcr-1-positive colistin-resistant E. coli from poultry environments in Bangladesh. 2020;22:546–52. doi: 10.1016/j.jgar.2020.03.028 [DOI] [PubMed] [Google Scholar]
- 48.Talukdar PK, Rahman M, Rahman M, Nabi A, Islam Z, Hoque MM, et al. Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS One. 2013;8(4):e61090. Epub 2013/04/11. doi: 10.1371/journal.pone.0061090 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mobarak-Qamsari M, Ashayeri-Panah M, Eftekhar F, Feizabadi MM. Integron mediated multidrug resistance in extended spectrum β-lactamase producing clinical isolates of Klebsiella pneumoniae. Braz J Microbiol. 2013;44(3):849–54. Epub 2014/02/12. doi: 10.1590/s1517-83822013000300028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jafari M, Ansari-Pour N. Why, When and How to Adjust Your P Values? Cell journal. 2019;20(4):604–7. Epub 2018/08/21. doi: 10.22074/cellj.2019.5992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei T, Simko V, Levy M, Xie Y, Jin Y, Zemla J. corrplot: Visualization of a correlation matrix. R package version 073. 2013;230(11). [Google Scholar]
- 52.Rezazadeh M, Baghchesaraei H, Peymani A. Plasmid-mediated quinolone-resistance (qnr) Genes in clinical isolates of Escherichia coli collected from several hospitals of Qazvin and Zanjan provinces, Iran. Osong Public Health Res Perspect. 2016;7(5):307–12. Epub 2016/11/05. doi: 10.1016/j.phrp.2016.08.003 53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Casarez EA, Pillai SD, Mott JB, Vargas M, Dean KE, Di Giovanni GD. Direct comparison of four bacterial source tracking methods and use of composite data sets. J Appl Microbiol. 2007;103(2):350–64. Epub 2007/07/26. doi: 10.1111/j.1365-2672.2006.03246.x [DOI] [PubMed] [Google Scholar]
- 54.Hasan J, Rahman MH, Ullah MR, Mredul MMH. Availability of aqua drugs and their uses in semi intensive culture farms at Patuakhali district in Bangladesh. Arch Agric Env Sci. 2020;5(3):368–76. [Google Scholar]
- 55.Sharmin A. Water and wastewater in Bangladesh, current status and a design of a decentralized solution. 2016. [Google Scholar]
- 56.Yan L, Liu D, Wang XH, Wang Y, Zhang B, Wang M, et al. Bacterial plasmid-mediated quinolone resistance genes in aquatic environments in China. Sci Rep. 2017;7:40610. Epub 2017/01/18. doi: 10.1038/srep40610 57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Osińska A, Harnisz M, Korzeniewska E. Prevalence of plasmid-mediated multidrug resistance determinants in fluoroquinolone-resistant bacteria isolated from sewage and surface water. Env Science Pollut Res Int. 2016;23(11):10818–31. Epub 2016/02/20. doi: 10.1007/s11356-016-6221-4 58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zurfluh K, Abgottspon H, Hächler H, Nüesch-Inderbinen M, Stephan R. Quinolone resistance mechanisms among extended-spectrum β-lactamase (ESBL) producing Escherichia coli isolated from rivers and lakes in Switzerland. PLoS One. 2014;9(4):e95864. Epub 2014/04/24. doi: 10.1371/journal.pone.0095864 59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tao Y, Zhou K, Xie L, Xu Y, Han L, Ni Y, et al. Emerging co-existence of three PMQR genes on a multiple resistance plasmid with a new surrounding genetic structure of qnrS2 in E. coli in China. Antimicrob Resist Infect Control. 2020;9(1):52. doi: 10.1186/s13756-020-00711-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fortini D, Fashae K, García-Fernández A, Villa L, Carattoli A. Plasmid-mediated quinolone resistance and β-lactamases in Escherichia coli from healthy animals from Nigeria. J Antimicrob Chemother. 2011;66(6):1269–72. Epub 2011/03/12. doi: 10.1093/jac/dkr085 [DOI] [PubMed] [Google Scholar]
- 61.Seo MR, Park YS, Pai H. Characteristics of plasmid-mediated quinolone resistance genes in extended-spectrum cephalosporin-resistant isolates of Klebsiella pneumoniae and Escherichia coli in Korea. Chemother. 2010;56(1):46–53. Epub 2010/03/06. doi: 10.1159/000290972 [DOI] [PubMed] [Google Scholar]
- 62.Goudarzi M, Azad M, Seyedjavadi SS. Prevalence of plasmid-mediated quinolone resistance determinants and OqxAB efflux pumps among extended spectrum β-lactamase producing Klebsiella pneumoniae isolated from patients with nosocomial urinary tract infection in Tehran, Iran. Scientifica (Cairo). 2015;2015:518167-. Epub 08/02. doi: 10.1155/2015/518167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cambray G, Guerout AM, Mazel D. Integrons. Annu Rev Genet. 2010;44:141–66. Epub 2010/08/17. doi: 10.1146/annurev-genet-102209-163504 . [DOI] [PubMed] [Google Scholar]



