Abstract
The pro-inflammatory cytokine, tumor necrosis factor-alpha (TNF-α), has been suggested to be a key factor in the induction of obesity-associated metabolic dysfunction. However, the role that macrophage-derived TNF-α has on regulating metabolic perturbations in obesity is not completely understood. Therefore, we utilized the TNF-αFlox/Flox(F/F), LyzMcre± mouse model to determine the impact that macrophage TNF-α deletion has on the development of high-fat diet (HFD)-induced obesity. At 10 weeks of age, male littermates were randomly assigned to 1 of 4 groups: TNF-αF/F low-fat diet (TNF-αF/F LFD), TNF-αF/F, LyzMCre LFD, TNF-αF/F HFD, or TNF-αF/F, LyzMCre HFD (n = 16–28/group) and were fed their respective diets for 18 weeks. Body weight was assessed throughout the course of the experiment. Body composition, hepatic lipid accumulation, and metabolic outcomes were also examined. A microarray gene expression experiment was performed from RNA isolated from epididymal adipose tissue of the HFD-fed groups (n = 10/group) and results were verified via qRT-PCR for all groups. Macrophage-derived TNF-α deletion significantly reduced adipose tissue TNF-α gene expression and circulating TNF-α and downregulated genes linked to the toll-like receptor (TLR) and NFκB signaling pathways. However, macrophage TNF-α deletion had no effect on hindering the development of obesity, hepatic lipid accumulation, or improving glucose metabolism or insulin sensitivity. In conclusion, macrophage-derived TNF-α is not a causative factor for the induction of obesity-associated metabolic dysfunction.
Keywords: insulin resistance, macrophage, metabolism, obesity, TNF-α
1 |. INTRODUCTION
It is well established that obesity has become a global epidemic.1 The obese phenotype is often characterized by a state of chronic low-grade inflammation and metabolic dysfunction. However, the extent to which these features are linked and causative of one another still remains unresolved. It is clear, however, that infiltrating macrophages are the primary mediators of adipose tissue inflammation.2 Furthermore, arguably the most infamous pro-inflammatory cytokine thought to link adipose tissue inflammation to metabolic dysfunction is adipose-tissue-derived tumor necrosis factor-alpha (TNF-α).
TNF-α is translated as a 26-k Da protein prior to undergoing proteolytic cleavage by the TNF-α converting enzyme (TACE) resulting in a soluble 17-k Da protein.3 In the 1990s, Hotamisligil and Spiegelman’s group, through the use of neutralizing antibodies and genetic ablation of TNF-α and its receptors, provided evidence of a central role of TNF-α to induce insulin resistance in obesity.4–7 However, the extent to which TNF-α regulates insulin resistance in obesity remains controversial. For instance, Salles et al. found that total body TNF-α knockout (KO) improved metabolic outcomes with high-fat diet (HFD) feeding,8 whereas Chen et al. found that whole-body deletion of TNF-α exacerbated obesity-related metabolic disorders.9 Furthermore, utilizing TNF-α receptor KO mice, a study by Schreyer et al. suggested that TNF-α, reacting through its receptors, is not a major contributor to obesity-associated insulin resistance.10 Additionally, because previous studies have implored whole-body KO mice to probe the role of TNF-α to regulate metabolic processes during obesity, it remains unknown which cell type is most responsible for TNF-α production. It is known that in an obese state both adipocytes and macrophages may produce TNF-α.11 Nonetheless, it has been shown that cells of the adipose tissue stromal vascular fraction (SVF), the majority of which are macrophages, have the capacity to produce significantly more TNF-α than adipocytes.12
Given the inconsistencies and limitations of previous studies, the purpose of our study was to examine the impact that macrophage-derived TNF-α has on regulating insulin resistance and adipose tissue inflammation in an HFD-induced obese mouse model. We hypothesized that macrophage TNF-α deletion would improve HFD-induced metabolic dysfunction.
2 |. MATERIALS AND METHODS
2.1 |. Animals
TNF-αFlox/Flox mice13 on a C57BL/6 background were a kind gift from Dr Sergei Nedospasov (Engelhardt Institute of Molecular Biology) via Dr Sergei Grivennikov (Fox Chase Cancer Center). LyzMCre mice on a C57BL/6J background were originally purchased from Jackson Laboratories (Bar Harbor, Maine). In order to produce TNF-αFlox/Flox, LysMCre± and TNF-αFlox/Flox, LyzMCre−/− littermate controls, TNF-αFlox/Flox, LyzMcre± were bred with TNF-αFlox/Flox mice. It should be noted that a limitation of the LyzMCre model is that cre activity is not limited to macrophages, but rather all myeloid lineage cells.14 This should be taken into consideration when examining the results of the study. We focused our study in male mice as female mice do not show as robust response with respect to adipose tissue inflammation resulting from HFD consumption.15
2.2 |. Lipopolysaccharide administration
A subset of TNF-αFlox/Flox, LyzMcre−/− (TNF-αF/F) and TNF-αFlox/Flox, LyzMcre± (TNF-αF/F,LyzMCre) mice were administered 100ng of lipopolysaccharide (LPS) (Sigma Aldrich, St. Louis, MO) intraperitoneally and were subsequently sacrificed 1hour after LPS administration. Whole blood was collected via the inferior vena cava and plasma was assessed for circulating TNF-α utilizing a mouse TNF-α ELISA kit (BioLegend, San Diego, CA).
2.3 |. Diets
At 10 weeks of age, male offspring were randomly assigned to 1 of 4 groups: TNF-αF/F low-fat diet (TNF-αF/F LFD), TNF-αF/F,LyzMCre LFD, TNF-αF/F HFD, or TNF-αF/F,LyzMCre HFD (n = 16–28/group). The LFD utilized in this experiment was the open-source, purified AIN-76A diet (3.77 kcal/g). The HFD was a custom-made purified HFD (4.57 kcal/g) comprised of 47%, 40%, and 13% of total calories from carbohydrate, fat, and protein, respectively, with saturated fat making up 12% of total calories in order to mimic the standard American diet (Bio-Serv, Frenchtown, NJ). Details and previous use of this diet are provided elsewhere.16–23 Mice were housed, 3–5/cage, maintained on a 12:12-h light-dark cycle in a low-stress environment (22°C, 50% humidity, low noise), and given food and water ad libitum. All methods were in accordance with the American Association for Laboratory Animal Science, and the Institutional Animal Care and Usage Committee of the University of South Carolina approved all experiments.
2.4 |. Body weight & body composition
Body weight was monitored on a weekly basis throughout the study. Body composition was assessed after 16 weeks of diet in order to use lean mass as the basis for the dose of glucose and insulin administration for glucose and insulin tolerance tests, respectively. For this procedure, mice were briefly anesthetized via isoflurane inhalation, and lean mass, fat mass, and percent body fat were assessed by dual-energy X-ray absorptiometry (Lunar PIXImus).
2.5 |. Circulating TNF-α measurement
At sacrifice, whole blood was collected via the inferior vena cava and plasma was assessed for TNF-α utilizing a mouse TNF-α ELISA kit (BioLegend, San Diego, CA). In order to detect the low levels of circulating TNF-α, 200 uL of plasma was used for the ELISA instead of the manufacturer’s recommended 100 μL.
2.6 |. Metabolism
Fasting blood glucose and insulin levels were assessed after 12 and 17 weeks of dietary treatment. After a 5-hour fast, blood samples were collected from the tip of the tail. A glucometer (Bayer Contour, Mishawaka, IN) was used to determine blood glucose concentrations in whole blood. Collected blood was centrifuged at 4,000 rpm for 10 minutes at 4°C. Plasma insulin concentrations were analyzed according to the manufacturer’s instructions using a mouse insulin ELISA kit (Mercodia, Winston Salem, NC). Glucose and insulin tolerances tests (GTTs and ITTs, respectively) were performed after 17 and 18 weeks of dietary treatment, respectively. For these procedures, mice were fasted for 5 hours, and glucose or insulin was administered intraperitoneally at 1g/kg or 0.75 U/kg lean mass, respectively. A glucometer (Bayer Contour) was used to measure blood glucose concentrations (tail sampling) intermittently over a 2-h period (0, 15, 30, 60, 90, and 120 minutes) for GTTs and intermittently over a 1-h period (0, 15, 30, 45, and 60 minutes) for ITTs. Area under the curve (AUC) was calculated using the trapezoidal rule. Blood was collected from the tip of the tail during the GTT (0, 15, 30, and 60 minutes) in order to assess the insulin response to the GTT for a subset of mice from each group (n = 9–15). Fasting serum was collected (using non-heparinized capillary tubes) for free fatty acid (FFA) analysis at the 0- and 30-min time points of the ITT to assess insulin’s ability to inhibit lipolysis. FFAs were analyzed using a commercially available kit according to the manufacturer’s instructions (Wako Diagnostics, Richmond, VA).
2.7 |. Tissue collection
After 18 weeks of dietary treatment, mice were euthanized via isoflurane inhalation for tissue collection. Epididymal, mesenteric, and perirenal fat pads, as well as the liver, and skeletal muscle (gastrocnemius and quadriceps) were removed, weighed, and immediately snap-frozen in liquid nitrogen and stored at −80°C or fixed in 4% formalin until analysis.
2.8 |. Hepatic lipid content
Lipids were isolated from the liver from each mouse utilizing a modified Folch extraction method and quantified gravimetrically, as previously described.20
2.9 |. Adipose tissue histology
Formalin-fixed tissues were embedded in paraffin blocks and sectioned (Instrumentation Resource Facility at the University of South Carolina). Epididymal fat was stained with hematoxylin and eosin (Biocare, Pacheco, CA). Representative images were taken at 40x (Revel Echo, San Diego, CA).
2.10 |. Microarray experiment
An EZNA Total RNA Kit (Omega Bio-Tek, Norcross, GA) was used to isolate RNA from epididymal adipose tissue. RNA quantity for n = 10 each for the TNF-αF/F HFD and TNF-αF/F,LyzMCre HFD groups was assessed using an Agilent 2100 Bioanalyzer and RNA Integrity Numbers (RIN) ranged from 7 to 8.5. For the gene expression experiments, total RNA samples were amplified and biotinylated using GeneChip WT PLUS Reagent Kit (Thermo Fisher Scientific, Cat. No. 902930), according to the manufacturer’s recommendations. Briefly, 100 ng of total RNA was reverse transcribed into ds-cDNA using NNN random primers that also contained the T7 RNA polymerase promoter sequence. Subsequently, T7 RNA polymerase was added to cDNA samples to amplify RNA molecules. Later, RNA was copied to ss-D NA and subsequently removed using RNase H. In the next step, ss-DNA molecules were fragmented and terminally labeled with biotin. Amplified/labeled samples were hybridized to Clariom S, Mouse Arrays (Thermo Fisher Scientific, Cat. No. 902930) during 16hours at 45°C using a GeneChip Hybridization Oven 645 (Thermo Fisher Scientific, Cat. No. 00–0331). Hybridized arrays were washed and stained using the kit mentioned above and GeneChip Fluidics Stations 450 (Thermo Fisher Scientific, Cat. No. 00–0079). Arrays were scanned with a GeneChip Scanner 3000 7G system (00–0218) using Affymetrix GeneChip Command Console Scan Control 4.0 software. At the same time, data were extracted from images and the resulting probe cell intensity (CEL) files (20 total) were imported into Transcriptome Analysis Console 4.0.2.15. CEL files were processed at the gene level using Clariom_S_Mouse.r1.na36.mm10.a1.transcript. csv annotation file and the Signal Space Transformation—Robust Multichip Analysis (SST-RMA) algorithm to generate CHP files. Data sets are available upon request to the corresponding author. After confirming data quality, phenotype-specific transcriptional responses were determined using one-way between-subject analysis of variance (ANOVA) with empirical Bayes correction. Differentially expressed genes with p values smaller than 0.05 and fold change higher than 1.5 and lower than −1.5 were used for further bioinformatics analysis. GO Enrichment Analysis of the differentially expressed gene set was performed with the enrichGO function (Using BP subontology) from package clusterProfiler v3.0.4 and mouse genome annotation file org. Mm.eg.db in R software.
2.11 |. Quantitative real-time PCR
An EZNA Total RNA Kit (Omega Bio-Tek, Norcross, GA) was used to isolate RNA from epididymal adipose tissue and skeletal muscle (gastrocnemius). In order to confirm changes in a subset of genes identified by the microarray, we performed qRT-P CR. Bio-Rad reverse transcription reagents and probe assays (Bio-Rad, Hercules, CA) were used to reverse transcribe and analyze the expression of the following genes in adipose tissue: EMR1, CD11c, CD206, MCP-1, TNFα, IL-10, IL-1β, MyD88, CD19, CD4, CD8b, and skeletal muscle: EMR1 and TNF-α. Potential reference genes (HPRT, 18s, GAPDH, β-Actin, HMBS, TBP, H2AFV, and B2 M) were analyzed for stability using Qbase+software (Biogazelle, Ghent, Belgium) for each tissue analyzed. The optimal number of reference genes was determined by Qbase+, and the geometric mean of these genes was used as the normalization factor for each analysis: epididymal adipose tissue (GAPDH, B2 M, and H2AFV), skeletal muscle (GAPDH, TBP, and H2AFV). Gene expression was quantified using the ΔΔCT method and Qbase+software.24
2.12 |. Statistical analysis
Data were analyzed using commercially available statistical software: Prism 6 (GraphPad Software, La Jolla, CA) and SigmaStat (Systat Software, San Jose, CA). A two-tailed student’s t test was used when only two groups were compared (LPS experiment). A two-way ANOVA (diet x genotype) followed by a Newman-Keuls post hoc analysis was used for the majority of outcomes analyzed. A two-way (time ×group) repeated-measures ANOVA followed by a Newman-Keuls post hoc test was also used for ITT analysis to determine differences in blood glucose levels from baseline (0 minutes) over the duration of the test. Any statistical test that did not pass the equal-variance test (Bartlett’s test for equal variances) was log-transformed and then reanalyzed. For microarray statistical analysis please see the microarray section. Data are presented as means ±SE, and the level of significance was set at P < .05.
3 |. RESULTS
3.1 |. TNF-αF/F,LyzMCre mice display significantly reduced circulating TNF-α when challenged with LPS
In order to assess the validity of LysMCre driven TNF-α deletion, we challenged a subset TNF-αF/F,LyzMCre, and TNF-αF/F mice with LPS. Similar to the results of others,13 mice with LyzMCre deletion of TNF-α displayed a significantly reduced TNF-α response to the LPS challenge (P < .05) (Figure S1).
3.2 |. Macrophage deletion of TNF-α reduces circulating TNF-α, but this has no effect at mitigating HFD-induced obesity
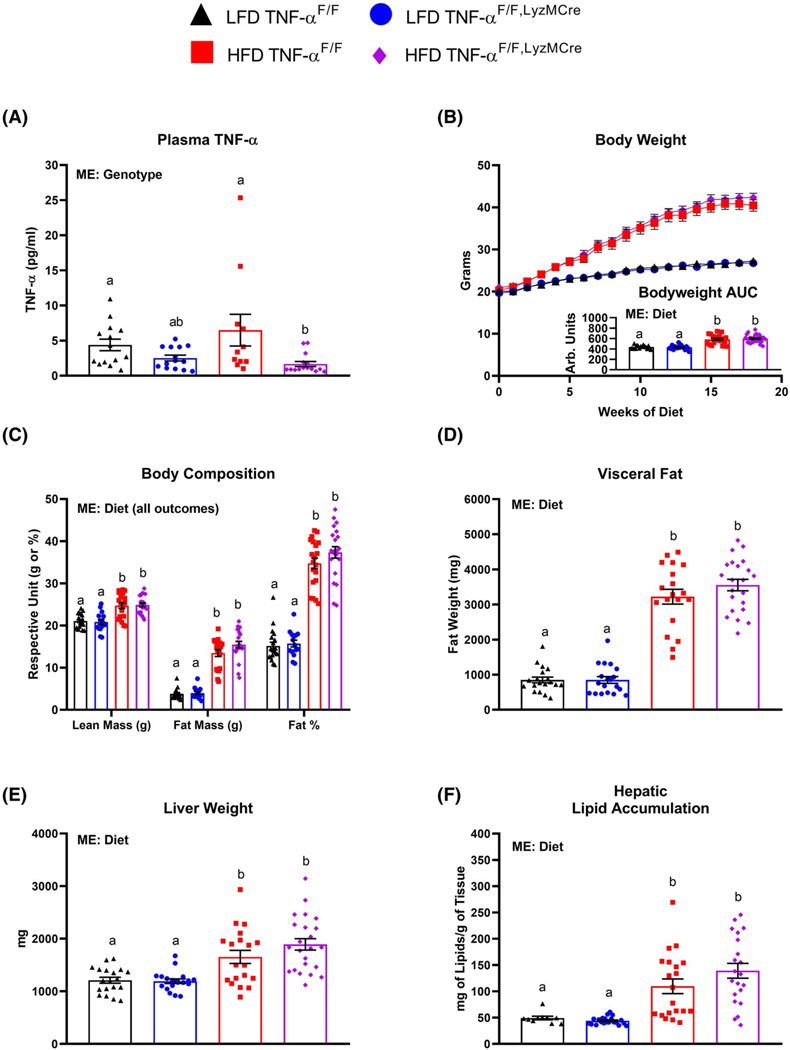
Although we did not find an increase in circulating TNF-α with HFD-feeding, there was a main effect of genotype to decrease circulating TNF-α in the LyzMCre mice (Figure 1A). However, independent of genotype, 18 weeks of HFD feeding resulted in body weight gain (Figure 1B) as well as increased lean mass, total fat mass, percent body fat (Figure 1C), visceral fat (Figure 1D), liver weight, (Figure 1E) and hepatic lipid accumulation (Figure 1F) compared to the LFD-fed mice (P < .05).
FIGURE 1.
Macrophage TNF-α deficiency does not protect against high-fat diet-induced (HFD) obesity. Male TNF-αF/F and TNF-αF/F,LyzMCre consumed either a low-fat diet (LFD) or HFD for 18 weeks. A, Plasma TNF-α (n = 11–1 5/group), B, body weight and body weight area under the curve (AUC), C, body composition (analyzed via DEXA after ≈16 weeks of dietary treatment), D, visceral fat, E, liver weight, and F, hepatic lipid accumulation (n = 16–28/group). Data are presented as mean ±SE. Bar graphs not sharing a common letter are significantly different from one another (P < .05). ME = Main Effect
3.3 |. LyzMCre-driven deletion of TNF-α does not dramatically impact metabolic processes
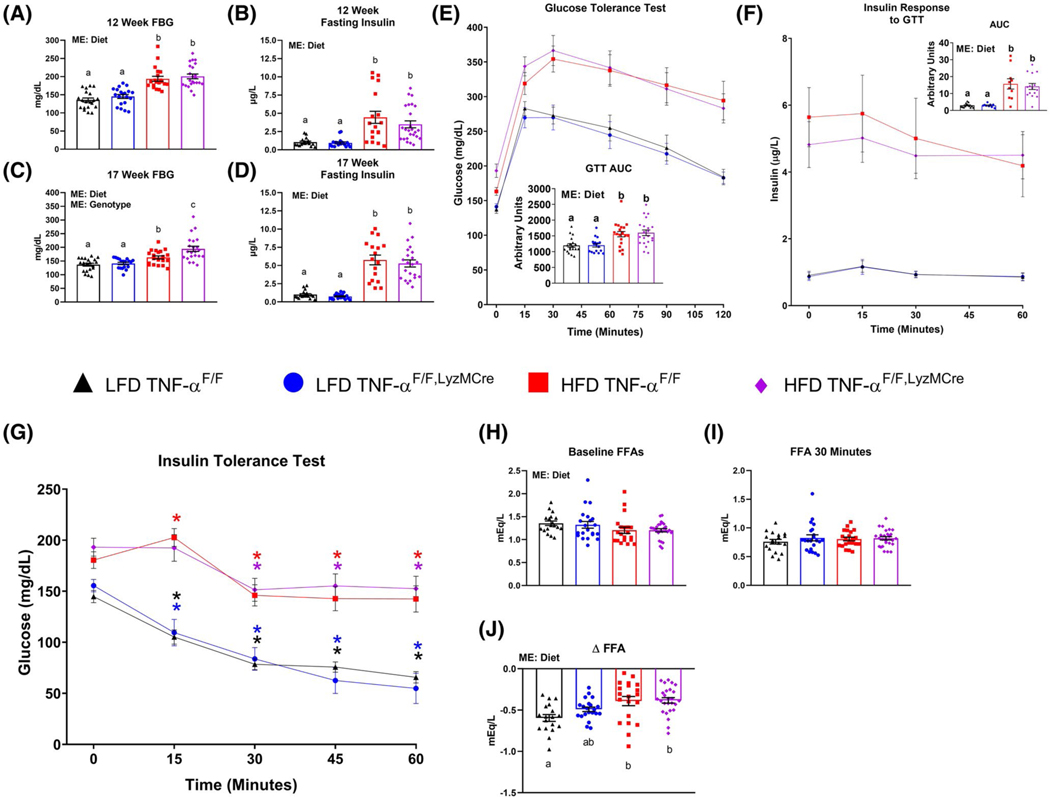
A main effect of diet was found to increase circulating levels of fasting blood glucose and insulin in the HFD-fed mice after 12 and 17 weeks of dietary treatment as well as impaired glucose tolerance compared to the LFD groups (P < .05) (Figure 2A–F). However, a main effect of genotype was found for the TNF-αF/F, LyzMCre mice to display elevated fasting blood glucose levels relative to TNF-αF/F mice after 17 weeks of dietary treatment (P < .05). For the ITT, only the LFD groups displayed lower blood glucose levels at 15 minutes, which continued throughout the remainder of the test relative to baseline levels (P < .05) (Figure 2F). Both HFD groups displayed lower blood glucose levels compared to baseline starting at 30 minutes post insulin administration (P < .05). However, only the TNF-αF/F HFD mice displayed an increase in blood glucose levels 15minutes after insulin administration compared to baseline levels (P < .05). Regarding blood concentration of FFAs, there was a main effect of diet to lower FFA levels in HFD-fed mice compared to LFD-fed mice at timepoint 0 of the ITT (Figure 2H) (P < .05). No difference was detected in blood FFAs among the groups 30 minutes into the ITT (Figure 2I). However, when examining the ΔFFAs levels between the two timepoints, a main effect of diet was found as the LFD-fed mice displayed the greatest drop in FFA levels compared to the HFD-fed mice (Figure 2J) (P < .05). However, when comparing between the groups, only the TNF-αF/F LFD mice exhibited a statistically significant drop in FFA levels compared to the HFD groups (P < .05).
FIGURE 2.
Macrophage TNF-α deficiency fails to improve obesity-linked metabolic dysfunction. Male TNF-αF/F and TNF-αF/F,LyzMCre consumed either a low-fat diet (LFD) or HFD for 18 weeks. A, 12-week fasting (5 hours) blood glucose (FBG), B, 12-week fasting (5 hours) plasma insulin, C, 17-week FBG (5 hours), D, 17-week fasting (5 hours) plasma insulin, E, glucose tolerance test (GTT) performed after ≈17 weeks of dietary treatment, F, insulin response to GTT, G, insulin tolerance test (ITT) performed after ≈18 weeks of dietary treatment, H, and I, serum free-fatty acid (FFA) concentration following a 5-hour fast measured at 0 and 30 minutes of the ITT, respectively, J, change in serum FFAs from 0 to 30 minutes of the ITT (n = 16–28/group). Data are presented as mean ±SE. Bar graphs not sharing a common letter are significantly different from one another (P < .05). Colored asterisks* represent statistically significant differences from the respective group’s baseline glucose levels (P < .05). ME = Main Effect
3.4 |. LyzMCre-driven deletion of TNF-α in the setting of obesity downregulates several inflammatory pathways
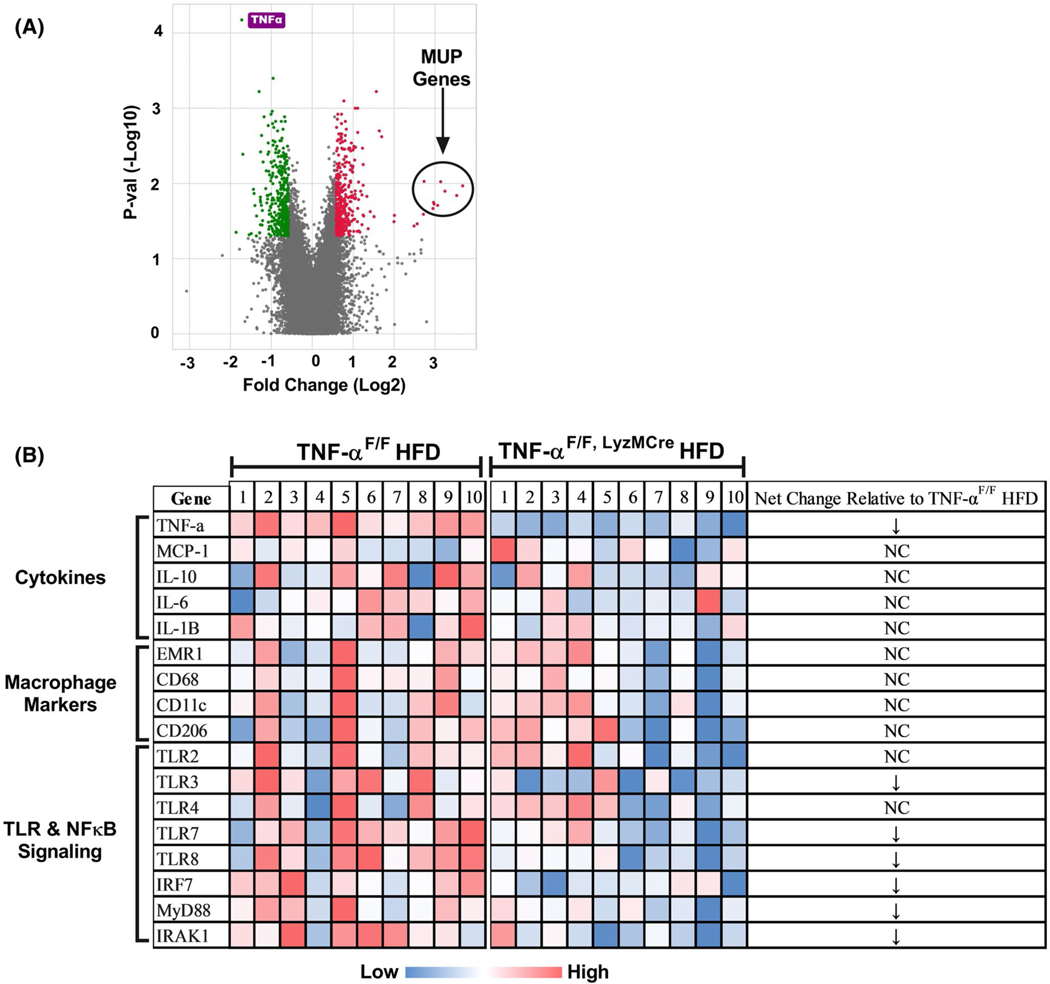
The Microarray experiment found a total of 145 differentially expressed genes that were ±1.5-f old changed in epididymal adipose tissue of TNF-αF/F, LyzMCre HFD vs TNF-αF/F HFD mice (P < .05) (Figure 3). Of the 145 genes that were significantly changed, TNF-α gene expression (−3.8x) was found to be the most statistically significantly changed (P = 7.67E-08) gene as a result of LyzMCre driven deletion (Figure 3A). Surprisingly, the genes found to be most up-regulated with TNF-α macrophage deletion were major urinary proteins (MUPs). Mup1, Mup2, Mup7, Mup8, Mup9, Mup10, Mup12, Mup13, and Mup19 were all found to be more than 15-fold increase in TNF-αF/F, LyzMCre HFD vs TNF-αF/F HFD mice (Figure 3A) (P < .05). Pathways analysis found several altered pathways as a result of LyzMCre mediated TNF-α deletion that piqued our interest: TNF-α superfamily cytokine production, toll-like receptor (TLR) signaling pathway, Iκβ Kinase/NFκB signaling, and regulation of TNF-α production (P < .05) (Table 1). Interestingly, we also found pathways regulating nucleotide processes to be changed as a result of LyzMCre mediated TNF-α deletion. Additionally, we did not see any differences in any macrophage (Figure 3B), B-Cell, or T-Cell markers.
FIGURE 3.
Microarray data from adipose tissue of TNF-αF/F and TNF-αF/F,LyzMCre HFD-fed mice. A, Volcano: a total of 145 genes were ±1.5x changed in epididymal adipose tissue of TNF-αF/F,LyzMCre HFD vs TNF-αF/F HFD mice (n = 10/group) (P < .05). The green and red colors signify genes that were −1.5x and +1.5x downregulated and upregulated, respectively, in the TNF-αF/F,LyzMCre HFD vs TNF-αF/F HFD mice (P < .05). TNF-α (labeled in purple) was found to be the most statistically significant down-regulated gene as a result of macrophage TNF-α deletion (P < .05). Major urinary proteins (MUPs) were found to be highly up-regulated (>15x) macrophage TNF-α deficient mice (P < .05). B, Clustering of gene expression profiles for each genotype
TABLE 1.
Adipose tissue gene pathway changes as a result of macrophage TNF-α deficiency
| Pathway | P-value | Gene # Changed | Up | Down | Net |
|---|---|---|---|---|---|
| Purine nucleotide metabolic process | .001056 | 35 | 24 | 11 | 13 |
| Purine ribonucleotide metabolic process | .001598 | 32 | 22 | 10 | 12 |
| Purine-containing compound metabolic process | .001598 | 36 | 24 | 12 | 12 |
| Ribose phosphate metabolic process | .001598 | 33 | 23 | 10 | 13 |
| Generation of precursor metabolites and energy | .001598 | 36 | 25 | 11 | 14 |
| Ribonucleotide metabolic process | .001631 | 32 | 22 | 10 | 12 |
| Nucleotide metabolic process | .014057 | 35 | 23 | 12 | 11 |
| Tumor necrosis factor production | .017636 | 19 | 5 | 14 | −9 |
| Nucleoside phosphate metabolic process | .017852 | 35 | 22 | 13 | 9 |
| Tumor necrosis factor superfamily cytokine production | .017852 | 19 | 5 | 14 | −9 |
| Pyruvate metabolic process | .017852 | 15 | 11 | 4 | 7 |
| Toll-like receptor signaling pathway | .018458 | 14 | 3 | 11 | −8 |
| I-kappa B kinase/NF-kappa B signaling | .025883 | 20 | 7 | 13 | −6 |
| Regulation of tumor necrosis factor production | .025962 | 18 | 5 | 13 | −8 |
| Regulation of I-kappa B kinase/ NF-kappa B signaling | .026226 | 18 | 7 | 11 | −4 |
| Regulation of tumor necrosis factor superfamily cytokine production |
.026591 | 18 | 5 | 13 | −8 |
| Nucleobase-containing small molecule metabolic process | .032999 | 37 | 24 | 13 | 11 |
| Purine nucleotide biosynthetic process | .033123 | 17 | 9 | 8 | 1 |
| Coenzyme A metabolic process | .033123 | 5 | 4 | 1 | 3 |
Note: 19 pathways were found to be significantly changed (P < .05) as a result of macrophage TNF-α deletion.
3.5 |. LyzMCre-driven deletion of TNF-α does not significantly affect obesity-induced adipose tissue macrophage infiltration
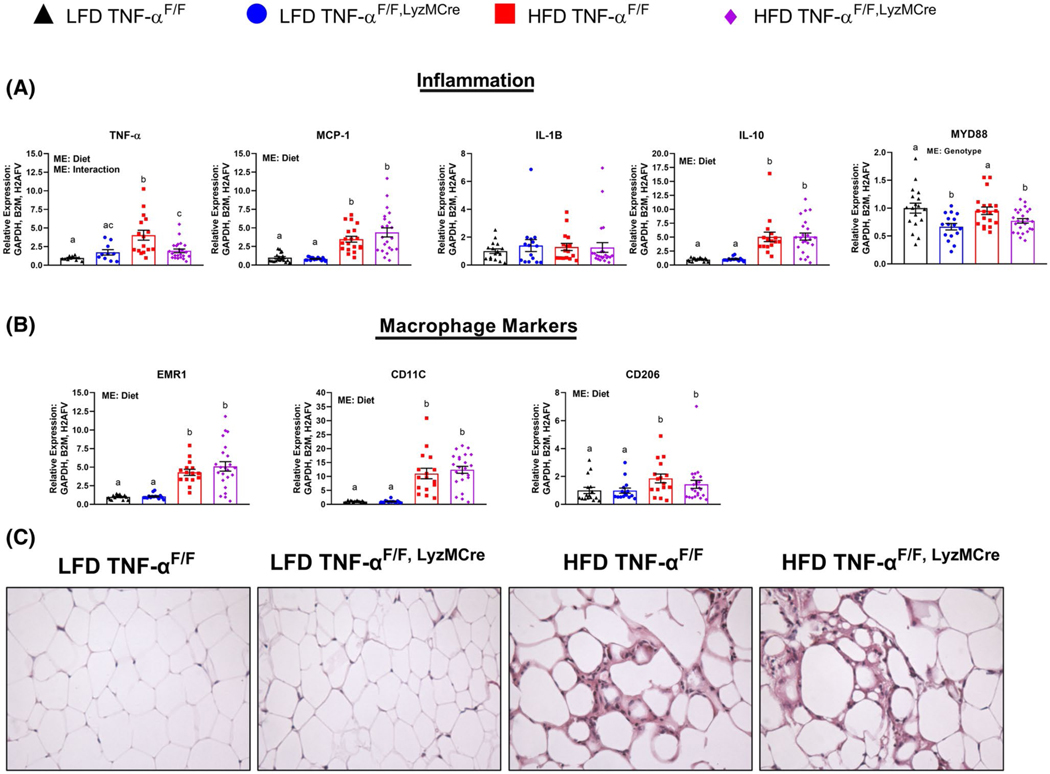
In order to corroborate the results of the microarray experiment, we performed qRT-PCR on all samples of each respective group—LFD and HFD. Not surprisingly, TNF-αF/F, LyzMCre HFD mice displayed significantly reduced levels of TNF-α gene expression in epididymal adipose tissue compared to TNF-αF/F HFD mice (P < .05) (Figure 4A). This lower TNF-α gene expression was similar to the levels exhibited by TNF-αF/F, LyzMCre LFD mice, but slightly elevated compared to TNF-αF/F LFD mice (P < .05). With respect to other cytokine gene expression, there was a diet effect for the HFD to increase adipose tissue gene expression of MCP-1 and IL-10, but not IL-1β (P < .05) (Figure 4A). We chose to measure mRNA expression of MYD88 as it is a common gene in the TLR and NFκB signaling pathways. Independent of diet, there was a significant effect of genotype (macrophage TNF-α deletion) to reduce MYD88 expression (P < .05). Similar to the findings of the microarray experiment, there was no difference in macrophage markers, EMR1 (total macrophage marker), CD11c (M1, pro-inflammatory macrophage marker), and CD206 (M2, anti-inflammatory macrophage marker) between the HFD groups (Figure 4B). However, there was a statistically significant diet effect for HFD consumption to increases these macrophage markers (P < .05). We did not observe any substantial differences in adipose tissue immune cell infiltration between the HFD-groups when viewed histologically (n = 5/group), but there was clear evidence of increased immune cell infiltration with HFD feeding (Figure 4C). In addition to substantiating the macrophage data, we also wanted to confirm that macrophage-derived TNF-α did not impact the infiltration of other immune cells, such as B-cells and T-cells. We found no difference across groups with respect to the pan B-cell marker, CD19, or T-cell markers, CD4a or CD8b, suggesting that there was no change in the infiltration of these cells with macrophage TNF-α deletion (Figure S2).
FIGURE 4.
Adipose tissue inflammation. A, TNF-α, MCP-1, IL-1β, IL-10, and MYD88, B, EMR1, CD11c, and CD206 (n = 16–28/group), C, representative epididymal adipose tissue H&E images taken at 40x. Data are presented as mean ±SE. Bar graphs not sharing a common letter are significantly different from one another (P < .05). NC = No change
3.6 |. No effect of genotype to impact skeletal muscle EMR1 or TNF-α gene expression
As we wanted to investigate whether macrophage-derived TNF-α deletion impacted skeletal muscle macrophage content or skeletal muscle TNF-α expression, we examined the gene expression of EMR1 and TNF-α, respectively, in skeletal muscle. No difference was detected in skeletal muscle EMR1 gene expression among the groups (Figure S3A). Interestingly, we found a main effect of diet (HFD) to decrease skeletal muscle TNF-α mRNA expression (Figure S3B) (P < .05).
4 |. DISCUSSION
The impact that TNF-α deficiency has on metabolic function linked to the obese phenotype remains controversial. Preclinical studies have produced mixed results with respect to the role that TNF-α plays in regulating insulin resistance in obesity. Furthermore, clinical studies utilizing TNF-α neutralizing strategies have failed to improve insulin sensitivity in type-2 diabetic patients and individuals with metabolic syndrome.25–28 Given these controversies and inconsistencies, the purpose of our study was to use a preclinical mouse model to shed light on the role that macrophage-derived TNF-α plays in regulating metabolic dysfunction and adipose tissue inflammation in an obese setting. Surprisingly, although we found several adipose tissue inflammatory pathways altered as a result of macrophage-derived TNF-α deletion, this had no effect to improve obesity-associated metabolic dysfunction.
The majority of previous preclinical studies that have explored the role that TNF-α plays in regulating obesity-associated insulin resistance have utilized either global TNF-α7–9,29 or TNF-α receptor-deficient mice7,10,30 and have yielded conflicting outcomes. Because macrophages are the predominant inflammatory cell that invades adipose tissue in an obese setting, we utilized the LyzMCre mouse model to target TNF-α deletion in macrophages. In order to be concrete in our findings, we utilized littermate controls and large sample size (n = 16–28) in conjunction with purified LFD and HFDs. Although we found that LyzMCre-driven deletion of TNF-α dramatically reduced adipose tissue TNF-α gene expression in an obese setting and lowered the concentration of circulating TNF-α, this had no impact to positively influence global glucose metabolism, insulin resistance, adipose tissue insulin action, or adipose tissue macrophage infiltration. However, the significant decrease in adipose tissue TNF-α gene expression corroborated that macrophage-derived TNF-α is the primary source of TNF-α production in an obese setting.
Our findings with respect to lack of metabolic changes as a result of macrophage-derived TNF-α deletion are in parallel with a study performed by De Taeye et al. who found that TNF-α−/− bone marrow transplanted to wild-type mice and subsequently fed an HFD displayed no protection against insulin resistance nor changes in adipose tissue macrophage markers.31 It was only when TNF-α-“sufficient” (TNF-α+/+) bone marrow was transplanted to TNF-α−/− mice that the authors found an effect of TNF-α+/+ bone marrow to exacerbate insulin resistance. The authors concluded that host-derived TNF-α must play a role in these outcomes. We proceeded to examine gene expression of TNF-α in skeletal muscle to determine if the deletion of macrophage-derived TNF-α impacted skeletal muscle production of the cytokine. We found no genotype effect nor change in the gene expression of TNF-α between the two HFD groups, suggesting that there was no compensatory skeletal muscle TNF-α production. Because TNF-α deficiency has been linked to hepatic steatosis in an obese setting,8,31 we examined hepatic lipid accumulation. We only found a statistically significant diet effect for the HFD to increase hepatic lipid accumulation independent of genotype, suggesting that macrophage-derived TNF-α deletion does not impact hepatic steatosis. A limitation of our study is we did not examine TNF-α expression in liver macrophages (Kupffer cells). It has previously been shown that Kupffer cells play a role in obesity-associated insulin resistance.32–34 However, we have previously found that 16 weeks of HFD feeding with the same HFD and LFD utilized in this investigation does not elicit statistically significant increases in hepatic TNF-α gene expression.17,23 Thus, because we found circulating TNF-α to be reduced as a result of LyzMCre activity, we can conclude one of the following in the LyzMCre mice: 1) Kupffer cell TNF-α production was blunted, 2) Kupffer cells are not a source of circulating TNF-α in our model, or 3) more likely, the length of HFD feeding was not sufficient to induce an increase in TNF-α production as supported by our previous publications.17,23
TNF-α has been suggested to have many pleiotropic effects within adipose tissue, including roles in lipogenesis, adipogenesis, thermogenesis, and stimulation of lipolysis.11 In order to gain a better understanding of the adipose tissue processes regulated by macrophage-derived TNF-α in an obese setting, we performed an adipose tissue gene expression microarray analysis with an n = 10 for each of the HFD groups. Surprisingly, no pathways linked to lipogenesis, lipolysis, or adipogenesis were found to be changed as a result of macrophage-derived TNF-α deletion. However, we found macrophage-derived TNF-α deletion resulted in a significant downregulation of genes in the TLR and NFκB signaling pathways. The finding that macrophage TNF-α deletion resulted in a downregulation of genes involved in the NFκB and TLR pathways is not surprising as TNF-α is a known activator of the NFκB pathway35 and has previously been shown to increase TLR expression in various cells.36–38 Moreover, we did not expect to see such significant increases of expression in a number of MUP genes and pathways involved in purine nucleotide metabolic processes with macrophage-derived TNF-α deletion. MUPs are members of the lipocalin family and are known to be primarily expressed in the liver of mice before being excreted in the urine.39 MUP production is stimulated by androgens and are thus found in higher quantity in males rather than females.40 In addition to having several known roles in rodents, including, regulating social behavior, MUPs, specifically Mup1, have also been shown to regulate energy expenditure and glucose metabolism.41 However, it seems not much is known regarding the function of MUPs in adipose tissue physiology. Our data suggest that there is a link between macrophage-derived TNF-α and MUP gene induction. Future studies are needed to better understand the relationship between these genes and their importance in adipose tissue. Purine nucleotides not only serve as the building blocks for RNA and DNA, but they also are necessary for providing cellular energy for metabolic processes and intracellular signaling (eg, ATP & GTP).42 We did not observe any differences in epididymal fat weight nor in insulin’s ability to inhibit lipolysis between the TNF-αF/F, LyzMCre HFD vs TNF-αF/F HFD mice, suggesting that any changes in metabolism did not impact these outcomes. Because whole adipose tissue was used for the microarray analysis, it is uncertain which cell (eg, adipocyte, macrophage, etc) or combinations of cells is most responsible for exhibiting these gene expression changes. Perhaps, macrophage TNF-α deletion impacted immune cell metabolism which evidently did not impact global glucose metabolism and insulin resistance.
Based on our findings we conclude that macrophage-derived TNF-α does not contribute to the metabolic dysfunction attributed to HFD-induced obesity. This finding supports the clinical studies that have shown TNF-α neutralization to be ineffective at enhancing insulin sensitivity in type-2 diabetic and obese patients.25–28 It should be noted that we did not find a statistically significant HFD effect to increase circulating TNF-α. This may explain why macrophage TNF-α deficiency had no positive impact on glucose metabolism or insulin signaling. A more advanced stage of obesity and adipose tissue dysfunction is likely necessary to increase circulating TNF-α levels beyond what is exhibited in adipose tissue. Nonetheless, our results provide evidence that factors other than TNF-α are the driving force behind obesity-associated metabolic dysfunction. For instance, an often debated topic is whether insulin resistance is a byproduct of inflammation or vice versa. Turner et al. performed a comprehensive study in which they showed that ectopic lipid accumulation and certain lipid species are likely the primary driver behind insulin resistance rather than adipose tissue inflammation in HFD-fed mice.43 We have also shown that when food intake is controlled for, macrophage depletion using clodronate liposomes does not rescue obesity-induced metabolic dysfunction.20 Additionally, Skurski et al. recently showed that reductions in adipose tissue inflammation, as evidenced by decreased adipose tissue TNF-α production and macrophage infiltration, do not improve insulin sensitivity in an HFD-induced obese model.44 Furthermore, it is only within the last several years that Philipp Scherer’s group elegantly showed that adipose tissue inflammation is necessary for “healthy” adipose tissue expansion and suppressing adipocyte inflammation promotes insulin resistance.45,46 The authors found that the inhibition of a pro-inflammatory response in adipose tissue resulted in ectopic lipid deposition, aggravated systemic inflammation, and impaired glucose metabolism. One of the models the authors utilized was a novel adipose tissue (aP2 promoter-driven) dominant-negative TNF-α transgenic mouse model (dnTNF tg) which produces variant TNF proteins that form heterotrimers with native TNF, thus abolishing TNF signaling.45 To the authors’ surprise, and in contrast to the findings of others who have shown the deletion of TNF-α improves metabolic processes in obesity, the dnTNF tg mice weighed less and displayed metabolic and adipose tissue dysfunction compared to their littermate controls. These results suggest a necessary role of adipose tissue TNF-α to regulate adipose tissue and systemic metabolic homeostasis, however, this was not corroborated in our investigation. The discrepancies between our results and those of others who have manipulated TNF-α to study the cytokine’s effect on metabolic processes in obesity may be explained by many factors including, but not limited to, use of littermate controls, the model of obesity (eg, genetic or HFD-induced), the length of HFD feeding, and the differences in microbiomes attributed to different animal facilities.
It is evident is that our results paired with those of others warrant further investigations to better understand how adipose tissue inflammation and metabolism work in concert in the quest to maintain systemic equilibrium under hypercaloric conditions. Future studies utilizing inducible, tissue-specific transgenic animal models will enhance our understanding of the role that adipose tissue inflammation plays in regulating local and global metabolic processes.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to acknowledge the Functional Genomics Core, COBRE Center for Targeted Therapeutics, University of South Carolina, College of Pharmacy for their help with the microarray experiment and Dr Sergei Grivennikov and Dr Sergei Nedospasov for the TNF-αFlox/Flox mice.
Funding information
This work was supported by grants K01-AT010348 (NIH—N CCIH) and the University of South Carolina Advanced
Support for Innovative Research Excellence (ASPIRE) to RTE. KTV is supported by R00-AT009206 (NIH—N CCIH)
Abbreviations:
- FFA
free-fatty acid
- GTT
glucose tolerance test
- HFD
high-fat diet
- ITT
insulin tolerance test
- LFD
low-fat diet
- TNF-α
tumor necrosis factor-alpha
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Bluher M. Obesity: global epidemiology and pathogenesis. Nat Rev Endocrinol. 2019;15:288–298. [DOI] [PubMed] [Google Scholar]
- 2.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW Jr. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black RA, Rauch CT, Kozlosky CJ, et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385:729–733. [DOI] [PubMed] [Google Scholar]
- 4.Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. [DOI] [PubMed] [Google Scholar]
- 5.Hotamisligil GS, Arner P, Caro JF, Atkinson RL, Spiegelman BM. Increased adipose tissue expression of tumor necrosis factor-alpha in human obesity and insulin resistance. J Clin Invest. 1995;95:2409–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hotamisligil GS, Budavari A, Murray D, Spiegelman BM. Reduced tyrosine kinase activity of the insulin receptor in obesity-diabetes. Central role of tumor necrosis factor-alpha. J Clin Invest. 1994;94:1543–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uysal KT, Wiesbrock SM, Marino MW, Hotamisligil GS. Protection from obesity-induced insulin resistance in mice lacking TNF-alpha function. Nature. 1997;389:610–614. [DOI] [PubMed] [Google Scholar]
- 8.Salles J, Tardif N, Landrier JF, et al. TNFalpha gene knockout differentially affects lipid deposition in liver and skeletal muscle of high-fat-diet mice. J Nutr Biochem. 2012;23:1685–1693. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Gong Q, Wang CY, et al. High-Fat Diet Induces Distinct Metabolic Response in Interleukin-6 and Tumor Necrosis Factor-alpha Knockout Mice. J Interferon Cytokine Res. 2016;36:580–588. [DOI] [PubMed] [Google Scholar]
- 10.Schreyer SA, Chua SC Jr, LeBoeuf RC. Obesity and diabetes in TNF-alpha receptor-deficient mice. J Clin Invest. 1998;102:402–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cawthorn WP, Sethi JK. TNF-a lpha and adipocyte biology. FEBS Lett. 2008;582:117–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fain JN, Bahouth SW, Madan AK. TNFalpha release by the non-fat cells of human adipose tissue. Int J Obes Relat Metab Disord. 2004;28:616–622. [DOI] [PubMed] [Google Scholar]
- 13.Grivennikov SI, Tumanov AV, Liepinsh DJ, et al. Distinct and nonredundant in vivo functions of TNF produced by t cells and macrophages/neutrophils: protective and deleterious effects. Immunity. 2005;22:93–104. [DOI] [PubMed] [Google Scholar]
- 14.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. [DOI] [PubMed] [Google Scholar]
- 15.Bader J, Carson M, Enos R, et al. High-fat diet-fed ovariectomized mice are susceptible to accelerated subcutaneous tumor growth potentially through adipose tissue inflammation, local insulin-like growth factor release, and tumor associated macrophages. Oncotarget. 2020;11:4554–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enos RT, Velazquez KT, McClellan JL, Cranford TL, Walla MD, Murphy EA. Lowering the dietary omega-6: omega-3 does not hinder nonalcoholic fatty-liver disease development in a murine model. Nutr Res. 2015;35:449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enos RT, Velazquez KT, Murphy EA. Insight into the impact of dietary saturated fat on tissue-specific cellular processes underlying obesity-related diseases. J Nutr Biochem. 2014;25:600–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enos RT, Velazquez KT, McClellan JL, et al. High-fat diets rich in saturated fat protect against azoxymethane/dextran sulfate sodium-induced colon cancer. Am J Physiol Gastrointest Liver Physiol. 2016;310:G906–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Velazquez KT, Enos RT, Carson MS, et al. miR155 deficiency aggravates high-fat diet-induced adipose tissue fibrosis in male mice. Physiol Rep. 2017;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bader JE, Enos RT, Velazquez KT, et al. Repeated clodronate-liposome treatment results in neutrophilia and is not effective in limiting obesity-linked metabolic impairments. Am J Physiol Endocrinol Metab. 2019;316:E358–E372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cranford TL, Enos RT, Velazquez KT, et al. Role of MCP-1 on inflammatory processes and metabolic dysfunction following high-fat feedings in the FVB/N strain. Int J Obes (Lond). 2016;40:844–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enos RT, Davis JM, Velazquez KT, et al. Influence of dietary saturated fat content on adiposity, macrophage behavior, inflammation, and metabolism: composition matters. J Lipid Res. 2013;54:152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enos RT, Velazquez KT, Carson MS, et al. A Low Dose of Dietary Quercetin Fails to Protect against the Development of an Obese Phenotype in Mice. PLoS One. 2016;11:e0167979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wascher TC, Lindeman JH, Sourij H, Kooistra T, Pacini G, Roden M. Chronic TNF-alpha neutralization does not improve insulin resistance or endothelial function in “healthy” men with metabolic syndrome. Mol Med. 2011;17:189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ofei F, Hurel S, Newkirk J, Sopwith M, Taylor R. Effects of an engineered human anti-TNF-alpha antibody (CDP571) on insulin sensitivity and glycemic control in patients with NIDDM. Diabetes. 1996;45:881–885. [DOI] [PubMed] [Google Scholar]
- 27.Dominguez H, Storgaard H, Rask-M adsen C, et al. Metabolic and vascular effects of tumor necrosis factor-alpha blockade with etanercept in obese patients with type 2 diabetes. J Vasc Res. 2005;42:517–525. [DOI] [PubMed] [Google Scholar]
- 28.Paquot N, Castillo MJ, Lefebvre PJ, Scheen AJ. No increased insulin sensitivity after a single intravenous administration of a recombinant human tumor necrosis factor receptor: Fc fusion protein in obese insulin-resistant patients. J Clin Endocrinol Metab. 2000;85:1316–1319. [DOI] [PubMed] [Google Scholar]
- 29.Ventre J, Doebber T, Wu M, et al. Targeted disruption of the tumor necrosis factor-alpha gene: metabolic consequences in obese and nonobese mice. Diabetes. 1997;46:1526–1531. [DOI] [PubMed] [Google Scholar]
- 30.Uysal KT, Wiesbrock SM, Hotamisligil GS. Functional analysis of tumor necrosis factor (TNF) receptors in TNF-alpha-mediated insulin resistance in genetic obesity. Endocrinology. 1998;139:4832–4838. [DOI] [PubMed] [Google Scholar]
- 31.De Taeye BM, Novitskaya T, McGuinness OP, et al. Macrophage TNF-alpha contributes to insulin resistance and hepatic steatosis in diet-induced obesity. Am J Physiol Endocrinol Metab. 2007;293:E713–725. [DOI] [PubMed] [Google Scholar]
- 32.Jager J, Aparicio-Vergara M, Aouadi M. Liver innate immune cells and insulin resistance: the multiple facets of Kupffer cells. J Intern Med. 2016;280:209–220. [DOI] [PubMed] [Google Scholar]
- 33.Tencerova M, Aouadi M, Vangala P, et al. Activated Kupffer cells inhibit insulin sensitivity in obese mice. FASEB J. 2015;29:2959–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lanthier N, Molendi-Coste O, Horsmans Y, van Rooijen N, Cani PD, Leclercq IA. Kupffer cell activation is a causal factor for hepatic insulin resistance. Am J Physiol Gastrointest Liver Physiol. 2010;298:G107–116. [DOI] [PubMed] [Google Scholar]
- 35.Hayden MS, Ghosh S. Regulation of NF-kappaB by TNF family cytokines. Semin Immunol. 2014;26:253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang WS, Han NJ, Kim JJ, Lee MJ, Park SK. TNF-alpha Activates High-Mobility Group Box 1 - Toll-Like Receptor 4 Signaling Pathway in Human Aortic Endothelial Cells. Cell Physiol Biochem. 2016;38:2139–2151. [DOI] [PubMed] [Google Scholar]
- 37.Hermoso MA, Matsuguchi T, Smoak K, Cidlowski JA. Glucocorticoids and tumor necrosis factor alpha cooperatively regulate toll-like receptor 2 gene expression. Mol Cell Biol. 2004;24:4743–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sukkar MB, Xie S, Khorasani NM, et al. Toll-like receptor 2, 3, and 4 expression and function in human airway smooth muscle. J Allergy Clin Immunol. 2006;118:641–648. [DOI] [PubMed] [Google Scholar]
- 39.Szoka PR, Gallagher JF, Held WA. In vitro synthesis and characterization of precursors to the mouse major urinary proteins. J Biol Chem. 1980;255:1367–1373. [PubMed] [Google Scholar]
- 40.Charkoftaki G, Wang Y, McAndrews M, et al. Update on the human and mouse lipocalin (LCN) gene family, including evidence the mouse Mup cluster is result of an “evolutionary bloom”. Hum Genomics. 2019;13:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hui X, Zhu W, Wang Y, et al. Major urinary protein-1 increases energy expenditure and improves glucose intolerance through enhancing mitochondrial function in skeletal muscle of diabetic mice. J Biol Chem. 2009;284:14050–14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pedley AM, Benkovic SJ. A New View into the Regulation of Purine Metabolism: The Purinosome. Trends Biochem Sci. 2017;42:141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner N, Kowalski GM, Leslie SJ, et al. Distinct patterns of tissue-specific lipid accumulation during the induction of insulin resistance in mice by high-fat feeding. Diabetologia. 2013;56:1638–1648. [DOI] [PubMed] [Google Scholar]
- 44.Skurski J, Penniman CM, Geesala R, et al. Loss of iRhom2 accelerates fat gain and insulin resistance in diet-induced obesity despite reduced adipose tissue inflammation. Metabolism. 2020;106:154194. [DOI] [PubMed] [Google Scholar]
- 45.Wernstedt Asterholm I, Tao C, Morley TS, et al. Adipocyte inflammation is essential for healthy adipose tissue expansion and remodeling. Cell Metab. 2014;20:103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu Q, An YA, Kim M, et al. Suppressing adipocyte inflammation promotes insulin resistance in mice. Mol Metab. 2020;39:101010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.