Abstract
PCR-restriction fragment length polymorphism analysis (PRA) using the novel region of the rpoB gene was developed for rapid and precise identification of mycobacteria to the species level. A total of 50 mycobacterial reference strains and 3 related bacterial strains were used to amplify the 360-bp region of rpoB, and the amplified DNAs were subsequently digested with restriction enzymes such as MspI and HaeIII. The results from this study clearly show that most of the mycobacterial species were easily differentiated at the species level by this PRA method. In addition, species with several subtypes, such as Mycobacterium gordonae, M. kansasii, M. celatum, and M. fortuitum, were also differentiated by this PRA method. Subsequently, an algorithm was constructed based on the results, and a blinded test was carried out with more than 260 clinical isolates that had been identified on the basis of conventional tests. Comparison of these two sets of results clearly indicates that this new PRA method based on the rpoB gene is more simple, more rapid, and more accurate than conventional procedures for differentiating mycobacterial species.
Since the early 1980s there has been an increase in disease caused by organisms called nontuberculous mycobacteria (NTM), which is the generic name for mycobacteria other than Mycobacterium tuberculosis and M. leprae. They affect both immune-competent and immune-compromised persons, and patients with the human immunodeficiency virus are known to be especially vulnerable. The NTM most frequently involved in disease cases are M. avium complex, M. kansasii, M. chelonae, M. abscessus, M. xenopi, M. malmoense, M. scrofulaceum, M. marinum, M. ulcerans, and M. haemophilum (28). Clinical diagnosis and treatment of NTM infections are an increasingly frequent challenge to clinicians.
Currently, identification of clinical isolates of mycobacteria to the species level is primarily based on cultural characteristics and biochemical tests. These conventional tests take several weeks, and they sometimes fail to provide precise identification. The procedures for these tests are complex and laborious, and they are usually impeded by the slow growth of mycobacteria in clinical laboratories. Additional methods, such as high-performance liquid chromatography, gas-liquid chromatography, and thin-layer chromatography (5, 21, 36) as well as DNA sequence analysis (3, 4, 15–17, 19, 26, 31, 32), can differentiate mycobacteria to the species level, but these techniques are labor intensive and difficult to perform for routine use in clinical laboratories. Recently developed molecular techniques using amplified DNA and probe hybridization (6, 8–10, 18, 20, 23) are also useful for direct and rapid identification of NTM species, but their overall cost is high. In addition, currently available kits are limited to the identification of a few species. In contrast to the above-mentioned techniques, PCR-restriction fragment length polymorphism analysis (PRA) offers an easy, rapid, and inexpensive way to identify several mycobacterial species in a single experiment. PRA techniques have been developed to target mycobacterial genes which are present in all mycobacteria, such as hsp65 (7, 11, 25, 29, 30, 34, 35), the 16S rRNA gene (2, 14, 37), and dnaJ (33). However, the previous PRA techniques are still cumbersome since they require several enzyme digestions for species identification, and the results are not easy to interpret for species identification due to the limited size differences of DNA fragments after digestion.
Therefore, the aim of this study was to develop a new PRA method that is easier to perform and more precise for mycobacterial species identification than currently available PRA techniques. We chose as a target for our new PRA method rpoB, a conserved gene that encodes the β subunit of RNA polymerase. The information-rich nature of the rpoB gene has been recently used in differentiation of mycobacteria by DNA hybridization array (10) and by DNA sequence (16) analyses. However, the rpoB region used in these previous studies has limited sequence differences, making it difficult to generate diverse restriction fragment length polymorphism (RFLP) profiles that can be used for species identification of mycobacteria. In the present study, we extended the genetic knowledge of the rpoB gene to a region that is relatively polymorphic and thus suitable for PRA. In addition, this region of the rpoB gene is flanked by conserved sequences, making it suitable for PCR amplification with the same set of PCR primers.
A 360-bp region of rpoB was amplified from the DNA of 50 mycobacterial reference strains, representing 44 species and a total of 260 clinical isolates, to evaluate this method for mycobacterial species identification. The results of this study show that this novel PRA method based on the rpoB genes of mycobacteria generates clear and distinctive results for easy, rapid, and precise identification of mycobacterial species.
MATERIALS AND METHODS
Mycobacterial samples.
A total of 50 mycobacterial reference strains representing 44 mycobacterial species and 3 related species which belong to 2 different genera (Table 1) were used to develop the new PRA method in this study. Among them, 40 mycobacterial strains and 3 related species were obtained from the Korean Institute of Tuberculosis (KIT), the Korean National Tuberculosis Association in Seoul. Four species were obtained from the Korean Collection for Type Cultures at the Korean Research Institute of Bioscience and Biotechnology (KRIBB), and M. abscessus, which was recently separated from M. chelonae as an independent new species, was obtained from the Department of Clinical Pathology at Yonsei University Medical College. Five subtypes of M. kansasii were generously given by V. Vincent of the Laboratoire de Référence des Mycobactéries, Institut Pasteur, Paris, France.
TABLE 1.
Bacterial strains used in this study
| Species | Strain | Source |
|---|---|---|
| M. abscessus | Pettenkofer Institute | YUMCa |
| M. africanum | ATCC 25420 | KIT |
| M. arcinogenes | ATCC 35753 | KIT |
| M. asiaticum | ATCC 25276 | KIT |
| M. aurum | ATCC 23366 | KIT |
| M. austroafricanum | ATCC 33464 | KRIBB |
| M. avium | ATCC 25291 | KIT |
| M. bovis | ATCC 19210 | KIT |
| M. bovis BCG | French strain 1173P2 | KIT |
| M. celatum type I/II | ATCC 51130/ATCC 51131 | KIT |
| M. chelonae | ATCC 35749 | KIT |
| M. chitae | ATCC 19627 | KIT |
| M. fallax | ATCC 35219 | KIT |
| M. fortuitum type I/II | ATCC 6841/ATCC 49404 | KIT |
| M. gallinarum | ATCC 19710 | KRIBB |
| M. gastri | ATCC 15754 | KIT |
| M. genavense | ATCC 51233 | KIT |
| M. gilvum | ATCC 43909 | KIT |
| M. gordonae | ATCC 14470 | KIT |
| M. haemophilum | ATCC 29548 | KIT |
| M. intracellulare | ATCC 13950 | KIT |
| M. interjectum | ATCC 51457 | KIT |
| M. intermedium | ATCC 51848 | KIT |
| M. kansasii type I-V | Pasteur Institute | |
| M. malmoense | ATCC 29571 | KIT |
| M. marinum | ATCC 927 | KIT |
| M. moriokaense | ATCC 43059 | KRIBB |
| M. mucogenicum | ATCC 49650 | KIT |
| M. neoaurum | ATCC 25795 | KIT |
| M. nonchromogenicum | ATCC 19530 | KIT |
| M. parafortuitum | ATCC 19686 | KIT |
| M. peregrinum | ATCC 14467 | KIT |
| M. phlei | ATCC 11758 | KIT |
| M. pulveris | ATCC 35154 | KRIBB |
| M. scrofulaceum | ATCC 19981 | KIT |
| M. smegmatis | ATCC 19420 | KIT |
| M. szulgai | ATCC 35799 | KIT |
| M. terrae | ATCC 15755 | KIT |
| M. thermoresistibile | ATCC 19527 | KIT |
| M. triviale | ATCC 23292 | KIT |
| M. tuberculosis H37Rv | ATCC 27294 | KIT |
| M. ulcerans | ATCC 19423 | KIT |
| M. vaccae | ATCC 15483 | KIT |
| M. xenopi | ATCC 19250 | KIT |
| Nocardia brasiliense | ATCC 19296 | KIT |
| N. nova | ATCC 21197 | KIT |
| Rhodococcus equi | ATCC 10146 | KIT |
YUMC, Yonsei University Medical College.
Clinical isolates subjected to PRA for evaluation of the new method were obtained from KIT. All clinical isolates used in this study were identified on the basis of conventional techniques that included microbiological characteristics and biochemical tests. In some cases, strains were subjected to the conventional PRA method, based on the hsp65 gene (7, 35), to aid in their precise identification.
DNA preparation.
To prepare a DNA sample for PCR amplification, a loopful of a bacterial colony was taken from a Lowenstein-Jensen medium culture and resuspended in 400 μl of distilled water in a screw-cap microcentrifuge tube. The sample was then boiled for 5 min prior to being centrifuged for 5 min to settle cell debris, and about 10 μl of supernatant, containing the genomic DNA, was used for subsequent PCR amplification.
PCR amplification.
Amplification of the region of interest in rpoB with a primer set consisting of 5′-TCAAGGAGAAGCGCTACGA-3′ (RPO5′) and 5′-GGATGTTGATCAGGGTCTGC-3′ (RPO3′) resulted in 360-bp PCR products (bases 902 to 1261 and codons 302 to 420, based on the sequence of the rpoB gene of M. tuberculosis [GenBank accession no. P47766]). The primer sequences were selected from the region of the rpoB gene which have been previously identified from M. tuberculosis, M. leprae, and M. smegmatis (12, 13, 22). The primers amplified the region between the first variable region (V1) and second conserved region (C2) based on the genetic information for the rpoB gene of Escherichia coli. As a result, the PCR products included 171 bp of variable region and 189 bp of conserved region. The variable region was amplified in this experiment based on the theory that the polymorphic nature of this region might help in the clear distinction of each mycobacterial species by the new PRA method. The RPO5′ primer was derived from relatively conserved sequences found in the variable region of rpoB.
PCR was carried out in a final volume of 50 μl consisting of 10 μl of DNA supernatant containing approximately 10 ng of genomic DNA, 10 pmol of each primer, 2 mM MgCl2, 200 μM deoxynucleoside triphosphates, and 1 U of DyNAzyme II DNA polymerase (FINNZYMESY, Espoo, Finland). DNA samples were first denatured completely by incubation at 94°C for 5 min before the amplification cycle; then DNA was amplified by subjecting it to 35 cycles of (i) denaturation at 94°C for 1 min, (ii) primer annealing at 58°C for 1 min, and (iii) elongation at 72°C for 1 min, using a Thermocycler (model 9600, Perkin-Elmer). After the last amplification cycle, the samples were incubated further at 72°C for 7 min for complete elongation of the final PCR products. Positive and negative controls were always included in each PCR. The positive control was the PCR mixture with DNA of the reference strain, M. bovis, and the negative control was the PCR mixture without any DNA. After the PCR, the amplification results were visualized by performing 1.5% agarose gel electrophoresis and ethidium bromide staining.
RFLP.
After successful amplification of the 360-bp PCR products was confirmed, the PCR products were subjected to restriction enzyme digestion. Most of the time, 10 to 16 μl of PCR product (approximately 1 to 1.5 μg of DNA) was digested in a 20-μl reaction volume containing 5 U of MspI (Boehringer Mannheim Biochemicals, Mannheim, Germany) and 2 μl of the 10× reaction buffer supplied by manufacturer. Similarly, 10 to 16 μl of PCR product was digested in a 20-μl reaction volume containing 5 U of HaeIII enzyme (Takara Shuzo Co., Ltd., Shiga, Japan) and the corresponding enzyme buffer. After 2 h of incubation at 37°C, 4 μl of gel loading buffer (0.25% bromophenol blue, 40% sucrose in water) was added, and the samples were loaded onto a 4% Metaphore agarose gel (FMC BioProducts, Rockland, Maine). Then, enzyme-digested fragments were visualized under UV light after ethidium bromide staining.
For the interpretation of the PRA profile generated by each species, a 50-bp ladder DNA size marker (Boehringer Mannheim) and the PRA profile of M. bovis, which generates restriction fragments of about 175, 80, 60, and 40 bp, were used as internal size markers. Using these markers, the sizes of the restricted fragments of each species were determined, and an algorithm was constructed based on this information.
RESULTS
To develop a more rapid and precise yet simple and easy-to-use PRA method for mycobacterial species identification, we chose one of the more highly conserved genes, rpoB. Since the genetic information for the rpoB genes of some mycobacteria is available, sequences were aligned and searched for regions suitable for PRA. As a result, a set of PCR primers was selected to amplify a 360-bp sequence of rpoB which contains a polymorphic region flanked by conserved regions.
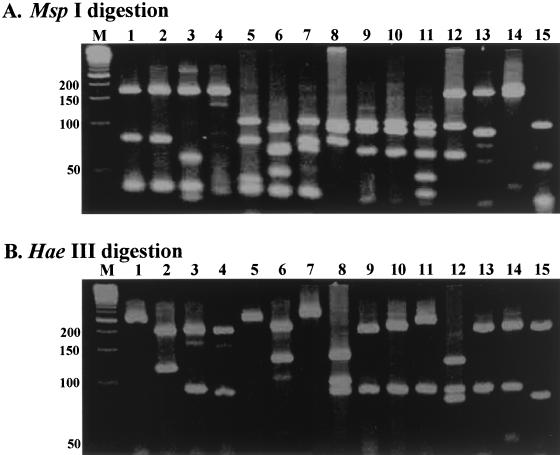
A total of 50 mycobacterial reference strains and 3 related bacterial strains that belong to the same class as mycobacteria (Actinomycetes) were used to amplify the 360-bp region of the rpoB gene (Table 1). The results showed that a conserved rpoB gene present in all mycobacteria and in some other bacteria, such as Nocardia and Rhodococcus species, was amplified. Subsequently, PCR products were subjected to two sets of restriction enzyme digestion, with MspI and HaeIII being used individually. These two enzymes were selected on the basis of sequence information for the rpoB genes of M. tuberculosis, M. leprae, and M. smegmatis (12, 13, 22). Based on this information, PCR products were subsequently subjected to RFLP analysis (Fig. 1). In short, this analysis showed that the RFLP profiles of PCR products from each mycobacterial species were distinctive. M. kansasii can be easily differentiated from M. gastri, which has much in common with nonpigmented variants of M. kansasii. In addition, M. abscessus, which had been classified as a subgroup of M. chelonae and was not easily differentiated from it by conventional biochemical tests, was also differentiated from the latter. Furthermore, for some species, such as M. fortuitum, M. celatum, M. gordonae, and M. kansasii, that are known to contain several subtypes, each subtype generated distinctive restriction profiles. Therefore, the results clearly indicated that this new PRA method could differentiate mycobacteria at the species and even the subspecies level.
FIG. 1.
Results of PRA for mycobacterial reference strains. Primer pair RPO5′ and RPO3′ was used. Amplified DNA was digested with MspI (A) or HaeIII (B) restriction enzyme and run on a 4% Metaphore agarose gel. Lanes: M, DNA size marker (50-bp ladder; positions are indicated on left [in base pairs]); 1, M. gordonae type IV; 2, M. szulgai; 3, M. kansasii type I; 4, M. gallinarum; 5, M. avium; 6, M. scrofulaceum; 7, M. asiaticum; 8, M. chelonae; 9, M. moriokaese; 10, M. phlei; 11, M. pulveris; 12, M. fortuitum type I; 13, M. austroafricanum; 14, M. smegmatis; 15, M. marinum.
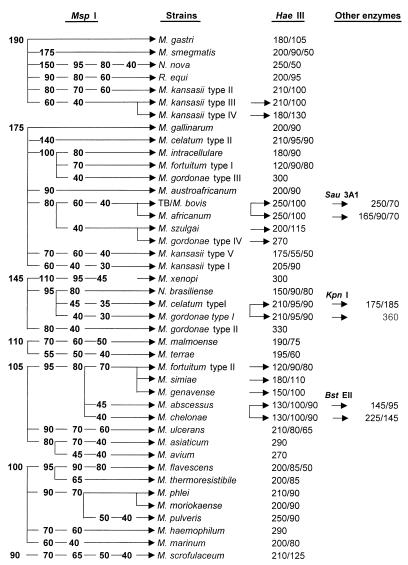
On the basis of the above PRA results, an algorithm was constructed (Fig. 2). In the algorithm, restriction fragments smaller than 40 bp were omitted in order to reduce confusion with primer-dimer bands. The different sizes of fragments were clearly separated from each other, making interpretation of results easier. In brief, the algorithm clearly shows that most mycobacterial species and other related bacterial species can be differentiated at the subspecies level by PRA using restriction enzymes MspI and HaeIII. In fact, except for a few mycobacterial species, most of these organisms can be identified by using a single enzyme, MspI, thus making this new method more useful for mycobacterial species identification in the clinical laboratory.
FIG. 2.
An algorithm was constructed based on the results of PRA with 40 mycobacterial reference strains and 3 other related bacterial strains. For algorithm conciseness, the PRA results of 10 other mycobacterial reference strains are not listed in this figure. TB, M. tuberculosis. The numbers indicate sizes (in base pairs) of resulting fragments.
For those strains that were not differentiated by two enzyme digestions, a third enzyme digestion was useful. For example, even though the members of M. tuberculosis complex (M. tuberculosis, M. bovis, and M. africanum) were not differentiated by using MspI and HaeIII, a third enzyme, Sau3AI, could be used to differentiate M. africanum from other members of M. tuberculosis complex. As another example, HincII can be used to differentiate M. gordonae type I from M. celatum type I (Fig. 2).
The different RFLP profiles generated for the PCR products strongly suggested to us that the rpoB region amplified by PCR in this study has a polymorphic nature. The next question was whether these differing RFLP profiles were species and, possibly, strain specific. If strains of a certain species also have polymorphic RFLP profiles, the use of this method for mycobacterial species identification will be very complex. Therefore, in a blinded test, clinical isolates that had been identified on the basis of conventional tests were subjected to PRA to determine their species based on the algorithm constructed during this study. The results of this experiment clearly show that there are no differences in the profiles of the various among clinical isolates that belong to the same species (data not shown). Subsequently, a substantial number of clinical isolates that had been identified on the basis of conventional tests were subjected to PRA. In this experiment, a total of 260 clinical isolates were analyzed, including M. tuberculosis, M. avium, M. intracellulare, M. fortuitum, M. chelonae, M. abscessus, M. terrae, M. gordonae, and M. szulgai specimens (Table 2). For easy interpretation of the PRA profiles generated by each clinical isolate, a 50-bp ladder size marker was used as a standard size marker and the PRA profile of M. bovis was used as an internal size marker (Fig. 3). Results from the PRA of clinical isolates were evaluated with the help of an algorithm generated on the basis of PRA profiles of reference strains. Most of the PRA results were consistent with conventional test results, while the PRA profiles of a few strains were not present in the reference algorithm. Based on the results of conventional tests and on molecular biological sequence analysis, some of these strains were determined to be M. terrae complex.
TABLE 2.
Clinical isolates of mycobacteria subjected to species identification by the new PRA method
| Species tested | No. of clinical isolates tested |
|---|---|
| M. tuberculosis | 40 |
| M. avium | 40 |
| M. intracellulare | 50 |
| M. gordonae | 25 |
| M. szulgai | 10 |
| M. fortuitum | 25 |
| M. chelonae | 15 |
| M. abscessus | 15 |
| M. kansasii | 20 |
| M. terrae | 20 |
| Total | 260 |
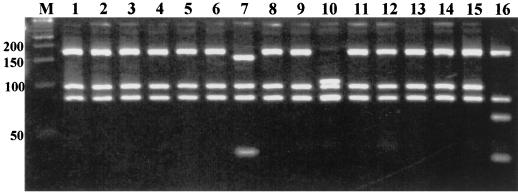
FIG. 3.
Application of rpoB-based PRA for species identification of mycobacterial isolates in the clinical laboratory. Clinical isolates were amplified using primers RPO5′ and RPO3′, digested with MspI, and run on a 4% Metaphore agarose gel. For each test, DNA size markers (lane M; 50-bp ladder (positions are indicated on the left; [in base pairs]) was employed, as were the PRA results for M. bovis, which were used as internal size markers (lane 16). Using the algorithm shown in Fig. 2, these clinical isolates were determined to be M. intracellulare (lanes 1 to 6, 8, 9, and 11 to 15), M. gordonae type II (lane 7), and M. abscessus (lane 10).
DISCUSSION
Mycobacterial identification to the species level not only is of academic interest but also is important because it provides a great deal of useful information on the epidemiology and pathogenesis of the organism, suggesting potential intervention strategies, including successful treatment of patients on a clinical basis. It is therefore important to develop methods that are rapid and simple but yet precise and cost-effective for use in a wide variety of clinical laboratories around the world. Currently available methods for the differentiation of mycobacteria to the species level are time-consuming evaluations based on phenotypic and biochemical tests or laborious procedures requiring expensive equipment.
The PRA technique certainly fits these requirements better than other molecular methods. It is rapid and precise since it employs PCR, and it is simple and cost-effective since it does not require any expensive equipment or laborious processes and can differentiate numerous species of mycobacteria within a single experiment. Owing to these advantages, several PRA methods based on different genes of mycobacteria have been developed (2, 7, 11, 14, 25, 29, 30, 33–35, 37). However, most of those methods require the use of more than two enzymes to differentiate mycobacteria at the species level, as well as a computer-assisted software program to differentiate restriction fragments since the profiles of certain mycobacterial species are not distinctive enough for bare-eyed interpretation.
The new PRA method developed through this investigation has more advantages than the previous ones. As presented in Fig. 1, it is apparent that most of the species have unique PRA profiles. Unlike other PRA profiles, which may require computer-assisted analysis and interpretation of the gels, ours does not face problems in terms of resolution of all patterns obtained during the experiments. Furthermore, problems such as gel-to-gel variation and confusion with regard to the sizes of the restriction fragments were limited by the use of the 50-bp size marker and of the M. bovis PRA profile as an internal size marker.
The restriction analysis of a 360-bp fragment within this gene after single MspI digestion is highly effective for differentiating most mycobacteria even at the species level. Only a few species require digestion with an additional enzyme, such as HaeIII, Sau3AI, or HincII. For some species, such as M. gordonae, M. kansasii, M. fortuitum, and M. celatum, the discrimination was even to the subtype level. For M. kansasii, this subdivision was clearly linked to genetic divergence observed previously by other investigators (1, 24, 27). It is therefore possible that by using this PRA method, discrimination at the subgroup level for other species could be similarly linked to bacteriological and clinical specificities. Currently, we are investigating several clinical isolates, with PRA profiles distinct from those of any of our reference strains, which also exhibit distinctive microbiological and biochemical characteristics.
On the other hand, the four members of the M. tuberculosis complex, which are difficult to differentiate by methods such as sequence analysis and high-performance liquid chromatography of mycolic acids, were also undistinguishable by our PRA method, confirming that they are genetically similar. However, unlike other methods, this new PRA method can further differentiate M. africanum from the other M. tuberculosis complex members with the added step of Sau3AI digestion. Therefore, in cases in which a clinical isolate exhibits M. tuberculosis complex profiles, PCR products can be further processed with Sau3AI to differentiate M. africanum from other M. tuberculosis complex constituents. In addition, M. tuberculosis and M. bovis can be differentiated by PCR amplification using PCR primers derived from the esat-6 gene, which is known to be present only in the genome of M. tuberculosis.
The fact that the species M. terrae contains strains of diverse genotype is consistent with previous observations by other investigators (35). Currently, we are investigating the genetic nature of these strains by sequence analysis and their phenotypic specificities by conventional tests. The data from these studies will be used for establishing a new mycobacterial taxonomy. In this regard, PRA seems to be a very effective method for identifying new mycobacterial species or subgroups of mycobacteria from clinical samples.
Currently, a substantial number of mycobacterial clinical isolates in our laboratory have now been identified by our new PRA method in parallel with other reference methods, including conventional tests and molecular biological methods such as PRA based on the hsp65 gene and sequence analysis based on the rpoB gene. From our experimental data it can be concluded that this new PRA is a rapid, cost-effective, and efficient method for the identification of mycobacteria in a clinical microbiology laboratory. The whole procedure can be done in 2 days when a culture is used. Successful PRA has been achieved with a loopful of culture taken from solid medium or 100 μl of a liquid culture such as MGIT for mycobacterial species identification. Both of the systems work well even when the genomic DNA is simply boiled for 5 min.
REFERENCES
- 1.Abed Y, Bollet C, de Micco P. Demonstration of Mycobacterium kansasii species heterogeneity by the amplification of the 16S-23S spacer region. J Med Microbiol. 1995;43:156–158. doi: 10.1099/00222615-43-2-156. [DOI] [PubMed] [Google Scholar]
- 2.Avaniss-Aghajani E, Jones K, Holtzman A, Aronson T, Glover N, Boian M, Froman S, Brunk C F. Molecular technique for rapid identification of mycobacteria. J Clin Microbiol. 1996;34:98–102. doi: 10.1128/jcm.34.1.98-102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böddinghaus B, Rogall T, Flohr T, Blöcker H, Böttger E C. Detection and identification of mycobacteria by amplification of rRNA. J Clin Microbiol. 1990;28:1751–1759. doi: 10.1128/jcm.28.8.1751-1759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosne S, Lévy-Frébault V. Mycobactin analysis as an aid for the identification of Mycobacterium fortuitum and Mycobacterium chelonae subspecies. J Clin Microbiol. 1992;30:1225–1231. doi: 10.1128/jcm.30.5.1225-1231.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler W R, Jost K C, Jr, Kilburn J O. Identification of mycobacteria by high-performance liquid chromatography. J Clin Microbiol. 1991;29:2468–2472. doi: 10.1128/jcm.29.11.2468-2472.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devallois A, Picardeau M, Goh K S, Sola C, Vincent V, Rastogi N. Comparative evaluation of PCR and commercial DNA probes for detection and identification to species level of Mycobacterium avium and Mycobacterium intracellulare. J Clin Microbiol. 1996;34:2756–2759. doi: 10.1128/jcm.34.11.2756-2759.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devallois A, Goh K S, Rastogi N. Rapid identification of mycobacteria to species level by PCR-restriction fragment length polymorphism analysis of the hsp65 gene and proposition of an algorithm to differentiate 34 mycobacterial species. J Clin Microbiol. 1997;35:2969–2973. doi: 10.1128/jcm.35.11.2969-2973.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiss E, Chehab F, Brooks G. DNA amplification and reverse dot blot hybridization for detection and identification of mycobacteria to the species level in the clinical laboratory. J Clin Microbiol. 1992;30:1220–1224. doi: 10.1128/jcm.30.5.1220-1224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fries J, Patel R, Piessens W, Wirth D. Genus- and species-specific DNA probes to identify mycobacteria using the polymerase chain reaction. Mol Cell Probes. 1990;4:87–105. doi: 10.1016/0890-8508(90)90011-n. [DOI] [PubMed] [Google Scholar]
- 10.Gingeras T R, Ghandour G, Wang E, Berno A, Small P M, Drobniewski F, Alland D, Desmond E, Holokniy M, Drenkow J. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic mycobacterium DNA arrays. Genome Res. 1998;8:435–448. doi: 10.1101/gr.8.5.435. [DOI] [PubMed] [Google Scholar]
- 11.Hance A J, Grandchamp B, Lévy-Frébault V, Lecossier D, Rauzier J, Bocart D, Gicquel B. Detection and identification of mycobacteria by amplification of mycobacterial DNA. Mol Microbiol. 1989;3:843–849. doi: 10.1111/j.1365-2958.1989.tb00233.x. [DOI] [PubMed] [Google Scholar]
- 12.Hetherington S V, Watson A S, Patrick C C. Sequence and analysis of the rpoB gene of Mycobacterium smegmatis. Antimicrob Agents Chemother. 1995;39:2164–2166. doi: 10.1128/aac.39.9.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Honore N T, Bergh S, Chanteau S, Doucet-Populaire F, Eiglmeier K, Garnier T, Geroges C, Launois P, Limpaiboon T, Newton S, Niang K, Del Portillo P, Ramesh G R, Reddi P, Ridel P R, Sittisombut N, Wu-Hunter S, Cole S T. Nucleotide sequence of the first cosmid from the Mycobacterium leprae genome project: structure and function of the Rif-Str regions. Mol Microbiol. 1993;7:207–214. doi: 10.1111/j.1365-2958.1993.tb01112.x. [DOI] [PubMed] [Google Scholar]
- 14.Hughes M S, Skuce R A, Beck L-A, Neill S D. Identification of mycobacteria from animals by restriction enzyme analysis and direct DNA cycle sequencing of polymerase chain reaction-amplified 16S rRNA gene sequences. J Clin Microbiol. 1993;31:3216–3222. doi: 10.1128/jcm.31.12.3216-3222.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapur V, Li L-L, Hamrick M R, Plikaytis B B, Shinnick T M, Telenti A, Jacobs W R, Banerjee A, Cole S, Yuen K Y, Clarridge J E, Kreiswirth B N, Musser J M. Rapid Mycobacterium species assignment and unambiguous identification of mutations associated with antimicrobial resistance in Mycobacterium tuberculosis by automated DNA sequencing. Arch Pathol Lab Med. 1995;119:131–138. [PubMed] [Google Scholar]
- 16.Kim B-J, Lee S-H, Lyu M-A, Kim S-J, Bai G-H, Kim S-J, Chae G-T, Kim E-C, Cha C-Y, Kook Y-H. Identification of mycobacterial species by comparative sequence analysis of the RNA polymerase gene (rpoB) J Clin Microbiol. 1999;37:1714–1720. doi: 10.1128/jcm.37.6.1714-1720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirschner P, Springer B, Vogel U, Meier A, Wrede A, Kiekenbeck M, Bange F-C, Böttger E C. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J Clin Microbiol. 1993;31:2882–2889. doi: 10.1128/jcm.31.11.2882-2889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kusunoki S, Ezaki T, Tamesada M, Hatanaka Y, Asano K, Hashimoto Y, Yabuuchi E. Application of colorimetric microdilution plate hybridization for rapid genetic identification of 22 mycobacterium species. J Clin Microbiol. 1991;29:1596–1603. doi: 10.1128/jcm.29.8.1596-1603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lévy-Frébault V, Daffé M, Goh K S, Lanéelle M-A, Asselineau C, David H L. Identification of Mycobacterium fortuitum and Mycobacterium chelonae. J Clin Microbiol. 1983;17:744–752. doi: 10.1128/jcm.17.5.744-752.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mabilat C, Desvarenne S, Panteix G, Machabert N, Bernillon M-H, Guardiola G, Cros P. Routine identification of Mycobacterium tuberculosis complex isolates by automated hybridization. J Clin Microbiol. 1994;32:2702–2705. doi: 10.1128/jcm.32.11.2702-2705.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marks J, Szulga T. Thin-layer chromatography of mycobacterial lipids as an aid to classification; technical procedures; Mycobacterium fortuitum. Tubercle. 1965;46:400–411. [Google Scholar]
- 22.Miller L P, Crawford J T, Shinnick T M. The rpoB gene of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1994;38:805–811. doi: 10.1128/aac.38.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Musial C E, Tice L S, Stockman L, Roberts G D. Identification of mycobacteria from culture by using the Gen-Probe rapid diagnostic system for Mycobacterium avium complex and Mycobacterium tuberculosis complex. J Clin Microbiol. 1988;26:2120–2123. doi: 10.1128/jcm.26.10.2120-2123.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Picardeau M, Prod'hom G, Raskine L, LePennec M P, Vincent V. Genotypic characterization of five subspecies of Mycobacterium kansasii. J Clin Microbiol. 1997;35:25–32. doi: 10.1128/jcm.35.1.25-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plikaytis B B, Plikaytis B D, Yakrus M A, Butler W R, Woodley C L, Silcox V A, Shinnick T M. Differentiation of slowly growing Mycobacterium species, including Mycobacterium tuberculosis, by gene amplification and restriction fragment length polymorphism analysis. J Clin Microbiol. 1992;30:1815–1822. doi: 10.1128/jcm.30.7.1815-1822.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogall T, Flohr T, Böttger E. Differentiation of mycobacterial species by direct sequencing of amplified DNA. J Gen Microbiol. 1990;136:1915–1920. doi: 10.1099/00221287-136-9-1915. [DOI] [PubMed] [Google Scholar]
- 27.Ross B C, Jackson K, Yang M, Sievers A, Dwyer B. Identification of a genetically distinct subspecies of Mycobacterium kansasii. J Clin Microbiol. 1992;30:2930–2933. doi: 10.1128/jcm.30.11.2930-2933.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shinners D, Yeager H., Jr . Nontuberculous mycobacterial infection: clinical syndromes and diagnosis: overview. In: Schlossberg D, editor. Tuberculosis and nontuberculous mycobacterial infections. 4th ed. Philadelphia, Pa: W. B. Saunders Co.; 1999. pp. 341–350. [Google Scholar]
- 29.Shinnick T M. The 65-kilodalton antigen of Mycobacterium tuberculosis. J Bacteriol. 1987;169:1080–1088. doi: 10.1128/jb.169.3.1080-1088.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shinnick T M, Vodkin M H, Williams J C. The Mycobacterium tuberculosis 65-kilodalton antigen is a heat shock protein which corresponds to common antigen and to the Escherichia coli GroEL protein. Infect Immun. 1988;56:446–451. doi: 10.1128/iai.56.2.446-451.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soini H, Böttger E C, Viljanen M K. Identification of mycobacteria by PCR-based sequence determination of the 32-kilodalton protein gene. J Clin Microbiol. 1994;32:2944–2947. doi: 10.1128/jcm.32.12.2944-2947.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Springer B, Stockman L, Teschner K, Roberts G D, Böttger E C. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J Clin Microbiol. 1996;34:296–303. doi: 10.1128/jcm.34.2.296-303.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takewaki S-I, Okuzumi K, Manabe I, Tanimura M, Miyamura K, Nakahara K-I, Yazaki Y, Ohkubo A, Nagai R. Nucleotide sequence comparison of the mycobacterial dnaJ gene and PCR-restriction fragment length polymorphism analysis for identification of mycobacterial species. Int J Syst Bacteriol. 1994;44:159–166. doi: 10.1099/00207713-44-1-159. [DOI] [PubMed] [Google Scholar]
- 34.Taylor T B, Patterson C, Hale Y, Safranek W W. Routine use of PCR-restriction fragment length polymorphism analysis for identification of mycobacteria growing in liquid media. J Clin Microbiol. 1997;35:79–85. doi: 10.1128/jcm.35.1.79-85.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Telenti A, Marchesi F, Balz M, Bally F, Böttger E C, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993;31:175–178. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsang A Y, Drupa I, Goldberg M, McClatchy J K, Brennan P J. Use of serology and thin-layer chromatography for the assembly of an authenticated collection of serovars within the Mycobacterium avium-Mycobacterium intracellulare-Mycobacterium scrofulaceum complex. Int J Syst Bacteriol. 1983;33:285–292. [Google Scholar]
- 37.Vaneechoutte M, Beenhouwer H D, Claeys G, Verschraegen G, Rouk A D, Paepe N, Elaichouni A, Portaels F. Identification of Mycobacterium species by using amplified ribosomal DNA restriction analysis. J Clin Microbiol. 1993;31:2061–2065. doi: 10.1128/jcm.31.8.2061-2065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]