Abstract
The spike protein of SARS-CoV-2 and the host ACE2 receptor plays a vital role in the entry to the cell. Among which the hotspot residue 501 is continuously subjected to positive selection pressure and induces unusual virulence. Keeping in view the importance of the hot spot residue 501, we predicted the potentially emerging structural variants of 501 residue. We analyzed the binding pattern of wild type and mutants (Spike RBD) to the ACE2 receptor by deciphering variations in the amino acids’ interaction networks by graph kernels along with evolutionary, network metrics, and energetic information. Our analysis revealed that N501I, N501T, and N501V increase the binding affinity and alter the intra and inter-residue bonding networks. The N501T has shown strong positive selection and fitness in other animals. Docking results and repeated simulations (three times) confirmed the structural stability and tighter binding of these three variants, correlated with the previous results following the global stability trend. Consequently, we reported three variants N501I, N501T, and N501V could worsen the situation further if they emerged. The relations between the viral fitness and binding affinity is a complicated game thus the emergence of high affinity mutations in the SARS-CoV-2 RBD brings up the question of whether or not positive selection favours these mutations or not?
Keywords: SARS-CoV-2, Emerging variants, Modeling, Simulation, Free energy
1. Introduction
Coronaviruses, which jolted the world in the 21st century with its perpetual appearance and phenomenal penetration, are placed within four genera alpha [(α), beta (β), gamma (γ) & delta (δ)] of subfamily Orthocoronavirinae of the family Coronaviridae [1,2]. The devastating epidemics of 2003 (SARS-CoV), 2012 (MERS-CoV), and 2019 (SARS-CoV-2) caused by the members of the beta (β) coronaviruses inflicted heavy losses to human health, wealth and social fabrics in the world [3,4]. These epidemics are attributed to the ability of the beta (β) coronaviruses to transmit from humans to humans [[5], [6], [7]]. The spike protein and angiotensin-converting enzyme (ACE2) receptors establish a link, which is instrumental for the transmission and pathogenesis of the virus [8]. Therefore, spike protein is an important and promising target for conducting and devising antiviral studies [9]. So far, the total number of infected person tally reached 140.45 million, whereas the death toll reached 3.05 million worldwide and 3% case fatality ratio (CFR) was reported for the SARS-CoV-2, which is comparatively lower to the 10%, and 35%, CFR caused by SARS-CoV-1 and MERS-CoV, respectively [5,10].
The epidemic caused by SARS-CoV-2 has been prolonged with the appearance of more contagious and lethal variants. Four variants like B.1.1.7 (UK), B.1.351 (South Africa), Delta (Indian), and P.1 (Brazil) are termed as variants of concern (VOCs) due to their higher contagious nature, severity, and possibility to evade the immune response [[11], [12], [13], [14]]. These VOCs harbour numerous mutations in the RBD domain, which includes N501Y, E484K and K417 N/T [[15], [16], [17]]. These variants, such as B.1.17 (Alpha variant) harbouring a mutation N501Y reported in the UK, D614G variant, lineage B.1.1.207 with P681H replacement, lineage B.1.351 (Beta) with E484K mutation reported in South Africa (B.1.351) and P.1 variant reported in Brazil [18]. Among the others, the cluster 5 variant, B.1.258Δ, B.1.1.28 or B.1.195, P.2, P.3, B.1.168, B.1.617, B.617.2, CAL.20C and the Delta variants are among the notable variants reported in Nigeria, Japan, Philippines, California, Denmark and Czech Republic aggravated the situation further [19,20]. They are reported to increase the binding affinity towards the host cellular receptor and also act as immune escaping variants [15,21]. These variants have also reduced the efficacy of different vaccines.
Proteomic-based solutions for the control of COVID-19 mainly depend on the understanding of the proteome of SARS-CoV-2. For instance, we previously reported that the E484K variant could be more devastating than anyone else and thus emerged as a Delta variant which is more lethal and contagious [15]. Therefore, predicting potentially emerging variants and estimating the impact on the binding of spike RBD to host receptor may help manage any devastating variant. The role of 501 residue in all the VOCs is indispensable in the tighter binding and could be an ideal target for researchers in devising antiviral therapies against SARS-CoV-2. A thorough, step-by-step and conclusive study is essential for unearthing the role of N501 substitutions on the binding of spike protein with ACE2 receptors and how it brings structure and functional changes. Hence, in the current research, protein-protein docking and biophysical methods were utilized to demonstrate variations in the protein's structure that brings the binding variations between RDB and ACE2 receptors due to predicted hypothetical variants and increase the transmissibility. This analysis will also provide an insight to understand these structural changes in the RBD domain for future repercussions.
2. Methods
2.1. Data retrieval and in silico mutagenesis of residue 501
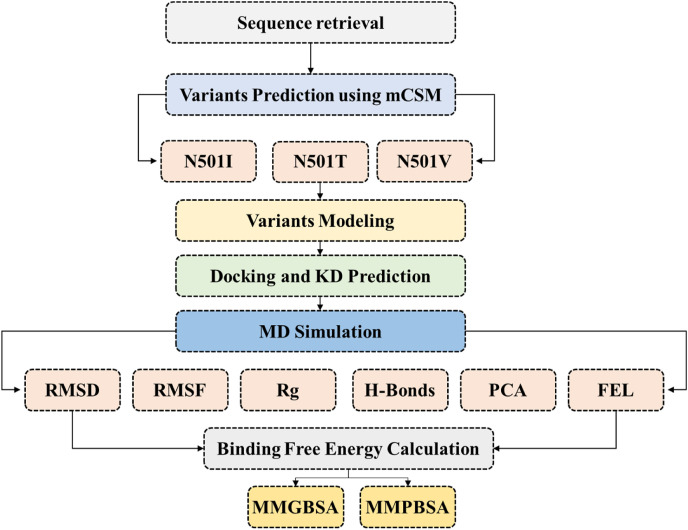
The role of spike protein in the attachment and pathogenesis of coronavirus infection is indispensable, and for recent variants a high transmission due to the mutations harbored by the RBD domain has been reported [[21], [22], [23], [24]]. In almost all the variants the residue at 501 was reported to favour the positive selection pressure and plays a vital role in the enhanced pathogenesis [15]. It is well observed that this residue at 501 position changed when SARS-CoV-2 evolved from SARS-CoV. In the SARS, this residue was Phenylalanine (501). However, this residue remained conserved between MERS and SARS-CoV-2 (Wuhan strain). With the emergence of new variants in the UK, South African and Brazil, this residue evolved as Tyrosine. A study reported that the binding of RBD-ACE2 significantly changed with this mutation and eventually led to higher pathogenesis. Keeping in view the structural and functional significance of this residue at position 501 in the spike protein in human pathogenesis, we used in silico mutagenesis approach to predict emerging hypothetical structural variants and understand its impact on the binding and potential role in pathogenesis. Using machine learning-enabled method known as graph-based structural signatures (mCSM) deployed as mCSM-PPI [25] to predict the most lethal variants of this residue. For this purpose, a recently reported structure of the spike protein of SARS-CoV-2 was retrieved from RCSB using an accession number 6M0J [26]. A mutation list excluding Phe, Asn, and Tyr was prepared and submitted to mCSM-PPI to predict the impact of the remaining 17 amino acids on the binding and stability of RBD. The 3D structures of mutations (N501I, N501T, N501V), which were predicted to tempt higher stability change and consequently increases the affinity by manifolds, were modeled using Modeller v15.2 [27] integrated in Chimera software [28]. The structures were minimized, prepared, and superimposed on the wild-type spike protein structure (6M0J) to determine the structural differences as RMSD (root mean square deviation). The overall flow of the work is given in Fig. 1 .
Fig. 1.
The overall workflow of the work, which includes mainly six steps.
2.2. Protein-protein docking and determination of dissociation constant (KD)
The binding variations between the wild type and the predicted hypothetical emerging mutants (N501I, N501T, N501V) were explored through HADDOCK (high ambiguity-driven protein-protein docking) algorithm [29]. Hydrogen bonding network, salt-bridges, and non-bonded contacts were analyzed through PDBsum. Subsequently, the dissociation constant (KD) was calculated to provide more authentic information about the docked complex between the spike protein and ACE2 receptor of both wild and mutants. It is an important parameter to determine the strength of a biological macromolecular complex. The online server PRODIGY (PROtein binDIng energy) [30] was used to compute the KD and the binding affinity for the wild type and mutant RBD-ACE2 complexes.
2.3. Molecular dynamics simulation (MDS) of the top complexes
The AMBER20 was used to perform the dynamic behaviour of wild type complex, N501I, N501T and N501V, through MD simulation using FF19SB force field [31]. The water molecules (TIP3P box) were added to solvate each complex while Na + ions were supplied for neutralization [32]. Gentle minimization of the complexes was achieved by employing steepest descent algorithm for 6000 cycles while the conjugate gradient algorithm for 3000 cycles were used respectively [33]. Equilibration at 1atm (constant pressure) with weak restraint and heating for 300°K was performed. Thereafter, the MDS in triplicate for each complex for a time scale of 200ns using PME (Particle mesh Ewald) and SHAKE algorithms for long-range and covalent interactions [31,34]. PTRAJ and CPPTRAJ integrated modules of AMBER20 were used for GPU accelerated simulation trajectories analysis [35].
2.4. Binding free energy calculations
The MM-GBSA and MM-PBSA approaches were used to analyze the actual binding energy of wild type complex, N501I, N501T and N501V [36]. The MM-GBSA is the most suitable approach used by different studies for the estimation of various biological complexes, such as protein-protein/DNA/RNA [[37], [38], [39], [40], [41]]. The total free energy was calculated by using the script MMPBSA.py [36].
The following equation (i) was used for free energy calculation:
| equation (i) |
Each component of the total free energy was estimated using the following equation (ii):
| equation (ii) |
2.5. Unsupervised clustering of MD trajectories and free energy landscape
To cluster the MD trajectories and understand the principal motions during the simulation time, Principal Component Analysis (PCA) [42,43]was computed by using CPPTRAJ package. Positional covariance matrix (PCM) of each eigenvector and their atomic coordinates were estimated. Exploiting the orthogonal coordinate transformation to retrieve the diagonal matrix of eigenvalues. Eigenvectors and eigenvalues of each principal components were calculated the motion of the simulation trajectories were mapped [44,45]. Principal components i.e. PC1 and PC2, were used further to calculate the free energy landscape (FEL) in the following equation (iii).
| equation (iii) |
where X specifies the response of the two PCs, KB represents a Boltzmann constant, and P(X) is the probability dispersion of the framework's first two principal components.
3. Results and discussion
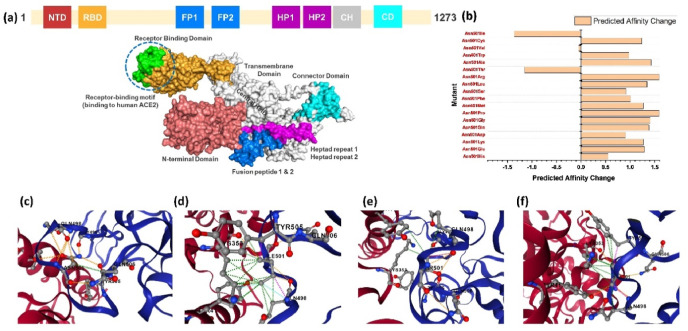
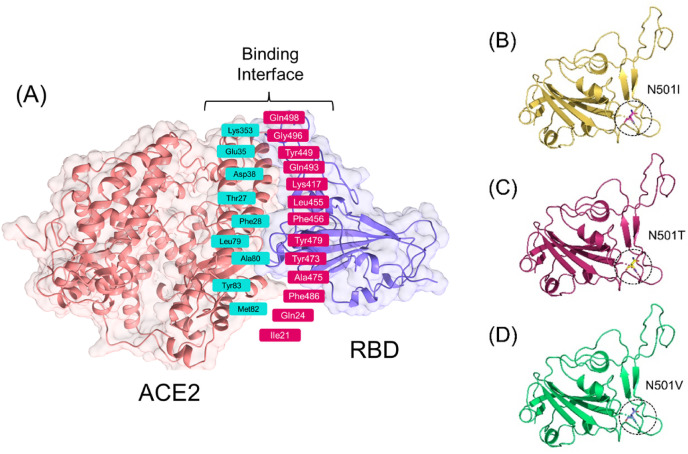
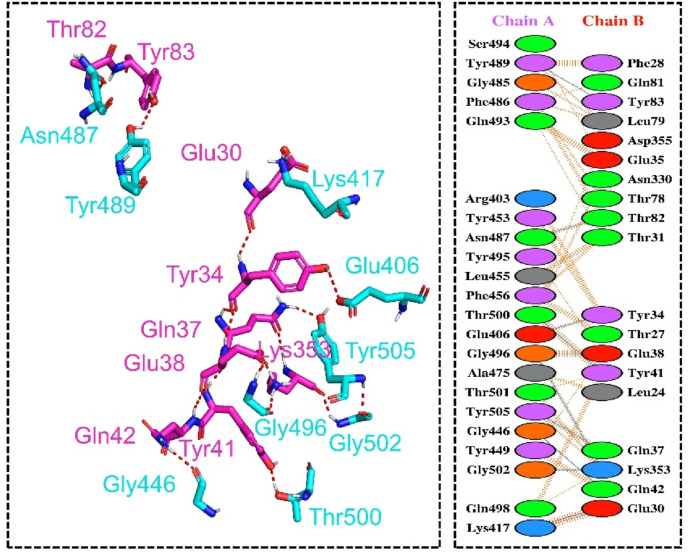
Spike glycoprotein is a multi-domain (Fig. 2 a) trimeric protein that initiates the SARS-CoV-2 infection by recruiting the host receptor ACE2. An interplay between the RBD domain of the spike protein and the host ACE2 receptor initiates a cascade of reactions that stabilize the binding and provides a platform for fusion and entry to the cell. Recently reported mutations in the spike glycoprotein unique to the RBD domain induces a different role and results in enhanced pathogenesis. Among the reported mutations, N501Y, E484K, K417 N, E484Q and L452R elevate the pathogenicity scale. The latest report explored that the hot spot residue 501 is continuously subjected to positive selection pressure and play an important role in the enhanced pathogenesis [15]. Keeping in view the importance of the hot spot residue 501 in the stronger attachment to ACE2 and induction of pathogenesis, we predicted the structural variants of 501 residue and analyzed the binding pattern of wild type and mutants (Spike RBD) to the ACE2 receptor. Recently our group also reported that E484K mutation alone in the RBD domain may increase Spike variant binding to ACE2 and hence enhance virus infectivity and transmissibility [15]. This speculation has come true and is now circulating and destroyed the health in India. We determined the impact of 17 different substitutions by deciphering variations in the amino acids’ interaction networks by graph kernels along with evolutionary, network metrics, and energetic information. The mCSM-PPI2 server predicted three residues which increase the binding affinity upon the mutation, while 14 residues were reported to decrease the binding affinity. Among the three residues, isoleucine was reported to change the binding affinity by −1.36 kcal/mol, threonine by −1.14 kcal/mol, while valine slightly altered the affinity by −0.049 kcal/mol. The binding affinity of each residue is shown in Fig. 2b. Further understanding was established by exploring the inter and intra residues bonding network. As given in Fig. 2c, the wild type (Asn501) of RBD forms a polar interaction with the Tyr41 of ACE2 while multiple polar bonds with neighboring residues of RBD and one hydrophobic interaction with Gln506. On the other hand, the non-polar and aliphatic isoleucine formed multiple hydrophobic and polar interactions with Tyr41 and Lys353. Comparatively, the polar threonine amino acid formed significant hydrophobic interactions with Tyr41 and Lys353. The intra-residue network of threonine 501 was significantly enriched with polar interactions. Intriguingly the valine at 501 formed mixed polar and hydrophobic interactions with the host receptor while the inter residues model was enriched with clashes and polar interactions. It can be seen that the wild type is enriched with polar contacts, while in the three predicted variants, the polar contents are reduced. This point is validated by a previous study that reported that reduced polar contents at the RBD site enhance the binding affinity. Recently a study based on sequence and structure-based predictions of the impact of mutations reported that the mutation N501I is a stabilizing mutation and affect the RBD binding by manifold [46]. Therefore these predicted mutations may significantly alter the affinity [47]. The intra and inter residues bonding networks of ile501, Thr501 and Val501 are given in Fig. 2d, e and 2f. Next, to uncover the binding mechanism of the predicted variants (N501I, N501T and N501V), we used HADDOCK to bring off the protein-protein (ACE2-RBD) docking. Prior to protein-protein docking homology modelling of the predicted mutants was performed using 6M0J as a template. The structures were minimized and prepared for docking using Chimera. The binding interface is given in Fig. 3 A, and the structures of N501I, N501T and N501V mutants are given in Fig. 3B, C, 3D.
Fig. 2.
Structure of the spike glycoprotein and mutations information. (a) Domain organization of spike glycoprotein and representation of RBD domain. (b) The predicted mutations' scores by mCSM-PPI2. (c) Represent the intra and inter residues bonding network of wild type while (d) (e) and (f) represent the intra and inter residues bonding network of the three mutants N501I, N501T and N501V.
Fig. 3.
(A) Structural representation of the spike glycoprotein (PDB ID:6M0J) and the receptor-binding domain of the SARS-CoV-2. (B) (C) and (D) shows different mutations N501I, N501T and N501V.
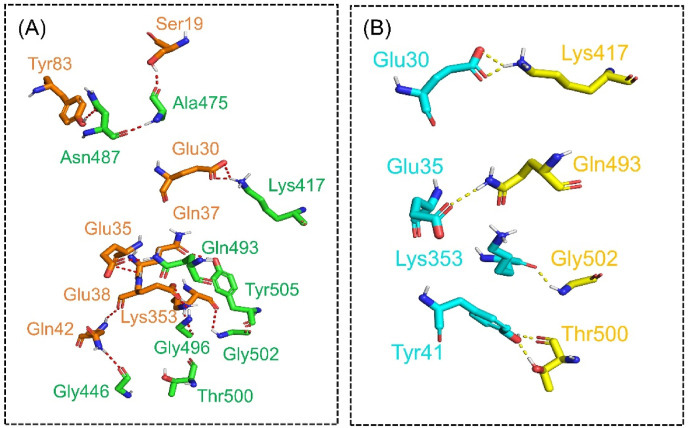
HADDOCK predicted the docking score −127.23 ± 1.2 for the ACE2-N501I complex. PDBsum interaction analysis revealed that both ACE2 and N501I structures form 10 hydrogen bonds and one salt bridge, while the non-bonded interaction between the two complexes were 121. The hydrogen bonds formed by the ACE2-N501I complex includes Gln493-Glu35, Asn487-Tyr83, Gly446-Gln42, Ala475-Ser19, Lys417-Glu30, Thr500-Tyr41, Gly496- Lys353, Gly496- Glu38, Gly502-Lys353 and Tyr505-Gln37 (Fig. 4 a). However, the salt bridge was reported between Glu30 and Lys417, which is conserved and reported in the wild type and previous new variants [15]. Previously published study also reported continued interactions between Gln93-Glu35 and Gln498-Glu38 [48]. This shows our consistent results of all the complexes. Additionally, Lys417 established an essential contact with the Asp30, which is also preserved here. Moreover, Verma and Subbarao also reported that the key Lys353 establish a hydrogen bond with Gly502 which shows our more accurate predictions [46]. The docking score for ACE2-N501T mutant was reported to be −125.52 ± 2.1 kcal/mol. The binding affinity of N501T was comparable with the N501I mutant with a total of one salt bridge, 10 hydrogen bonds and 112 non-bonded contacts. The salt bridge was formed between Lys417 and Glu30 residues. Among the hydrogen bonds Tyr489-Tyr83, Asn487-Thr82, Thr500-Tyr41, Glu406-Tyr34, Gly496-Glu38, Gly496-Gln37, Tyr505-Gln37, Gly446-Gln42, Gly502-Lys353 and Lys417-Glu30 residues are involved (Fig. 4b). The HADDOCK docking score for N501V (ACE2-spike RBD) was reported to be −123.65 ± 3.2 kcal/mol. Through molecular interaction analysis, it has been revealed that the substituted residue N501V decreases the binding of ACE2 with the spike RBD domain as compared to the N501I mutants. A total of one salt bridge, 5 hydrogen bonds and 100 non-bonded contacts were reported (Fig. 5 ). Among the hydrogen bonds, Lys417-Glu30, Thr500-Tyr41, Gln493-Glu35 and Gly502-Lys353 residues are involved. Furthermore, HDOCK predicted the docking scores for each complex was wild type (−302.84 kcal/mol), N501I (−317.32 kcal/mol), N501T (−315.66 kcal/mol), while for N501V, the docking score was reported to be −308.02 kcal/mol. Similarly, the electrostatic interactions substantially increased the binding. Previous research have found that evaluating the binding of SARS-CoV-2 and SARS-CoV RBDs with ACE2 indicated stronger hydrogen bonds and electrostatic interactions in SARS-CoV-2 [26,49]. Thus, this shows that the emergence of these variants could possibly induce higher infectivity than the first strain reported in Wuhan. Our predictions are supported by an experimental study conducted in Ferrets. The researchers found N501T SARS-CoV-2 variants in 11/11 experimentally infected ferrets, with a rising fraction of the virome demonstrated over time, indicating significant positive selection in ferrets [50]. On the other hand, the N501V are reported to stabilize the interactions [51]. Hence, our results are corroborated with these previous findings as N501T shows strong positive fitness while the N501V failed in the experimental validation, which is also reported as a weak binding mutation. This further confirms that our results interesting and needs further exploration. All the docking and KD results are shown in Table 1 . The KD scores are validated that the two mutations N501I and N501T particularly are strong binder than the wild type. However, the results of wild type and N501V are comparable.
Fig. 4.
Docking representation of N501I and N501T mutant complexes. The figure represents the binding interface of N501I (A) and N501T (B) complexes along with its stick representation of the key hydrogen interactions.
Fig. 5.
Docking representation of N501V mutant complexes. The figure represents the binding interface of N501V complex along with its stick representation of the key hydrogen interactions. While the right panel shows the binding interface with key hydrogen bonding interactions of the N501V mutant.
Table 1.
HADDOCK predicted docking scores and KD predicted dissociation constants for each complex.
| Docking | Wild Type | N501I | N501T | N501V |
|---|---|---|---|---|
| HADDOCK score | −111.8 ± 4.6 | −127.23 ± 1.2 | −125.52 ± 2.1 | −123.65 ± 3.2 |
| HDOCK Scores | −302.84 | −317.32 | −315.66 | −308.02 |
| KD (M) at 38.0 °C | 3.4E−09 | 1.4E−09 | 6.1E−10 | 2.0E−09 |
| [15] | This study | This study | This study |
3.1. Root Mean Square Deviations (RMSD) analysis
The simulation trajectories were analyzed first by RMSD to look for structural alterations in the docked Spike-ACE2 complex under dynamic conditions. RMSD is a statistical parameter to superimpose all MD frames over the initial reference and measure distance between corresponding atoms. Herein we measured the distance between carbon alpha atoms of the systems. Mathematically, RMSD is
where.
di is the difference of position between atoms and i refers to the original and superimposed structure.
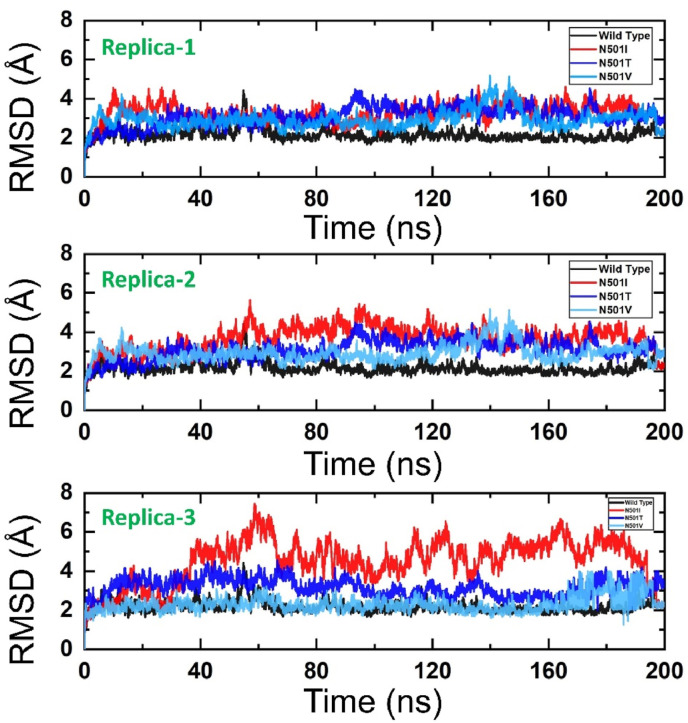
In all triplicates, the wild-type complex is structurally more stable, shown consistent intermolecular affinity and firm chemical interactions profile as compared to the N501I, N501T, and N501V variants. Except for small window deviations between 50 and 60 ns, RMSD plots of the wild type are very smooth and gain equilibrium after 5 ns. The average stability of the wild type plots is ∼2 Å, while the sharp deviation discussed above reaches 4 Å. These findings of the wild type RBD-ACE2 complex is in coherence with the previous finding and concluded almost similar RMSD trend. The N501I variant RMSD behaves differently in all three runs; the second run is more stable than the first and third. In the second run, RMSD steadily increased till 35 ns touching RMSD value of 5 Å, followed by decreasing RMSD to 3 Å. This trend was noticed till 90 ns, and the system then entered into another phase of constant mini deviations ∼4 Å. Towards the end time, the complex appears to gain structural stability after 180 ns though the RMSD is still high with RMSD of ∼4 Å. The first run of N501I variant, unlike the second run, is more uniform up to 50 ns, then sharply reaches to ∼6 Å, then suddenly stable to ∼2.5 Å, followed by fluctuating RMSD >4 Å until it became stable at 140 ns, which was seen till the simulation end. The third run of N501I variant among all is the most unstable, with maximum RMSD reaching 8 Å. Throughout the simulation period, the RMSD of this run is in continuous oscillation and faces significant perturbations. This can be inferred as not well docked intermolecular conformation with continuous formation and breakage of chemical interaction in an attempt to get the proper binding mode of the interacting molecules. As stability in the intermolecular docked pose is not achieved in the performed simulation, extending the simulation time might be needed until the system is converged. Superimposition of N501I three replica snapshots picked at different nanoseconds (20 ns,35 ns,55 ns,74 ns,115 ns,125 ns and 160 ns) over the control revealed an RMSD of 1.271 Å. This indicates no conformational modifications in the structures and structural deviations as noticed in the RMSD plot is the outcome of flexible loops of both Spike and RBD. In addition, a recent study based on in silico analysis also reported that the N501I mutation remained stable during the MD simulation in complex with ACE2 [46]. Nevertheless, these loop-mediated fluctuations do not confer any impact on the intermolecular binding and complex formation. Like N501I, N501T variant behaves in different dynamics in the triplicate run. The third run of N501T variant, in particular, is concluded as highly unstable complex and behaves very flexibly, especially after 40 ns. Till 75 ns, the system RMSD remained constant (∼4 Å), afterwards lower RMSD of the inconsistent pattern was seen till 165 ns. Towards the end, some equilibrium of the system was noticed that pointed docked conformation stability. In case of N501T third run, snapshots at different nanoseconds like 25 ns, 40 ns, 145 ns, and 180 ns revealed an RMSD of 1.129 Å after superimposition. It was again observed that the deviations of RMSD plots is due to flexible loops, which don't affect the intermolecular complex conformation. The first and second run of the N501T variant is somewhat similar in terms RMSD deviations and considerably more stable with minor deviations until 85 ns. Both the systems then suffer from increasing RMSD that continues till 180 ns and then get equilibrated as the simulation proceeds. These findings reported previously confirms that N501T may produce destabilizing effect, however long run simulations (∼microseconds) can confirm the findings in more details [46]. The first and second run N501V RMSD highly resemble each other, and the complexes are structurally highly stable till 140 ns without experiencing any major deviation. Two shorts but bit higher deviations were revealed between 140 and 150 ns with RMSD above 4 Å. At the simulation end, the systems are super stable with no deviations plotted. The third run of this variant, in contrast to the first and second run, is the most stable, with no deviations reported at all with consistent RMSD ∼2 Å. Our findings are strongly correlated with the RBD stability trend induced by mutations which consequently increases the ACE2-binding affinity [47]. Furthermore, earlier studies show a close link between RBD stability and affinity, with mutations that enhances stability also increases in binding affinity [52,53]. It can be further witnessed as a destabilizing mutation C432D has been reported to reduce the affinity for ACE2 and thus entry to the cell [47]. In the recently reported mutations, including B.1.1.7 (N501Y), B.1.351, P.1, B.1.617 and B.1.618, increased in the stability increased was strongly correlated with a stable evolution of the new variants and tighter binding [15]. Thus, our finding shows that N501I, N501T and N501V possess stable dynamics and may evolve stably subjected to fitness and their adaptive significance, which may further increase the unusual virulence consequently but the impact of different environmental conditions i.e. pH, temperature may affect the virulence. The RMSDs of all the complexes in triplicate are shown in Fig. 6 while the mean RMSDs and standard deviation of all the triplicates are given in Table 2 . The SARS-CoV-2 variants with mutations at 501 are reported to have reduced susceptibility to antibodies neutralization in immunized mice. Further, the neutralization resistance reported for 501 variants unveiled compromised efficacy of vaccines and monoclonal antibodies [54].
Fig. 6.
The figure represents the RMSDs of all the complexes in triplicate. The RMSDs of the wild type is shown in black colour while the other mutants are given in different colours.
Table 2.
mean RMSDs and standard deviation of all the replicates of different systems.
| Complex | Replicate 1 |
Replicate 2 |
Replicate 3 |
|||
|---|---|---|---|---|---|---|
| Mean RMSD | Standard Deviation | Mean RMSD | Standard Deviation | Mean RMSD | Standard Deviation | |
| Wild Type | 2.38 | 0.34 | 2.38 | 0.34 | 2.38 | 0.34 |
| N501I | 3.25 | 0.47 | 3.67 | 0.63 | 4.55 | 1.11 |
| N501T | 3.82 | 0.62 | 3.09 | 0.5 | 5.14 | 2.68 |
| N501V | 3.53 | 0.58 | 2.94 | 0.44 | 4.48 | 3.01 |
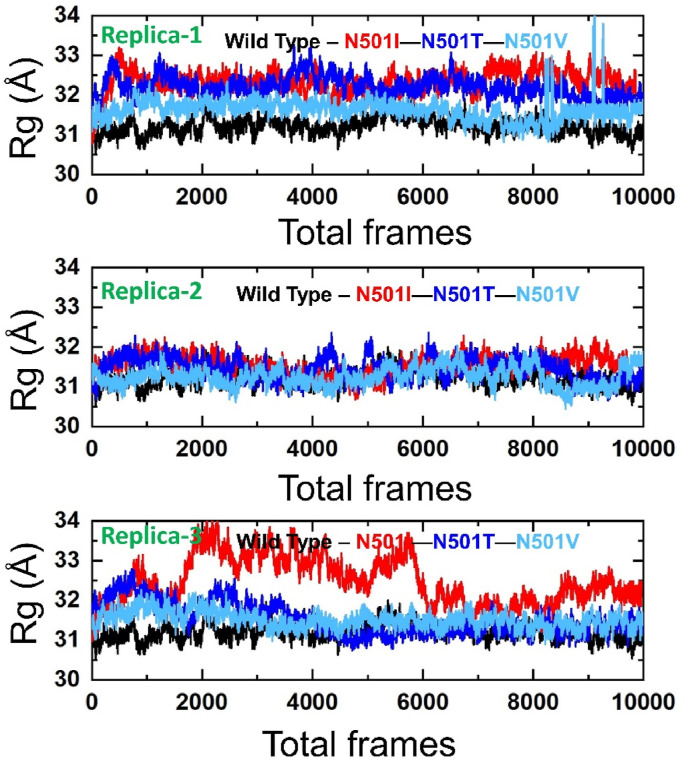
3.2. Radius of Gyration (RoG) analysis
The compact nature of the variants with reference to the wild type was elucidated through RoG analysis. From a general perspective, the RoG results are more in homology to the RMSD and almost inferred the same dynamics. As analyzed in the RMSD, the wild type compact nature remained the same in all three runs, and its average RoG is ∼31 Å. On the other side, variants RoG is not the same in triplicate as captured earlier in the RMSD. The N501I RoG in the third run is facing high structure deviations due to its non-compact nature. As pointed in the RMSD, this may explain the unstable docked conformation of the RBD with the ACE2 and attempting to get more stable interactions by breaking some and forming others. These conformation deviations events are more pronounced between frame 1500 to 6000. The second run of the variant is more compact and is not subject to any major structural alterations. The first, however, showed some deviations, particularly at the start, followed by a stable, compact nature. N501T second run RoG appears to be more compact than the first and third run as predicted by RMSD. The first run interpreted the complex as less compact at the start till 7–8 ns, and then the system is a somewhat compact exception to more minor deviation throughout the length of simulation time. The third run RoG acts inconsistently till 40 ns, and then reaches static till the end. Compared to the above-mentioned variants, all three runs of the N501V variant produced considerably more stable RoG with few minor fluxes. The Rg(s) of all the complexes in triplicate are shown in Fig. 7 .
Fig. 7.
The figure represents the Rg(s) of all the complexes in triplicate. The Rg(s) of the wild type is shown in black colour while the other mutants are given in different colours.
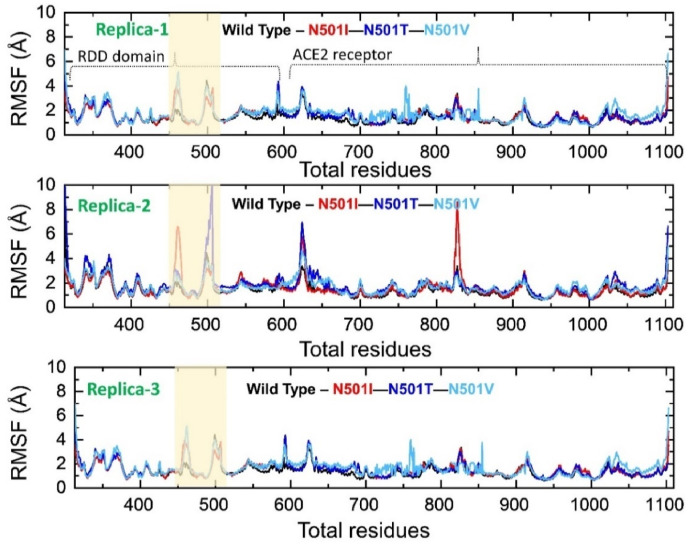
3.3. Root Mean Square Fluctuations (RMSF) analysis
An insight into the residue level fluctuations of the wild and the variants was further accomplished as such local level flexibility confers strength to intermolecular binding, negatively impact molecular recognition and can potentially influence the overall function of the biological molecule. Higher and lower RMSF implies flexible and stable regions, respectively. Usually, loop regions are more unstable due to no fixed secondary structure and therefore correspond to higher RMSF. The wild type in triplicate demonstrated stable residues fluctuations, and all the systems agree with respect to each, as noted before in RMSD and RoG. N501I residues range: 170–175, 300–305,505-515, and residues at the C and N-terminal regions are more flexible in the first run, while these residues range were found more stable in the second and third run except the C- and N-terminal. Same observations were made for the N501T in the first, where the regions highlighted for the first run of N501I follow the same pattern of fluctuations. The N501V variant residues are comparatively more stable than other variants in the first run but depicting more fluctuations, especially in the regions of 100–200 400–460. Similar findings where the three loops in the RBD domain γ1, γ2 and γ3 correspond to position 474–485, 488–490 and 494–505 have been reported to be essential for enhanced binding in the other variants. Similarly conformational dynamics of various mutations in the RBD reported in a previous study revealed that the N501I mutations particularly (at 501 position) increases the flexibility [46]. Our findings also showed that region 469–505 corresponds (highlighted) possess higher fluctuation in the mutant complexes. From these findings, it can be observed the Spike protein is undergoing structural adjustments to bind efficiently to the ACE2 receptor and consequently increases the affinity for the host-receptor thus enhanced entry to the host cells. The RMSFs of all the complexes in triplicate are shown in Fig. 8 .
Fig. 8.
This figure represents the residual flexibility (RMSF) index of the wild type and mutant complexes. The shaded region represents the three important loops γ1 (474–485), γ2 (488–490) and γ3 (494–505) that are crucial for binding with ACE2.
3.4. Hydrogen bonds analysis
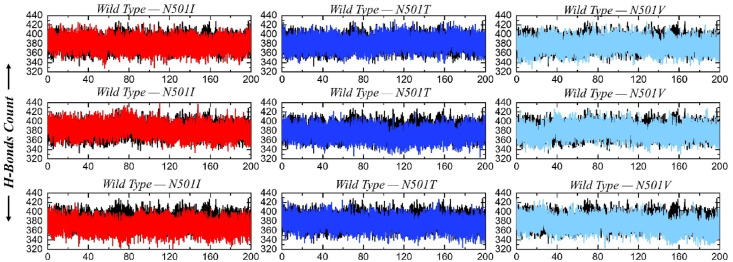
Hydrogen bonds analysis was further done to disclose the variants specificity for the ACE2 receptor as a result of biochemical events steered by hydrogen bonding. This is also key to deduce stable interactions allowing the intermolecular contact for a longer time, and carry out functional roles. As can be inferred from the hydrogen bond plots, the wild Spike and variants are associated with the ACE2 receptor via hundreds of hydrogen bonds in each frame of MD simulations. This further affirms the above MD simulation analysis and classified the systems as highly stable. The average number of hydrogen bonds in each complex of the triplicate run was calculated. In the wild type, the average hydrogen bonds were 382 while in N501I 387, 385, 381, in N501T 382, 386, 383 and in N501V 382, 381 and 382.This finding shows that the mutations in these three variants have altered their hydrogen-bonding network and may use a different strategy if they emerged as potential variants. All the H-bond results are presented in Fig. 9 .
Fig. 9.
This figure represents the total number of hydrogen bonds in all the replicates during the 200ns simulation.
3.5. MM/GBSA and MM/PBSA binding energies
Binding free energy prediction of small molecules to a larger biological macromolecule by MM/GBSA and MM/PBSA is arguably the most widely used approach to re-investigate docking conformation, determining structural stability and predicting interacting hotspots and binding affinities. Both aforementioned methods are computationally less expensive than the extensive alchemical free energy methods and are categorized as more accurate than conventional scoring functions. Taking into account the high significance of these methods, they were applied herein to shed light on the structure, function and interaction impact of N501I, N501T and N501V mutations on Spike RBD binding to ACE2 receptor. The different energy components estimated by both MM/GBSA and MM/PBSA methods for wild and variants complexes are tabulated in Table 3 and Table 4 , respectively. Each energy component is presented in triplicate to get full confidence in the data. As per MM/GBSA, the variants have shown inconsistent binding energy results; however, the net binding energy for the wild type system repeatedly found the same. The N501I variant among all is the highly stable variant with respect to ACE2, followed by N501T and N501V. The average net MM/GBSA binding energy of N501I is −64.52 kcal/mol, while for N501T and N501V, the net energy is −55.64 kcal/mol and −45.74 kcal/mol, respectively. The low net energy value demonstrates the high intermolecular complex formation by N501I with ACE2, which in turn indicates better interaction of the N501I with the host cells allowing the variant to spread rapidly compared to the wild type. The N501T and N501V can also be interpreted as more infectious based on the net MM/GBSA binding energy considering the wild type net binding energy as reference. Decomposing the net energies into respective energy components, the net electrostatic binding energy was found to dominate the variants binding to ACE2. The van der Waal interactions seem to also play a favorable contribution in binding. Previously for the N501Y substitution reported in B.1.1.7 variant the total binding energy was also reported to have increased than the wild type [22]. Herein, a direct comparison revealed that the N510Y possess higher total binding energy than the reported variants here but the results here are extracted from long-run simulation cannot be compared with the previous report which performed only 100ns. Thus, to compare the B.1.1.7 with the predicted variants long-run repeated simulations are required. To overall binding, the polar solvation energy played a non-favorable contribution in contrast to good contribution from non-polar. The charge distribution in the dynamic's environment appears to interact with itself less frequently, thus accounting for non-significant contribution in binding. In short, the solvation energy is non-favorable in variants binding to ACE2, whereas gas-phase energy is key to the strong binding of the variants to ACE2.
Table 3.
The TBE employing MM-GBSA method for the wild type and three predicted variants (kcal/mol).
| MM/GBSA (replicate 1) | |||||
|---|---|---|---|---|---|
| Complex | vdW | Elec | EGB | ESURF | Total Binding Energy |
| Wild Type | −87.61 ± 0.14 | −616.56 ± 0.64 | 672.19 ± 0.62 | −12.74 ± 0.016 | −44.72 ± 0.193 |
| N501I | −100.38 ± 0.18 | −602.41 ± 1.03 | 651.66 ± 0.59 | −14.05 ± 0.012 | −65.18 ± 1.154 |
| N501T | −88.86 ± 0.12 | −639.22 ± 0.92 | 685.09 ± 0.71 | −12.77 ± 0.019 | −55.77 ± 1.620 |
| N501V | −84.19 ± 0.13 | −581.68 ± 0.73 | 632.98 ± 0.55 | −12.34 ± 0.021 | −45.23 ± 1.002 |
| N501Y | −105.84 ± 0.21 | −623.69 ± 1.14 | 668.42 ± 0.57 | −11.89 ± 0.018 | −73.00 ± 0.48 [22] |
| MM/GBSA (replicate 2) | |||||
| Complex | vdW | Elec | EGB | ESURF | Total Binding Energy |
| Wild Type | −87.61 ± 0.14 | −616.56 ± 0.64 | 672.19 ± 0.62 | −12.74 ± 0.016 | −44.72 ± 0.193 |
| N501I | −88.83 ± 0.16 | −615.37 ± 0.95 | 655.00 ± 0.64 | −12.04 ± 0.015 | −61.24 ± 1.186 |
| N501T | −87.41 ± 0.11 | −590.31 ± 0.95 | −629.85 ± 0.42 | −9.97 ± 0.011 | −57.84 ± 1.021 |
| N501V | −92.44 ± 0.91 | −617.64 ± 0.75 | 675.90 ± 0.93 | −12.95 ± 0.013 | −47.14 ± 1.019 |
| MM/GBSA (replicate 3) | |||||
| Complex | vdW | Elec | EGB | ESURF | Total Binding Energy |
| Wild Type | −87.61 ± 0.14 | −616.56 ± 0.64 | 672.19 ± 0.62 | −12.74 ± 0.016 | −44.72 ± 0.193 |
| N501I | −87.69 ± 0.15 | −620.33 ± 0.52 | 652.34 ± 0.58 | −11.46 ± 0.015 | −67.14 ± 1.166 |
| N501T | −75.57 ± 0.10 | −607.87 ± 0.89 | 641.97 ± 0.95 | −11.83 ± 0.013 | −53.31 ± 1.139 |
| N501V | −84.85 ± 0.11 | −611.72 ± 0.81 | 663.42 ± 0.89 | −12.18 ± 0.013 | −44.85 ± 1.016 |
Table 4.
The TBE employing MM-PBSA method for the wild type and three predicted variants (kcal/mol).
| MM/PBSA (replicate 1) | ||||
|---|---|---|---|---|
| Complex | vdW | Elec | EPB | Total Binding Energy |
| Wild Type | −87.61 ± 0.14 | −616.56 ± 0.64 | 649.01 ± 45.23 | −4.89 ± 0.56 |
| N501I | −100.38 ± 0.18 | −602.41 ± 1.03 | 624.66 ± 41.54 | −8.83 ± 1.28 |
| N501T | −88.86 ± 0.12 | −639.22 ± 0.92 | 655.85 ± 42.02 | −6.25 ± 0.93 |
| N501V | −84.19 ± 0.13 | −581.68 ± 0.73 | 609.00 ± 39.19 | −4.67 ± 0.44 |
| MM/PBSA (replicate 2) | ||||
| Complex | vdW | Elec | EPB | Total Binding Energy |
| Wild Type | −87.61 ± 0.14 | −616.56 ± 0.64 | 649.01 ± 45.23 | −4.89 ± 0.56 |
| N501I | −88.83 ± 0.16 | −615.37 ± 0.95 | 633.56 ± 43.67 | −7.62 ± 1.11 |
| N501T | −87.41 ± 0.11 | −590.31 ± 0.95 | 615.03 ± 40.05 | −6.92 ± 0.98 |
| N501V | −92.44 ± 0.91 | −617.64 ± 0.75 | 640.12 ± 44.61 | −5.20 ± 0.68 |
| MM/PBSA (replicate 3) | ||||
| Complex | vdW | Elec | EPB | Total Binding Energy |
| Wild Type | −87.61 ± 0.14 | −616.56 ± 0.64 | 649.01 ± 45.23 | −4.89 ± 0.56 |
| N501I | −87.69 ± 0.15 | −620.33 ± 0.52 | 637.58 ± 42.27 | −9.44 ± 1.43 |
| N501T | −75.57 ± 0.10 | −607.87 ± 0.89 | 622.35 ± 41.67 | −6.16 ± 1.04 |
| N501V | −84.85 ± 0.11 | −611.72 ± 0.81 | 638.30 ± 43.54 | −4.66 ± 0.42 |
Analogous to the MM/GBSA results, MM/PBSA reflected the same trend of binding energy for the variants and wild type. The net binding energy of N501I is −8.63 kcal/mol, which is stable than N501T (−6.44 kcal/mol) and N501V (−4.84 kcal/mol). According to these values, the N501I and N501T are strong binders of ACE2 than N501V, which is comparable to the wild type. In MM/PBSA the non-polar solvation energy is much lower than that observed in MM/GBSA. The difference in the net binding energy of MM/GBSA and MM/PBSA may be due to the more sensitive nature of the latter for the investigated systems. These findings are consistent with the previous findings which reported that mutations at 501 position particularly N501I increased the binding free energy [46]. Therefore, the systems require an extra length of MD simulations time to achieved convergence in the net energy values.
3.6. Principle Component Analysis (PCA)
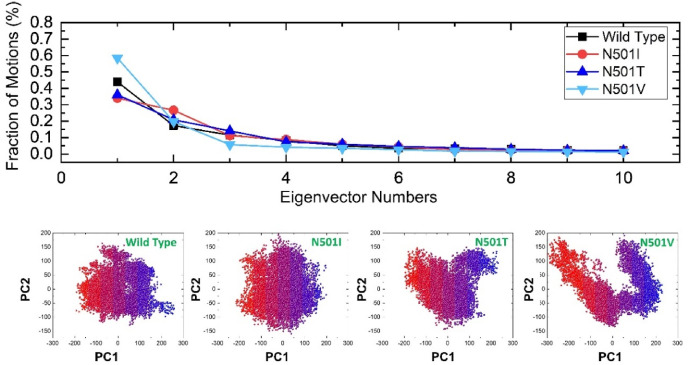
We further study the dynamic behaviour of ACE2-RBD (wild and mutant) binding, using PCA to identify collective motions of each complex. Principal component analysis (PCA) is a statistical technique for reducing the size of a data collection without compromising important information. Ample fluctuations were shown by the first three eigenvectors, while localized fluctuations were displayed by the remaining eigenvectors in each complex, as shown in Fig. 9. In the case of the wild ACE2-RBD complex, the first three eigenvectors accounted for 43% of the overall observed motion, while N501I accounted for 33%, N501T for 36%, and N501V for 56%. This behaviour may explain the structural rearrangement due to the mutations, and thus it may empirically be proposed that the RBD interaction may stabilize the protein by reducing the dynamics of the active regions to a lower level, particularly in N501I and N501T complexes. The current findings also signify the previous reports with higher magnitude of motion in the RBD-ACE2 complexes and directions identical to the wild type [46]. Additionally, the first two PCs were chosen to investigate the projection of their paths during phase space simulations (Fig. 10 ). The structural alterations throughout the simulation are depicted by the blue to red dots. By achieving a stable structural state red colour, these conformational levels were shown to be closer to each other, notably in wild type, N501I, and N501T, and were regarded an energetically stable conformational state.
Fig. 10.
Principal component analysis (PCA) of all the complexes i.e. wild type, N501I, N501T and N501V. The first PC1 and second PC2 from the PCA of the backbone carbon were used.
3.7. Free Energy Landscape (FEL)
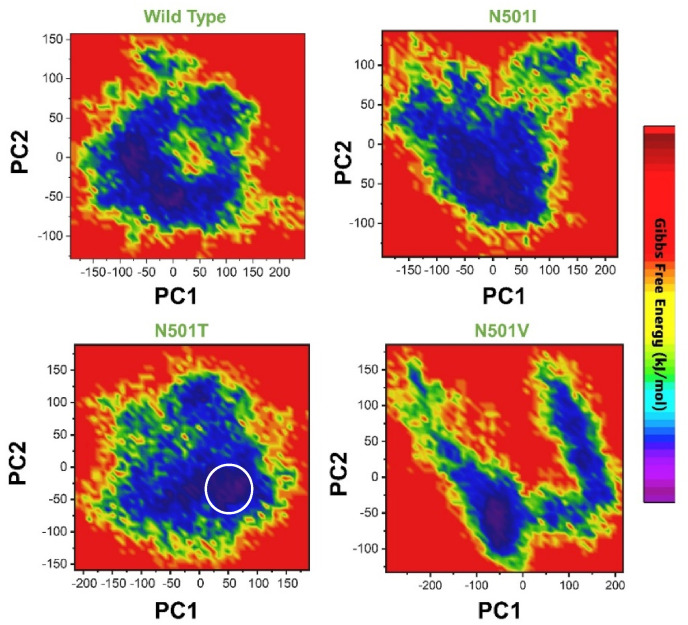
Furthermore, a free energy landscape (FEL) was created to extract the distinguishing features and dynamics properties. To extract the lowest energy conformation based on the given data and finally to link the switches between these minima, FEL was used. Fig. 11 represent the FEL of all the complexes i.e. wild type, N501I, N501T and N501V. The wild type shows two minima separated by a small subspace, while the mutant complexes N501I, N501T and N501V reached only one energy minima, thus demonstrates the global conformational differences accustomed by the mutant complexes in response to mutations.
Fig. 11.
Free Energy Landscape (FEL) of all the complexes i.e., wild type, N501I, N501T and N501V. The first PC1 and second PC2 from the PCA of the backbone carbon were used.
4. Conclusions
Though the study is quite helpful in deciphering mutations induced SARS-CoV-2 mechanisms of interactions and binding conformation with host ACE receptor, which hold significant importance from a therapeutic perspective and COVID-19 management yet this methodology has several limitations. We predicted the potential structural variants of 501 residue, which enforce a more robust interaction response and infectivity. Our results yield that some variants that have shown fitness in other animals like Ferrets may emerge and further exacerbate the situation. The docking predictions are often misleading because the docking tools cannot correctly infer entropy and solvation effects. In this study, docking limitations are overcome by the long run of MD simulations and downward sophisticated computational analysis. However, still we are not clear whether the discussed outcomes are real and therefore required experimental follow-up. MD simulations force fields are not well refined, and the systems simulation requires a lengthier time scale to converge and achieved more accuracy in results. The MMPB/GBSA approaches are highly accepted endpoint techniques in estimating binding free energies; the role of ligand-water interactions and protein-water interactions are often skipped. We discovered that N501I and N501T might potentially cause unusual virulence if they emerged in the near future. The relations between the viral fitness and binding affinity is a complicated game thus the emergence of high affinity mutations in the SARS-CoV-2 RBD brings up the question of whether or not positive selection favours these mutations or not?
Funding
Dong-Qing Wei is supported by grants from the Key Research Area Grant 2016YFA0501703 of the Ministry of Science and Technology of China, the National Science Foundation of China (Grant No. 32070662, 61832019, 32030063), the Science and Technology Commission of Shanghai Municipality (Grant No.: 19430750600), the Natural Science Foundation of Henan Province (162300410060), as well as SJTU JiRLMDS Joint Research Fund and Joint Research Funds for Medical and Engineering and Scientific Research at Shanghai Jiao Tong University (YG2017ZD14).
Availability of data
All the data is available on RCSB, UniProt and any simulation data will be provided on demand.
Declaration of competing interest
Declared None.
Acknowledgments
The computations were partially performed at the Center for High-Performance Computing, Shanghai Jiao Tong University. We acknowledge their help.
References
- 1.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. New England Journal of Medicine; 2019. 2020. A Novel Coronavirus from Patients with Pneumonia in China. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan W.-j., Ni Z.-y., Hu Y., Liang W.-h., Ou C.-q., He J.-x., Liu L., Shan H., Lei C.-l., Hui D.S. MedRxiv; 2020. Clinical Characteristics of 2019 Novel Coronavirus Infection in China. [Google Scholar]
- 4.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiss S.R., Leibowitz J.L. Elsevier; 2011. Coronavirus Pathogenesis, Advances in Virus Research; pp. 85–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Z., Lian X., Su X., Wu W., Marraro G.A., Zeng Y. From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir. Res. 2020;21:1–14. doi: 10.1186/s12931-020-01479-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., Qin C. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11:59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khan A., Khan S., Saleem S., Nizam-Uddin N., Mohammad A., Khan T., Ahmad S., Arshad M., Ali S.S., Suleman M. Immunogenomics guided design of immunomodulatory multi-epitope subunit vaccine against the SARS-CoV-2 new variants, and its validation through in silico cloning and immune simulation. Comput. Biol. Med. 2021:104420. doi: 10.1016/j.compbiomed.2021.104420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haque S.M., Ashwaq O., Sarief A., Azad John Mohamed A.K. A comprehensive review about SARS-CoV-2. Future Virol. 2020;15:625–648. doi: 10.2217/fvl-2020-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Challen R., Brooks-Pollock E., Read J.M., Dyson L., Tsaneva-Atanasova K., Danon L. medRxiv; 2021. Increased Hazard of Mortality in Cases Compatible with SARS-CoV-2 Variant of Concern 202012/1-a Matched Cohort Study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A., Russell T.W., Tully D.C., Washburne A.D. Estimated transmissibility and impact of SARS-CoV-2 lineage B. 1.1. 7 in England. Science. 2021:372. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galloway S.E., Paul P., MacCannell D.R., Johansson M.A., Brooks J.T., MacNeil A., Slayton R.B., Tong S., Silk B.J., Armstrong G.L. Emergence of SARS-CoV-2 b. 1.1. 7 lineage—United States, december 29, 2020–january 12, 2021. MMWR (Morb. Mortal. Wkly. Rep.) 2021;70:95. doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Challen R., Brooks-Pollock E., Read J.M., Dyson L., Tsaneva-Atanasova K., Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ (Clinical research ed.) 2021:372. doi: 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan A., Zia T., Suleman M., Khan T., Ali S.S., Abbasi A.A., Mohammad A., Wei D.-Q. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J. Cell. Physiol. 2021 doi: 10.1002/jcp.30367. n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang J.W., Tambyah P.A., Hui D.S. Emergence of a new SARS-CoV-2 variant in the UK. J. Infect. 2020 doi: 10.1016/j.jinf.2020.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toovey O.T., Harvey K.N., Bird P.W., Tang J.W.-T.W.-T. Introduction of Brazilian SARS-CoV-2 484K. V2 related variants into the UK. J. Infect. 2021 doi: 10.1016/j.jinf.2021.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirby T. New variant of SARS-CoV-2 in UK causes surge of COVID-19. Lancet Respir. Med. 2021;9:e20–e21. doi: 10.1016/S2213-2600(21)00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirotsu Y., Omata M. Detection of R. 1 lineage severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with spike protein W152L/E484K/G769V mutations in Japan. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirotsu Y., Omata M. Discovery of a SARS-CoV-2 variant from the P. 1 lineage harboring K417T/E484K/N501Y mutations in Kofu, Japan. J. Infect. 2021;82:276–316. doi: 10.1016/j.jinf.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.A. Khan, D.-Q. Wei, K. Kousar, J. Abubaker, S. Ahmad, J. Ali, F. Al-Mulla, S.S. Ali, N. Nizam-Uddin, A.M. Sayaf, Preliminary structural data revealed that the SARS-CoV-2 B. 1.617 variant's RBD binds to ACE2 receptor stronger than the wild type to enhance the infectivity, Chembiochem. [DOI] [PMC free article] [PubMed]
- 22.Khan A., Zia T., Suleman M., Khan T., Ali S.S., Abbasi A.A., Mohammad A., Wei D.Q. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J. Cell. Physiol. 2021;236:7045–7057. doi: 10.1002/jcp.30367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan A., Gui J., Ahmad W., Haq I., Shahid M., Khan A.A., Shah A., Khan A., Ali L., Anwar Z. The SARS-CoV-2 B. 1.618 variant slightly alters the spike RBD–ACE2 binding affinity and is an antibody escaping variant: a computational structural perspective. RSC Adv. 2021;11:30132–30147. doi: 10.1039/d1ra04694b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan A., Khan T., Ali S., Aftab S., Wang Y., Qiankun W., Khan M., Suleman M., Ali S., Heng W. SARS-CoV-2 new variants: characteristic features and impact on the efficacy of different vaccines. Biomed. Pharmacother. 2021:112176. doi: 10.1016/j.biopha.2021.112176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodrigues C.H., Myung Y., Pires D.E., Ascher D.B. mCSM-PPI2: predicting the effects of mutations on protein–protein interactions. Nucleic Acids Res. 2019;47:W338–W344. doi: 10.1093/nar/gkz383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 27.Eswar N., Webb B., Marti‐Renom M.A., Madhusudhan M., Eramian D., Shen M.y., Pieper U., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protocols Protein Sci. 2007;50:2.9. 1–2.9. 31. doi: 10.1002/0471140864.ps0209s50. [DOI] [PubMed] [Google Scholar]
- 28.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 29.Dominguez C., Boelens R., Bonvin A.M. HADDOCK: a protein− protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 30.Xue L.C., Rodrigues J.P., Kastritis P.L., Bonvin A.M., Vangone A. PRODIGY: a web server for predicting the binding affinity of protein–protein complexes. Bioinformatics. 2016;32:3676–3678. doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- 31.Salomon‐Ferrer R., Case D.A., Walker R.C. An overview of the Amber biomolecular simulation package. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2013;3:198–210. [Google Scholar]
- 32.Price D.J., Brooks C.L., III A modified TIP3P water potential for simulation with Ewald summation. J. Chem. Phys. 2004;121:10096–10103. doi: 10.1063/1.1808117. [DOI] [PubMed] [Google Scholar]
- 33.Watowich S.J., Meyer E.S., Hagstrom R., Josephs R. A stable, rapidly converging conjugate gradient method for energy minimization. J. Comput. Chem. 1988;9:650–661. [Google Scholar]
- 34.Kräutler V., Van Gunsteren W.F., Hünenberger P.H. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 2001;22:501–508. [Google Scholar]
- 35.Roe D.R., Cheatham T.E., III PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theor. Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 36.Hou T., Wang J., Li Y., Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011;51:69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khan A., Junaid M., Li C.-D., Saleem S., Humayun F., Shamas S., Ali S.S., Babar Z., Wei D.-Q. Dynamics insights into the gain of flexibility by Helix-12 in ESR1 as a mechanism of resistance to drugs in breast cancer cell lines. Front. Mol. Biosci. 2020;6:159. doi: 10.3389/fmolb.2019.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.A. Khan, W. Heng, Y. Wang, J. Qiu, X. Wei, S. Peng, S. Saleem, M. Khan, S.S. Ali, D.-Q. Wei, In Silico and in Vitro Evaluation of Kaempferol as a Potential Inhibitor of the SARS-CoV-2 Main Protease (3CLpro), Phytotherapy research: PTR. [DOI] [PMC free article] [PubMed]
- 39.Khan A., Rehman Z., Hashmi H.F., Khan A.A., Junaid M., Sayaf A.M., Ali S.S., Hassan F.U., Heng W., Wei D.-Q. An integrated systems biology and network-based approaches to identify novel biomarkers in breast cancer cell lines using gene expression data. Interdiscipl. Sci. Comput. Life Sci. 2020:1–14. doi: 10.1007/s12539-020-00360-0. [DOI] [PubMed] [Google Scholar]
- 40.Khan A., Khan M., Saleem S., Babar Z., Ali A., Khan A.A., Sardar Z., Hamayun F., Ali S.S., Wei D.-Q. Phylogenetic analysis and structural perspectives of RNA-dependent RNA-polymerase inhibition from SARs-CoV-2 with natural products. Interdiscipl. Sci. Comput. Life Sci. 2020:1–14. doi: 10.1007/s12539-020-00381-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khan A., Khan M.T., Saleem S., Junaid M., Ali A., Ali S.S., Khan M., Wei D.-Q. Structural Insights into the mechanism of RNA recognition by the N-terminal RNA-binding domain of the SARS-CoV-2 nucleocapsid phosphoprotein. Comput. Struct. Biotechnol. J. 2020 doi: 10.1016/j.csbj.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wold S., Esbensen K., Geladi P. Principal component analysis. Chemometr. Intell. Lab. Syst. 1987;2:37–52. [Google Scholar]
- 43.Pearson K. LIII. On lines and planes of closest fit to systems of points in space. London Edinburgh Dublin Philosophical Magazine J. Sci. 1901;2:559–572. [Google Scholar]
- 44.Balsera M.A., Wriggers W., Oono Y., Schulten K. Principal component analysis and long time protein dynamics. J. Phys. Chem. 1996;100:2567–2572. [Google Scholar]
- 45.Ernst M., Sittel F., Stock G. Contact-and distance-based principal component analysis of protein dynamics. J. Chem. Phys. 2015;143 doi: 10.1063/1.4938249. 12B640_641. [DOI] [PubMed] [Google Scholar]
- 46.Verma J., Subbarao N. Insilico study on the effect of SARS-CoV-2 RBD hotspot mutants' interaction with ACE2 to understand the binding affinity and stability. Virology. 2021;561:107–116. doi: 10.1016/j.virol.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310. doi: 10.1016/j.cell.2020.08.012. e1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ali A., Vijayan R. Dynamics of the ACE2–SARS-CoV-2/SARS-CoV spike protein interface reveal unique mechanisms. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-71188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020 doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sawatzki K., Hill N.J., Puryear W.B., Foss A.D., Stone J.J., Runstadler J.A. Host barriers to SARS-CoV-2 demonstrated by ferrets in a high-exposure domestic setting. Proc. Natl. Acad. Sci. Unit. States Am. 2021;118 doi: 10.1073/pnas.2025601118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xue T., Wu W., Guo N., Wu C., Huang J., Lai L., Liu H., Li Y., Wang T., Wang Y. Single point mutations can potentially enhance infectivity of SARS-CoV-2 revealed by in silico affinity maturation and SPR assay. bioRxiv. 2020 doi: 10.1039/d1ra00426c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davenport Thaddeus M., Gorman J., Joyce M.G., Zhou T., Soto C., Guttman M., Moquin S., Yang Y., Zhang B., Doria-Rose Nicole A., Hu S.-L., Mascola John R., Kwong Peter D., Lee Kelly K. Somatic hypermutation-induced changes in the structure and dynamics of HIV-1 broadly neutralizing antibodies. Structure. 2016;24:1346–1357. doi: 10.1016/j.str.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ovchinnikov V., Louveau J.E., Barton J.P., Karplus M., Chakraborty A.K. Role of framework mutations and antibody flexibility in the evolution of broadly neutralizing antibodies. Elife. 2018;7 doi: 10.7554/eLife.33038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Q., Nie J., Wu J., Zhang L., Ding R., Wang H., Zhang Y., Li T., Liu S., Zhang M. SARS-CoV-2 501Y. V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184:2362–2371. doi: 10.1016/j.cell.2021.02.042. e2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data is available on RCSB, UniProt and any simulation data will be provided on demand.