Abstract
We developed a dual-mode surface-enhanced Raman scattering (SERS)-based aptasensor that can accurately diagnose and distinguish severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and influenza A/H1N1 at the same time. Herein, DNA aptamers that selectively bind to SARS-CoV-2 and influenza A/H1N1 were immobilized together on Au nanopopcorn substrate. Raman reporters (Cy3 and RRX), attached to the terminal of DNA aptamers, could generate strong SERS signals in the nanogap of the Au nanopopcorn substrate. Additionally, the internal standard Raman reporter (4-MBA) was immobilized on the Au nanopopcorn substrate along with aptamer DNAs to reduce errors caused by changes in the measurement environment. When SARS-CoV-2 or influenza A virus approaches the Au nanopopcorn substrate, the corresponding DNA aptamer selectively detaches from the substrate due to the significant binding affinity between the corresponding DNA aptamer and the virus. As a result, the related SERS intensity decreases with increasing target virus concentration. Thus, it is possible to determine whether a suspected patient is infected with SARS-CoV-2 or influenza A using this SERS-based DNA aptasensor. Furthermore, this sensor enables a quantitative evaluation of the target virus concentration with high sensitivity without being affected by cross-reactivity. Therefore, this SERS-based diagnostic platform is considered a conceptually new diagnostic tool that rapidly discriminates against these two respiratory diseases to prevent their spread.
Keywords: Surface-enhanced Raman scattering, Aptasensor, Au Nanopopcorn substrate, Duplex assays, SARS-CoV-2, Influenza virus
1. Introduction
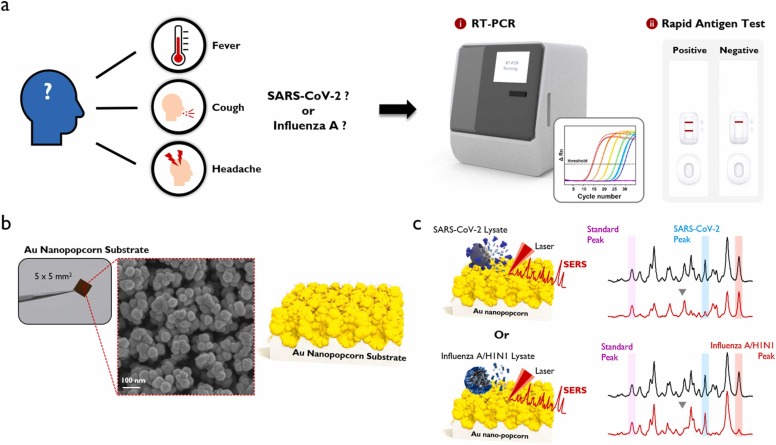
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and influenza virus have many clinical similarities [1], [2], [3]. Both viruses are transmitted through the respiratory tract and show mild symptoms in the initial infection state but lead to death when the virus reaches a severe condition. In addition, they are primarily airborne and have similar symptoms such as fever, cough, fatigue, and muscle pain [4], [5], [6]. Therefore, if the SARS-CoV-2 pandemic situation is prolonged and favorable conditions for influenza virus are created due to the seasonal characteristics, we cannot exclude the possibility for the simultaneous outbreaks of SARS-CoV-2 and influenza virus. Pharmaceutical companies such as Pfizer and Moderna, which have successfully developed an RNA vaccine against SARS-CoV-2 [7], [8], [9], have also started developing a vaccine that can act simultaneously against these two respiratory infectious diseases. However, it is urgent to develop a diagnostic technology rapidly discriminating these two respiratory diseases to prevent their spread. In preparation for such a situation, dual reverse-transcription polymerase chain reaction (RT-PCR) reagents to simultaneously diagnose SARS-CoV-2 and influenza virus from clinical nasopharyngeal samples were already commercialized [10], [11]. Furthermore, dual-mode rapid immunodiagnostic kits for SARS-CoV-2 and influenza virus using antibody-antigen interactions were also developed [12], [13]. Scheme 1a shows the symptoms that appear when infected with SARS-CoV-2 or influenza A and the RT-PCR and Rapid antigen kit that can diagnose them.
Scheme 1.
(a) Symptoms that appear when infected with SARS-CoV-2 or influenza A, and RT-PCR and rapid antigen kit for their diagnosis. (b) Photograph and SEM image of Au nanopopcorn substrate. (c) Working principle of dual aptamer-immobilized Au nanopopcorn substrate for virus assays.
As reported so far, RT-PCR shows high sensitivity and specificity, but it takes a long time for diagnosis since it needs a viral RNA extraction and 20–40 PCR amplification steps. Therefore, the research into shortening the diagnosis time using isothermal PCR or gene scissors is actively underway in RT-PCR diagnosis [14], [15], [16]. In the case of an immunoassay-based rapid kit, the diagnostic accuracy of the initial or asymptomatic infection patient is very low due to the limit of its detection sensitivity. As a result, the false-negative diagnosis of infected patients is recognized as the most severe problem [17], [18]. To resolve this problem, many researchers have made efforts to improve the detection sensitivity of a rapid kit through optical measurements such as fluorescence and chemiluminescence [19], [20], [21], [22], [23]. Our research group has been trying to improve its diagnostic sensitivity and accuracy by using a surface-enhanced Raman scattering (SERS)-based detection different from the fluorescence or chemiluminescence detections used in conventional molecular diagnostics or immunoassays [24], [25], [26], [27], [28]. The SERS detection is a method of measuring the high-sensitivity Raman scattering signals of incident light amplified by the localized surface plasmon effects of molecules present in the nanogaps of the Au nanocomposite [29], [30], [31], [32]. We recently reported a novel SERS-based DNA aptasensor platform that can detect SARS-CoV-2 [33] or influenza virus [34] with high sensitivity. Herein, Au nanopopcorn substrates were used as a susceptible and reproducible SERS platform for the virus assays. Additionally, specific DNA aptamers [34], [35], [36], [37], [38], [39] that selectively bind to the virus target protein biomarkers were used as the receptors (Scheme 1b).
To quickly distinguish whether a suspected patient with similar symptoms is infected with SARS-CoV-2 or influenza virus, we developed a dual-mode virus assay platform that rapidly and accurately differentiates the virus type in this study. Herein, SARS-CoV-2 and influenza A aptamers were simultaneously immobilized on the same Au nanopopcorn substrate and then selectively reacted according to the kind of virus approaching the substrate (Scheme 1c). This simultaneous dual-mode DNA aptasensor provides an efficient diagnostic tool that can accurately determine which virus is infected and respond quickly when a patient has cold symptoms during the changing seasons.
2. Experimental section
2.1. Reagent and materials
Ethanol (99.5%), tris(2-carboxyethyl) phosphine hydrochloride (TCEP), 6-mercapto-1-hexanol (MCH), 4-mercaptobenzoic acid (4-MBA), saline-sodium citrate (SSC) buffer (pH 7.0), and magnesium chloride were purchased from Sigma-Aldrich (St. Louis, MO, USA). Phosphate-buffered saline (PBS) (10 ×, pH 7.4) was purchased from Invitrogen Corporation (Carlsbad, CA, USA). Ultrapure water (0.055 μs/cmc) was obtained from a laboratory water system (Goettingen, Germany). Capture DNAs and aptamer probes oligonucleotides were purchased from Integrated DNA Technologies, Inc. (Coralville, IA, USA), having the sequences of 5′-SH-(CH2O)3(CH2)3-TGT CCA TTA ACG CCC-3′, and capture DNA 2: 5′-SH-(CH2O)3(CH2)3-TGC GCC GAG GTG ATG-3′. Aptamer probe 1: 5′-CAG CAC CGA CCT TGT GCT TTG GGA GTG CTG GTC CAA GGG CGT TAA TGG ACA- Cy3–3′ [33], [35] and aptamer probe 2: 5′-TAC TGC ACA CGA CAC CGA CTG TCA CCA TCA CCT CGG CGC A-RRX-3′ [39]. SARS-CoV-2 lysate was prepared at Korea Research Institute of Bioscience and Biotechnology (KRIBB), (Daejeon, South Korea). Influenza A/H1N1, influenza A/H3N2 and influenza B viruses were purchased from Microbix (Mississauga, Ontario, Canada). Chung-Ang University Hospital provided virus-free clinical nasopharyngeal samples. Various concentrations of virus samples were prepared by spiking SARS-CoV-2 lysate or influenza A/H1N1 lysate into transport media containing virus-free nasopharyngeal samples to detect each virus under conditions similar to actual clinical tests. The research protocol for clinical samples was approved by the Institutional Review Board (IRB) of the Chung-Ang University Hospital. All chemicals used in this study were of analytical reagent grade and were used without further purification.
2.2. Instrumentation
The Raman images and SERS spectra were acquired using an in Via Renishaw Raman microscope system (Renishaw, New Mills, UK). A He–Ne laser operated at 632.8 nm was used as the excitation source. The Raman spectral data were collected using a charge-coupled device (CCD) camera. The Raman mapping images were obtained by using a 20 × (NA 0.4) objective lens with a diffraction limit of around 0.9 µm. The baseline correction of Raman spectra was performed using the WiRE V 4.0 software (Renishaw, New Mills, UK). The spectral analysis was performed using the Spectragryph software and Origin Pro V8 software (OriginLab Corporation, Northants, USA).
2.3. Fabrication and characterization of SERS-based DNA aptasensors
The fabrication procedure of the Au nanopopcorn surface has been previously reported [33], [34]. Before the functionalization, this plasmonic substrate was cleaned with ethanol and deionized water. Before the hybridization, equal molar concentrations of the capture DNA 1 and the aptamer probe 1 were mixed and heated to 90 °C for 10 min to unfold their strain and then cooled to room temperature. The identical process was performed for the capture DNA 2 and the aptamer probe 2. Then the DNAs were treated with TCEP to activate the thiol groups at the end of capture DNA sequences at pH 4 for 1 h. PBS buffer solution was added to adjust the concentration of the mixing two aptamer probes to be 2 μM. Subsequently, the plasmonic substrate was incubated for 2 h in the mixing aptamers DNA solution and then immersed in 2 mM MCH containing a 0.1 mM 4-MBA solution at room temperature for 2 h. After the functionalization on the surface of the Au nanopopcorn, the aptasensors were dried with nitrogen gas. The Raman image of the fabricated SERS-based aptasensor (5 mm × 5 mm size) was measured using a mapping tool with a 20 × objective lens. A computer-controlled microscope stage was used to acquire 36 Raman spectra with a step size of 20 µm (total size 120 µm ×120 µm). All spectra were measured with an exposure time of 5 s. Raman mapping images for five random areas on the same Au substrate were measured to investigate spot-to-spot fluctuations of Raman signal intensity. In addition, Raman mapping images for 6 different substrates were also measured to investigate the substrate-to-substrate fluctuations due to the change of the substrate.
2.4. Single viral assays for SARS-CoV-2 and influenza virus A and cross-reactivity tests
For the single viral assays, 5 μL of sample solution (SARS-CoV-2, 0–5000 PFU/mL) or (H1N1, 0–2016 HAU/mL) was dropped onto the single aptamer-immobilized Au nanopopcorn substrate, followed by incubation for 15 min in a humid chamber at room temperature. Then the aptasensors were instantly rinsed with a washing buffer to remove non-specific binding species between the specific aptamers and target proteins. After being dried with nitrogen gas, the aptasensors were measured using the Raman mapping technique. For the cross-reactivity tests, 5 μL of sample solution containing (SARS-CoV-2, 0.32–200 PFU/mL) or (H1N1, 0.13–80.6 HAU/mL) was dropped onto the duplex SERS-based aptasensor, followed by incubation for 15 min in a humid chamber at room temperature. After the aptasensors were instantly rinsed with a washing buffer, they were dried with nitrogen gas. Then corresponding Raman mapping images were measured and analyzed.
2.5. Simultaneous dual-mode assays of SARS-CoV-2 and influenza virus A and selectivity tests
For the dual assays, 5 sample solutions containing (SARS-CoV-2, 0–200 PFU/mL) and (H1N1, 0–80.6 HAU/mL) were mixed with different molar ratios, and then 5 μL of sample solution was dropped onto the duplex SERS-based aptasensor, followed by incubation for 15 min in a humid chamber at room temperature. The Raman mapping images were measured after the aptasensors were instantly rinsed with a washing buffer and dried with nitrogen gas. Meanwhile, 6 mixture sample solutions were prepared and then measured with the same method. For the specificity tests, 5 samples: blank, SARS-CoV-2 (200 PFU/mL), influenza A/H3N2 (1000 HAU/mL), influenza A /H1N1 (80 HAU/mL), and influenza B (500 HAU/mL) were prepared, and then their Raman mapping images were measured and analyzed.
3. Results and discussion
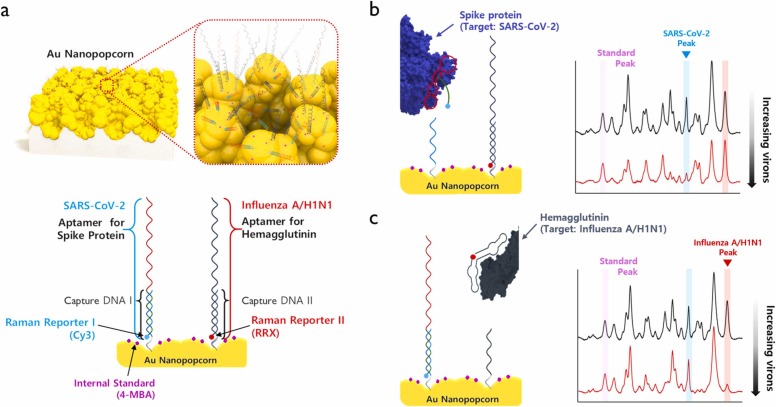
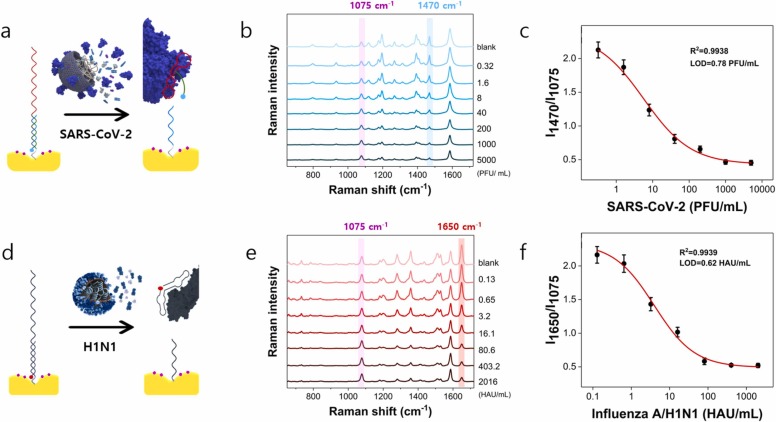
Fig. 1 shows the operating principle of the SERS aptasensor, which can quickly classify and quantitatively analyze SARS-CoV-2 virus and influenza A/H1N1 virus. As shown in Fig. 1a, spike protein DNA aptamer was immobilized on Au nanopopcorn substrate through the hybridization with capture DNA 1 for SARS-CoV-2 detection. At the same time, hemagglutinin DNA aptamer was simultaneously immobilized on the same Au nanopopcorn substrate through the hybridization with capture DNA 2 to detect influenza A/H1N1 virus (Fig. S1). Strong SERS signals for both double-strand DNA aptamers were observed because the Cy3 reporter bound to the spike protein DNA aptamer and the RRX reporter attached to the hemagglutinin DNA aptamer is located very close to the Au nanopopcorn substrate. As shown in Fig. 1b, when SARS-CoV-2 lysates are approaching the Au nanopopcorn substrate, the DNA aptamer corresponding to SARS-CoV-2 binds to the spike protein of SARS-CoV-2 lysate. As a result, the DNA aptamer including Cy3 moves away from the Au nano popcorn surface, and the Raman peak intensity decreases. When influenza virus A/H1N1 approaches Au nanopopcorn surface, the Raman peak intensity for the DNA aptamer, including RRX, drops by the identical mechanism. Therefore, it is possible to quantify the amounts of SARS-CoV-2 and influenza A/H1N1 simultaneously by monitoring the decrease in characteristic Raman peak intensities (1470 cm−1 for Cy3 and 1650 cm−1 for RRX) of both Raman reporter molecules. To correct the variations of Cy3 or RRX SERS signals under different measurement conditions, another Raman reporter 4-MBAs were labeled on the surface of Au nanopopcorn for the use of an internal standard. As shown in Fig. 1b and c, the characteristic Raman peak intensity of MBA at 1075 cm−1 has a constant peak intensity regardless of the presence of viruses. As the concentration of SARS-CoV-2 increases, the number of DNA aptamers that escape from the Au nanopopcorn substrate increases, so the characteristic Raman peak intensity of Cy3 at 1470 cm−1 decreases. As the concentration of influenza A/H1N1 increases, the peak intensity of RRX at 1650 cm−1 decreases by the same principle. Therefore, it is possible to quantify SARS-CoV-2 and influenza A/H1N1 concentrations by monitoring changes in Raman peak intensity of Cy3 and RRX.
Fig. 1.
Schematic illustration for the quantitative evaluation of influenza A/H1N1 and SARS-CoV-2 using the dual-mode SERS aptasensor. (a) Two Raman reporters-labeled DNA aptamers are hybridized with capture DNAs on the Au nanopopcorn substrate. The internal standard 4-MBAs are immobilized along with aptamer DNAs on the Au nanopopcorn substrate. Recognition of target protein induces the aptamer’s conformational change, leading to decreased corresponding Raman signal intensities for (b) SARS-CoV-2, 0 PFU/mL (black) to 1000 PFU/mL (red) and (c) Influenza A/H1N1, 0 HAU/mL (black) to 403 HAU/mL(red).
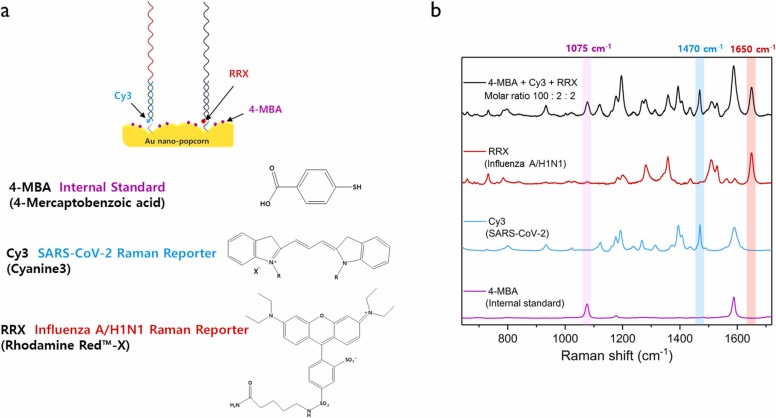
Fig. 2a shows the molecular structures of three Raman reporters, 4-MBA (internal standard), Cy3 (SARS-CoV-2), and RRX (influenza A/H1N1), used in this work. Among the Raman reporter molecules used here, 4-MBA has a much smaller Raman cross-section than Cy3 or RRX, so the enhancement factor is small when the same concentration of molecules is used for measurement. Therefore, we used a much higher molar concentration of 4-MBA than that of Cy3 or RRX for SERS measurements. Fig. 2b shows the SERS spectra for the 100:2:2 molar concentration ratio of 4-MBA, Cy3, and RRX attached to the Au nanopopcorn substrate. At this time, it is possible to quantitatively analyze SASR-CoV-2 and influenza A/H1N1 by monitoring the Raman peak intensity change for each Raman reporter molecule at 1075 cm−1 (4-MBA), 1470 cm−1 (Cy3), and 1650 cm−1 (RRX).
Fig. 2.
(a) Molecular structures of three Raman reporters. (b) Average SERS spectra of 0.1 mM 4-MBA, 2 μM Cy3, 2 μM RRX and their 1:1:1 molar mixtures on Au nanopopcorn substrate. Characteristic Raman peaks of Cy3 at 1470 cm− 1) and RRX at 1650 cm− 1 were used for the quantitative evaluation of SARS-CoV-2 and influenza A/H1N1, respectively. The Raman peak of 4-MBA at 1075 cm− 1 was used as an internal standard.
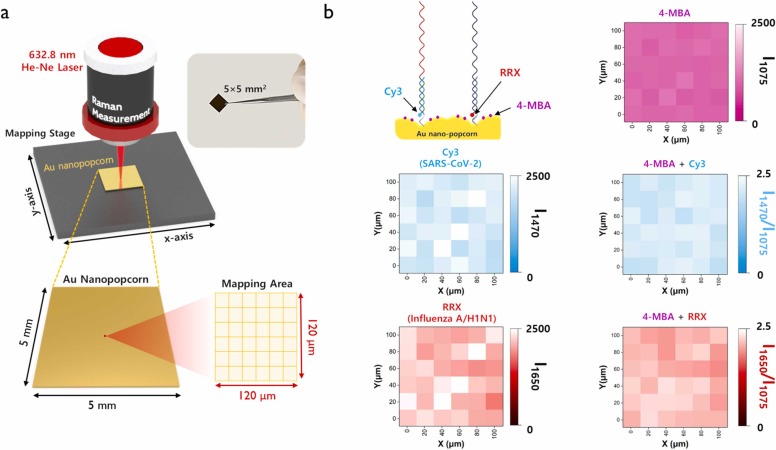
Fig. 3a shows the photograph of an Au nanopopcorn substrate and Raman mapping method used in this study. A 5 × 5 mm2 Au nanopopcorn substrate was used as the substrate for a dual-mode SERS-based aptasensor. A 120 × 120 µm2 area was randomly selected on the substrate, and its Raman mapping image was measured at 20 µm intervals (Fig. 3a). The upper figure of Fig. 3b is a Raman mapping image for Raman peak intensity at 1075 cm−1 of 4-MBA used as an internal standard. The figure on the right is a color decoding bar representing Raman peak intensity distributions. The middle and lower figures are Raman mapping images showing the peak intensity changes for Cy3 and RRX. Both images show more uniform intensity distributions when the corresponding intensities are corrected with the Raman mapping intensity value of the internal standard 4-MBA (I1470/I1075 and I1650/I1075). It shows that the reproducibility of each Raman reporter molecule is improved when each Raman intensity is corrected using the peak of the internal standard 4-MBA. When we used the internal standard, the relative standard deviations (RSDs) were improved from 8.0% to 5.2% for SARS-CoV-2 and 9.1% to 5.0% for Influenza A, respectively.
Fig. 3.
Data collections using the Raman mapping technique. (a) Photograph of an Au nanopopcorn substrate and Raman mapping method. Raman mapping images were obtained with an excitation laser wavelength of 632.8 nm, scanned over a 120 µm (x-axis) and 120 µm (y-axis) range using a computer-controlled microscopic stage. (1 pixel=20 µm×20 µm). (b) Raman mapping image measured with the Raman peak intensity at 1075 cm−1 (4-MBA, pink), 1470 cm−1 (Cy3, blue), and 1650 cm−1 (RRX, red). Images on the right are the Raman mapping images for normalized Raman peaks intensity ratios (I1470/I1075, blue and I1650/I1075, red). The scale bars show color decoding bars representing Raman peak intensity distributions.
Herein, it needs to evaluate the substrate-to-substrate reproducibility since many pieces of Au nanopopcorn substrates are used in actual virus assays. Fig. S2a and b show photographs of six different substrate pieces and the average Raman spectra for 36 point pixels of each substrate, respectively. Fig. S2c shows histograms of the RSDs for their normalized Raman peak intensity ratios. This figure shows that the substrate-to-substrate reproducibility is excellent since RSDs for Cy3 and RRX are 1.7% and 2.1%, respectively. In addition, we also evaluated spot-to-spot reproducibility for the same substrate. As shown in Fig. S3a, five areas were selected on the same substrate, and their corresponding Raman spectra were measured for six random points chosen for each location (Fig. S3b). According to our experimental data, the spot-to-spot reproducibility is also nice because the RSDs for the five testing areas were estimated to be 9.7% (I1470) and 11.9% (I1650), respectively.
Fig. 4a shows the process of assaying SARS-CoV-2 lysate after immobilizing 4-MBA and spike protein DNA aptamers on the Au nanopopcorn substrate. Fig. 4b displays the corresponding Raman spectral changes in the concentration range 0–5000 PFU/mL of SARS-CoV-2 lysate. This figure also shows that the characteristic Raman peak at 1075 cm−1 of the internal standard 4-MBA shows a constant intensity. On the other hand, the Raman peak intensity of the Cy3 Raman reporter at 1470 cm−1, attached to the DNA aptamer terminal, decreases concomitantly as the concentration increases. This is because the spike protein of SARS-CoV-2 lysate induces the DNA aptamer to detach from the substrate and reduces its Raman intensity. Fig. 4c shows the calibration curve for the I1470/I1075 ratio for the increase of SARS-CoV-2 concentration. The Raman peak intensity ratio for each concentration was determined from the average value of Raman peak intensities for 36 mapping area pixels, which showed a good correlation of R2 = 0.9938 and LOD= 0.78 PFU/mL for SARS-CoV-2 in Fig. 4c. Fig. 4d, e and f also show the process of assaying influenza A/H1N1 after immobilizing DNA aptamers for hemagglutinin, the change in Raman spectra in the 0–2016 HAU/mL concentration range, and corresponding calibration curve, respectively. At this time, the LOD and R2 were estimated to be 0.62 HAU/mL and 0.9939, respectively.
Fig. 4.
Evaluation of Raman peak intensity variations with increasing concentration of the target viruses using single DNA aptamer-immobilized SERS sensors. The internal standard Raman reporter 4-MBA was immobilized on the Au nanopopcorn substrate. (a) Addition of SARS-CoV-2 lysate to a spike protein aptamer-immobilized Au popcorn substrate. (b) Average Raman spectra in the presence of various SARS-CoV-2 concentrations (0−5000 PFU/mL) and (c) the corresponding calibration curve to show a variation of the normalized Raman peak intensity ratio of I1470 /I1075 as a function of the SARS-CoV-2 lysate concentration. LOD= 0.78 PFU/mL and R2 = 0.9938. (d) Addition of influenza A/H1N1 lysate to a hemagglutinin DNA aptamer-immobilized Au popcorn substrate. (e) Average Raman spectra in the presence of various influenza A/H1N1 concentrations (0−2016 HAU/mL) and (f) the corresponding calibration curve to show a variation of the normalized Raman peak intensity ratio I1650 /I1075 as a function of the influenza A/H1N1 lysate concentration. LOD= 0.68 HAU/mL and R2 = 0.9939.
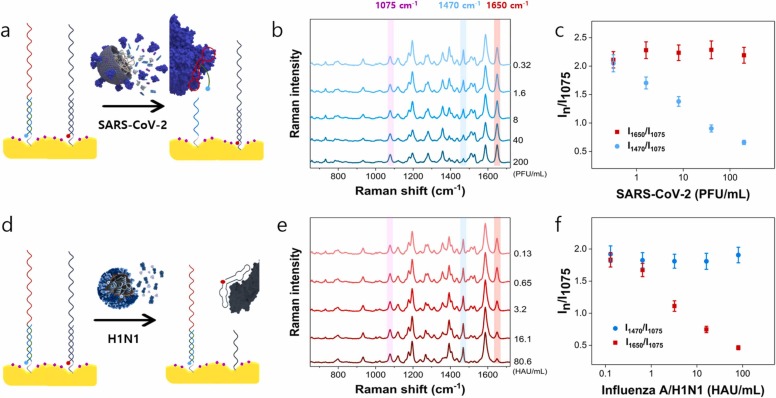
Fig. 5 shows the results of evaluating the cross-reactivity between SARS-CoV-2 and influenza A/H1N1 using a dual-mode SERS-based aptasensor. The internal standard Raman reporter 4-MBA was immobilized on the Au nanopopcorn substrate. Then two different DNA aptamers binding to spike protein and hemagglutinin were immobilized on the substrate with the same molar concentrations. The bindings between DNA aptamers and target proteins were induced by adding various concentrations of SARS-CoV-2 or influenza A/H1N1 lysate to the substrate. When the SARS-CoV-2 lysate was added to Au nanopopcorn substrate (Fig. 5a), only the spike protein DNA aptamer selectively binds to SARS-CoV-2, and the characteristic Raman peak intensity of Cy3 at 1470 cm−1 decreases (Fig. 5b). On the other hand, the characteristic Raman peak intensity at 1650 cm−1 of RRX attached to the hemagglutinin DNA aptamer is maintained at a constant intensity regardless of the concentration of SARS-CoV-2 (Fig. 5c). Conversely, as shown in Fig. 5d, when influenza A/H1N1 lysate was added to Au nanopopcorn substrate, only hemagglutinin DNA aptamers selectively combined with influenza A/H1N1 and the characteristic Raman peak intensity of RRX at 1650 cm−1 decreases (Fig. 5e). In this case, the Raman peak intensity at 1470 cm−1 is kept constant (Fig. 5 f). From these experimental results, we can conclude that the spike protein- and hemagglutinin-compatible DNA aptamers, fixed together on the Au nanopopcorn substrate, have low cross- reactivity against SARS-CoV-2 and influenza A/H1N1 without any interference. Therefore, it is possible to quickly and accurately distinguish SARS-CoV-2 and influenza A/H1N1 using this dual-mode DNA aptasensor platform.
Fig. 5.
Evaluation of the cross-reactivity between SARS-CoV-2 and influenza A/H1N1 using a dual-mode SERS-based aptasensor. The internal standard Raman reporter 4-MBA was immobilized on the Au nanopopcorn substrate. (a) Addition of SARS-CoV-2 lysate to a dual aptamer-immobilized Au popcorn substrate. (b) Average Raman spectra in the presence of various SARS-CoV-2 lysate (0.32−200 PFU/mL) concentrations and (c) the variations of the I1470/I1075 and I1650 /I1075. (d) Addition of influenza A/H1N1 lysate to a dual aptamer-immobilized Au popcorn substrate. (e) Average Raman spectra in the presence of various influenza/H1N1 lysate (0.13−80.6 HAU/mL) concentrations and (f) the variations of the I1470/I1075 and I1650 /I1075.
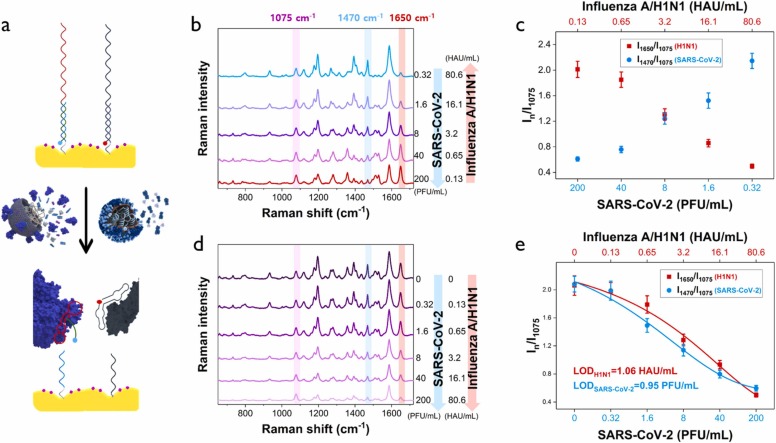
When the concentrations of SARS-CoV-2 and influenza A/H1N1 were relatively changed, corresponding variations in SERS signal intensity were also measured ( Fig. 6a, b). In Fig. 6c, the relative changes in SERS signal intensity were plotted when the concentrations of SARS-CoV-2 and influenza A/H1N1 lysates ranged in 0.32–200 PFU/mL and 80.6–0.13 HAU/mL, respectively. When the concentration ratio of H1N1 and SARS-CoV-2 was 0.13/200, the SERS peak intensity of H1N1 was relatively strong but when it was 3.2/8, they had almost the same intensity. However, when the ratio is 80.6/0.32, the SERS intensity of SARS-CoV-2 was relatively strong. SERS spectra were also measured with the increase of both concentrations of SARS-CoV-2 and influenza A/H1N1 equally in the ranges of 0.2–200 PFU/mL and 0–80.6 HAU/mL (Fig. 6d). As the concentration of each virus increased, the corresponding SERS intensity for both viruses concomitantly decreased in a similar pattern (Fig. 6e). Thus, the concentration of each virus can be accurately determined using those calibration curves.
Fig. 6.
Simultaneous evaluations of SARS-CoV-2 and influenza A/H1N1 viruses using a dual-mode SERS-based aptasensor. (a) Addition of different concentration ratios of SARS-CoV-2 and influenza A/H1N1 lysates to a dual-mode Au nanopopcorn substrate. (b) Average Raman spectra for different concentration ratios of SARS-CoV-2 and influenza A/H1N1 when the concentration of SARS-CoV-2 lysate decreases but that of influenza A/H1N1 increases (200 PFU/mL/0.13 HAU/mL to 0.32 PFU/mL/80.6 HAU/mL) and (c) the corresponding variations of the I1470/I1075 and I1650 /I1075 ratios. (d) Average Raman spectra for different concentration ratios of SARS-CoV-2 and influenza A/H1N1 when both concentrations increase (0 PFU/mL/0 HAU/mL to 200 PFU/mL/80.6 HAU/mL) and (e) the corresponding variations of the I1470/I1075 and I1650 /I1075 ratios.
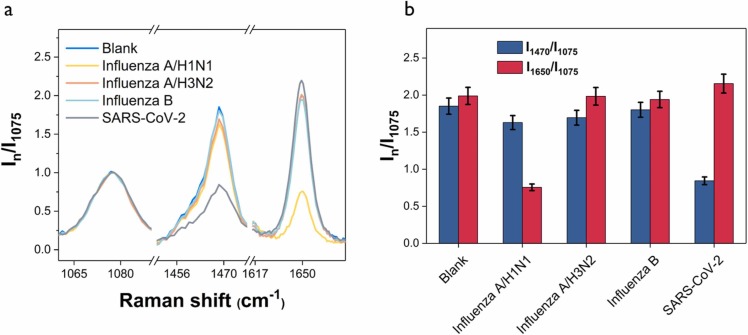
To test the specificity of the dual-mode SERS-based aptasensor, four different viruses, SARS-CoV-2 (200 PFU/mL), Influenza A/H3N2 (1000 HAU/mL), Influenza A/ H1N1 (80 HAU/mL), and Influenza B (500 HAU/mL), were added to the substrate and corresponding SERS spectra were measured. Fig. 7a shows the normalized Raman spectra showing only the characteristic Raman peaks of 4-MBA, Cy3, and RRX for each virus. The full Raman spectra for related viruses are shown in Fig. S4. Fig. 7b shows the changes in relative SERS peak intensities I1470/I1075 and I1650/I1075 for each virus. Compared to the SERS intensity of the blank, the I1650/I1075 and I1470/I1075 show a considerable decrease only for influenza A/H1N1 and SARS-CoV-2. On the other hand, both peak intensity ratios show similar values for other viruses. Therefore, we can conclude that the spike protein and hemagglutinin DNA aptamers have good selectivity only for SARS-CoV-2 and influenza A/H1N1.
Fig. 7.
Evaluation of assay specificity of the dual-mode SERS-based aptasensor. (a) Normalized average Raman spectra for blank, SARS-CoV-2 (200 PFU/mL), influenza A/H3N2 (1000 HAU/mL), influenza A /H1N1 (80 HAU/mL), and influenza B (500 HAU/mL). (b) Corresponding histograms for the normalized Raman peak intensity ratios (I1470/I1075) and (I1650/I1075) collected from the SERS peak intensities in (a).
4. Conclusion
There is a possibility that SARS-CoV-2 and influenza A will coincide in autumn or winter. Since both respiratory infectious diseases have similar symptoms, it is critical to differentiate them through a prompt and accurate diagnosis. This study developed a conceptually new SERS-based aptasensor that can accurately distinguish them using Au nanopopcorn substrates co-immobilized with the spike protein and hemagglutinin DNA aptamers. When SARS-CoV-2 approaches the substrate, only the corresponding spike protein DNA aptamer binds to the SARS-CoV-2 virus and moves away from the substrate, and the Raman peak intensity of Cy3 at 1470 cm−1 decreases. When the influenza A/H1N1 virus approaches the substrate, the hemagglutinin DNA aptamer that binds to the influenza/H1N1 virus moves away from the substrate. As a result, the characteristic Raman peak intensity of RRX at 1650 cm−1 decreases. The characteristic Raman peak intensity at 1075 cm−1 of 4-MBA was used as an internal standard to reduce errors caused by changes in the measurement environment. It was possible to measure very minute changes in the concentrations of SARS-CoV-2 or influenza A/H1N1 without the influence of cross-reactivity using the SERS-based aptasensor developed in this study. Therefore, this SERS-based aptasensor is expected to be used as a new diagnostic tool that can quickly and accurately determine which virus is infected when a patient shows a symptom of infection.
CRediT authorship contribution statement
Hao Chen: Conceptualization, Validation, Formal analysis, Investigation. Sung-Kyu Park: Funding acquisition, Methodology, Investigation. Younju Joung: Validation, Formal analysis, Investigation. Taejoon Kang: Funding acquisition, Conceptualization, Methodology, Visualization. Mi-Kyung Lee: Clinical Validation, Conceptualization, Methodology. Jaebum Choo: Funding acquisition, Conceptualization, Methodology, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Research Foundation of Korea (Grant nos. 2019R1A2C3004375, 2020R1A5A1018052, 2021M3H4A1A02051048, and 2021M3E5E3080379), the government-wide Research and Development Fund for the research of infectious diseases in Korea (Grant no. HG18C0062), and Technology Development Program for Biological Hazards Management in Indoor Air through Korea Environment Industry & Technology Institute (KEITI) funded by Ministry of Environment (ME) of Korea (2021003370003). This work was also supported by Nanomedical Devices Development Project of NNFC (CSM2105M101) and the KRIBB Research Initiative Program (1711134081).
Biographies
Hao Chen: Received his B.S. degree in 2011 from the Department of Chemistry at Shandong University, China. He is currently studying for his Ph.D. degree in the Department of Chemistry at Chung-Ang University in South Korea.
Sung-Kyu Park: Received his Ph.D. degree in 2010 from the Department of Chemical Engineering at KAIST, South Korea. He is currently a Senior Researcher at the Korea Institute of Materials Science (KIMS) in South Korea.
Younju Joung: Received her B.S. degree in 2019 from the Department of Bionano Engineering at Hanyang University, South Korea. She is currently studying for her Ph.D. degree in the Department of Chemistry at Chung-Ang University in South Korea.
Taejoon Kang: Received his Ph.D. degree in 2010 from the Department of Chemistry at KAIST, South Korea. He is currently a Senior Researcher at the Korea Research Institute of Bioscience and Biotechnology (KRIBB) in South Korea.
Mi-Kyung Lee: Received her Ph.D. degree in 1999 from Medical School at Chung-Ang University, South Korea. She is currently a Professor in the Clinical Laboratory at Chung-Ang University Hospital in South Korea.
Jaebum Choo: Received his Ph.D. degree in 1994 from the Department of Chemistry at Texas A&M University, U.S.A. Now, he is a Distinguished Professor in the Department of Chemistry at Chung-Ang University in South Korea.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.snb.2021.131324.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Cuadrado-Payan E., Montagud-Marrahi E., Torres-Elorza M., Bodro M., Blasco M., Poch E., Soriano A., Pineiro G.J. SARS-CoV-2 and influenza virus co-infection. Lancet. 2020;395 doi: 10.1016/S0140-6736(20)31052-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petersen E., Koopmans M., Go U., Hamer D.H., Petrosillo N., Castelli F., Storgaard M., Al Khalili S., Simonsen L. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 2020;20:e238–e244. doi: 10.1016/S1473-3099(20)30484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khorramdelazad H., Kazemi M.H., Najafi A., Keykhaee M., Zolfaghari Emameh R., Falak R. Immunopathological similarities between COVID-19 and influenza: investigating the consequences of Co-infection. Microb. Pathog. 2021;152 doi: 10.1016/j.micpath.2020.104554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hashemi S.A., Safamanesh S., Zadeh-moghaddam H.G., Ghafouri M., Azimian A. High prevalence of SARS-CoV-2 and influenza A virus (H1N1) coinfection in dead patients in Northeastern Iran. J. Med. Virol. 2021;93:1008–1012. doi: 10.1002/jmv.26364. [DOI] [PubMed] [Google Scholar]
- 5.Kong W.-H., Li Y., Peng M.-W., Kong D.-G., Yang X.-B., Wang L., Liu M.-Q. SARS-CoV-2 detection in patients with influenza-like illness. Nat. Microbiol. 2020;5:675–678. doi: 10.1038/s41564-020-0713-1. [DOI] [PubMed] [Google Scholar]
- 6.Cheng Y., Ma J., Wang H., Wang X., Hu Z., Li H., Zhang H., Liu X. Co-infection of influenza A virus and SARS-CoV-2: a retrospective cohort study. J. Med. Virol. 2021;93:2947–2954. doi: 10.1002/jmv.26817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mullin R. Pfizer, Moderna ready vaccine manufacturing networks. Chem. Eng. News. 2020;98:12. [Google Scholar]
- 8.Skowronski D.M., De Serres G. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. New Engl. J. Med. 2021;384:1576–1578. doi: 10.1056/NEJMc2036242. [DOI] [PubMed] [Google Scholar]
- 9.Deva Priya S.A., Kavitha S., Venugopal P., Sriram D.K., George M. Can mRNA vaccines turn the tables during the COVID-19 pandemic? Current status and challenges. Clin. Drug Investig. 2021;41:499–509. doi: 10.1007/s40261-021-01022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noerz D., Hoffmann A., Aepfelbacher M., Pfefferle S., Luetgehetmann M. Clinical evaluation of a fully automated, laboratory-developed multiplex RT-PCR assay integrating dual-target SARS-CoV- 2 and influenza A/B detection on a high-throughput platform. J. Med. Microbiol. 2021;70 doi: 10.1099/jmm.0.001295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung H.-Y., Jian M.-J., Chang C.-K., Lin J.-C., Yeh K.-M., Chen C.-W., Chiu S.-K., Wang Y.-H., Liao S.-J., Li S.-Y., Hsieh S.-S., Tsai S.-H., Perng C.-L., Yang J.-R., Liu M.-T., Chang F.-Y., Shang H.-S. Novel dual multiplex real-time RT-PCR assays for the rapid detection of SARS-CoV-2, influenza A/B, and respiratory syncytial virus using the BD MAX open system. Emerg. Microbes Infect. 2021;10:161–166. doi: 10.1080/22221751.2021.1873073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C., Yang X., Zheng S., Cheng X., Xiao R., Li Q., Wang W., Liu X., Wang S. Development of an ultrasensitive fluorescent immunochromatographic assay based on multilayer quantum dot nanobead for simultaneous detection of SARS-CoV-2 antigen and influenza A virus. Sens. Actuators B. 2021;345 doi: 10.1016/j.snb.2021.130372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C., Zheng T., Wang H., Chen W., Huang X., Liang J., Qiu L., Han D., Tan W. Rapid one-pot detection of SARS-CoV‑2 based on a lateral flow assay in clinical samples. Anal. Chem. 2021;93:3325–3330. doi: 10.1021/acs.analchem.0c05059. [DOI] [PubMed] [Google Scholar]
- 14.Ganguli A., Mostafa A., Berger J., Aydin M.Y., Sun F., de Ramirez S.A.S., Valera E., Cunningham B.T., King W.P., Bashir R. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 2020;117:22727–22735. doi: 10.1073/pnas.2014739117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pang B., Xu J., Liu Y., Peng H., Feng W., Cao Y., Wu J., Xiao H., Pabbaraju K., Tipples G., Joyce M.A., Saffran H.A., Tyrrell D.L., Zhang H., Le X.C. Isothermal amplification and ambient visualization in a single tube for the detection of SARS-CoV‑2 using loop-mediated amplification and CRISPR technology. Anal. Chem. 2020;92:16204–16221. doi: 10.1021/acs.analchem.0c04047. [DOI] [PubMed] [Google Scholar]
- 16.Mukhopadhyay C.D., Sharma P., Sinha K., Rajarshib K. Recent trends in analytical and digital techniques for the detection of the SARS-Cov-2. Biophys. Chem. 2021;270 doi: 10.1016/j.bpc.2020.106538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berger1 A., Nsoga1 M.T.N., Perez-Rodriguez F.J., Aad Y.A., Sattonnet-Roche P., Gayet-Ageron A., Jaksic C., Torriani G., Boehm E., Kronig I., Sacks J.A., de Vos M., Bausch F.J., Chappuis F., Renzoni A., Kaiser L., Schibler M., Eckerle I. Diagnostic accuracy of two commercial SARS-CoV-2 antigen-detecting rapid tests at the point of care in community-based testing centers. PLoS One. 2021;16 doi: 10.1371/journal.pone.0248921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guglielmi G. Rapid coronavirus tests: a guide for the perplexed. Nature. 2021;590:202–205. doi: 10.1038/d41586-021-00332-4. [DOI] [PubMed] [Google Scholar]
- 19.Chen Z., Zhang Z., Zhai X., Li Y., Lin L., Zhao H., Bian L., Li P., Yu L., Wu Y., Lin G. Rapid and sensitive detection of anti-SARS-CoV-2 IgG using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal. Chem. 2020;92:7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 20.Wang C., Cheng X., Liu L., Zhang X., Yang X., Zheng S., Rong Z., Wang S. Ultrasensitive and simultaneous detection of two specific SARS-CoV-2 antigens in human specimens using direct/enrichment dual-mode fluorescence lateral flow immunoassay. ACS Appl. Mater. Interfaces. 2021;13:40342–40353. doi: 10.1021/acsami.1c11461. [DOI] [PubMed] [Google Scholar]
- 21.Liu D., Ju C., Han C., Shi R., Chen X., Duan D., Yan J., Yan X. Nanozyme chemiluminescence paper test for rapid and sensitive detection of SARS-CoV-2 antigen. Biosens. Bioelectron. 2021;173 doi: 10.1016/j.bios.2020.112817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suleman S., Shukla S.K., Malhotra N., Bukkitgar S.D., Shetti N.P., Pilloton R., Narang J., Tan Y.N., Aminabhavi T.M. Point of care detection of COVID-19: advancement in biosensing and diagnostic methods. Chem. Eng. J. 2021;414 doi: 10.1016/j.cej.2021.128759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asif M., Xu Y., Xiao F., Sun Y. Diagnosis of COVID-19, vitality of emerging technologies and preventive measures. Chem. Eng. J. 2021;423 doi: 10.1016/j.cej.2021.130189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Z., Choi N., Wang R., Lee S., Moon K.C., Yoon S.-Y., Chen L., Choo J. Simultaneous detection of dual prostate specific antigens using SERS-based immunoassay for accurate diagnosis of prostate cancer. ACS Nano. 2017;11:4926–4933. doi: 10.1021/acsnano.7b01536. [DOI] [PubMed] [Google Scholar]
- 25.Wang X., Choi N., Cheng Z., Ko J., Chen L., Choo J. Simultaneous detection of dual nucleic acids using a SERS-based lateral flow biosensor. Anal. Chem. 2017;89:1163–1169. doi: 10.1021/acs.analchem.6b03536. [DOI] [PubMed] [Google Scholar]
- 26.Wang X., Park S.-G., Ko J., Xiao X., Gianini V., Maier S.A., Kim D.-H., Choo J. Sensitive and reproducible immunoassay of multiple mycotoxins using surface-enhanced Raman scattering mapping on three-dimensional plasmonic nanopillar arrays. Small. 2018;14 doi: 10.1002/smll.201801623. [DOI] [PubMed] [Google Scholar]
- 27.Dang H., Park S.-G., Wu Y., Choi N., Yang J.-Y., Lee S., Joo S.-W., Chen L., Choo J. Reproducible and sensitive plasmonic sensing platforms based on Au-nanoparticle-internalized nanodimpled substrate. Adv. Funct. Mater. 2021;31 [Google Scholar]
- 28.Das A., Kim K., Park S.-G., Choi N., Choo J. SERS-based serodiagnosis of acute febrile diseases using plasmonic nanopopcorn microarray platforms. Biosens. Bioelectron. 2021;192 doi: 10.1016/j.bios.2021.113525. [DOI] [PubMed] [Google Scholar]
- 29.Baumberg J.J., Aizpurua J., Mikkelsen M.H., Smith D.R. Extreme nanophotonics from ultrathin metallic gaps. Nat. Mater. 2019;18:668–678. doi: 10.1038/s41563-019-0290-y. [DOI] [PubMed] [Google Scholar]
- 30.Chen S., Zhang Y., Shih T.-M., Yang W., Hu S., Hu X., Li J., Ren B., Mao B., Yang Z., Tian Z. Plasmon-induced magnetic resonance enhanced Raman spectroscopy. Nano Lett. 2018;18:2209–2216. doi: 10.1021/acs.nanolett.7b04385. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z., Zong S., Wu L., Zhu D., Cui Y. SERS-activated platforms for immunoassay: probes, encoding methods, and applications. Chem. Rev. 2017;117:7910–7963. doi: 10.1021/acs.chemrev.7b00027. [DOI] [PubMed] [Google Scholar]
- 32.Wu K., Li T., Schmidt M.S., Rindzevicius T., Boisen A., Ndoni S. Gold nanoparticles sliding on recyclable nanohoodoos-engineered for surface-enhanced Raman spectroscopy. Adv. Funct. Mater. 2018;28 [Google Scholar]
- 33.Chen H., Park S.-G., Choi N., Kwon H.-J., Kang T., Lee M.-K., Choo J. Sensitive detection of SARS-CoV-2 using SERS-based aptasensor. ACS Sens. 2021;6:2378–2385. doi: 10.1021/acssensors.1c00596. [DOI] [PubMed] [Google Scholar]
- 34.Chen H., Park S.-G., Choi N., Moon J.-I., Dang H., Das A., Lee S., Kim D.-G., Chen L., Choo J. SERS imaging-based aptasensor for ultrasensitive and reproducible detection of influenza virus A. Biosens. Bioelectron. 2020;167 doi: 10.1016/j.bios.2020.112496. [DOI] [PubMed] [Google Scholar]
- 35.Song Y., Song J., Wei X., Huang M., Sun M., Zhu L., Lin B., Shen H., Zhu Z., Yang C. Discovery of aptamers targeting the receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal. Chem. 2020;92:9895–9900. doi: 10.1021/acs.analchem.0c01394. [DOI] [PubMed] [Google Scholar]
- 36.Liu R., He L., Hu Y., Luo Z., Zhang J.A. Serological aptamer-assisted proximity ligation assay for COVID-19 diagnosis and seeking neutralizing aptamers. Chem. Sci. 2020;11:12157–12164. doi: 10.1039/d0sc03920a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Lai B.S., Juhas M. Recent advances in aptamer discovery and applications. Molecules. 2019;24:941. doi: 10.3390/molecules24050941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasegawa H., Savory N., Abe K., Ikebukuro K. Methods for improving aptamer binding affinity. Molecules. 2016;21:421. doi: 10.3390/molecules21040421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhardwaj J., Chaudhary N., Kim H., Jang J. Subtyping of influenza A H1N1 virus using a label-free electrochemical biosensor based on the DNA aptamer targeting the stem region of HA protein. Anal. Chim. Acta. 2019;1064:94–103. doi: 10.1016/j.aca.2019.03.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material