Abstract
Copper(II) carboxylate complexes [Cu2(OOCR)4L2] (1) and [Cu2(OOCR`)4OCO(R`)CuL2]n (2), where L = 2-methyl pyridine, R = 2-chlorophenyl acetate and R` = 2-fluorophenyl acetate were synthesized and characterized by FT-IR spectroscopy and single crystal X-ray analysis. Complex 1 exhibits the typical paddlewheel array of a dinuclear copper(II) complex with carboxylate ligands. In complex 2, this scaffold is further extended into a polymeric arrangement based on alternate paddlewheel and square planar moieties with distinct coordination spheres. The complexes showed better 2,2-diphenyl-1-picrylhydrazyl (DPPH) and hydroxyl radical scavenging activities and have been found to be more potent antileishmanial agents than their corresponding free ligand acid species. UV-Vis absorption titrations revealed good DNA binding abilities {Kb = 9.8 × 104 M–1 (1) and 9.9 × 104 M–1 (2)} implying partial intercalation of the complexes into DNA base pairs along with groove binding. The complexes displayed in vitro cytotoxic activity against malignant glioma U-87 (MG U87) cell lines. Computational docking studies further support complex-DNA binding by intercalation. Molecular docking investigations revealed probable interactions of the complexes with spike protein, the nucleocapsid protein of SARS-CoV-2 and with the angiotensin converting enzyme of human cells.
Keywords: Copper(II) complex, DNA interaction, Anticancer, Antioxidant, SARS-CoV-2
Graphical abstract
.
1. Introduction
Dinuclear copper(II) complexes with four bridging carboxylate ligands have been widely studied due to their facile preparation and diverse applications. The coordination chemistry of copper(II) carboxylates in general as well as in combination with additional donating ligands such as pyridine derived ligands is of particular interest to us in a medicinal-chemical context [1,2]. Such complexes have the general formula [Cu2(OOCR)4L2], where RCOO–is a syn bridging carboxylate ligand at equatorial positions stabilizing the charge of copper(II) ions and L is a pyridine derived N-donor ligand at the axial positions of the paddlewheel structural arrangement. A literature search of the Cambridge Crystallographic Database shows more than 1500 structurally characterized examples for dinuclear paddlewheel copper complexes and those with aromatic N-donor ligands complementing the respective coordination spheres are still well over 300 [3]. In the last two years (2018/2019) alone, almost thirty respective structures have been reported [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17]. The apical nitrogen donor ligands normally saturate the coordination spheres of the copper centers and thereby result in the commonly observed discrete di-nuclear molecular units [2], [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17]. Less common polymeric paddlewheel structures may result in cases where carboxylates act as bridging bidentate ligands in the apical positions [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34]. The formation of di- or polymeric structures may be attributed mainly to both steric as well as electronic effects of the employed ligands. Here we show that varying the electronic structures of the comparatively small phenyl acetic acids by halogen substituent based modulation together with appropriate pyridine co-ligands, can lead to the formation of either of such complex types selectively. The distinct combinations of carboxylates with N-donor ligands not only result in various complexes with a range of geometries but also (fine-)tunes their chemical thermal, magnetic and biological properties [2,5,35,36].
Two further copper(II) carboxylate complexes were synthesized and structurally characterized in course of our continued investigations into these species’ redox active natures, their capability to interact with biomolecules and, hence, their biomedical relevance. The complexes were tested in vitro for their anticancer, antioxidant, antileishmanial activity and DNA interaction potential. Since the outbreak of the coronavirus disease in 2019 (COVID-19), much attention has been paid to developing effective and safe antiviral agents [37] along with vaccination efforts. The potential of inorganic complexes being used as alternatives to conventional small organic molecules or antibody-based therapies is an area of immense recent interest [38,39]. Well defined three-dimensional shapes of the coordination compounds which are used to bind the target protein(s) are prerequisite for an informed and detailed evaluation of their specific biological activities [40]. Thus, the synthesized complexes of this study were characterized structurally and then screened computationally for their interaction ability with the spike protein (protein that mediates the entry of coronavirus into host cells), with the Nucleocapsid protein of SARS-CoV-2 (a target for vaccine development), and with the angiotensin converting enzyme of human cells (host for SARS-CoV-2) as potential sites for successful corona inhibition.
2. Experimental
2.1. Materials and methods
Copper(II) chloride dihydrate, 2-chlorophenylacetic acid (HL1), 2-florophenylacetic acid (HL2), 2-methylpyridine, ascorbic acid, 2,2-diphenyl-1-picrylhydrazyl radical (DPPH), iron(II) sulfate, sulfuric acid and hydrogen peroxide were purchased from Sigma-Aldrich and used without further purification. All solvents were of analytical grade and used without further purification. Distilled water was used after one time distillation. The melting points were measured using a Digital Electro-Thermal Melting Point Apparatus. The copper percentage was determined by using a Perkin Elmer Atomic Absorption Spectrometer A Analyst 700. The FT-IR Spectra (4000–400 cm−1) were recorded on a Nicolet-6700 FTIR spectrophotometer, Thermoscientific, USA using attenuated total reflectance (ATR) technique. The electronic spectra of the complexes for DNA interaction studies were recorded using a Perkin Elmer UV/VIS Spectrophotometer Lambda 25 in aqueous dimethyl sulfoxide (DMSO).
2.2. Synthesis of copper complexes
2.2.1. [Tetrakis(μ-(2-chlorophenyl)acetato-κ2O,O')bis(2-methylpyridine)dicopper(II){Cu‒Cu} (1)
For the synthesis of complex 1, a solution of HL1 (0.50 g, 2.93 mmol) in methanol (10 mL) was added drop wise to the constantly stirring solution of copper(II) chloride dihydrate (0.25 g, 1.46 mmol) in methanol (10 mL). The reaction mixture was refluxed for 3 h followed by the addition of 2-methylpyridine (0.145 mL, 1.46 mmol). The resulting reaction mixture was refluxed for further 3 h. The reaction was stopped and the mixture was then cooled to room temperature to afford dark green colored crystals of the product after a few days. These crystals were separated from the solution and analyzed by different analytical and spectroscopic techniques.
Yield: 80%; M.p. 178 °C; Anal. Calcd. for C44H38Cl4Cu2N2O8(%): C, 53.29; H, 3.86; N, 2.82. Found: C, 53.19; H, 3.81; N, 2.79; FT-IR (cm−1): 1632s v(COO)asym, 1389s v(COO)sym , 243 Δv, 1601 v(C=N), 1476 v(C=C), 740 s pyridyl ring vibration, 685 s v(C—Cl), 473 v(Cu—N), 428 v(Cu—O). AAS Anal. calcd for Cu(%): 12.82. Found: 12.80. UV–VIS [MeOH; λnm]: 726 (d-d transition), 262 (charge transfer transition).
2.2.2. Catena-poly[Tetrakis(μ-(2-fluorophenyl)acetato‒κ2O,O')dicopper(II){Cu‒Cu}(μ2–2-fluorophenyl)acetato-1-O:2-O)(bis-2-methylpyridine)Cu(II)(μ2–2-fluorophenyl)acetato-1-O:2-O) (2)
For the synthesis of complex 2, the same experimental procedure as used in the preparation of complex 1 was followed. HL2 (0.45 g, 2.93 mmol) was used instead of HL1. Yield: 70%; M.p. 198 °C; Anal. Calcd. for C60H50Cu3F6N2O12(%): C, 55.62; H, 3.89; N, 2.16. Found: C, 55.54; H, 3.80; N, 2.10; FT-IR (cm−1): 1644s, 1547s v(OCO)asym, 1395s, 1380s v(OCO)sym , 249, 167 Δv, 1611 v(C = N), 1492 v(C = C), 1228 v(C-F), 750 s pyridyl ring vibration, 461 v(Cu-N), 434 v(Cu-O). AAS Anal. calcd for Cu(%): 14.71. Found: 14.68. UV–VIS [MeOH; λnm]: 738 (d-d transition), 263 (charge transfer transition).
2.3. Crystal structure determination
For the X-ray single crystal analyses appropriate sized crystals of the synthesized complexes (1 and 2) were mounted on a glass fiber. The corresponding structural data were collected using a STOE-IPDS II diffractometer equipped with a normal-focus, 2.4 kW sealed-tube X-ray source with graphite-monochromated Mo Kα radiation (λ= 0.71,073 Å). The single crystal of 1 was measured at room temperature and that of 2 at low temperature (170 K). The program X-Area was used for the integration of diffraction profiles. Numerical absorption corrections were carried out with the programs X-Shape and X-Red32 (all from STOE ©). The structures were solved by direct methods with SHELXT-16 and refined by full-matrix least-squares methods using SHELXL-16 [41, 42]. All non-hydrogen atoms were refined anisotropically and hydrogen atoms were refined isotropically at calculated positions using a riding model (U isovalues constrained to 1.5 Ueq of their pivot atoms for methyl groups and 1.2 Ueq of their pivot atoms for all other groups). The fluorine substituent of one carboxylic acid in the asymmetric unit of complex 2 is disordered over two positions (76% vs. 24%). The hydrogen atom in these two positions was not split but refined with full occupancy on the carbon atom (C4), to which the 24% fluorine atom is bound. The respective C—H distance was fixed (DFIX) and the displacement parameter constrained. Other than that, no constraints or restraints were applied for modelling this disorder. Crystallographic data were deposited with the Cambridge Crystallographic Data Centre, CCDC, 12 Union Road, Cambridge CB21EZ, UK. These data can be obtained free of charge on quoting the depository numbers CCDC 2031623 (1) and 2031624 (2) by FAX (+44-1223-336-033), email (deposit@ccdc.cam.ac.uk) or their web interface (at http://www.ccdc.cam.ac.uk).
2.4. DNA binding study
Salmon sperm DNA (SS-DNA) (10 mg) was dissolved in distilled water (pH=7.0) with continuous stirring for 24 h. The absorbance ratio (A260/A280) of this solution was found to be 1.8, indicating that the DNA solution was sufficiently free of protein impurities [43]. The SS-DNA {ε = 6600M−1cm−1(260 nm)} concentration was determined via a UV absorption measurement [44] and was found to be 0.6 × 10−4 M. Solutions of the test compounds {200 µM} were prepared in aqueous DMSO (1:4). UV absorption titrations were carried out with the test compound of fixed concentration and with SS-DNA solution of varying concentrations. Equivalent solutions of SS-DNA were added to the test compound and reference solutions, to eliminate the absorbance of DNA itself. Test compound-DNA solutions were allowed to incubate for an hour at room temperature before absorption measurements. Spectra were recorded using 1 cm path length cuvettes at room temperature (25±1 °C).
2.5. Antioxidant activities
2.5.1. DPPH free radical scavenging activity
Carboxylic acids (HL1 and HL2) and copper complexes (1 and 2) were assessed in vitro for their DPPH free radical scavenging potentials following a reported procedure [45]. Different concentrations (15, 30, 60, 120 and 240 µM) of the test compounds were mixed with 1000 µL ethanolic solution of DPPH (255 µM). The solution mixtures were shaken and incubated in the dark for half an hour at room temperature. The absorbances of the solution mixtures were recorded at 518 nm. Ascorbic acid at the same concentrations as the test compounds was used as a positive control. The experiments were carried out in triplicates. The percentage inhibition by the test compounds was calculated using the formula:
Ac and As constitute the observed absorbance values of the control and the sample, respectively.
2.5.2. Hydroxyl radical scavenging activity
The hydroxyl radical scavenging activities of the acids (HL1 and HL2) and complexes (1 and 2) were assessed according to a reported method [46]. Different concentrations (15, 30, 60, 120 and 240 µM) of the test compounds were mixed with 200 µL of 3.7 mM O-phenanthroline, 1000 µL of 0.4 M phosphate buffer (pH 6.6), 100 µL of 7.5 mM iron(II) sulfate and 100 µL of H2O2 (0.1%). The mixtures were diluted up to 3000 µL with distilled water and then incubated for half an hour at room temperature. The absorbance of each mixture was recorded at 510 nm. Ascorbic acid was used as a standard.
The scavenging power was calculated using the formula:
As, Ac and Ab constitute the observed absorbance values of the sample, control and blank, respectively.
2.6. Antileishmanial activity
The antileishmanial activity test was performed with a slight modification of the previously reported method [47]. Leishmania tropica promastigotes (1 × 105 parasites/well) and test compounds were dispensed in a flat-bottom 96-well microtiter plate such that the final concentration of each test compound was 1000, 500 and 250 µg/mL in DMSO. Amphotericin-B was used as a positive control while growth media with DMSO were used as blanks. The plates were then incubated at 25 ± 1 °C for 72 h. After the incubation period was over, 100 µL of 3-(4,5-methylthiazol-2-yl)−2, 5-diphenyltetrazolium bromide (MTT) solution was added to each well and the plate was re-incubated for 4 h at 31 °C. 40 µL of DMSO were then added to each well to dissolve the formazan crystals of MTT. The plates were stirred gently for 15 min. The optical absorbance of each plate was measured by the microplate reader (Bio Tek™ ELx800) at 570 nm. Percentage inhibition was then calculated using the following formula.
2.7. Anticancer activity
The malignant glioma U-87 (MG U87) cell lines were cultured as previously described [48]. The cell lines were revived from cryopreserved vials placed in the vapor phase of a liquid nitrogen storage and immediately cultured in an incubator at 37 °C in presence of 5% CO2 supply. The cells were grown as adherent cultures in Dulbecco's Modified Eagle Medium and Ham's F12 nutrient mixture media (DMEM:F12) in 1:1 concentration, supplemented with 10% Fetal bovine serum, 100 U/mL penicillin, and 100 µg/mL streptomycin in T25 cell culture flasks. Upon reaching a confluency of >80%, the cells were split into further passages by plating 50% into double surface area flasks to increase the number of cells.
The growth inhibition assay was conducted as previously described [49]. The malignant glioma U87 cell lines were plated into 96-well plates, with each well containing 5000 cells per well. The cell count was done with a hemocytometer followed by culturing in DMEM:F12 medium for 24 h in a CO2 incubator with 5% CO2 supply. Following 24 h incubation, different concentrations of the test compounds (HL1, HL2, 1 and 2) were prepared in DMSO. The cell lines were treated with test compounds at different serial concentrations ranging from 0 to 250 µg (0 µg/mL (untreated control), 7.812 µg/mL, 15.625 µg/mL, 31.25 µg/mL, 62.5 µg/mL, 125 µg/mL and 250 µg/mL). All test compounds were analyzed at each concentration in triplicates. The plates’ patterns were repeated three times, each with 24 h of treatment. Following the specified time of drug treatments, cells were fixed in 4% formalin, followed by staining with 0.1% crystal violet for 10 min [50,51]. Each plate was washed three times using 1x phosphate buffered saline and air-dried, followed by addition of 200 µL of acetic acid into each well. The absorbance of each plate (well) was recorded at 630 nm using a 96-well plate spectrophotometer reader.
2.8. Molecular docking
The three dimensional (3D) structures of the newly synthesized Cu-complexes were drawn with the Molecular Operating Environment (MOE-2016) software [52]. The hydrogen atoms were added to the synthesized compounds by 3D protonation followed by energy minimization with MOE. The crystal structures of the Salmon sperm-DNA (PDB id: 1BNA), angiotensin converting enzyme of human (PDB id: 1R42), spike protein of SARS-CoV-2 (PDB id: 6CS2), and nucleocapsid protein of SARS-CoV-2 (PDB id: 6M3M) were retrieved from the protein databank [www.rcsb.org/pdb]. The 3D protonation and energy minimizations of the retrieved DNA and proteins were carried out with the MOE software using its default parameters. The macromolecules (DNA, protein) were allowed to dock to the synthesized compounds with MOE and default parameters, i.e., Placement: Triangle Matcher, Rescoring: London dG for DNA and GBVI/WSA dG for proteins. For each test substance ten conformations were generated. The top-ranked conformation of each compound was used for further analysis.
3. Results and discussion
3.1. Synthesis
The complexes were synthesized by the reaction of copper(II) chloride dihydrate with 2-chlorophenylacetic acid (HL1) or 2-florophenylacetic acid (HL2) in the presence of 2-methylpyridine as co-ligand in methanol under reflux. The complexes are dark green crystalline solids and were obtained in good yields. Both complexes were soluble in methanol and DMSO.
3.2. FR-IR spectroscopy
The comparative analysis of FT-IR spectra of the synthesized complexes (1 and 2) with the corresponding free ligand acids (HL1 and HL2) spectra confirmed the complex formations. The most vivid difference was the absence of broader bands in the complex spectra in the range 3400–2400cm–1 which appear due to OH vibrations of the COOH moiety in the free ligand acids [2]. The presence of coordinated deprotonated carboxylate ligands in the complex spectra was also supported by the appearance of new medium intensity bands at 428 and 434 cm–1 for complexes 1 and 2, respectively. The coordination of 2-methyl pyridine to copper centers is reflected by the medium intensity bands at 592 and 593 cm–1 in the spectra of complexes 1 and 2, respectively, owing to the newly established Cu-N bonds and concomitant decrease in the vibration frequencies of the vC=N bond [2,36]. The presence of 2-methyl pyridine was also supported by the strong pyridyl ring vibration at 740 and 750 cm–1 in complexes 1 and 2, respectively [2]. Such notable difference in the value for this vibration in the two complexes suggests a considerably distinct environment of the 2-methyl pyridine ring which was further confirmed by the X-ray single crystal analyses of both the complexes. The strong bands due to C=O (1706, 1689 cm−1) and C—O (1233, 1287 cm−1) vibrations in the free 2-chlorophenylacetic acid and 2-flurophenylacetic acid were replaced by sets of new peaks due to antisymmetric and symmetric vibrations of the COO– moiety in the complex spectra. In complex 1, antisymmetric and symmetric COO– vibrations appeared as strong bands at 1632 and 1389 cm−1 with a Δv value of 243, suggesting a bridging bidentate mode of binding of the carboxylate ligand in complex 1 as also confirmed by the single crystal analysis [53]. Strong two bands each were observed for antisymmetric (1644, 1547) and symmetric vibrations (1395, 1380) of the COO– moiety in complex 2 with Δv values of 249 and 167, respectively, suggesting bridging bidentate modes of coordination of the carboxylate ligand but in two different spatial environments as also confirmed by the single crystal structure of complex 2. Spectra of complexes 1 and 2 exhibit strong bands for the Ar-Cl and Ar-F bonds at 685 and 1228 cm−1, respectively.
3.3. X-ray single crystal analysis
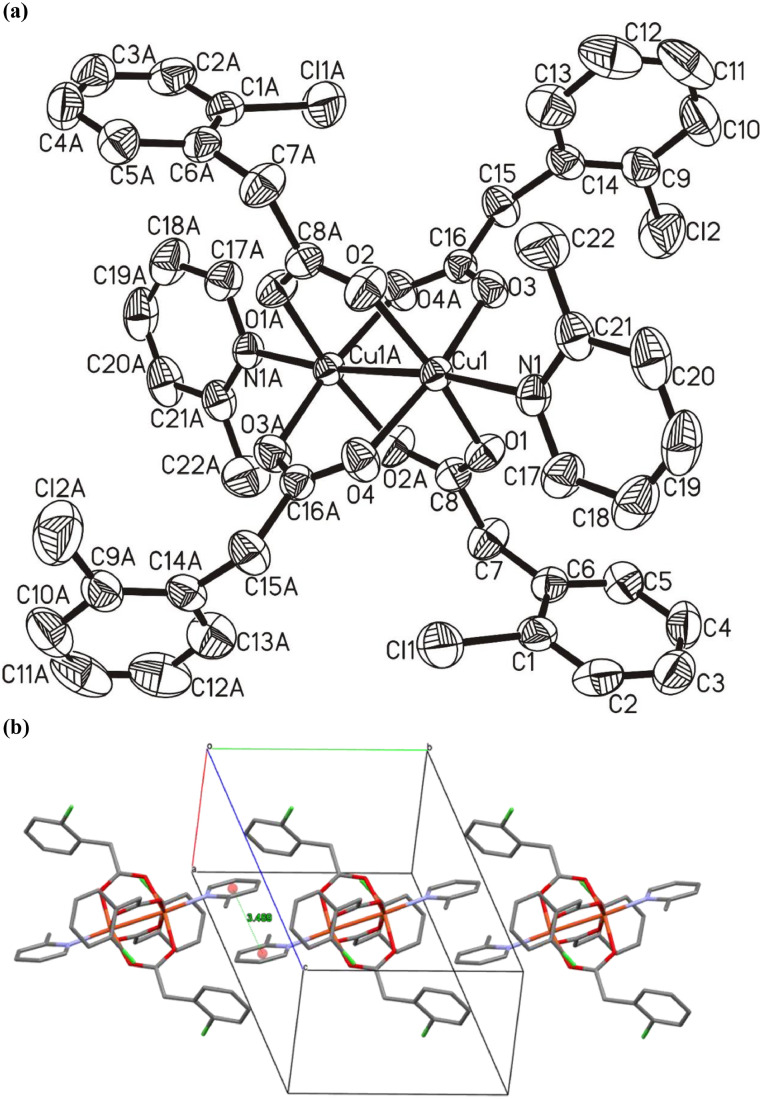
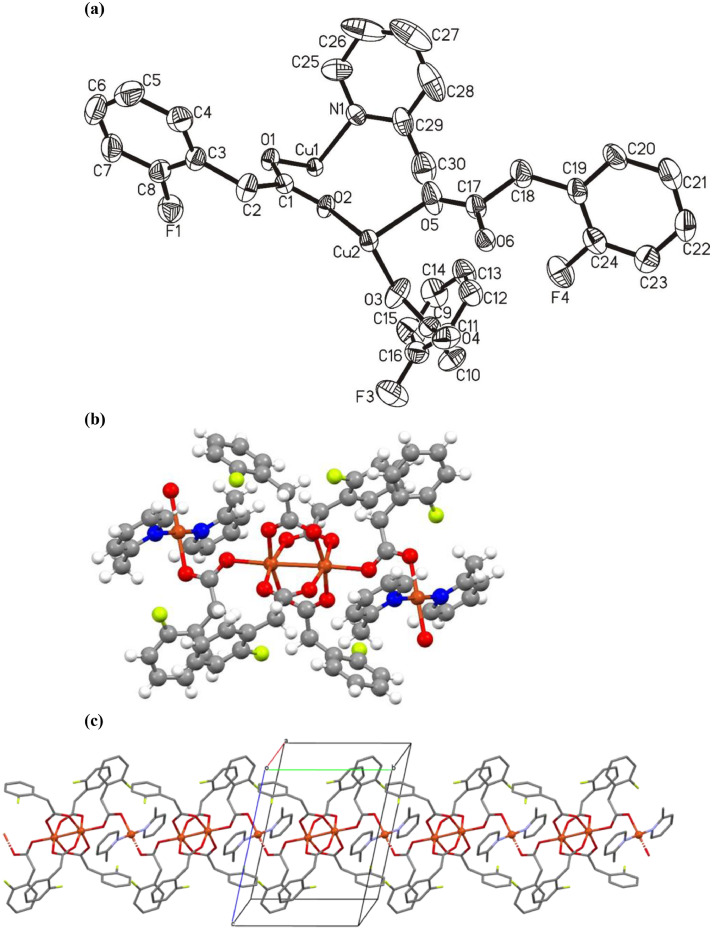
The two copper complexes reported here were synthesized by the reaction of HL1 and HL2 with [CuCl2].2H2O in the presence of 2-methyl pyridine. Crystals of 1 and 2 suitable for X-ray analysis were grown from methanol and the molecular structures are shown in Figs. 1 a and 2 a, respectively. The crystal and refinement data for both complexes are summarized in Table 1 .
Fig. 1.
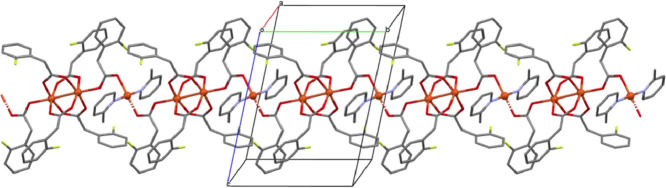
(a) Molecular structure of complex 1 (ellipsoids at the 50% level; H-atoms are omitted for clarity) (b) Crystal packing diagram of complex 1 (H-atoms are omitted for clarity) illustrating π-π-interactions (centroid-centroid distance: 3.489 Å) in the crystal lattice forming indefinite chains along the crystallographic b-axis.
Fig. 2.
(a) Molecular structure of the asymmetric unit of complex 2 (ellipsoids at the 50% level; H-atoms are omitted for clarity). (b) Molecular structure of complex 2 with complete coordination spheres generated by symmetry operations showing the two types of copper centers which alternately form the coordination polymer. (c) Crystal packing diagram of complex 2 emphasizing the coordination polymeric structure with two types of copper centers, which protrudes along the crystallographic b-axis (H-atoms are omitted for clarity).
Table 1.
Crystal and refinement data of complexes 1 and 2.
| Complex | 1 | 2 |
|---|---|---|
| Empirical formula | C44H38Cl4Cu2N2O8 | C60H50Cu3F6N2O12 |
| Formula mass | 991.64 | 1295.64 |
| Temperature (K) | room tepmerature | 170 |
| Wavelength (Å) | Mo Kα radiation, 0.71073 | Mo Kα radiation, 0.71073 |
| Crystal system, space group | Triclinic, P | Triclinic, P |
| Unit cell dimensions |
a= 8.3204 (17) Å b = 10.874 (2) Å c = 12.726 (3) Å α = 76.29 (3)° β = 75.33 (3)° γ = 83.76 (3)° |
a = 8.7723 (18) Å b = 12.318 (3) Å c = 13.833 (3) Å α = 101.77 (3)° β = 105.28 (3)° γ = 104.70 (3)° |
| Volume (Å3) | 1080.7 (4) | 1334.7 (6) |
| Crystal size (mm) | 0.30 × 0.30 × 0.26 | 0.26 × 0.17 × 0.13 |
| Z, Calculated density (g•cm−3) | 1, 1.524 | 1, 1.612 |
| Absorption coefficient (mm−1) | 1.29 | 1.27 |
| F(000) | 506 | 661 |
| Θ range for data collection | 6.4–59.1° | 6.3–59.0° |
| Limiting indices | h = −11→11, k = −14→14, l = −13→16 | h = −12→12, k = −15→16, l = −16→19 |
| Measured reflections | 10,754 | 14,996 |
| Independent reflections | 5327 | 7268 |
| Reflections with I > 2σ(I) | 4045 | 4531 |
| Rint | 0.032 | 0.049 |
| Goodness-of-fit on F2 | 1.008 | 0.945 |
| Final R indexes [I>=2σ (I)] | R1 = 3.51%, wR2 = 0.089 | R1 = 4.71%, wR2 = 0.126 |
| Refinement method | Full-matrix least-squares on F2 | Full-matrix least-squares on F2 |
| Data/restraints/ parameters | 5327/0/272 | 7268/1/390 |
| R[F2 > 2σ(F2)] | 0.035 | 0.047 |
| Δ〉max, Δ〉min (e Å−3) | 0.38, −0.54 | 1.60, −0.85 |
Complex 1 is a typical example of the copper carboxylate with paddlewheel arrangement of the type [Cu2(OOCR)4L2] (Fig. 1a). In complex 2, the paddlewheel array [Cu2(OOCR`)4] is extended further to a polymeric composition of the type [Cu2(OOCR`)4OCO(R`)CuL2]n by the axial bridging carboxylate ligands as shown in Fig. 2a. The 2-methyl pyridine ligand, which occupies the typical axial positions of the paddlewheel moiety in complex 1, is binding instead to a square planar mono-nuclear building block in complex 2. The square planar geometry is necessarily perfect as the mono-nuclear copper ion is located on an inversion center and is further evidenced by the geometry index, τ4 = 0 for β = α = 180°.
Despite the nearly identical ancillary ligands and identical reaction conditions, structurally quite distinct species are formed. The difference between the two reaction systems is the ortho substituted halogen atom on the carboxylate ligands. Accordingly, it has to be presumed that the inductive effect of a chloro versus a fluoro substituent in ortho position to the acid functional group has an impact on the formation of the resulting structural arrangement. The Cu—Cu (2.692 Å) bond length in complex 1 is different by 0.034 Å from the respective bond length in complex 2 (2.658 Å). Therefore, the ancillary ligands in the paddlewheel arrangement seemingly also affect the Cu—Cu bond lengths. The copper-copper distance in complex 2 is notably longer than in the closely related bis(2-fluorobenzoate) copper(II) C30.9H23.6Cu2F4O10.9 {Cu—Cu = 2.5882(13) Å} and in the 2,6-difluorobenzoate analogue [Cu2(C6H4F2)4(H2O)2] {Cu—Cu = 2.6139(2) Å} [54,55]. In the former example the exact same carboxylate is employed as in complex 2 but the axial ligands are alcohol derived O-donor ligands. The observed differences in the molecular structures including the Cu-Cu distances may therefore be attributed to the ligands located at the axial positions of the paddlewheel cores as well.
Selected bond lengths and bond angles for complex 1 are summarized in Table 2 . The carboxylate ligands bind in equatorial positions, presuming that the main molecular (i.e. vertical) axis runs through the two copper centers. Two of the phenyl rings of the carboxylate ligands point ‘upwards’ to one axial pyridine ligand while the two phenyl rings of the opposite carboxylates point ‘down’ towards the second axial pyridine ligand. The Cu—O bond lengths lie within the narrow range of 1.9647–1.9785 Å showing almost perfect symmetry in the geometry around copper. These bond lengths are comparatively shorter than the respective bond lengths reported for bis(2-fluorobenzoate) copper(II) {Cu—O = 1.949–2.126 Å} [55]. The Cu—N = 2.2009 (17) Å bond length for the axial ancillary ligand was found to be 0.05 Å longer than the one previously observed in [pyCu(µ2—OOCCH2C6H4R)4Cupy] with Cu—N = 2.157(2) Å [36]. The bond angles centered at the copper ions, i.e. O—Cu—O {O1—Cu1—O3 = 89.37 (8)°, O4—Cu1—O1 = 89.25 (8)°, O2—Cu1—O3 = 89.05 (8)° and O2—Cu1—O4 = 89.12 (9)°} highlight the near to perfection octahedral coordination geometry and are comparable to those in the closely related [Cu2(2-ClC6H4CH2COO)4(py)2] complex [16]. The entire molecule is generated from the asymmetric unit by a symmetry operation (inversion center in the center of the unit cell). The proximity of these angles to 90° is comparable to the already reported CuC14H8O4F2 complex which also exhibits rather regular O—Cu—O bond angles, i.e. 89.7o [55]. Distortions in the octahedral geometry arise from the O—Cu—N bond angles {O1—Cu1—N1 = 91.71 (7)°, O2—Cu1—N1 = 101.95 (7)°, O3—Cu1—N1 = 99.07 (7)°, O4—Cu1—N1 = 94.42 (7)°}. The torsion between the planes O1—Cu1—O2—O1—Cu1—O2 and the plane produced by the methyl pyridine C19—C20—C21—N1—C17—C18 is 34.98°. The rotational orientation of the phenyl rings is supported by the centroid interactions between the two methyl pyridine rings of the adjacent di-nuclear complexes with a centroid to centroid distance of 3.489 Å. These interactions are responsible for the generation of the crystal packing pattern that is shown in Fig. 1b. The Cu—Cu—O bond angles all lie within the range of 82.59–85.30° supporting a slight distortion in the octahedral geometry. A non-classical intramolecular hydrogen bond between Cl1 and H15B with a bond length of 3.09 Å further stabilizes the crystal structure of complex 1.
Table 2.
Selected bond lengths and bond angles of complex 1.
| Moiety | Bond Length, Å | Moiety | Bond length, Å |
|---|---|---|---|
| Cu1—O2 | 1.9647 (18) | O1—C8 | 1.250 (3) |
| Cu1—O4 | 1.9725 (17) | O2—C8i | 1.250 (3) |
| Cu1—O1 | 1.9780 (16) | O3—C16 | 1.248 (3) |
| Cu1—O3 | 1.9785 (17) | O4—C16i | 1.252 (3) |
| Cu1—N1 | 2.2009 (17) | N1—C21 | 1.329 (3) |
| Cu1—Cu1i | 2.6917 (8) | N1—C17 | 1.340 (3) |
| Moiety | Bond Angle, ° | Moiety | Bond Angle, ° |
| O2—Cu1—O4 | 89.12 (9) | O3—Cu1—Cu1i | 85.30 (5) |
| O2—Cu1—O1 | 166.33 (6) | N1—Cu1—Cu1i | 172.81 (5) |
| O4—Cu1—O1 | 89.25 (8) | C8—O1—Cu1 | 124.15 (14) |
| O2—Cu1—O3 | 89.05 (8) | C8i—O2—Cu1 | 123.48 (14) |
| O4—Cu1—O3 | 166.47 (6) | C16—O3—Cu1 | 121.16 (14) |
| O1—Cu1—O3 | 89.37 (8) | C16i—O4—Cu1 | 126.36 (15) |
| O2—Cu1—N1 | 101.95 (7) | C21—N1—C17 | 118.2 (2) |
| O4—Cu1—N1 | 94.42 (7) | C21—N1—Cu1 | 127.34 (17) |
| O1—Cu1—N1 | 91.71 (7) | C17—N1—Cu1 | 114.02 (16) |
| O3—Cu1—N1 | 99.07 (7) | O2i—C8—O1 | 125.5 (2) |
| O2—Cu1—Cu1i | 83.75 (6) | O2i—C8—C7 | 115.36 (19) |
| O4—Cu1—Cu1i | 81.18 (5) | O1—C8—C7 | 119.1 (2) |
| O1—Cu1—Cu1i | 82.59 (6) | ||
Symmetry code: (i) −x + 1, −y + 1, −z + 1.
In complex 2 [Cu2(OOCR)4OCO(R)CuL2]n, the square planar copper ions on either side of the paddlewheel moieties bear the 2-methylpyridine ligands plus oxygen donors from the bridging carboxylates in trans positions as shown in Fig. 2b. The methyl groups of the pyridine ligands point to opposite sides of the square planar geometry, to avoid any steric strain. A similar observation was reported previously for the copper complex with 6-methylpicolinic acid, having a geometry that was intermediate between square pyramidal and trigonal bipyramidal [56]. Due to the symmetry operations the two Cu—O bond lengths {Cu1—O1 = 1.979 (2) Å, Cu1—O1i = 1.979 (2) Å} as well as the two Cu-N bond lengths {Cu1—N1 = 1.972 (3) Å, Cu1—N1i = 1.972 (3) Å} of the square pyramidal building block are identical (Table 3 ). These distances are comparable with the respective bond lengths in a copper complex with 6-methylpicolinic acid {Cu1–O2 = 1.962(18) Å, Cu1–O3 = 1.961(19) Å, Cu1–N1 = 2.014(2) and Cu1–N2 = 2.031(2) Å} [56]. Most notably, the order of the bond lengths is reversed in complex 2 where the Cu-O bonds are elongated and the Cu-N distances are shorter. The relatively shorter distances of 2-methylpyridine may be attributed to the fact that the oxygen donor atoms are displaced from an ideal distance due to steric and spatial strain on the carboxylate ligand, while bridging the massive paddlewheel core and the mononuclear unit. Consequently the N-donor ligands move closer to the fourfold coordinated copper center to make up for the diminished electron density in the O—Cu—O axis. This reasoning is supported by the comparatively longer Cu—N and shorter Cu—O bond lengths in complex 1. The 2-Methylpyridine rings are twisted by 63.64° relative to the Cu1—O1—N1—O1—N1 plane. The twist in the 2-methylpyridine rings is enforced by an intermolecular C—H···F hydrogen bond as described in Table 4 {C26—H26···F1iii = 2.26 Å}. The paddlewheel structure, unlike in complex 1 comprises two types of coordinating carboxylates, i.e. one type acting as bridging ligands connecting the two paddlewheel copper ions and one connecting this moiety to the square planar copper centers via µ-Κ2-O:O´ and µ-O,O´ arrangements, respectively. The Cu—O bond lengths in the paddlewheel structure vary in the range of 1.95-1.97 Å {Cu2—O3 = 1.957 (3) Å, Cu2—O4ii = 1.962 (3) Å, Cu2—O5 = 1.960 (3) Å and Cu2—O6ii = 1.974 (2) Å}, and are thus, a bit more diverse than in complex 1. The axial carboxylate bonds, Cu2—O2 = 2.153 (2) Å are notably longer than the respective Cu—O bonds in equatorial positions and are in accordance with the shorter Cu-Cu distances within the paddlewheel relative to the distance between paddlewheel and mono-nuclear copper ions. The more flexible and weaker coordination of the axial donor ligands is also reflected in the bond angles of the paddlewheel copper centers. The O—Cu—O bond angles can be divided into two groups: (i) O—Cu—O bond angles formed by carboxylate coordination solely to the paddlewheel and (ii) O—Cu—O bond angles including the axial carboxylate. The equatorial angles O3—Cu2—O5 = 89.18 (14)°, O3—Cu2—O6ii = 90.88 (12)°, O4ii—Cu2—O6ii = 88.27 (11)°, and O5—Cu2—O4ii = 88.89 (13)° are all close to 90° representing only small distortions in the octahedral geometry around copper. With respect to the axial carboxylates the bond angles are deviating to a greater extent with O3—Cu2—O2 = 92.74 (10)°, O4ii—Cu2—O2 = 99.92 (10)°, O5—Cu2—O2 = 95.68 (10)° and O6ii—Cu2—O2 = 97.03 (9)°. The packing diagram as shown in Fig. 2c, displays hydrogen bonding interactions stabilizing the 3-dimensional array in the lattice, i.e. C2—H2B···O4ii = 2.34 Å and C2—H2A···O6ii = 2.63 Å (Table 4). The observed distortions of the ideal coordination geometries are possibly also based to some extent on the strong intermolecular hydrogen bonds.
Table 3.
Selected bond lengths and bond angles of complex 2.
| Moiety | Bond Length, Å | Moiety | Bond length, Å |
|---|---|---|---|
| Cu1—N1i | 1.972 (3) | Cu2—O2 | 2.153 (2) |
| Cu1—N1 | 1.972 (3) | Cu2—Cu2ii | 2.6586 (9) |
| Cu1—O1 | 1.979 (2) | C8—F1 | 1.337 (5) |
| Cu1—O1i | 1.979 (2) | F3—C16 | 1.358 (5) |
| Cu2—O3 | 1.957 (3) | F4—C24 | 1.357 (4) |
| Cu2—O5 | 1.960 (3) | N1—C25 | 1.344 (5) |
| Cu2—O4ii | 1.962 (3) | N1—C29 | 1.348 (5) |
| Cu2—O6ii | 1.974 (2) | O1—C1 | 1.269 (3) |
| O4—C9 | 1.253 (4) | O2—C1 | 1.248 (4) |
| O5—C17 | 1.251 (4) | O3—C9 | 1.247 (4) |
| O6—C17 | 1.243 (4) | ||
| Moiety | Bond Angle, ° | Moiety | Bond Angle, ° |
| N1i—Cu1—N1 | 180.0 | C1—O1—Cu1 | 112.85 (19) |
| N1i—Cu1—O1 | 89.89 (11) | C1—O2—Cu2 | 129.7 (2) |
| N1—Cu1—O1 | 90.11 (11) | C9—O3—Cu2 | 123.2 (2) |
| N1i—Cu1—O1i | 90.11 (11) | C9—O4—Cu2ii | 123.2 (2) |
| N1—Cu1—O1i | 89.89 (11) | C17—O5—Cu2 | 122.0 (2) |
| O1—Cu1—O1i | 180.0 | C17—O6—Cu2ii | 124.1 (2) |
| O3—Cu2—O5 | 89.18 (14) | O2—C1—O1 | 122.7 (3) |
| O3—Cu2—O4ii | 167.32 (10) | O2—C1—C2 | 118.6 (3) |
| O5—Cu2—O4ii | 88.89 (13) | O1—C1—C2 | 118.6 (3) |
| O3—Cu2—O6ii | 90.88 (12) | O3—C9—O4 | 125.9 (3) |
| O5—Cu2—O6ii | 167.28 (10) | O3—C9—C10 | 118.3 (3) |
| O4ii—Cu2—O6ii | 88.27 (11) | O3—Cu2—Cu2ii | 83.86 (8) |
| O3—Cu2—O2 | 92.74 (10) | O5—Cu2—Cu2ii | 84.78 (8) |
| O5—Cu2—O2 | 95.68 (10) | O4ii—Cu2—Cu2ii | 83.49 (8) |
| O4ii—Cu2—O2 | 99.92 (10) | O6ii—Cu2—Cu2ii | 82.58 (7) |
| O6ii—Cu2—O2 | 97.03 (9) | O2—Cu2—Cu2ii | 176.56 (7) |
Symmetry codes: (i) −x + 1, −y + 2, −z + 1; (ii) −x + 1, −y + 1, −z + 1.
Table 4.
Hydrogen-bond geometry of complex 2 (Å, °).
| D—H···A | D—H | H···A | D···A | D—H···A |
|---|---|---|---|---|
| C2—H2A···O6ii | 0.99 | 2.63 | 3.236 (4) | 120 |
| C2—H2B···O4ii | 0.99 | 2.34 | 3.166 (4) | 140 |
| C26—H26···F1iii | 0.95 | 2.26 | 3.212 (7) | 176 |
Symmetry codes: (ii) −x + 1, −y + 1, −z + 1; (iii) x − 1, y, z.
3.4. Antioxidant activities
3.4.1. DPPH free radical scavenging activity
A DPPH free radical scavenging assay was performed for the free ligand acids (HL1 and HL2) and their copper complexes (1 and 2) and the results are shown in Table 5 . A dose dependent response was exhibited by the samples with an activity order 1 > HL1 > 2 > HL2. Complexes 1 and 2, both have better radical scavenging activities than their corresponding free carboxylic acid ligands HL1 and HL2, respectively. The observed activity enhancement after complexation reflects the importance of the metal center as well as the N-donor ligands for the reaction with the radical in this assay. The copper complexes acquire additional superoxide-dismuting centers leading to superior antioxidant activity [57]. Notably, the chloro substituted phenylacetic acid (HL1) is a better DPPH radical scavenger than the fluoro substituted phenyl acetic acid (HL2). The higher activity of HL1 could be attributed to the ease with which free radical formation can take place through homolytic fission in the presence of the chloro substituent for subsequent reaction with the DPPH. On the other hand the polarity induced by the fluorine probably hinders the process of free radical formation in HL2. Complex 1 has shown comparable activity to the standard drug ascorbic acid (vitamin C) therefore it might be utilized in the future as DPPH scavenger after some further evaluation.
Table 5.
DPPH free radical scavenging activity of ligand acids and synthesized complexes.
| Compound | % Inhibition |
||||
|---|---|---|---|---|---|
| Dose Conc. (µM) | |||||
| 15 | 30 | 60 | 120 | 240 | |
| HL1 | 8.74 ± 1.45 | 12.3 ± 1.67 | 23.4 ± 2.02 | 38.12 ± 2.11 | 43.00 ± 1.34 |
| HL2 | 4.65 ± .04 | 7.41 ± 1.02 | 13.02 ± 1.32 | 19.76 ± 1.58 | 27.06 ± 2.21 |
| 1 | 17.34 ± 1.21 | 20.45 ± 2.10 | 49.76 ± 2.68 | 67.76 ± 2.97 | 78.56 ± 2.38 |
| 2 | 5.39 ± .68 | 8.10 ± 0.86 | 18.29 ± 1.42 | 26.7 ± 1.32 | 36.09 ± 2.13 |
| Vit C | 15.00 ± 1.34 | 32.88 ± 2.65 | 49.00 ± 1.89 | 72.00 ± 2.55 | 92.00 ± 1.91 |
3.4.2. Hydroxyl radical scavenging activity
The hydroxyl radical scavenging assay results of the tested compounds are shown in Table 6 . At lower concentrations essentially the same order of activity was observed as in the DPPH assay. Complex 1 exhibits the highest hydroxyl scavenging ability throughout, followed by its corresponding acid ligand, HL1. Complex 2 was more active than its corresponding acid ligand, HL2, at lower concentrations. However, the activity order reverses at the two highest concentrations where HL2 exhibits better activity than complex 2. Overall, the results point again at the role of the carbon halogen bond polarity in the radical scavenging activities.
Table 6.
Hydroxyl radical scavenging activity of ligand acids and synthesized complexes.
| Compound | % Inhibition |
||||
|---|---|---|---|---|---|
| Dose Conc. (µM) | |||||
| 15 | 30 | 60 | 120 | 240 | |
| HL1 | 10.00 ± 0.91 | 16.90 ± 2.21 | 22.70 ± 2.04 | 30.83 ± 2.53 | 42.87 ± 2.63 |
| HL2 | 1.70 ± 0.20 | 3.54 ± 0.02 | 6.09 ± 1.06 | 9.99 ± 1.01 | 13.89 ± 1.21 |
| 1 | 13.90 ± 1.01 | 20.78 ± 2.04 | 30.55 ± 2.56 | 44.99 ± 2.57 | 71.78 ± 2.48 |
| 2 | 2.95 ± 0.07 | 5.98 ± 0.97 | 8.09 ± 1.08 | 9.00 ± 1.78 | 11.02 ± 1.01 |
| Vit C | 17.56 ± 1.05 | 27.99 ± 2.45 | 32.76 ± 2.79 | 49.76 ± 2.87 | 76.78 ± 2.78 |
3.5. Antileishmanial activity
The antileishmanial activity data, summarized in Table 7 , show dose dependent antileishmanial activities for all investigated species. In particular, the free carboxylic acid ligands give rise to an exponential increase at 1000 ppm concentration. The complexes exhibit higher activities than their free acid ligands (HL1 and HL2) at all concentrations. However, the difference was more pronounced at lower concentrations. The enhanced activities of the complexes (1 and 2) compared to the free acid ligands reflect the significance of the metal center as well as the N-donor ligands for the antileishmanial activity and can be explained on the basis of their ability to interact with DNA of parasite through intercalation [58]. At lower concentrations the two complexes displayed significant differences in their activities. Comparable activities of the complexes were observed at higher concentration (1000 ppm). Among the tested compounds, complex 1 was found to be the most potent antileishmanial agent at all concentrations. In the applied concentration range, its activity was essentially dose independent within the error margin suggesting that even lower doses should be considered. Complex 1 is a strong candidate for further and more intense studies regarding its application as a potential antileishmanial agent. Amphotericin-B was used as a positive control in this assay and reached 100% inhibition.
Table 7.
Antileishmanial activity of ligand acids and synthesized complexes.
| Compound | % Inhibition |
||
|---|---|---|---|
| Dose Conc. (µM) | |||
| 250 | 500 | 1000 | |
| HL1 | 17.04 ± 1.12 | 25.11 ± 1.89 | 85.13 ± 1.56 |
| HL2 | 14.53 ± 1.23 | 16.38 ± 0.97 | 84.97 ± 1.32 |
| 1 | 89.91 ± 2.77 | 91.13 ± 2.78 | 91.99 ± 2.34 |
| 2 | 61.66 ± 2.53 | 76.90 ± 2.11 | 91.03 ± 2.71 |
3.6. Anticancer activity
The acid ligands (HL1 and HL2) and synthesized complexes (1 and 2) were screened in vitro for their anticancer potential against malignant glioma U-87 (MG U87) cell lines and the results are summarized in Table 8 . The tested compounds exhibit dose dependent cytotoxic responses. As the concentrations of the test compounds were increased sharp increases in the cytotoxic effects were observed by an exponential decrease in the survival fractions (%) of the target cells. However, at elevated concentrations (62.5-250 µg/mL) the decreases in the survival fraction (%) of the target cells became less and less steep. Surprisingly, a further increase in the concentration of the test compounds above 250 µg/mL resulted in a lower cytotoxic effect of the test compounds, most notably for the complexes 1 and 2. The decrease in the cytotoxic effect of the test compounds at higher concentration (500 µg/mL) may be attributed to auto association/interaction of the drug molecules (e.g. intermolecular H-bonding for the complexes) which decrease their ability to interact with the active sites of the target cells. Alternatively, the higher concentrations, in particular those of the complexes might trigger defense responses of the cells leading to better survival. Up to 500 µg/mL concentrations, no significant activity differences were found among the four tested compounds.
Table 8.
Cytotoxic effect of ligand acids and complexes on malignant glioma U-87 cell lines.a
| Compound | Survival fraction (%) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Dose Conc. (µg/mL) | ||||||||
| 0 | 7.812 | 15.625 | 31.25 | 62.5 | 125 | 250 | 500 | |
| HL1 | 100 | 75.74 ± 0.06 | 50.70 ± 0.04 | 36.66 ± 0.04 | 23.84 ± 0.02 | 15.69 ± 0.01 | 16.98 ± 0.00 | 19.92 ± 0.01 |
| HL2 | 100 | 83.84 ± 0.09 | 58.94 ± 0.04 | 37.29 ± 0.04 | 20.34 ± 0.03 | 15.51 ± 0.01 | 15.04 ± 0.02 | 18.97 ± 0.05 |
| 1 | 100 | 84.13 ± 0.09 | 60.27 ± 0.04 | 45.09 ± 0.03 | 25.05 ± 0.03 | 20.92 ± 0.03 | 17.52 ± 0.02 | 61.44 ± 0.21 |
| 2 | 100 | 82.28 ± 0.02 | 49.55 ± 0.11 | 34.02 ± 0.07 | 19.41 ± 0.02 | 15.86 ± 0,02 | 14.21 ± 0.01 | 54.65 ± 0.18 |
a = The cell lines were treated with different concentrations of the test compounds in vitro for 24 h.
3.7. DNA interaction study
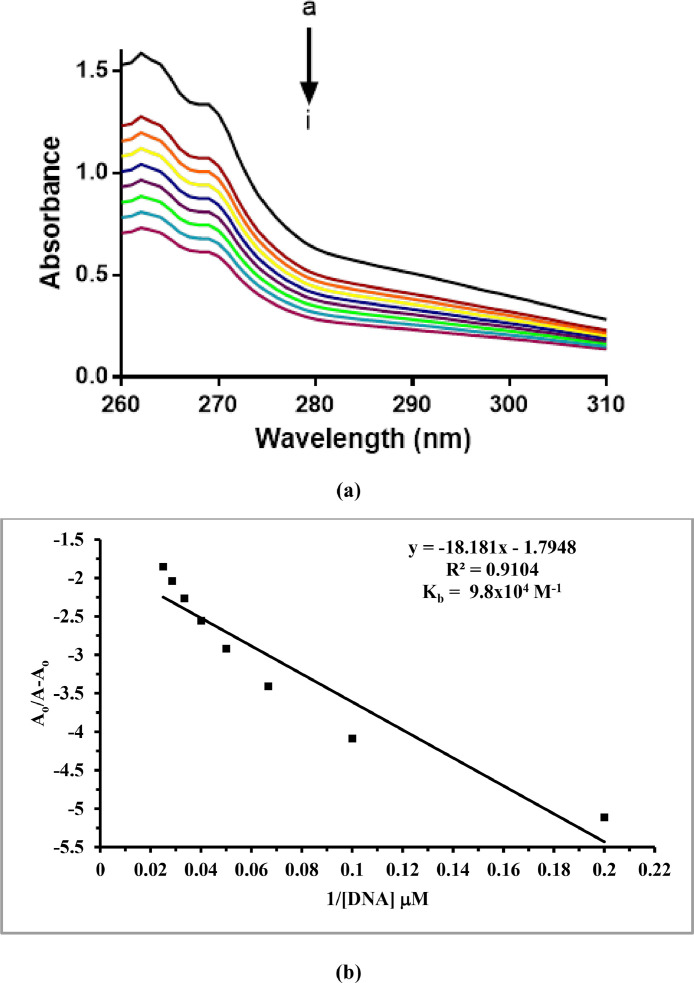
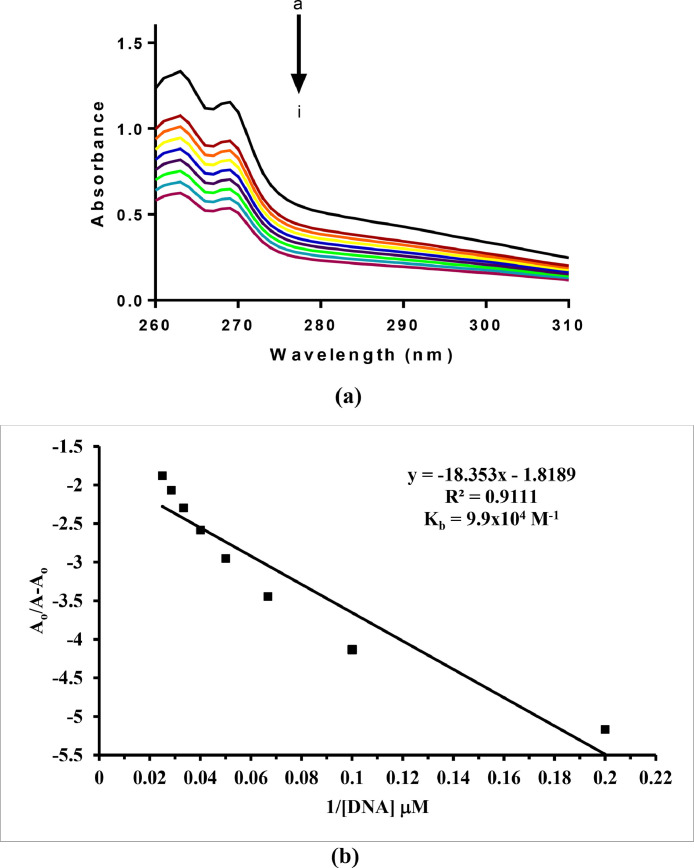
Preliminary DNA binding data of the synthesized complexes 1 and 2 were determined by electronic absorption measurements. UV-Vis absorption titrations were performed with a fixed concentration of complex solutions (200 µM) and varying concentrations (0-40 µM) of salmon sperm DNA (SSDNA). Before measurements, complex-DNA adduct solutions were kept at room temperature for at least one hour to allow any possible interactions to develop. The experiments led to the same results when repeated with the identical solutions after 24 h, emphasizing the stability of the respective complex-DNA adducts. The results of the absorption measurements for complexes 1 and 2, are shown in Figs. 3 and 4 , respectively. Both complexes show a pronounced hypochromism after addition of DNA with no red/blue shifts, pointing towards a mixed binding mode comprising partial intercalation of the complexes into DNA base pairs as well as groove binding [2].
Fig. 3.
(a) Absorption spectrum of complex 1 in the absence (a) and presence of 5 µM (b), 10 µM (c), 15 µM (d), 20 µM (e), 25 µM (f), 30 µM (g), 35 µM (h) and 40 µM (i) DNA. The arrow direction indicates increasing concentrations of DNA. (b) Plot of A/(A-A) vs. 1/[DNA] for binding constant determination of the complex 1-DNA adduct.
Fig. 4.
(a) Absorption spectrum of complex 2 in the absence (a) and presence of 5 µM (b), 10 µM (c), 15 µM (d), 20 µM (e), 25 µM (f), 30 µM (g), 35 µM (h) and 40 µM (i) DNA. The arrow direction indicates increasing concentrations of DNA. (b) Plot of A/(A-A) vs. 1/[DNA] for binding constant determination of the complex 2-DNA adduct.
The calculated binding constant (Kb) values using the Benesi–Hildebrand equation were 9.8 × 104 and 9.9 × 104 M−1 for complexes 1 and 2, respectively [2]. The reported complexes are relatively better DNA binders compared to the previously reported copper(II) complexes with similar carboxylate ligands [2,59,60]. Since in a dinuclear complex the two adjacent Lewis acid metal sites are favorably poised to bind or interact with the donor sites of double-stranded DNA and by tuning the groups on the dimetal system one can even effectively control the nature of interaction with DNA [61]. The response of both complexes towards DNA in terms of interaction mode and binging strength was found to be very similar. This observation reflects that varying the halogen (F or Cl) on the coordinated ligand has no effect on the DNA binding ability of the complexes as long as the complexes have other structural or spacial geometric similarities. However, the position of the halogen substituent on the phenyl acetate ligand, considering previously reported studies, was found to affect the DNA binging ability, i.e. decreasing it in the order 2-Cl > 4-Cl > 3-Cl [2,60]. Notably, the copper complexes with the paddlewheel arrangement are relatively more active DNA binders compared to the complexes with similar phenyl acetate ligands and bidentate nitrogen donor ligands in a different structural environment [2,60].
3.8. Molecular docking studies
3.8.1. Complex-DNA interaction
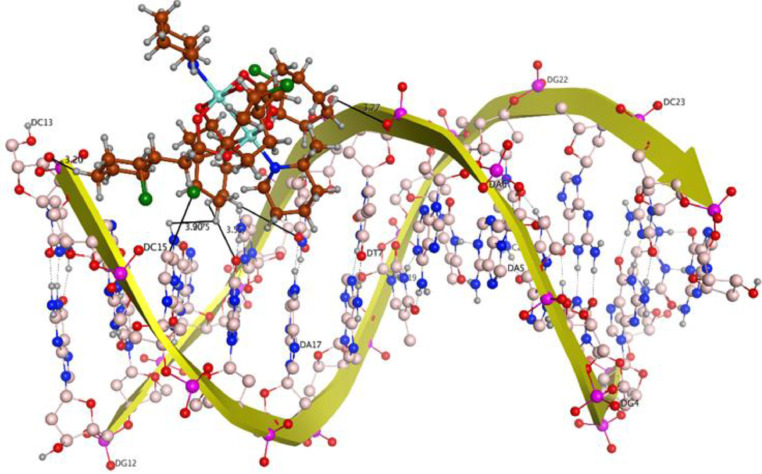
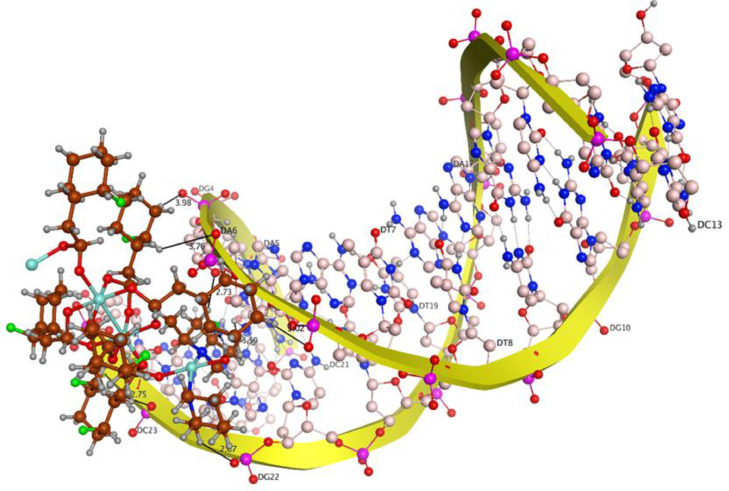
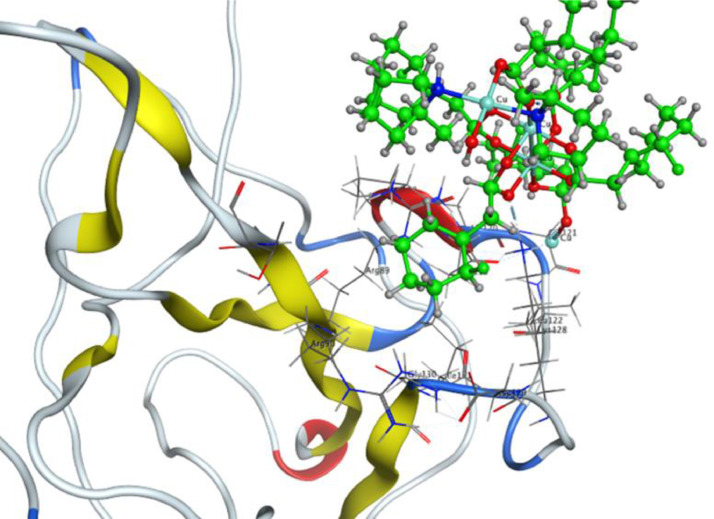
The theoretical DNA binding mode of the synthesized copper complexes was investigated by molecular docking. The docking results show that the complexes prefer to bind to DNA by intercalation. From the computed docking conformation of complex 1 (docking score = −8.9371), it was observed that this complex establishes six polar interactions with the active residues (DC 13, DG 14, DC 15, DG 16, DA 17 and DG 22) of DNA as shown in Fig. 5 . Complex 2 (docking score = −9.1065), which in addition to the paddlewheel also comprises the mono-nuclear copper center, formed seven polar interactions with the active site residues (DG 4, DA 5, DA 6, DC 21, DG 22, DC 23) of DNA (Fig. 6 ). An excellent arrangement was obtained as the best docked pose, which showed important binding features that are predominantly based on interactions of various interacting moieties of the complexes and phosphate derived functional groups of the DNA backbone.
Fig. 5.
Docking orientation of complex 1 inside the active site of DNA.
Fig. 6.
Docking orientation of complex 2 inside the active site of DNA.
3.8.2. Complexes-SARS-CoV-2 interaction
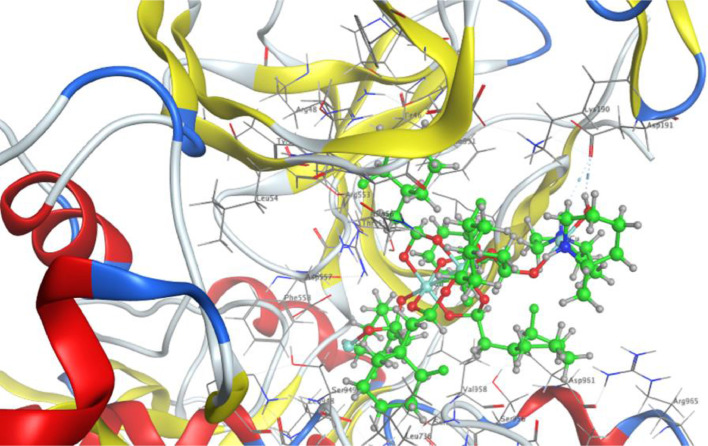
Molecular docking investigations were performed to find any possible complex and SARS-CoV-2 interactions. For the docking studies a spike protein, the nucleocapsid protein of SARS-CoV-2 and the angiotensin converting enzyme of the human cell (host for SARS-CoV-2) were selected as target sites [62,63]. The results of this theoretical study show that the synthesized complexes 1 and 2 do indeed bind to spike protein, nucleocapsid protein of SARS-CoV-2 and that they can also inhibit the angiotensin converting enzyme of the human cell. Complex 1 (docking score= −6.5162) showed one notable H-donor interaction with GLY 638 of the spike protein and 2 (docking score= −8.4282) showed two particular H-donor interactions with ASP 191 of the spike protein. Interestingly, no such interactions with spike protein were observed in case of triorganotin(IV) carboxylates [64]. The interaction of complex 1 with the nucleocapsid protein of SARS-CoV-2 attached to RNA (docking score= −5.2152) includes one H-Acceptor interaction with the active site residue GLY 165 of the nucleocapsid protein, whereas complex 2 (docking score= −5.1609) exhibited three interactions. Two are H-Acceptor types and one is a metal ion interaction with the active sites LYS 128, GLU 119 of the nucleocapsid protein. The docking scores reflect that complex 2 can interact more efficiently with spike and nucleocapsid proteins of SARS-CoV-2 as shown in Figs. 7 and 8 , respectively. Complexes 1 and 2 were also found to engage in one interaction each with GLN 531 and ASP 67 of the human angiotensin converting enzyme with docking scores of −5.2022 and −5.4059, respectively. Since, free copper is toxic, its metal complexes are of particular importance in biological applications. Docking studies of copper complex-SARS-CoV-2 interactions are rare [65] and the complexes could be used as potential corona virus inhibitors.
Fig. 7.
Docking orientation of complex 2 inside the active site of spike protein.
Fig. 8.
Docking orientation of complex 2 inside the active site of nucleocapsid protein.
4. Conclusion
The successful synthesis of copper(II) carboxylate complexes by the condensation reaction of copper(II) chloride dihydrate with 2-chlorophenyl acetic acid (HL1) and 2-fluorophenyl acetic acid (HL2) in the presence of 2-methyl pyridine co-ligand was verified by FT-IR and X-ray single crystal analyses. While the reaction with the chloro-substituted acid led to a typical discrete paddlewheel structure, the fluoro-analog resulted in a coordination polymer with a di-nuclear paddlewheel moiety plus a square planar mono-nuclear building block. The complexes exhibit better antioxidant and antileishmanial activity compared to the free acid ligands thus reflecting the importance of the synergistic effect of the metal ion center, carboxylate and nitrogen donor ligands for the activity. A mixed DNA interaction mode consisting of partial intercalation and groove binding was established by UV-Vis absorption spectroscopy. A dose dependent cytotoxic response against malignant glioma U-87 (MG U87) cell lines was observed at relatively low concentrations. The decrease in the cytotoxic effect at higher concentration suggests a detrimental influence of intermolecular H-bonding on the cytotoxic activity of the complexes or that they trigger defense mechanisms of the cell. A theoretical docking study puts the two complexes forward as potential corona virus inhibitors.
CRediT authorship contribution statement
: Investigation, Validation. Niaz Muhammad: Supervision, Resources, Writing – review & editing. Ishaq N. Khan: . Zafar Ali: . Mohammad Ibrahim: . Shaukat Shujah: . Saqib Ali: . Muhammad Ikram: . Sadia Rehman: . Gul Shahzada Khan: . Abdul Wadood: . Awal Noor: . Carola Schulzke: .
Declaration of Competing Interest
The author(s) declare(s) that there is no conflict of interest regarding the publication of this paper.
Acknowledgment
Viola (PIN No. 315-17243-2PS3-132) and Dr. Niaz Muhammad are thankful to the Higher Education Commission, Pakistan for the financial support through project number 20-3093/NRPU/R&D/HEC/2013.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2021.132308.
Appendix. Supplementary materials
References
- 1.A.Crawford C., Day E.F., E.Streib W., Huffman J.C., Christou G. One-dimensional polymerization of M2(OAc)4 (M = Cu, Rh) units using 2-(aminomethyl)pyridine: preparation and characterization of [Rh2(OAc)4(amp)]n and [Cu4(OAc)8(amp)2]n. Polyhedron. 1994;13(21):2933–2942. [Google Scholar]
- 2.Deka K., Sarma R.J., Baruah J.B. Nitrogen–oxygen bond formation during oxidative reactions of copper(II)benzoate complexes having 3,5-dimethylpyrazole. Inorg. Chem. Commun. 2006;9(9):931–934. [Google Scholar]
- 3.Adonin S.A., Novikov A.S., Sokolov M.N., Fedin V.P. Heteroleptic Cu(II) iodoacetate complex: appearance of halogen bonding in solid state. Inorg. Chem. Commun. 2019;105:221–224. [Google Scholar]
- 4.Afanas'ev I.B., Ostrakhovitch E.A., Mikhal'chik E.V., Ibragimova G.A., Korkina L.G. Enhancement of antioxidant and anti-inflammatory activities of bioflavonoid rutin by complexation with transition metals. Biochem. Pharmacol. 2001;61:677–684. doi: 10.1016/s0006-2952(01)00526-3. [DOI] [PubMed] [Google Scholar]
- 5.Ali T., Muhammad N., Ali Z., Samad A., Ibrahim M., Ikram M., Rehman S., Shujah S., Khan G.S., Wadood A., Ali S., Schulzke C. Syntheses, crystal structures, antioxidant, in silico DNA and SARS-CoV-2 interaction studies of triorganotin(IV) carboxylates. J. Mol. Struct. 2021;1234 doi: 10.1016/j.molstruc.2021.132308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altürk S., Avcı D., Başoğlu A., Tamer Ö., Atalay Y., Dege N. Copper(II) complex with 6-methylpyridine-2-carboxyclic acid: experimental and computational study on the XRD, FT-IR and UV–Vis spectra, refractive index, band gap and NLO parameters. Spectrochim. Acta Part A. 2018;190:220–230. doi: 10.1016/j.saa.2017.09.041. [DOI] [PubMed] [Google Scholar]
- 7.Bregman H., Carroll P.J., Meggers E. Rapid access to unexplored chemical space by ligand scanning around a ruthenium center: discovery of potent and selective protein kinase inhibitors. J. Am. Chem. Soc. 2006;128(3):877–884. doi: 10.1021/ja055523r. [DOI] [PubMed] [Google Scholar]
- 8.Bromley R., Davey R., Oliver L., Harvie R., Baldock C. A preliminary investigation of cell growth after irradiation using a modulated x-ray intensity pattern. Phys. Med. Biol. 2006;51(15):3639–3651. doi: 10.1088/0031-9155/51/15/003. [DOI] [PubMed] [Google Scholar]
- 9.Bruno I.J., Cole J.C., Edgington P.R., Kessler M., Macrae C.F., McCabe P., Pearson J., Taylor R. New software for searching the Cambridge structural database and visualizing crystal structures. Acta Crystallogr. Sect. B. 2002;B58:389–397. doi: 10.1107/s0108768102003324. [DOI] [PubMed] [Google Scholar]
- 10.Butcher R.J., Overman J.W., Sinn E. Bi- and polynuclear antiferromagnetic metal complexes (copper,nickel) with salicylaldimines and acetate-type ligands. New modes of acetate bridging. J. Am. Chem. Soc. 1980;102(9):3276–3278. [Google Scholar]
- 11.Chen S.M., Lu C.Z., Xia C.K., Xu X.J., Zhai Q.G. Reactivity of 1,4-Bis[2-(5-phenyloxazoly)]benzene toward Cu salts under different reaction conditions. Cryst. Growth Des. 2005;5(4):1485–1490. [Google Scholar]
- 12.Chen Z., Liu L., Wang Y., Zou H., Zhang Z., Liang F. A two-dimensional homospin Cu(II) ferrimagnet featuring S-shaped hexanuclear secondary building blocks. Dalton Trans. 2014;43(22):8154–8157. doi: 10.1039/c4dt00375f. [DOI] [PubMed] [Google Scholar]
- 13.Chifotides H.T., Dunbar K.R. Interactions of metal−metal-bonded antitumor active complexes with DNA fragments and DNA. Acc. Chem. Res. 2005;38(2):146–156. doi: 10.1021/ar0302078. [DOI] [PubMed] [Google Scholar]
- 14.Choi C.W., Kim S.C., Hwang S.S., Choi B.K., Ahn H.J., Lee M.Y., Park S.H., Kim S.K. Antioxidant activity and free radical scavenging capacity between Korean medicinal plants and flavonoids by assay-guided comparison. Plant Sci. 2002;163(6):1161–1168. [Google Scholar]
- 15.Chung Y.H., Wei H.H., Liu Y.H., Lee G.H., Wang Yu. Reinvestigation of the crystal structure and cryomagnetic behaviour of copper(II) propionates. Polyhedron. 1998;17(4):449–455. [Google Scholar]
- 16.Cirri D., Pratesi A., Marzo T., Messori L. Metallo therapeutics for COVID-19. Exploiting metal-based compounds for the discovery of new antiviral drugs. Expert Opin. Drug Discovery. 2021;16:39–46. doi: 10.1080/17460441.2020.1819236. [DOI] [PubMed] [Google Scholar]
- 17.Ackermann H., Geiseler G., Harms K., Leo R., Massa W., Weller F., Dehnicke K. Triethylphosphanimin-Komplexe der Acetate von Kupfer(II) und Zink. Kristallstrukturen von [Zn(O2C–CH3)2(HNPEt3)], [Cu5(O2C–CH3)10(HNPEt3)2] und [Cu(O2C–CH3)2(HNPEt3)2] Z. Anorg. Allg. Chem. 1999;625(9):1500–1506. [Google Scholar]
- 18.Devaux C.A., Rolain J.M., Colson P., Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents. 2020;55(5) doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dey S., Sarkar S., Paul H., Zangrando E., Chattopadhyay P. Copper(II) complex with tridentate N donor ligand: synthesis, crystal structure, reactivity and DNA binding study. Polyhedron. 2010;29(6):1583–1587. [Google Scholar]
- 20.Giese A., Rief M.D., Loo M.A., Berens M.E. Determinants of human astrocytoma migration. Cancer Res. 1994;54(14):3897–3904. [PubMed] [Google Scholar]
- 21.Gogoleva N.V., Aleksandrov G.G., Pavlov A.A., Kiskin M.A., Sidorov A.A., Eremenko I.L. Effect of the structure of pyridine ligands and the substituent in the carboxylate anion on the geometry of transition metal complexes [M2(O2CR)4L2] Russ. J. Coord. Chem. 2018;44(2):91–102. [Google Scholar]
- 22.Hassouna I., Sperling S., Kim E., Schulz-Schaeffer W., Rave-Fränk M., Hasselblatt M., Jelkmann W., Giese A., Ehrenreich H. Erythropoietin augments survival of glioma cells after radiation and temozolomide. Int. J. Radiat. Oncol. Biol. Phys. 2008;72(3):927–934. doi: 10.1016/j.ijrobp.2008.06.1923. [DOI] [PubMed] [Google Scholar]
- 23.Hinz A., Köckerling M. A new mixed-valent copper cyanido complex and a new copper(II) acetato complex, prepared with an ionic liquid. Z. Anorg. Allg. Chem. 2015;641(7):1347–1351. [Google Scholar]
- 24.Hökelek T., Mert Y., Ünalerouglu C. catena-Poly[[tetrakis([mu]-3,5-dinitrobenzoato-O:O')dicopper(II)]bis[([mu]-3,5-dinitrobenzoato-O:O')(methanol-O)copper(II)]] Acta Crystallogr. Sect. C. 1998;C54(3):310–313. [Google Scholar]
- 25.Iqbal M., Ahmad I., Ali S., Muhammad N., Ahmed S., Sohail M. Dimeric ‘’paddle-wheel’’ carboxylates of copper(II): synthesis, crystal structure and electrochemical studies. Polyhedron. 2013;50:524–531. [Google Scholar]
- 26.Iqbal M., Ali S., Muhammad N., Sohail M. Synthesis, crystal structures and electrochemical characterization of dinuclear paddlewheel copper(II) carboxylates. Polyhedron. 2013;57:83–93. [Google Scholar]
- 27.Iqbal M., Ali S., Tahir M.N., Muhammad N., Shah N.A., Sohail M., Pandarinathan V. Synthesis, crystal structure and electrochemical and DNA binding studies of oxygen bridged-copper(II) carboxylate. J. Mol. Struct. 2015;1093:135–143. [Google Scholar]
- 28.Iqbal M., Karim A., Ali S., Tahir M.N., Sohail M. Synthesis, characterization, structural elucidation, electrochemistry, DNA binding study, micellization behaviour and antioxidant activity of the Cu(II) carboxylate complexes. Polyhedron. 2020;178 [Google Scholar]
- 29.Islam Z., Kouli P.K., Baruah S., Chakrabortty M., Bepari R.A., Das BK. Dimeric copper(II) tetracarboxylates as catalysts in the selective epoxidation of styrene. Polyhedron. 2018;155:351–358. [Google Scholar]
- 30.Kani Y., Tsuchimoto M., Ohba S., Matsushima H., Tokii T. catena-Poly[[tetra-kis-(μ-3,3-di-methylbutyrato-O:O')dicopper(II)]-μ-3,3-di-methylbutyrato-O:O'-[bis-(2,6-di-methylpyri dine-N)copper(II)]-μ-3,3-di-methylbutyrato-O:O'] and tetra-kis-(μ-2,2-di-methylpropanoato-O:O')bis-[(2,6-di-methylpyridine-N)copper(II)] Acta Crystallogr. Sect. C. 2000;C56(8):923–925. [Google Scholar]
- 31.Karges J., Cohen S.M. Metal complexes as antiviral agents for SARS-CoV-2. ChemBioChem. 2021;22:2600–2607. doi: 10.1002/cbic.202100186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan I., Baeesa S., Bangash M., Schulten H.J., Alghamdi F., Qashqari H., Madkhali N., Carracedo A., Saka M., Jamal A., Al-Maghrabi J., AlQahtani M., Al-Karim S., Damanhouri G., Saini K., Chaudhary A., Abuzenadah A., Hussein D. Pleomorphism and drug resistant cancer stem cells are characteristic of aggressive primary meningioma cell lines. Cancer Cell Int. 2017;17:72. doi: 10.1186/s12935-017-0441-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li P., Huo L., Su W., Lu R., Deng C., Liu L., Deng Y., Guo N., Lu C., He C. Free radical-scavenging capacity, antioxidant activity and phenolic content of Pouzolzia zeylanica. J. Serb. Chem. Soc. 2011;76(5):709–717. [Google Scholar]
- 34.Lim E.K., Teoh S.G., Teh J.B.J., Fun H.K., Yuen C.W. Tetra-kis(μ-2,6-di-fluoro-benzoato-κ2O:O′)bis-[aqua-copper(II)] Acta Crystallogr. Sect. E Crystallogr. Commun. 2007;E63(4):m991–m993. [Google Scholar]
- 35.Molecular operating environment MOE . Inc.; Montreal Quebec Canada: 2016. C.C.G.I.M, Quebec Canada, MOE-Dock, Chemical Computing Group. [Google Scholar]
- 36.Muhammad N., Ikram M., Perveen F., Ibrahim M., Ibrahim M., Abel V., Rehman S., Shujah S., Khan W., Shams D.F., Schulzke C. Syntheses, crystal structures and DNA binding potential of copper(II) carboxylates. J. Mol. Struct. 2019;1196:771–782. [Google Scholar]
- 37.Mushtaq A., Ali S., Tahir Nawaz M., Ismail H., Mirza B., Saadiq M., Abdul M.H., Iqbal M. New bioactive heteroleptic copper(II) carboxylates: structure, enzymatic and DNA-binding studies. Acta Chim. Slov. 2017;64:397–408. doi: 10.17344/acsi.2017.3250. [DOI] [PubMed] [Google Scholar]
- 38.Nakamoto K. Wiley; New York: 1986. Infrared and Raman Spectra of Inorganic and Coordinative Compounds. [Google Scholar]
- 39.Navarro M., Cisneros-Fajardo E.J., Sierralta A., Fernández-Mestre M., Silva P., Arrieche D., Marchán E. Design of copper DNA intercalators with leishmanicidal activity. JBIC J. Biol. Inorg. Chem. 2003;8:401–408. doi: 10.1007/s00775-002-0427-2. [DOI] [PubMed] [Google Scholar]
- 40.Neels A., Stoeckli-Evans H., Escuer A., Vicente R. Ten-membered rings of coppers interconnected by 2,5-Bis(2-pyridyl)pyrazine and acetate groups: synthesis, crystal structure, and magnetic properties of the two-dimensional polymer catena-(Octakis(.mu.2-acetato)[2,5-bis(2-pyridyl)pyrazine]tetracopper(II)) Inorg. Chem. 1995;34(7):1946–1949. [Google Scholar]
- 41.Njogu E.M., Omondi B., Nyamori V.O. Crystal structure of tetrakis(µ2-acetato-κ2O:O′)-bis{[(E)-2,6-diisopropyl-N-(pyridin-3-ylmethylene)aniline]copper(II)}, C44H56 Cu2N4O8. Z. Kristallogr NCS. 2018;233(3):373–375. [Google Scholar]
- 42.Paiva R.E.F., Neto A.M., Santos I.A., Jardim A.C.G., Corbi P.P., Bergamini F.R.G. What is holding back the development of antiviral metallodrugs? A literature overview and implications for SARS-CoV-2 therapeutics and future viral outbreaks. Dalton Trans. 2020;49:16004–16033. doi: 10.1039/d0dt02478c. [DOI] [PubMed] [Google Scholar]
- 43.Pal M., Musib D., Roy M. Transition metal complexes as potential tools against SARS-CoV-2: an in silico approach. New J. Chem. 2021;45:1924–1933. [Google Scholar]
- 44.Perlepes S.P., Libby E., Streib W.E., Folting K., Christou G. The reactions of Cu2(O2CMe)4(H2O)2 with 2,2′-bipyridine (bpy): Influence of the Cu: bpy ratio, and the structure of a linear polymer comprising two alternating types of Cu2 units. Polyhedron. 1992;11(8):923–936. [Google Scholar]
- 45.Premkumar M., Kaleeswaran D., Kaviyarasan G., Prasanth D.A., Venkatachalam G. Mono and dinuclear Cu(II) carboxylate complexes with pyridine and 1-methylimidazole as co–ligands: synthesis, structure, antibacterial activity and catalytic nitroaldol reactions. Chem. Select. 2019;4(25):7507–7511. [Google Scholar]
- 46.Ren-Shu W., Jing F., Ming-Lei L., De-Shun K., Yi-Zhu L., Kai-Yi S. Two Cu(II) complexes with hydrolyzed nicotinamide ligand: crystal structures and bioactivities. Chin. J. Struct. Chem. 2019;38(8):1297–1310. [Google Scholar]
- 47.Sánchez-Férez F., Bayés L., Font-Bardia M., Pons J. Solvent dependent formation of Cu(II) complexes based on isonicotinamide ligand. Inorg. Chim. Acta. 2019;494:112–122. [Google Scholar]
- 48.Sánchez-Férez F., Guerrero M., Ayllón JA., Calvet T., Font-Bardia M., Planas J.G., Pons J. Reactivity of homoleptic and heteroleptic core paddle wheel Cu(II) compounds. Inorg. Chim. Acta. 2019;487:295–306. [Google Scholar]
- 49.Sánchez-Férez F., Soldevila-Sanmartín J., Ayllón JA., Calvet T., Font-Bardia M., Pons J. Synthesis and characterization of three new Cu(II) paddle-wheel compounds with 1, 3-benzodioxole-5-carboxylic acid. Polyhedron. 2019;164:64–73. [Google Scholar]
- 50.Sanchez-Sala M., Pons J., Alvarez-Larena A., Bayés-García L., Font-Bardia M., Ayllón J.A. Cu(II) 4-phenoxybenzoate dimers and monomer coordinated by pyridines: synthesis and crystal structures. Polyhedron. 2018;151:545–553. [Google Scholar]
- 51.Sarma R., Baruah J.B. A mixed carboxylate polymer as an intermediate during synthesis of mononuclear bis -3, 5-dimethylpyrazole copper(II) benzoates. J. Coord. Chem. 2008;61(20):3329–3335. [Google Scholar]
- 52.Sastri C.V., Eswaramoorthy D., Giribabu L., Maiya B.G. DNA interactions of new mixed-ligand complexes of cobalt(III) and nickel(II) that incorporate modified phenanthroline ligands. J. Inorg. Biochem. 2003;94(1–2):138–145. doi: 10.1016/s0162-0134(02)00622-0. [DOI] [PubMed] [Google Scholar]
- 53.Segľa P., Kuchtanin V., Tatarko M., Švorec J., Moncol J., Valko M. Structural study and magnetic properties of copper(II) thiophene-2-carboxylate with 4-pyridinemethanol and isonicotinamide. Chem. Pap. 2018;72(4):863–876. [Google Scholar]
- 54.Shahid M., Mazhar M., O'Brien P., Afzaal M., Raftery J. catena-Poly[diethyl(2-hydroxyethyl)-ammonium[[tetra-μ-acetato-κ8O:O′-dicuprate (II) (Cu—Cu)]-μ-acetatoκ2O:O′] dichloromethane solvate] Acta Crystallogr. Sect. E Struct. Rep. 2009;E65(2):m163–m164. doi: 10.1107/S1600536808044048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sheldrick G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C. 2015;C71(1):3–8. doi: 10.1107/S2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sheldrick G.M. SHELXT – integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A. 2015;A71(1):3–8. doi: 10.1107/S2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soldevila-Sanmartín J., Sanchez-Sala M., Calvet T., Font-Bardia M., Ayllón J.A., Pons J. [Cu(μ-MeCO2)2(4-Bzpy)]2 (4-Bzpy = 4-benzylpyridine): study of the intermolecular C H⋯O hydrogen bonds at two temperatures. J. Mol. Struct. 2018;1171:808–814. [Google Scholar]
- 58.Sun P., Lu X., Xu C., Sun W., Pan Bo. Understanding of COVID-19 based on current evidence. J. Med. Virol. 2020;92:548–551. doi: 10.1002/jmv.25722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Timco G.A., Fernandez A., Kostopoulos A.K., Charlton J.F., Lockyer S.J., Hailes T.R., Adams R.W., McInnes E.J.L., Tuna F., Vitorica-Yrezabal I.J., Whitehead G.F.S., Winpenny R.E.P. Hybrid organic–inorganic rotaxanes, including a hetero-hybrid [3]rotaxane featuring two distinct heterometallic rings and a molecular shuttle. Angew. Chem. Int. Ed. 2018;57(34):10919–10922. doi: 10.1002/anie.201805439. [DOI] [PubMed] [Google Scholar]
- 60.Viola N.M., Ikram M., Rehman S., Ali S., Akhtar M.N., AlDamen MA., Schulzke C. A paddle wheel dinuclear Copper(II) carboxylate: crystal structure, thermokinetic and magnetic properties. J. Mol. Struct. 2019;1196:754–759. [Google Scholar]
- 61.Wein AN., Cordeiro R., Owens N., Olivier H., Hardcastle KI., Eichler JF. Synthesis and characterization of Cu(II) paddlewheel complexes possessing fluorinated carboxylate ligands. J. Fluor. Chem. 2009;130(2):197–203. [Google Scholar]
- 62.Wojciechowski K., Bitner A., Bernardinelli G., Brynda M. Azacrown ether–copper(II)–hexanoate complexes. From monomer to 1-D metal organic polymer. Dalton Trans. 2009;(7):1114–1122. doi: 10.1039/b811374b. [DOI] [PubMed] [Google Scholar]
- 63.Youngme S., Cheansirisomboon A., Danvirutai C., Pakawatchai C., Chaichit N. Polynuclear paddle-wheel copper(II) propionate with di-2-pyridylamine or 1,10-phenanthroline: preparation, characterization and X-ray structure. Inorg. Chem. Commun. 2008;11(1):57–62. [Google Scholar]
- 64.Zghair K.H., Saheb E.J., Al-Qadhi B.N. Antiparasitic effect of carbonnanotubes on leishmania donovani in vitro. Iraqi J. Sci. 2016;57(4B):2641–2649. [Google Scholar]
- 65.Zhang J., Zhang C., Yu X., Qin Y., Zhang S. Two novel trinuclear cluster-based coordination polymers with 2,6-Di-imidazol-1-yl-pyridine: solvothermal syntheses, crystal structures, properties and Hirshfeld surface analysis. Supramoecular Chem. 2016;28(3-4):231–238. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.