Abstract
Scientists rely on high-throughput screening tools to identify promising small-molecule compounds for the development of biochemical probes and drugs. This study focuses on the identification of promiscuous bioactive compounds, which are compounds that appear active in many high-throughput screening experiments against diverse targets but are often false-positives which may not be easily developed into successful probes. These compounds can exhibit bioactivity due to nonspecific, intractable mechanisms of action and/or by interference with specific assay technology readouts. Such “frequent hitters” are now commonly identified using substructure filters, including pan assay interference compounds (PAINS). Herein, we show that mechanistic modeling of small-molecule reactivity using deep learning can improve upon PAINS filters when modeling promiscuous bioactivity in PubChem assays. Without training on high-throughput screening data, a deep learning model of small-molecule reactivity achieves a sensitivity and specificity of 18.5% and 95.5%, respectively, in identifying promiscuous bioactive compounds. This performance is similar to PAINS filters, which achieve a sensitivity of 20.3% at the same specificity. Importantly, such reactivity modeling is complementary to PAINS filters. When PAINS filters and reactivity models are combined, the resulting model outperforms either method alone, achieving a sensitivity of 24% at the same specificity. However, as a probabilistic model, the sensitivity and specificity of the deep learning model can be tuned by adjusting the threshold. Moreover, for a subset of PAINS filters, this reactivity model can help discriminate between promiscuous and nonpromiscuous bioactive compounds even among compounds matching those filters. Critically, the reactivity model provides mechanistic hypotheses for assay interference by predicting the precise atoms involved in compound reactivity. Overall, our analysis suggests that deep learning approaches to modeling promiscuous compound bioactivity may provide a complementary approach to current methods for identifying promiscuous compounds.
Graphical Abstract
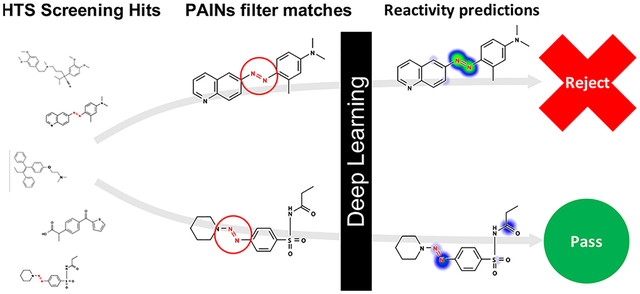
INTRODUCTION
Identifying high-quality, small-molecule probes with specific bioactivity combined with useful mechanisms of action has become a critical component of both basic science research and early drug discovery efforts.1 To this end, many high-throughput screening (HTS) technologies have been developed to rapidly screen libraries of tens of thousands of compounds for modulation of specific biomolecules or production of certain cellular phenotypes. Light-based readouts, including absorbance, fluorescence, luminescence, and resonance energy transfer (e.g., FRET), are commonly used to enable higher throughputs.2 These assays and other technologies are susceptible to technology-related and generalized sources of compound-mediated assay interference.3,4 We and others have noted the importance of this distinction.5,6 Technology-related interferences occur when compounds interfere with some aspect of signal transmission related to a given assay technology and do not represent actual target modulation. Common mechanisms of technology-related interference include light-based interferences (auto-fluorescence, quenching, inner-filter effects, light scattering),7 capture reagent disruption,8,9 luciferase reporter modulation,10 and singlet oxygen quenching and scavenging.11 Generalized interference represents on-target but often poorly tractable mechanisms of bioactivity. Compounds causing generalized interference modulate the target protein as desired, but also exhibit other undesirable properties such as promiscuous activity against off-target proteins, an inconsistent dose–response relationship, or activity that is highly variable with assay conditions. Common mechanisms of generalized interference can include aggregation,12 nonspecific reactivity,13 redox activity,14 chelation,15 membrane perturbation,16 and metal contaminants.17 We note that there can be overlap between these two categories in those cases where the interference source acts upon biological targets and the assay technology, such as when nonspecific compound reactivity may modulate a biological target but also a key reagent.18 Given the implications of each interference category, it is critical to consider these distinctions as well as overlaps when analyzing assay interference and bioassay promiscuity.
The increasing use of HTS to identify small-molecule probes and drugs, combined with a lack of thorough control and validation studies, has led to a proliferation of low-quality probes in the literature that exhibit nonspecific activity against a variety of targets upon further testing.1,19–21 The popularity of virtual HTS techniques has compounded this problem by allowing scientists without experimental chemistry or biological assay expertise to perform screening experiments in silico.22 While many problematic compounds can be recognized by an experienced medicinal chemist and pharmacologists, it can be difficult for nonexperts to recognize potentially undesirable properties. In addition, some interference com-pounds can be difficult to identify a priori based on their chemical structures without performing appropriate follow-up experiments. Clearly, both scientists and publishers need effective tools to flag potentially problematic compounds to guide appropriate orthogonal assays, counter-screens, and controls.
There are several types of interference counter-screens considered the gold-standard for derisking compound-mediated assay interference. For nonspecific thiol reactivity, an often significant source of assay interference and nonspecific bioactivity, such assays can include incubating compounds with thiol-based probes such as glutathione (GSH), with compound-thiol adducts detected by fluorometry or mass spectrometry (MS).23 Advanced techniques for identifying thiol-reactivity include ALARM NMR and protein MS.24 However, some of these methods can require substantial expertise and instrumentation and may be difficult to implement in certain academic settings.
One current in silico approach to this problem uses filters to match chemical substructures known to be problematic. One commonly used filter set, known as Pan Assay Interference Compounds (PAINS) filters, was designed by analyzing a set of six AlphaScreenassays, a luminescence-based assay technology. While it is thought nonspecific reactivity is the main contribution to assay interference in this study, PAINS filters may capture multiple assay interference mechanisms. Since these filters were created in a data-driven (observational) process, some of these filters flag substructures that do not have a clear interference mechanism. In addition, not all compounds matching a PAINS filter are promiscuously active and PAINS filters cannot discriminate these compounds. Indeed, it has been previously noted that PAINS filters match FDA approved drugs.25,26 Some of these drugs are cytotoxic agents, used for treating serious diseases such as cancers and infectious disease.26 Importantly, promiscuous activity alone does not necessarily prevent a drug from being effective.27 The specific mechanism of a compound’s bioactivity, whether promiscuous or not, more directly determines its viability as a drug.
We note that the general applicability of PAINS filters has been called into question for a number of reasons.6,25 It is important to consider that these filters were developed using data from a single screening methodology (AlphaScreen) and may or may not reflect promiscuity in other assay technologies. Furthermore, these assays were carried out at a single compound concentration between 10 and 30 μM, which may not necessarily reflect interference or bioassay promiscuity at alternative compound concentrations. Finally, it has been noted that the primary data and some of the study targets are not publicly available, which can limit independent studies.6 One study reported that PAINS filters generalize poorly to other assay technologies used in screening data deposited in the PubChem database, and many PAINS filters match so-called “dark chemical matter”,28 which are compounds that appear inert in most bioassays.25 An analysis by Jasial and colleagues revealed similar observations.29 Besides overly strict filters, these observations could also be caused by varying compound screening concentrations, library curation practices, definitions of bioactivity, and capture reagents. While this may be true of certain chemotypes that may interfere with singlet oxygen transmission (e.g., quenching, scavenging), capture reagent disruption, or light transmission, one hypothesis is that such compounds should have interfered in many of the original PAINS screens independent of the six targets.30 Notably, these screens were run using different compound concentrations, and the magnitude (“end effect”) of singlet oxygen interference on the final readout in any of these original assays may also depend on compound concentration and other experimental conditions such as buffer composition, background intensity, signal intensity, and assay time. Therefore, further studies may be useful to better characterize the consequences of compound concentration on singlet oxygen interference in AlphaScreenassays. When applying PAINS filters, it is equally important to consider both the PAINS structure and the chemical context in which it appears and implement appropriate follow-up experiments when indicated, points that have been expressed repeatedly since the original PAINS publication.31–33
In this work, we investigate how a deep learning model of small-molecule, covalent reactivity with scavenger probes (cyanide and GSH) and biological macromolecules (DNA and protein) can be used to identify promiscuously active small-molecules.34,35 While this model only captures a single mode of assay interference, it can provide mechanistic predictions that suggest why a particular molecule may be reactive, thereby enabling those involved in HTS triage to make more informed decisions to mitigate nonspecific reactive behaviors. Data-driven modeling of compound interference mechanisms should provide three key benefits: such models should enable (1) better selection of compounds for screening libraries, (2) nonexperts to better identify problematic compounds, and (3) publishers to flag potentially questionable chemical matter for closer scrutiny.
Toward this goal, we studied the utility of reactivity modeling in enhancing prediction and mechanistic understanding of compound-mediated assay interference and bioassay promiscuity. PAINS filters were benchmarked for their ability to identify promiscuous bioactive compounds tested in large PubChem screening assays, followed by a similar analysis using our reactivity model. Next, we studied whether our reactivity model could distinguish promiscuous and nonpromiscuous compounds that match the same PAINS filter, and investigated how this reactivity model could be used to identify chemical mechanisms of reactivity among PAINS filter matches. We then developed a simplified scoring model which combines reactivity scores from our model into a single promiscuity prediction, followed by a combined model incorporating reactivity scores with PAINS filters to flag promiscuous bioactive compounds.
DATA AND METHODS
PubChem BioAssay.
The complete PubChem BioAssay database was downloaded via the NCBI FTP service (data accessed April seventh, 2016).36 We focused on HTS assays in PubChem and excluded assays imported from ChEMBL, another bioassay database. To focus on applicability to HTS, analyses were restricted to bioassays testing at least 1000 compounds, and those compounds tested in at least 100 discrete bioassays, resulting in a data set of 384 328 unique compounds (Figure 1A). Compounds were defined as active if the value of the PUBCHEM_ACTIVITY_OUTCOME field was set as “Active” and were defined as inactive otherwise. We note these activity outcomes are based on the activity selection criteria of individual assay authors and are not uniform. Both cell-based and target-based assays were included in the data set.
Figure 1.
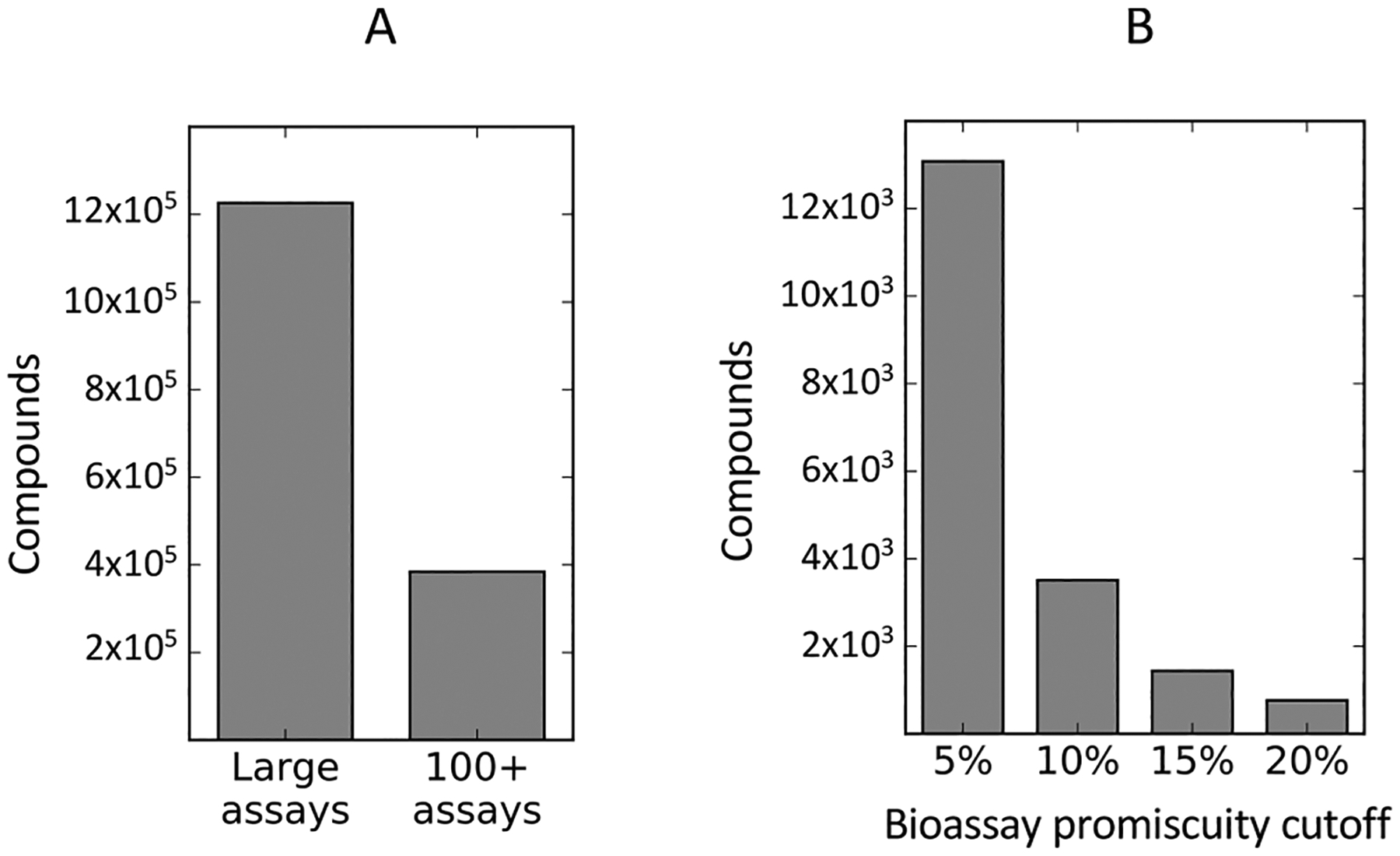
Assay data from PubChem reveals a large number of potential bioassay promiscuous compounds. (A) Analysis of all non-ChEMBL PubChem assays testing greater than 1000 compounds. This study restricted analysis to compounds tested in greater than 100 bioassays (384 328 compounds, from an initial 1 226 075). (B) Compounds were defined as promiscuous bioactives if they were active above a fixed percentage of tested bioassays. Cutoffs of 5, 10, 15, and 20% were initially considered. Promiscuous activity of compounds follows an approximate power distribution (Figure S1). At a promiscuity cutoff of 5%, approximately 3.40% of compounds in the data set were considered promiscuous. Note, many compounds were active in more than 20% of tested assays.
An initial analysis demonstrated both the number of compounds tested in PubChem and the activity rates of compounds follow an approximate power law distribution, with the majority of compounds having been tested in fewer than 100 assays and active in fewer than ten assays (Figure S1). These data appear similar to a recent PubChem analysis of bioassay promiscuity.29 We defined promiscuous compounds as those compounds exhibiting activity above a fixed percentage of assays. Because the classification methods used in this study are binary classifiers, this definition of promiscuous compounds enables a straightforward, systematic comparison between promiscuous actives, PAINS filters and our reactivity models. While this facilitates our analyses, we note that binary classifiers and other data binning have some potential shortfalls, such as the potential to inflate correlations.37 To address this, we chose these cutoffs prior to running our classifiers, and we show data from multiple cutoffs for some of our experiments to demonstrate that the data have similar interpretation regardless of the choice of cutoff. We initially experimented with cutoffs of 5%, 10%, 15% and 20% (Figure 1B). A cutoff of 5% marked only 3.4% of compounds as promiscuously active, with exponentially fewer compounds exhibiting pan-assay activity at higher cutoffs. A complete list of compounds and their promiscuity labels can be found in the supplementary files.
A relationship between lipophilicity and promiscuous activity of drugs has been previously noted in the literature.38 Specifically, drugs with logP greater than three were observed to have substantially higher rates of promiscuity. We additionally analyzed the relationship between solubility (octanol/water partition coefficient, logP), molecular weight, and bioassay activity rates among those compounds meeting our filtering criteria (Figure S2). The majority of compounds (90%) conform to a variant of the Lipinski rule of five criteria39 for these metrics: molecular weight between 180 and 500 Da and logP between −0.4 and 5.6. In this data set, no apparent relationship was observed between logP and bioactivity or molecular weight and bioactivity.
DrugBank.
The complete DrugBank database in XML format was downloaded from www.drugbank.ca (accessed April 20th, 2016). FDA-approved, small-molecule drugs were identified by entries annotated with the “small molecule” type and the “approved” group. Corresponding PubChem Compound IDs (CIDs) for each DrugBank small-molecule were identified from the DrugBank XML records, and any drugs not having an associated CID in our filtered PubChem data set were not considered for further analyses.
PAINS Filters.
PAINS filters were acquired in SMARTS format from the RDKit GitHub repository (Table S7, accessed May 25th, 2016). These filters were compiled from work translating the original SLN format40 PAINS filters to SMARTS format, which was published by the original authors of the PAINS filters.41 The following RDKit pipeline was implemented for applying pains filters: (1) PAINS filters were loaded using RDKit SMARTS parser with the mergeHs flag set, (2) input test molecules were read in SDF format with the sanitize flag set, (3) existing hydrogen atoms were removed and then explicit hydrogen atoms added to the molecule, (4) detection algorithms were invoked for aromaticity, conjugation, and hybridization, and (5) pains filter matches were identified using the RDKit substructure matching algorithm. This procedure was performed in accordance with the RDKit author’s guidelines for applying PAINS filters.
SMARTS substructure queries are known to behave differently on different software platforms. Besides using RDKit, we also experimented with applying the PAINS filters using CDK, a java-based cheminformatics toolkit. Interesting, we found that a substantial number of compounds matched PAINS filters using the RDKit implementation but did not match when using the CDK implementation and vice versa (Figure S3). However, we found that this discrepancy did not affect the overall sensitivity and specificity of PAINS filters on our data set (Figures S4C and F), and subsequent analyses utilized the RDKit implementation.
For enrichment calculations and predictive analysis, PAINS filters were grouped by the common functional groups they targeted, as indicated by their text names. For example, there are four filters targeting the quinone functional group: quinone_A(370), quinone_B(5), quinone_C(2), and quinone_D(2), which were all included in the filter group “quinone”.
A complete list of filters used in this study, along with the groups to which they were assigned, are provided as Supporting Information (Table S7). In addition, a complete list of compounds and their matched PAINS filters is provided as a supplementary file.
Lilly MedChem Filters.
Lilly MedChem filtering software42 was acquired from that project’s github page (https://github.com/IanAWatson/Lilly-Medchem-Rules). The filtering software was run with default options, with the flags -expert and -B to write molecules flagged by filtering to a separate file. Any molecule flagged by the filtering software was classified as a potential promiscuous bioactive and those not flagged were classified as nonpromiscuous bioactives.
Small Molecule Reactivity Model.
Compounds were analyzed using a previously developed model of small-molecule reactivity with trapping agents GSH and cyanide as well as biological macromolecules DNA and protein.34,35 Briefly, a convolutional neural network model was trained using literature-derived data extracted from the Accelrys Metabolite Database and other sources. The model was trained on a set of 2803 molecules encompassing nonreactive molecules, as well as molecules reactive with each of the modeled substrates: cyanide, GSH, DNA and protein. The model was constructed and trained in two stages (Figure 2A). First, atoms and molecules in the training data were described numerically. Topology-based descriptors were computed for each atom in the training set, including 154 valence, ring membership, aromaticity, and the number of nearby heteroatoms (Table S9). These vectors formed the input to an atom-level neural network, which was then trained to predict sites (discrete atoms) at which nucleophilic attack by GSH, cyanide, DNA and/or protein may occur. Next, for each molecule (including those without a known site of reactivity), the top five reactive-site predictions obtained from the site-level neural network were combined with 15 molecule-level descriptors including molecular weight, span, total polar surface area to form a set of 20 molecule level descriptors (Table S8). The resulting vector formed the input to a second neural network that was trained to predict which molecules underwent conjugation reactions with GSH, cyanide, DNA, and/or protein.
Figure 2.
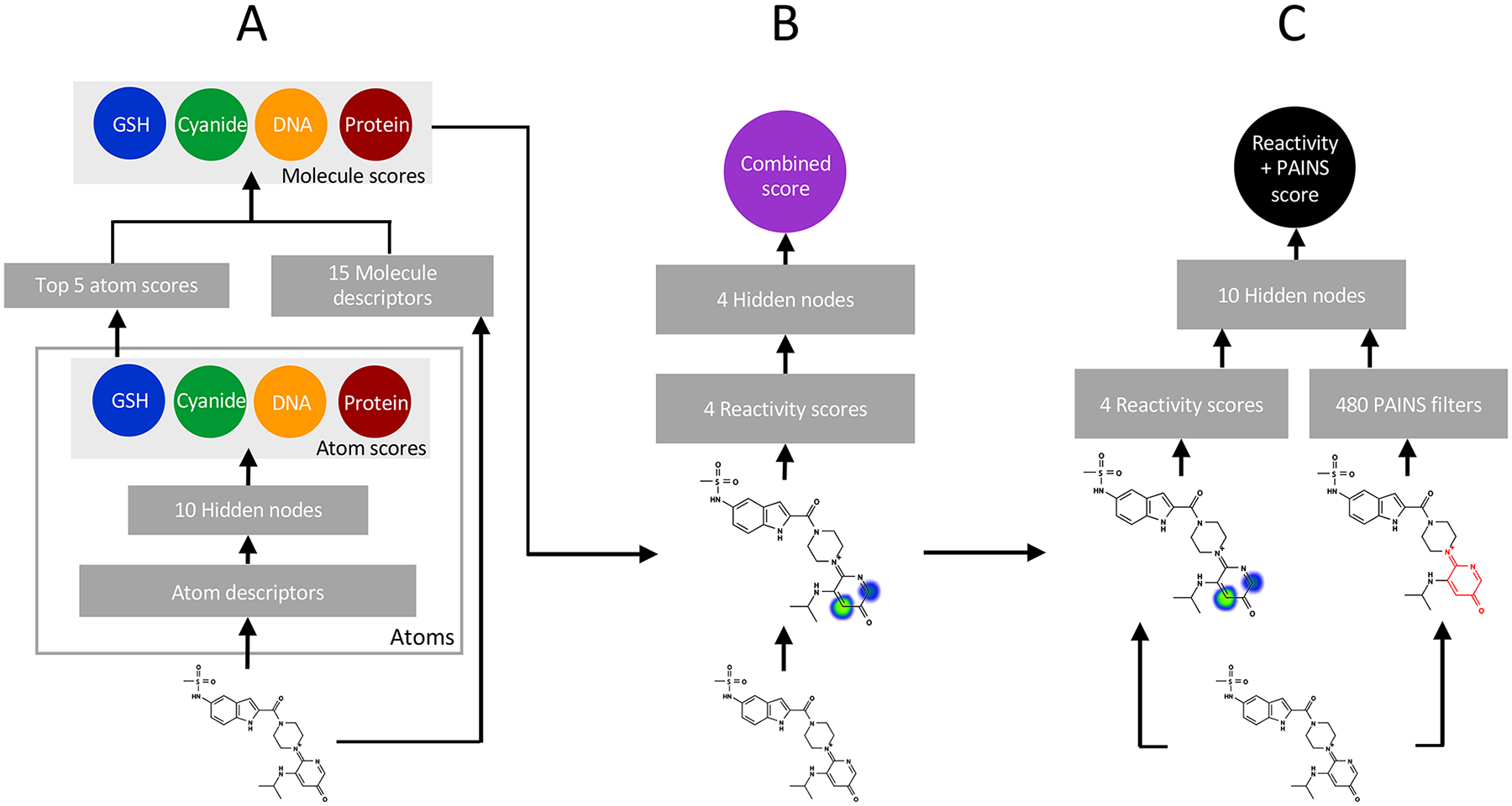
Schematic of deep convolutional neural network models for predicting small-molecule reactivity and bioassay promiscuity. (A) Atoms in a test compound are represented as rows of numerical descriptors in a data matrix. These data are input to a neural network with one hidden layer of ten units. This neural network calculates four atom reactivity scores, each score predicts nucleophilic attack at that atom by GSH, cyanide, DNA, or protein. The top five atom reactivity scores in each category are then combined with molecule descriptors and are then used to calculate four molecule reactivity scores. Each molecule level reactivity score is then trained to predict conjugation of the input molecule to either GSH, cyanide, DNA, or protein.34,35 (B) Molecule-level reactivity scores are further combined with another neural network to produce a single integrated reactive promiscuity score. This network can then be trained to predict promiscuous bioactivity in HTS data sets. (C) A hybrid model combines molecule-level reactivity scores with binary indicators for PAINS substructure filter matches. A single hidden layer neural network is then trained to predict promiscuous behavior in HTS data sets.
Compounds were also analyzed by two additional models of bioassay promiscuity. In the first model, four reactivity scores for a molecule (from GSH, cyanide, DNA, protein scores) were combined into a single promiscuity score via a single hidden-layer neural network with four hidden nodes (Combined Score, Figure 2B). In the second model, the four reactivity scores were combined with 480 binary indicator variables that identified whether a match to a particular PAINS filter was present in the molecule. These inputs were combined by a single hidden layer neural network with ten hidden nodes to produce a single bioactivity promiscuity score (React +PAINS Score). Reactivity scores for all compounds and all six models used in this study are available as a supplementary file.
RESULTS AND DISCUSSION
Substructure Filters Identify Promiscuous Actives.
PAINS filters proved to be effective at screening promiscuously active compounds in PubChem. Using the RDKit chemistry toolkit, we applied PAINS filters to the promiscuity annotated PubChem data set. At the 5% cutoff, 13% of PAINS filter matches were marked as promiscuous compounds, which represents an enrichment of 3.85-fold (Figure 3A). The sensitivity of PAINS filters for promiscuous compounds at the 5% cutoff was 20.3% and specificity was 95.5% (Figure 3B and C). This balance of specificity and sensitivity is acceptable for a screening test. The sensitivity and specificity of PAINS filters was not substantially affected by the choice of promiscuity cutoff (Figures S4A and D).
Figure 3.
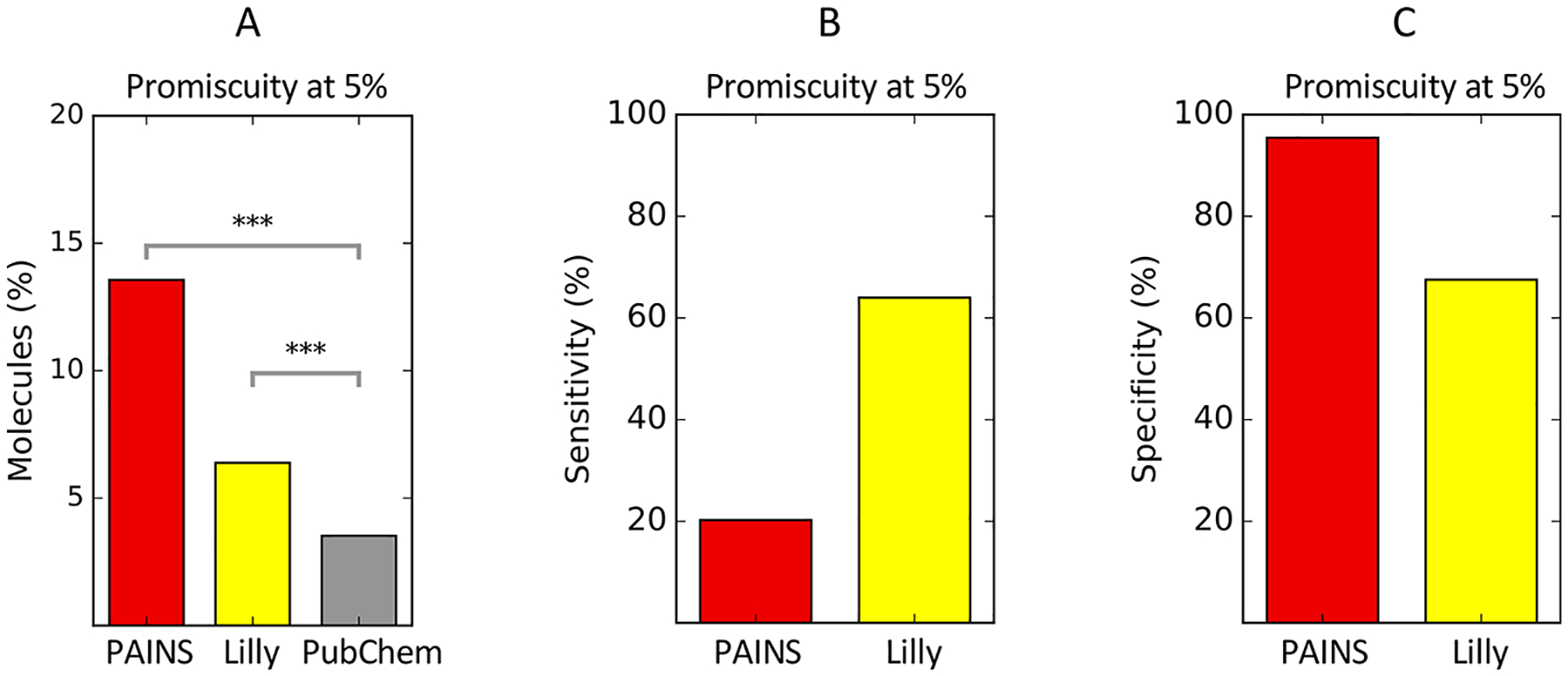
Substructure filter-based methods for flagging promiscuous and/or assay interference compounds detecting promiscuous bioactives in PubChem. (A) At the 5% promiscuity cutoff, PAINS filters (red) are enriched 3.85-fold for promiscuous bioactives (p < 10−10, χ2 test) and Lilly MedChem filters (yellow) are enriched 1.85-fold for promiscuous bioactives (p < 10−10, χ2 test). (B) PAINS filters have a lower sensitivity than the Lilly MedChem filters for promiscuous bioactive compounds in PubChem. (C) However, PAINS filters have a 95% specificity for promiscuous actives, while Lilly MedChem filters have 67.5% specificity. ***: p < 0.0001.
In addition to PAINS filters, we also applied the Lilly MedChem filters to the PubChem data set.42 Lilly MedChem is designed to identify not only compounds that interfere with biological assays but also those compounds that are unlikely to become drugs due to bioavailability or toxicity issues. Lilly MedChem achieved a sensitivity of 64% and a specificity of 67.5% at the 5% promiscuity cutoff (Figure 3B and C). These filters identify problematic structures not necessarily associated with HTS interference, which may contribute to their low specificity and make the Lilly MedChem filters less useful in this context. However, the MedChem filters do gain substantial sensitivity with increasing promiscuity cutoffs, which suggests that this filter set is optimized for identifying some highly problematic compounds (Figures S4B and E).
Analysis of PAINS Filters and Predicted Compound Reactivity.
While PAINS filters were designed with principles of medicinal chemistry in mind, many of these filters have unclear and/or unconfirmed chemical mechanisms of assay interference, and compounds flagged by PAINS filters can demonstrate a spectrum of promiscuity profiles.25,29 Compound-mediated assay interference can be a complex phenomenon, and minor chemical changes may significantly affect interference. Additional confounding factors include random errors (e.g., false-positives, false-negatives), assay precision, library biases, compound stability, definitions of bioactivity, and experimental protocols, to name a few.
With these limitations in mind, we hypothesized that improvements in flagging interference compounds can be made by directly modeling assay interference mechanisms in a data-driven manner. We therefore compared the performance of the conventional PAINS filters to a small-molecule reactivity model previously developed in our lab.34,35 We initially focused on modeling compound reactivity because (1) it represents a significant source of nonspecific bioactivity versus many biological targets and (2) it likely represents a significant source of interference in the original PAINS training set, given detergent and decoy proteins were included in assay buffer to mitigate aggregation. The reactivity model provides mechanistic predictions of reactivity, often pinpointing the precise atom at which a covalent bond with a biological nucleophile is formed. Furthermore, this model can predict reactivity with diverse biological molecules including GSH, cyanide, DNA, and proteins. Importantly, the model distinguishes between reactive and nonreactive molecules containing the same chemotypes, such as epoxides35 and sulfur oxides.43 This is a key advantage which separates deep-learning approaches from substructure-based methods. Our model was constructed using deep convolutional neural networks to compute both site (atom)-level reactivity scores and molecule-level reactivity scores. These scores are scaled between zero and one and are well-calibrated probabilities.44 A well-calibrated probabilistic model outputs scores which are proportional to the ratio of positives to negatives among all examples in the training set assigned the same score. For example, among training examples assigned scores close to 0.3, 30% are positive examples.
Many of the PAINS chemotypes are hypothesized to interfere via nonspecific reactivity. Accordingly, PAINS matches in PubChem were assigned higher GSH reactivity scores than non-PAINS by our model (p = 2.06 × 10−7, Mann–Whitney U-test, Figure 4A). In contrast, FDA approved drugs, whether PAINS matches or not, were assigned low GSH reactivity scores, similar to non-PAINS in PubChem. Compounds active in more than 5% of tested assays had substantially higher reactivity scores than nonpromiscuous compounds (Figure 4B). In addition, compounds matching multiple PAINS filters exhibited higher rates of bioassay promiscuity and were assigned higher reactivity scores than compounds matching fewer filters (Figure S13).
Figure 4.
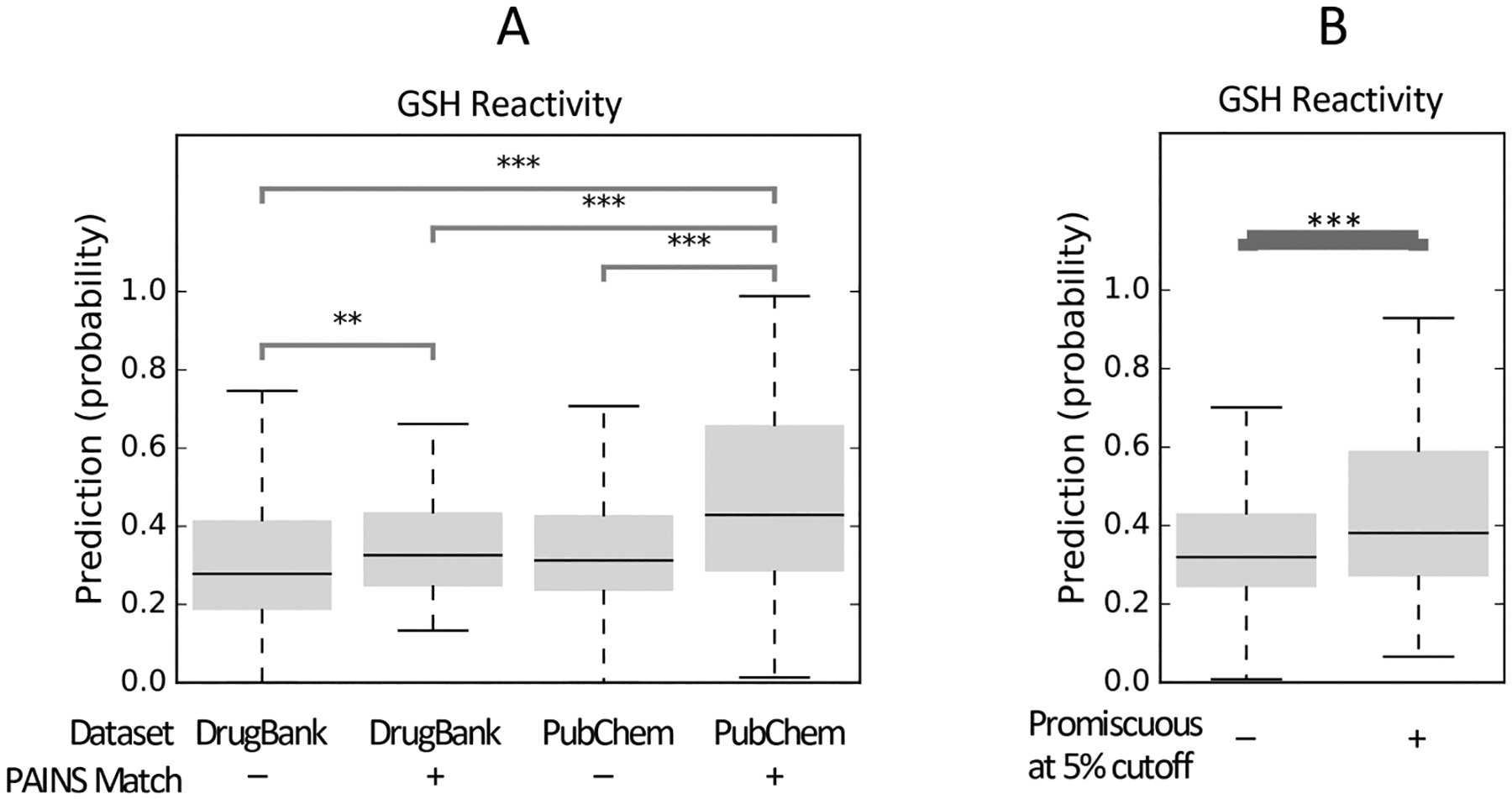
GSH reactivity predictions for compounds in DrugBank and PubChem. PAINS filters are associated with increased GSH reactivity scores in PubChem, but reactivity is not increased among FDA-approved drugs. (A) Reactivity scores for FDA-approved drugs in DrugBank are comparable between PAINS and non-PAINS, whereas reactivity scores of PubChem PAINS matches are substantially elevated compared to non-PAINS and compared to DrugBank (p = 2.06 × 10−7, Mann–Whitney U-test). While some FDA-approved drugs act via a reactive mechanism, the majority of FDA-approved drugs are not explicitly reactive and not found to be promiscuous bioactives. Outliers are not shown. (B) Compounds active in more than 5% of tested assays in PubChem have substantially higher reactivity scores than nonpromiscuous compounds (p < 10−10, Mann–Whitney U-test). Outliers are not shown. **: p < 0.001. ***: p < 0.0001.
PAINS matches in DrugBank are predictive of promiscuous activity in PubChem assays but are not associated with increased predicted reactivity. Out of 926 FDA-approved small-molecule drugs in DrugBank that have assay data in PubChem, 65 were found to be PAINS filter matches. We found that 235 drugs were promiscuous at the 5% cutoff, with many drugs exhibiting higher degrees of promiscuity (e.g, 100 drugs were active in at least 10% of tested assays, and the antineoplastic agent bortezomib was active in 50% of tested assays, most of which were annotated by bioassay ontology45 as cell-based assays). Among DrugBank PAINS matches, 41 of the 65 were also promiscuous actives, which represents a 2.48-fold enrichment for promiscuous activity (Figure S6A). However, reactivity modeling of these drugs showed that there is only a small 13% difference in average reactivity scores between PAINS matches and nonmatches (p = 0.0012, Mann–Whitney U-test). Furthermore, there is only a small 8.2% difference in average reactivity score between drugs which display promiscuous activity and those that do not (p = 0.0043, Mann–Whitney U-test, Figure S6B). In the in vivo context, reactive compounds may give rise to a host of undesirable off-target effects including toxicity, and as such, reactive compounds are traditionally less common among FDA-approved drugs. We note drugs, including certain reactive compounds, may be enriched in bioassays by virtue of their known bioactivity and that drugs may be assayed at concentrations that are not physiologically or therapeutically relevant. These factors may confound interpretation of this data.
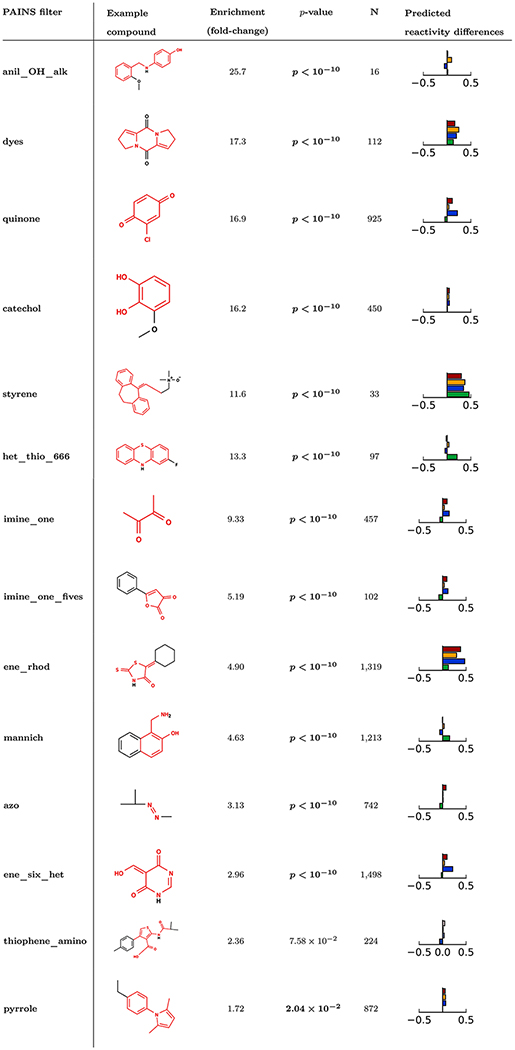
Our analyses identified 45 PAINS filter groups that exhibited a statistically significant enrichment for promiscuous actives (p < 0.05, Bonferroni-corrected χ2 test). Several of the most enriched PAINS filters were also associated with increased reactivity (Tables 1 and S1). Among these filters, quinones,46 rhodanines,47 Mannich bases,48 and styrenes49,50 are all associated with covalent reactivity.
Table 1.
Selected PAINS Filter Groups That Are Enriched for Promiscuous Bioactives Compared to the Backgrounda
|
Enrichment scores in bold indicate statistically-significant enrichment for promiscuous bioactives above the background rate of 3.84% (p < 0.05, Bonferroni-corrected χ2 test). Example molecules shown are the lowest molecular weight match in PubChem to the given filter group. Substructures matched by the filter group are shown in red. In addition, the change in mean reactivity score between molecules matching the PAINS filter and those not matching is shown: cyanide (blue), GSH (green), DNA (orange), protein (red). A complete list of filter groups significantly enriched for promiscuous actives is provided in the supplementary material (Table S1). Filter groups are sorted by promiscuous activity enrichment fold change.
Some known reactive motifs are still useful pharmacophores. For example, quinones are used in numerous drugs51 and are present in electron transport cofactor molecules for photosynthesis and cellular respiration.52 They are most likely identified by PAINS filters because of their tendency to form reactive oxygen species in situ (redox active)46,53 and exhibit strong, nonspecific reactivity with a variety of biological nucleophiles.46 Unsurprisingly, quinones are strongly enriched for promiscuous activity, and our reactivity model predicts an increase in reactivity with both GSH and protein.
Some reactive motifs have been reported extensively in the literature yet have never been incorporated into a successful drug. Our analyses identified reactive rhodanine-containing compounds such as those matched by the ene_rhod filter group. Certain rhodanines and related compounds such as thiazolidinediones can react with thiols (e.g., those that contain an exocyclic unsaturated bond), but that even some rhodanines and related compounds with unsaturated exocyclic bonds do not show gross reactivity in certain experimental conditions or bioassay promiscuity.54 The ultimate utility of these com-pounds is part of an ongoing discussion in the medicinal chemistry community.47,54,55 Our reactivity model assigns high scores for reactivity of this class with all the modeled substrates, and based on current evidence and an abundance of caution, we would recommend flagging these compounds for additional interference characterization such as protein reactivity.
In addition to filters with known reactive liabilities, we also identified several enriched PAINS filters which were not associated with increases in predicted reactivity. For example, the PAINS filter group most strongly enriched for promiscuous actives is the anil_OH_alk filter group. These compounds are not predicted to be reactive by our model. However, these compounds contain anilines, which can interfere with certain assay technologies such as AlphaScreendue to potential physical or chemical quenching of singlet oxygen species.11,56
Furthermore, catechols are also enriched for promiscuous bioactivity. However, our reactivity model does not suggest substantial increases in reactivity for these chemical species. In addition to being prone to nonspecific reactivity,24 catechols can be redox active or chelate metals.57 These alternative assay interference mechanisms may better account for the observed enrichment for promiscuous behavior.
These studies show that our reactivity model assigns higher reactivity scores to compounds which match PAINS filters and are also promiscuous bioactives in PubChem. Numerous PAINS filters enriched for promiscuous bioactives in PubChem also receive higher reactivity scores. Reactivity models provide additional confirmation of a reactive interference mechanism for some filters, and identify some filters with potential nonreactive interference mechanisms. Furthermore, our reactivity model does not assign higher scores to FDA-approved drugs matching PAINS filters, leading us to hypothesize that it may be able to discriminate promiscuous bioactives among compounds matching the same PAINS filter. We further investigate this hypothesis in the following sections.
Reactivity Scores Predict Promiscuity.
Despite only modeling one of many potential assay interference mechanisms, our reactivity model identifies many promiscuous bioactives. To further evaluate the utility of our model as a screening tool, we performed receiver operator characteristic curve analysis.59 At the 5% bioactivity cutoff, promiscuous actives are identified with area under the receiver operator curve (AUC) of 64%, 62%, 62%, and 56% for our GSH, protein, DNA, and cyanide molecule reactivity scores, respectively (Figure 5). Our GSH reactivity score approaches 16% sensitivity at the same specificity as PAINS filters, which have a sensitivity of 20.3%. This is notable, considering that PAINS filters identify compounds that are not specifically reactive.
Figure 5.
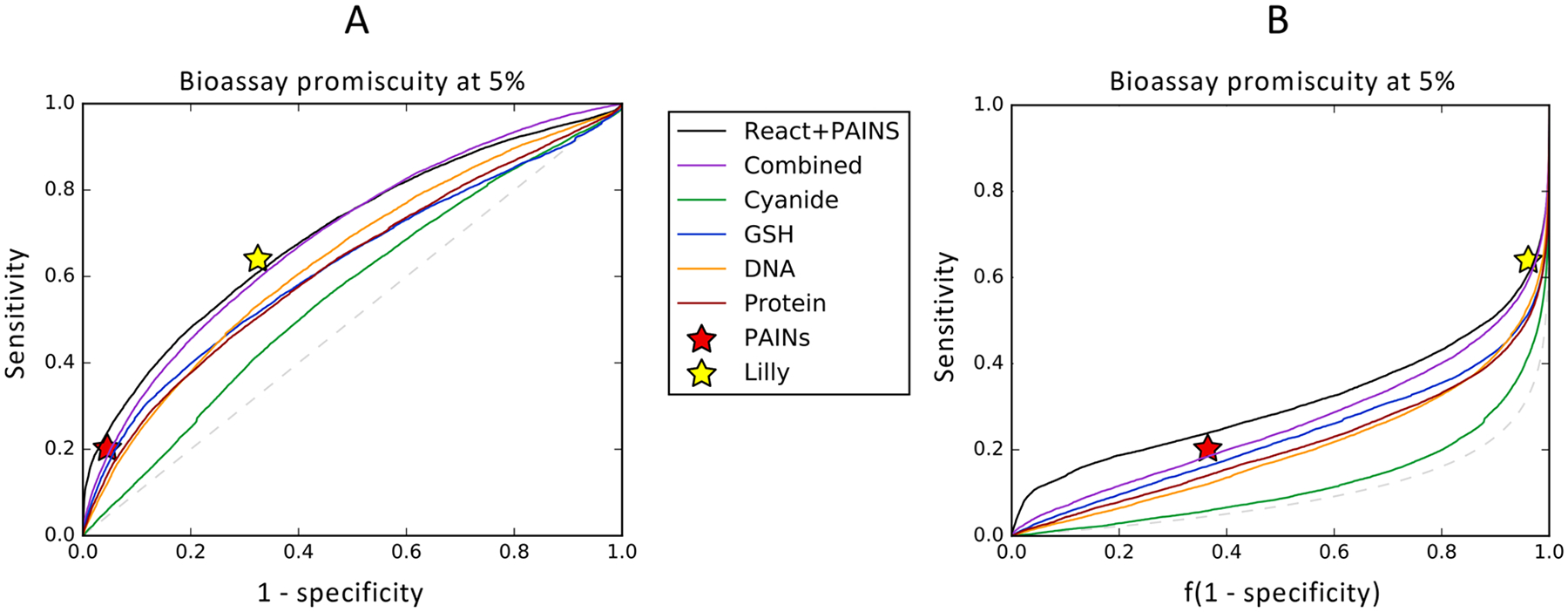
Reactivity scores are predictive of promiscuous bioactivity. (A) Model scores of small-molecule reactivity with DNA (AUC 63.8%), GSH (AUC 62.1%), and protein (AUC 62.1%) are all modestly predictive of promiscuous behavior at the 5% bioactivity cutoff. Predictions of GSH reactivity achieve similar sensitivity and specificity to PAINS filters, while Lilly MedChem filters have a higher sensitivity for promiscuous actives but lower specificity. Combining the four reactivity scores into a single integrated score via a small neural network achieves a 100-fold cross validated AUC of 69.1%. Including PAINS filter matches with reactivity scores in a similar manner achieves a 100-fold cross validated AUC of 69.5%. (B) CROC curves58 with the exponential transform (α = 10) show a substantial increase in early recall for the combined PAINS and reactivity model, with a 4% increase in sensitivity compared to PAINS filters at the same specificity.
GSH reactivity scores can predict promiscuous bioactivity among PAINS filter matches, and also among compounds not matching any PAINS filters. Since PAINS filters match chemical substructures associated with reactivity, it is important to ask whether PAINS filters capture all potentially reactive compounds, and whether they may indiscriminately flag compounds as reactive that may ultimately be tractable, less-promiscuous compounds.
Toward this end, we collected compounds flagged by at least one PAINS filter. This filtered data set consisted of 2967 promiscuous compounds and 19 928 nonpromiscuous com-pounds using the 5% bioactivity cutoff in PubChem. GSH reactivity scores predict promiscuous actives among these compounds at the 5% cutoff with an AUC of 64%, comparable to the AUC for the whole data set (Figure S7 A). Early recall, the left portion of the curve corresponding to bioactives with high reactivity scores, is diminished, suggesting that PAINS filters already capture some of the most highly reactive chemical groups. Conversely, we analyzed those compounds that did not match any PAINS filter. This filtered data set contained 11 682 promiscuous compounds and 364 444 nonpromiscuous compounds. GSH reactivity score predicted promiscuous bioactivity among these compounds at the 5% bioactivity cutoff with an AUC of 60% (Figure S7B). This notable residual predictive power suggests that PAINS filters do not capture all possibly reactive compounds and that our model can provide additional predictive power independent of PAINS filters. One explanation for this observation may be that multiple notoriously reactive chemotypes were purposefully excluded from the screening library from which the PAINS were derived and would therefore not be included in the PAINS training set.
We then hypothesized that combining reactivity scores into a single comprehensive reactive promiscuity score can improve predictions of promiscuous behavior. Each reactivity score provides some predictive power to identify promiscuously bioactive compounds, and each score predicts the reactivity of compounds with different biological nucleophiles. For example, compounds reactive with cyanide and DNA tend to be reactive with other “hard” nucleophiles, while those reacting with GSH tend to be reactive with other “soft” nucleophiles.60–65 This information represents independent predictive power for bioassay promiscuity due to nonspecific compound reactivity. To maximize the sensitivity of our model, we combined the four reactivity scores into a single comprehensive score using a simple neural network classifier (Figure 2B). The neural network takes as input the output score of each of the four reactivity scores. The network was trained to predict whether a molecule demonstrated bioassay promiscuity at the 5% bioassay activity cutoff. The network has only 25 trainable parameters. We then tested our model in 100-fold cross validated experiments on the PubChem data set and computed a receiver operator curve for the collected predictions from each fold (Figure 5). This combined model achieves an AUC of 69.1%, with only a 1.8% decrease in sensitivity compared to PAINS filters at the same specificity.
Given the comparability of our combined reactivity model with conventional PAINS substructure filtering, we then hypothesized that combining PAINS filters and our reactivity model could further improve performance compared to either method individually. We thereby constructed a combined neural network classifier using information from both methods (Figure 2C). For each test compound, this classifier took as input the four reactivity scores from our reactivity model and a 480-bit binary vector indicating which of the PAINS filters matched the molecule. This network was then trained to predict the probability that a given molecule was promiscuous at the 5% bioactivity cutoff. This model achieved an AUC of 69.5% in 100-fold cross validated experiments (Figure 5). While this AUC is not substantially different from the combined reactivity model (i.e., 69.1%), early recall is improved. As a result, the sensitivity of this model is 3.7% greater than PAINS filters at the same specificity. In practical terms, since large HTS primary screens can identify hundreds of actives, our models may identify a substantial number of additional actives as nonspecifically reactive. Furthermore, our models could be used to identify and eliminate particularly problematic compounds from large screening libraries.
Reactivity Model Improves PAINS Filters.
Many PAINS filters match chemical groups known to interfere with HTS assays by nonspecific covalent reactivity. However, many of these same chemical groups can also be found among drugs. Importantly, not all molecules with a given reactive group will be necessarily reactive or interfere with assays. PAINS filters were created by identifying chemical moieties enriched for promiscuous behavior, without further workup of the underlying mechanisms of compound-mediated assay interference including chemical reactivity. For example, while specific sites of chemical reactivity can often be identified by trained scientists, the PAINS filters themselves do not indicate which part of a molecule forms a covalent bond with proteins in an assay solution. In addition, some PAINS filters are correlated to other common chemical groups, which may in turn be mechanistically linked to interference.
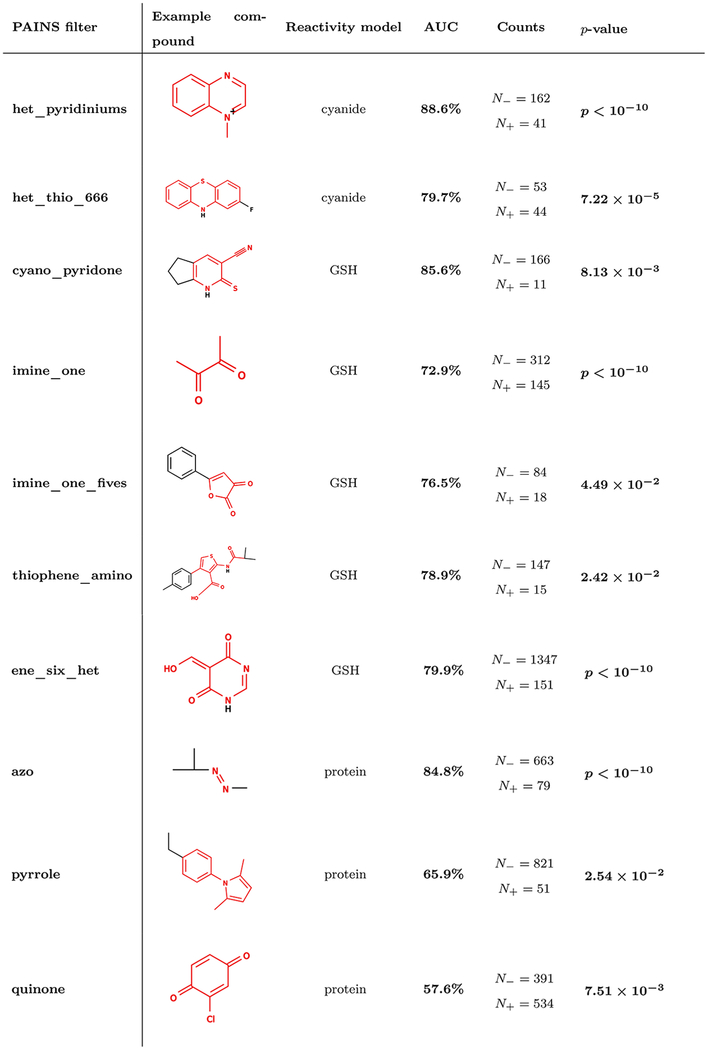
As a potential useful add-on to existing filtering techniques, our reactivity model can help predict reactive and nonreactive molecules within the same compound class, and can also predict the precise atoms that are susceptible to nucleophilic attack. Such capability could be useful for discriminating between compounds flagged as PAINS that may otherwise prove tractable. We computed AUCs and statistical significance using a Bonferroni-corrected Mann–Whitney U-test for each pair of reactivity score and PAINS filter group. Fifteen unique filter groups were identified for which reactivity scores provided statistically significant predictive power to discriminate promiscuous actives at the 5% bioactivity cutoff among compounds matching the PAINS filter group (Tables S2–S5). Promiscuity in some filter groups could be predicted by more than one reactivity score. For cyanide, GSH, DNA, and protein scores, respectively six, seven, six, and seven PAINS filter groups were identified. Statistically significant AUC values ranged from very strong predictive power (88.6% for the het_pyridinium class) to weak predictive power (57.6% for the quinone class) (Table 2).
Table 2.
Selected PAINS filter Groups Enhanced by Reactivity Modelinga
|
AUCs shown in bold indicate statistical significance with p < 0.05 using a Mann-Whitney U test and a Bonferroni correction for the number of tested filter groups. Example compounds are the lowest molecular weight match in PubChem to the given filter group. Filtered substructures are shown in red. The number of promiscuous (N+) and non-promiscuous (N−) molecules in each class are shown. P-values were calculated from AUC values by the normal approximation to the Mann–Whitney U-test. A complete list of filter groups for which reactivity modeling improves bioassay promiscuity predictions is provided (Tables S2–S5).
Reactivity Model Provides Mechanistic Hypotheses of Interference.
PAINS filters were originally constructed by an observational process intended to identify chemical substructures associated with promiscuous bioactivity using AlphaScreentechnology and have subsequently been generalized to other HTS technologies. The individual filters themselves may be recognized by scientists with medicinal chemistry expertise as being associated with particular mechanisms of assay interference. However, in cases where PAINS filters are actually identifying reactive compounds, the filters cannot explicitly indicate the reactive atoms. In contrast, our reactivity model provides mechanistic predictions, and can identify both the site of covalent interaction and the probability of that interaction.
Critically, our reactivity model provides additional mechanistic interpretations for PAINS filter group matches. To demonstrate this point, we discuss (1) four cases of PAINS filters that have potential reactive mechanisms directly related to the matched substructure and supported by reactivity modeling and (2) six cases where reactivity modeling identifies additional potentially reactive structural elements not otherwise identified by a given PAINS filter group.
PAINS Filters with Proposed Reactive Mechanisms Supported by Reactivity Modeling.
Some PAINS filters are associated with covalent reactivity as identified in literature studies11,18 and match common reactive motifs, such as Michael-acceptors. Some have undergone rather extensive studies but such studies are resource-intensive and are fewer in number.18 To demonstrate the utility of reactive site prediction by our model, we present three examples of PAINS filters that match potentially reactive substructures that are also predicted by our reactivity model.
The reactivity model performs best at discriminating compounds matching the het_pyridiniums PAINS filters. The het_pyridiniums filter group is already enriched 5.94-fold for promiscuous actives in PubChem (p < 10−10, χ2 test, Table 1). Our cyanide reactivity score discriminates promiscuous compounds within this class with an AUC of 88.6% (p < 10−10, Mann–Whitney U-test, Table 2). Carbons adjacent to the charged pyridinium nitrogen present in these compounds can react with cyanide.66,67
Some PAINS filter groups directly match a known reactive motif that is also identified by our reactivity model. For example, azo nitrogen can react with certain biological macromolecules,68,69 and the azo PAINS filter group is enriched 313% for promiscuous activity in PubChem (p = 1.60 × 10−43, Bonferroni-corrected χ2 test, Figure S9 A). We extracted azo nitrogen site-level reactivity scores from PubChem compounds using a SMARTS query (Table S6 Pattern 7). We found that these site-level scores were sufficient to predict promiscuous bioactivity with an AUC of 84.5% (Figure S9B). Furthermore, this azo site-level ROC curve is indistinguishable from that based on the molecule-level protein reactivity score (p = 0.490, ROC Z-test70).
Reactivity scores can help identify distinct chemical mechanisms of reactivity-based assay interference and bioassay promiscuity for individual filters in a group. For example, the cyano_pyridone PAINS filter group consists of a core pyridone ring with one cyano substituent. This filter group does not substantially enrich for promiscuous bioactive compounds in the PubChem data set at the 5% bioactivity cutoff (p = 1.00, Bonferroni-corrected χ2 test). Furthermore, this class of compounds is predicted to have marginally greater reactivity with GSH compared to other compounds in PubChem (p = 0.0226, Bonferroni-corrected Mann–Whitney U test), which indicates a weak association between this filter group and reactivity-mediated promiscuous behavior (Figure S10). Nevertheless, GSH reactivity scores predict promiscuity of this compound class with an AUC of 85.6% (p = 8.13 × 10−3, Bonferroni-corrected Mann–Whitney U-test). On further inspection, two distinct classes of promiscuous bioactive compounds matching this filter group were observed. The first group contained a Michael-acceptor motif and was predicted to have appreciable reactivity, while the second group did not contain this motif and was predicted to have moderate-to-low reactivity (Figure 6A, B). Interestingly, the reactive class was active in a larger percentage of assays, on average, compared to the nonreactive class (Figure 6C). Notably, in the original AlphaScreenassays used to generate these filters, the same pattern of activity was observed for these two classes.11
Figure 6.
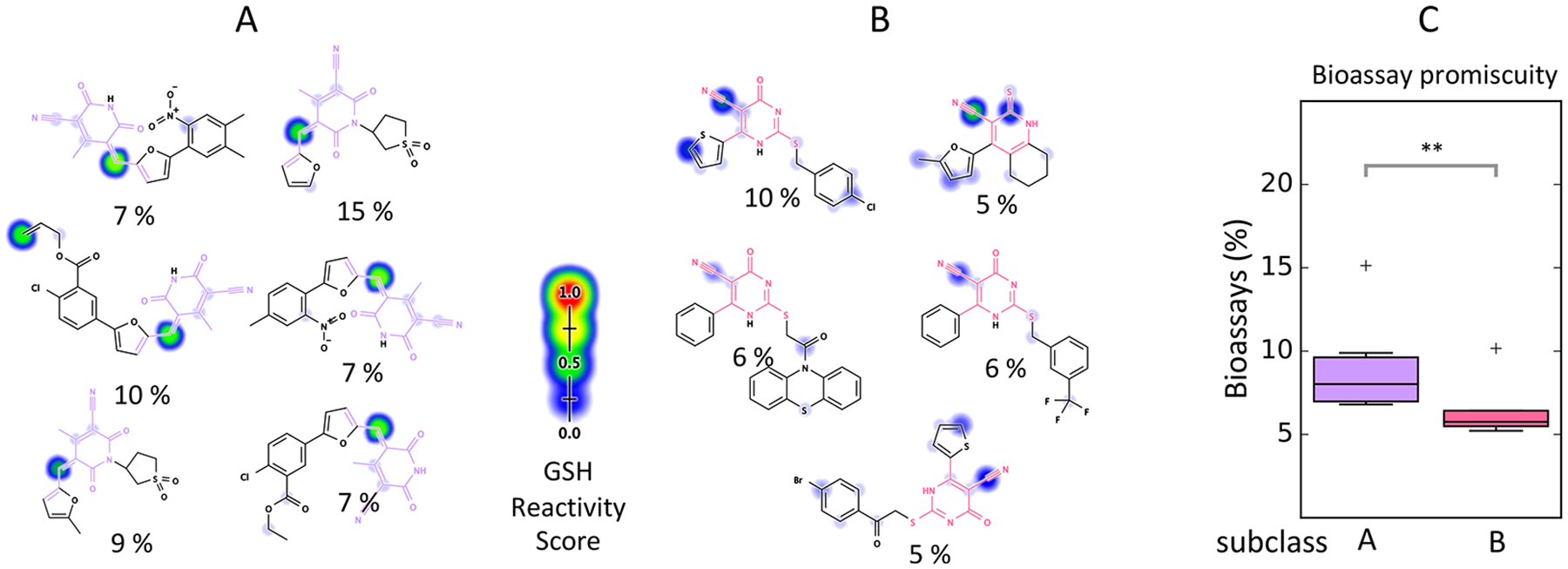
Reactivity scores flag subclasses of chemotypes with predicted enriched biological reactivity. The cyano_pyridone PAINS filter group consists of a core pyridone ring with one cyano substituent. (A) Note six of the 11 promiscuous compounds matched by this filter group are predicted to be reactive at an sp2 carbon within a Michael-acceptor-like motif located meta to the cyano group (purple). Reactivity modeling predicts that GSH attacks this electron-deficient region. The percentage of biological assays in which each compound was active are noted. (B) The other five promiscuous compounds within this filter group correspond to variants of the cyano_pyridone filter group not containing a traditional Michael-acceptor (pink), which are not predicted to be strongly reactive with GSH. (C) These predicted less-reactive compounds are active in a smaller percentage of biological assays (p = 0.0001, Mann–Whitney U test). **: p < 0.001.
PAINS Filters with Proposed Alternative Reactive Mechanisms.
To further demonstrate the potential additional utility of incorporating reactivity site prediction, we analyze in-depth six PAINS filter groups in which the reactive sites are predicted to be separate from the chemotypes flagged by a PAINS filter group. Chemical mechanisms of interference were proposed for some PAINS chemotypes in the original report, but follow-up investigations were not performed in that study.11 We note that some PAINS and related chemotypes have been more extensively studied in follow-up studies.18 However, interference with the underlying AlphaScreentechnology remains to be explicitly investigated for most, if not all, of the PAINS chemotypes. Furthermore, we note some PAINS filters do not have obvious mechanisms that could be suggested for their biological activity when they were originally identified. Compound libraries are often reflective of the underlying synthetic and combinatorial chemistry, and this may lead to correlations between substructures within compounds in a library. Subsequently, in some cases PAINS filters may only identify a substructure correlated with promiscuous behavior, but not mechanistically linked to that behavior.
Some PAINS filters are associated with adjacent or overlapping reactive chemotypes, such as Michael-acceptors. Michael-acceptors are electrophilic motifs with α,β-unsaturated carbonyl groups.71 The imineone, thiophene_amino, and ene_six_het PAINS filter groups are all associated with a nearby or overlapping Michael-acceptor motif. In the case of the ene_six_het filter group, a Michael-acceptor motif is part of the filter itself, while the imineone and thiophene_amino filter groups are correlated with Michael-acceptor motifs because they include carbonyl functional groups. In the case of all three filters, reactivity scores at these Michael-acceptor motifs account for a substantial part of the reactivity model’s power to discriminate promiscuous bioactives among com-pounds matching these filter groups.
The imineone filter group matches molecules containing either adjacent diones or adjacent imine and ketone groups. This filter group is enriched 9.33-fold for bioassay promiscuity (p < 10−10, Bonferroni-corrected χ2 test, Table 1), and predicted GSH reactivity discriminates promiscuous actives in this filter group with an AUC of 72.9 (p < 10−10, Bonferroni-corrected Mann–Whitney U-test Table 2). Many imine_one compounds also contained a Michael-acceptor motif adjacent to, and overlapping with, a PAINS filter motif (Figure 7A). We then searched for compounds containing Michael-acceptor motifs among com-pounds matching the imine_one filter group using a SMARTS query (Table S6, Pattern 1). This search identified 185 compounds, which were enriched 3.99-fold for promiscuous bioactive compounds compared to the imine_one filter alone (p = 9.41 × 10−22, χ2 test, Figure 7C). In addition, compounds containing the queried Michael-acceptor motif were assigned 74% higher GSH reactivity scores than those matching only the imine_one filter (p < 10−10, Figure S11A). This suggests that the imine_one filter is enriched for promiscuous bioactivity, perhaps because it is associated with other reactive motifs. It should be noted that this combination of filters will also match ortho-quinones, which are likely to share undesirable bioactivity with paraquinones, and both moieties are also matched by the quinone PAINS filter group.
Figure 7.
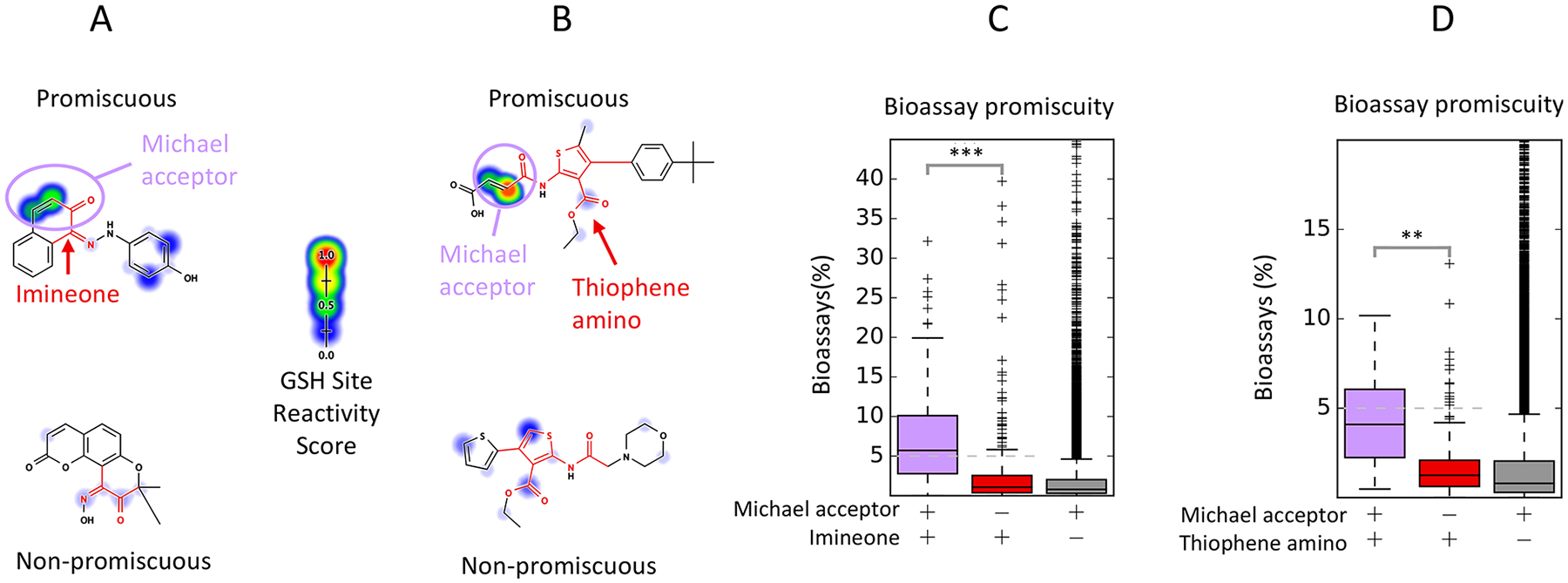
Many PAINS filters are associated with a nearby or overlapping reactive Michael-acceptor motif. (A) The imineone filter group matches a chemical motif with adjacent imine and ketone groups, as well as diones. Among 457 compounds matching this filter (red), 185 compounds (40%) possess a Michael-acceptor motif that overlaps with the motif matched by this filter group (purple). Michael-acceptor motifs are well-known electrophiles assigned high reactivity scores by our model.71 (B) The thiophene_amino filters match various substituted thiophene rings. Among 224 compounds matching this filter group, 28 (13%) contain a Michael-acceptor motif adjacent to the amide and outside the motif matched by the filter group (purple). (C, D) Compounds with this Michael-acceptor are enriched 3.99-fold for promiscuous actives among compounds matching the imineone filter (p < 10−10, χ2 test), while compounds matching the thiophene amino filter group and the Michael-acceptor motif are enriched 3.30-fold for promiscuous actives (p = 7.97 × 10−3, χ2 test). Compounds with Michael-acceptor motifs not matching the imineone or thiophene amino filters are not strongly enriched for promiscuous bioactivity. **: p < 0.001, ***: p < 0.0001.
The thiophene_amino is another Michael-acceptor associated PAINS filter group for which reactivity predictions can discriminate promiscuous actives. Thiophenes are five-membered, sulfur-containing aromatic rings that may undergo S-oxidation to form reactive compounds.72 The thiophene_amino filters match various substituted thiophenes. This filter group is enriched 2.36-fold for promiscuous actives (not significant, p = 0.075, χ2 test). GSH reactivity predicts promiscuity among compounds matching this filter group with an AUC of 78.9% (p = 2.42 × 10−2, Bonferroni-corrected Mann–Whitney U-test). We observed that a number of compounds matching this filter group also contained an adjacent Michael-acceptor motif that shares the amide carbonyl with the PAINS filter (Figure 7B). We identified 28 compounds containing this Michael-acceptor motif among those matching the thiophene_amino filter group using a SMARTS query (Table S6, Pattern 3). These compounds are enriched 3.30-fold for promiscuous actives compared to those just matching the thiophene_amino filter group, and they are assigned much higher reactivity scores (Figures 7D and S11B).
In yet another example of Michael-acceptor associated PAINS filters, the ene_six_het PAINS filter group matches heteroatom rings containing a Michael-acceptor motif (Figure S8A). This filter group is enriched 2.96-fold for promiscuous actives (p < 10−10, Bonferroni-corrected χ2 test). GSH reactivity scores predict bioassay promiscuity among com-pounds matching this filter group with an AUC of 79.9% (p < 10−10, Bonferroni-corrected Mann–Whitney U-test). Since this compound also contains a Michael-acceptor, we extracted site-level GSH reactivity prediction at the β carbon of the Michael-acceptor using a SMARTS query (Table S6, Pattern 4). Site-level reactivity at this position achieved an AUC of 67.8% for predicting promiscuous bioactive compounds within this class (p < 10−10, Mann–Whitney U-test, Figure S8B). Furthermore, the early recall portion of the ROC is perfectly recovered using only this information. We also identified compounds in this class with additional conjugated pi bonds adjacent to the Michael-acceptor motif. We extracted the maximum site-level reactivity score among the atoms in these systems using a SMARTS query (Table S6, Pattern 5). We combined this reactivity score with the former site-level score by choosing the maximum score between the two, and then constructed a receiver operator curve (Figure S8B). Incorporating this information recovers additional predictive power from the original model. While the ene_six_het PAINS filter group captures some key information about the reactive mechanism of this class of compounds, integrating additional information from the whole molecule may enhance the characterization of certain promiscuous bioactives.
Since many Michael-acceptor-like motifs contain ketone, sulfonyl, cyano, or other electron-donating groups, the correlation of Michael-acceptors with ketones may seem obvious. However, correlations between reactive motifs and PAINS filters include cases without a clear structural overlap. The imine_one_fives filter group matches a five-membered ring motif containing both imine and ketone groups. This filter group is enriched 5.19-fold for promiscuous bioactives (p < 10−10, Bonferroni-corrected χ2 test, Table 1), and predicted GSH reactivity discriminates actives in this filter class with an AUC of 76.5% (p = 4.49 × 10−2, Bonferroni-corrected Mann–Whitney U-test, Table 2). We noted that many of these compounds also contain a thioamide group conjugated to the aromatic ring system, but such compounds were not flagged by the PAINS filter group (Figure 8A). We subsequently searched for compounds containing this thioamide group among compounds matching the imine_one_fives filter group using a SMARTS query (Table S6, Pattern 2), which identified 28 compounds. Compounds matching this thioamide group were enriched 3.31-fold for promiscuous bioactives compared to those matching only the imine_one_fives filter group, and these compounds are also assigned higher reactivity scores (Figure 8B and C). Within this filter group, atoms matching the filter group were commonly assigned lower reactivity scores than those matching other parts of the molecule (Figure S12). This analysis suggests that the imine_one filter group is also enriched for promiscuous bioactivity because of its correlation with other reactive functional groups.
Figure 8.
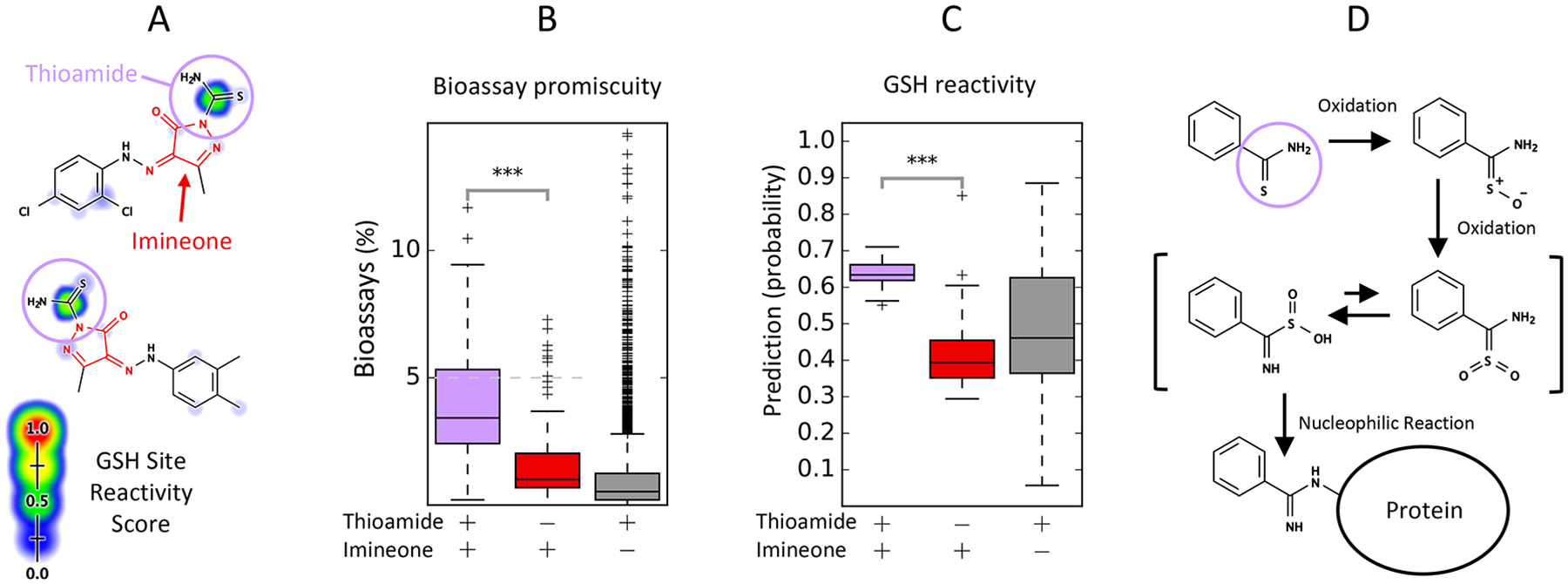
Some PAINS filters may be associated with other, unrelated reactive motifs. (A) The imine_one_fives filter group matches a five-membered ring motif containing both imine and ketone groups. Among 102 compounds matching this filter group (red), 28 (27%) contain a thioamide group conjugated to the ring motif (purple). (B) Compounds matching the filter group and this thioamide group are enriched 3.31-fold for promiscuous actives compared to compounds matching only the filter group (p = 7.97 × 10−3, χ2 test). (C) Compounds containing the thioamide group are assigned higher reactivity scores than compounds matching only the filter group (p < 10−10, Mann–Whitney U-test). (D) Oxidation of the thioamide group is known to form a reactive intermediate that can conjugate to proteins in rat hepatocytes.73 **: p < 0.001. ***: p < 0.0001.
The promiscuous bioactivity of compounds flagged by certain PAINS substructures may not have a clear connection to the filter structure a priori. We note pyrrole-containing compounds were identified in the original PAINS publication, but only anecdotal evidence of chemical instability was provided to hypothesize a chemical mechanism of interference.11 There has been a subsequent report of certain pyrroles decomposing to form interfering polymers.75 In spite of the supposed idiosyncratic effect, we found that pyrrole-containing compounds were still enriched 1.72-fold for promiscuous bioactive compounds in PubChem (p = 2.04 × 10−2, Bonferroni-corrected χ2 test). Even more surprising, reactivity modeling predicted pyrrole promiscuity with an AUC of 65.3% (p = 2.54 × 10−2, Bonferroni-corrected, Mann–Whitney U-test). We observed that many promiscuous pyrrole compounds contained a predicted reactive double bond adjacent to the motif matched by this filter group (Figure 9). In 7 of 51 (14%) promiscuous compounds, this double bond was part of the ene_rhod PAINS filter group. However, an additional 15 promiscuous compounds with the double bond did not match any other PAINS filter. We then extracted the protein site-level reactivity score at this bond using a SMARTS query (Table S6, Pattern 6). Compounds not matching the query were assigned a score of zero. The score at this double bond predicts promiscuous behavior of pyrroles with an AUC of 60.6%, which is a decrease of only 4.7% compared to the molecule-level protein reactivity score (p = 0.19, ROC Z-test70). Our model suggests that pyrroles may be enriched for promiscuity because they are correlated with other promiscuous substructures. Additional studies would be useful to clarify the nature of pyrrole structure-interference relationships including the determinants of potential chemical instability as well as reactivity.
Figure 9.
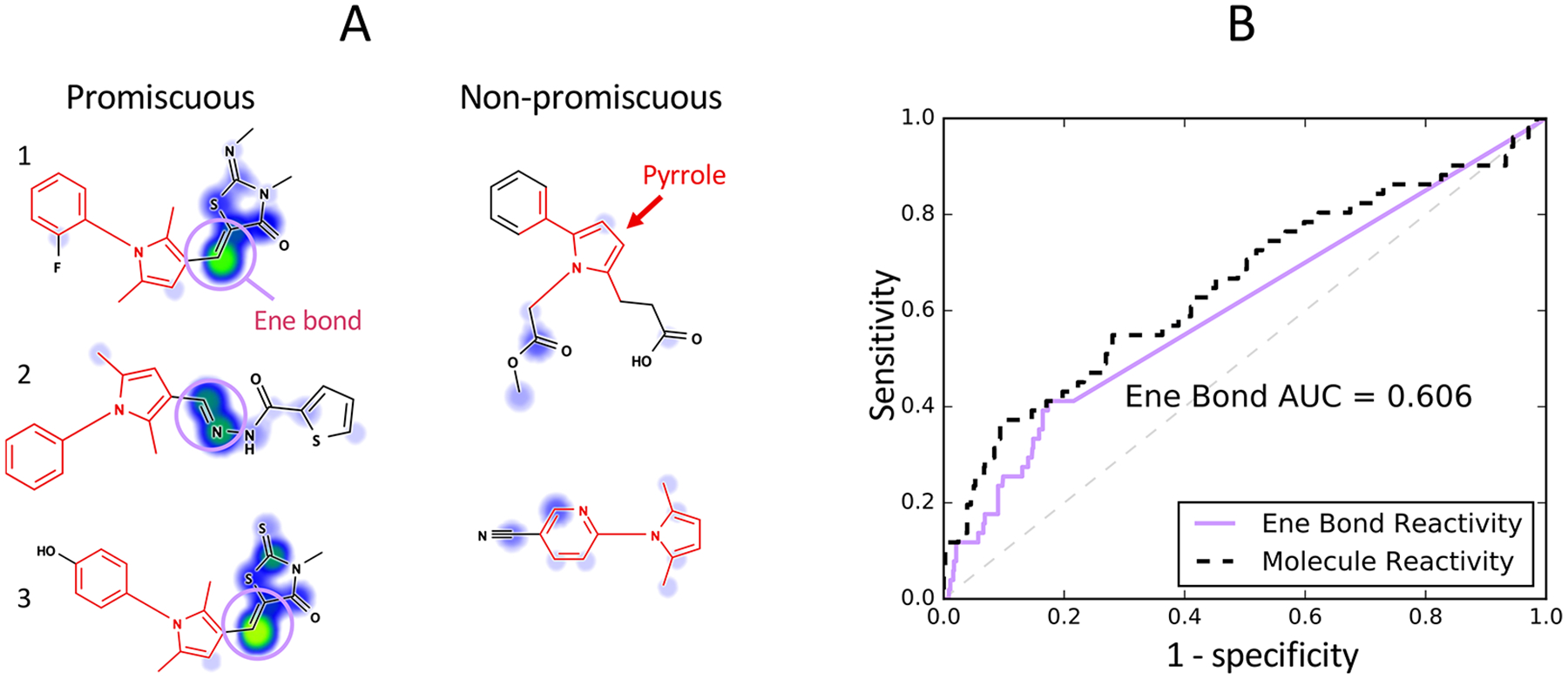
Reactivity scores suggest mechanisms of promiscuity for PAINS filters without a known mechanism. (A) The pyrrole filter group matches compounds containing the five-membered nitrogen aromatic ring pyrrole. Many promiscuous bioactive compounds contain a reactive double bond motif (purple). Compounds 1 and 3 also match the ene_rhod PAINS filter group, but compound 2 does not match another PAINS filter. Compound 2 is a hydrazone, which may tautomerize to form a reactive azo compound68,74 (B) Predicted atom-level GSH reactivity at this double bond was used to construct a ROC curve. Compounds matching the filter group but not containing an adjacent double bond motif received a score of 0. Molecule-level protein reactivity predicts pyrrole promiscuity (AUC = 65.3%). GSH reactivity scoring of the double bond achieves an AUC of 60.6%, which accounts for the majority of the predictive power of this model (difference not statistically significant, p = 0.19, ROC Z-test70). The dashed line denotes the expected ROC of a random model.
In some cases, motifs with high predicted reactivity are not obviously reactive, and not associated with a PAINS filter. The het_thio_666 PAINS filter group consists of tricyclic, six-membered, heteroaromatic, sulfur-containing compounds. Compounds matching these filters are enriched 13.34-fold for promiscuous activity at the 5% cutoff (p < 10−10, χ2 test, Table 1). Our cyanide reactivity scores are predictive of promiscuous activity among compounds matching these filters, with an AUC of 79.4% (p < 10−10, Bonferroni-corrected Mann–Whitney U-test, Table 2). However, site-level cyanide reactivity scores show that only atoms outside the region matched by the filter group are predicted to be reactive (Figure 10 B). We noted that tertiary nitrogen-containing rings such as piperidines, piperazines, and pyrrolidines were common in the side chains of compounds matching this filter group (Figure 10 A). We searched for compounds containing these ring structures among het_thio_666 matches using SMARTS queries (Table S6, Patterns 8–10). Promiscuous bioactive compounds are enriched 1.94-fold among these amine ring-containing compounds compared to other het_thio_666 filter group matches at the 5% promiscuity cutoff (p = 0.004, χ2 test). These compounds can be bioactivated by oxidation to iminium ions that are reactive with cyanide (Figure 10C).66,67
Figure 10.
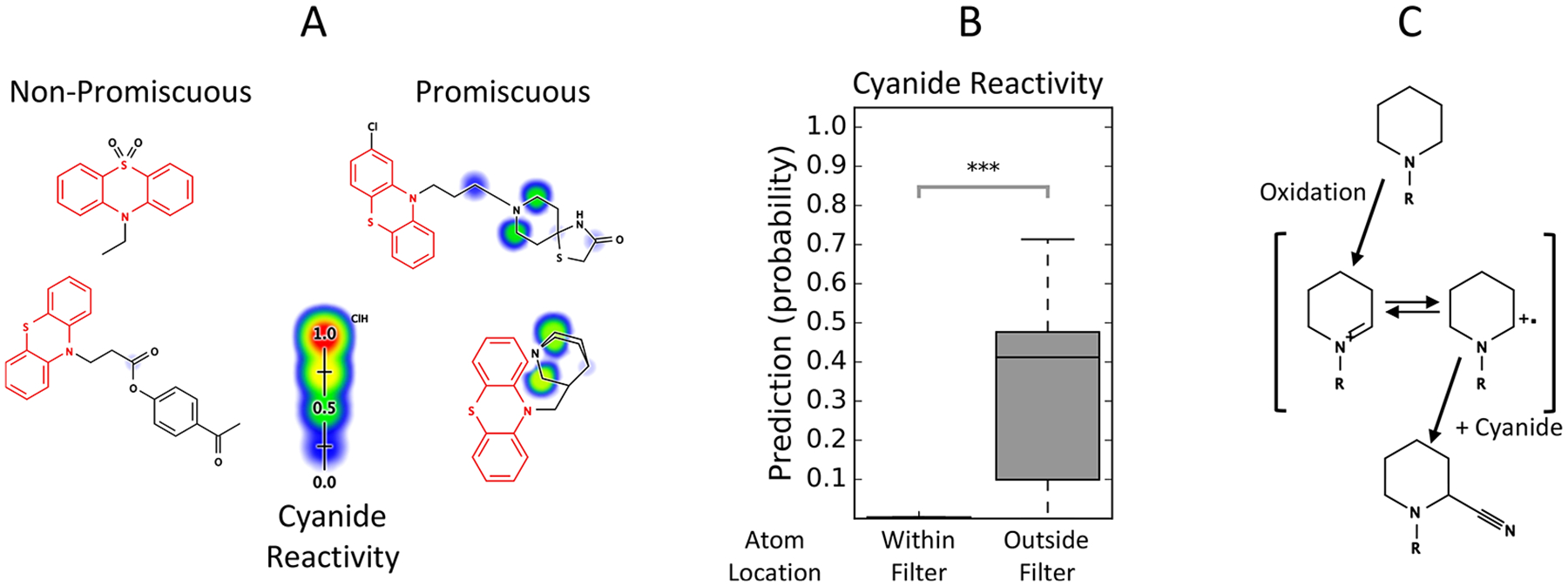
Reactivity scores identify non-obvious, reactive motifs not captured by PAINS filters. (A) The het_thio_666 PAINS filter group consists of tricyclic, heteroaromatic, sulfur-containing compounds. Twenty-four of 44 bioassay promiscuous compounds matching this filter group in PubChem also contain tertiary amine rings such as piperidines, piperazines, or pyrrolidines. (B) Among compounds matching this PAINS filter group, site-level cyanide reactivity scores are nonzero only on the atoms not matched by the filter group, suggesting a reactive mechanism unrelated to the motifs in this filter group. (C) Cyclic tertiary amines are known to be oxidized in vivo by Cytochrome P450 enzymes.66,67 This oxidation leads to the formation of an iminium ion intermediate that can react with cyanide or biological substrates. ***: p < 0.0001.
While our reactivity model generally provided more informative predictions than PAINS filters for known reactivity mechanisms, there were exceptions. For example, quinone species are a common, electrophilic, reactive species that can covalently bind to diverse biological molecules.46,53 We identified 925 compounds containing the quinone moiety in our PubChem data set. Since covalent reactivity is common among these species, we expected the reactivity model to be able to distinguish between promiscuous and nonpromiscuous quinone species. However, we found that protein reactivity scores only predicted quinone bioassay promiscuity with an AUC of 57.6% (p = 7.51 × 10−3, Bonferroni-corrected Mann–Whitney U-test). This low AUC may be a consequence of an alternative mechanism of promiscuous activity among certain quinones. Some quinone species are capable of generating abundant reactive oxygen species by redox cycling in the presence of DTT or other reducing agents commonly used in HTS assays.46 Reactive oxygen species can cause nonspecific modulation of biological systems in biochemical and cell-based assays. Quinones and other electron-accepting chemical species may also contribute to the oxidation of key reagents and/or biological targets (e.g., oxidation of cysteine thiols) which may further interfere with assay readouts by disrupting certain assay technologies or broadly modulating biological macromolecules. These orthogonal mechanisms of assay interference are not addressed by our reactivity model and may be a confounding factor.
Case Study of Histone Acetyltransferase Inhibitors.
High-quality chemical probes can enable unique analyses of complex biological systems and can be complementary and even orthogonal to analogous genetic perturbations. Unfortunately, many probes reported in the literature actually have nonspecific activity due to indiscriminate reactivity.4 Recently, the majority of 23 reported inhibitors of histone acetyltransferases (HAT) were shown to react nonspecifically with biological nucleophiles such as GSH, CoA, and the human La antigen. Many of compounds displayed nonspecific cellular readouts indistinguishable from prototypical thiol-reactive and redox-active compounds.19
In this context, our reactivity model may help identify nonspecific, reactive chemical matter among reported chemical probes. We applied conventional PAINS filters and our combined reactive promiscuity model to analyze 23 reported HAT inhibitors and four inactive control substances. We compared predictions of PAINS filters and our reactivity model to the results of GSH and coenzyme A (CoA) reactivity counterscreen results. PAINS had sensitivities of 70.0% and 75.0% for the GSH and CoA thiol-reactivity counterscreens, respectively, and specificities of 66.7% and 63.6%, respectively. The combined reactivity model could predict the outcomes of both GSH and CoA thiol-reactivity counterscreens with the same sensitivity as PAINS filters, but perfect (100%) specificity (Figure 11A and B). Furthermore, our site-level GSH reactivity score provides mechanistic predictions for each potential reactive compound including C646, gossypol, and MB-3 (Figure 11C).77–79 Interestingly, we found that our model predicted sites of thiol-reactivity to be within a PAINS filter group in only 5 of 12 thiol-reactive compounds.
Figure 11.
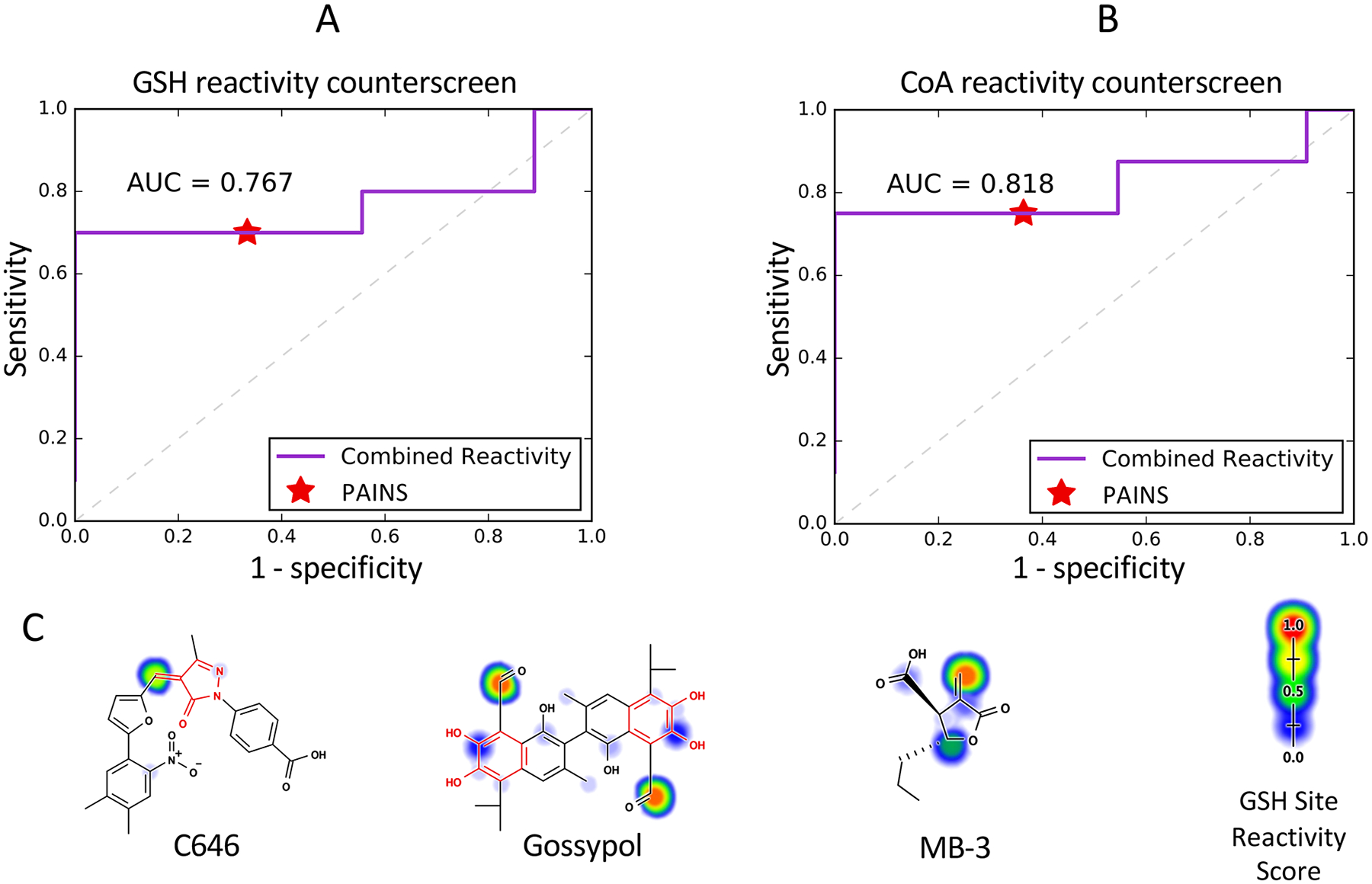
A reactivity analysis of literature reported HAT inhibitors identifies likely sites of nonspecific reactivity. (A) The combined reactive promiscuity model predicts the results of their GSH adduct formation counterscreen with the same sensitivity (75%) as PAINS filters, but with enhanced specificity (100% versus 63.6%, respectively). (B) The reactivity model also predicts the results of their CoA adduct formation counterscreen with the same sensitivity (66.7%) as PAINS filters, but with enhanced specificity (100% versus 70%, respectively). (C) Example reported HAT inhibitors and respective GSH reactivity predictions. From left to right: C646 contains an ene_rhod PAINS filter group match, a common reactive motif. Our GSH reactivity score predicts that the mechanism of nonspecific thiol reactivity involves nucleophilic attack at the β carbon of a Michael-acceptor contained within the motif matched by this filter group. The catechol groups of gossypol match the catechol PAINS filter group, though our model predicts the aldehyde substituents as thiol-reactive. We note gossypol can undergo redox-activity and form quinones under certain conditions, and it is confirmed which adduct(s) are formed under any given assay condition.76 While MB-3 is not flagged by PAINS filters, it contains a reactive terminal olefin group. Dashed line denotes the expected ROC of a random model.
Limitations of Reactivity Modeling.
The application of our reactivity model has some notable limitations. The reactivity model is trained on in vitro and in vivo data on covalent reactivity and conjugate formation, which may be affected by cellular metabolic processes such as metabolism by cytochrome P450s.62 This model may be quite useful in the cell-based assay context, but may have more false-positives in cell-free assays that do not model such complex biological processes. An improved reactivity model for the cell-free assay context could be produced by removing examples of implicit metabolism from the training data. Regardless, the reactivity model has been demonstrated to identify promiscuous actives in both cell-based and target-based assays included in our data set. Second, the model is intended to identify promiscuous bioactive compounds if they are covalently reactive. Many other mechanisms of promiscuity exist, and by some estimates, covalent reactivity may not represent the most common cause of promiscuous biological activity in certain biochemical assays.4,24 Third, for some known reactive motifs such as quinones, we have observed that our reactivity model does not substantially improve promiscuity predictions compared to PAINS filters. This may be due to an alternate mechanism of interference for these PAINS, or our model may need to be improved to discriminate these highly reactive species more accurately. Fourth, the reactivity model assumes the chemical structure of the bioactive substance is “as drawn”. However, for some compounds such as azo-phenols (Figure 7A), there could be additional tautomers based on substituent effects and experimental conditions. Future work may benefit from modeling tautomers explicitly. However, many compounds may undergo chemical transformations in situ or in storage.33,80–82 Related, bioactivity may be due to contaminants, impurities, or incorrectly annotated compounds. Future work will seek to experimentally and prospectively validate compound reactivity, especially for chemotypes with unconfirmed mechanisms of interference and those chemotypes with a wide spectrum of bioassay promiscuity.
CONCLUSIONS
In this study, we show that a model of small-molecule reactivity with several biological substrates predicts promiscuous activity in HTS with similar sensitivities and specificities as the popular PAINS substructure filters. PAINS filters may capture many mechanisms of promiscuous bioactivity and/or assay interference (e.g., thiol reactivity, aggregation, light-based interference, singlet oxygen interference). Interestingly, the aforementioned reactivity model achieves nearly equivalent performance (by sensitivity and specificity metrics) by modeling only a single mechanism: reactivity of compounds with biological nucleophiles. When combining PAINS substructure filters and reactivity scores into a single model, the hybrid model is able to achieve a sensitivity of 24% while maintaining the same specificity, a potentially useful improvement over PAINS filters alone in the context of HTS triage. Furthermore, PAINS filters differentiate between promiscuous and nonpromiscuous compounds matching the same PAINS filter group. In contrast, we demonstrated that our reactivity model could be used to differentiate these compounds for 15 PAINS filter groups. The model may enhance HTS triage by flagging specific reactive sites not otherwise specified by traditional substructure-based filters. As support of principle, we demonstrate that this reactivity model can flag nonspecific thiol-reactive compounds among a series of reported HAT inhibitors with nonspecific thiol reactivity.
While PAINS filters have useful predictive utility for flagging potentially problematic compounds from HTS experiments that may interfere with assay readouts and/or modulate targets nonspecifically, we show that data-driven modeling of small-molecule reactivity using deep learning can enhance such analyses. Additional mechanistic models of other interference phenomena such as light-based interferences, luciferase interference, aggregation, redox cycling, and cytotoxicity may enable more robust identification of promiscuous bioactive and/or interference compounds with poor tractability.
Future studies may need to investigate the contributions of various subsets of PAINS filters, notably the three subsets (A, B, C). For example, the “A” subset, which is based on the most analogs and the most robust experimental evidence, may be essential for broadly screening bioassay promiscuity. By contrast, some of the “C” subset, which is derived from far fewer analogs, may need to be modified or culled depending on additional evidence.
Such tools should enhance drug and chemical probe discovery and development by derisking compounds for interference and promiscuity and flagging potentially problematic compounds for triage or appropriate counterscreens.
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to the developers of the open-source cheminformatics tools Open Babel, RDKit, and the Chemistry Development Kit. Research reported in this publication was supported by the National Library of Medicine of the National Institutes of Health under Award Numbers R01LM012222 and R01LM012482, and by the National Institutes of Health under Award Number GM07200. The content is the sole responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Computations were performed using the facilities of the Washington University Center for High Performance Computing, which were partially funded by NIH grant nos. 1S10RR022984-01A1 and 1S10OD018091-01. We also thank both the Department of Immunology and Pathology at the Washington University School of Medicine, the Washington University Center for Biological Systems Engineering, and the Washington University Medical Scientist Training Program for their generous support of this work.
ABBREVIATIONS
- AUC
area under curve
- PAINS
pan assay interference compounds
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jcim.8b00104.
PubChem_annotations_reactivity_pains_promiscuity.txt: Tab-separated format table of PubChem CIDs and annotated information for compounds included in this study. Reactivity scores for GSH, Cyanide, DNA, Protein, Combined, and React+PAINS models; matched PAINS filters; promiscuity labels for 5, 10, 15, and 20% cutoffs (ZIP)
Supplementary note, supplementary Figures S1–S13, supplementary Tables S1–S9 (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Arrowsmith CH; Audia JE; Austin C; Baell J; Bennett J; Blagg J; Bountra C; Brennan PE; Brown PJ; Bunnage ME; Buser-Doepner C; Campbell RM; Carter AJ; Cohen P; Copeland RA; Cravatt B; Dahlin JL; Dhanak D; Edwards AM; Frederiksen M; Frye SV; Gray N; Grimshaw CE; Hepworth D; Howe T; Huber KVM; Jin J; Knapp S; Kotz JD; Kruger RG; Lowe D; Mader MM; Marsden B; Mueller-Fahrnow A; Müller S; O’Hagan RC; Overington JP; Owen DR; Rosenberg SH; Ross R; Roth B; Schapira M; Schreiber SL; Shoichet B; Sundström M; Superti-Furga G; Taunton J; Toledo-Sherman L; Walpole C; Walters MA; Willson TM; Workman P; Young RN; Zuercher WJ The promise and peril of chemical probes. Nat. Chem. Biol 2015, 11, 536–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Inglese J; Johnson RL; Simeonov A; Xia M; Zheng W; Austin CP; Auld DS High-throughput screening assays for the identification of chemical probes. Nat. Chem. Biol 2007, 3, 466–479. [DOI] [PubMed] [Google Scholar]
- (3).Shoichet BK Screening in a spirit haunted world. Drug Discovery Today 2006, 11, 607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Thorne N; Auld DS; Inglese J Apparent activity in high-throughput screening: origins of compound-dependent assay interference. Curr. Opin. Chem. Biol 2010, 14, 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Xie Y; Dahlin JL; Oakley AJ; Casarotto MG; Board PG; Baell JB Reviewing hit discovery literature for difficult targets: glutathione transferase omega-1 as an example. J. Med. Chem 2018, DOI: 10.1021/acs.jmedchem.8b00318. [DOI] [PubMed] [Google Scholar]
- (6).Kenny PW Comment on The Ecstasy and Agony of Assay Interference Compounds. J. Chem. Inf. Model 2017, 57, 2640–2645. [DOI] [PubMed] [Google Scholar]
- (7).Simeonov A; Jadhav A; Thomas CJ; Wang Y; Huang R; Southall NT; Shinn P; Smith J; Austin CP; Auld DS; Inglese J Fluorescence Spectroscopic Profiling of Compound Libraries. J. Med. Chem 2008, 51, 2363–2371. [DOI] [PubMed] [Google Scholar]
- (8).Brenke JK; Salmina ES; Ringelstetter L; Dornauer S; Kuzikov M; Rothenaigner I; Schorpp K; Giehler F; Gopalakrishnan J; Kieser A; Gul S; Tetko IV; Hadian K Identification of Small-Molecule Frequent Hitters of Glutathione S-Transferase–Glutathione Interaction. J. Biomol. Screening 2016, 21, 596–607. [DOI] [PubMed] [Google Scholar]
- (9).Falk H; Connor T; Yang H; Loft KJ; Alcindor JL; Nikolakopoulos G; Surjadi RN; Bentley JD; Hattarki MK; Dolezal O; Murphy JM; Monahan BJ; Peat TS; Thomas T; Baell JB; Parisot JP; Street IP An efficient high-throughput screening method for MYST family acetyltransferases, a new class of epigenetic drug targets. J. Biomol. Screening 2011, 16, 1196–1205. [DOI] [PubMed] [Google Scholar]
- (10).Auld DS; Southall NT; Jadhav A; Johnson RL; Diller DJ; Simeonov A; Austin CP; Inglese J Characterization of Chemical Libraries for Luciferase Inhibitory Activity. J. Med. Chem 2008, 51, 2372–2386. [DOI] [PubMed] [Google Scholar]
- (11).Baell JB; Holloway GA New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem 2010, 53, 2719–2740. [DOI] [PubMed] [Google Scholar]
- (12).McGovern SL; Caselli E; Grigorieff N; Shoichet BK A Common Mechanism Underlying Promiscuous Inhibitors from Virtual and High-Throughput Screening. J. Med. Chem 2002, 45, 1712–1722. [DOI] [PubMed] [Google Scholar]
- (13).Rishton GM Reactive compounds and in vitro false positives in HTS. Drug Discovery Today 1997, 2, 382–384. [Google Scholar]
- (14).Johnston PA; Soares KM; Shinde SN; Foster CA; Shun TY; Takyi HK; Wipf P; Lazo JS Development of a 384-Well Colorimetric Assay to Quantify Hydrogen Peroxide Generated by the Redox Cycling of Compounds in the Presence of Reducing Agents. Assay Drug Dev. Technol 2008, 6, 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Schorpp K; Rothenaigner I; Salmina E; Reinshagen J; Low T; Brenke JK; Gopalakrishnan J; Tetko IV; Gul S; Hadian K Identification of small-molecule frequent hitters from AlphaScreen high-throughput screens. J. Biomol. Screening 2014, 19, 715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Ingólfsson HI; Thakur P; Herold KF; Hobart EA; Ramsey NB; Periole X; de Jong DH; Zwama M; Yilmaz D; Hall K; Maretzky T; Hemmings HC; Blobel C; Marrink SJ; Koçer A; Sack JT; Andersen OS Phytochemicals Perturb Membranes and Promiscuously Alter Protein Function. ACS Chem. Biol 2014, 9, 1788–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hermann JC; Chen Y; Wartchow C; Menke J; Gao L; Gleason SK; Haynes N-E; Scott N; Petersen A; Gabriel S; Vu B; George KM; Narayanan A; Li SH; Qian H; Beatini N; Niu L; Gan Q-F Metal impurities cause false positives in high-throughput screening campaigns. ACS Med. Chem. Lett 2013, 4, 197–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Dahlin JL; Nissink JWM; Strasser JM; Francis S; Higgins L; Zhou H; Zhang Z; Walters MA PAINS in the Assay: Chemical Mechanisms of Assay Interference and Promiscuous Enzymatic Inhibition Observed during a Sulfhydryl-Scavenging HTS. J. Med. Chem 2015, 58, 2091–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Dahlin JL; Nelson KM; Strasser JM; Barsyte-Lovejoy D; Szewczyk M; Shrimp JH; Meier JL; Arrowsmith CH; Brown PJ; Baell JB; Walters MA; et al. Assay interference and off-target liabilities of reported histone acetyltransferase inhibitors. Nat. Commun 2017, 8, 1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Nelson KM; Dahlin JL; Bisson J; Graham J; Pauli GF; Walters MA The Essential Medicinal Chemistry of Curcumin. J. Med. Chem 2017, 60, 1620–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Bisson J; McAlpine JB; Friesen JB; Chen S-N; Graham J; Pauli GF Can Invalid Bioactives Undermine Natural Product-Based Drug Discovery? J. Med. Chem 2016, 59, 1671–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Dahlin JL; Walters MA The essential roles of chemistry in high-throughput screening triage. Future Med. Chem 2014, 6, 1265–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Dahlin JL; Baell J; Walters MA Assay interference by chemical reactivity; Eli Lilly & Company and the National Center for Advancing Translational Sciences, 2015. [PubMed] [Google Scholar]
- (24).Huth JR; Mendoza R; Olejniczak ET; Johnson RW; Cothron DA; Liu Y; Lerner CG; Chen J; Hajduk PJ ALARM NMR: A Rapid and Robust Experimental Method To Detect Reactive False Positives in Biochemical Screens. J. Am. Chem. Soc 2005, 127, 217–224. [DOI] [PubMed] [Google Scholar]
- (25).Capuzzi SJ; Muratov EN; Tropsha A Phantom PAINS: Problems with the Utility of Alerts for Pan-Assay INterference CompoundS. J. Chem. Inf. Model 2017, 57, 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Senger MR; Fraga CA; Dantas RF; Silva FP Filtering promiscuous compounds in early drug discovery: is it a good idea? Drug Discovery Today 2016, 21, 868–872. [DOI] [PubMed] [Google Scholar]
- (27).Anighoro A; Bajorath J; Rastelli G Polypharmacology: Challenges and Opportunities in Drug Discovery. J. Med. Chem 2014, 57, 7874–7887. [DOI] [PubMed] [Google Scholar]
- (28).Wassermann AM; Lounkine E; Hoepfner D; Le Goff G; King FJ; Studer C; Peltier JM; Grippo ML; Prindle V; Tao J; Schuffenhauer A; Wallace IM; Chen S; Krastel P; Cobos-Correa A; Parker CN; Davies JW; Glick M Dark chemical matter as a promising starting point for drug lead discovery. Nat. Chem. Biol 2015, 11, 958–966. [DOI] [PubMed] [Google Scholar]
- (29).Jasial S; Hu Y; Bajorath J How Frequently Are Pan-Assay Interference Compounds Active? Large-Scale Analysis of Screening Data Reveals Diverse Activity Profiles, Low Global Hit Frequency, and Many Consistently Inactive Compounds. J. Med. Chem 2017, 60, 3879–3886. [DOI] [PubMed] [Google Scholar]
- (30).Baell JB; Nissink JWM Seven Year Itch: Pan-Assay Interference Compounds (PAINS) in 2017-Utility and Limitations. ACS Chem. Biol 2018, 13, 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Baell JB Observations on screening-based research and some concerning trends in the literature. Future Med. Chem 2010, 2, 1529–1546. [DOI] [PubMed] [Google Scholar]
- (32).Baell JB; Ferrins L; Falk H; Nikolakopoulos G PAINS: Relevance to tool compound discovery and fragment-based screening. Aust. J. Chem 2013, 66, 1483–1494. [Google Scholar]
- (33).Dahlin JL; Walters MA How to Triage PAINS-Full Research. Assay Drug Dev. Technol 2016, 14, 168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Hughes TB; Miller GP; Swamidass SJ Site of Reactivity Models Predict Molecular Reactivity of Diverse Chemicals with Glutathione. Chem. Res. Toxicol 2015, 28, 797–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Hughes TB; Dang NL; Miller GP; Swamidass SJ Modeling Reactivity to Biological Macromolecules with a Deep Multitask Network. ACS Cent. Sci 2016, 2, 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Wang Y; Xiao J; Suzek TO; Zhang J; Wang J; Zhou Z; Han L; Karapetyan K; Dracheva S; Shoemaker BA; Bolton E; Gindulyte A; Bryant SH PubChem’s BioAssay Database. Nucleic Acids Res. 2012, 40, D400–D412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Kenny PW; Montanari CA Inflation of correlation in the pursuit of drug-likeness. J. Comput.-Aided Mol. Des 2013, 27, 1–13. [DOI] [PubMed] [Google Scholar]
- (38).Waring MJ Lipophilicity in drug discovery. Expert Opin. Drug Discovery 2010, 5, 235–248. [DOI] [PubMed] [Google Scholar]
- (39).Ghose AK; Viswanadhan VN; Wendoloski JJ A Knowledge-Based Approach in Designing Combinatorial or Medicinal Chemistry Libraries for Drug Discovery. 1. A Qualitative and Quantitative Characterization of Known Drug Databases. J. Comb. Chem 1999, 1, 55–68. [DOI] [PubMed] [Google Scholar]
- (40).Ash S; Cline MA; Homer RW; Hurst T; Smith GB SYBYL Line Notation (SLN): A Versatile Language for Chemical Structure Representation†. J. Chem. Inf. Model 1997, 37, 71–79. [Google Scholar]
- (41).Saubern S; Guha R; Baell JB KNIME Workflow to Assess PAINS Filters in SMARTS Format. Comparison of RDKit and Indigo Cheminformatics Libraries. Mol. Inf 2011, 30, 847–850. [DOI] [PubMed] [Google Scholar]
- (42).Bruns RF; Watson IA Rules for Identifying Potentially Reactive or Promiscuous Compounds. J. Med. Chem 2012, 55, 9763–9772. [DOI] [PubMed] [Google Scholar]
- (43).Dang NL; Hughes TB; Miller GP; Swamidass SJ Computational Approach to Structural Alerts: Furans, Phenols, Nitroaromatics, and Thiophenes. Chem. Res. Toxicol 2017, 30, 1046–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Niculescu-Mizil A; Caruana R Predicting good probabilities with supervised learning. Proc. 22nd Int. Conf. Machine Learning 2005, 625–632. [Google Scholar]
- (45).Vempati UD; Przydzial MJ; Chung C; Abeyruwan S; Mir A; Sakurai K; Visser U; Lemmon VP; Schürer SC Formalization, Annotation and Analysis of Diverse Drug and Probe Screening Assay Datasets Using the BioAssay Ontology (BAO). PLoS One 2012, 7, e49198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Graham DG; Tiffany SM; Bell WR; Gutknecht WF Autoxidation versus covalent binding of quinones as the mechanism of toxicity of dopamine, 6-hydroxydopamine, and related compounds toward C1300 neuroblastoma cells in vitro. Mol. Pharmacol 1978, 14, 644–653. [PubMed] [Google Scholar]
- (47).Tomašič T; Peterlin Mašič L Rhodanine as a scaffold in drug discovery: a critical review of its biological activities and mechanisms of target modulation. Expert Opin. Drug Discovery 2012, 7, 549–560. [DOI] [PubMed] [Google Scholar]
- (48).Gardner PD; Rafsanjani HS; Rand L Reaction of phenolic Mannich base methiodides and oxides with various nucleophiles. J. Am. Chem. Soc 1959, 81, 3364–3367. [Google Scholar]
- (49).Olaj OF; Kauffmann HF; Breitenbach JW Spectroscopic measurements on spontaneously polymerizing styrene, 2. The estimation of the reactivity of the two Diels-Alder-isomers towards polymer radicals. Makromol. Chem 1977, 178, 2707–2717. [Google Scholar]
- (50).Walling C; Briggs ER; Wolfstirn KB; Mayo FR Copolymerization. X. The effect of meta-and para-substitution on the reactivity of the styrene double bond. J. Am. Chem. Soc 1948, 70, 1537–1542. [Google Scholar]
- (51).Bolton JL; Trush MA; Penning TM; Dryhurst G; Monks TJ Role of quinones in toxicology. Chem. Res. Toxicol 2000, 13, 135–160. [DOI] [PubMed] [Google Scholar]
- (52).Sun I; Sun E; Crane F; Morre D; Lindgren A; Löw H Requirement for coenzyme Q in plasma membrane electron transport. Proc. Natl. Acad. Sci. U. S. A 1992, 89, 11126–11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Bova MP; Mattson MN; Vasile S; Tam D; Holsinger L; Bremer M; Hui T; McMahon G; Rice A; Fukuto JM The oxidative mechanism of action of ortho-quinone inhibitors of protein-tyrosine phosphatase α is mediated by hydrogen peroxide. Arch. Biochem. Biophys 2004, 429, 30–41. [DOI] [PubMed] [Google Scholar]
- (54).Mendgen T; Steuer C; Klein CD Privileged Scaffolds or Promiscuous Binders: A Comparative Study on Rhodanines and Related Heterocycles in Medicinal Chemistry. J. Med. Chem 2012, 55, 743–753. [DOI] [PubMed] [Google Scholar]
- (55).Kaminskyy D; Kryshchyshyn A; Lesyk R Recent developments with rhodanine as a scaffold for drug discovery. Expert Opin. Drug Discovery 2017, 12, 1233–1252. [DOI] [PubMed] [Google Scholar]
- (56).Young RH; Brewer D; Kayser R; Martin R; Feriozi D; Keller RA On the mechanism of quenching by amines: a new method for investigation of interactions with triplet states. Can. J. Chem 1974, 52, 2889–2893. [Google Scholar]
- (57).Dietrich LE; Teal TK; Price-Whelan A; Newman DK Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science 2008, 321, 1203–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Swamidass SJ; Azencott C-A; Daily K; Baldi P A CROC stronger than ROC: measuring, visualizing and optimizing early retrieval. Bioinformatics 2010, 26, 1348–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Beck JR; Shultz EK The use of relative operating characteristic (ROC) curves in test performance evaluation. Arch. Pathol. Lab. Med 1986, 110, 13–20. [PubMed] [Google Scholar]
- (60).Srivastava A; Maggs J; Antoine D; Williams D; Smith D; Park B Adverse Drug Reactions; Springer, 2010; pp 165–194. [Google Scholar]
- (61).Park BK; Kitteringham NR; Maggs JL; Pirmohamed M; Williams DP The role of metabolic activation in drug-induced hepatotoxicity. Annu. Rev. Pharmacol. Toxicol 2005, 45, 177–202. [DOI] [PubMed] [Google Scholar]
- (62).Attia SM Deleterious effects of reactive metabolites. Oxid. Med. Cell. Longevity 2010, 3, 238–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).LoPachin RM; Gavin T Molecular mechanisms of aldehyde toxicity: a chemical perspective. Chem. Res. Toxicol 2014, 27, 1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Gerberick GF; Vassallo JD; Bailey RE; Chaney JG; Morrall SW; Lepoittevin J-P Development of a peptide reactivity assay for screening contact allergens. Toxicol. Sci 2004, 81, 332–343. [DOI] [PubMed] [Google Scholar]
- (65).Dennehy MK; Richards KA; Wernke GR; Shyr Y; Liebler DC Cytosolic and nuclear protein targets of thiol-reactive electrophiles. Chem. Res. Toxicol 2006, 19, 20–29. [DOI] [PubMed] [Google Scholar]
- (66).Argoti D; Liang L; Conteh A; Chen L; Bershas D; Yu C-P; Vouros P; Yang E Cyanide Trapping of Iminium Ion Reactive Intermediates Followed by Detection and Structure Identification Using Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS). Chem. Res. Toxicol 2005, 18, 1537–1544. [DOI] [PubMed] [Google Scholar]
- (67).Kalgutkar A; Gardner I; Obach R; Shaffer C; Callegari E; Henne K; Mutlib A; Dalvie D; Lee J; Nakai Y; O’Donnell J; Boer J; Harriman S A Comprehensive Listing of Bioactivation Pathways of Organic Functional Groups. Curr. Drug Metab 2005, 6, 161–225. [DOI] [PubMed] [Google Scholar]
- (68).Boulègue C; Löweneck M; Renner C; Moroder L Redox Potential of Azobenzene as an Amino Acid Residue in Peptides. ChemBioChem 2007, 8, 591–594. [DOI] [PubMed] [Google Scholar]
- (69).Hultin T Reactions of C14-labeled carcinogenic azo dyes with rat liver proteins. Exp. Cell Res 1957, 13, 47–59. [DOI] [PubMed] [Google Scholar]
- (70).Hanley JA; McNeil BJ The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982, 143, 29–36. [DOI] [PubMed] [Google Scholar]
- (71).Attia SM Deleterious Effects of Reactive Metabolites. Oxid. Med. Cell. Longevity 2010, 3, 238–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Dansette P; Thang DC; Mansuy HEAD Evidence for thiophene-s-oxide as a primary reactive metabolite of thiophene in vivo: Formation of a dihydrothiophene sulfoxide mercapturic acid. Biochem. Biophys. Res. Commun 1992, 186, 1624–1630. [DOI] [PubMed] [Google Scholar]
- (73).Ikehata K; Duzhak TG; Galeva NA; Ji T; Koen YM; Hanzlik RP Protein Targets of Reactive Metabolites of Thiobenzamide in Rat Liver in Vivo. Chem. Res. Toxicol 2008, 21, 1432–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Hihara T; Okada Y; Morita Z Photo-oxidation of pyrazolinylazo dyes and analysis of reactivity as azo and hydrazone tautomers using semiempirical molecular orbital PM5 method. Dyes Pigm. 2006, 69, 151–176. [Google Scholar]
- (75).Zhu W; Groh M; Haupenthal J; Hartmann RW A Detective Story in Drug Discovery: Elucidation of a Screening Artifact Reveals Polymeric Carboxylic Acids as Potent Inhibitors of RNA Polymerase. Chem. - Eur. J 2013, 19, 8397–8400. [DOI] [PubMed] [Google Scholar]
- (76).Kovacic P Mechanism of drug and toxic actions of gossypol: focus on reactive oxygen species and electron transfer. Curr. Med. Chem 2003, 10, 2711–2718. [DOI] [PubMed] [Google Scholar]
- (77).Bowers EM; Yan G; Mukherjee C; Orry A; Wang L; Holbert MA; Crump NT; Hazzalin CA; Liszczak G; Yuan H; Larocca C; Saldanha SA; Abagyan R; Sun Y; Meyers DJ; Marmorstein R; Mahadevan LC; Alani RM; Cole PA Virtual ligand screening of the p300/CBP histone acetyltransferase: identification of a selective small molecule inhibitor. Chem. Biol. (Oxford, U. K.) 2010, 17, 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Biel M; Kretsovali A; Karatzali E; Papamatheakis J; Giannis A Design, Synthesis, and Biological Evaluation of a Small-Molecule Inhibitor of the Histone Acetyltransferase Gcn5. Angew. Chem., Int. Ed 2004, 43, 3974–3976. [DOI] [PubMed] [Google Scholar]
- (79).Sorum AW; Shrimp JH; Roberts AM; Montgomery DC; Tiwari NK; Lal-Nag M; Simeonov A; Jadhav A; Meier JL Microfluidic mobility shift profiling of lysine acetyltransferases enables screening and mechanistic analysis of cellular acetylation inhibitors. ACS Chem. Biol 2016, 11, 734–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Olson ME; Abate-Pella D; Perkins AL; Li M; Carpenter MA; Rathore A; Harris RS; Harki DA Oxidative reactivities of 2-furylquinolines: ubiquitous scaffolds in common high-throughput screening libraries. J. Med. Chem 2015, 58, 7419–7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Matson SL; Chatterjee M; Stock DA; Leet JE; Dumas EA; Ferrante CD; Monahan WE; Cook LS; Watson J; Cloutier NJ; Ferrante MA; Houston JG; Banks MN Best practices in compound management for preserving compound integrity and accurately providing samples for assays. J. Biomol. Screening 2009, 14, 476–484. [DOI] [PubMed] [Google Scholar]
- (82).Engeloch C; Schopfer U; Muckenschnabel I; Le Goff F; Mees H; Boesch K; Popov M Stability of screening compounds in wet DMSO. J. Biomol. Screening 2008, 13, 999–1006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


