Abstract
Limbal stem cell deficiency (LSCD) and corneal disorders are among the top global threats for human vision. Emerging therapies that integrate stem cell transplantation with engineered hydrogel scaffolds for biological and mechanical support are becoming a rising trend in the field. However, methods for high-throughput fabrication of hydrogel scaffolds, as well as knowledge of the interaction between limbal stem/progenitor cells (LSCs) and the surrounding extracellular matrix (ECM) are still much needed. Here, we employed digital light processing (DLP)-based bioprinting to fabricate hydrogel scaffolds encapsulating primary LSCs and studied the ECM-dependent LSC phenotypes. The DLP-based bioprinting with gelatin methacrylate (GelMA) or hyaluronic acid glycidyl methacrylate (HAGM) generated microscale hydrogel scaffolds that could support the viability of the encapsulated primary rabbit LSCs (rbLSCs) in culture. Immunocytochemistry and transcriptional analysis showed that the encapsulated rbLSCs remained active in GelMA-based scaffolds while exhibited quiescence in the HAGM-based scaffolds. The primary human LSCs (hLSCs) encapsulated within bioprinted scaffolds showed consistent ECM-dependent active/quiescent statuses. Based on these results, we have developed a novel bioprinted dual ECM ‘Yin-Yang’ model encapsulating LSCs to support both active and quiescent statues. Our findings provide valuable insights towards stem cell therapies and regenerative medicine for corneal reconstruction.
Keywords: DLP-based bioprinting, Limbal stem cell, Stem cell quiescence, Endogenous stem cell, Hyaluronic acid, Regenerative medicine
1. Introduction
Corneal epithelium is a transparent nonkeratinized epithelium that contributes to the refractive power of eye and serves as the first protective barrier against the outside world [1,2]. Limbal stem/progenitor cells (LSCs) are endogenous stem cells that reside at the limbus, the periphery of the cornea [3]. LSCs are responsible for the homeostasis of corneal epithelium, thus, facilitating optical clarity and light transmission [2,3]. Worldwide, there are over 5 million individuals affected by corneal blindness and limbal stem cell deficiency (LSCD) being a common etiology [4-6]. Conventional LSCD treatments employ surgical repair interventions using such sources as amniotic membrane (AM) as substrate or scaffold combined with keratolimbal autografts, or allografts [7]. These treatment approaches are limited by the lack of standardized preparation of AM, risk of developing iatrogenic LSCD and immunologic rejection [8-10].
Recent advances in regenerative medicine and tissue engineering have facilitated the development of novel transplantation approaches using advanced biomaterials for the treatment of LSCD [11]. Hydrogel scaffolds based on collagen, gelatin, hyaluronic acid (HA), and synthetic polymers have been investigated as LSC carriers for transplantation [12-15]. Among the various approaches of hydrogel scaffold fabrication, digital light processing (DLP)-based bioprinting stands out as a high-throughput platform allowing fabrication of hydrogel scaffolds that support the encapsulation of numerous types of stem cells including retinal progenitor cells, conjunctival stem cells, mesenchymal stem cells, neural progenitor cells and cancer stem cells [16-21]. The spatiotemporal control of light exposure afforded by DLP-based bioprinting also enables the stiffness tunability within desired regions of the fabricated hydrogel scaffolds, thus allowing one to manipulate the phenotypes of the encapsulated cells [19,22-25]. Moreover, DLP-based bioprinting enables the use of multiple extracellular matrix (ECM) components and multiple cell types during hydrogel fabrication to better recapitulate the complex native microenvironment of stem cells [16,17,26].
Biological and biomechanical interactions between stem cells and their ECM have been shown to affect cell fate and phenotype [27-29]. Biomechanical factors such as substrate stiffness have been shown to regulate the activities of LSCs and the corneal regeneration under physiological and pathological conditions [30]. Stem cells can also interact with the scaffolds in a composition-dependent way as various types of cell surface receptors responding to the ECM by triggering downstream intracellular signaling pathways that dynamically and comprehensively manipulate cell programming [12,31-35]. The delicate balance between activation and quiescence of endogenous stem cells, including LSCs, is critical for the system homeostasis under varying healthy, aging, and diseased circumstances [36-38]. Recent studies have showed that engineered scaffolds are able to tune the transition of activation/quiescence in LSCs [33,39]. Therefore, understanding how the different ECM compositions regulate LSCs in a 3D microenvironment is important for developing novel transplantable LSC scaffolds.
In this study, we present a bioprinting approach in fabricating primary LSC-encapsulated microscale hydrogel scaffolds to study the ECM-dependent LSC activities. With the customized DLP-based bioprinting system, we fabricated microscale hydrogel scaffolds with gelatin methacrylate (GelMA) and hyaluronic acid glycidyl methacrylate (HAGM) that supported the encapsulation and cell viability of primary rabbit LSCs (rbLSCs). Next, we analyzed the different phenotypes of encapsulated rbLSCs at mRNA and protein levels. In addition, we extended the study on primary human LSCs (hLSCs) from different individuals with bioprinting. Furthermore, we performed multimaterial bioprinting and fabricated a dual ECM ‘Yin-Yang’ model encapsulating primary rbLSCs in active/quiescent status. Overall, we developed an innovative DLP-based bioprinting approach for LSC engineering while broadening the understanding of ECM-dependent LSCs phenotypes, which is a meaningful step towards the development of regenerative medicine for LSCD and other severe ocular surface diseases.
2. Materials and Methods
2.1. Primary rabbit, human LSCs isolation and culture
The rabbit tissues from 10-12 weeks old New Zealand White rabbit eyes (Oryctolagus Cuniculus) were acquired from Sierra for Medical Science, Inc. (Whittier, CA). The human corneoscleral rims were acquired from One Legacy or Saving Sight eye banks. Consent was obtained by the eye banks for the tissues to be used for research. Experimentation on human tissue adhered to the tenets of the Declaration of Helsinki. The protocol for human corneal tissue collection and dissection was evaluated and exempted by the University of California, Los Angeles (UCLA) Institutional Review Boards (IRB#12-000363). The overall procedure was approved by University of California San Diego Institutional Biosafety Committee.
For rabbit LSCs (rbLSCs), rabbit eyeballs were washed in Dulbecco's phosphate-buffered saline (DPBS) and Dulbecco’s Modified Eagle Medium (DMEM, ThermoFisher Scientific) with penicillin-streptomycin, respectively, and the corneoscleral rims were isolated for further dissection. Human LSCs (hLSCs) were harvested from donor corneoscleral rims stored in Optisol-GS. Corneoscleral rims from three different donors with no significant history of corneal diseases were used in this study. The isolation of both rbLSCs and hLSCs was performed as previously described [40]. Briefly, limbal epithelium with underlying stroma was excised circumferentially and minced using Vannas scissors. Type IV collagenase (0.2%, Sigma Aldrich) was used for digestion at 37°C with constant shaking at 120 rpm for 1-1.5 hr. Following the incubation, cells were pelleted and washed with PBS. Following a 10 min digestion with 0.25% Trypsin-EDTA (Sigma Aldrich) digestion, the cells were filtered through a 70 μm cell strainer (Corning) to obtain single cells. The cells were seeded onto Collagen I coated plates (ThermoFisher Scientific). The culture medium used was composed of DMEM /F-12 (3:1, ThermoFisher Scientific) with 10% Fetal Bovine Serum (ThermoFisher Scientific), penicillin-streptomycin (ThermoFisher Scientific), 400 ng/ml hydrocortisone (Sigma Aldrich), 1x insulin-transferrin-selenium (Corning), 2 nM reverse T3 (Sigma Aldrich), and 0.1 nM cholera toxin (Sigma Aldrich), 10 ng/ml epidermal growth factor (EGF, R&D System), and 10 μM Y27632 (Tocris Bioscience).
2.2. Material synthesis and photocrosslinkable bioink preparation
The synthesis process of GelMA and HAGM was performed following previously established protocols [16,17,19,26,41]. Briefly, for GelMA, a 10% (w/v) gelatin solution was prepared by dissolving porcine skin gelatin type A (Sigma Aldrich) in a 0.25 M carbonate-bicarbonate (3:7) buffer at pH 9 while stirring at 50°C. Methacrylic anhydride (Sigma Aldrich) was then mixed in a dropwise fashion to the gelatin solution to reach 100 μl methacrylic anhydride per gram of gelatin. Then, following 1 hr of continuous stirring at 50°C, the product was subjected to overnight fluid dynamic dialysis using 13.5 kDa dialysis tubes (Repligen). Lyophilization for three days was then conducted to produce GelMA powder which was then stored at −80°C. The degree of methacrylation of the resultant GelMA is approximately 95% [17].
To synthesize HAGM, 1.0 g of Sodium Hyaluronate (Lifecore Biomedical) was dissolved in 100 mL water: acetone solution (1:1 ratio) and stirred at room temperature overnight to prepare a 1% (w/v) HA solution. The flask was subjected to vacuum for 3 sec or until the solution boils then flooded with Argon. This step was repeated twice and the solution was stirred overnight protected from light. On the next day, 7.2 ml triethylamine (Sigma Aldrich) 20-fold in excess was slowly added to the reaction flask until thoroughly mixed. The reaction was then flooded with argon gas, then immediately sealed, and mixed for 30 min. Using a syringe, 7.2 mL of glycidyl methacrylate (GM, Sigma Aldrich) in 20-fold excess was added dropwise to the reaction. Afterwards, the reaction was flooded with Argon, sealed, and stirred overnight at room temperature. The resulting material was precipitated using acetone and vacuum filtration was used to collect the precipitate which was dissolved in DI water. The dissolved material was then dialyzed, lyophilized, and stored at −80°C until further use. The degree of methacrylation of the resultant HAGM is approximately 35% [17].
For photopolymerization, lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) was used as a photoinitiator and synthesized per previously published protocols [16,19]. Briefly, dimethyl phenylphosphonite (18 mmol, Sigma Aldrich) was added dropwise to an equimolar 2,4,6-trimethylbenzoyl chloride (Acros Organics). The reaction was constantly stirred for 18 hr at room temperature. A solution of lithium bromide (6.1g, Sigma Aldrich) in 100 ml of 2-butanone (Sigma Aldrich) was then mixed into the reaction. Following a 10-minute stirring at 50°C, the mixture was incubated overnight at room temperature. Filter-washing with 2-butanone for three times was carried out to remove unreacted lithium bromide. The LAP solids that resulted from the reaction were crushed into powder and stored under argon in the dark at 4 °C.
8% (w/v) GelMA with 0.25% (w/v) LAP and 4% (w/v) HAGM with 0.25% (w/v) LAP were dissolved in warm DPBS, filtered using a 0.22 μm syringe and used as prepolymer solutions for DLP-based bioprinting with or without LSCs. The cells were detached from the culture plates with 0.25% trypsin-EDTA, and then neutralized with a pre-made culture medium. The cell solution was then filtered with a 70 μm cell strainer and the cell concentration was measured with a hemocytometer. The bioink containing 1-2×107 cells/mL LSCs and GelMA/HAGM prepolymer solution was prepared right before printing.
2.3. Bioprinting of GelMA and HAGM hydrogel scaffolds
Our in-house DLP-based bioprinting system was used for the rapid biofabrication of hydrogel scaffolds. The system is composed of a 365 nm light source (Hamamatsu), a projection optics assembly, a motion-controlled stage (Newport) and a digital micromirror device (DMD, Texas Instruments.) used for patterning the light. We generated digital patterns using Adobe photoshop which were imported into the custom operation software that controls the DMD chip to modulate light projection depending on the imported pattern. For the bioprinting setup, two polydimethylsiloxane (PDMS) spacers with thickness of 250 μm were set between a PDMS base that is attached to a glass slide and a methacrylated coverslip. This creates a gap of desired thickness where the prepolymer bioink was loaded. Then, photopolymerization was performed with the DLP bioprinter and the printed constructs were immediately moved to a 24-well plate and washed in pre-warmed DPBS to remove the excess bioink materials. The DPBS was then substituted with warmed culture medium and the bioprinted constructs were incubated in 5% CO2 at 37°C.
2.4. Immunofluorescence staining
Primary LSCs cultured on Millicell EZ slides (Millipore Sigma) were washed twice with DPBS to prepare for 2D cell staining. The cells were fixed at room temperature for 20 min with 4% (w/v) paraformaldehyde (FUJIFILM Wako), followed by three washes with DPBS, each for 10 min. Then, the samples underwent blocking and permeabilization for 1 hr using 5% bovine serum albumin (Sigma Aldrich) and 0.3% triton X-100 (Sigma Aldrich) in DPBS at room temperature. Primary antibody incubation was done at 4°C overnight followed by three DPBS washes for 10 min each. Afterwards, the cells were incubated with secondary antibodies (Alexa Fluor-conjugated, Invitrogen) for 1 hr at room temperature. The samples were further washed with DPBS and nuclear staining was done with 1:500 DAPI (4',6-Diamidino-2-Phenylindole; ThermoFisher Scientific) in DPBS for 10 min. After a final DPBS wash the samples were left to air-dry for 30 sec and mounted with Fluoromount-G™ Mounting Medium (ThermoFisher Scientific). Hydrogel cells staining was performed with the exact same procedure, except without mounting, where the samples were left in DPBS to be imaged. The samples were all imaged within 48 hr of staining to preserve clarity. Further information on details of antibodies and dilution rates is provided in Supplementary Table 1.
2.5. Mechanical properties characterization
A micromechanical testing machine (Microsquisher, CellScale) was used to determine the Young’s modulus of the bioprinted scaffolds based on GelMA and HAGM. Cylindrical test specimens (500 μm diameter, 500 μm height) printed with 8% GelMA or 4% HAGM were fabricated and incubated at 37°C for overnight. We followed the manufacturer’s instructions when measuring the compressive modulus. The sample’s hysteresis was removed using two cycles of predetermined compression. During mechanical testing, the samples were compressed at a 10% strain with a strain rate of 2 μm/s. After the force and displacement data was collected from the Microsquisher, we used custom MATLAB script to calculate the compressive Young's modulus.
2.6. Viability evaluation
The viability of the LSCs encapsulated in the hydrogels were studied with the Viability/Cytotoxicity kit (Thermo Fisher), also known as Live/Dead™ staining. They were incubated with 2 μM calcein acetoxymethyl ester, along with 4 μM ethidium homodimer in DPBS, for 30 min at 37°C. Fluorescent imaging was done with a Leica microscope (DMI 6000-B). The viability test was carried out in triplicates.
2.7. RNA isolation, reverse transcription, and real time quantitative PCR
For RNA extraction, a TRIzol® reagent (Ambion Thermo Fisher) was continuously pipetted into the pelleted 2D-cultured cells. For the encapsulated LSCs in GelMA- and HAGM-based bioprinted hydrogel scaffolds, the constructs were stripped off their coverslips using a scalpel and subjected to enzymatic digestion with 0.2% Type IV collagenase (Sigma Aldrich) and 1kU/ml hyaluronidase (STEMCELL Technologies), respectively, at 37°C for 15 min. The resulting cell solution was pelleted with centrifugation immediately followed by addition of TRIzol® reagent to the pellet. The lysate was then used directly or stored in −80°C. Direct-zol™ RNA Purification kit (Zymo Research) was used for the extraction of RNA following the manufacturer's protocol. NanoDrop™ 2000 (Thermo Fisher Scientific) was used to quantify the purified RNA. The RNA was then used for cDNA synthesis and reverse transcription using the iScript™ cDNA Synthesis Kit (Bio-Rad) with thermal cycler StepOne™ Real-Time PCR System (Thermo Fisher Scientific). Luna® Universal qPCR Master Mix was used for Real-Time quantitative polymerase chain reaction (qPCR). The primer details used in the qPCR can be found in Supplementary Table 2.
2.8. Flow cytometry
For flow cytometry, GelMA- and HAGM-based bioprinted scaffolds were enzymatically digested to isolate the encapsulated LSCs. Following the enzymatic digestion, the cells extracted from the scaffolds and 2D-cultured cells were digested with 0.25% trypsin-EDTA and filtered with a 70 μm cell strainer. After centrifugation, cell pellets were resuspended and fixed with Cytofix™ Fixation Buffer (BD) for 20 min followed by three 5-minute wash with Cell Staining Buffer (Biolegend) supplemented with 0.2% triton X-100. Primary antibodies were diluted with Cell Staining Buffer and applied for 20 min. Following a wash, secondary antibodies were diluted with Cell Staining Buffer and applied for 20 min. All the antibody incubations were performed at room temperature. Cell solutions were then kept on ice in the dark until use. The propidium iodide (Biolegend) viability staining was performed following the manufacturer’s procedures. Briefly, 10 μl per million cells of the propidium iodide solution was added to the cell suspension. The solution was then incubated for 15 minutes at 4 °C avoiding light before analysis. Flow cytometry was performed using BD Accuri™ C6 flow cytometer following the instructions of the manufacturer. The data was collected from at least 100,000 events for each group and processed using FlowJo.
2.9. Imaging and processing
Confocal and brightfield/regular fluorescence imaging of the samples were taken using SP8 Confocal and DMI 6000-B Leica microscopes, respectively. ImageJ and LAS X were used to further process the images.
2.10. Statistical analysis
Data obtained from the experiments were processed with Microsoft Excel and presented in a way of mean ± standard deviations Student’s t-test (two tailed) or one-way ANOVA were applied to determine statistical significance which was denoted on the figures with an asterisk where appropriate (*: P < 0.05; **: P < 0.01; ***: P < 0.001.).
3. Results
3.1. Bioprinted GelMA and HAGM hydrogel scaffolds supported the viability of encapsulated primary rbLSCs
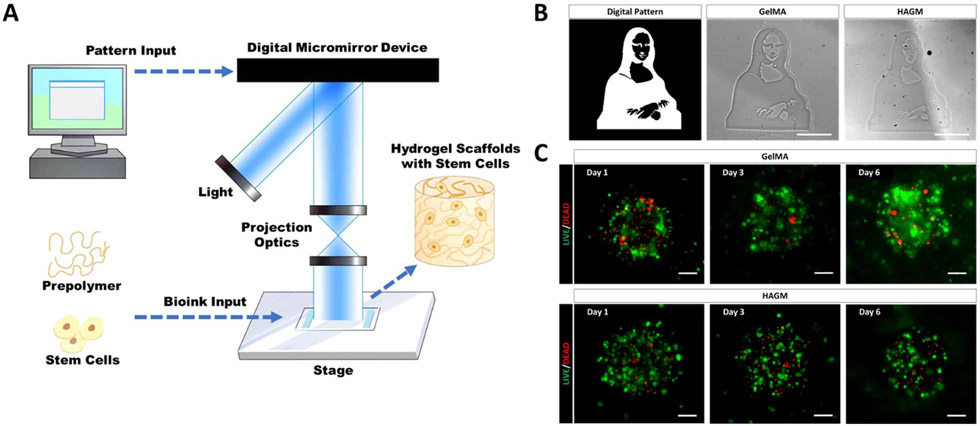
Our customized DLP-based bioprinting system can spatially manipulate light based on user-defined input designs, allowing for precise photopolymerization-based patterning of cellularized hydrogel constructs containing different material compositions (Figure 1A) [18,19,24]. GelMA is a photocrosslinkable gelatin that has been extensively studied as a bioink for the bioprinting of stem cells including conjunctival stem cells and mesenchymal stem cells [19,42]. HAGM as another photocrosslinkable bioink, was found to support the encapsulation of retinal progenitor cells and cancer stem cells [16,17]. Using DLP-based bioprinting techniques, we were able to fabricate GelMA- or HAGM-based hydrogel scaffolds with a complex pattern and microscale resolution within a matter of seconds (Figure 1B). To encapsulate LSCs in 3D scaffolds, we isolated and expanded the primary rbLSCs from fresh rabbit limbal tissues and characterized with immunofluorescence staining of various LSC markers (Supplementary Figure S1 A). To test biocompatibility, we fabricated GelMA- or HAGM-based bioprinted scaffolds encapsulating primary rbLSCs. The Live/Dead™ staining confirmed that both types of bioprinted scaffolds were able to support the viability of the encapsulated rbLSCs after 7 days of culture (Figure 1C). We have also quantified the viability of the encapsulated rbLSC in both types of bioprinted scaffolds by flow cytometry with propidium iodide staining (Supplementary Figure S1B). Based on the results, the live cell ratios were 86.7±1.6% in GelMA scaffolds and 92.1±0.8 % in HAGM scaffolds after 7 days of culture. In brief, we were able to fabricate both GelMA- and HAGM-based scaffolds encapsulating viable primary rbLSCs using our DLP-based bioprinting system.
Figure 1.
Bioprinting of GelMA- or HAGM-based scaffolds encapsulating primary rbLSCs. (A) Schematic of DLP-based bioprinting workflow; (B) representative bright field images of bioprinted Mona Lisa with acellular GelMA or HAGM; (C) representative images of Live/Dead™ staining of rbLSCs encapsulated with GelMA- or HAGM-based bioprinted scaffolds at Day 1, Day 3 and Day 6 of culture (scale bars: 100 μm).
3.2. Encapsulated primary rbLSCs displayed active status in GelMA-based bioprinted scaffolds while exhibiting quiescence in HAGM-based bioprinted scaffolds
While both GelMA- and HAGM-based bioprinted scaffolds maintained viable encapsulated primary rbLSCs, the cells displayed different behaviors depending on which scaffold they were cultured in. More cell aggregates or colonies were observed in GelMA-based bioprinted scaffolds, while rbLSCs encapsulated with HAGM-based scaffolds largely remained as single-cells after 6 days of culture (Supplementary Figure S1C). These results suggest that the interaction between rbLSCs and their surrounding ECM in the different bioprinted scaffolds influenced the stem cell status following the encapsulation. To further explore the effect of the scaffold matrix material on LSC-ECM interaction, we first needed to control the stiffness. Our DLP-based bioprinting system enables us to control the mechanical properties of the fabricated hydrogel scaffolds via spatiotemporal regulation of light exposure [23,24]. Mechanical testing of GelMA-and HAGM-based bioprinted scaffolds indicated positive linear correlation between the Young’s modulus and the light exposure time in our printing system (Figure 2A, Supplementary Figure S1D). Based on the results, GelMA and HAGM scaffolds had a similar Young’s modulus with the light exposure time set to 25 seconds which was adopted as the primary bioprinting parameters for subsequent experiments.
Figure 2.
Encapsulated primary rbLSCs displayed different status in GelMA- or HAGM-based bioprinted scaffolds. (A) Linear plot of compressive modulus of the GelMA- or HAGM-based scaffolds versus the light exposure time (mean ± sd, n ⩾ 3); (B) representative images of immunofluorescence staining of proliferation marker KI67 and LSC lineage marker PAX6 on rbLSCs encapsulated in GelMA- or HAGM-based scaffolds after 2 days of culture (scale bars: 50 μm); (C)KI67-positive and ΔNP63-positve populations of the primary rbLSCs cultured in 2D and encapsulated in GelMA- or HAGM-based scaffolds after 2 days of culture as determined by flow cytometry (mean ± sd, n = 3); (D) heatmap of real-time qPCR data showing relative mRNA expression of LSC markers (KRT14, P63, PAX6, BMI1), LSC quiescent markers (CD200, P27KIP1), corneal epithelium differentiation marker (KRT3), canonical WNT signaling pathway marker (CTNNB1) and non-canonical WNT signaling pathway markers (WNT5A, VANGL1) on rbLSCs on 2D surface or encapsulated in GelMA- or HAGM-based scaffolds after 2 days of culture.
To investigate the behavior of LSCs in the different scaffolds, we examined the expression of various LSC markers. Immunofluorescence staining showed the expression of PAX6, an ocular lineage marker, in both GelMA- and HAGM-encapsulated rbLSCs while the expression of proliferation marker, KI67, was present only in the GelMA-based scaffolds (Figure 2B). Consistently, flow cytometry identified significantly smaller percentage of KI67 positive rbLSCs encapsulated in HAGM-based bioprinted scaffolds compared to the population encapsulated in GelMA, while the positive ratio of stemness marker, ΔNP63, remained identical in both scaffolds (Figure 2C, Supplementary Figure S2A). The decreased KI67 positive population of rbLSCs encapsulated in HAGM-based bioprinted scaffolds can be reversed by releasing the cells from scaffolds (Supplementary Figure S2B). We have also performed transcriptional analysis with real-time qPCR to compare rbLSCs in 2D culture or encapsulation with GelMA- or HAGM-based bioprinted scaffolds (Figure 2D). We found up-regulated mRNA expression of PAX6 and BMI1 in the HAGM group compared to the 2D control, while P63 was up-regulated in the GelMA group. In addition, the expression of two previously reported LSC quiescence markers, CD200 and P27KIP1, were up-regulated in the HAGM group [43-45]. The expression of corneal epithelium differentiation marker, KRT3, was downregulated in the GelMA group and showed no significant change in the HAGM group in comparison with the 2D control. Furthermore, mRNA expression of markers of non-canonical WNT pathway, VANGL1 and WNT5A, were up-regulated in the HAGM group but down-regulated in the GelMA group. Meanwhile, the expression of marker of canonical WNT pathway, CTNNB1, remained unchanged among the three groups. These results indicated the potential participation of non-canonical WNT pathway in the LSCs-ECM interactions [46-49]. Collectively, these results demonstrated an active status of the rbLSCs encapsulated in GelMA-based scaffolds and quiescent characteristics of rbLSCs in the HAGM-based scaffolds.
3.3. Encapsulated primary hLSCs were viable but displayed different status in GelMA- or HAGM-based scaffolds
Based on the ECM-dependent response of rbLSCs in the GelMA- or HAGM-based scaffolds, we further explored the LSC-ECM interaction in human LSCs. Primary hLSCs were isolated and expanded from human corneoscleral rims of three different donors and subjected to bioprinting with GelMA and HAGM, respectively. Similar to the rbLSCs, Live/Dead™ staining showed that most of the encapsulated hLSCs remained viable in both types of bioprinted scaffolds during culture (Figure 3A). Consistent with rbLSCs, aggregated colonies of hLSCs were largely found in the GelMA-based scaffolds but rarely observed in the HAGM-based scaffolds (Figure 3B). Real-time qPCR showed that the hLSCs encapsulated in HAGM-based scaffolds had significantly higher expression of PAX6, CD200 and P27KIP1, while the expression of KI67 was significantly down-regulated compared to the 2D control and the GelMA group (Figure 3C). In addition, KRT14 expression was significantly up-regulated in both bioprinted groups comparing with the 2D control. These results reinforce the observation that LSCs respond differently (e.g., exhibiting active proliferation or quiescence) to the surrounding ECM composition, and appears to be consistent whether the LSCs are isolated from rabbits or humans, suggesting that this may be highly valuable for future clinical studies.
Figure 3.
Encapsulated primary hLSCs remained viable and displayed different status in GelMA- or HAGM-based scaffolds. (A) Representative images of Live/Dead™ staining of hLSCs encapsulated with GelMA- or HAGM-based scaffolds at Day 2 and Day 5 of culture (scale bars: 100 μm); (B) representative bright field images of bioprinted scaffolds with GelMA/HAGM encapsulating primary hLSCs after 5 days of culture (scale bars: 100 μm); (C) real-time qPCR data showing relative mRNA expression of proliferation marker (KI67), LSC markers (KRT14, PAX6), LSC quiescent markers (CD200, P27KIP1) of the primary hLSCs on 2D surface or encapsulated in GelMA- or HAGM-based scaffolds after 2 days of culture (mean ± sd, n = 3).
3.4. DLP-based bioprinting of dual ECM ‘Yin-Yang’ LSC model
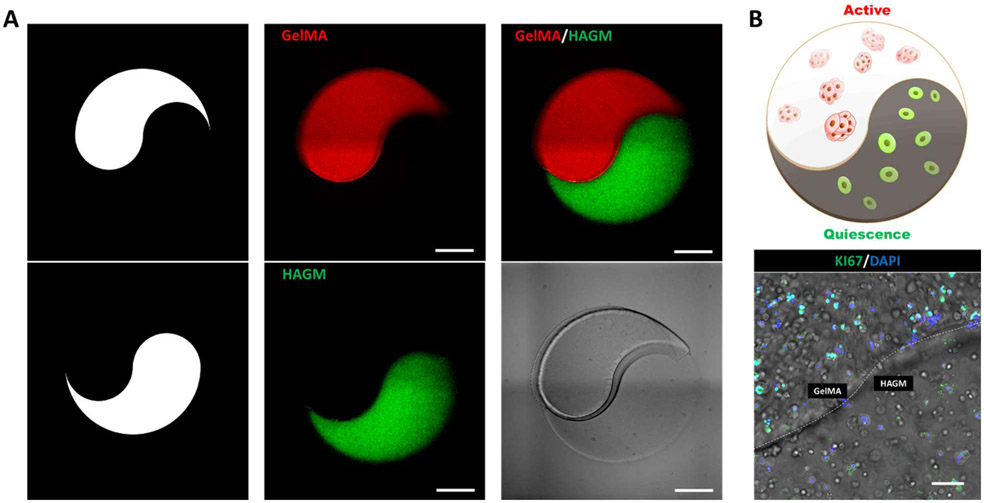
After ascertaining the ECM-dependent active/quiescent status of encapsulated LSCs in GelMA- and HAGM-based scaffolds, we aimed to build a dual in vitro ECM model that could facilitate these differential statuses of cells within the same hydrogel, thus coming closer to recapitulating native LSC niches where cells in both activated/quiescent states coexist [3]. For this, we chose to utilize a ‘Yin-Yang’ pattern that allows for the placement of GelMA and HAGM distinctly separate yet spatially close regions. To demonstrate the feasibility of our design, we first printed the ‘Yin-Yang’ pattern in microscale with GelMA and HAGM mixed with fluorescence microspheres (Figure 4A). Fluorescent microscopic imaging showed the precise patterning of the acellular hydrogel materials matching our design specification. In follow-up prints, we replaced fluorescent microspheres with primary rbLSCs and verified the status of the encapsulated rbLSCs in different parts of the dual ECM model (Figure 4B). Immunofluorescence staining showed the positive expression of KI67 in the GelMA-based region while few KI67 positive cells were found in the HAGM-based region (Figure 4B). Therefore, with our bioprinting method, we were able to fabricate the dual ECM ‘Yin-Yang’ model whose separate ECM-portions induced active/quiescent statuses for the LSC.
Figure 4.
Bioprinting of a dual ECM ‘Yin-Yang’ model encapsulating rbLSCs. (A) Illustration of the design patterns and bright field images of the acellular dual ECM ‘Yin-Yang’ models encapsulating fluorescent microspheres (scale bars: 500 μm); (B) illustration and a representative image of immunofluorescence staining of KI67 on dual ECM ‘Yin-Yang’ model encapsulating primary rbLSCs after 2 days of culture (scale bars: 100 μm).
4. Discussion
With the recent technological advances in tissue engineering and regenerative medicine, stem cell therapies based on hydrogel scaffolds have become popular for the treatment of LSCD [12]. However, cost effective approaches for the high-throughput fabrication of hydrogel scaffolds encapsulating primary LSCs remains an active area of research. Furthermore, behavior that LSCs exhibit in response to different 3D matrices presents an attractive opportunity in formulating scaffolds that can effectively recapitulate the native microenvironment of the LSC niche. Our study demonstrated a novel engineering approach applying DLP-based bioprinting for the fabrication of hydrogel scaffolds encapsulating both rabbit and human primary LSCs. We successfully printed GelMA- and HAGM-based microscale hydrogel scaffolds that maintained the viability of the encapsulated primary rbLSCs. The cells exhibited ECM-dependent phenotypes with an active status in GelMA- and quiescent status in HAGM-based scaffolds. We repeated the bioprinting experiments with hLSCs and confirmed the consistency of the ECM-dependent phenotype in primary human cells. Moreover, we applied DLP-based bioprinting to build a dual ECM ‘Yin-Yang’ model encapsulating LSCs in active and quiescent status within the same culture.
3D engineered tissues are 3D fabricated biomimetic systems consisted of the corresponding cells from the target tissue and organ, as well as the hydrogel scaffolds that mimic ECM[18,50]. As the construct geometry and the cell distribution can be manipulated to recapitulate the physiological microenvironment, 3D engineered tissues could have better performance in applications like drug screening, disease modeling, and regenerative medicine compared to the 2D monolayer cell model[17,19,51]. Tissue regeneration using endogenous stem cell is a promising solution for many medical conditions [2,28,52,53]. As the essential endogenous epithelial stem cell contributing to corneal regeneration, LSCs have been explored in various approaches in combination with hydrogel scaffolds for corneal epithelium reconstruction [54,55]. DLP-based bioprinting has been instrumental in tissue engineering as it facilitated the fabrication of high-throughput hydrogel scaffolds encapsulating various types of stem cells [18]. We used DLP-based bioprinting to produce GelMA- or HAGM-based hydrogel scaffolds encapsulating primary rbLSCs and hLSCs. DLP-based bioprinting maintained the viability and stemness of the encapsulated LSCs in both materials. With flexible and precise control over morphological structures, the microscale hydrogel scaffolds encapsulating LSCs can be optimized by our bioprinting system to serve various therapeutic purposes including minimally invasive injectable stem cell transplantation [56]. In addition, the translucent nature of the GelMA and HAGM scaffolds not only enabled facile monitoring of cell morphology and behaviors, but also makes ideal candidates for corneal tissue-on-a-chip in vitro disease modeling. Although the bioprinted hydrogel scaffolds in this study were 2.5D constructs that have consistent pattern along the thickness direction, they were able to provide insightful information for disease modeling[17,26]. In the future, we could use the DLP-based bioprinting system to fabricate true 3D constructs such as a corneal tissue with complex 3D features for various biomedical applications[57].
The ECM-dependent regulation and reprogramming of epithelial stem cell fate have been indicated as prevalent mechanisms in different tissues including epidermis, lung, intestine, colon and cornea [30,58-61]. By controlling the matrix stiffness with our bioprinting system, we were able to compare the influence of ECM component on the encapsulated LSCs. As a result, we found that primary rbLSCs and hLSCs actively proliferated and formed aggregated colonies in the GelMA-based scaffolds while showed inhibited proliferation and aggregation in the HAGM-based scaffolds. Further analysis showed the active/quiescent status of encapsulated LSCs by comparing proliferation and stemness markers. The quiescence of LSCs in HAGM-based scaffolds can potentially be mediated by the HA-specific cell adhesion excluding integrins [62-64]. We also found the HAGM-encapsulated rbLSCs presented proliferative status after being released from scaffolds and cultured for a week, indicating that the HAGM-encapsulated rbLSCs were reversibly quiescent. Non-canonical WNT signaling pathways (planar cell polarity) have been reported to modify the activation/quiescence in multiple endogenous stem cells [48,49,65]. Notably, we found upregulated mRNA expression of markers related to non-canonical WNT signaling pathways in primary LSCs encapsulated in HAGM-based scaffolds, which is consistent with the previously reported ECM-response of primary LSCs cultured on engineered HA scaffolds [39,66]. As a proof-of-concept, we further combined these findings with multimaterial bioprinting to fabricate a dual ECM ‘Yin-Yang’ model simultaneously encapsulating primary LSCs in active/quiescent status. The dual ECM model can be an attractive platform for drug screening since it reproduced stem cell quiescence that was correlated to drug-resistance and recapitulated the stem cells in heterogeneous status that could react to drugs differently [67-69].
5. Conclusions
We applied DLP-based bioprinting to fabricate engineered microscale hydrogel scaffolds based on GelMA and HAGM. These scaffolds supported not only the viability of encapsulated primary rbLSCs and hLSCs, but also exhibited differential regulation. LSCs were found to display an ECM-dependent active/quiescent status as they actively proliferated in the GelMA-based scaffolds and took on quiescent characteristics in the HAGM-based scaffolds. A bioprinted dual-ECM ‘Yin-Yang’ model encapsulating both active and quiescent LSCs were fabricated based on these findings. Together, these results illustrated an innovative engineering approach for disease modeling, drug screening and the development of an LSC-based regenerative therapy for the treatment of LSCD and related ocular diseases. Future studies exploring other types of biomaterials or integrating different cell types would be valuable to investigate.
Supplementary Material
Footnotes
Competing Interests
The authors declare no competing financial interests.
Data Availability
The raw/processed data required to reproduce these findings can be shared by the authors upon request.
References
- [1].Dhouailly D, Pearton DJ, Michon F, The vertebrate corneal epithelium: From early specification to constant renewal, Dev. Dyn 243 (2014) 1226–1241. 10.1002/dvdy.24179. [DOI] [PubMed] [Google Scholar]
- [2].Ouyang H, Xue Y, Lin Y, Zhang X, Xi L, Patel S, Cai H, Luo J, Zhang M, Zhang M, Yang Y, Li G, Li H, Jiang W, Yeh E, Lin J, Pei M, Zhu J, Cao G, Zhang L, Yu B, Chen S, Fu XD, Liu Y, Zhang K, WNT7A and PAX6 define corneal epithelium homeostasis and pathogenesis, Nature. 511 (2014) 358–361. 10.1038/nature13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dziasko MA, Daniels JT, Anatomical Features and Cell-Cell Interactions in the Human Limbal Epithelial Stem Cell Niche, Ocul. Surf 14 (2016) 322–330. 10.1016/j.jtos.2016.04.002. [DOI] [PubMed] [Google Scholar]
- [4].Bourne RRA, Flaxman SR, Braithwaite T, Cicinelli MV, Das A, Jonas JB, Keeffe J, Kempen J, Leasher J, Limburg H, Naidoo K, Pesudovs K, Resnikoff S, Silvester A, Stevens GA, Tahhan N, Wong T, Taylor HR, Ackland P, Arditi A, Barkana Y, Bozkurt B, Wormald R, Bron A, Budenz D, Cai F, Casson R, Chakravarthy U, Congdon N, Peto T, Choi J, Dana R, Palaiou M, Dandona R, Dandona L, Shen T, Dekaris I, Del Monte M, Deva J, Dreer L, Frazier M, Ellwein L, Hejtmancik J, Frick K, Friedman D, Javitt J, Munoz B, Quigley H, Ramulu P, Robin A, Tielsch J, West S, Furtado J, Gao H, Gazzard G, George R, Gichuhi S, Gonzalez V, Hammond B, Hartnett ME, He M, Hirai F, Huang J, Ingram A, Joslin C, Khanna R, Stambolian D, Khairallah M, Kim J, Lambrou G, Lansingh VC, Lanzetta P, Lim J, Mansouri K, Mathew A, Morse A, Musch D, Nangia V, Battaglia M, Yaacov F, Raju M, Rossetti L, Saaddine J, Sandar M, Serle J, Shetty R, Sieving P, Silva JC, Sitorus RS, Tejedor J, Tsilimbaris M, van Meurs J, Varma R, Virgili G, Volmink J, Xing Y, Wang NL, Wiedemann P, Zheng Y, Magnitude, temporal trends, and projections of the global prevalence of blindness and distance and near vision impairment: a systematic review and meta-analysis, Lancet Glob. Heal (2017). 10.1016/S2214-109X(17)30293-0. [DOI] [PubMed] [Google Scholar]
- [5].Deng SX, Borderie V, Chan CC, Dana R, Figueiredo FC, Gomes JAP, Pellegrini G, Shimmura S, Kruse FE, Global consensus on definition, classification, diagnosis, and staging of limbal stem cell deficiency, Cornea. 38 (2019) 364–375. 10.1097/ICO.0000000000001820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Le Q, Xu J, Deng SX, The diagnosis of limbal stem cell deficiency, Ocul. Surf 16 (2018) 58–69. 10.1016/j.jtos.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Meller D, Pires RTF, Mack RJS, Figueiredo F, Heiligenhaus A, Park WC, Prabhasawat P, John T, McLeod SD, Steuhl KP, Tseng SCG, Amniotic membrane transplantation for acute chemical or thermal burns, Ophthalmology. (2000). 10.1016/S0161-6420(00)00024-5. [DOI] [PubMed] [Google Scholar]
- [8].Hopkinson A, McIntosh RS, Tighe PJ, James DK, Dua HS, Amniotic membrane for ocular surface reconstruction: Donor variations and the effect of handling on TGF-β content, Investig. Ophthalmol. Vis. Sci 47 (2006) 4316–4322. 10.1167/iovs.05-1415. [DOI] [PubMed] [Google Scholar]
- [9].Ramuta TŽ, Starčič Erjavec M, Kreft ME, Amniotic Membrane Preparation Crucially Affects Its Broad-Spectrum Activity Against Uropathogenic Bacteria, Front. Microbiol 11 (2020) 469. 10.3389/fmicb.2020.00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Deng SX, Kruse F, Gomes JAP, Chan CC, Daya S, Dana R, Figueiredo FC, Kinoshita S, Rama P, Sangwan V, Slomovic AR, Tan D, Global Consensus on the Management of Limbal Stem Cell Deficiency, Cornea. 39 (2020) 1291–1302. 10.1097/ICO.0000000000002358. [DOI] [PubMed] [Google Scholar]
- [11].Mahdavi SS, Abdekhodaie MJ, Mashayekhan S, Baradaran-Rafii A, Djalilian AR, Bioengineering Approaches for Corneal Regenerative Medicine, Tissue Eng. Regen. Med 17 (2020) 567–593. 10.1007/s13770-020-00262-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nguyen KN, Bobba S, Richardson A, Park M, Watson SL, Wakefield D, Di Girolamo N, Native and synthetic scaffolds for limbal epithelial stem cell transplantation, Acta Biomater. 65 (2018)21–35. 10.1016/j.actbio.2017.10.037. [DOI] [PubMed] [Google Scholar]
- [13].Chen D, Qu Y, Hua X, Zhang L, Liu Z, Pflugfelder SC, Li DQ, A hyaluronan hydrogel scaffold-based xeno-free culture system for ex vivo expansion of human corneal epithelial stem cells, Eye. 31 (2017) 962–971. 10.1038/eye.2017.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sanie-Jahromi F, Eghtedari M, Mirzaei E, Jalalpour MH, Asvar Z, Nejabat M, Javidi-Azad F, Propagation of limbal stem cells on polycaprolactone and polycaprolactone/gelatin fibrous scaffolds and transplantation in animal model, BioImpacts. 10 (2020) 44–54. 10.15171/bi.2020.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Soman P, Lee JW, Phadke A, Varghese S, Chen S, Spatial tuning of negative and positive Poisson’s ratio in a multi-layer scaffold, Acta Biomater. (2012). 10.1016/j.actbio.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang P, Li X, Zhu W, Zhong Z, Moran A, Wang W, Zhang K, Chen S, 3D bioprinting of hydrogels for retina cell culturing, Bioprinting. (2018). 10.1016/j.bprint.2018.e00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tang M, Xie Q, Gimple RC, Zhong Z, Tam T, Tian J, Kidwell RL, Wu Q, Prager BC, Qiu Z, Yu A, Zhu Z, Mesci P, Jing H, Schimelman J, Wang P, Lee D, Lorenzini MH, Dixit D, Zhao L, Bhargava S, Miller TE, Wan X, Tang J, Sun B, Cravatt BF, Muotri AR, Chen S, Rich JN, Three-dimensional bioprinted glioblastoma microenvironments model cellular dependencies and immune interactions, Cell Res. (2020). 10.1038/s41422-020-0338-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yu C, Schimelman J, Wang P, Miller KL, Ma X, You S, Guan J, Sun B, Zhu W, Chen S, Photopolymerizable Biomaterials and Light-Based 3D Printing Strategies for Biomedical Applications, Chem. Rev 120 (2020) 10695–10743. 10.1021/acs.chemrev.9b00810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhong Z, Deng X, Wang P, Yu C, Kiratitanaporn W, Wu X, Schimelman J, Tang M, Balayan A, Yao E, Tian J, Chen L, Zhang K, Chen S, Rapid bioprinting of conjunctival stem cell micro-constructs for subconjunctival ocular injection, Biomaterials. (2021). 10.1016/j.biomaterials.2020.120462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Qu X, Zhu W, Huang S, Li YS, Chien S, Zhang K, Chen S, Relative impact of uniaxial alignment vs. form-induced stress on differentiation of human adipose derived stem cells, Biomaterials. (2013). 10.1016/j.biomaterials.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Soman P, Tobe BTD, Lee JW, Winquist AM, Singec I, Vecchio KS, Snyder EY, Chen S, Three-dimensional scaffolding to investigate neuronal derivatives of human embryonic stem cells, Biomed. Microdevices (2012). 10.1007/s10544-012-9662-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, Engler AJ, Interplay of matrix stiffness and protein tethering in stem cell differentiation, Nat. Mater (2014). 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yu C, Ma X, Zhu W, Wang P, Miller KL, Stupin J, Koroleva-Maharajh A, Hairabedian A, Chen S, Scanningless and continuous 3D bioprinting of human tissues with decellularized extracellular matrix, Biomaterials. 194 (2019) 1–13. 10.1016/j.biomaterials.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ma X, Yu C, Wang P, Xu W, Wan X, Lai CSE, Liu J, Koroleva-Maharajh A, Chen S, Rapid 3D bioprinting of decellularized extracellular matrix with regionally varied mechanical properties and biomimetic microarchitecture, Biomaterials. (2018). 10.1016/j.biomaterials.2018.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hribar KC, Choi YS, Ondeck M, Engler AJ, Chen S, Digital plasmonic patterning for localized tuning of hydrogel stiffness, Adv. Funct. Mater (2014). 10.1002/adfm.201400274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ma X, Qu X, Zhu W, Li YS, Yuan S, Zhang H, Liu J, Wang P, Lai CSE, Zanella F, Feng GS, Sheikh F, Chien S, Chen S, Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting, Proc. Natl. Acad. Sci. U. S. A (2016). 10.1073/pnas.1524510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gouveia RM, Connon CJ, Biomechanical modulation therapy—a stem cell therapy without stem cells for the treatment of severe ocular burns, Transl. Vis. Sci. Technol 9 (2020) 1–11. 10.1167/tvst.9.12.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xia H, Li X, Gao W, Fu X, Fang RH, Zhang L, Zhang K, Tissue repair and regeneration with endogenous stem cells, Nat. Rev. Mater 3 (2018) 174–193. 10.1038/s41578-018-0027-6. [DOI] [Google Scholar]
- [29].Ramos T, Scott D, Ahmad S, An Update on Ocular Surface Epithelial Stem Cells: Cornea and Conjunctiva, Stem Cells Int. (2015). 10.1155/2015/601731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gouveia RM, Lepert G, Gupta S, Mohan RR, Paterson C, Connon CJ, Assessment of corneal substrate biomechanics and its effect on epithelial stem cell maintenance and differentiation, Nat. Commun (2019). 10.1038/s41467-019-09331-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chakraborty A, Dutta J, Das S, Datta H, Comparison of ex vivo cultivated human limbal epithelial stem cell viability and proliferation on different substrates, Int. Ophthalmol 33 (2013) 665–670. 10.1007/s10792-013-9765-z. [DOI] [PubMed] [Google Scholar]
- [32].Bonnans C, Chou J, Werb Z, Remodelling the extracellular matrix in development and disease, Nat. Rev. Mol. Cell Biol 15 (2014) 786–801. 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gesteira TF, Sun M, Coulson-Thomas YM, Yamaguchi Y, Yeh LK, Hascall V, Coulson-Thomas VJ, Hyaluronan rich microenvironment in the limbal stem cell niche regulates limbal stem cell differentiation, Investig. Ophthalmol. Vis. Sci 58 (2017) 4407–4421. 10.1167/iovs.17-22326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cho IJ, Lui PPW, Obajdin J, Riccio F, Stroukov W, Willis TL, Spagnoli F, Watt FM, Mechanisms, Hallmarks, and Implications of Stem Cell Quiescence, Stem Cell Reports. 12 (2019) 1190–1200. 10.1016/j.stemcr.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cooper J, Giancotti FG, Integrin Signaling in Cancer: Mechanotransduction, Stemness, Epithelial Plasticity, and Therapeutic Resistance, Cancer Cell. 35 (2019) 347–367. 10.1016/j.ccell.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cho IJ, Lui PPW, Obajdin J, Riccio F, Stroukov W, Willis TL, Spagnoli F, Watt FM, Mechanisms, Hallmarks, and Implications of Stem Cell Quiescence, Stem Cell Reports. 12 (2019) 1190–1200. 10.1016/j.stemcr.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang YAZ, Plane JM, Jiang P, Zhou CJ, Deng W, Concise review: Quiescent and active states of endogenous adult neural stem cells: Identification and characterization, Stem Cells. 29 (2011) 907–912. 10.1002/stem.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sagga N, Kuffová L, Vargesson N, Erskine L, Collinson JM, Limbal epithelial stem cell activity and corneal epithelial cell cycle parameters in adult and aging mice, Stem Cell Res. 33 (2018) 185–198. 10.1016/j.scr.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chen S-Y, Han B, Zhu Y-T, Mahabole M, Huang J, Beebe DC, Tseng SCG, HC-HA/PTX3 Purified From Amniotic Membrane Promotes BMP Signaling in Limbal Niche Cells to Maintain Quiescence of Limbal Epithelial Progenitor/Stem Cells, Stem Cells. 33 (2015) 3341–3355. 10.1002/stem.2091. [DOI] [PubMed] [Google Scholar]
- [40].Ouyang H, Xue Y, Lin Y, Zhang X, Xi L, Patel S, Cai H, Luo J, Zhang M, Zhang M, Yang Y, Li G, Li H, Jiang W, Yeh E, Lin J, Pei M, Zhu J, Cao G, Zhang L, Yu B, Chen S, Fu XD, Liu Y, Zhang K, WNT7A and PAX6 define corneal epithelium homeostasis and pathogenesis, Nature. (2014). 10.1038/nature13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Shirahama H, Lee BH, Tan LP, Cho NJ, Precise tuning of facile one-pot gelatin methacryloyl (GelMA) synthesis, Sci. Rep (2016). 10.1038/srep31036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhu W, Cui H, Boualam B, Masood F, Flynn E, Rao RD, Zhang ZY, Zhang LG, 3D bioprinting mesenchymal stem cell-laden construct with core-shell nanospheres for cartilage tissue engineering, Nanotechnology. 29 (2018). 10.1088/1361-6528/aaafa1. [DOI] [PubMed] [Google Scholar]
- [43].Vattulainen M, Ilmarinen T, Koivusalo L, Viiri K, Hongisto H, Skottman H, Modulation of Wnt/BMP pathways during corneal differentiation of hPSC maintains ABCG2-positive LSC population that demonstrates increased regenerative potential, Stem Cell Res. Ther 10 (2019) 236. 10.1186/s13287-019-1354-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bojic S, Hallam D, Alcada N, Ghareeb A, Queen R, Pervinder S, Buck H, Amitai Lange A, Figueiredo G, Rooney P, Stojkovic M, Shortt A, Figueiredo FC, Lako M, CD200 Expression Marks a Population of Quiescent Limbal Epithelial Stem Cells with Holoclone Forming Ability, Stem Cells. 36 (2018) 1723–1735. 10.1002/stem.2903. [DOI] [PubMed] [Google Scholar]
- [45].Barbara V, Testa A, Di Iorio E, Mavilio F, Pellegrini G, De Luca M, C/EBPδ regulates cell cycle and self-renewal of human limbal stem cells, J. Cell Biol (2007). 10.1083/jcb.200703003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mentink RA, Rella L, Radaszkiewicz TW, Gybel T, Betist MC, Bryja V, Korswagen HC, The planar cell polarity protein VANG-1/Vangl negatively regulates Wnt/β-catenin signaling through a Dvl dependent mechanism, PLoS Genet. 14 (2018). 10.1371/journal.pgen.1007840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Katoh M, Canonical and non-canonical WNT signaling in cancer stem cells and their niches: Cellular heterogeneity, omics reprogramming, targeted therapy and tumor plasticity (Review), Int. J. Oncol 51 (2017) 1357–1369. 10.3892/ijo.2017.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Chavali M, Klingener M, Kokkosis AG, Garkun Y, Felong S, Maffei A, Aguirre A, Non-canonical Wnt signaling regulates neural stem cell quiescence during homeostasis and after demyelination, Nat. Commun 9 (2018) 1–17. 10.1038/s41467-017-02440-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sugimura R, He XC, Venkatraman A, Arai F, Box A, Semerad C, Haug JS, Peng L, Zhong XB, Suda T, Li L, Noncanonical Wnt signaling maintains hematopoietic stem cells in the niche, Cell. 150 (2012) 351–365. 10.1016/j.cell.2012.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].ET V, J D, W L, B L, L S, Three-dimensional cell culture models for anticancer drug screening: Worth the effort?, J. Cell. Physiol 233 (2018) 2993–3003. 10.1002/JCP.26052. [DOI] [PubMed] [Google Scholar]
- [51].Ma X, Liu J, Zhu W, Tang M, Lawrence N, Yu C, Gou M, Chen S, 3D bioprinting of functional tissue models for personalized drug screening and in vitro disease modeling, Adv. Drug Deliv. Rev 132 (2018) 235–251. 10.1016/J.ADDR.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].He L, Nguyen NB, Ardehali R, Zhou B, Heart regeneration by endogenous stem cells and cardiomyocyte proliferation: Controversy, fallacy, and progress, Circulation. 142 (2020) 275–291. 10.1161/CIRCULATIONAHA.119.045566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lin H, Ouyang H, Zhu J, Huang S, Liu Z, Chen S, Cao G, Li G, Signer RAJ, Xu Y, Chung C, Zhang Y, Lin D, Patel S, Wu F, Cai H, Hou J, Wen C, Jafari M, Liu X, Luo L, Zhu J, Qiu A, Hou R, Chen B, Chen J, Granet D, Heichel C, Shang F, Li X, Krawczyk M, Skowronska-Krawczyk D, Wang Y, Shi W, Chen D, Zhong Z, Zhong S, Zhang L, Chen S, Morrison SJ, Maas RL, Zhang K, Liu Y, Lens regeneration using endogenous stem cells with gain of visual function, Nature. (2016). 10.1038/nature17181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Mobaraki M, Abbasi R, Vandchali SO, Ghaffari M, Moztarzadeh F, Mozafari M, Corneal repair and regeneration: Current concepts and future directions, Front. Bioeng. Biotechnol 7 (2019) 135. 10.3389/fbioe.2019.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wright B, Mi S, Connon CJ, Towards the use of hydrogels in the treatment of limbal stem cell deficiency, Drug Discov. Today 18 (2013) 79–86. 10.1016/j.drudis.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Selver ÖB, Yağci A, Eğrilmez S, Gürdal M, Palamar M, Çavuşoğlu T, Ateş U, Veral A, Güven Ç, Wolosin JM, Limbal stem cell deficiency and treatment with stem cell transplantation, Turkish J. Ophthalmol (2017). 10.4274/tjo.72593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].HH H, S Y, X M, L K, G V, N L, X W, H. S, W Z, S C, High throughput direct 3D bioprinting in multiwell plates, Biofabrication. 13 (2020). 10.1088/1758-5090/AB89CA. [DOI] [PubMed] [Google Scholar]
- [58].Daley WP, Peters SB, Larsen M, Extracellular matrix dynamics in development and regenerative medicine, J. Cell Sci 121 (2008) 255–264. 10.1242/jcs.006064. [DOI] [PubMed] [Google Scholar]
- [59].Yui S, Azzolin L, Maimets M, Pedersen MT, Fordham RP, Hansen SL, Larsen HL, Guiu J, Alves MRP, Rundsten CF, Johansen JV, Li Y, Madsen CD, Nakamura T, Watanabe M, Nielsen OH, Schweiger PJ, Piccolo S, Jensen KB, YAP/TAZ-Dependent Reprogramming of Colonic Epithelium Links ECM Remodeling to Tissue Regeneration, Cell Stem Cell. 22 (2018) 35–49.e7. 10.1016/j.stem.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Chermnykh E, Kalabusheva E, Vorotelyak E, Extracellular matrix as a regulator of epidermal stem cell fate, Int. J. Mol. Sci 19 (2018). 10.3390/ijms19041003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Meran L, Baulies A, Li VSW, Intestinal Stem Cell Niche: The Extracellular Matrix and Cellular Components, Stem Cells Int. 2017 (2017). 10.1155/2017/7970385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kim Y, Kumar S, CD44-mediated adhesion to hyaluronic acid contributes to mechanosensing and invasive motility, Mol. Cancer Res 12 (2014) 1416–1429. 10.1158/1541-7786.MCR-13-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lam J, Truong NF, Segura T, Design of cell-matrix interactions in hyaluronic acid hydrogel scaffolds, Acta Biomater. 10 (2014) 1571–1580. 10.1016/j.actbio.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Frith JE, Mills RJ, Cooper-White JJ, Lateral spacing of adhesion peptides influences human mesenchymal stem cell behaviour, J. Cell Sci 125 (2012) 317–327. 10.1242/jcs.087916. [DOI] [PubMed] [Google Scholar]
- [65].Zheng M, Tian C, Fan T, Xu B, Fibronectin regulates the self-renewal of rabbit limbal epithelial stem cells by stimulating the Wnt11/Fzd7/ROCK non-canonical Wnt pathway, Exp. Eye Res 185 (2019) 107681. 10.1016/j.exer.2019.05.021. [DOI] [PubMed] [Google Scholar]
- [66].Tseng SCG, Chen SY, Mead OG, Tighe S, Niche regulation of limbal epithelial stem cells: HC-HA/PTX3 as surrogate matrix niche, Exp. Eye Res 199 (2020) 108181. 10.1016/j.exer.2020.108181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Chen W, Dong J, Haiech J, Kilhoffer MC, Zeniou M, Cancer stem cell quiescence and plasticity as major challenges in cancer therapy, Stem Cells Int. 2016 (2016). 10.1155/2016/1740936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].De Angelis ML, Francescangeli F, La Torre F, Zeuner A, Stem cell plasticity and dormancy in the development of cancer therapy resistance, Front. Oncol 9 (2019) 626. 10.3389/fonc.2019.00626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Rehman SK, Haynes J, Collignon E, Brown KR, Wang Y, Nixon AML, Bruce JP, Wintersinger JA, Singh Mer A, Lo EBL, Leung C, Lima-Fernandes E, Pedley NM, Soares F, McGibbon S, He HH, Pollet A, Pugh TJ, Haibe-Kains B, Morris Q, Ramalho-Santos M, Goyal S, Moffat J, O’Brien CA, Colorectal Cancer Cells Enter a Diapause-like DTP State to Survive Chemotherapy, Cell. 184 (2021) 226–242.e21. 10.1016/j.cell.2020.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw/processed data required to reproduce these findings can be shared by the authors upon request.