Abstract
Cryptosporidiosis is an important disease in neonatal calves, causing watery diarrhoea, loss of appetite, and production losses. Dehydration from diarrhoea often results in the calf requiring rehydration or veterinary treatment to prevent calf mortality. Transmission of Cryptosporidium to calves still has some major knowledge gaps, such as the initial source of oocysts ingested by calves and how these oocysts can persist between calving periods. Some studies have examined the role of adult cattle in the transmission of Cryptosporidium oocysts, although these have yielded inconclusive results. In this study, highly sensitive oocyst extraction from faeces and detection techniques, sensitive to 5 oocysts per gram using a 50 g sample, were used to genotype faecal samples from adult cattle and their calves to determine if adult cattle could be a source of Cryptosporidium infection for their calves. On a dairy farm, faecal samples from adult cattle were collected twice per week for 0–3 weeks before calving and from their calves three times per week until they reached 3 weeks of age followed by twice per week until they reached 6 weeks of age. On a beef farm, samples were collected from both adults and calves at a single time point. Faecal samples were examined to compare species and multilocus genotypes of Cryptosporidium parvum. Results show that C. parvum was the most prevalent species on both the dairy and beef farms. The calves within each herd appear to have one predominant single multilocus genotype, whereas adult cattle have multiple distinct genotypes. Adult cattle on the dairy farm, tested before calving, in the majority of cases had a multilocus genotype that is different from that detected in their calves. On the beef farm, where samples were taken at the same time, the majority of adult cattle matched the multilocus genotype of their calves. This study shows that adult cattle display a higher diversity of C. parvum genotypes on both farms compared to the calves. The data also represent a detailed longitudinal prevalence study of the shedding profiles and genotype of Cryptosporidium parasites detected in dairy calves from birth to 6 weeks of age.
Keywords: Cryptosporidiosis, Cryptosporidium parvum, Transmission, Cattle, Longitudinal study
Graphical abstract
Highlights
-
•
On a dairy farm, the majority of adult cattle had a multilocus genotype that was different to those in calves.
-
•
On the beef farm, the majority of adult cattle matched the multilocus genotype of their calves.
-
•
Calves on the dairy farm sampled longitudinally exhibited intermittent shedding of Cryptosporidium oocysts.
-
•
Increased C. parvum genotype diversity was observed in the adult cattle compared to the calves.
1. Introduction
Cryptosporidiosis is caused by the protozoan parasites of the genus Cryptosporidium and is an important disease on many farms worldwide (Ryan et al., 2016). Despite there being over 44 recognised species to date (Holubová et al., 2020), Cryptosporidium parvum is the only species that is known to cause clinical disease in naturally infected calves (Thomson et al., 2017) although some evidence exists detailing reduced milk yield in adult cattle shedding Cryptosporidium andersoni (see Esteban & Anderson, 1995; Anderson, 1998). The symptoms of cryptosporidiosis include watery diarrhoea, loss of appetite, depression, and dehydration, which can become severe enough to cause calf mortality (Thomson et al., 2017). Oocyst shedding starts around 2–6 days of age and lasts between 7 and 15 days (Tzipori et al., 1983; Uga et al., 2000). During this time, each infected calf will shed millions of oocysts, ranging from 3.89 × 1010 in a 6–12 day-old calf to 3.8 × 107 in a 50–56 day-old calf (Nydam et al., 2001). This results in huge environmental contamination and a risk to other young calves vulnerable to transmission and infection from these oocysts (Nydam et al., 2001).
Cryptosporidiosis has been reported as being one of the leading causes of neonatal calf diarrhoea worldwide (Thomson et al., 2017). Young calves between 7 and 14 days post-infection have a significant reduction in growth rate (Klein et al., 2008), and this reduction is maintained over six months (Shaw et al., 2020). It has been shown that severe clinical cryptosporidiosis early in life results in an average difference in weight gain of 34 kg at six months of age, potentially costing 130 GBP per infected calf (Shaw et al., 2020). Cryptosporidiosis is difficult to tackle on-farm as the parasite oocyst possesses a strong outer shell, rendering it resistant to many of the common disinfectants used on farms. It requires a very low dose of oocysts to infect and cause disease in neonatal livestock (de Graaf et al., 1999; Zambriski et al., 2013, Zambriski et al., 2013).
Cryptosporidium is transmitted via the faecal-oral route, and the potential of adult cattle as a source of oocysts to infect naïve calves is disputed in the literature. Young calves tend to show clinical signs of infection in the second week of life (Faubert & Litvinsky, 2000) and so this would suggest that infection occurs very soon after birth, due to the 5–7 day incubation period of the parasite (Tzipori et al., 1983). Prevalence of C. parvum in adult cattle ranges from 0 to 86.2% (Lorenzo et al., 1993; Atwill & Pereira, 2003; Wells et al., 2015; Thomson et al., 2019) but different detection methods were used, with only the two most recent studies using a very sensitive method developed to detect Cryptosporidium oocysts in 50 g of faeces (Wells, et al., 2015; Thomson, et al., 2019). The consensus from the older papers is that adult cattle do not play a significant role in transmission to other cattle or environmental contamination due to the low number of C. parvum-positive samples obtained. However, these studies did not use the most sensitive techniques for oocyst detection and so prevalence may have been under-reported. Not only this but it has been documented that as few as 17 oocysts are required to infect a calf (Zambriski et al., 2013, Zambriski et al., 2013) and so even excretion in low concentrations could pose a risk, especially as adult cattle produce much larger faecal pats compared to the calves and therefore the total number of oocysts shed into the environment by adult cattle should be considered when looking at sources of transmission.
The main issue with previous studies is the difficulty in analysing faecal samples from adult cattle for Cryptosporidium oocysts. The larger sample volume, reduced oocyst number, and fibrous nature of the faeces makes laboratory detection from adult faecal samples difficult (Wells et al., 2016). A recent report described a highly sensitive technique for detecting Cryptosporidium oocysts, using 50 g of faeces in an acid flocculation and salt flotation technique, which effectively deals with extraction from adult faeces. This method has a sensitivity of 5 oocysts per gram and allows for 50 g to be analysed rather than the 2–4 g used in previous techniques (Wells et al., 2016). Higher prevalences of C. parvum in adult cattle were found in the studies using this technique compared to those which used alternative techniques (Lorenzo et al., 1993; Atwill & Pereira, 2003). Two studies conducted using this sensitive extraction technique found that the predominant species in adult cattle was C. parvum (Wells et al., 2015; Thomson et al., 2019), and not C. andersoni as reported by older studies using previous detection methods. Previous work described in all of the aforementioned studies either lacked direct pairings between the dam and calf or did not genotype at multiple loci.
This study aims to examine multilocus genotypes of both adult cattle and their calves to further explore the genetic diversity of C. parvum. This information will be vital in understanding potential sources of infection for calves. At present, very few studies describe the shedding profile of individual calves on a longitudinal basis and only one of these has been conducted in Scotland (Thomson et al., 2019). This information is valuable when it comes to controlling this parasite on the farm. This study provides detailed information on the species and genotypes of Cryptosporidium parasites shed over six weeks in calves from birth on a Scottish dairy farm.
2. Materials and methods
2.1. Sample collection
In Study 1 (Dairy farm), adult cattle faecal samples were mixed and 125 ml collected using a plastic collection pot following observation of defecation. To reduce environmental contamination, only the freshly deposited faeces were collected, avoiding any straw bedding. Samples were collected from 79 adult Holstein cattle twice a week between 0 and 3 weeks before calving. These were all the adult cattle that were due to calf during the study period. Due to the nature of this collection method, it was not always possible to collect a sample from each individual at every time point. Samples were stored at 4 °C until processed.
Once the adults had given birth, female calves were removed from their mothers within 12 h, placed in individual pens, and fed from pooled colostrum. Male calves were not kept on-farm and so could not be included in the study. Faecal samples were collected from the female calves three times per week (n = 38), for 3 weeks, followed by twice per week for weeks 4–6. The entire motion was collected following observation of each individual, inside a plastic bag. It was not always possible to gain a sample from all calves at every sampling point.
In Study 2 (Beef farm), adult cattle faecal samples were mixed and 125 ml collected using a plastic collection pot following observation of defecation. These samples were collected at a single time point from all adult Belgian Blue female cattle (n = 27) and 23 of their Belgian Blue × Limousin, mixed-gender calves, which were all housed together and allowed to suckle colostrum and milk from their mothers. These calves ages ranged from 0 to 4 weeks. The entire motion was collected following observation of each individual, inside a plastic bag, avoiding bedding material. Five of the calves died during the study. All were positive for cryptosporidiosis although other factors were involved. Therefore not every adult has a matched calf sample for comparison.
2.2. Sample processing
All faecal samples from adult cattle were processed according to the protocol described in Wells et al. (2016), which had a detection limit of 5 oocysts per gram of faeces. This included using 50 g of starting material in an acid flocculation, followed by a salt flotation on the pellet. Faecal samples from calves were thoroughly mixed before 3 g of faeces was removed and added to 1 ml TE buffer (10 mM Tris-HCl, 0.5 mM EDTA). Both adult and calf samples underwent 10× freeze/thaw cycles in liquid nitrogen before DNA was extracted using a modified Macherey Nagal tissue kit protocol (NZ740952250, Macherey-Nagel, Duren, Germany), which is described in Wells et al. (2016).
2.3. 18S nssm PCR
The species of Cryptosporidium present was determined using a multiplex nested species-specific PCR, which amplified the 18S region (Thomson et al., 2019) and allowed the identification of the common cattle species C. parvum, Cryptosporidium bovis, Cryptosporidium ryanae, and C. andersoni. Modifications to this PCR protocol were applied during the second round of PCR, where first-round PCR products were not diluted as a template for the second round PCR when testing the adult cattle samples. Calf DNA samples underwent the PCR as previously described (Thomson et al., 2019). All DNA samples were run in triplicate with a negative control, a DNA extraction control, and a positive control. Cycling conditions were 3 min at 94 °C, followed by 35 cycles of 45 s at 94 °C, 45 s at 56 °C, and 1 min at 72 °C. The final extension was 7 min at 72 °C. Secondary PCR products were visualized following electrophoresis on a 1.5% agarose gel stained with GelRed™ (Biotium) on an AlphaImager 2000. This multiplex PCR method was tested against known positive samples for the four common cattle species during its development (Thomson et al., 2019). A proportion of positive samples from this study was also confirmed via sequencing by submitting positive samples to MWG Eurofins.
2.4. gp60 PCR
The 450 bp fragment of the gp60 gene was amplified from all C. parvum-positive samples using a previously described nested PCR (Brook et al., 2009) and sequenced by GATC Biotech (Köln, Germany). Genotype for the gp60 was assigned according to Sulaiman et al. (2005).
2.5. Multi-locus genotyping
Four microsatellite markers (MM5, MM18, MM19 and TP14) described previously (Mallon et al., 2003; Morrison et al., 2008), were used to determine amplicon length for each allele at each locus and therefore assign a multilocus genotype (MLG) across the four markers to each sample. Amplicon lengths and locus assignments are shown in Table 1. This reaction included 10× PCR buffer (45 mM Tris-HCl pH 8.8, 11 mM (NH4)2SO4, 4.5 mM MgCl2, 4.4 μM EDTA, 113 μg/ml BSA, 1 mM each of the four deoxyribonucleotide triphosphates) (Burrells et al., 2013), 0.5 units of Bioline BioTaq, and 10 μM of forward and reverse primers. The PCR protocol was modified to the one described by removing the 1:100 dilution of the primary product for adult cattle samples. A positive control for each fragment size of all four loci was included in the PCR, as was a negative (no template) control.
Table 1.
Amplicon length (bp) for each of the MLG alleles described
| Allele | MM5 | MM18 | MM19 | TP14 |
|---|---|---|---|---|
| 1 | 262 | 288 | 298 | 296 |
| 2 | 235 | 294 | 304 | 304 |
| 3 | 225 | 318 | 292 | 252 |
| 4 | 412 | 316 | ||
| 5 | 270 | |||
| 6 | 253 | |||
| 7 | 281 | |||
| 8 | 285 | |||
| 9 | 495 | |||
| 10 | 222 |
PCR products underwent fragment analysis (Applied Biosystems; University of Dundee, UK) using Genescan ROX500 and the results were analysed using STRand (https://www.vgl.ucdavis.edu/informatics/strand.php). MLG visualisation was achieved using Phyloviz (https://www.phyloviz.net) in which the allele sizes for loci gp60, MM5, MM18, MM19, and TP14 were inputted.
3. Results
3.1. Species present in adult cattle in Study 1
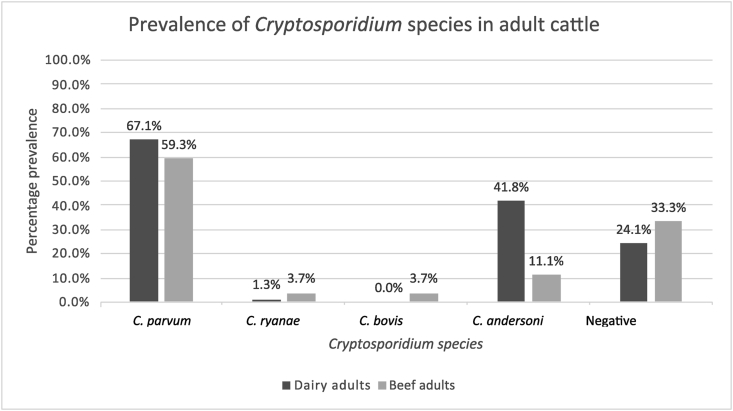
Adult dairy cattle in Study 1 had a C. parvum prevalence of 67.1% (±10.4%). This includes cattle shedding C. parvum only (n = 26) and C. parvum as a mixed infection with other species (n = 27). Twenty-six of these mixed infections were with C. andersoni and one with C. ryanae. The only other species detected in the dairy cattle as a single infection was C. andersoni (8.9% (±6.3%)). The rest of the cattle, 24.1% (±9.5%) were negative for the four most common species of Cryptosporidium found in cattle (Fig. 1).
Fig. 1.
Prevalence of Cryptosporidium species in adult cattle on both a dairy (Study 1) and a beef (Study 2) farm. Mixed infections are accounted for by including them in each of the species categories, therefore the total will exceed 100%
3.2. Species present in adult cattle in Study 2
Of the 27 samples collected from adult beef cattle, 59.3% (±18.6%) contained C. parvum. One of these was a mixed infection with C. andersoni, C. bovis, and C. ryanae. The only other species detected in adult beef cattle as a single species was C. andersoni, detectable in 7.4% (±7.4%) of cattle. No detectable Cryptosporidium species were present in 33.3% (±17.7%) of cattle (Fig. 1).
3.3. Species present in calves in Study 1
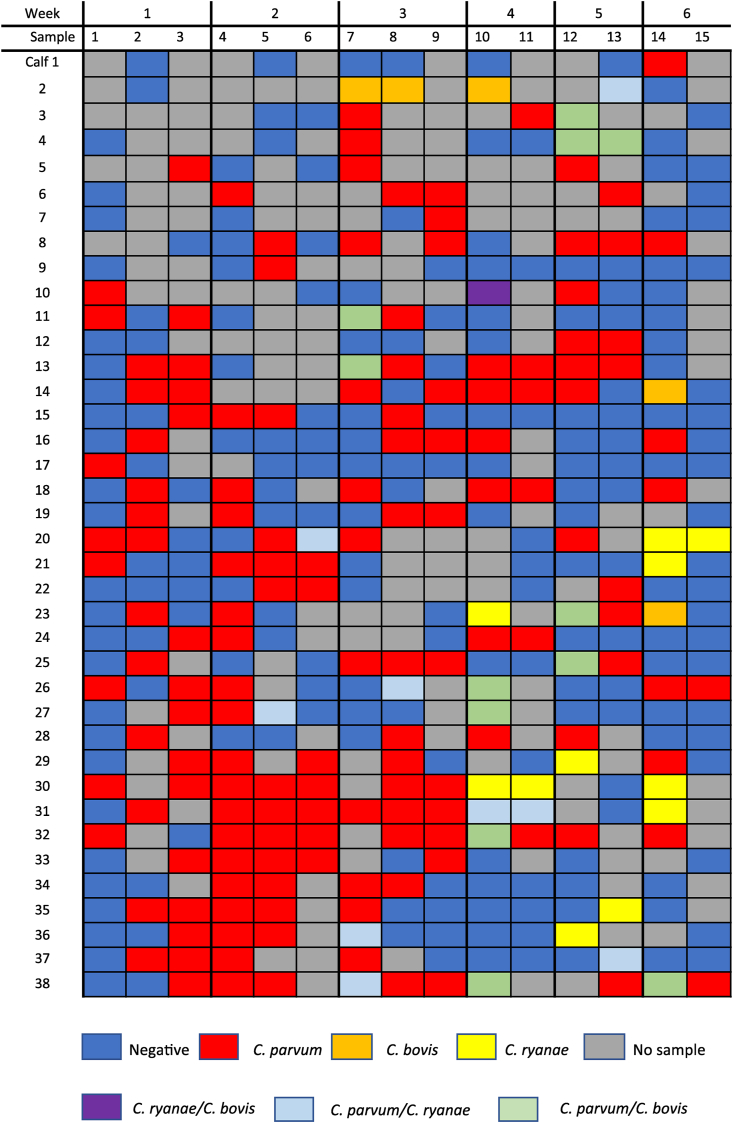
Female calves on the dairy farm (n = 38) were tested for Cryptosporidium species from birth until they reached 6 weeks of age (Fig. 2). A positive C. parvum result from any of the samples that week was counted as C. parvum-positive. If multiple species were found during that week, either in a single sample or between samples within a week, it was counted as a mixed infection.
Fig. 2.
Longitudinal prevalence data for Cryptosporidium species in dairy calves
In total, there were 15 sampling points for each calf which totalled 400 faecal samples. The average number of samples obtained was 10.4 per calf but this ranged between 6 and 15 samples. Of these 400 samples, 180 were positive for C. parvum giving a sample prevalence of 45% (±4.9%). For individual calves, the prevalence of C. parvum in all collected faecal samples ranged from 8.3 to 90.9%. The lowest C. parvum prevalence was observed in the final sampling week at 8.3% and the highest observed at sampling timepoint three which occurred in the first week of life at 68.2%.
Results also detail longitudinal species prevalence data for the 38 individual calves, which are shown in Fig. 2. Of the 38 calves, 25 showed intermittent shedding of C. parvum oocysts. In the majority of these cases, the gap was only observed in a single sample (n = 15) but one calf had four gaps in C. parvum shedding. Two calves had four negative samples between C. parvum-positive samples. No calves remained negative for the full 6-week period, with every calf testing positive for C. parvum at some point over the sampling period (Fig. 2). Calves tended to be positive for C. parvum in the first two weeks of life, although C. parvum was detected at all sampling time points, whereas C. bovis and C. ryanae tended to be detected in weeks 3–6. In two instances (calves 9 and 17), C. parvum was only detected once early in infection and no Cryptosporidium was detected thereafter. This could suggest that infection in these calves was effectively controlled. While the overall pattern also indicates control of C. parvum, several calves (e.g. calf 16, 18 and 26) were shedding Cryptosporidium from week 1 to week 6. While a few calves were positive at all sampling time points during a week (e.g. calves 30–33 in week 2), in the majority of cases there was variable detection of parasites in samples taken during a week and between weeks, indicating variable and intermittent shedding of oocysts, for all three Cryptosporidium species. For most calves, a parasite species detected once was also detected on a further occasion, indicating infection establishment and persistence and providing reassurance that the experimental setup was robust.
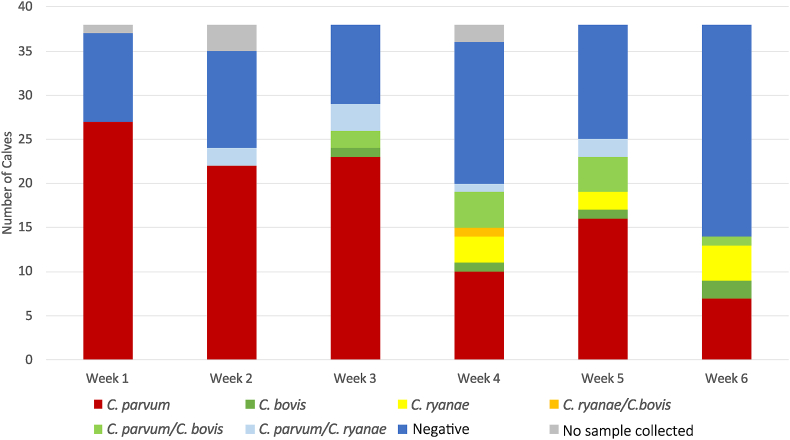
Cryptosporidium species present in dairy calves from birth over a 6-week period are summarised in Fig. 3. In week 1, 73% (±14.3%) were positive for C. parvum, with the remainder having no detectable Cryptosporidium. In week 2, 62.9% (±16%) were positive for C. parvum with two calves having a C. parvum + C. ryanae mixed infection. In week 3, 61% (±15.5%) were positive for C. parvum with two calves showing a C. parvum + C. bovis mixed infection and three showing a C. parvum + C. ryanae mixed infection. One calf was positive for C. bovis only. In week 4, 27.8% (±14.6%) were positive for C. parvum. One calf was positive for C. bovis and three calves for C. ryanae. One calf was positive for C. ryanae + C. bovis as a mixed infection, four calves had a C. parvum + C. bovis mixed infection, and one a C. parvum + C. ryanae mixed infection. At 5 weeks, 42.1% (±15.7%) were positive for C. parvum, one for C. bovis, and two calves for C. ryanae. Four calves had a C. parvum and C. bovis mixed infection and two calves a C. parvum + C. ryanae mixed infection. In week 6, 18.4% (±12.3%) were positive for C. parvum, two for C. bovis, and four for C. ryanae. There was also one C. parvum + C. bovis mixed infection. No samples were able to be collected for one calf in week 1, three calves in week 2 and two calves in week 4.
Fig. 3.
Species of Cryptosporidium present in dairy calves (Study 1) in the first six weeks of life
3.4. Species present in calves in Study 2
On the beef farm, 23 calves were tested, and all of them were positive for C. parvum. No other species was found either alone or as a mixed infection.
3.5. gp60 subtyping in Study 1
The adult cattle samples that were positive for C. parvum underwent sequence typing at the gp60 gene. The genotypes present were IIaA15R1 (n = 26), IIaA15G2R1 (n = 22), IIcA5G3 (n = 2), and IIaA17G1R1 (n = 1). We were unable to sequence two samples at the gp60 gene. The first sample containing C. parvum oocysts from each of the 38 calves was chosen for gp60 subtyping. All of the 38 C. parvum-positive calves had the gp60 genotype IIaA15G2R1.
3.6. gp60 subtyping in Study 2
In Study 2, 16 adult cattle and 23 calves gave a positive test for C. parvum on the beef farm and these underwent gp60 genotyping. All except for one adult cow were positive on amplification of this gene. The predominant gp60 genotype was IIaA17G1R1, which was present in 63% (10/16) of the adult cattle and 100% (23/23) of the C. parvum-positive calves. IIaA15R1, IIaA19G2R1, IIaA16G3R1 were found in 19% (3/16), 6% (1/16), and 6% (1/16) of the adult cattle respectively.
3.7. Multilocus genotyping
3.7.1. Study 1
Those adult cattle that were the mothers of the 38 calves in this study were chosen for microsatellite analysis. The sample selected for adult cattle was the C. parvum-positive sample collected closest to the time of calving, as this was the most likely genotype to be shed at birth. The calf sample chosen was the first detected C. parvum-positive in the first two weeks of life to characterise the primary infection source to the calf. Of the 38 adults, 28 were positive for C. parvum and underwent further genotyping. Amplification at all five alleles was successful for 19 out of 28 adults. Of the 38 calves, 34 were positive for C. parvum in the first two weeks of life. For the calf samples, 32 out of 34 were successfully assigned an MLG by amplifying at all five loci.
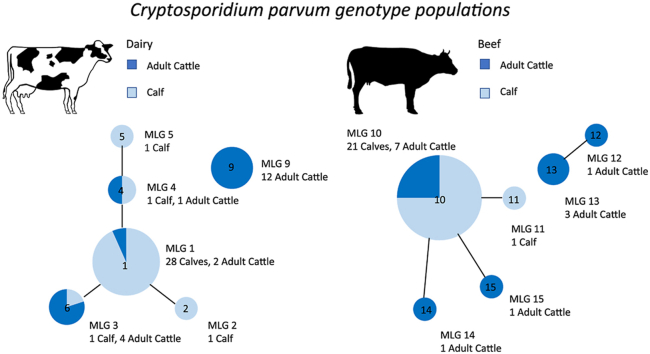
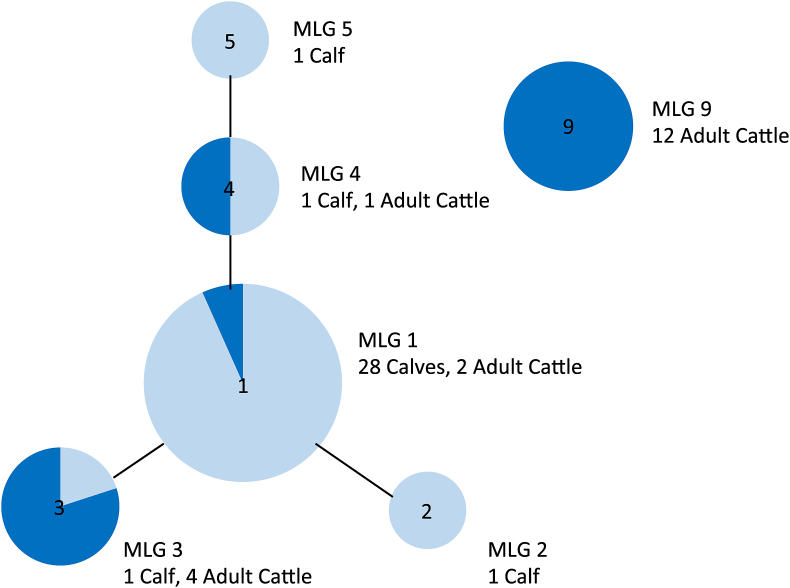
Results show that 28 calves and 2 adult cattle had MLG 1, although only one of these was a matched dam and calf pair. A single calf had MLG 2. One calf had MLG 3 which was shared by four of the adult cattle, none of which was the dam of that calf. One calf and one adult had MLG 4 and they were also unrelated. One calf was shedding MLG 5 and the majority of adult cattle (n = 12) were shedding MLG 9. The multilocus genotype results for adult cattle and their matched calves are provided in Table 2. The relationship between these MLGs as a whole dataset is visualised in Fig. 4 showing the relatedness between the MLGs. Those MLGs which are joined by a line share 4 out of the 5 alleles and so there is a smaller difference between these two multilocus genotypes compared to those which are not joined by a line. This information is vital as it shows how related the genotypes are.
Table 2.
Multilocus genotypes between matched calves and their mothers for C. parvum on a dairy farm (Study 1)
| Calf | Calf gp60 | Calf MM5 | Calf MM18 | Calf MM19 | Calf TP14 | Calf MLG | Dam gp60 | Dam MM5 | Dam MM18 | Dam MM19 | Dam TP14 | Dam MLG |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | × | × | × | × | × | × | IIaA15R1/IIaA15G2R1 | 2 | 1 | 3 | 1 | 9 |
| 2 | × | × | × | × | × | × | IIaA15R1/IIaA15G2R1 | 2 | 1 | 3 | 1 | 9 |
| 3 | × | × | × | × | × | × | IIaA15R1 | 2 | 1 | 3 | 1 + 2 | 9 |
| 4 | IIaA15G2R1 | 2 | 1 | 3 + 1 | 2 | 4 | IIaA15G2R1 | 2 | 1 | 1 | 2 | 1 |
| 5 | IIaA15G2R1 | 2 | 2 | 3 + 1 | 2 | 5 | × | × | × | × | × | × |
| 6 | IIaA15G2R1 | 2 | 1 | 1 | 1 | 3 | IIaA15R1 | 2 | 1 | 3 | 1 | 9 |
| 7 | IIaA15G2R1 | 2 | 2 | 1 | 2 | 2 | × | × | × | × | × | × |
| 8 | IIaA15G2R1 | 2 | 1 | 1 | 2 | 1 | IIaA15R1 + IIcA5G3 | 2 | 1 | 3 | 1 | 9 |
| 9 | IIaA15G2R1 | 2 | 1 | 1 | 2 | 1 | IIaA17G1R1 | × | × | 1 | 1 | × |
| 10 | IIaA15G2R1 | 2 | 1 | 1 | 2 | 1 | IIaA15R1 | 2 | 1 | 3 | 1 | 9 |
| 11 | IIaA15G2R1 | 2 | 1 | 1 | 2 | 1 | IIaA15R1 | 2 | 1 | 3 | 1 | 9 |
| 12 | × | × | × | × | × | × | × | × | × | × | × | × |
| 13 | IIaA15G2R1 | 2 | 1 | 1 | 2 | 1 | IIaA15G2R1 | 2 | 1 | 1 | 1 | 3 |
| 14 | IIaA15G2R1 | 2 | 1 | 1 | 2 | 1 | IIaA15R1 | 2 | 1 | 3 | 1 | 9 |
| 15 | IIaA15G2R1 | 2 | 1 | 1 | 2 | 1 | × | × | × | × | × | × |
| 16 | IIaA15G2R1 | 2 | × | × | 2 | × | IIaA15G2R1 | × | × | × | 2 | × |
| 17 | llaA15G2R1 | × | × | × | × | × | × | × | × | × | × | × |
| 18 | llaA15G2R1 | 2 | 1 | 1 | 2 | 1 | IIaA15G2R1 | × | × | × | × | × |
| 19 | llaA15G2R1 | 2 | 1 | 1 | 2 | 1 | × | × | × | × | × | × |
| 20 | llaA15G2R1 | 2 | 1 | 1 | 2 | 1 | IIaA15G2R1 | 2 | 1 | 3 | 2 | 4 |
| 21 | llaA15G2R1 | 2 | 1 | 1 | 2 | 1 | × | × | × | × | × | × |
| 22 | llaA15G2R1 | 2 | 1 | 1 | 2 | 1 | × | × | × | × | × | × |
| 23 | llaA15G2R1 | 2 | 1 | 1 | 2 | 1 | IIaA15R1/IIaA15G2R1 | 2 | 1 | 3 | 1 | 9 |
| 24 | llaA15G2R1 | 2 | 1 | 1 | 2 | 1 | IIaA15G2R1 | 2 | 1 | 1 | 1 | 3 |
| 25 | llaA15G2R1 | 2 | 1 | 1 | 2 + 1 | 1 | IIaA15R1 | 2 | 1 | 3 | 1 | 9 |
| 26 | llaA15G2R1 | 2 | 1 | 1 | 2 | 1 | IIaA17G2R1 | × | × | × | 1 | × |
| 27 | llaA15G2R1 | 2 | 1 | 1 | 2 + 1 | 1 | IIaA15G2R1 | × | × | × | 1 | × |
| 28 | llaA15G2R1 | 2 | 1 | 1 | 2 | 1 | × | × | × | × | × | × |
| 29 | llaA15G2R1 | 2 | 1 | 1 | 2 | 1 | IIaA16G1R1 | × | × | × | 1 | × |
| 30 | llaA15G2R1 | 2 | 1 | 1 | 2 | 1 | llaA15R1 | × | × | × | × | × |
| 31 | llaA15G2R1 | 2 | 1 | 1 | 2 + 1 | 1 | IIaA15R1 | × | × | × | 1 | × |
| 32 | llaA15G2R1 | 2 | 1 | 1 | 2 | 1 | IIaA15R1 | × | × | × | 1 | × |
| 33 | llaA15G2R1 | 2 | 1 | 1 | 2 | 1 | IIaA15G2R1/IIaA15R1 | 2 | 1 | 1 | 1 | 3 |
| 34 | llaA15G2R1 | 2 | 1 | 1 | 2 | 1 | × | × | × | × | × | × |
| 35 | llaA15G2R1 | 2 | 1 | 1 | 2 | 1 | IIaA15R1 | 2 | 1 | 3 | 1 | 9 |
| 36 | llaA15G2R1 | 2 | 1 | 1 | 2 | 1 | IIaA15G2R1 | 2 | 1 | 1 | 1 | 3 |
| 37 | llaA15G2R1 | 2 | 1 | 1 | 2 | 1 | IIaA15R1/IIaA15G2R1 | 2 | 1 | 3 | 1 | 9 |
| 38 | llaA15G2R1 | 2 | 1 | 1 | 2 | 1 | IIaA15G2R1/IIaA17G1R1 | 2 | 1 | 1 | 2 | 1 |
Notes: An ‘×’ denotes that a genotype was not obtained at this locus, either because the sample was not C. parvum-positive or due to insufficient PCR amplification. Final MLG assignment is presented in bold. Matched calf and dam pairs have their MLG underlined.
Fig. 4.
Multilocus genotypes (MLGs) present in calves and their mothers on a dairy farm (Study 1). Each node represents a single MLG. The size of the node represents the number of animals with that MLG. Nodes are connected by a line if they share 4 out of 5 tested alleles. Dark blue represents adult cattle and light blue represents the calves
3.7.2. Study 2
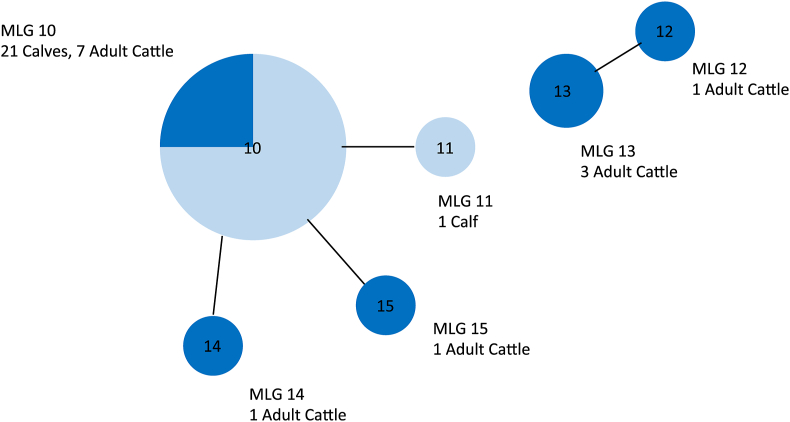
All C. parvum-positive cows and calves (16 and 23, respectively), underwent microsatellite analysis. Of the 16 positive adult cattle samples, 13 were amplified at all 5 alleles and were assigned an MLG. Positive calf samples (22 out of 23) were successfully amplified at all 5 alleles and were assigned an MLG. Results show that 21 calves and 7 adult cattle had MLG 10, and all 7 adult cattle shedding MLG 10 had the same MLG as their calves. A single calf had MLG 11, one adult was shedding MLG 15, and another MLG 14, which is different from MLG 10 only by a single allele. One adult was shedding MLG 12 and another adult was shedding MLG 13. The matched pairs between calves and their mothers are shown in Table 3 and the relationship between these MLGs as a whole dataset is visualised in Fig. 5.
Table 3.
Multilocus genotypes between matched calves and their mothers for C. parvum on a beef farm (Study 2)
| Calf | Calf gp60 | Calf MM5 | Calf MM18 | Calf MM19 | Calf TP14 | Calf MLG | Dam gp60 | Dam MM5 | Dam MM18 | Dam MM19 | Dam TP14 | Dam MLG |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IIaA17G1R1 | 2 | 1 | 3 | 1 | 10 | × | × | × | × | × | × |
| 2 | IIaA17G1R1 | 2 | 1 | 3 | 1 | 10 | × | × | × | × | 1 | × |
| 3 | IIaA17G1R1 | 2 | 1 | 3 | 1 | 10 | IIaA17G1R1 | 2 | 1 + 2 | 3 | 1 | 10 |
| 4 | IIaA17G1R1 | 2 | 1 | 10 + 3 | 1 | 11 | IIaA15R1 | 2 | 1 + 2 | 8 | 1 | 13 |
| 5 | IIaA17G1R1 | 2 | 1 | 3 + 9 | 1 | 10 | × | × | × | × | × | × |
| 6 | IIaA17G1R1 | 2 | 1 | 3 | 1 | 10 | × | × | × | × | × | × |
| 7 | IIaA17G1R1 | 2 | 1 | 3 | 1 | 10 | IIaA15R1 | 2 | 1 | 8 | 1 | 13 |
| 8 | IIaA17G1R1 | 2 | 1 | 3 | 1 | 10 | × | × | × | × | × | × |
| 9 | IIaA17G1R1 | 2 | 1 | 3 | 1 | 10 | IIaA17G1R1 | 2 | 1 | 3 | 1 | 10 |
| 10 | IIaA17G1R1 | 2 | 1 | 3 | 1 | 10 | IIaA17G1R1 | × | × | × | × | × |
| 11 | IIaA17G1R1 | 2 | 1 | 3 + 9 | 1 | 10 | IIaA17G1R1 | 2 | 1 | 3 | 1 | 10 |
| 12 | IIaA17G1R1 | 2 | 1 | 3 + 9 | 1 | 10 | IIaA17G1R1 | 2 | 4 | 3 | 1 | 14 |
| 13 | IIaA17G1R1 | 2 | 1 | 3 | 1 | 10 | × | × | × | × | × | × |
| 14 | IIaA17G1R1 | 2 | 1 | 3 | 1 | 10 | IIaA17G1R1 | 2 | 1 | 3 | 1 | 10 |
| 15 | IIaA17G1R1 | 2 | 1 | 3 | 1 | 10 | IIaA16G3R1 | 2 + 3 | 1 | 3 | 1 | 15 |
| 16 | IIaA17G1R1 | 2 | 1 | 3 | 1 | 10 | × | × | × | × | × | × |
| 17 | IIaA17G1R1 | 2 | 1 | 3 + 9 | 1 | 10 | IIaA19G2R1 | × | × | × | 1 | × |
| 18 | IIaA17G1R1 | 2 | 1 | 3 + 9 | 1 | 10 | IIaA19G2R1 | 2 | 1 | 8 | 1 | 12 |
| 19 | IIaA17G1R1 | 2 | 1 | 3 + 9 | 1 | 10 | IIaA17G1R1 | 2 + 3 | 1 | 3 | 1 | 10 |
| 20 | IIaA17G1R1 | 2 | 1 | 3 | 1 | 10 | IIaA17G1R1 | 2 | 1 | 3 | 1 | 10 |
| 21 | IIaA17G1R1 | 2 | 1 | 3 + 9 | 1 | 10 | × | × | × | × | × | × |
| 22 | IIaA17G1R1 | 2 | 1 | 3 + 9 | 1 | 10 | IIaA17G1R1 | 2 | 1 | 3 | 1 | 10 |
| 23 | Calf died | IIaA17G1R1 | × | × | × | 1 | × | |||||
| 24 | Calf died | IIaA15R1 | 2 | 1 | 8 | 1 | 13 | |||||
| 25 | IIaA17G1R1 | × | × | × | × | × | × | × | × | × | × | × |
Notes: An ‘×’ denotes that a genotype was not obtained at this locus, either because the sample was not C. parvum-positive or due to insufficient PCR amplification. Final MLG assignment is presented in bold. Matched calf and dam pairs have their MLG underlined.
Fig. 5.
Multilocus genotypes (MLGs) present in calves and their mothers for a beef farm (Study 2). Each node represents a single MLG. The size of the node represents the number of animals with that MLG. Nodes are connected by a line if they share four out of five tested alleles. Dark blue represents adult cattle and light blue represents the calves
4. Discussion
Cryptosporidium is a parasite of great importance on farms as it is one of the leading causes of neonatal calf diarrhoea. The role that adult cattle play in potentially being a source of C. parvum oocysts to their calves is currently unknown due to the difficulty of concentrating and detecting oocysts in adult faecal samples (Wells et al., 2016). Overall, C. parvum was the most prevalent species found in pre-weaned calves on both farms in this study, which is consistent with previous research conducted in Scotland, England, and Wales (Smith et al., 2014; Wells et al., 2015; Thomson et al., 2019). Cryptosporidium species of cattle tend to follow an infection pattern that depends on the age of the host animal, with neonatal and pre-weaned calves mostly shedding C. parvum, post-weaned calves with C. bovis and C. ryanae, and adult cattle with C. andersoni (Robertson et al., 2014; Thomson et al., 2019). Cryptosporidium ryanae was detected in one calf in the second week of life. Although uncommon, this species has previously been found in pre-weaned calves (Murakoshi et al., 2013; Åberg et al., 2020).
Longitudinal sampling results from birth to six weeks (Fig. 2) show that calves intermittently shed C. parvum oocysts over multiple sampling periods. Despite difficulties in collecting a sample at every time point for every calf, the data indicate that calves can shed C. parvum in their faeces (or the other Cryptosporidium species) on two occasions with negative results in-between. There are very few studies looking at longitudinal shedding profiles of calves and so these data provides evidence that calves shedding Cryptosporidium oocysts can do so intermittently during the pre-weaned stage. Existing studies in calves state that shedding increases, peaks, and then decreases rather than being shed intermittently (Nydam et al., 2001; Tiranti et al., 2011; Zambriski et al., 2013, Zambriski et al., 2013). However, two of these only examined the calves for 21 days (Nydam et al., 2001; Zambriski et al., 2013, Zambriski et al., 2013) and none of them reported calf data on an individual basis. A study by Thomson et al. (2019) did collect data from calves on an individual basis although these data was not shared in the publication. Results from that study showed intermittent shedding in individual calves and this has also been seen in experimentally infected animals (F. Katzer, unpublished observations). To the best of our knowledge, this is the first study to report Cryptosporidium shedding for calves on an individual basis from birth to six weeks. Intermittent shedding has, however, been reported for natural C. parvum infection in humans (Chappell et al., 1996). Our data indicate that there is variation in shedding profiles across the calves, with some calves shedding oocysts for longer periods and in consecutive sampling, others being intermittently positive, and a few calves for which oocysts were only detected on one occasion. Understanding the reasons behind this variation, and the relative contribution of infectious dose size, time of exposure, nutrition, maternal antibodies or genetics, would be of significant interest concerning patterns of disease in calves, and these data highlight the value of longitudinal genotyping studies.
Cryptosporidium parvum was detected as the most prevalent species on both farms in adult cattle, with 67.1% prevalence on the dairy farm (Study 1) and 59.3% on the beef farm (Study 2). The finding that C. parvum is the most prevalent species in adult cattle is consistent with previous studies using the same sensitive techniques designed to work with adult cattle faeces and molecular typing tools (Wells et al., 2015; Thomson et al., 2019).
In this study, a high prevalence of C. parvum in adult cattle was found both on its own or as a mixed infection with other species. C. andersoni was present in 41.8% of the adult cattle either alone or as a mixed infection with C. parvum in Study 1 and in 11.1% of the adult cattle in Study 2. When these techniques were used in previous studies, similar results were shown; a study looking at the species of Cryptosporidium present in livestock in a catchment in the Cairngorms, Scotland, found that C. parvum was the predominant species in adult cattle, present throughout three sampling time points (Wells et al., 2015). A further study examining the shedding of Cryptosporidium in calves and dams found that 86.2% of the adult cattle were shedding C. parvum (see Thomson et al., 2019). This present study builds on this work by continuing the research on alternative farms and typing multiple loci and examining paired samples from dams and their calves.
The gp60 genotype discovered in the calves in Study 1 was exclusively IIaA15G2R1. This appears to be the most common genotype in calves in the UK (Brook et al., 2009; Smith et al., 2014; Wells et al., 2015) and in Europe (Holzhausen et al., 2019, Holzhausen et al., 2019). In Study 2, however, the calves had exclusively IIaA17G1R1, which is less common although has been found in the UK, Germany, Hungary, Sweden, and Poland (Plutzer & Karanis, 2007; Silverlås & Blanco-Penedo, 2013; Smith et al., 2014; Kaupke & Rzezutka, 2015; Holzhausen et al., 2019, Holzhausen et al., 2019). The studies by Silverlås and Blanco-Penedo (2013) and Smith et al. (2014) were conducted in dairy cattle but unfortunately, the other studies do not state whether these were from dairy or beef calves. Cryptosporidium parvum subtype IIaA15G2R1 was not found in any of the animals in Study 2, but IIaA17G1R1 was detected in one adult cow in Study 1. This might suggest that IIaA15G2R1 may be associated with increased pathogenicity when compared to IIaA17G1R1 and indeed the clinical disease in Study 1 was notably higher than in Study 2 when considering the whole group (our unpublished observations). Pathogenicity of different C. parvum genotypes has been shown to differ according to the corresponding gp60 genotype (Sayed et al., 2016), and work looking at the pathogenicity of C. parvum showed that different isolates differ in their ability to alter host cell monolayers (Holzhausen et al., 2019, Holzhausen et al., 2019). It has been suggested that C. parvum with the gp60 genotype IIaA15G2R1 may be one of the most virulent to infect calves and lambs (Feng et al., 2013).
Species identification was achievable using the present protocols when used for adult cattle samples; however, the protocol had to be adapted when amplifying at the MM5, MM18, MM19, and TP14 gene to remove dilution steps. Multiple PCR repeats were also required to ensure adult cattle samples were amplified at all loci. This could suggest that either there is a reduced number of C. parvum oocysts present in adult cattle samples, that the ratio of faeces to oocysts is greater resulting in reduced sensitivity, or the presence of PCR inhibitors has underestimated the prevalence in adult cattle (Moreira et al., 2020). The 18S target sequence is a multi-copy target while the other alleles used in the study are single-copy targets of the genome. An effective qPCR to quantify the C. parvum DNA would be useful although this would also be subject to PCR inhibition. An alternative could be to separate oocysts using immunomagnetic separation (IMS) and quantify using microscopy. Future work should include improving detection rates in adult cattle faeces by optimising methodology in order to increase the reliability of prevalence data in adult cattle.
Despite previous studies finding C. parvum in adult cattle, many of these have not been genotyped or have only been genotyped at the gp60 locus, making it difficult to conclude if the genotype in the adults matches the genotype in the calves, especially if the IIaA15G2R1 genotype is found as this is the most common genotype in calves and humans within the UK. When further genotyping is done at multiple loci, as in the present study, the results show a greater variety of genotypes present in the adult cattle compared to their calves. In the dairy farm (Study 1), the majority of the adult cattle had a genotype that differs by more than one of the five tested loci. Furthermore, only one matched calf and dam shed the same genotype on the dairy farm. This would suggest that the main genotype found in adult cattle is different from the main genotype found in the calves. However, the genotype which is present in the calves may be present within the adult cattle, but at levels below the detection threshold. This result is mirrored in the multilocus genotypes found in adult cattle and calves in a Scottish catchment, where the majority of adult cattle had an MLG that differed from that of the calves, although calves in this study were not matched pairs (Wells et al., 2015).
In the beef farm (Study 2), the majority of adult cattle did have a multilocus genotype that matches the calves (54%) although it must be noted that these animals were sampled at the same time. When it came to matched calf and dam pairs, there were seven out of a possible 25 matches. This match cannot directly confirm transmission between the two; however, it does suggest that adult cattle may be a potential source of oocysts to their calves. This is not an unexpected result as these adults and calves are housed together in the same shed and therefore this result could reflect also that adult cattle may become infected from oocysts that have originated from the calves. The way these cattle are housed together is likely to account for the difference in genotypes shed in matched dam-calf pairs observed when compared to the dairy farm. The dairy calves were removed from their dams within 12 h of birth whereas the beef calves were housed alongside their dams. Further work is required to determine whether these matched genotypes in the adult cattle could pose a transmission risk to calves. This would involve a detailed look at how oocysts are able to move from the adult cattle, through the environment to then become a source of infection to the calves. Future work should include testing the adult cattle before the birth of the calves, to see if a similar pattern is observed.
Results from the dairy farm (Study 1) show that adult cattle and their calves predominantly have different multilocus genotypes with only one matched pair. The beef farm presented with seven matched pairs which show multilocus genotypes shed by the calves can also be shed by the adult cattle. Therefore adult cattle may potentially be a source of oocysts to calves, but this is more likely on a beef farm where they are housed together than on a dairy farm where they are separated. Overall the difficulty in amplification indicated a low level of oocyst shedding in both the adult dairy and beef cattle, although further method optimisation is required to confirm this. Nevertheless, as little as 17 oocysts have been shown to infect calves and result in diarrohea and oocyst shedding (Zambriski et al., 2013, Zambriski et al., 2013), and therefore low-level shedding within a larger sample volume in adult cattle may still contribute a substantial number of oocysts and therefore be responsible for maintaining C. parvum between calving seasons.
As cattle on the dairy farm calved all year around, it was impossible to determine how genotypes evolve over a calving period. This information would be useful as it would allow for the development of risk forecasting and provide future opportunities to discover if oocysts persist between calving seasons. Therefore future work could involve similar studies on farms, which have set calving periods, to see how genotypes evolve and persist.
5. Conclusions
The results of this study have shown that when using the most sensitive technique available for the processing of adult cattle faecal samples, C. parvum is the most prevalent species in both adult cattle and calves on both a dairy and beef farm. The dairy farm study (Study 1) is one of the first studies to examine longitudinal shedding profiles on an individual calf basis from birth to six weeks and it revealed intermittent shedding of Cryptosporidium oocysts by the calves. The difficulty in multilocus amplification of C. parvum in adult cattle and the requirement to alter the published protocols suggests that further work is required to optimise the detection of oocysts in adult cattle faeces. This study indicates that adult cattle do have the ability to shed the same genotype of C. parvum as their calves and so with the low infectious dose and increased sample size, could potentially be a source of oocysts to calves. Further work would be required to confirm their potential role in transmission by analysing oocyst movement and survival in the environment.
Funding
This study was funded by the Agricultural and Horticultural Development Board and the Scottish Governmentʼs Rural and Environment Science and Analytical Services Division. Liam Morrison and the Roslin Institute were also supported by a core grant from the BBSRC (BBS/E/D/20231762; BBS/E/D/20002173).
Ethical approval
The study was carried out with the permission of the farmers and all positive animals were experiencing a natural infection with Cryptosporidium which was confirmed using 18S nssm PCR. Both farms had a history of clinical cryptosporidiosis.
CRediT author statement
FK, EI, LM and HS conceived and designed the study. HS collected the samples, performed the laboratory analysis, genotyping analysis and drafted the manuscript. CA and KU aided in sample collection and processing. FK, EI, LM edited the manuscript. All authors read and approved the final manuscript.
Declaration of competing interests
The authors declare that they have no competing interests.
Editorial disclosure
Given his role as Co-Editor, Frank Katzer had no involvement in the peer-review of this article and has no access to information regarding its peer-review. Full responsibility for the editorial process of this article was delegated to Aneta Kostadinova.
Acknowledgements
The authors would like to thank the farmers for their permission and help provided during the sampling periods and Sarah Thomson for her guidance with sample collection.
Contributor Information
Hannah Jade Shaw, Email: hshaw@harper-adams.ac.uk.
Frank Katzer, Email: frank.katzer@moredun.ac.uk.
References
- Åberg M., Emanuelson U., Troell K., Björkman C. A single-cohort study of Cryptosporidium bovis and Cryptosporidium ryanae in dairy cattle from birth to calving. Vet. Parasitol. X: Reg. Stud. Rep. 2020;100400 doi: 10.1016/j.vprsr.2020.100400. [DOI] [PubMed] [Google Scholar]
- Anderson B.C. Cryptosporidiosis in bovine and human health. J. Dairy Sci. 1998;81:3036–3041. doi: 10.3168/jds.S0022-0302(98)75868-0. [DOI] [PubMed] [Google Scholar]
- Atwill E., Pereira M.G.C. Lack of detectable shedding of Cryptosporidium parvum oocysts by periparturient dairy cattle. J. Parasitol. 2003;89:1234–1236. doi: 10.1645/GE-3192RN. [DOI] [PubMed] [Google Scholar]
- Brook E.J., Anthony Hart C., French N.P., Christley R.M. Molecular epidemiology of Cryptosporidium subtypes in cattle in England. Vet. J. 2009;179:378–382. doi: 10.1016/j.tvjl.2007.10.023. [DOI] [PubMed] [Google Scholar]
- Burrells A., Bartley P.M., Zimmer I.A., Roy S., Kitchener A.C., Meredith A., et al. Evidence of the three main clonal Toxoplasma gondii lineages from wild mammalian carnivores in the UK. Parasitology. 2013;140:1768–1776. doi: 10.1017/S0031182013001169. [DOI] [PubMed] [Google Scholar]
- Chappell C.L., Okhuysen P.C., Sterling C.R., DuPont H.L. Cryptosporidium parvum: Intensity of infection and oocyst excretion patterns in healthy volunteers. J. Infect. Dis. 1996;173:232–236. doi: 10.1093/infdis/173.1.232. [DOI] [PubMed] [Google Scholar]
- de Graaf D.C., Vanopdenbosch E., Ortega-Mora L.M., Abbassi H., Peeters J.E. A review of the importance of cryptosporidiosis in farm animals. Int. J. Parasitol. 1999;29:1269–1287. doi: 10.1016/s0020-7519(99)00076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban E., Anderson B.C. Cryptosporidium muris: Prevalence, persistency, and detrimental effect on milk production in a drylot dairy. J. Dairy Sci. 1995;78:1068–1072. doi: 10.3168/jds.S0022-0302(95)76723-6. [DOI] [PubMed] [Google Scholar]
- Faubert G., Litvinsky Y. Natural transmission of Cryptosporidium parvum between dams and calves on a dairy farm. J. Parasitol. 2000;86:495–500. doi: 10.1645/0022-3395(2000)086[0495:NTOCPB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Feng Y., Torres E., Li N., Wang L., Bowman D., Xiao L. Population genetic characterisation of dominant Cryptosporidium parvum subtype IIaA15G2R1. Int. J. Parasitol. 2013;43:1141–1147. doi: 10.1016/j.ijpara.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Holubová N., Tůmová L., Sak B., Hejzlarová A., Konečný R., McEvoy J., Kváč M. Description of Cryptosporidium ornithophilus n. sp. (Apicomplexa: Cryptosporidiidae) in farmed ostriches. Parasit. Vectors. 2020;13:340. doi: 10.1186/s13071-020-04191-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzhausen I., Lendner M., Daugschies A. Bovine Cryptosporidium parvum field isolates differ in cytopathogenicity in HCT-8 monolayers. Vet. Parasitol. 2019;273:67–70. doi: 10.1016/j.vetpar.2019.08.006. [DOI] [PubMed] [Google Scholar]
- Holzhausen I., Lendner M., Göhring F., Steinhöfel I., Daugschies A. Distribution of Cryptosporidium parvumgp60 subtypes in calf herds of Saxony, Germany. Parasitol. Res. 2019;118:1549–1558. doi: 10.1007/s00436-019-06266-1. [DOI] [PubMed] [Google Scholar]
- Kaupke A., Rzezutka A. Emergence of novel subtypes of Cryptosporidium parvum in calves in Poland. Parasitol. Res. 2015;114:4709–4716. doi: 10.1007/s00436-015-4719-1. [DOI] [PubMed] [Google Scholar]
- Klein P., Kleinova T., Volek Z., Simunek J. Effect of Cryptosporidium parvum infection on the absorptive capacity and paracellular permeability of the small intestine in neonatal calves. Vet. Parasitol. 2008;152:53–59. doi: 10.1016/j.vetpar.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo M.L., Ares-Mazas E., De Maturana I.V.M. Detection of oocysts and IgG antibodies to Cryptosporidium parvum in asymptomatic adult cattle. Vet. Parasitol. 1993;47:9–15. doi: 10.1016/0304-4017(93)90171-i. [DOI] [PubMed] [Google Scholar]
- Mallon M.E., MacLeod A., Wastling J.M., Smith H., Tait A. Multilocus genotyping of Cryptosporidium parvum Type 2: Population genetics and sub-structuring. Infect. Genet. Evol. 2003;3:207–218. doi: 10.1016/s1567-1348(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Moreira N.C., Cabrine-Santos M., de Oliveira-Silva M.B. Evaluation of different oocyst DNA extraction methods for Cryptosporidium spp. research in environmental samples. Acta Parasitol. 2020;65:995–998. doi: 10.1007/s11686-020-00235-w. [DOI] [PubMed] [Google Scholar]
- Morrison L.J., Mallon M.E., Smith H.V., MacLeod A., Xiao L., Tait A. The population structure of the Cryptosporidium parvum population in Scotland: A complex picture. Infect. Genet. Evol. 2008;8:121–129. doi: 10.1016/j.meegid.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi F., Tozawa Y., Inomata A., Horimoto T., Wada Y., Kato K. Molecular characterization of Cryptosporidium isolates from calves in Ishikari District, Hokkaido, Japan. J. Vet. Med. Sci. 2013;75:837–840. doi: 10.1292/jvms.12-0435. [DOI] [PubMed] [Google Scholar]
- Nydam D.V., Wade S.E., Schaaf S.L., Mohammed H.O. Number of Cryptosporidium parvum oocysts or Giardia spp. cysts shed by dairy calves after natural infection. Am. J. Vet. Res. 2001;62:1612–1615. doi: 10.2460/ajvr.2001.62.1612. [DOI] [PubMed] [Google Scholar]
- Plutzer J., Karanis P. Genotype and subtype analyses of Cryptosporidium isolates from cattle in Hungary. Vet. Parasitol. 2007;146:357–362. doi: 10.1016/j.vetpar.2007.02.030. [DOI] [PubMed] [Google Scholar]
- Robertson L.J., Björkman C., Axén C., Fayer R. In: Cryptosporidium: Parasite and disease. Cacciò S.M., Widmer G., editors. Springer Vienna; Vienna: 2014. Cryptosporidiosis in farmed animals; pp. 149–235. [DOI] [Google Scholar]
- Ryan U., Paparini A., Monis P., Hijjawi N. Itʼs official - Cryptosporidium is a gregarine: What are the implications for the water industry? Water Res. 2016;105:305–313. doi: 10.1016/j.watres.2016.09.013. [DOI] [PubMed] [Google Scholar]
- Sayed F.G., Hamza A.I., Galal L.A., Sayed D.M., Gaber M. Virulence of geographically different Cryptosporidium parvum isolates in experimental animal model. Ann. Parasitol. 2016;62:221–232. doi: 10.17420/ap6203.56. [DOI] [PubMed] [Google Scholar]
- Shaw H.J., Innes E.A., Morrison L.J., Katzer F., Wells B. Long-term production effects of clinical cryptosporidiosis in neonatal calves. Int. J. Parasitol. 2020;50:371–376. doi: 10.1016/j.ijpara.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverlås C., Blanco-Penedo I. Cryptosporidium spp. in calves and cows from organic and conventional dairy herds. Epidemiol. Infect. 2013;141:529–539. doi: 10.1017/S0950268812000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.P., Clifton-Hadley F.A., Cheney T., Giles M. Prevalence and molecular typing of Cryptosporidium in dairy cattle in England and Wales and examination of potential on-farm transmission routes. Vet. Parasitol. 2014;204:111–119. doi: 10.1016/j.vetpar.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulaiman I.M., Hira P.R., Zhou L., Al-Ali F.M., Al-Shelahi F.A., Shweiki H.M., et al. Unique endemicity of cryptosporidiosis in children in Kuwait. J. Clin. Microbiol. 2005;43:2805–2809. doi: 10.1128/JCM.43.6.2805-2809.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson S., Hamilton C.A., Hope J.C., Katzer F., Mabbott N.A., Morrison L.J., et al. Bovine cryptosporidiosis: Impact, host-parasite interaction and control strategies. Vet. Res. 2017;48:42. doi: 10.1186/s13567-017-0447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson S., Innes E.A., Jonsson N.N., Katzer F. A multiplex PCR test to identify four common cattle-adapted Cryptosporidium species-CORRIGENDUM. Parasitol. Open. 2019;5 doi: 10.1017/pao.2016.2. [DOI] [Google Scholar]
- Tiranti K., Larriestra A., Vissio C., Picco N., Alustiza F., Degioanni A., Vivas A. Prevalence of Cryptosporidium spp. and Giardia spp., spatial clustering and patterns of shedding in dairy calves from Córdoba, Argentina. Rev. Bras. Parasitol. Vet. 2011;20:140–147. doi: 10.1590/s1984-29612011000200009. [DOI] [PubMed] [Google Scholar]
- Tzipori S., Smith M., Halpin C., Angus K., Sherwood D., Campbell I. Experimental cryptosporidiosis in calves: Clinical manifestations and pathological findings. Vet. Rec. 1983;112:116–120. doi: 10.1136/vr.112.6.116. [DOI] [PubMed] [Google Scholar]
- Uga S., Matsuo J., Kono E., Kimura K., Inoue M., Rai S., Ono K. Prevalence of Cryptosporidium parvum infection and pattern of oocyst shedding in calves in Japan. Vet. Parasitol. 2000;94:27–32. doi: 10.1016/s0304-4017(00)00338-1. [DOI] [PubMed] [Google Scholar]
- Wells B., Shaw H., Hotchkiss E., Gilray J., Ayton R., Green J., et al. Prevalence, species identification and genotyping Cryptosporidium from livestock and deer in a catchment in the Cairngorms with a history of a contaminated public water supply. Parasit. Vectors. 2015;8:66. doi: 10.1186/s13071-015-0684-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells B., Thomson S., Innes E., Katzer F. Development of a sensitive method to extract Cryptosporidium oocysts from adult cattle faecal samples. Vet. Parasitol. 2016;227:26–29. doi: 10.1016/j.vetpar.2016.07.018. [DOI] [PubMed] [Google Scholar]
- Zambriski J.A., Nydam D.V., Bowman D.D., Bellosa M.L., Burton A.J., Linden T.C., et al. Description of fecal shedding of Cryptosporidium parvum oocysts in experimentally challenged dairy calves. Parasitol. Res. 2013;112:1247–1254. doi: 10.1007/s00436-012-3258-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambriski J.A., Nydam D.V., Wilcox Z.J., Bowman D.D., Mohammed H.O., Liotta J.L. Cryptosporidium parvum: Determination of ID50 and the dose-response relationship in experimentally challenged dairy calves. Vet. Parasitol. 2013;197:104–112. doi: 10.1016/j.vetpar.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]