Abstract
The nucleoside reverse transcriptase inhibitor abacavir (ABC) is used commonly to treat young children with HIV infection and is a component of the fixed-dose-combination Triumeq®. ABC can trigger a severe hypersensitivity reaction in people who are homozygous or heterozygous for HLA-B*57:01. Testing for HLA-B*57:01 before ABC initiation is standard-of-care in high-resource settings, but current tests are costly or difficult to access in resource-limited settings. To address these gaps, we developed an inexpensive simple-to-use rapid assay to detect HLA-B*57:01. We designed and optimized a multiplexed polymerase chain reaction (PCR) to amplify HLA-B*57 subtypes and the human beta-globin gene; employed probes and ligation to specifically tag the HLA-B*57:01 allele with biotin. Tagged-ligated products were detected by immunocapture in an enzyme-linked immunosorbent assay plate or lateral flow strip. Cell lines with known HLA genotypes were used to optimize the assay. The optimized assay was then compared with genotypes of clinical specimens (n = 60) determined by sequencing, with specimens enriched for individuals with HLA-B*57:01. The optimized assay utilizes 40-min 35-cycle multiplex PCR for B*57 and beta-globin; 20-min ligation reaction; and 15-min detection. Evaluation of the HLA-B*57:01 oligonucleotide ligation assay using clinical specimens had a sensitivity of 100% (n = 27/27 typed as B*57:01) and specificity of 100% (n = 33/33 typed as non-B*57:01) by visual interpretation of lateral flow strips. The cost is US$5.96/specimen. This rapid and economical assay accurately detects HLA-B*57:01 in clinical specimens. Use of this assay could expand access to HLA-B*57:01 genotyping and facilitate safe same-day initiation of ABC-based treatment.
Keywords: oligonucleotide ligation assay, HLA-B*57:01, abacavir, hypersensitivity reactions, pediatric ART, genetic screening
Introduction
Abacavir (ABC) is a nucleoside reverse transcriptase inhibitor used in combination therapy to treat HIV.1–3 Despite high tolerability in most patients, ABC triggers a hypersensitivity reaction in 5%–8% of people within 6 weeks of treatment initiation, characterized by fever, rash, nausea, and respiratory distress. Treatment continuation or drug reinitiation can be fatal.4 The risk of developing a hypersensitivity reaction to ABC is strongly associated with the HLA-B*57:01 allele.5 The allele frequency of HLA-B*57:01 varies by ethnicity; ranging from 0% to 3% in African and South American populations, 6%–10% in Caucasian populations and up to 19% in Southwest Asian populations.6 Screening for HLA-B*57:01 has become standard-of-care before ABC use in high-resource communities.2
Current tests for ABC hypersensitivity detect the HLA-B*57:01 subtype by allele-specific polymerase chain reaction (PCR) or sequencing. Allele-specific PCR uses primers7,8 to specifically amplify the HLA-B*57:01 allele, requiring a standard thermocycler and electrophoresis equipment7 (cost estimated at US$1,500), or real-time fluorescence instruments9 (cost estimated at US$15,000–50,000). Sanger sequencing or next-generation sequencing, completed in high-resource settings7,10 requires even more expensive equipment (>US$100,000). Owing to high-cost instruments, most HLA-B*57:01 testing is performed at centralized laboratories, which requires shipping of specimens. Slow turnaround times of 3 days to weeks can delay treatment initiation. Tests that could be performed inexpensively without the need for sequencing technology could increase the safe use of ABC in settings where screening for HLA-B*57:01 is otherwise cost-prohibitive.
In this study, we report the development and validation of a lateral flow-based oligonucleotide ligation assay (OLA) that detects a polymorphism differentiating HLA-B*57:01 from other B*57 subtypes. The B*57 region is amplified from genomic DNA followed by annealing to two probes that bisect the HLA-B*57:01 allele. A ligase joins a genotype-specific probe and a biotin-tagged detector probe only when the probes match the genotype of the amplified product at the ligation site. The ligated product is detected by capture on a single-use lateral flow strip or enzyme-linked immunosorbent assay plate. This assay only requires a standard thermocycler and microcentrifuge (cost estimated <US$1,000), which are relatively inexpensive and common in small laboratories, allowing use in near point-of-care settings.
Materials and Methods
Study design and population
PCR primers were chosen to amplify the region of chromosomal DNA encoding HLA-B*57 subtypes.7 OLA probes were designed to detect HLA-B*57:01 but not other HLA-B*57 subtypes. The PCR for HLA-B*57 was multiplexed with primers for human beta-globin as a positive control for DNA extraction, amplification, and ligation. Assay optimization used DNA from five cell lines (LADA, MYE 2004, FH18, 32/32, and DBB; from the Fred Hutchinson International Histocompatibility Working Group, Seattle, WA) including HLA-B*57:01, B*57:02, B*57:03, and a B*57 negative control. Optimization of assay parameters was completed using a plate-based OLA before clinical validation using a paper-based lateral flow OLA. Clinical validation used 60 banked specimens, including 52 Mexican10 and 8 Seattle Primary Infection Cohort11 DNA specimens; all collected following protocols approved by respective review boards for research on human subjects, and de-identified for this project.
The Mexican specimens were enriched for HLA-B*57:01 and other common B*57 subtypes (i.e., B*57:02/03) compared with the Mexican population, with genotypes previously determined using next-generation sequencing [Trusight HLA v2 Sequencing Panel on MiniSeq platform (Illumina, San Diego, CA), and Assign v2.0 software was used to assign HLA subtypes] at the Centre for Research in Infectious Diseases (CIENI) of the National Institute of Respiratory Diseases (INER) in Mexico City. Studies collecting these samples and those from the Seattle Primary Infection Cohort were approved by respective Ethics Committees. De-identified DNA from Mexican specimens were sent to Seattle for testing. After blinded testing by the HLA-B*57:01 OLA assay, the OLA results were compared with the genotypes by sensitivity and specificity analysis.
Sample preparation
DNA was extracted from buffy coat (200 μL) or PBMCs (peripheral blood mononuclear cells, ∼6 million cells) processed from whole blood using the QIAmp DNA Blood Mini Kit (Qiagen, Valencia; CA) according to manufacturer's specifications. Extracted DNA was loaded directly into the PCR reaction mix at a concentration of 2–4 ng/μL.
Primers and PCR
The primer sets coamplify a fragment comprising 161 nucleotides of the human beta-globin gene (forward primer: 5′-GGGATCTGTCCACTCCTGATGCTGT; reverse primer: 5′-ATCCACGTGCAGCTTGTCACAGTG) and a region (179 bp) specific to B*57 subtypes using a modification of primers previously published7 (forward primer: 5′-CCAGGGTCTCACATCATCCAGGT; reverse primer: 5′-CGCCTCCCACTTGCGCTGGG) (Integrated DNA Technology, Coralville, Iowa). The PCR was performed in a 25-μL reaction containing 0.625 U Terra PCR Direct Polymerase (Takara Bio, Inc., Shiga, Japan), 0.4 μM of each primer, 12.5 μL Terra buffer, and 2.5 μL of extracted DNA (5–10 ng total DNA).
Two-step PCR conditions are as follows: 30 s at 95°C, 35 cycles with denaturation for 15 s at 95°C followed by annealing for 30 s at 68°C, with a final extension at 68°C for 5 min.
Oligonucleotide ligation assay
Oligonucleotide probes for HLA-B*57:01 target a cytosine nucleotide at position 1094 of the B*57:01 allele (reference sequence HLA-B*57:01:01, GenBank Accession number AJ458991), but not other B*57 types7: B*57-probe-1: 5′-FAM-TCCTCC GCGGGCATGACCAGTC; B*57-probe-2: 5′-Phosphorylation-YGCCTACGACGGCAAGGATTACA–Biotin. Beta-globin probes ligate when annealed to the beta-globin amplicon and serve as an amplification and ligation control: beta-globin-probe-1: 5′-Digoxigenin-CTCGGTGCCTTTAGTGATGGC CTG; beta-globin-probe-2: 5′-Phosphorylation-GCTCACCTGGACAACCTCAAGGG–Biotin.
Two microliters of amplicon from the multiplex PCR are added directly into 20 μL of ligation mix containing 1.33 U Ampligase DNA Ligase (Lucigen, Middleton, WI), 12.5 mM KCl, 1 mM NAD, 1 × ligase buffer (20 mM Tris-HCl, 10 mM MgCl2, 1 mM DDT), 0.08% Triton™X-100, 16.7 nM B*57 probes, 4.2 nM beta-globin-probe-1 and 16.7 nM beta-globin-probe-2. The ligation reaction conditions are 10 cycles with denaturation at 94°C for 30 s followed by ligation for 30 s at 50°C.
Detection of ligation products
Plate-based OLA
Ligated products are captured on a streptavidin-coated 96-well plate (Sigma-Aldrich, St Louis, MO) for 1 h at room temperature and then denatured by washing with dilute NaOH (0.01 N NaOH and 0.05% Tween®20), followed by incubation with antifluorescein and antidigoxigenin antibodies (Sigma-Aldrich). Secondary reporters are added sequentially for B*57:01 and beta-globin, and the optical density (OD) was measured using a Spectramax i3x plate reader (Molecular Devices, San Jose, CA) at 405 and 450 nm, respectively.12 Samples with high signal (OD >1.0) at both wavelengths were classified as B*57:01 positive. Samples with low (OD <0.15) signal at 405 nm and high signal at 450 nm were classified as negative.
Paper-based OLA
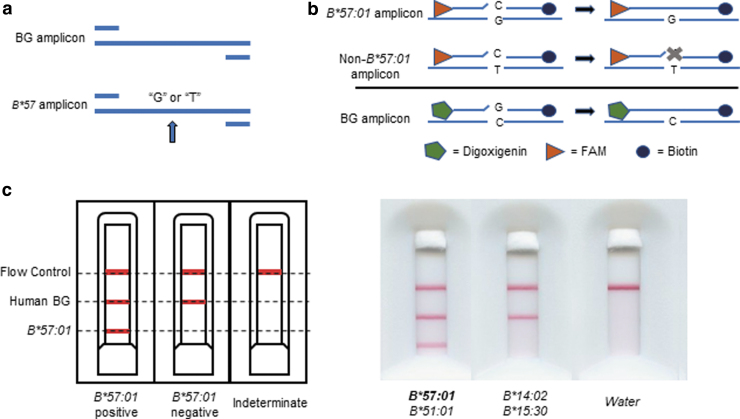
Unlabeled oligonucleotides with sequences identical to those used in the plate-based assay were added at 40-fold higher concentration and denatured at 90°C for 30 s. The 26.8-μL ligation reaction is then added onto lateral flow detection strips and after 5 min is chased by 43-μL buffer containing antibiotin antibodies conjugated gold nanoparticles (OD = 0.7 at 520 nm). Ten minutes after the addition of the gold buffer, the strips were visually inspected by a single laboratory technician for appearance of precipitated bands to determine genotype or by scanner and in-house software (Fig. 1). Indeterminate results, indicated by either the lack of a precipitated band for the flow control or beta-globin, would be reprocessed from extracted DN. Any sample that is indeterminate on repeat testing would be interpreted as having inhibitor contamination or low DNA.
FIG. 1.
OLA for detection of HLA-B*57:01 by lateral flow strip. (a) Amplification: Extracted DNA is added to the PCR reaction mixture for multiplexed amplification of human BG and B*57 gene fragments. (b) Ligation: labeled B*57:01 probes are selectively ligated together in the presence of amplicons with the B*57:01 allele. BG probes ligate in the presence of the amplified BG region of the somatic genome and serve as a (+) control. (c) Lateral flow detection: chip layout and scanned images of participant sample assays. Appearance of a red line at “Flow Control” indicates adequate flow of reagents on the single-use cartridge; a red line at “Human BG” indicates presence of human BG amplicon, and confirms adequate DNA extraction, PCR amplification and ligation; a red line at “B*57:01” indicates detection of B*57:01, either a specimen that is homozygous or heterozygous for the B*57:01 subtype. HLA-B* types determined by Illumina sequencing are shown for two participants' samples tested by OLA. BG, beta-globin; OLA, oligonucleotide ligation assay; PCR, polymerase chain reaction. Color images are available online.
Preparation of lateral flow test
Antifluorescein (Southern Biotech, Birmingham, AL) and antidigoxigenin (Novus Biologicals, Centennial, CO) antibodies capture ligated products complementary to HLA-B*57:01 and beta-globin, respectively, in the lateral flow strips. BSA-conjugated biotin (Sigma-Aldrich) was used as a flow control as previously described.13–15
Automated assay interpretation by in-house software
To avoid human error, the lateral flow strips can be analyzed using in-house Python software, based on an algorithm previously described.13 The software automatically detects bands in the paper strips and quantifies signal intensity by subtracting brightness within band and adjacent background regions.
Results
Development of the HLA-B*57:01 OLA assay
The assay, described in Methods, co-amplifies the beta-globin gene and the B*57 allele and uses a paper-based OLA to detect HLA B57:01 for clinical management and beta-globin to verify the input of sufficient human DNA and the absence of PCR inhibitors (Fig. 1). The B*57:01 probe concentrations and ligation conditions were optimized to maximize signal-to-noise ratio in a 96-well ELISA-based OLA using cell lines with known B*57:01/02/03 subtypes or no B*57 variants. The OLA was then adapted to a lateral flow strip format with intensity suitable for visual reading of results. In addition, an in-house software was adapted to quantify band signal intensities from scanned images of the lateral flow strip.
Testing of 60 human specimens detected 100% of specimens previously genotyped as B*57 positive by paper-based detection (details presented below, see Table 1). The costs per specimen were DNA extraction reagents US$3.36, PCR amplification reagents and plasticware US$1.09, and paper-based ligation US$1.51.
Table 1.
HLA-B*57:01 Typing Results by Lateral Flow OLA and Illumina Sequencing in a Mexican Population Enriched for HLA-B*57 and Seattle Primary Infection Cohort
| HLA-B typing results | ||||
|---|---|---|---|---|
| HLA-B type by Illumina sequencing | No. of specimens | Expected outcome by OLA | Correct OLA outcome by visual interpretation | Correct OLA outcome by in-house software |
| B*57:01 | 27 | + | 27 | 27 |
| B*57:02 | 1 | − | 1 | 1 |
| B*57:03 | 9 | − | 9 | 9 |
| Non-B*57a | 23 | − | 23 | 23 |
Non-B*57 types tested included—B*07, 08, 13, 14, 15, 18, 35, 38, 39, 40, 41, 42, 44, 45, 48, 49, 51, 52.
OLA, oligonucleotide ligation assay.
Validation of OLA for detection of HLA-B*57:01 subtypes using clinical specimens with interpretation by visual inspection or in-house software
The lateral flow strips read by visual inspection, reported earlier, were also classified as positive or negative for both beta globin and B*57:01 by automated software. All samples were classified as positive for the human beta globin control, with no indeterminate results. Compared with HLA typing by sequencing, both visual inspection and the software classified as positive all 27 B*57:01 specimens (all heterozygotes) [100% sensitivity (95% confidence interval: 87.2%–100%)] (Table 1). The remaining 33 specimens were correctly classified as B*57:01 negative by both visual inspection and the software [100% specificity (95% confidence interval: 89.4%–100%)].
Discussion
We developed and evaluated a relatively rapid and inexpensive assay that accurately detects the HLA-B*57:01 subtype, associated with a hypersensitivity reaction to ABC, and discriminates it from similar common B*57 subtypes (i.e., B*57:02 and B*57:03). The assay requires minimal supplies and uses inexpensive equipment and delivers results in <2 h (i.e., 30-min DNA extraction, 40-min PCR, 20-min ligation, and 15-min detection). Reagents and plasticware cost US$5.96/specimen, and the thermocycler can be purchased for as little as US$550. Results are simple to analyze by the unaided eye or using a scanner and our in-house software.14,15 Given these characteristics, this assay stands out compared with other assays7–10 as better suited for near point-of-care small laboratories, as it offers the potential for rapid turnaround times that could expedite the timely initiation of antiretrovirals.
This assay showed high sensitivity and specificity for detection of HLA-B*57:01 in clinical samples compared with HLA typing by sequencing. Performance of the OLA was similar to other nonsequencing methods such as allele-specific PCR.7,9,16
This study was limited by a relatively small number of clinical samples tested; however, it was enriched to include all with the B*57:01 subtype from a HIV cohort of ∼1,800 individuals in Mexico City (calculated prevalence 1.5%). A sample panel from more diverse populations would allow a more comprehensive validation of primers and probes and discrimination of B*57:01 from other HLA subtypes.
The primers and probes used in this assay are complementary at the site of ligation to sequences described for two other alleles (B*57:08 and B*55:14).7 However, peak frequencies of B*57:08 and B*55:14 are both <0.1%; in contrast the frequency of B*57:01 ranges from 1% to 15% globally in large studies.17–19 Given these frequencies and target locations for this assay, such as Mexico and South Asia, the assay is likely to have few false positive reactions (notably, in Mexico B*57:08 and B*55:14 were not identified among the 1,800 tested).
In this study, we only tested DNA extracted from blood samples as we did not have access to other sample types. The use of an inhibitor-resistant polymerase for PCR may facilitate direct input from fingerstick blood or buccal swab, thus eliminating the need for DNA extraction and/or venipuncture.20 A change in specimen type and use of direct-to-PCR samples would reduce total reagent costs to US$2.60/specimen. To increase ease of use, test reagents can be lyophilized for single-use aliquots.14,21 These improvements would simplify the procedure, shorten assay time, which would aid in implementation in near point-of-care settings.
Conclusion
ABC is recommended as first-line HIV treatment in children by multiple experts,2,22 and is a component of a once daily fixed-dose-combination, Triumeq®. However, the safe use of these antiretrovirals is limited by the high cost and limited access to genotypic assays to detect B*57:01, which increases the risk of a severe hypersensitivity reactions.4,5 Our simple, rapid, and inexpensive B*57:01 OLA described in this study should allow testing for this HLA subtype in low-resource settings, which could facilitate safer same-day initiation of ABC-based treatment.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the funders.
Authors' Contributions
J.J.W. developed and optimized the assay and performed blinded sample testing. I.A.B. and N.P. developed the assay platform and provided writing and project guidance. N.P. and P.S.R. manufactured the tests and performed software analysis for the project. H.V.-P., M.S.-N., and S.Á.-R. collected and provided samples for blinded testing and sequencing data. B.R.L., S.Á.-R., and L.M.F. oversaw the project and provided guidance and feedback.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
Support for this project came from the National Institutes of Health (NIH) including R01 AI110375 (L.M.F.); R01 AI145486 (B.R.L.); the Clinical and Retrovirology Research Core and the Molecular Profiling and Computational Biology Core of the University of Washington Fred Hutch Center for AIDS Research [P30 AI027757]; and the International Maternal Pediatric Adolescent AIDS Clinical Trials Network (IMPAACT). IMPAACT is funded by the National Institute of Allergy and Infectious Diseases (NIAID) with co-funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) and the National Institute of Mental Health (NIMH), all components of NIH under Award Numbers UM1AI068632 (IMPAACT LOC), UM1AI068616 (IMPAACT SDMC), and UM1AI106716 (IMPAACT LC), and by NICHD contract number HHSN275201800001I. The study was supported in part by funds from the Canadian Institutes of Health Research (grants PJT-148621 and PJT-159625), received by S.Á.-R., as well as funds from the Mexican Government (Comisión de Equidad y Género de las Legislaturas LX-LXI y Comisión de Igualdad de Género de la Legislatura LXII de la H. Cámara de Diputados de la República Mexicana), received by S.Á.-R.
References
- 1. Staszewski S, Katlama C, Harrer T, et al. : A dose-ranging study to evaluate the safety and efficacy of abacavir alone or in combination with zidovudine and lamivudine in antiretroviral treatment-naive subjects. AIDS 1998;12:197–202. [DOI] [PubMed] [Google Scholar]
- 2. Update of Recommendations on First- and Second-Line Antiretroviral Regimens. World Health Organization, Geneva, Switzerland, 2019. [Google Scholar]
- 3. Moyle GJ, DeJesus E, Cahn P, et al. : Abacavir once or twice daily combined with once-daily lamivudine and efavirenz for the treatment of antiretroviral-naive HIV-infected adults: Results of the Ziagen Once Daily in Antiretroviral Combination Study. J Acquir Immune Defic Syndr 2005;38:417–425. [DOI] [PubMed] [Google Scholar]
- 4. Hetherington S, McGuirk S, Powell G, et al. : Hypersensitivity reactions during therapy with the nucleoside reverse transcriptase inhibitor abacavir. Clin Ther 2001;23:1603–1614. [DOI] [PubMed] [Google Scholar]
- 5. Mallal S, Nolan D, Witt C, et al. : Association between presence of HLA-B* 5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet 2002;359:727–732. [DOI] [PubMed] [Google Scholar]
- 6. Martin MA, Klein TE, Dong BJ, et al. : Clinical pharmacogenetics implementation consortium guidelines for HLA-B genotype and abacavir dosing. Clin Pharmacol Ther 2012;91:734–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martin AM, Nolan D, Mallal S: HLA-B*5701 typing by sequence-specific amplification: Validation and comparison with sequence-based typing. Tissue Antigens 2005;65:571–574. [DOI] [PubMed] [Google Scholar]
- 8. Stocchi L, Cascella R, Zampatti S, Pirazzoli A, Novelli G, Giardina E: The pharmacogenomic HLA biomarker associated to adverse Abacavir reactions: Comparative analysis of different genotyping methods. Curr Genomics 2012;13:314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cesbron-Gautier A, Simon P, Achard L, Cury S, Follea G, Bignon JD: Luminex technology for HLA typing by PCR-SSO and identification of HLA antibody specificities. Ann Biol Clin 2004;2004:93–98. [PubMed] [Google Scholar]
- 10. Soto-Nava M, Avila-Ríos S, Valenzuela-Ponce H, et al. : Weaker HLA footprints on HIV in the unique and highly genetically admixed host population of Mexico. J Virol 2018;92:e01128-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schacker T, Collier AC, Hughes J, Shea T, Corey L: Clinical and epidemiologic features of primary HIV infection. Ann Intern Med 1996;125:257–264. [DOI] [PubMed] [Google Scholar]
- 12. Beck IA, Crowell C, Kittoe R, et al. : Optimization of the oligonucleotide ligation assay, a rapid and inexpensive test for detection of HIV-1 drug resistance mutations, for non-North American variants. J Acquir Immune Defic Syndr 2008;48:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Panpradist N, Beck IA, Chung MH, Kiarie JN, Frenkel LM, Lutz BR: Simplified paper format for detecting HIV drug resistance in clinical specimens by oligonucleotide ligation. PLoS One 2016;11:e0145962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Panpradist N, Beck IA, Vrana J, et al. : OLA-Simple: A software-guided HIV-1 drug resistance test forlow-resource laboratories. EBioMedicine 2019;50:34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Panpradist N, Beck I, Ruth P, et al. : Near point-of-care, point-mutation test to detect drug resistance in HIV-1: A validation study in a Mexican cohort. AIDS 2020;34:1331–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Darke C, Corbin SA: External quality assessment of patient HLA-B*57:01 testing prior to abacavir prescription. Int J Immunogenet 2014;41:277–280. [DOI] [PubMed] [Google Scholar]
- 17. Hurley CK, Kempenich J, Wadsworth K, et al. : Common, intermediate and well-documented HLA alleles in world populations: CIWD version 3.0. 0. HLA 2020;95:516–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gragert L, Madbouly A, Freeman J, Maiers M: Six-locus high resolution HLA haplotype frequencies derived frommixed-resolution DNA typing for the entire US donor registry. Hum Immunol 2013;74:1313–1320. [DOI] [PubMed] [Google Scholar]
- 19. Gonzalez-Galarza FF, McCabe A, Santos EJ, et al. : Allele frequency net database (AFND) 2020 update: Gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res 2020;48:D783–D788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Komatsu H, Tsunoda T, Inui A, et al. : Successful use of saliva without DNA extraction for detection of macrolide-resistant Mycoplasma pneumoniae DNA in children using LNA probe-based real-time PCR. J Infect Chemother 2013;19:1087–1092. [DOI] [PubMed] [Google Scholar]
- 21. Panpradist N, Wang Q, Ruth PS, et al. : Simpler and faster Covid-19 testing: Strategies to streamline SARS-CoV-2 molecular assays. EBioMedicine 2021;64:103236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Panel on Antiretroviral Therapy and Medical Management of Children Living with HIV: Guidelines for the use of antiretroviral agents in pediatric HIV infection. Available at http://aidsinfo.nih.gov/content-files/lvguidelines/pediatricguidelines.pdf accessed March 10, 2021.