Abstract
Background
Tylocinum Y.C. Li & Zhu L. Yang 2016 is a Boletaceae genus belonging in subfamily Leccinoideae. It was described in 2016 from China and, prior to this study, it contained only one species, T.griseolum Y.C. Li & Zhu L. Yang 2016. During our survey of Boletaceae from Thailand, we collected some specimens that could be identified as a Tylocinum species, different from T.griseolum.
New information
The bolete specimens, collected in forests dominated by Dipterocarpaceae and Fagaceae in northern Thailand, are described as Tylocinumbrevisporum Raghoonundon & Raspé sp. nov. Macroscopic and microscopic descriptions with illustrations are provided, as well as a 3-gene phylogeny, which confirms the new taxon’s position in Tylocinum. Tylocinumbrevisporum differs from the only other known Tylocinum species (T.griseolum) by its brownish-grey colour, greyish-orange to brownish-orange colour change in the hymenophore when bruised, smaller pores (≤ 0.5 mm), longer tubes (up to 6 mm long), shorter and narrower basidiospores, longer and broader basidia and longer pleurocystidia relative to cheilocystidia. T.brevisporum is the second species from the genus Tylocinum and the only one to be found outside China thus far.
Keywords: new species, Boletaceae, Leccinoideae, molecular phylogeny, taxonomy, Thailand
Introduction
Tylocinum Y.C. Li & Zhu L. Yang 2016, is a monotypic genus of ectomycorrhizal (ECM) boletes (Boletaceae, Boletales, Agaricomycetes, Basidiomycota, Fungi). Typical characters of the genus are its dark scabrous stipe surface, white to pallid unchanging context in the pileus and stipe, white to pallid hymenophore, trichodermium pileipellis and smooth basidiospores (Wu et al. 2016). The type species Tylocinumgriseolum Y.C. Li & Zhu L. Yang 2016, was originally described from China and was the only species known from this genus at the time. The phylogenetic analyses by Wu et al. (2016) showed that Tylocinum forms a separate clade from all other generic clades in the subfamily Leccinoideae.
The plant family Dipterocarpaceae includes many species of large trees that are often dominant in the tropical and subtropical lowlands of Southeast Asia, where the species diversity of Dipterocarpaceae is highest (Ashton 1982, Hamilton et al. 2019). Many Dipterocarpaceae are well known to be ECM, symbiotically associating with various ECM fungi, including mushroom-forming species (Watling et al. 2002, Yuwa-Amornpitak et al. 2006, Brearley 2012). Several new genera and species of boletes have recently been documented from tropical dipterocarp forest (Desjardin et al. 2009, Neves et al. 2012, Hosen et al. 2013, Halling et al. 2014, Raspé et al. 2016, Wu et al. 2016, Vadthanarat et al. 2019, Chuankid et al. 2019). Members of the Fagaceae, which also form ECM associations, co-occur with dipterocarps in Southeast Asia (Smith et al. 2008), which promotes higher mycodiversity and ECM colonisation in those tropical forest ecosystems (Corrales et al. 2018).
In this study, we describe a new species of Tylocinum from dry dipterocarp forests of northern Thailand, with description, illustrations and molecular phylogenetic analyses of a multi-gene DNA sequence dataset (atp6, tef1 and rpb2).
Materials and methods
Specimens collected
Fresh basidiomata were collected during the rainy season (2019) from Chiang Mai and Chiang Rai Provinces, orthern Thailand. The basidiomata were photographed on-site and wrapped in aluminium foil. The descriptions of the macroscopic features were made on the same day, after which the basidiomata were dried in an electric drier at 45–50°C. Specimens were deposited in the Mae Fah Luang University (MFLU) or CMUB Herbaria.
Ecological, morphological and taxonomic study
The habitat, locality information and macro-chemical reactions on fresh basidiomata were recorded. Spore prints were taken for each collection. Colour codes were given using Kornerup and Wanscher (1978) as a guide. Microscopic characters were studied in the dried specimens. The following mounting solutions were used to observe the tissues: 10% aqueous potassium hydroxide (KOH) or 28–30% ammonium hydroxide (NH4OH) solutions or 1% ammoniacal Congo red solution. The microscopic structures were studied at magnifications of 60× and 100×, photographed with a calibrated Nikon Y-TV55 camera, fitted to a Nikon DIC microscope. A total of 60 basidiospores, 30 basidia, 30 pleurocystidia, 30 cheilocystidia and 30 terminal cells and 30 hyphae for both the pileipellis and stipitipellis were measured. The dimensions of the microscopic features are presented in the following format: (a–) b–c–d (−e), in which c represents the average, b the 5th percentile, d the 95th percentile and a and e the minimum and maximum values, respectively. Q, the length/width ratio for the spores, is presented in the same format. All microscopic features were drawn by free hand, using a drawing tube. Faces of Fungi (Jayasiri et al. 2015) and MycoBank numbers are provided for the new species.
DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted from CTAB-preserved tissues or dry specimens (ca. 10 mg) using a CTAB isolation procedure, adapted from Doyle and Doyle (1990). The atp6, tef1 and rpb2 gene regions were amplified by polymerase chain reaction (PCR). For amplification of atp6, the primers ATP6-1M40F and ATP6-2M were used (Raspé et al. 2016). EF1-983F and EF1-2218R (Rehner and Buckley 2005) were used to amplify tef1 and bRPB2-6F and bRPB2-7.1R (Matheny 2005) were used to amplify rpb2. The PCR amplification, purification and sequencing of atp6, rpb2 and tef1 were used following the procedure from Raspé et al. (2016).
Sequence alignment and phylogenetic analysis
The sequences were assembled using Geneious 8 (Biomatters). The Basic Local Alignment Search Tool (BLAST) (https://blast.ncbi.nlm.nih.gov/Blast.cgi) from GenBank was used to find the closest matches to the sequences. Reference sequences (Table 1) were downloaded and aligned using MAFFT v. 7 (Katoh and Standley 2013; http://mafft.cbrc.jp/alignment/server/). Then, the concatenated three-gene matrix was prepared.
Table 1.
List of collections used for DNA analyses, with origin, GenBank accession numbers and reference(s).
| Species | Voucher | Origin | atp 6 | tef 1 | rpb 2 | References |
| Baorangiamajor | OR0209 | Thailand | MG897421 | MG897431 | MG897441 | Phookamsak et al. (2019) |
| Baorangiapseudocalopus | HKAS75739 | China | – | KJ184570 | KM605179 | Wu et al. (2015) |
| Baorangiarufomaculata | BOTH4144 | USA | MG897415 | MG897425 | MG897435 | Phookamsak et al. (2019) |
| Borofutusdhakanus | OR0345 | Thailand | MH614660 | MH614709 | MH614755 | Vadthanarat et al. (2018) |
| Ionosporuslongipes | LEE1180 | Malaysia | MT085461 | MT085471 | MH712031 | Khmelnitsky et al. (2019) |
| Lanmaoaasiatica | OR0228 | China | MH614682 | MH614730 | MH614777 | Vadthanarat et al. (2019) |
| Lanmaoacarminipes | BOTH4591 | USA | MG897419 | MG897429 | MG897439 | Phookamsak et al. (2019) |
| Lanmaoapallidorosea | BOTH4432 | USA | MG897417 | MG897427 | MG897437 | Phookamsak et al. (2019) |
| Leccinummonticola | HKAS76669 | China | – | KF112249 | KF112723 | Wu et al. (2014) |
| Leccinumquercinum | HKAS63502 | China | – | KF112250 | KF112724 | Wu et al. (2014) |
| Leccinumscabrum | RW105a | Belgium | KT823979 | KT824045 | KT824012 | Raspé et al. (2016) |
| Leccinumscabrum | VDKO0938 | Belgium | MG212549 | MG212593 | MG212635 | Vadthanarat et al. (2018) |
| Leccinumschistophilum | VDKO1128 | Belgium | KT823989 | KT824055 | KT824022 | Raspé et al. (2016) |
| Leccinumvariicolor | HKAS57758 | China | – | KF112251 | KF112725 | Wu et al. (2014) |
| Leccinumvariicolor | VDKO0844 | Belgium | MG212550 | MG212594 | MG212636 | Vadthanarat et al. (2018) |
| Leccinellumaff.crocipodium | HKAS76658 | China | – | KF112252 | KF112728 | Wu et al. (2014) |
| Lecinellumcf.intusrubens | OR0082 | Thailand | MZ803019 | MZ803024 | MZ824749 | This study |
| Leccinellumcrocipodium | VDKO1006 | Belgium | KT823988 | KT824054 | KT824021 | Raspé et al. (2016) |
| Leccinellumcremeum | HKAS90639 | China | – | KT990781 | KT990420 | Wu et al. (2016) |
| Leccinellum sp. | HKAS53427 | China | – | KF112253 | KF112727 | Wu et al. (2014) |
| Leccinellum sp. | OR0711 | Thailand | MH614685 | MH614733 | MH614780 | Vadthanarat et al. (2019) |
| Octavianiahesperi | KPM-NC 17793 | Japan | KC552150 | JN378422 | – | Orihara et al. (2016) |
| Octavianiajaponimontana | KPM-NC 17797 | Japan | KC552151 | JN378425 | – | Orihara et al. (2016) |
| Octavianianonae | KPM-NC 17748 | Japan | KC552143 | JN378403 | – | Orihara et al. (2016) |
| Octavianiatasmanica | MEL 2341996 | Australia | KC552156 | JN378436 | – | Orihara et al. (2012), Orihara et al. (2016) |
| Octavianiazelleri | MES270 | USA | KC552161 | JN378440 | – | Orihara et al. (2012), Orihara et al. (2016) |
| Pseudoaustroboletuscf.valens | OR0477 | China | MZ803020 | MZ803025 | MZ824750 | This study |
| Retiboletusbrevibasidiatus | OR0570 | Thailand | MT085469 | MT085476 | MT085479 | Chuankid et al. (2021) |
| Retiboletusbrunneolus | HKAS 52680 | China | – | KF112179 | KF112690 | Wu et al. (2014) |
| Retiboletusfuscus | OR0231 | China | MG212556 | MG212600 | MG212642 | Vadthanarat et al. (2018) |
| Retiboletusfuscus | OR0738 | Thailand | MT085462 | MT085472 | MT085477 | Chuankid et al. (2021) |
| Retiboletusgriseus | MB03-079 | USA | KT823964 | KT824030 | KT823997 | Raspé et al. (2016) |
| Retiboletuskauffmanii | OR0278 | China | MG212557 | MG212601 | MG212643 | Vadthanarat et al. (2018) |
| Retiboletusnigrogriseus | BC0179 | Thailand | MT085464 | MT085474 | MT085478 | Chuankid et al. (2021) |
| Retiboletusnigrogriseus | OR049 | Thailand | KT823967 | KT824000 | KT824033 | Raspé et al. (2016) |
| Retiboletusornatipes | MBsn | USA | MT219514 | MT219516 | MT219515 | Chuankid et al. (2021) |
| Rhodactinarostratispora | SV170 | Thailand | MG212560 | MG212605 | MG212645 | Vadthanarat et al. (2018) |
| Rossbeeveraeucyanea | TUMH-40252 | Japan | KC552116 | KC552069 | – | Orihara et al. (2016) |
| Rossbeeveragriseovelutina | TUMH-40266 | Japan | KC552121 | KC552073 | – | Orihara et al. (2016) |
| Rossbeeveravittatispora | A.W. Claridge 2137 | Australia | KC552105 | KC552063 | – | Orihara et al. (2016) |
| Spongiformathailandica | DED7873 | Thailand | MG212563 | KF030436 | MG212648 |
Nuhn et al. (2013), Vadthanarat et al. (2018) |
| Spongisporatemasekensis | ACMF5 | Singapore | MZ803018 | MZ803023 | MZ824748 | This study |
| Turmalineamesomorphasubsp.mesomorpha | KPM-NC 18012 | Japan | KC552139 | KC552090 | – | Orihara et al. (2016) |
| Turmalineapersicina | KPM-NC 18001 | Japan | KC552130 | KC552082 | – | Orihara et al. (2016) |
| Turmalinea sp. | Muroi361 | USA | DQ218885 | DQ219224 | DQ219046 | Orihara et al. (2016) |
| Tylocinumgriseolum | HKAS50281 | China | – | KF112284 | KF112730 | Wu et al. (2014) |
| Tylocinumbrevisporum | OR622 | Thailand | MZ803021 | – | MZ824751 | This study |
All analyses were done on the CIPRES Science Gateway (https://www.phylo.org; Miller et al. 2012). Maximum Likelihood (ML) phylogenetic tree inference was done using RAxML-HPC2 v.8.2.10 (Stamatakis 2006), using the GTRCAT model of sequence evolution with 25 categories. Three Lanmaoa species and three Baorangia species were selected as outgroup. Four partitions were defined: atp6, tef1 exons, rpb2 exons and introns. Statistical support of the clades was obtained using 1,000 rapid bootstrap replicates.
Using jModeltest2 (Darriba et al. 2012) on XSEDE via the CIPRES Science Gateway, the best-fit model of substitution for analysis in MrBayes was estimated for each gene, based on the Bayesian Information Criterion (BIC). GTR + I + G for atp6 and introns, SYM + I + G for tef1 exons and K80 + I + G for rpb2 exons were selected as the best fit models. Partitioned Bayesian analysis was performed with MrBayes 3.2.7a (Ronquist et al. 2012). Two runs of four cold and one heated chains were run for 1,000,000 generations and sampled every 200 generations. The average standard deviation of split frequencies was 0.005106 at the end of the runs. The burn-in phase (25%) was estimated by checking the stationarity in the plot generated by the sump command.
Taxon treatments
Tylocinum brevisporum
Raghoonundon & Raspé sp. nov.
64D99B61-7573-598D-8BE9-02D0AC73258B
MB841102
FoF 10255
Materials
Type status: Holotype. Taxon: kingdom: Fungi; phylum: Basidiomycota; class: Agaricomycetes; order: Boletales; family: Boletaceae; genus: Tylocinum; specificEpithet: brevisporum; taxonRank: species; Location: country: Thailand; stateProvince: Chiang Rai Province, Chang Wat, Doi Pui; verbatimElevation: 730 m; verbatimCoordinates: 19°48'50"N, 99°51'57"E; Identification: identifiedBy: Bhavesh Raghoonundon; Event: eventDate: 20 August 2019; Record Level: institutionID: MFLU 21-0144; institutionCode: Mae Fah Luang University Herbarium; collectionCode: BR137
Type status: Other material. Taxon: kingdom: Fungi; phylum: Basidiomycota; class: Agaricomycetes; order: Boletales; family: Boletaceae; genus: Tylocinum; specificEpithet: brevisporum; taxonRank: species; Location: country: Thailand; stateProvince: Chiang Mai Province, Mueang District; verbatimElevation: 450 m; verbatimCoordinates: 18°48'40"N, 98°56'31"E; Identification: identifiedBy: Olivier Raspé; Event: eventDate: 18 May 2015; Record Level: institutionID: CMU-B OR622; collectionID: OR622; institutionCode: Chiang Mai University Herbaria
Description
Basidiomata pileo-stipitate, small to medium-sized (Fig. 1). Pileus (1.5–)2.0–2.5 cm in diameter, convex when young, becoming plano-depressed with age; margin deflexed to uplifted, surface finely tomentose, dull and dry, at first brown (7E4) to greyish-brown (8E3–8F4), becoming paler (8D3) near the margin with age; context 3–5 mm thick halfway to the margin, soft and fleshy, off-white, slightly browning on exposure. Stipe central, cylindrical, (3.4–)4.9–6.5 cm × 0.6–1.3 cm, surface even, dull and dry, scabrous, covered with granular squamules (dotted-verrucose), brownish-grey (7E2–8E2) when young to reddish-brown (8E5) to dark brown (8F5) with age, no colour change when bruised, basal mycelium off-white; context solid, fleshy, off-white, reddish-brown to dark brown near the stipe base (8F7) and in worm wounds, slightly browning on exposure. Hymenophore tubulate, subventricose, adnexed, slightly depressed around apex of the stipe, greyish-orange to brownish-orange when bruised. Tubes 3–6 mm long halfway to the margin, off-white, easily separable from one another. Pores ≤ 0.5 mm wide at mid-radius, regularly arranged, angular, off-white, turning brown to dark brown (8E5–8F5) when bruised. Odour fungoid. Taste bitter. Spore print not obtained.
Figure 1.
Photograph of Tylocinumbrevisporum sp. nov. a, b Basidioma of specimen OR622; c Basidioma of the holotype (BR 137).
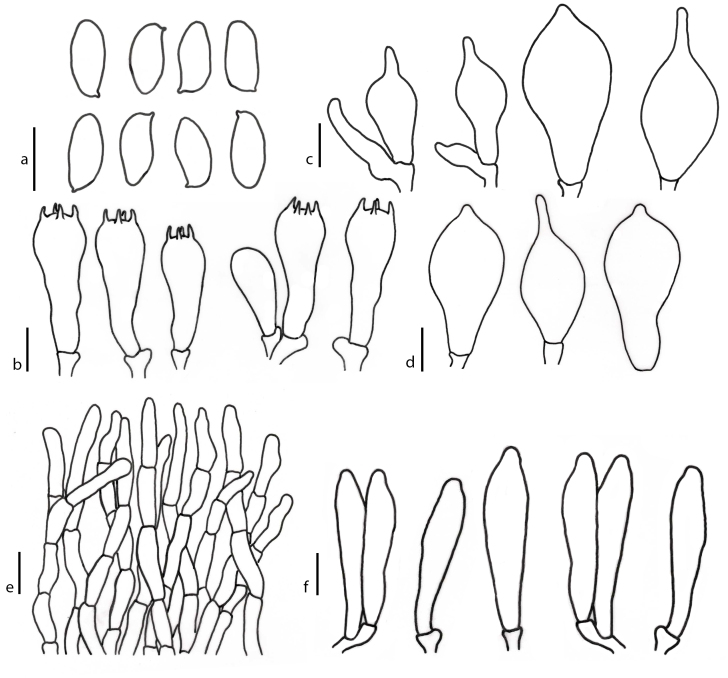
Basidiospores (6.7–)7.5–10–11.7(–11.8) × (3.1–)3.5–4.7–5.8(–5.9) µm (n = 50) Q = (1.7–) 1.79–2.15–2.5 (–2.61), ellipsoid in central view, oblong to subcylindrical in side view, smooth under light microscope, yellowish to brownish in KOH (Fig. 2). Basidia 4-spored, (27–)27–37.4–54(–54) × (9–)9–12.3–19(–19) µm, clavate, yellowish to brownish in KOH, sterigmata up to 3 µm long. Cheilocystidia (19–)19.3–25.5–33(–35) × (4–)4.1–6–8.2(–8.5) µm, frequent, fusiform, thin-walled, yellowish to brownish hyaline in KOH and NH4OH. Pleurocystidia (40–)41–53–69(–70) × (8–)7.4–12–16.6(–17) µm, thin-walled, fusiform to broadly fusiform with a long pedicel and sharp apex, occasionally containing yellowish inclusions, yellowish to brownish hyaline in KOH and NH4OH. Hymenophoral trama boletoid, elements smooth, cylindrical, hyaline, 5–10 µm wide. Pileipellis a trichodermium, hyphae terminations with 3–4 cells that are 5–11 µm wide and terminal cells 31–48 µm × 6–10 µm, colourless to slightly brownish in KOH. Pileus trama composed of interwoven hyaline hyphae 5–9 µm wide. Stipitipellis a disrupted hymeniderm with hyphae 3.7–7.4 µm wide, colourless to slightly brownish in KOH and caulocystidia (24–)24.5–35–47(–48) × (9–)9.2–12.4–16.9(–17) µm, thin-walled, clavate to broadly clavate with a sharp apex, yellowish to brownish hyaline in KOH and NH4OH. Stipe trama composed of cylindrical, hyaline, interwoven hyphae 3.7–7.4 µm wide. Clamp connections absent.
Figure 2.
Microscopic features of Tylocinumbrevisporum; a Basidiospores; b Basidia; c, d Caulocystidia; e Pleurocystidia; f Cheilocystidia; g Pileipellis. Scale bars: a, b, c, d, f = 10 µm, e = 20 µm, g = 50 µm.
Diagnosis
This species is distinguished from Tylocinumgriseolum by its greyish-brown colour, greyish-orange to brownish-orange colour change in the hymenophore when bruised, smaller pores (≤ 0.5 mm) and longer tubes (up to 6 mm long). Additionally, the basidiospores are shorter and narrower compared to T.griseolum and the basidia are slightly longer and broader. Furthermore, the pleurocystidia of Tylocinumbrevisporum are longer than its cheilocystidia.
Etymology
Epithet “brevisporum”; from the Latin words brevi (short) and sporae (spores), referring to the shorter spores of this species compared to Tylocinumgriseolum.
Distribution
Thus far known only from northern Thailand.
Ecology
Solitary, in tropical forest dominated by Dipterocarpaceae (Dipterocarpus spp. and Shorea spp.), with some Fagaceae (Quercus spp., Lithocarpus spp. and Castanopsiscalathiformis).
Notes
Morphologically, Tylocinumbrevisporum is similar to Tylocinumgriseolum, with which it shares the overall grey colour of the basidiomata and dark scabrous stipe surface. However, Tylocinumbrevisporum is more brownish as compared to the grey Tylocinumgriseolum. In addition, Wu et al. (2016) mentioned no discolouration in the context of Tylocinumgriseolum. The context of Tylocinumbrevisporum becomes slightly brown when bruised. The hymenophore of T.brevisporum changes to greyish-orange to brownish-orange when bruised as compared to the unchanging hymenophore of T.griseolum. Moreover, T.griseolum has relatively larger pores (up to 1.5 mm) than that of T.brevisporum (< 0.5 mm). The tubes in T.griseolum are also shorter than those of T.brevisporum.
The basidiospores of Tylocinumbrevisporum [(6.7–)7.5–10–11.7(–11.8) × (3.1–)3.5–4.7–5.8(–5.9) µm, Q = (1.7–)1.79–2.15–2.5(–2.61)] are shorter and narrower than those of Tylocinumgriseolum [(11)12.0–14.5(16) × 4.5–5.5 µm Q = 2.60–3.22] from China. The basidia of T.brevisporum [(27–)27–37.4–54(–54) × (9–)9–12.3–19(–19) µm] are also slightly longer and broader than T.griseolum [30–45 × 10–12 µm]. Wu et al. (2016) reported that, for T.griseolum, the pleurocystidia and cheilocystidia are similarly-sized. In T.brevisporum, the pleurocystidia are longer than the cheilocystidia. Phylogenetically, T.brevisporum clusters with T.griseolum, together forming a well-supported clade (MLB/BPP = 93/1.00) i.e. the genus Tylocinum.
Analysis
Phylogenetic analysis
The concatenated gene dataset comprised 47 terminals. The final alignment contained 121 sequences (38 for atp6, 46 for tef1, 37 for rpb2) and was 2,676 characters long, including gaps. Both ML and Bayesian analyses produced the same tree topology; thus, only the ML tree is shown with both Maximum Likelihood Bootstrap (MLB) and Bayesian Posterior Probabilities (BPP) values. In the analyses, the new species Tylocinumbrevisporum shared a sister relationship with the type species Tylocinumgriseolum (Fig. 3), providing strong statistical support (MLB = 93 and BPP = 1.00) for the genus Tylocinum (Leccinoideae). The atp6 sequence of the holotype (BR 137) was 100% identical to OR622.
Figure 3.
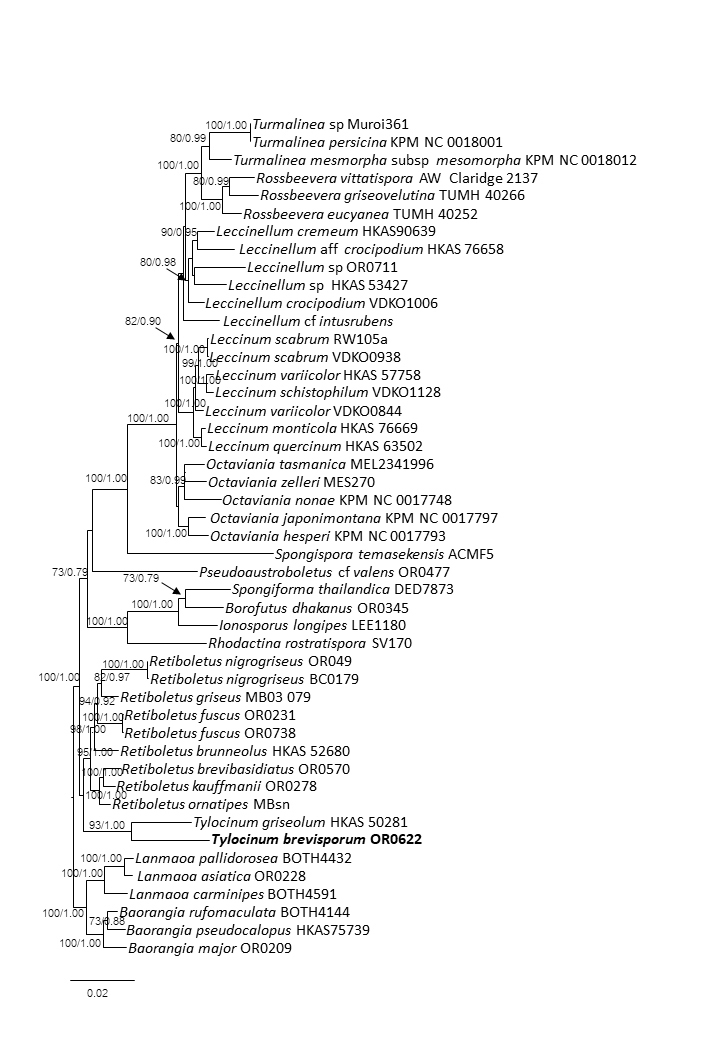
Maximum Likelihood phylogenetic tree inferred from the three-gene dataset (atp6, rpb2, tef1). The three Lanmaoa and three Baorangia species were used as outgroup taxa. Maximum Likelihood Bootstrap (MLB, left) ≥ 70% and Bayesian Posterior Probabilities (BPP, right) ≥ 0.95 are shown above supported branches. The new species is in bold.
Discussion
Boletales is a globally-distributed order of fungi, comprising morphologically diverse groups (Binder and Hibbett 2006, Wu et al. 2016), with ECM, ligninolytic, saprobic and mycoparasitic members (Binder and Hibbett 2006, Kirk et al. 2008). Thorough morphological and phylogenetic analyses of the order has led to the discovery of new genera and other taxa (e.g. Binder and Bresinsky 2002, Wu et al. 2014, Zhu et al. 2015, Wu et al. 2016, Orihara et al. 2016, Vadthanarat et al. 2019, Zhang et al. 2019). Boletaceae Chevall. 1826 is a morphologically diverse family currently comprising of 94 genera distributed amongst seven subfamilies (Binder and Hibbett 2006, Wu et al. 2014, Wu et al. 2016). The subfamily Leccinoideae was revealed by the phylogenetic analyses of Wu et al. (2014). Currently, this subfamily comprises fifteen genera, viz. Binderoboletus T.W. Henkel & M.E. Sm. 2016, Borofutus Hosen & Z.L. Yang 2012, Chamonixia Rolland 1899, Ionosporus Khmeln. 2018, Kaziboletus Iqbal Hosen & Zhu L. Yang 2021, Leccinum Gray 1821, Lecinellum Bresinsky & Manfr. Binder 2003, Pseudoaustroboletus Y.C. Li & Zhu L. Yang 2014, Octavania, Retiboletus Manfr. Binder & Bresinsky 2002, Rossbeevera T. Lebel & Orihara 2012, Rhodactina Pegler & T.W.K. Young 1989, Spongiforma Desjardin, Manfr. Binder, Roekring & Flegel 2009, Spongispora G. Wu, S.M.L. Lee, E. Horak & Z.L. Yang 2018, Turmalinea Orihara & N. Maek. 2015 and Tylocinum. Only ten of these genera are stipitate-pileate.
Our survey on the diversity of boletes in northern Thailand led to the discovery of a second species of Tylocinum (the focus of the present study), being found in tropical forests dominated by Dipterocarpaceae, which have been reported as ECM hosts for Boletaceae (Desjardin et al. 2009, Halling et al. 2014, Wu et al. 2018, Vadthanarat et al. 2019). According to Wu et al. (2016), the white to dirty white hymenophore of Tylocinum is similar to that of Tylopilus Karst. 1881 when young, while the verrucose stipe surface is similar to Leccinum. The stipe surface of Tylocinum is dotted-verrucose, which may give a more or less rough touch, but it does not produce markedly projecting scabers like in Leccinum. Tylocinum is also similar to Tylopilus, but there are some morphological differences between the two genera. Tylopilus species usually produce larger basidiomata and have minutely and densely tomentose to dotted-tomentose, but never dotted-verrucose, stipitipellis. Moreover, some Tylopilus species have reticulate stipe, whereas, in Tylocinum, the stipe is at most longitudinally venose near the apex. As the diversity of Boletaceae in Thailand is high and remains understudied (e.g. Vadthanarat et al. 2021), further studies may uncover additional species of Tylocinum or related taxa.
Supplementary Material
Acknowledgements
B. Raghoonundon appreciates the kind support given by the Mushroom Research Foundation (MRF), Thailand. The authors thank the Thailand Science Research and Innovation (TSRI) (Grant No. DBG6280009) entitled “Macrofungi diversity research from the Lancang-Mekong Watershed and surrounding areas”. The authors are also grateful to Amy Choong for loaning her collection of Spongisporatamasekensis, Changlin Zhao for sequencing of Tylocinum specimens and to the reviewers for their comments.
References
- Ashton P. Dipterocarpaceae . The Hague, The Netherlands: Martinus-Nijhoff Publishers.; 1982. [Google Scholar]
- Binder M., Bresinsky A. Derivation of a polymorphic lineage of Gasteromycetes from boletoid ancestors. Mycologia. 2002;94:85–98. doi: 10.1080/15572536.2003.11833251. [DOI] [PubMed] [Google Scholar]
- Binder M, Hibbett D. S. Molecular systematics and biological diversification of Boletales. Mycologia. 2006;98:971–981. doi: 10.1080/15572536.2006.11832626. [DOI] [PubMed] [Google Scholar]
- Brearley F. Q. Ectomycorrhizal associations of the Dipterocarpaceae. Biotropica. 2012;44(5):637–648. doi: 10.1111/j.1744-7429.2012.00862.x. [DOI] [Google Scholar]
- Chuankid B, Vadthanarat S, Hyde K. D., Raspé Olivier, et al. Three new Phylloporus species from tropical China and Thailand. Mycological Progress. 2019;18(5):603–614. doi: 10.1007/s11557-019-01474-6. [DOI] [Google Scholar]
- Chuankid B, Vadthanarat S, Thongbai B, Stadler Marc, et al. Retiboletus (Boletaceae) in Northern Thailand: one novel species and two first records. Mycoscience. 2021;62:1–10. doi: 10.47371/mycosci.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales A, Henkel T. W., Smith M. E. Ectomycorrhizal associations in the tropics–biogeography, diversity patterns and ecosystem roles. New Phytologist. 2018;220(4):1076–1091. doi: 10.1111/nph.15151. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada G. L., Doallo, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods. 2012;9(8):772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardin D, Binder M., Roekring S., Flegel T. Spongiforma, a new genus of gasteroid boletes from Thailand. Fungal Diversity. 2009;37:1–8. [Google Scholar]
- Doyle J. J, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Halling R, Desjardin D, Fechner N, Arora D, et al. New porcini (Boletussect.Boletus) from Australia and Thailand. Mycologia. 2014;106:830–834. doi: 10.3852/13-340. [DOI] [PubMed] [Google Scholar]
- Hamilton R., Hall T., Stevenson J., Penny D. Distinguishing the pollen of Dipterocarpaceae from the seasonally dry and moist tropics of south-east Asia using light microscopy. Review of Palaeobotany and Palynology. 2019;263:117–133. doi: 10.1016/j.revpalbo.2019.01.012. [DOI] [Google Scholar]
- Hosen MI, Feng B, Wu G, Zhu XT, et al. Borofutus, a new genus of Boletaceae from tropical Asia: phylogeny, morphology and taxonomy. Fungal Diversity. 2013;58:215–226. doi: 10.1007/s13225-012-0211-8. [DOI] [Google Scholar]
- Jayasiri SC, Hyde KD, Ariyawansa HA, Bhat J, et al. The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts. https://www.facesoffungi.org/ Fungal Diversity. 2015;74(1):3–18. doi: 10.1007/s13225-015-0351-8. [DOI] [Google Scholar]
- Katoh K, Standley DM. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelnitsky O, Davoodian N, Singh P, et al. Ionosporus: a new genus for Boletuslongipes (Boletaceae), with a new species, I.australis, from Australia. Mycological Progress. 2019;18(3):439–451. doi: 10.1007/s11557-018-01463-1. [DOI] [Google Scholar]
- Kirk P. M., Cannon P. F., Minter D. W., Stalpers J. A. Dictionary of the Fungi. 10th edition. CABI International, Wallingford.; 2008. [Google Scholar]
- Kornerup A, Wanscher J. H. Methuen Handbook of Colour. Eyre Methuen; 1978. [Google Scholar]
- Matheny P. B. Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Molecular Phylogenetics and Evolution. 2005;35:1–20. doi: 10.1016/j.ympev.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Miller Mark A., Pfeiffer W, Schwartz T. The CIPRES science gateway: enabling high-impact science for phylogenetics researchers with limited resources. XSEDE '12: Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the eXtreme to the campus and beyond. 2012;39:1–8. doi: 10.1145/2335755.2335836. [DOI] [Google Scholar]
- Neves M. A., Binder M., Halling R., Hibbett D, et al. The phylogeny of selected Phylloporus species inferred from NUC–LSU and ITS sequences, and descriptions of new species from the Old World. Fungal Diversity. 2012;55(1):109–123. doi: 10.1007/s13225-012-0154-0. [DOI] [Google Scholar]
- Nuhn M. E., Binder M., Taylor A. F.S, Halling R. E, et al. Phylogenetic overview of the Boletineae. Fungal Biology. 2013;117:479–511. doi: 10.1016/j.funbio.2013.04.008. [DOI] [PubMed] [Google Scholar]
- Orihara T, Smith M. E, Shimomura N., Iwase K, et al. Diversity and systematics of the sequestrate genus Octaviania in Japan: two new subgenera and eleven new species. Persoonia. 2012;28:85–112. doi: 10.3767/003158512X650121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orihara T., Lebel T., Ge Z. W., Smith M. E, et al. Evolutionary history of the sequestrate genus Rossbeevera (Boletaceae) reveals a new genus Turmalinea and highlights the utility of ITS minisatellite–like insertions for molecular identification. Persoonia. 2016;37:173–198. doi: 10.3767/003158516X691212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phookamsak R, Hyde K. D, Jeewon R, et al. Fungal diversity notes 929-1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Diversity. 2019;95:1–273. doi: 10.1007/s13225-019-00421-w. [DOI] [Google Scholar]
- Raspé O., Vadthanarat S., Kesel A. De, Degreef J., et al. Pulveroboletusfragrans, a new Boletaceae species from Northern Thailand, with a remarkable aromatic odor. Mycological Progress. 2016;15(38):38. doi: 10.1007/s11557-016-1179-7. [DOI] [Google Scholar]
- Rehner S. A, Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97:84–98. doi: 10.1080/15572536.2006.11832842. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Mark P Van Der, Ayres DL, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. E, Read D. J, Smith S. E, Read D. Structure and development of ectomycorrhizal roots. Mycorrhizal Symbiosis. 2008;3:191–268. doi: 10.1002/9781118951446.ch4. [DOI] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Vadthanarat S, Raspé O, Lumyong S. Phylogenetic affinities of the sequestrate genus Rhodactina (Boletaceae), with a new species, R.rostratispora from Thailand. MycoKeys. 2018;29:63–80. doi: 10.3897/mycokeys.29.22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadthanarat S, Saisamorn L., Raspé O. Cacaoporus, a new Boletaceae genus, with two new species from Thailand. MycoKeys. 2019;54:1–29. doi: 10.3897/mycokeys.54.35018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadthanarat S, Halling R. E, Amalfi M., Lumyong S. An unexpectedly high number of new Sutorius (Boletaceae) species from Northern and Northeastern Thailand. Frontiers in Microbiology. 2021;12:643505. doi: 10.3389/fmicb.2021.643505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watling R, Lee S. S, Turnbull E. The occurrence and distribution of putative ectomycorrhizal basidiomycetes in a regenerating South-East Asian rain forest. Tropical Mycology. 2002;1:25–43. [Google Scholar]
- Wu G, Feng B, Xu J, et al. Molecular phylogenetic analyses redefine seven major clades and reveal 22 new generic clades in the fungal family Boletaceae. Fungal Diversity. 2014;69:93–115. doi: 10.1007/s13225-014-0283-8. [DOI] [Google Scholar]
- Wu G, Zhao K, Li Y. C, et al. Four new genera of the fungal family Boletaceae. Fungal Diversity. 2015;81:1–24. doi: 10.1007/s13225-015-0322-0. [DOI] [Google Scholar]
- Wu G, Li Y. C., Zhu X. T. One hundred noteworthy boletes from China. Fungal Diversity. 2016;81:25–188. doi: 10.1007/s13225-016-0375-8. [DOI] [Google Scholar]
- Wu G, Lee S. M.L, Horak E, Yang Z. L. Spongisporatemakensis, a new boletoid genus and species from Singapore. Mycologia. 2018;1105:919–929. doi: 10.1080/00275514.2018.1496387. [DOI] [PubMed] [Google Scholar]
- Yuwa-Amornpitak T., Vichitsoonthonkul T., Tanticharoen M., Cheevadhanarak S., et al. Diversity of ectomycorrhizal fungi on Dipterocarpaceae in Thailand. Journal of Biological Sciences. 2006;6:1059–1064. doi: 10.3923/jbs.2006.1059.1064. [DOI] [Google Scholar]
- Zhang M., Li T. H., Wang C. Q., Zeng N. K., et al. Phylogenetic overview of Aureoboletus (Boletaceae, Boletales), with descriptions of six new species from China. MycoKeys. 2019;61:111–145. doi: 10.3897/mycokeys.61.47520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X. T, Wu G, Zhao K., Halling R. E, et al. Hourangia, a new genus of Boletaceae to accommodate Xerocomuscheoi and its allied species. Mycological Progress. 2015;14:37. doi: 10.1007/s11557-015-1060-0. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.