Abstract
Objectives
Identification of patients with coronavirus disease 2019 (COVID‐19) at risk for deterioration after discharge from the emergency department (ED) remains a clinical challenge. Our objective was to develop a prediction model that identifies patients with COVID‐19 at risk for return and hospital admission within 30 days of ED discharge.
Methods
We performed a retrospective cohort study of discharged adult ED patients (n = 7529) with SARS‐CoV‐2 infection from 116 unique hospitals contributing to the National Registry of Suspected COVID‐19 in Emergency Care. The primary outcome was return hospital admission within 30 days. Models were developed using classification and regression tree (CART), gradient boosted machine (GBM), random forest (RF), and least absolute shrinkage and selection (LASSO) approaches.
Results
Among patients with COVID‐19 discharged from the ED on their index encounter, 571 (7.6%) returned for hospital admission within 30 days. The machine‐learning (ML) models (GBM, RF, and LASSO) performed similarly. The RF model yielded a test area under the receiver operating characteristic curve of 0.74 (95% confidence interval [CI], 0.71–0.78), with a sensitivity of 0.46 (95% CI, 0.39–0.54) and a specificity of 0.84 (95% CI, 0.82–0.85). Predictive variables, including lowest oxygen saturation, temperature, or history of hypertension, diabetes, hyperlipidemia, or obesity, were common to all ML models.
Conclusions
A predictive model identifying adult ED patients with COVID‐19 at risk for return for return hospital admission within 30 days is feasible. Ensemble/boot‐strapped classification methods (eg, GBM, RF, and LASSO) outperform the single‐tree CART method. Future efforts may focus on the application of ML models in the hospital setting to optimize the allocation of follow‐up resources.
Keywords: clinical prediction model, COVID‐19, discharge planning, emergency department, machine learning, prognosis, readmissions, SARS‐CoV‐2
1. INTRODUCTION
1.1. Background
As of December 2020, there were an estimated 97 million symptomatic cases of coronavirus disease 2019 (COVID‐19) resulting in 4.1 million hospitalizations, and >535,000 attributed deaths in the United States. 1 Limitation of inpatient hospital resources has been linked with excess patient mortality and may contribute to disparities in regional outcomes. 2 , 3 Although some patients with COVID‐19 may present in severe distress and clearly require hospital admission, the majority will present with mild symptoms and appear well enough to be discharged from the emergency department (ED). However, within this discharged cohort, a potentially important but understudied fraction will experience disease progression and deterioration requiring a return ED visit with hospitalization. 4 Clinical uncertainty exists around predicting the likelihood of disease progression and deterioration in this cohort.
1.2. Importance
This study provides the first predictive model from a national sample of patients with COVID‐19 discharged from the ED to identify those most at risk for disease progression and most likely to benefit from outpatient case management services and support after discharge. In practice, such a model has the potential to aid in the allocation of scarce case management and home health equipment, such as pulse oximeters, during a pandemic and may have the potential to alleviate hospital crowding during COVID surges and improve patient outcomes.
1.3. Goals of this investigation
To develop and compare clinical prediction models that identify patients at risk for return hospital admission within 30 days following COVID‐19 diagnoses and discharge from the ED.
2. METHODS
2.1. Study design and setting
We performed a retrospective cohort study of adult (aged ≥18 years) patients discharged from the ED with polymerase chain reaction (PCR)–confirmed severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection. Data were obtained from the National Registry of Suspected COVID‐19 in Emergency Care (RECOVER) network, a large ED‐based COVID‐19 registry with patient data from 116 EDs within 40 hospital systems across 27 US states. 5 , 6 Data from 80,176 patient visits beginning the first week of February until the final week of October 2020 were downloaded from the registry on March 15, 2021. The RECOVER registry protocol was reviewed by the institutional review board (IRB) at each contributing site. The transparency in reporting of a multivariate prediction model for individual diagnosis and prognosis guidelines were adhered to during the database design, data collection, recording, and data analysis phases of the project. 7 This was a preplanned analysis.
2.2. Selection of participants
Adult patients from the RECOVER network discharged from EDs with positive molecular RNA tests for SARS‐CoV‐2 infection were included. Patients who were asymptomatic who received SARS‐CoV‐2 testing for automated or administrative purposes (eg, preoperative testing) were excluded. Patients were enrolled only once.
2.3. Measures
The RECOVER case report form (CRF) included 204 questions with 360 answers capturing demographics, past medical history, home medications, vital signs, test results, 28 symptoms from the International Severe Acute Respiratory and Emerging Infection Consortium (ISARIC)/World Health Organization's Clinical Characterization Protocol for Severe Infectious Diseases, and 14 contact exposure risks. 8 Selection of potential covariates from the original RECOVER CRF occurred using a modified Delphi method before all analyses and was based on published literature and expert author consensus regarding risk factors for moderate to severe COVID‐19 illness. Furthermore, candidate variables were chosen based on availability of data that could be used in real time to predict downstream need for healthcare use. The final model input selection reduced candidate variables to 128 demographic, social, and clinical features.
2.4. Outcomes
The primary outcome for this analysis was return ED visits resulting in hospital admission (return hospital admission) within 30 days of the index visit. Standard instructions were given to site investigators to search their electronic medical records and data available via Care Everywhere or other shared health information exchanges or data marts across health systems for any ED or hospital encounters in the 30 days after the index ED visit.
2.5. Analyses
Descriptive statistics and model development were performed with RStudio version 1.3.1093, running R version 4.0.3. 9 , 10 , 11 Covariates with zero variability or >15% missingness were excluded from the analyses. Missing values were imputed. 12 The distribution of all features for each cohort was visualized, and summary statistics were calculated. 13 , 14 Vital signs were winsorized setting outliers to the 0.5% and 99% percentiles to improve the model robustness by minimizing the influence of outlier continuous values. 15 Model cut‐points were established to achieve a 95% negative predictive value (NPV). 16 This threshold was chosen given a low tolerance for incorrect negative predictions from the model given its planned utility.
We developed 3 machine‐learning (ML) models using gradient boosting machine (GBM), random forest (RF), and least absolute shrinkage and selection operator (LASSO) regression. 17 , 18 , 19 A classification and regression tree (CART) reference model was also derived. 20 Hyperparameter tuning was performed. 21 Data were randomly partitioned into a 70% training set and a 30% test set balanced by the outcome variable. All models were trained using 10‐fold cross‐validation.
The Bottom Line.
Identification of patients with coronavirus disease 2019 (COVID‐19) at risk for deterioration after discharge from the emergency department (ED) remains a clinical challenge that might be aided by specific prediction models. Using a retrospective cohort of >7500 discharged adult ED patients with SARS‐CoV‐2 infection from 116 unique hospitals contributing to the National Registry of Suspected COVID‐19 in Emergency Care (RECOVER), 4 machine‐learning statistical models were employed to predict patients with COVID‐19 who might be at low risk for discharge without 30‐day return hospitalization. The models performed similarly, and 571 (7.6%) patients returned for hospital admission within 30 days. Predictive variables, including lowest oxygen saturation, temperature, or history of hypertension, diabetes, hyperlipidemia, or obesity, were common to all models. Machine learning aided predictive models identifying adult ED patients with COVID‐19 at risk for return for return hospital admission within 30 days of ED discharge is feasible.
The prediction performance of each model was evaluated by calculating the area under the receiver operating characteristic curve (AUC) and calculating statistical measures of classification performance (ie, sensitivity, specificity, positive predictive value [PPV], and NPV) at the aforementioned prespecified cut point. Permutation‐based model‐specific variable importance plots (VIPs) were used to examine the contribution of each predictor in the RF and GBM ML models using the vip package in R. 22 These are determined by the normalized mean value of difference between the prediction accuracy of the out‐of‐the‐bag estimation and that of the same measure after permuting each predictor. Regression coefficients were calculated for the LASSO model.
3. RESULTS
3.1. Characteristics of study subjects
From the original cohort of 80,176 patients, 72,323 adults (aged ≥18 years) were identified, with 16,791 patients found to have SARS‐CoV‐2 by PCR (Figure 1). The analysis group consisted of 7529 patients discharged from the ED with known ED return visit outcome data. Within this cohort, 571 returned to the ED and were subsequently admitted to the hospital within 30 days of ED discharge for an event rate of ≈7.6%. Univariate comparisons of patient cohort demographics, social history, and major comorbid diagnoses are listed in Table 1 and Table S1.
FIGURE 1.
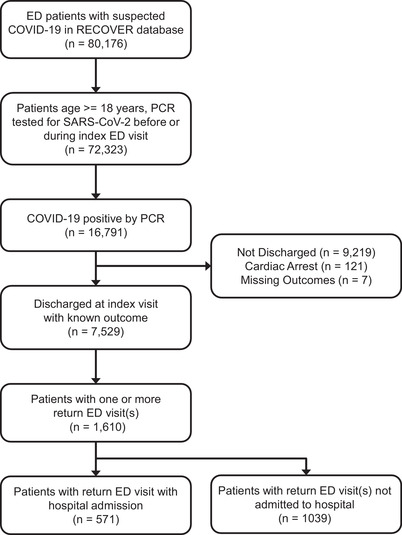
CONSORT flow diagram identifying adult patients in the National Registry of Suspected COVID‐19 in Emergency Care data set with polymerase chain reaction (PCR)–confirmed severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) who had a return hospital admission within 30 days of discharge from their index emergency department (ED) visit. Models were designed to distinguish the characteristics of the patients in the third box from the bottom (n = 7529) that predicted (or were associated with) their appearance in the final box at the bottom (n = 517). COVID‐19, coronavirus disease 2019
TABLE 1.
Demographic and clinical characteristics
| No return, n = 6958 | Return with admission, n = 571 | Total, n = 7529 | |
|---|---|---|---|
| Sex | |||
| Female, n (%) | 3586 (51.5) | 278 (48.7) | 3864 (51.3) |
| Age | |||
| Mean (SD) | 46.4 (17.2) | 54.925 (16.4) | 47.074 (17.3) |
| Range | 18–120 | 18–97 | 18–120 |
| Race/ethnicity, n (%) | |||
| Hispanic | 2037 (29.3) | 153 (26.8) | 2190 (29.1) |
| Non‐Hispanic Black | 2351 (33.8) | 231 (40.5) | 2582 (34.3) |
| Non‐Hispanic White | 1252 (18.0) | 132 (23.1) | 1384 (18.4) |
| Unknown/other | 1318 (18.9) | 55 (9.6) | 1373 (18.2) |
| O2 saturation at triage | |||
| Mean (SD) | 97.5 (2.6) | 96.5 (2.9) | 97.5 (2.6) |
| Range | 80–100 | 80–100 | 80–100 |
| Minimum O2 saturation in ED | |||
| Mean (SD) | 96.8 (3.5) | 95.2 (3.9) | 96.7 (3.5) |
| Range | 62–100 | 62–100 | 62.000–100 |
| Temperature | |||
| Mean (SD) | 37.2 (0.7) | 37.5 (0.9) | 37.2 (0.8) |
| Range | 35.7–39.6 | 35.7–39.6 | 35.7–39.6 |
| Diabetes, n (%) | 731 (10.5) | 162 (28.4) | 893 (11.9) |
| Hypertension, n (%) | 1419 (20.4) | 254 (44.5) | 1673 (22.2) |
| Obesity, n (%) | 1161 (16.7) | 194 (34.0) | 1355 (18.0) |
| Hyperlipidemia, n (%) | 528 (7.6) | 136 (23.8) | 664 (8.8) |
| Statins, n (%) | 381 (5.5) | 107 (18.7) | 488 (6.5) |
| Smoking, n (%) | 588 (8.5) | 33 (5.8) | 621 (8.2) |
| Chest X‐ray performed, n (%) | 3753 (53.9) | 454 (79.5) | 4207 (55.9) |
| Systolic blood pressure | |||
| Mean (SD) | 132.7 (19.5) | 133.2 (21.4) | 132.8 (19.6) |
| Range | 83–197 | 83–197 | 83–197 |
| Diastolic blood pressure | |||
| Mean (SD) | 80.3 (12.9) | 79.9 (13.3) | 80.3 (13.0) |
| Range | 46–122 | 46–122 | 46–122 |
Abbreviation: SD, standard deviation.
3.2. Model performance
Receiver operator curves displaying the discrimination of the models predicting return hospital admission (Figure 2) yielded an AUC of 0.59 (95% confidence interval [CI], 0.55–0.63) for the CART model, whereas RF, GBM, and LASSO had similar AUCs of 0.75 (95% CI, 0.71–0.78). The predicted probabilities of hospital admission generated by each model relative to the true outcomes of the test set are displayed in box plots (Figure 3). The Yates discrimination slopes for the 3 models were 0.071, 0.072, and 0.076 for the GBM, LASSO, and RF models, respectively. 23 Using probability cutoff thresholds to achieve a 95% NPV, the sensitivity, specificity, PPV, positive likelihood ratio, and negative likelihood ratio for each model were calculated (Table 2).
FIGURE 2.
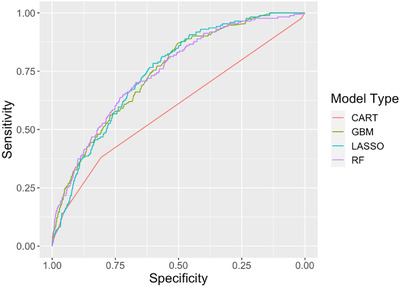
Receiver operator characteristic curves for models predicting return hospital admission of patients who are positive for coronavirus disease 2019 infection and discharged from the emergency department. CART, classification and regression tree; GBM, gradient boosted machine; LASSO, least absolute shrinkage and selection; RF, random forest
FIGURE 3.
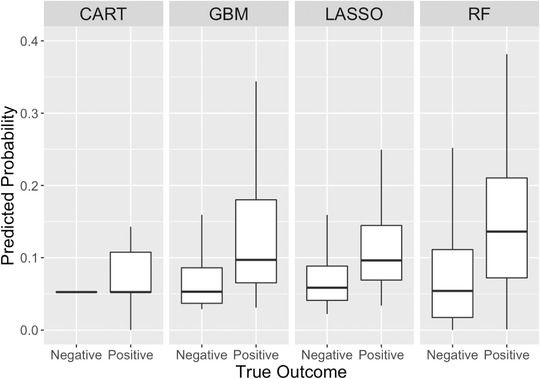
Predicted probabilities of return hospital admission compared with true outcome for the 30% test set (n = 1669). CART, classification and regression tree; GBM, gradient boosted machine; LASSO, least absolute shrinkage and selection; RF, random forest
TABLE 2.
Comparison of model performance using cut point 95% NPV
| Model | AUC (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | NPV (95% CI) | PPV (95% CI) | PLR (95% CI) | NLR (95% CI) |
|---|---|---|---|---|---|---|---|
| CART | 0.586 (0.546–0.627) | 0.380 (0.307–0.457) | 0.798 (0.780–0.815) | 0.940 (0.919–0.946) | 0.133 (0.122–0.175) | 1.880 (1.525–2.318) | 0.777 (0.690–0.875) |
| GBM | 0.747 (0.711–0.783) | 0.474 (0397–0.551) | 0.818 (0.801–0.835) | 0.950 (0.933–0.955) | 0.176 (0.160–0.226) | 2.608 (2.174–3.130) | 0.643 (0.557–0.742) |
| RF | 0.747 (0.710–0.784) | 0.462 (0.386–0.540) | 0.838 (0.821–0.853) | 0.950 (0.933–0.955) | 0.189 (0.172–0.241) | 2.844 (2.355–3.435) | 0.642 (0.558–0.739) |
| LASSO | 0.747 (0.714–0.781) | 0.491 (0.414–0.569) | 0.792 (0.774–0.809) | 0.950 (0.933–0.955) | 0.162 (0.148–0.209) | 2.362 (1.985–2.811) | 0.642 (0.553–0.745) |
Abbreviations: AUC, area under the receiver operating characteristic curve; CART, classification and regression tree; CI, confidence interval; GBM, gradient boosted machine; LASSO, least absolute shrinkage and selection; NLR, negative likelihood ratio; NPV, negative predictive value; PLR, positive likelihood ratio; PPV, positive predictive value; RF, random forest.
3.3. GBM and RF model variable importance
Variable importance plots were used to visualize the contribution of the 15 most important features of the GBM and RF models across all feature values (Figure 4). Several model input features, including age, statin use, comorbid conditions (ie, diabetes, hyperlipidemia, hypertension, and obesity) and physical exam findings (ie, lowest documented O2 saturation, temperature, and heart rate), were important predictors of return hospital admission in both the GBM and RF models.
FIGURE 4.
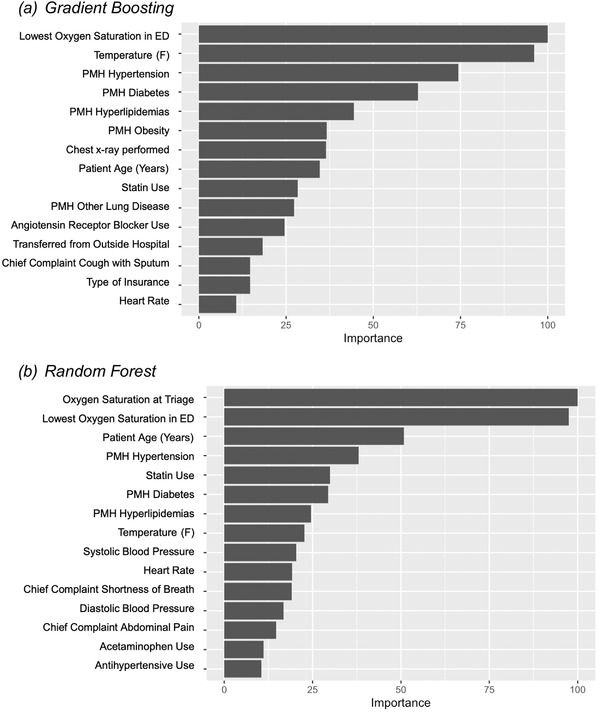
Variable importance plots for (A) gradient boosting machine and (B) random forest models. ED, emergency department
3.4. LASSO model coefficients
The LASSO regression model selected many of the features prominent within the aforementioned tree‐based models, including age, comorbid conditions (ie, diabetes, hyperlipidemia, hypertension, and obesity) and physical exam findings (ie, temperature, lowest documented O2 saturation; Table 3).
TABLE 3.
Logistic odds ratios of training set using LASSO selected inputs
| Variable | Odds ratio | Lower 95% CI | Upper 95% CI | Pr(>|Z|) |
|---|---|---|---|---|
| Transferred from outside hospital | 2.30 | 1.24 | 4.28 | 8.32E‐03 |
| Age | 1.02 | 1.01 | 1.03 | 4.87E‐08 |
| Cough with sputum | 1.64 | 1.15 | 2.33 | 6.12E‐03 |
| Lowest O2 saturation | 0.98 | 0.94 | 1.03 | 4.50E‐01 |
| O2 saturation at triage | 0.97 | 0.92 | 1.03 | 2.83E‐01 |
| Temperature | 1.53 | 1.34 | 1.74 | 1.70E‐10 |
| History of diabetes | 1.65 | 1.25 | 2.19 | 4.21E‐04 |
| History of hypertension | 1.31 | 1.01 | 1.72 | 4.55E‐02 |
| History of obesity | 1.94 | 1.51 | 2.49 | 2.13E‐07 |
| History of hyperlipidemia | 1.17 | 0.77 | 1.80 | 4.57E‐01 |
| History of other lung disease | 2.46 | 1.11 | 5.45 | 2.69E‐02 |
| Statin | 1.16 | 0.73 | 1.84 | 5.22E‐01 |
| Angiotensin receptor blocker | 1.63 | 1.05 | 2.55 | 3.04E‐02 |
| Chest X‐ray performed | 1.69 | 1.30 | 2.19 | 1.01E‐04 |
Abbreviations: CI, confidence interval; LASSO, least absolute shrinkage and selection; PR, probability.
3.5. Limitations
This study has several limitations that introduce the potential for bias from several important sources. First, model validation was performed on retrospective data and lacks prospective external validation, although a held‐out validation cohort was reserved for internal validation. Second, the RECOVER network included some smaller hospital systems or single medical centers where ED visits may not have been tracked across multiple sites. Such a limitation would likely bias the database toward an underestimate of the true rate of return hospital admission. It is likely that this would have a minimal impact on our model validity as these cases are likely to be missing at random. The RECOVER data set provided us with limited information regarding the exact timing and reason for return hospital admission. Also, there was significant heterogeneity of sampling procedures at RECOVER sites for inclusion into the registry. This limitation is offset by the size and number of health systems participating in the registry. For example, much of the data collection occurred early in the pandemic with variable, and often limited, access to SARS‐CoV‐2 testing. This may have resulted in a cohort that was more ill and could influence the test characteristics of the models. Laboratory values were excluded because of the high degree of missingness within the cohort of discharged patients. This missingness might reflect resource limitations at some sites during regional COVID‐19 surges where laboratory results may not have been ordered for patients who were likely to be discharged. As a result, potentially useful data were excluded from the models. Conversely, this means that the models do not depend on advanced testing that may be limited in certain settings. Finally, the development and broad adoption of safe and effective outpatient COVID‐19 therapies such as monoclonal antibodies and oral antivirals has the potential to reduce the specificity of our model prediction as their use is likely to be initially focused on patients with many of the high‐risk features identified in our model VIPs.
4. DISCUSSION
4.1. Principal results
This study represents one of the first systematic attempts to stratify patients according to their risk of return ED visit with hospitalization in a geographically diverse sample of adults discharged from the ED with laboratory‐confirmed COVID‐19. 24 Using a well‐described derivation population from the national RECOVER ED network of 116 hospitals, we applied ML approaches to derive models that predict the probability of return hospital admission within a 30‐day horizon. After patient discharge, this model is designed to identify patients with COVID‐19 at highest risk for delayed clinical deterioration. Such information could be used administratively to target potentially scarce hospital resources, including telehealth, home health equipment such as pulse oximeters, and home visits within that discharged population. Although not intended as a real‐time clinical decision tool, a quantitative estimate of deterioration risk could be used to reduce ED disposition uncertainty for well‐appearing patients with apparent risk factors for severe COVID‐19 disease.
Each of the model development techniques used performed similarly well as evidenced by overlapping AUC and other test characteristic CIs, with the exception of the CART model. This is not surprising given the small number of nodes in the CART model representing significantly fewer prediction variables. Furthermore, the VIP plots illustrate that several variables have comparable importance in driving the prediction, which is difficult to capture in a CART model with a small number of nodes.
4.2. Comparison with prior work
Although a number of studies have examined the risk of adverse events after diagnosis of COVID‐19, these have generally focused on serious short‐term adverse events such as intubation or death in a predominantly admitted cohort 25 or readmissions after hospital discharge. 26 , 27 The overall rate of 30‐day ED return after discharge index visit discharge in our study was 21.4% (1610/7529), which is significantly higher than the 14.6% rate reported in a smaller study of patients who were symptomatic and discharged from a mix of EDs, immediate care centers, and drive‐through testing facilities within a single health system. 28 This difference is likely reflective an overall lower patient acuity within that mixed‐site study. Our overall hospital readmission rate at 30 days of 7.6% (571/7529) is surprisingly high compared with the rate of 8.5% reported in a population of patients discharged from the ED or inpatient care with COVID‐19 pneumonia on supplemental oxygen. 29 However, it should be noted that in that study, patients received longitudinal nursing telephone follow‐up as needed, which might have played a role in reducing readmissions.
The predictors identified in our study are quite consistent with those most frequently reported by many inpatient prognostic models, including age, comorbidities, vital signs, and imaging features. In contrast to several inpatient prognostic models, gender was not predictive of return for hospital admission in our data set. 25 We advance this literature by focusing on a different patient population, specifically, those determined appropriate for discharge from the ED.
Our results are also consistent with the limited number of published models focused on identifying high‐risk adults discharged from the ED with the diagnosis of COVID‐19. One early research letter from a single health system of 1419 patients diagnosed with COVID‐19 within 7 days of an index ED visit identified 4 dichotomized covariates associated with a 72‐hour return for admission or observation, including age >60 years, hypoxia (SpO2 <95% on room air) upon presentation, fever (temperature >38°C), or abnormal chest radiograph. 30 Our approach identified a similar collection of important covariates and offers more generalizability as it draws from a larger sample across multiple hospital systems.
In contrast to the study by Kilaru et al, 30 we used a large number of candidate variables, allowing us to investigate more subtle or unexpected drivers of poor outcome. The larger number of candidate variables typically allows for better risk prediction given a more robust description of the cohort, as evidenced in this study by the relatively high AUCs described. Similar ML approaches have been successfully used to predict hospital admission and return ED visits in patients with no COVID‐19 infection. 31 , 32 However, ML approaches come with 2 drawbacks worth considering. The first drawback of this approach is that mathematically complex ML models are not amenable to rapid dissemination as they are best operationalized through time‐ and cost‐intensive integration with an electronic health record (EHR) system. Until such integrations become more seamless and routine, this limits the implementation of our study. Second, and more general to the concept of model development, the large number of candidate variables always raises the possibility of model overfitting. This situation was mitigated by using methods offering lower variance solutions such as LASSO, which reduces variance through regularization techniques, and RF, which uses an averaging technique known as bagging and random variable selection. 33 In addition, methods such as cross‐validation provided a mechanism to reduce overfitting incurred during the model derivation steps.
4.3. Clinical implications
Model performance in terms of sensitivity, specificity, NPV and PPV, and likelihood ratios are influenced by the choice of cut points used to convert predicted probabilities of the outcome into binary classifications. We have presented model performance using a cut point that would have a 95% NPV; alternative methods could be used to establish alternative cut points such as the Youden index, which seeks to maximize the sum of sensitivity and specificity. Although these cut points may increase sensitivity compared with the 95% NPV cut point, as the specificity of the model decreases, so does the number of false positive outcomes that predicted. The choice of where to establish a cut point may vary from health system to health system based on the availability of outpatient follow‐up and remote patient monitoring equipment.
It is worth considering the potential clinical implications and application of any of these models, with the exception of CART given its poor characteristics. As mentioned previously, all of these models would need to be incorporated into an EHR given their mathematical complexity. To function in real time, based on the candidate variables selected, symptom inventories using discrete data fields would have to be used requiring active maintenance of problem and medication lists, a recurrent problem in clinical settings, particularly the ED. 34 However, it is worth noting that these data were derived from real medical records with those limitations built in to the candidate variables. Furthermore, we did not apply natural language processing to these data, so data obtained from unstructured text or clinical notes not completed in real time could represent a complication to real‐time implementation. An alternative approach is to apply the tool in a post hoc manner to identify recently discharged patients with the highest risk of clinical decompensation. Although this might mitigate some of the problems surrounding real‐time data entry or notes completed after a patient's disposition is already determined, the other aforementioned data quality issues remain.
In summary, using a large geographically diverse national sample of patients diagnosed with COVID‐19 and discharged from the ED, we developed, cross‐validated, and tested multiple predictive models using a number of ML techniques with GBM, RF, and LASSO performing similarly well in predicting a return ED visit with hospitalization. These models may be of value in the distribution of limited outpatient resources to those at highest risk for clinical decompensation or in the identification of patient cohorts with near‐zero risk for return or decompensation.
CONFLICTS OF INTEREST
Samuel A. McDonald declares grant funding as site coinvestigator for “Predictors of Severe COVID Outcomes” funded and sponsored by Verily Life Sciences, and the Innovative Support for Patients with SARS‐COV2 Infections Registry (INSPIRE) funded by the Centers for Disease Control and Prevention, “Modeling of Infectious Network Dynamics for Surveillance, Control and Prevention Enhancement (MINDSCAPE).”
AUTHOR CONTRIBUTIONS
David G. Beiser conceived the study; developed analysis plan; assisted with data models and analyses; and wrote, edited, and approved this article. Zachary J. Jarou conceived the study, developed data models; and wrote, edited, and approved this article. Alaa A. Kassir assisted with data collection, data models, analyses, and writing and edited and approved this article. Michael A. Puskarich conceived the study, developed analysis plan, assisted with data models and analyses, and edited and approved this article. Marie C. Vrablik contributed to study design, assisted with data analysis, and edited and approved this article. Elizabeth D. Rosenman assisted with data analysis and edited and approved this article. Samuel A. McDonald assisted with data models and analyses and edited and approved this article. Andrew C. Meltzer contributed to the study design, collected data, and edited and approved this article. D. Mark Courtney and Christopher Kabrhel edited and approved this article. Jeffrey A. Kline conceived the RECOVER database, organized sites, obtained funding, collected data, and edited and approved this article.
Supporting information
Supporting information.
ACKNOWLEDGMENTS
David G. Beiser acknowledges the invaluable assistance of Ross Han, BS, and Aswathy Ajith, MS, with retrieval and preparation of the UChicago data. This project was funded by the Department of Emergency Medicine at the Indiana University School of Medicine. The funder had no role in the review or approval of the article. Alaa A. Kassir is supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number TL1TR002388. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Biography
David Beiser, MD, is an Associate Professor of Emergency Medicine who specializes in therapeutic hypothermia and ultrasound at The University of Chicago in Chicago, IL.

Beiser DG, Jarou ZJ, Kassir AA, et al. Predicting 30‐day return hospital admissions in patients with COVID‐19 discharged from the emergency department: A national retrospective cohort study. JACEP Open. 2021;2:e12595. 10.1002/emp2.12595
David G. Beiser and Zachary J. Jarou contributed equally to this study
This project was funded by the Department of Emergency Medicine at the Indiana University School of Medicine. The funder had no role in the review or approval of the manuscript. A.A.K. is supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under Award TL1TR002388.
Supervising Editor: Chadd Kraus, DO, DrPH.
REFERENCES
- 1. Centers for Disease Control and Prevention . COVID Data Tracker. Centers for Disease Control and Prevention; 2021. [Google Scholar]
- 2. Ji Y, Ma Z, Peppelenbosch MP, Pan Q. Potential association between COVID‐19 mortality and health‐care resource availability. Lancet Glob Heal 2020;8(4):e480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Karan A, Wadhera RK. Healthcare system stress due to COVID‐19: evading an evolving crisis. J Hosp Med. 2021;16(2):127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wood F, Roe T, Newman J, Wood L. Risk factors for deterioration in mild COVID‐19 remain undefined. Emerg Med J. 2021;38(5):406. [DOI] [PubMed] [Google Scholar]
- 5. Kline JA, Pettit KL, Kabrhel C, Courtney DM, Nordenholz KE, Camargo CA. Multicenter registry of United States emergency department patients tested for SARS‐CoV‐2. J Am Coll Emerg Physicians Open. 2020;1(6):1341–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kline JA, Camargo CA, Courtney DM, et al. Clinical prediction rule for SARS‐CoV‐2 infection from 116 U.S. emergency departments. PLoS One. 2021;16(3):e0248438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moons KGM, Altman DG, Reitsma JB, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162(1):W1–W73. [DOI] [PubMed] [Google Scholar]
- 8. https://isaric.net/ccp/
- 9. RStudio Inc . RStudio Team: Integrated Development for R. Boston, MA: RStudio Inc; 2020. [Google Scholar]
- 10. R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2018. [Google Scholar]
- 11. Wickham H, Averick M, Bryan J, et al. Welcome to the tidyverse. J Open Source Softw. 2019;4(43):1686. [Google Scholar]
- 12. Stekhoven DJ, Bühlmann P. Missforest‐non‐parametric missing value imputation for mixed‐type data. Bioinformatics. 2012;28(1):112–118. [DOI] [PubMed] [Google Scholar]
- 13. Krasser R. explore: Simplifies Exploratory Data Analysis. 2020. https://cran.r‐project.org/
- 14. Heinzen E, Sinnwell J, Atkinson E, et al. Arsenal: An Arsenal of “R” Functions for Large‐Scale Statistical Summaries. Vienna, Austria: The Comprehensive R Archive Network (CRAN); 2020. [Google Scholar]
- 15. Royston P, Sauerbrei W. Improving the robustness of fractional polynomial models by preliminary covariate transformation: a pragmatic approach. Comput Stat Data Anal. 2007;51(9):4240–4253. [Google Scholar]
- 16. López‐Ratón M, Rodríguez‐Álvarez MX, Cadarso‐Suárez C, Gude‐Sampedro F. Optimalcutpoints: an R package for selecting optimal cutpoints in diagnostic tests. J Stat Softw. 2014;61(8):1–36. [Google Scholar]
- 17. Greenwell B, Boehmke B, Cunningham J. gbm: Generalized Boosted Regression Models. 2020. https://cran.r-project.org/
- 18. Wright MN, Ziegler A. Ranger: a fast implementation of random forests for high dimensional data in C++ and R. J Stat Softw. 2017;77(1):1–17. 10.18637/jss.v077.i01 [DOI] [Google Scholar]
- 19. Taddy M. One‐step estimator paths for concave regularization. J Comput Graph Stat. 2017;26(3):525–536. [Google Scholar]
- 20. Therneau T, Atkinson B. rpart: Recursive Partitioning and Regression Trees. 2019. https://cran.r-project.org/
- 21. Kuhn M. caret: Classification and Regression Training. 2020. https://cran.r-project.org/
- 22. Greenwell B, Boehmke B. Variable importance plots—An introduction to the vip package. R J. 2020;12(1):343. [Google Scholar]
- 23. Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: a framework for some traditional and novel measures. Epidemiology. 2010;21(1):128–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wynants L, Van Calster B, Collins GS, et al. Prediction models for diagnosis and prognosis of covid‐19: systematic review and critical appraisal. BMJ. 2020;369:m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sharp AL, Huang BZ, Broder B, et al. Identifying patients with symptoms suspicious for COVID‐19 at elevated risk of adverse events: the COVAS score. Am J Emerg Med. 2020;46:489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Atalla E, Kalligeros M, Giampaolo G, Mylona EK, Shehadeh F, Mylonakis E. Readmissions among patients with COVID‐19. Int J Clin Pract. 2020:75(3):1–10. [DOI] [PubMed] [Google Scholar]
- 27. Donnelly JP, Wang XQ, Iwashyna TJ, Prescott HC. Readmission and death after initial hospital discharge among patients with COVID‐19 in a large multihospital system. JAMA. 2020;325(3):2020–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haag A, Dhake SS, Folk J, et al. Emergency department bounceback characteristics for patients diagnosed with COVID‐19. Am J Emerg Med. 2021;47:239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Banerjee J, Canamar CP, Voyageur C, et al. Mortality and readmission rates among patients with COVID‐19 after discharge from acute care setting with supplemental oxygen. JAMA Netw Open. 2021;4(4):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kilaru AS, Lee K, Snider CK, et al. Return hospital admissions among 1419 COVID‐19 patients discharged from five U.S. emergency departments. Acad Emerg Med. 2020;27(10):1039–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Raita Y, Goto T, Faridi MK, Brown DFM, Camargo CA, Hasegawa K. Emergency department triage prediction of clinical outcomes using machine learning models. Crit Care. 2019;23(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hong WS, Haimovich AD, Taylor RA. Predicting 72‐hour and 9‐day return to the emergency department using machine learning. JAMIA Open. 2019;2(3):346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. New York, NY: Springer; 2001. [Google Scholar]
- 34. Wang ECH, Wright A. Characterizing outpatient problem list completeness and duplications in the electronic health record. J Am Med Inform Assoc. 2020;27(8):1190–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.


