Abstract
Natural killer (NK) cells and T cells are key effectors of anti-tumor immune responses and major targets of checkpoint inhibitors. In multiple cancer types, we find that the expression of Wnt signaling potentiator R-spondin genes (e.g. RSPO3) is associated with favorable prognosis and is positively correlated with gene signatures of both NK cells and T cells. While endothelial cells and cancer-associated fibroblasts comprise the R-spondin3-producing cells, NK cells and T cells correspondingly express the R-spondin3 receptor LRG6 within the tumor microenvironment. Exogenous expression or intratumor injection of R-spondin3 in tumors enhanced the infiltration and function of cytotoxic effector cells, which led to tumor regression. NK cells and CD8+ T cells independently and cooperatively contributed to R-spondin3-induced control of distinct tumor types. The effect of R-spondin3 was mediated in part through upregulation of MYC and ribosomal biogenesis. Importantly, R-spondin3 expression enhanced tumor sensitivity to anti-PD1 therapy, thereby highlighting new therapeutic avenues.
SIGNIFICANCE
Our study identifies novel targets in enhancing anti-tumor immunity and sensitizing immune checkpoint inhibition, which provides a rationale for developing new immunotherapies against cancers. It also offers mechanistic insights on Wnt signaling-mediated modulation of anti-cancer immunity in the TME and implications for a putative R-spondin-LGR6 axis in regulating NK-cell biology.
Keywords: R-spondin, LGR6, tumor microenvironment, NK cells and T cells, immune checkpoint inhibitor
INTRODUCTION
Natural killer (NK) cells are essential innate immune effector cells that can recognize and rapidly kill oncogenically transformed target cells. More importantly, NK cell interactions with other immune cells in the tumor microenvironment (TME), such as dendritic cells (DC) and T cells, are crucial to magnify the overall immune response against the cancer[1, 2]. Supporting this notion, elevated numbers and enhanced functionality of NK cells are associated with better responses to immune checkpoint blockade therapies that largely target T cells[2], and emerging evidence indicates that direct targeting on NK cells may also exist[3, 4]. However, NK cells typically exhibit poor capacity to infiltrate tumors and frequently become functionally exhausted within tumors due to various immunosuppressive facets of the TME[5]. Thus, the identification of molecular targets and developing therapeutic strategies to promote infiltration and maintain or restore anti-tumor functions of NK cells in the TME have been an outstanding clinical priority to improve cancer outcomes.
Wnt-signaling pathways control a wide range of cellular processes and are delicately regulated by a variety of positive or negative regulators with temporospatial specificity[6]. Several recent studies highlight an important role of Wnt signaling in regulating NK-cell anti-tumor functionality[7]. Activation of the Wnt/β-catenin pathway via inhibition of GSK3β enhanced the maturation and function of NK cells[8]. Cancer cell secretion of the Wnt signaling antagonists Dickkopf 1/2 (DKK1/2) facilitated the evasion of NK cell-mediated anti-tumor responses in certain contexts[9, 10]. On the other hand, hyperactivation of Wnt-β catenin pathway is often a hallmark of cancer cells and crucial in tumor formation. And evidence is also showing up for an immune cell exclusion phenotype associated with tumor cell-intrinsic aberrant β-catenin signaling activation across cancers[11, 12]. Thus, the TME is controlled by an intricate interplay of Wnt agonists, antagonists, and anti-antagonists, and there could be certain components in the Wnt signaling pathway that play critical roles in tuning the activity and infiltration of NK cells in the TME.
The R-spondin gene family, RSPO1 thru RSPO4, encode four evolutionary conserved secreted proteins. R-spondins can potentiate canonical Wnt signaling in the low dose of Wnt following binding to the leucine-rich repeat-containing G-protein coupled receptors (LGR) LGR4, LGR5, and LGR6 with high affinity[13, 14]. Previous studies have described functions for R-spondins mainly in embryonic development, adult stem cell maintenance, and tumorigenesis[15, 16]. However, the roles of R-spondins in modulating anti-tumor immunity remain ill-defined and largely unexplored.
LGR6 shares a similar structural basis with LGR4/5 and together they belong to the B-type LGR subfamily that is characterized by a long ectodomain containing 17 leucine-rich repeats (LRR)[16]. These three LGRs were considered as obligate high-affinity receptors for R-spondins[13]. While LGR-independent enhancement of Wnt signaling has also been reported recently for RSPO2 and RSPO3[17, 18]. LGR6 was shown to have a unique expression pattern and has been extensively reported to mark distinct types of adult stem cells in actively self-renewing tissues, such as epidermis and mammary glands[19-22]. However, the expression and function of LGR6 in cell types other than the stem/progenitor cell populations remains unspecified.
Here we identify R-spondin family members R-spondin3 and R-spondin1, which are mainly expressed by endothelial cells (EC) and cancer-associated fibroblasts (CAF) in the TME, as potential modulators of anti-tumor immunity in cancers. Exogenous expression of R-spondin3 in the TME promotes tumor suppression largely through NK cells, as well as CD8+ T cells. The mechanism of R-spondin3 enhancement of effector cell responses involves enhanced expression of the Wnt target gene MYC in NK cells. Importantly, R-spondin3 and PD-1 blockade therapy cooperatively enhance immune control of tumors. These findings provide molecular and mechanistic insights on Wnt signaling components in modulating anti-tumor immunity and a strong rationale for developing novel anti-cancer immunotherapeutic strategies utilizing R-spondins.
RESULTS:
RSPO3 and RSPO1 levels positively correlate with anti-cancer immune-cell signatures and better prognosis in multiple cancers.
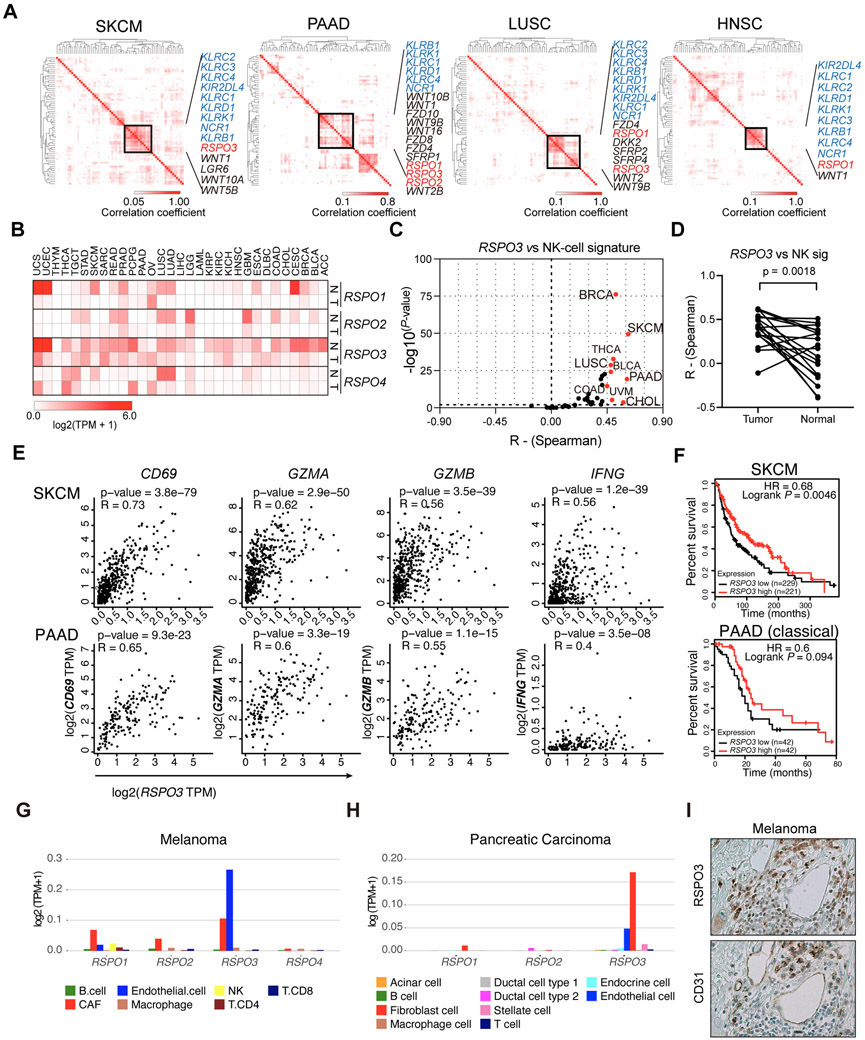
To explore Wnt signaling components that may regulate the anti-tumor immune responses of NK cells in the TME, we generated correlation matrices with The Cancer Genome Atlas (TCGA) datasets using 60 genes encoding Wnt signaling components (Supp Table 1) and 9 NK-cell signature genes (KIR2DL4, NCR1, KLRD1, KLRC1, KLRC2, KLRC3, KLRC4, KLRB1, KLRK1)[23]. These genes are enriched in NK cells and can be used to indirectly infer the abundance of NK-cells within tumor tissue. RSPO1 and RSPO3 recurrently showed positive associations with the NK-cell signature genes in four cancer types: melanoma (SKCM), pancreatic adenocarcinoma (PAAD), lung squamous carcinoma (LUSC), and head and neck squamous carcinoma (HNSC) (Figure 1A). Notably, all four members of the R-spondin family are downregulated in the majority of cancer types compared to matched normal tissues (Figure 1B), suggesting a potential for a shared underlying mechanistic role of R-spondin in different cancers. We then performed further correlation analyses specifically for the NK-cell signature genes with RSPO3 or RSPO1 in a total of 33 cancer types from TCGA datasets. Nine cancer types showed positive correlations (R>0.45, P<0.05) for RSPO3, with SKCM, PAAD, breast invasive carcinoma (BRCA), and cholangiocarcinoma (CHOL) showing strong correlations (R>0.5) (Figure 1C). Four cancer types showed positive correlations (R>0.45, P<0.05) for RSPO1, with PAAD and CHOL showing strong correlations (R>0.5) (Supp Figure 1A). In contrast, RSPO2 or RSPO4 did not show positive correlations with the NK-cell signature in the TCGA datasets (R>0.45, P<0.05) (Supp Figure 1B-C). Together, these data suggest positive correlations between the expression of RSPO3 or RSPO1 with NK-cell signature across different cancers, particularly strong in SKCM, PAAD, BRCA, CHOL, and LUSC.
Figure 1. EC- and CAF-derived R-spondins correlate with anti-cancer immune cell signatures and prognosis in multiple cancers.
(A) Hierarchically clustered Kendall’s correlation matrices using the indicated datasets from TCGA database based on components of Wnt signaling pathway and NK-cell signature genes. (B) Heatmap visualization of the gene expression levels of RSPO1, RSPO2, RSPO3, and RSPO4 in different cancer tissues (T) and the matched normal tissues (N) in the TCGA datasets. Data are shown as log2 (TPM+1). (C) Results of Spearman’s rank correlation analyses of RSPO3 with NK-cell signature genes using TCGA datasets plotted with coefficient R and −log10 (P-value). Cancer types with correlation P-value<0.01 and R>0.45 are marked red. (D) Overall comparison of Spearman’s rank correlation coefficients for RSPO3 with NK-cell signature for a total of 19 types of tumors and normal tissues. Wilcoxon tests were performed. (E) Spearman’s rank correlation plots for RSPO3 with CD69, GZMA, GZMB, and IFNG in SKCM and PAAD of TCGA datasets. (F) Kaplan-Meier curves showing the prognostic impact of RSPO3 expression levels for the overall survival of patients with SKCM and classical PAAD of TCGA datasets. Hazard ratios (HR) and P values of Log-rank tests are shown. Patients were grouped by the median expression levels of RSPO3. (G-H) The expression of the detectable RSPO genes in the single-cell RNA-seq datasets of melanoma (G) and pancreatic carcinoma (H) patients reported previously. (I) Representative pictures of human melanoma tissues with immunohistochemical staining of RSPO3 and CD31 in serial sections. Pictures are shown as 200×. Abbreviations for the cancer types are listed in Supp Table 1.
To further investigate whether the correlations for RSPO3 or RSPO1 with NK-cell signature are unique in tumor tissues relative to normal tissues, we analyzed the available TCGA normal tissues or The Genotype-Tissue Expression (GTEx) dataset. Results showed no or weaker positive correlations were observed in the counterpart normal tissues in the majority of cancer types analyzed, except for bladder urothelial carcinoma (BLCA) and thyroid carcinoma (THCA) (Supp Figure 1D-E). A generally stronger correlation with NK-cell signature in tumor tissues relative to counterpart normal tissues was observed for RSPO3, while not for RSPO1 (Figure 1D, Supp Figure 1F). The impact of confounding clinical factors on these association was assessed by multivariant analysis, including age, gender, history of neoadjuvant therapy, tumor grade, and tumor stage. The results showed that RSPO3 level remained as an independent factor that correlated with the expression of NK-cell signature in both SKCM and PAAD (Supp Figure 1G). These data suggest a positive correlation between the expression of RSPO3 and NK-cell signature in tumor tissues.
NK cells exhibit tight interactions with DC and T cells and subsequently affect the overall anti-cancer immune responses[1]. To further explore whether the positive correlation seen for R-spondin genes with NK-cell signature could also be observed for DC or T-cell signature, we did correlation analyses for RSPO3 with a conventional type 1 dendritic cell (cDC1) signature (KIT, CCR7, BATF3, FLT3, ZBTB46, IRF8, BTLA, MYCL, CLEC9A, XCR1)[24] and a T-cell signature (CD8A, CD8B, and CD3E). Fourteen cancer types showed positive correlations (R>0.45, P<0.05) between RSPO3 and DC1 signature, while 10 cancer types showed positive correlations between RSPO3 and T-cell gene signatures (Supp Figure 2A-B). We next checked whether RSPO3 or RSPO1 correlated with the expression of immune cell activation marker (CD69) or cytotoxic functional genes (GZMA, GZMB, IFNG) in the TME. Strong positive correlations (R>0.55, P<0.05) with RSPO3 were observed for CD69, GZMA, and GZMB in SKCM and PAAD (Figure 1E). Overall positive correlations (R=0.3-0.75, P<0.1) were observed for these markers with RSPO3 in CHOL, BRCA, and LUSC (Supp Figure 2C) and with RSPO1 in PAAD and CHOL (Supp Figure 2D). Together, these data suggest that high expression levels of RSPO3 or RSPO1 are associated with better anti-cancer immunity in the TME.
Importantly, survival analyses revealed that patients with higher levels of RSPO3 had a better prognosis in SKCM, PAAD (classical subtype), CHOL, and BRCA (non-luminal, triple-negative, Her2+) (Figure 1F, Supp Figure 2E). Patients with higher levels of RSPO1 had better survival in CHOL (Supp Figure 2F). Collectively, this body of evidence suggests that expression of RSPO3 or RSPO1 are associated with better NK and T cell activity as well as improved prognosis in cancers.
RSPO3 is expressed by ECs and CAFs in the TME.
To explore the source of R-spondins in the TME, we analyzed available scRNAseq datasets of human melanoma and pancreatic carcinoma[25, 26]. Results showed that endothelial cells (ECs) and cancer-associated fibroblasts (CAFs) were the major cell populations that expressed RSPO1 and RSPO3 in the TME, with RSPO3 being much more abundantly expressed (Figure 1G-H). Low to negligible expression levels of RSPO2 and RSPO4 were observed in the TME of the two cancer types (Figure 1G-H). Further, immunohistochemistry staining of R-spondin3 and EC marker CD31 showed an abundant protein level of R-spondin3 in the melanoma TME with regional overlap with CD31 staining (Figure 1I). These data indicate that EC and CAF are among the major cell sources of R-spondin3 in the TME.
R-spondin receptor LGR6 is prominently expressed by human NK cells.
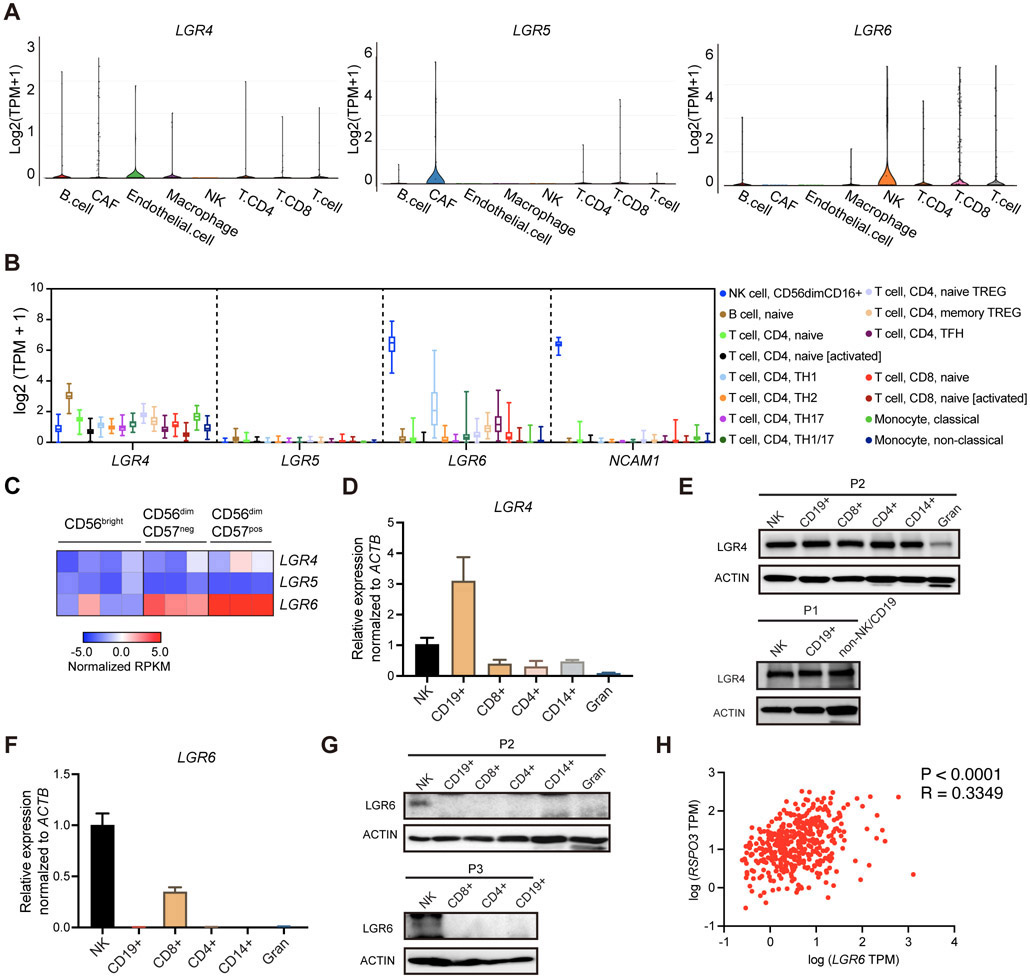
We next interrogated the expression of R-spondin receptors in the TME. LGR4, LGR5, and LGR6 are considered the obligate high-affinity receptors for R-spondins 1-4[13, 17]. By analyzing the single-cell RNA-seq dataset of human melanoma[25], we found that NK cells were the predominant cell population that expressed LGR6 in the TME, while ECs and CAFs express an appreciable amount of LGR4 and LGR5, respectively (Figure 2A). To further investigate the expression pattern of LGRs 4-6 in normal NK cells and other immune cells, we analyzed the Database of Immune Cell Expression (DICE), which covers 15 immune cell subtypes that include NK cells, B cells, T cells, and monocytes from 91 healthy donors[27]. Among these cell populations, NK cells showed the most pronounced expression of LGR6, whose transcript level (median Transcripts Per Million (TPM) = 88.55, 95% CI: 76.52-102.5) was comparable to that of NCAM1 (median TPM = 84.04, 95% CI: 81.15-86.93) which encodes for the canonical NK-cell marker CD56 (Figure 2B). Meanwhile, CD4+ Th1 cells displayed a varied but overall lower level of LGR6 compared to NK cells (Figure 2B). For the other two receptors, appreciable expression of LGR4 could be observed in B cells and LGR5 was not abundantly expressed by any of these immune cells (Figure 2B). Another dataset from the Primary Cell Atlas in BioGPS Dataset Library[28, 29], a meta-analysis of microarray datasets of 745 human primary cell samples, also showed that NK cells and, to a lesser extent, CD8+ T cells are the two cell populations that had prominent expression levels of LGR6 (Supp Figure 3). The other two known R-spondin receptors – LGR4 and LGR5 – are mainly expressed by embryonic stem cells and mesoderm-mesenchymal stem cell-derived cell lineages (Supp Figure 3). Interestingly, analysis of a transcriptome dataset of human peripheral blood NK cell subsets[30] suggested that the mature and cytotoxic subset of NK cells (CD56dimCD57+) had higher levels of LGR6 compared to the less mature and less cytotoxic subsets (CD56bright) (log2FC = 5.69, P<0.0001) (Figure 2C). Together, these evidence consistently suggest that NK cells have a high transcriptional level of LGR6.
Figure 2. LGR6 is prominently expressed by human NK cells.
(A) The expression of LGR4, LGR5, LGR6 in the single-cell RNA-seq dataset of human melanoma reported previously accessed from Broad Institute’s Single Cell Portal. (B) Visualization of LGR4, LGR5, LGR6, and NCAM1 expression in human immune cell subtypes in DICE datasets. (C) Heatmap visualization of the gene expression levels of LGR4, LGR5, and LGR6 in bulk RNA-seq dataset of human circulating NK cell subsets reported previously. (D) Quantitative RT-PCR analysis of LGR4 expression for different immune cell types isolated from human peripheral blood pooled by two different donors. (E) Protein expression of LGR4 in different immune cell types isolated from human peripheral blood from two different donors. (F) Quantitative RT-PCR analysis of LGR6 expression for different immune cell types isolated from human peripheral blood pooled by two different donors. (G) Protein expression of LGR6 in different immune cell types isolated from human peripheral blood from two different donors. (H) Spearman’s rank correlation plot for RSPO3 with LGR6 using TCGA SKCM dataset. Data are shown as mean ± s.d. for panels D and F.
To validate the findings from these bioinfo-analyses, we purified human NK cells, CD19+ B cells, CD8+ T cells, CD4+ T cells, CD14+ monocytes, and granulocytes from the peripheral blood of healthy donors and performed RT-qPCR or WB assays. Supporting the results shown in Figure 2B, RT-qPCR analysis revealed over 3-fold greater level of LGR4 in B cells compared to that in the rest of the peripheral blood cell populations (Figure 2D). Protein level of LGR4 showed no dramatic differences between the peripheral blood mononuclear cell fractions but showed a much lower level in the granulocytes (Figure 2E). Importantly, while LGR6 mRNA was detected in NK cells and, to a less extent, by CD8+ T cells, the LGR6 protein was substantially expressed by NK cells but not by any other peripheral blood mononuclear cells with a decent amount (Figure 2F-G). These results suggest LGR6 is prominently expressed by human NK cells. Consistent with these data, a moderate positive correlation (R=0.33, P<0.05) was observed for LGR6 with RSPO3 in TCGA dataset of melanoma (Figure 2H), indicating active R-spondin ligands-LGR6 interactions possibly existed within the tumor tissue. Collectively, these data suggest that LGR6 is highly expressed by human NK cells and the R-spondin3/LGR6 axis may serve as a signaling axis in the TME to regulate NK-cell mediated anti-tumor immunity in human cancers.
Interestingly in mouse NK cells, as revealed by the immGen project dataset[31], a higher level of Lgr6, as well as Lgr4, was observed in the mature and cytotoxic subset (CD11b+CD27−) relative to the less mature and less cytotoxic subset (CD11b-CD27+) (Lgr6: log2FC = 4.09, P<0.05, Lgr4: log2FC = 3.03, P<0.05) (Supp Figure 4A), suggesting a conserved regulatory mechanism for the expressions of R-spondin receptors that may exist between mouse and human NK cells. Notably, unlike in human samples, the overall transcription level of mouse Lgr6 in bulk NK cells, as revealed by the Haemopedia RNA-seq datasets[32], was not as abundant as that of mouse NK cell marker genes, such as Ncr1, Klrb1c (Supp Figure 4B), implying differential functional significance for the LGR6-mediated signaling pathway in the associated biological processes between the two species.
Exogenous expression of R-spondin3 in the TME inhibits tumor progression
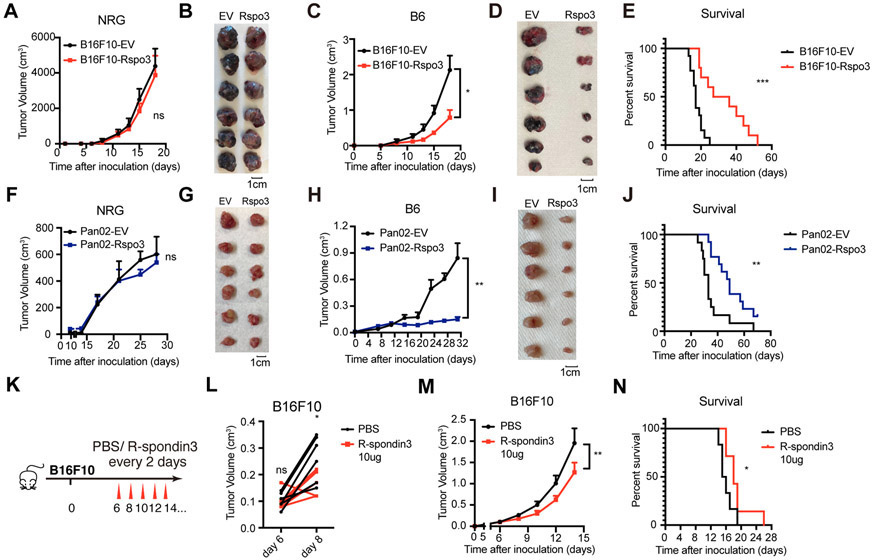
To investigate whether an increased level of R-spondin3 in the TME affects the tumor progression, we generated mouse melanoma tumor cell line B16F10 overexpressing R-spondin3. The endogenous expression level of RSPO3 in B16F10 was low compared to several other tumor cell lines analyzed (Supp Figure 5A). B16F10 cells were transduced with empty vector (B16F10-EV) or vector expressing R-spondin3 (B16F10-Rspo3) (Supp Figure 5B-C). The two lines showed marginal growth difference in vitro (Supp Figure 5D) and no growth difference in vivo in the immunodeficient NRG mice (NOD-Rag1null IL2rgnull, NOD rag gamma) that lack both innate and adaptive immunity (Figure 3A-B, Supp Figure 5E). Notably, in the immune-competent syngeneic mice, B16F10-Rspo3 group showed substantially impaired tumor progression (Figure 3C-D, Supp Figure 5F) and prolonged overall survival (Figure 3E) relative to the B16F10-EV group. Similar effects were recapitulated in a mouse pancreatic carcinoma cell line Pan02, in which the overexpression of R-spondin3 (Supp Figure 5G-H) showed marginal effects on in vitro growth (Supp Figure 5I) and in vivo growth in NRG mice (Figure 3F-G, Supp Figure 5J), but inhibited the tumor progression in syngeneic B6 mice (Figure 3H-J, Supp Figure 5K). The tumors of Pan02-Rspo3 were also more movable and with clearer boundaries from surrounding tissues compared to Pan02-EV tumors, indicating less invasiveness. To further confirm the role of R-spondin3 in inhibiting tumor progression, we performed intra-tumor injections of R-spondin3 protein to B16F10 syngeneic tumor models (Figure 3K). The treatment was effective in suppressing tumor growth and extending survival (Figure 3L-N). Importantly, in the Lgr6−/− mice, the exogenously expressed R-spondin3-mediated tumor suppression could still be observed in both tumor models, while the effect was diminished in the Pan02 model as shown by an enhanced tumor progression of Pan02-Rspo3 tumors in the Lgr6−/− mice relative to that in the wild type mice (Supp Figure 5L-O), suggesting LGR6 partially mediated the tumor suppression caused by enhanced R-spondin3 levels in the TME in a tumor-specific fashion.
Figure 3. Exogenous expression of R-spondin3 in the TME inhibits tumor progression.
(A-B) Growth curves of B16F10-EV and B16F10-Rspo3 in NRG mice (n = 8 mice per group) (A) and representative pictures of tumors dissected on 18 days after inoculation (B). (C-D) Growth curves of B16F10-EV and B16F10-Rspo3 in syngeneic B6 mice (n = 8 mice per group) (C) and representative pictures of tumors dissected on 18 days after inoculation (D). (E) Survival curves and result of the Log-rank test for growth of B16F10-EV and B16F10-Rspo3 in syngeneic B6 mice (n = 11 for each group). (F-G) Growth curves of Pan02-EV and Pan02-Rspo3 in NRG mice (n = 8 mice per group) (F) and representative pictures of tumors dissected on 33 days after inoculation (G). (H-I) Growth curves of Pan02-EV and Pan02-Rspo3 in syngeneic B6 mice (n = 8 mice per group) (H) and representative pictures of tumors dissected on 33 days after inoculation (I). (J) Survival curves and result of the Log-rank test for growth of Pan02-EV and Pan02-Rspo3 in syngeneic B6 mice (n = 12 for each group). (K) Experimental design for panels L-N. Recombinant mouse R-spondin3 (10μg) or the same volume of PBS were intratumorally injected to B16F10 tumors in syngeneic B6 mice (n = 6 mice per group). (L) Tumor volumes measured before and 2 days after the 1st dose of R-spondin3 i.t. therapy were compared. Two-way ANOVA test with Sidak’s multiple comparisons. (M) Tumor growth curves. (N) Survival curves and result of the Log-rank test. For panels A, C, F, H, and M, two-way ANOVA test with or without Tukey's multiple comparisons tests were performed. Data are shown as mean ± s.e.m or individual sample results. Data are representative of at least two independent experiments. P < 0.05 is considered as statistically significant. ns, not statistically significant, *P < 0.05, **P < 0.01, ***P < 0.001.
Exogenous expression of R-spondin3 in the TME enhances anti-tumor immunity.
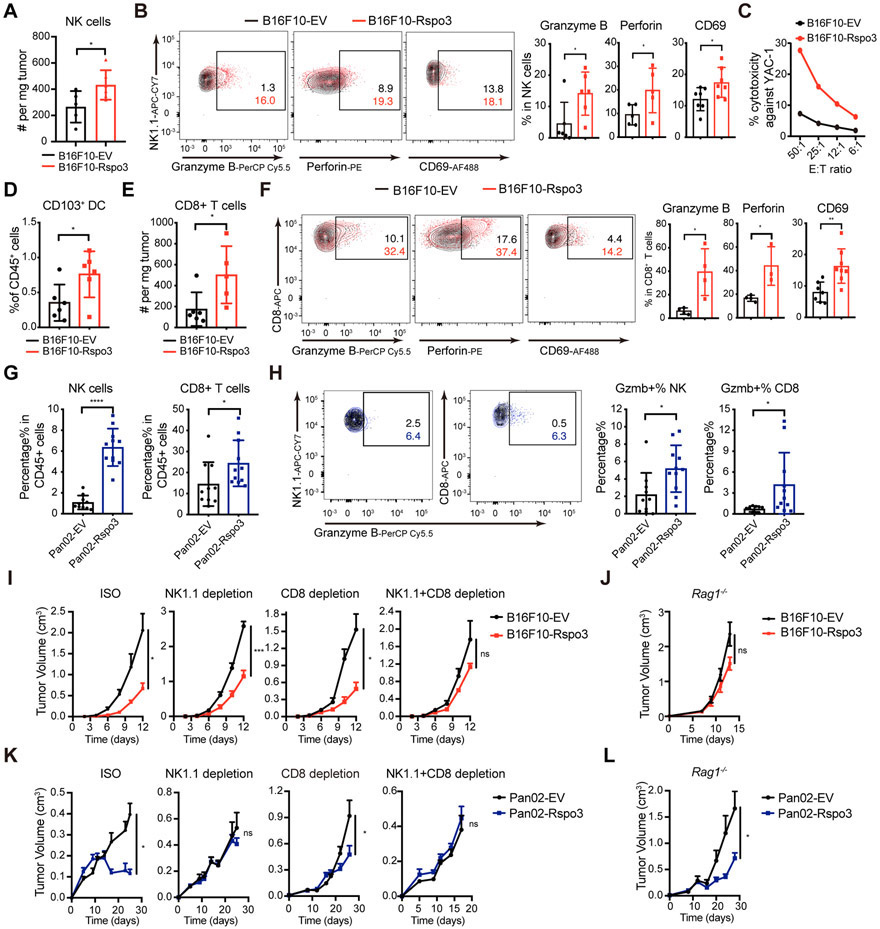
To investigate whether R-spondin3 in the TME affects the NK-cell and overall anti-tumor immunity, we analyzed the tumor-infiltrating immune cells from both B16F10-EV and B16F10-Rspo3 tumors. An increased infiltration of CD45+ immune cells and NK cells was observed by both flow cytometry analysis and immunohistochemistry (Figure 4A, Supp Figure 6A-C). In addition, enhanced expressions of cytotoxic molecules granzyme B and perforin and activation marker CD69 were observed in the NK cells from B16F10-Rspo3 tumors (Figure 4B). In vitro killing capacity against B16F10 cells or YAC-1 cells, an NK-cell sensitive MHC-I low-expressing lymphoma cell line, of the NK cells derived from B16F10-Rspo3 tumors were stronger compared to those derived from the B16F10-EV tumors (Figure 4C, Supp Figure 6D), indicating a better NK cell functionality. Furthermore, in the B16F10-Rspo3 tumors, we observed an increased proportion of CD103+ cDC1s (Figure 4D, Supp Figure 6E) and CD8+ T cells (Figure 4E, Supp Figure 6F-G), whose expressions of granzyme B, perforin, CD69, and IFN-γ were also increased (Figure 4F, Supp Figure 6H) compared to those in the B16F10-EV tumors, indicating a better overall anti-cancer immunity. Similarly, in the Pan02 pancreatic cancer model, increased percentages of NK cells and CD8+T cells in the CD45+ immune cell population, with enhanced expressions of granzyme B, were detected in the Pan02-Rspo3 group relative to the Pan02-EV group (Figure 4G-H), although enhanced infiltrations could not be observed when measured as absolute infiltrating cell number per tumor weight (Supp Figure 6I-J), which could be related with an increased amount of stromal tissue observed in the Pan02-Rspo3 tumors (Supp Figure 6K). Collectively, these data indicate that increasing R-spondin3 levels in the TME enhances the NK-cell anti-tumor immunity and promotes better overall anti-tumor responses.
Figure 4. Exogenous expression of R-spondin3 in the TME enhances anti-tumor immunity.
(A) Absolute numbers of tumor-infiltrating NK cells (CD45+CD3−NK1.1+DX5+) in B16F10-EV and B16F10-Rspo3 tumors (n = 5-6 mice per group) by flow cytometry analysis. (B) Representative flow plots (left) and summary (right) of the percentages of tumor-infiltrating NK cells expressing Granzyme B, perforin, and CD69 in the B16F10-EV and B16F10-Rpso3 tumors (n = 5-7 mice per group). (C) Cytotoxicity of freshly isolated tumor-infiltrating NK cells from B16F10-EV and B16F10-Rspo3 tumors was measured with chromium (51Cr)-release assays using YAC-1 cells as target cells with the indicated E: T (effector: target ratios). Purified NK cells were pooled from four mice per group. (D) Percentages of CD103+ DC (lineage [CD90.2, CD45R, Ly6G, NK1.1]−, CD45+, Ly6C−, MHC-II+, F4/80−, CD24+) in the CD45+ cell population in the B16F10-EV and B16F10-Rpso3 tumors by flow cytometry analysis (n = 6 mice per group). (E) Absolute numbers of tumor-infiltrating CD8+ cells (CD45+CD3+CD8+) in B16F10-EV and B16F10-Rspo3 tumors by flow cytometry analysis (n = 5-6 mice per group). (F) Representative flow plots (left) and summary (right) of the percentages of tumor-infiltrating CD8+ cells expressing Granzyme B, perforin, and CD69 in the B16F10-EV and B16F10-Rpso3 tumors (n = 4-8 mice per group). (G) Percentages of tumor-infiltrating NK cells and CD8+ T cells in the CD45+ cell population by flow cytometry analysis of Pan02-EV and Pan02-Rspo3 tumors (n = 9-11 mice per group). (H) Representative flow plots (left) and summary (right) of the percentages of Granzyme B-expressing tumor-infiltrating NK cells and CD8+ T cells in the Pan02-EV and Pan02-Rpso3 tumors (n = 9-11 mice per group). (I) Growth curves of B16F10-EV and B16F10-Rspo3 cells in syngeneic B6 mice treated with isotype control, anti-NK1.1 depletion antibody, anti-CD8a depletion antibody, or both (n = 5-8 mice per group). (J) Growth curves of B16F10-EV and B16F10-Rspo3 cells in Rag1−/− mice (n = 6 mice per group). (K) Growth curves of Pan02-EV and Pan02-Rspo3 cells in syngeneic B6 mice treated with isotype control, anti-NK1.1 depletion antibody, anti-CD8a depletion antibody, or both (n = 5 - 8 mice per group). (L) Growth curves of Pan02-EV and Pan02-Rspo3 cells in Rag1−/− mice (n = 6 mice per group). Data are shown as mean ± s.e.m. and are representative of at least two independent experiments. For panels A, B, and D-H, Student’s t-tests or Welch’s t-tests were performed. For panels I-L, two-way ANOVA tests were performed. P < 0.05 is considered as statistically significant. ns, not statistically significant, *P < 0.05, **P < 0.01, ***P < 0.001.
To dissect the roles of different immune cells in the TME in the R-spondin3-meditated tumor suppression, we inoculated B16F10-EV and B16F10-Rspo3 tumors in mice lacking CD8+ T cells and/or NK cells by injecting the anti-CD8a and/or anti-NK1.1 depleting antibodies (Supp Figure 7A-C). Depletion of both CD8+ T and NK cells, but not by depletion of either cell type alone, abrogated the tumor-suppressive effect of R-spondin3 exogenous expression (Figure 4I, Supp Figure 7D), indicating that NK cells and CD8+ T cells are the major populations mediating tumor suppression and both can act at least partially independently on each other. Further functional dissection using Rag1−/− mice, which lacks mature B cells and T cells, also abrogated tumor suppression (Figure 4J, Supp figure 7E), suggesting that other T cell subsets or B cells may also play roles here. Interestingly, in the Pan02 pancreatic cancer model, depleting NK cells alone was sufficient to abrogate the tumor suppression, and depleting CD8+ T cells alone or growth in Rag1−/− mice could just modestly but noticeably reduced R-spondin3-mediated tumor suppression (Figure 4K-L, Supp Figure 7F-H). These results suggested that both NK cells and CD8+ T cells contributed to the R-spondin3-mediated tumor suppression in this pancreatic carcinoma tumor model and there existed a T-cell-independent effect. Together, these data indicate that both NK cells and CD8+ T cells mediate the tumor-suppressive effects of R-spondin3.
R-spondin3 promotes MYC expression of NK cells in the TME.
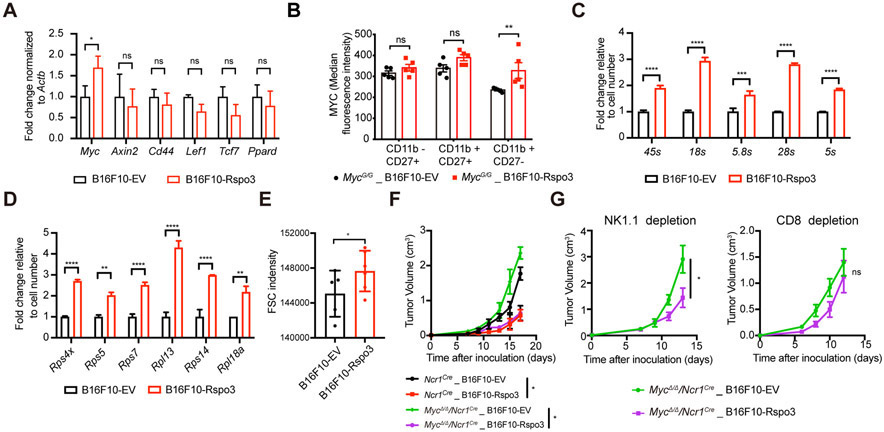
R-spondins are able to potentiate the canonical Wnt signaling activity, which upregulates the expressions of a series of target genes in a cell-type- and context-specific manner[6, 33]. To explore the mechanism by which the R-spondin3 promote the anti-tumor immunity in the TME, we sorted NK cells from the B16F10-EV and B16F10-Rspo3 tumors and measured the expression levels of several known Wnt target genes, including Myc, Axin2, Cd44, Lef1, Tcf7, Ppard, Mmp7, and Ccnd1. Among the detectable genes, Myc is significantly upregulated in the NK cells from the B16F10-Rspo3 tumor relative to the B16F10-EV tumor (Figure 5A). Using syngeneic recipients of MycG/G mice, in which the endogenous Myc locus has been modified to encode a GFP-MYC fusion protein enabling the measurement of MYC expression with GFP signal intensities, we further confirmed an increased MYC protein level in the tumor-infiltrating NK cells from the B16F10-Rspo3 tumors compared to B16F10-EV tumors (Figure 5B). Thus, R-spondin3 promotes MYC expression in NK cells in the TME.
Figure 5. R-spondin3 promotes MYC expression in NK cells in the TME.
(A) Quantitative RT-PCR results of Wnt target genes of flow-sorted CD11b+CD27− NK cells from tumor tissues (n = 4 for each group). (B) Flow analysis of tumor-infiltrating NK cells from B16F10-EV or B16F10-Rspo3 tumors inoculated subcutaneously to MycG/G mice (n = 5 for each group). Median fluorescence intensities of GFP for the indicated NK cell subpopulations are shown. (C-D) Quantitative RT-PCR results of rRNA (C) or ribosomal protein mRNA (D) of flow-sorted tumor-infiltrating NK cells from B16F10-EV or B16F10-Rspo3 tumor tissues. Data are shown as mean ± s.d. Expression levels were normalized by cell numbers sorted. (E) Median FSC intensities of flow analyzed tumor-infiltrating NK cells (CD45+CD3−NK1.1+DX5+) from B16F10-EV and B16F10-Rspo3 tumors (n = 4-5 per group). (F) Tumor growth curves of B16F10-EV and B16F10-Rspo3 in Ncr1Cre and MycΔ/Δ/Ncr1Cre mice (n = 4-5 per group). (G) Tumor growth curves of B16F10-EV and B16F10-Rspo3 cells in Ncr1Cre and MycΔ/Δ/Ncr1Cre mice depleted with NK cells (left) or CD8+ T cells (right) (n = 4-5 per group). For panels A-D, and F-G, two-way ANOVA with or without Sidak’s multiple comparisons tests were performed. For panel E, student’s t-test was performed. Data represent at least two independent experiments. Data are shown as mean ± s.e.m. unless otherwise noted. P < 0.05 is considered as statistically significant. ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001.
We next explored the functional impact of altered MYC expression of NK cells in the R-spondin3-mediated tumor suppression. Implications from a Myc conditional knock-out mouse model – MycΔ/Δ/Ncr1Cre mice – which depletes MYC expression in NK cells and restricted subsets of innate lymphoid cells[34], indicated that ribosome and NK cell-mediated cytotoxicity were the two most enriched pathways in the differentially down-regulated genes (FDR<0.05) by RNA-seq analysis of the isolated splenic NK cells from MycΔ/Δ/Ncr1Cre mice (Supp Figure 8A, Supp Table 2). Translation-associated gene set was also shown to be negatively enriched in the NK cells with Myc deletion (Supp Figure 8B). These results suggested impaired ribosomal biogenesis as a feature for the NK cells with MYC deficiency. To determine whether the tumor-infiltrating NK cells from B16F10-Rspo3 tumors, which have enhanced MYC expression, also have enhanced ribosomal biogenesis compared to that from B16F10-EV tumors, quantitative RT-PCR was performed using the NK cells sorted from these tumors. Results showed increased expressions of rRNAs and mRNA level of ribosomal proteins by the NK cells from B16F10-Rspo3 tumors compared to that from the B16F10-EV tumors (Figure 5C-D). An increased FSC intensity revealed in flow cytometric analysis, a phenomenon usually observed in cells with enhanced ribosomal biogenesis associated with larger cell sizes, was also seen for the tumor-infiltrating NK cells in B16F10-Rspo3 tumors relative to -EV tumors (Figure 5E). Together, these indicate a stronger ribosomal biogenesis capacity of the NK cells in TME with a higher level of R-spondin3.
To determine whether MYC expression in NK cells is required for the R-spondin3-mediated tumor suppression, B16F10 tumors were inoculated to MycΔ/Δ/Ncr1Cre mice and Ncr1Cre controls. While the R-spondin3-mediated tumor suppression could still be observed in the MycΔ/Δ/Ncr1Cre mice (Figure 5F), which corresponded to the results shown in Figure 4E that depleting NK cells alone was not enough to abrogate tumor suppression in wild type mice, we found the tumor suppression was abrogated in the MycΔ/Δ/Ncr1Cre mice with CD8+ T cell-depleted (Figure 5G), suggesting MYC expression in NK cells is required for the contribution of NK cells to the R-spondin3-mediated effects.
R-spondin3 sensitizes tumors to PD-1 blocking therapy
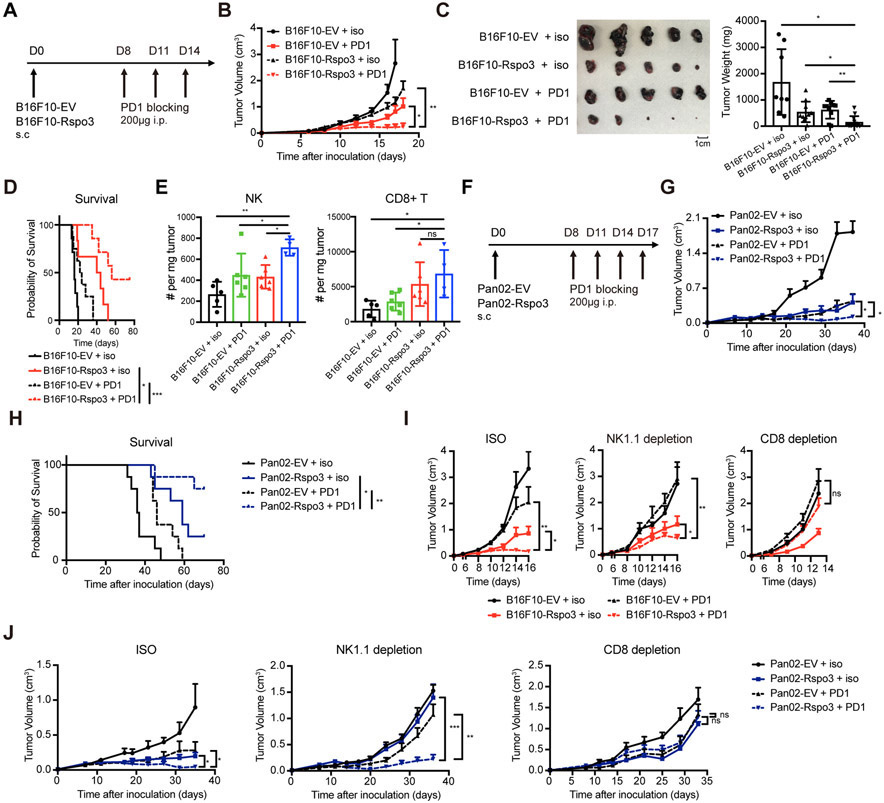
Given that NK cell activity and immune cell frequencies associate with patient responses to immune checkpoint inhibitors[2], we next tested the therapeutic efficacy of anti-PD1 antibody in the R-spondin3-overexpressing B16F10 tumors, whose parental line is known to be resistant to anti-PD1 antibody therapy (Figure 6A). Results showed a sensitization of B16F10 tumors with R-spondin3 overexpression to anti-PD1 antibody treatment (Figure 6B), with some B16F10-Rspo3 tumors got nearly completed rejected (Figure 6C). The response rate of B16F10-Rspo3 tumors to anti-PD1 therapy was significantly higher compared to B16F10-EV tumors (Supp Figure 9A-B). Survival of mice inoculated with B16F10-Rspo3 tumors with anti-PD1 therapy was substantially extended, with some achieved durable tumor remission (Figure 6D). Importantly, the percentages of tumor-infiltrating NK cells and CD8+ T cells were both increased in the B16F10-Rspo3 tumors with anti-PD1 antibody treatment compared to other groups (Figure 6E, Supp Figure 9C), indicating a better infiltration of cytotoxic cells. Further, therapeutic merits could also be observed in the Pan02-Rspo3 tumors with anti-PD1 antibody treatment (Figure 6F-H), indicating R-spondin3 and anti-PD1 therapy cooperatively enhance tumor control.
Figure 6. R-spondin3 sensitizes tumors to PD-1 blocking therapy.
(A) Experimental design for panels B-E. B16F10-EV or B16F10-Rspo3 cells (5×105 cells) were inoculated subcutaneously to B6 mice. 200ug anti-PD1 antibody or isotype antibody were intraperitoneally injected at days 8, 11, and 14. (B) Tumor growth curves of B16F10-EV and B16F10-Rspo3 cells with isotype or anti-PD1 therapy (n = 6-8 mice per group). (C) Representative tumor pictures (left) and summary of tumor weights (right) of B16F10-EV and B16F10-Rspo3 tumors with isotype or anti-PD1 therapy dissected 18 days after inoculation. (D) Survival curves and results of the Log-rank test of B16F10-EV and B16F10-Rspo3 tumors with isotype or anti-PD1 therapy (n = 6-8 mice for each group). (E) Absolute numbers of tumor-infiltrating NK cells (CD45+CD3−NK1.1+DX5+) and CD8+ cells (CD45+CD3+CD8+) in B16F10-EV and B16F10-Rspo3 tumors with isotype or anti-PD1 antibody therapy (n = 3-6 mice per group). (F) Experimental design for panels G-H. Pan02-EV or Pan02-Rspo3 cells (5×105 cells) were inoculated subcutaneously to B6 mice. 200ug anti-PD1 antibody or isotype antibody were intraperitoneally injected at days 8, 11, 14 and 17. (G) Tumor growth curves of Pan02-EV and Pan02-Rspo3 cells with isotype or anti-PD1 therapy (n = 6-8 mice per group). (H) Survival curves and results of the Log-rank test of B16F10-EV and B16F10-Rspo3 tumors with isotype or anti-PD1 therapy (n = 6-8 mice for each group). (I) Growth curves of B16F10-EV and B16F10-Rspo3 cells with or without anti-PD1 antibody therapy in mice treated with isotype control, anti-NK1.1 depletion antibody, or anti-CD8a depletion antibody (n = 5 - 8 mice per group) were shown. (J) Growth curves of Pan02-EV and Pan02-Rspo3 cells in mice treated with isotype control, anti-NK1.1 depletion antibody, or anti-CD8 depletion antibody (n = 6 - 8 mice per group). For panels B, G, I, and J, two-way ANOVA tests were performed. For panels C and E, two-way ANOVA tests with Tukey's multiple comparisons were performed. Data are representative of at least two independent experiments. Data are shown as mean ± s.e.m.. P < 0.05 is considered as statistically significant. ns, not statistically significant, *P < 0.05, **P < 0.01, and ***P < 0.001.
We next dissected the contribution of NK cells and CD8+ T cells to the combinatory effect of R-spondin3 with anti-PD1 therapy. Results showed that depletion of CD8+ T cells completely abrogated the exceptional outcome achieved by anti-PD1 treatment in -Rspo3 tumors in both tumor models (Figure 6I-J), suggesting a dependence on CD8+ T cells for the combinatory effects of anti-PD1 therapy with increased tumor R-spondin3 level. Importantly, although this combinatory effect could still be observed after NK cell depletion (Figure 6I-J), the strength was much diminished in the B16F10 model (Supp Figure 9D), suggesting NK cells also play a role here. Together, these data suggest a robust sensitization for PD-1 blocking therapy by enhanced R-spondin3 level in the TME.
DISCUSSION
In this study, we identified R-spondin3 and R-spondin1 derived from ECs/CAFs in the TME as critical regulators for anti-tumor immunity to affect cancer outcomes and sensitivity to immune checkpoint inhibitors. The expression of LGR6, a high-affinity receptor for R-spondins, is prominently expressed in human NK cells. Mechanistically, R-spondin3 enhances the MYC and ribosomal biogenesis gene expressions of NK cells in the tumor tissues.
Wnt signaling is delicately regulated by a variety of positive or negative regulators with temporospatial specificity. DKK1 is a secreted Wnt signaling negative regulator. Being a target gene of Wnt signaling, DKK1 is highly secreted by cancer cells with Wnt signaling aberrant activation[35]. On the other hand, previous literature has shown cancers with aberrant β-catenin activation present immune deserts lacking infiltration of immune cells[11, 12]. Interestingly, R-spondins synergize with Wnt proteins to activate canonical Wnt signaling with particular potency in the presence of DKK1[16]. These collectively suggest that the levels of R-spondins are reasonable with the capacity to modulate the activity of canonical Wnt signaling in the TME. This could particularly be the case for non-cancer cells in the TME including anti-tumor immune cells, whose activation of Wnt signaling are not like in cancer cells that are determined by intrinsic mutations or aberrant activation, but are affected, to a greater extent, by the alterations of signals in the TME. These may also partially explain why numerous drugs inhibiting Wnt/MYC signaling showed good efficacy in vitro, while compromised in vivo[36]. Therefore, our study provides new clues for these important questions.
The gene expressions of R-spondins are widely reduced across multiple cancers shown in TCGA data, while the underlying mechanism remains unclear. It is possible that R-spondin proteins are essential sustaining factors for immune cells in the TME and downregulating the expression levels of R-spondins may be a key mechanism for cancer cells to evade anti-tumor immunity. Our data showed positive correlations of RSPO3 and, to a less extent, RSPO1 with immune cell signatures and cancer outcomes, while we did not observe significant correlations for RSPO2 and RSPO4. This selectivity could be related to a more abundant expression of RSPO3 in tumor tissues compared to other RSPO genes (Figure 1B). Besides, although all four R-spondins are able to potentiate Wnt signaling, their expression patterns and phenotypes shown in knockout mice have striking differences[37], suggesting distinct roles for the four R-spondin members in modulating a variety of biological processes.
Our data showed a pronounced expression of LGR6 in human NK cells, particularly by the more mature and cytotoxic subsets. The finding is of notable interest given a wide belief that the three B-type LGRs (LGR4/5/6) mainly regulate embryonic development and adult stem cell self-renewing as determined by their unique expression patterns majorly observed in stem/progenitor cell populations[16]. Interestingly, in contrast to the neonatal lethality found in both Lgr4 null and Lgr5 null mutations mice, Lgr6 knockout mice are healthy and fertile[38], implying an essential difference for the biological functions of LGR6 from the other two LGRs. Future studies will be of great interests to clarify whether LGR6, like its expression in other tissues, marks specific NK cell populations with self-renewing capacities, such as memory NK cells; or whether it functions, like most other GPCRs that are highly specialized, to regulate certain NK-cell biological functions through G-proteins-mediated signaling, a general mechanism used by GPCRs while seems not yet identified to be used by the three B-type LGRs[13].
The R-spondins bind to LGR4/5/6 with high affinity through their furin-2 repeat, which allows the other furin repeat in R-spondin to interact with RNF43/ZNRF3, a membrane E3 ubiquitin ligase complex that removes Wnt receptors from cell surface. The subsequent endocytosis of the R-spondin-LGR-RNF43/ZNRF3 complex in turn leads to membrane clearance of the E3 ligases and persistence of Wnt receptors on the cell surface, thereby promoting Wnt signaling strength and duration[16, 39]. Of note, LGR-independent enhancement of Wnt signaling has also been reported recently for RSPO2 and RSPO3, which were determined by direct interaction of R-spondins with RNF43/ZNRF3[17, 18]. Interestingly, while RNF43/ZNRF3 homologs exist in invertebrates, the R-spondin/LGR/RNF43 module is considered as a relatively recent evolutionary ‘‘add-on’’ seen only in vertebrates and largely dedicated to adult stem cells[40]. In this regard, although the pronounced LGR6 expression in human NK cells is highly likely to mediate profound biological functions in human cancers, the R-spondin3-mediated tumor suppression revealed by our study in mouse tumor models is not necessarily to be mediated through LGR6, as shown in our data of a minor or no rescue of R-spondin3-mediated tumor suppression in Lgr6−/− mice (Supp Figure 5L-O). In contrast, LGR-independent signaling or LGR4/5 mediated signaling in other cell components in the TME may paly roles here. Our observation of a small peak expression bar shown in the mouse NK cells relative to other cell types by RNA-seq analysis (Supp Figure 4B) may provide one evidence for the evolution of this LGR-mediated exquisite modulation of Wnt signaling in an early phase shown in mice.
MYC, as the key target gene of canonical Wnt signaling pathway, is a master regulator controlling a variety of cellular processes, which includes ribosomal biogenesis that regulates mRNA translation. The expression of MYC has been shown essential for NK cell metabolism and functional status[41, 42] . Reduced MYC expression in the peripheral blood NK cells of patients with cancers was also reported[43]. Of note, one hallmark of NK cells is that they maintain abundant mRNA levels of cytotoxic molecules at rest, and a ready-to-go ribosomal biogenesis machinery that ensures prompt translation of cytotoxic molecules when encountering target cells is critically needed for their innate killing capacity[44]. Thus, our data that showed enhanced MYC and ribosomal biogenesis gene expressions in the NK cells with increased R-spondin3 in TME provides mechanistic insights on how R-spondin3 promotes anti-tumor immunity. Importantly, since the exact molecular basis of R-spondin-mediated signaling modulation in this context was unspecified, further studies may be performed to interrogate whether the enhanced MYC expression is a direct target of enhanced canonical Wnt signaling potentiated by R-spondin or a secondary consequence of an improved TME by R-spondin through activating other signaling pathways, such as non-canonical signaling. Given the use of a constitutive Ncr1Cre in our study that could result in an altered NK cell compartment and function, potential confounding factors could be involved to affect the observed phenotypes. Thus, an inducible Ncr1-iCreER allele that has been reported recently[45] could be a better tool to be used in the future to study the NK cell biology in cancers.
Inhibition of the PD-1/PD-L1 pathway has become a very powerful therapeutic strategy that remarkably improved the prognosis of patients with cancers. However, resistance remains a hurdle for a broader application, with multiple mechanisms proposed to contribute, which include an inadequate T or NK cell infiltration. Regarding this, in our tumor models, enhanced R-spondin3 in the TME promoted a better infiltration of both T cells and NK cells in tumor tissues, which is likely to be one mechanism for the sensitization to anti-PD1 therapy. Importantly, this combinatory effect is not only dependent on CD8+ T cells, depletion of NK cells also reduced the sensitivity to anti-PD1 antibody observed in the B16F10-Rspo3 tumor model, which could be due to a diminished tumor suppression by R-spondin3 and/or a consequence of immune checkpoint inhibitors to directly target NK cells[3, 4]. Further, depletion of CD8+ T cells completely eliminated any anti-tumor benefits observed in the B16F10-Rspo3 tumors, indicating complexity of the CD8+ T cell-independent mechanism by anti-PD1 antibody combined with R-spondin3. Future studies to investigate whether there is an altered expression of PD1 or PD-L1 on NK cells and CD8+ T cells will be of interest and informative to clarify underlying mechanisms.
Our study provides supports for a translational potential of R-spondin proteins as immunotherapeutic agents to treat cancers. While the safety of R-spondins being therapeutic agents should take into consideration of their roles in regulating the differentiation and proliferation of adult stem cells and tumorigenesis[46, 47]. Gene fusions involving RSPO3 or RSPO2 were previously identified in colon cancers, and anti-RSPO3 treatment was demonstrated to inhibit tumor growth in PTPRK-RSPO3-fusion-positive human tumor xenografts through mechanisms including regulating intestinal stem-cell function and promoting differentiation[48]. Of note, although transgenic overexpression of RSPO3 or RSPO3 fusion genes could result in adenomatous growth of the intestine, these alone were not sufficient to promote continued tumor growth[49, 50], supporting the observation that RSPO fusion genes always co-occur with either BRAF or KRAS mutation in colon cancers[47]. Therefore, while care should be taken, the strategy of utilizing R-spondins as immunotherapeutic agents remains promising.
To conclude, our study identified a novel role of R-spondins in promoting anti-tumor immunity in the TME (Supp Figure 10). Although R-spondin3 showed the broadest expression across tissue types, it warrants further studies to elucidate which R-spondin is the most robust one in providing therapeutic benefits. It also warrants further studies to determine the molecular or immunological features of the cancers that may benefit from R-spondin-based therapy. Future studies integrating nanoparticles or other bio-engineering technologies with the administration of R-spondin proteins will be valuable in advancing the translation of these new findings to clinical use in cancer therapies.
METHODS
Bio-info Analysis of Patient Transcriptome Data
Kendall’s correlation matrix analysis was performed based on a total of 60 components of Wnt signaling pathway (Supp Table 1) and NK cell signature of SKCM, PAAD, LUSC, HNSC, and visualized as heatmap after hierarchical clustering in Phantasus v1.5.1 (https://genome.ifmo.ru/phantasus). For the correlation analyses between genes or gene signatures, Spearman’s correlation analyses were performed in GEPIA2 using TCGA datasets[51] and summarized in Prism 8.0.1. Survival analyses were performed with TCGA datasets in GEPIA2. The gene expressions in the single-cell RNA-seq datasets of melanoma or pancreatic carcinoma patients were visualized with R or in Broad Institute’s Single Cell Portal (http://singlecell.broadinstitute.org/single_cell) using the previously reported datasets[52]. DICE dataset was downloaded from https://dice-database.org. Data presented with Primary Cell Atlas in BioGPS Dataset was obtained from BioGPS portal (http://biogps.org/#goto=welcome). Statistical significance of gene expressions between NK cell subsets was performed with EdgeR algorithm by galaxy tool[53]. Heatmaps were visualized with Phantasus v1.5.1.
Cell Lines
B16F10 cell line was purchased from American Type Culture Collection (ATCC). Pan02 cell line was purchased from the Division of Cancer Treatment and Diagnosis (DCTD), National Cancer Institute. Both cell lines were actively cultured for less than four months after purchase and not further authenticated. Mycoplasma testing was performed at least every two months by Universal Mycoplasma Detection Kit (ATCC, 30-1012K), with the latest testing date on Jan 5, 2021. The B16F10 cell line was cultured in DMEM (Thermo Fisher, 12430054) including 10% fetal bovine serum (Thermo Fisher, 16140-071) and 1× penicillin and streptomycin (Thermo Fisher, 15140-122). The Pan02 cell line was cultured in RPMI-1640 (Thermo Fisher, 21870-076) including 10% fetal bovine serum (Thermo Fisher, 16140-071) and 1× penicillin and streptomycin (Thermo Fisher, 15140-122). All cells were cultured at 37 °C, 5% CO2.
Transient Transfection and Retrovirus Infection
ORF clone of mouse Rspo3 (NM_028351.3) in the pcDNA3.1 vector was purchased from GenScript (Piscataway, NJ, USA). The full length of the ORF region was amplified with PCR using the primers: 5’-CTTGTCGACGCCACCATGCACTTGCGACTG-3’ (forward), and 5’-GTCGAGAATTCTTATCACTTATCGTCGTCATC-3’ (reverse) and cloned into the pMSCV-hpGK-GFP vector using the SalI and EcoRI restriction enzyme. Retroviruses were generated by calcium phosphate transient co-transfection of the retroviral vectors (MSCV-Rspo3-hpGK-eGFP or MSCV-hpGK-eGFP) with the packaging plasmids Gag and Eco-env into 293T cells. The supernatant was harvested at 48 hours and 72 hours and filtrated with a 0.45um filter. B16F10 or Pan02 cells were plated in a 6-well plate one day before the transduction. On the day of transduction, 1ml of the original media was kept and 2ml of the retroviral supernatant was added. Polybrene was used at the final concentration of 6 ug/ml. Cells were centrifuged at 800g for 90 minutes at RT. The GFP positive cells were sorted using flow cytometry two weeks after the transduction for further use.
Mice
All mice were bred and housed in specific pathogen-free conditions in the animal barrier facility at the Cincinnati Children’s Hospital Medical Center (CCHMC). All animal studies were conducted in accordance with an approved Institutional Animal Care and Use Committee protocol and federal regulations. Lgr6−/− mice (Jackson stock #016934), NRG mice (NOD-Rag1null IL2rgnull, NOD rag gamma), Rag1−/− mice (Jackson stock #002216), C57BL/6 mice (Jackson stock #000664), C57BL/6 congenic BoyJ mice were purchased from Jackson or Comprehensive Mouse and Cancer Core of CCHMC. All BoyJ mice used are confirmed with the expression of NKp46 by flow cytometry analysis with peripheral blood samples. The MycG/G mice were a kind gift from Dr. H. Leighton Grimes at Cincinnati Children’s Hospital Medical Center, OH, USA. Mycf/f mice and Ncr1Cre mice were backcrossed to C57BL/6 background at our lab. All mice used were 8 to 12 weeks old. Age and sex matching were performed for each independent experiment. The MycG/G mice, MycΔ/Δ/Ncr1Cre mice were born at the expected Mendelian ratios and showed normal WBC, hemoglobin, and platelet counts.
Syngeneic Mouse Tumor Models.
7- to 12-week-old mice were used to establish syngeneic mouse tumor models. Mice were subcutaneously (s.c.) inoculated with B16F10 or Pan02 lines (5×105 cells/mouse) into the right flank of the mouse. A caliper is used to measure the length and width of the tumor, and tumor volumes are estimated using the formula: [(length)x(width)x(width)]x0.52. The tumor volumes were monitored. Mice were killed before the tumor reached the maximum permitted size. Anti-PD1 antibody (29F.1A12) and isotype (2A3) were purchased from Bio X Cell, West Lebanon, NH, and administered 200μg/mouse intraperitoneally at the indicated time point as described. Recombinant carrier-free mouse R-spondin3 protein (R&D, 4120-RS-025/CF) was used for intra-tumor injection with the regimen indicated. For immune cell depletion studies, antibodies against CD8a (YTS 169.4, Bio X cell), NK1.1 (PK136, Bio X Cell) were used. CD8a depletion (400ug) was administered by intraperitoneal injection started on day −1, day 1, and was continued weekly for the duration of the experiment. NK1.1 depletion (100ug) was administered by intraperitoneal injection started on day 0 and was continued weekly for the duration of the experiment. Lymphocyte depletions were confirmed in peripheral blood lymphocytes and tumor-infiltrating lymphocytes by flow cytometry with the following antibodies: CD8a (53-6.7) and NKp46 (29A1.4).
Flow Cytometry and Cell Sorting
Flow cytometry analysis and cell sorting were performed with FACS Canto, LSR Fortessa, or FACSAria instruments (BD Biosciences). Single-cell suspensions of mouse peripheral blood, bone marrow, spleen, and lymph node were obtained by forcing of organs through 70 μm cell strainer. Single-cell suspensions of tumors were digested in HBSS buffer in the presence of Collagenase D (Sigma, 2mg/ml), Hyaluronidase (Sigma, 0.75mg/ml), and DNaseI (Sigma, 0.4mg/ml) for 45 min at 37°C before passing through the cell strainer. Erythrocytes were then eliminated by RBC lysis buffer. Single-cell suspensions were used for surface staining in phosphate-buffered saline PBS containing 2% fetal bovine serum (FBS) and followed by intracellular staining or secondary staining if necessary. Fixation/Permeabilization Solution Kit (BD Biosciences) was used for intracellular staining of perforin, granzyme B, and IFN-γ. Antibodies were purchased from Biolegend, BD Bioscience, eBioscience, or Thermo Fisher: CD3 (145-2C11 or 17A2), NK1.1 (PK136), CD49b (DX5), CD11b (M1/70), CD27 (LG.3A10), NKp46 (29A1.4), CD107a (1D4B), IFN-γ (XMG1.2), Ly6G (1A8), B220 (RA3-6B2), CD8 (53-6.7), CD4 (GK1.5), CD115 (AFS98), CD25 (PC61), CD11c (HL3), MHC-II (M5/114.15.2), CD19 (6D5), mouse CD45 (30-F11), Ly6C (HK1.4), CD24 (30-F1), F4/80 (BM8), CD103 (2E7), CD69 (H1.2F3), MYC (Y69), perforin (eBioOMAK-D), granzyme B (QA16A02), BV421 goat anti-rabbit IgG, Alexa Fluor 488 donkey anti-rabbit IgG (H+L), streptavidin. 7-AAD (BD Biosciences, 559925) or Zombie Aqua Fixable Viability Kit (Biolegend, 423101) was used to exclude dead cells during analysis. For measurement of absolute cell number of tumor-infiltrating immune cells per tumor weight, CountBright Absolute Counting Beads (Thermo Fisher, C36950) were used. Data were analyzed using FlowJo software. All flow cytometric data were acquired using equipments maintained by the Research Flow Cytometry Core in the Division of Rheumatology at Cincinnati Children’s Hospital Medical Center.
RNA Preparation and Real-Time qPCR
Bone marrow or spleen single-cell suspensions were prepared and stained as stated above before sorted into different populations using a FACSAria Cell Sorter (BD Biosciences). The purity of sorted cell populations was >95%. Sorted cells were lysed directly in RLT buffer from the RNeasy Micro kit (QIAGEN), and total RNA was extracted according to the manufacturer’s instructions. Amounts of total RNA were measured using NanoDrop according to the manufacturer’s instructions. cDNA was synthesized using the SuperScript III First-Strand Synthesis System for the RT-PCR Kit (Invitrogen). The cDNA was amplified using SYBR Green Master Mix (Life Technologies) with an Applied Biosystems Step One Plus thermal cycler (Applied Biosystems). Expression of target genes were determined using Atcb as internal control unless otherwise noted. Specific primers for each gene are shown in Supp Table 3.
RNA-seq and Data Analysis
CD3−NK1.1+DX5+ NK cells were sorted by flow cytometry from the splenic cells of three Mycf/f and two MycΔ/Δ/Ncr1Cre mice using FACSAria Cell Sorter (BD Biosciences). Total RNA was prepared as described above and submitted for RNA-seq analysis. Directional RNA-seq was performed by the Genomics, Epigenomics and Sequencing Core (GESC) at the University of Cincinnati. The RNA quality was determined by Bioanalyzer (Agilent, Santa Clara, CA). NEBNext Poly(A) mRNA Magnetic Isolation Module (New England BioLabs, Ipswich, MA) was used to isolate the polyA RNA. A total of 1 μg of good quality total RNA was used as input. The dUTP-based stranded library was prepared using the NEBNext Ultra II Directional RNA Library Prep Kit (New England BioLabs). The library was indexed and amplified under 8 PCR cycles. After library Bioanalyzer QC analysis and quantification, individually indexed and compatible libraries were proportionally pooled and sequenced using the Hiseq 1000 (Illumina, San Diego, CA). About 25 million pass filter reads per sample were generated under the sequencing setting of single read 1x51 bp.
Sequence reads were aligned to the reference genome using the TopHat aligner[54] and aligned reads to each known transcript were counted using Bioconductor packages and were used for further data analysis[55]. The analysis of differentially expressed genes between the Mycf/f and MycΔ/Δ/Ncr1Cre group was performed using the negative binomial statistical model of read counts as implemented in the edgeR Bioconductor package[56]. The pathway enrichment analysis was performed using The Database for Annotation, Visualization and Integrated Discovery (DAVID) gene functional classification tool[57]. Enrichment analysis of translation-associated gene sets (REACTOME_TRANSLATION) from MSigDB (Broad Institute, Inc., Massachusetts Institute of Technology, and Regents of the University of California) was performed using gene set enrichment analysis (GSEA)[58]. The number of permutations was 1,000. The signal-to-noise method was used. The raw RNA-seq sequencing data reported in this paper have been deposited in the Gene Expression Omnibus database under GSE142685.
Immunohistochemistry Staining
The formalin-fixed, paraffin-embedded tumor tissue sections were used for immunohistochemistry staining or hematoxylin and eosin (H&E) staining. For the former, samples were stained with anti-CD8 antibody (EPR20305, ab209775, Abcam, Cambridge, MA) or anti-NK1.1 antibody (ab197979, Abcam, Cambridge, MA). A Biotin Link was used as secondary antibody followed by streptavidin-peroxidase method, visualized with the DAB chromogen, and finally counter-stained with hematoxylin. The percentages of positive-staining cells were counted with at least four representative fields at 400 x magnification by two individual researchers independently. Scoring of tumor stroma area is based on methods reported before[59]. Human melanoma FFPE tissues were purchased from BioCore USA, and were stained with anti-RSPO3 (17193-1-AP, ProteinTech, Rosemont, USA) and anti-CD31 (ab28364, Abcam, Cambridge, MA).
Isolation of Lymphocytes
Human peripheral blood samples of healthy donors were obtained from the Cell Processing Core and studies were approved by Institutional Review Board at Cincinnati Children's Hospital Medical Center. Written, informed consent was obtained from each donor be included. Peripheral blood mononuclear cells and granulocytes were obtained by Ficoll (07801, STEMCELL, Cambridge, MA) processing based on the manufacture’s instruction. Lymphocytes were purified by magnetically labeling with human NK Cell Isolation Kit (130-092-657), human CD19 MicroBeads (130-050-301), human CD4+ T Cell Isolation Kit (130-096-533), and human CD8+ T Cell Isolation Kit (130-096-495) purchased from Miltenyi Biotech and sorted with an autoMACS Pro Separator.
Western Blotting
Human peripheral blood immune cells were purified with magnetic selection. The cell pellets were then lysed in sodium dodecyl sulfate (SDS) sample buffer containing 10 mM NaF, 10mM β-Glycerophosphate, 1mM phenylmethylsulfonyl fluoride, 0.2mM Na3VO4, 2.5mM dithiothreitol, 5% 2-mercaptoethanol, 1mM 4-Amidinophenylmethanesulfonyl Fluoride Hydrochloride, and proteinase inhibitors followed sonication. Samples were boiled at 95 °C for 5 minutes and loaded to SDS-polyacrylamide gel electrophoresis (PAGE). The separated proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Merck KGaA, Darmstadt, Germany) and blocked with 5% BSA in PBST for 1 h at room temperature. The membranes were further probed with the indicated primary antibodies overnight at 4°C. The following primary antibodies were used: anti-LGR6 antibody (ab126747, Abcam, Cambridge, MA), anti-RSPO3 antibody (17193-1-AP, ProteinTech, Rosemont, USA), and anti-β-actin antibody (ab49900, Abcam, Cambridge, MA). Horseradish peroxidase-conjugated antibody to rabbit (NA934V, GE Healthcare) was used to detect primary antibodies using the Super Signal West Dura Chemiluminescent Substrate (Pierce) was used for ECL detection. Band intensity quantification was determined using Image Lab (version 5.2.1) software. All images presented are representative of two to three independent experiments.
NK Cell Cytotoxicity Assay
Tumor tissues were digested into single-cell suspension as shown in the section of Flow Cytometry and Cell Sorting and further processed with Ficoll (07801, STEMCELL, Cambridge, MA) to remove dead cells. Tumor-infiltrating NK cells were isolated using CD49b (DX5) MicroBeads, mouse (Miltenyi Biotec, 130-052-501) according to the manufacturer’s instructions by an autoMACS Pro Separator (Miltenyi Biotec). B16F10 or YAC1 target cells were labeled for 2 hour with 2μCi of 51Cr per 1 × 104 target cells at 37 °C, 5% CO2. Washing procedures were performed to remove excess 51Cr. Labeled target cells were added to 96-well round-bottom plates (1 × 104 cells/well). Isolated NK cells were added to the plates with E:T ratios ranged between 50:1 and 6:1. The amount of 51Cr released, which corresponds to target cell death, was measured by a gamma scintillation counter. The percent cytotoxicity against target cells was calculated as: ((experimental lysis – spontaneous lysis)/(maximal lysis – spontaneous lysis)) × 100. To determine maximal lysis, 51Cr-labeled target cells were treated with 3% Triton X for 4 hours. To determine spontaneous release, target cells without effector cells were used for the assay.
Cell Viability Assays
Cells were seeded in 96-well plates in triplicate at a density of 4000 cells/100 μL/well. Cell viability was assayed with Cell Counting Kit-8 reagent (Dojindo, Japan) based on manufacture’s instruction, and the relative growth was calculated by normalizing to day 0 results.
Statistical Analysis.
Statistical analyses were performed using Prism 8.0.1 software. Selections of all statistical analysis methods meet the assumptions of the tests. Equality of variances between the groups was statistically compared. Student’s t-test or Welch’s t-test were used for comparisons of two groups. ANOVA with multiple comparisons was used for three or more groups and tumor growth profiles. The log-rank test was used to determine statistical significance for overall survival data. Multivariant regression analysis was used to determine whether RSPO3 level is an independent factor affecting NK-cell signature in TCGA datasets. Unless specifically noted, all data are representative of >2 independent experiments. Data are shown as mean ± s.d. unless otherwise noted. P < 0.05 was considered statistically significant. P-value is shown if 0.05<P<0.1.
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Freudiger, M. Rife, A. Woeste, P. Seig, L. Tilton, C. Sexton (Cincinnati Children’s Hospital Medical Center) for experiment assistance. We thank X. Pan for bioinfo analysis inquiry. We thank Dr. H. L. Grimes (Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA) for kindly providing the MycG/G mice. We thank Dr. J.S. Palumbo (Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA) for kindly providing the YAC-1 cell line. We thank Cincinnati Children’s Research Flow Cytometry Core (RFCC) in the Division of Rheumatology, Cincinnati Children’s Veterinary Services, and J. Bailey and V. Summey (Cincinnati Children’s Comprehensive Mouse and Cancer Core) for experiment assistance. RNA-Seq was conducted by Genomics, Epigenomics and Sequencing Core, Department of Environmental Health, University of Cincinnati. A part of RNA-Seq analyses was conducted by X. Zhang, J. Chen, and M. Medvedovic at Laboratory for Statistical Genomics and Systems Biology, Department of Environmental Health, University of Cincinnati. The results shown in this work are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
FINANCIAL SUPPORT:
This work was supported by National Institutes of Health (NIH) (R01DK105014 and 1R01CA248019 to G. Huang, and DA038017, AI148080, AR073228 to S.N. Waggoner), the CCTST Pilot Collaborative Studies (PCS) Grant to G. Huang, the Taub Foundation and EvansMDS Foundation to G. Huang, National Natural Science Foundation of China (81570196) to J. Zhou, the Arnold W. Strauss Fellow Award to Y. Tang, Pelotonia postdoctoral fellowship to Y. Tang, and postdoctoral fellowship from the American Heart Association to D.E. Ochayon.
Footnotes
CONFLICT OF INTEREST DISCLOSURE:
A patent application related to this work has been submitted to the U.S. Patent and Trademark Office; U.S. DEPARTMENT OF COMMERCE entitled ‘‘R-Spondins Enhance NK-cell Immunity’’ (application no. 63/034,010)
REFERENCE
- 1.Souza-Fonseca-Guimaraes F, Cursons J, Huntington ND. The Emergence of Natural Killer Cells as a Major Target in Cancer Immunotherapy. Trends Immunol 2019; 40(2):142–158. [DOI] [PubMed] [Google Scholar]
- 2.Barry KC, Hsu J, Broz ML, Cueto FJ, Binnewies M, Combes AJ, et al. A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat Med 2018; 24(8):1178–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu J, Hodgins JJ, Marathe M, Nicolai CJ, Bourgeois-Daigneault MC, Trevino TN, et al. Contribution of NK cells to immunotherapy mediated by PD-1/PD-L1 blockade. J Clin Invest 2018; 128(10):4654–4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong W, Wu X, Ma S, Wang Y, Nalin AP, Zhu Z, et al. The Mechanism of Anti-PD-L1 Antibody Efficacy against PD-L1-Negative Tumors Identifies NK Cells Expressing PD-L1 as a Cytolytic Effector. Cancer Discov 2019; 9(10):1422–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cozar B, Greppi M, Carpentier S, Narni-Mancinelli E, Chiossone L, Vivier E. Tumor-Infiltrating Natural Killer Cells. Cancer Discov 2021; 11(1):34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramakrishnan AB, Cadigan KM. Wnt target genes and where to find them. F1000Res 2017; 6:746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galluzzi L, Spranger S, Fuchs E, Lopez-Soto A. WNT Signaling in Cancer Immunosurveillance. Trends Cell Biol 2019; 29(1):44–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cichocki F, Valamehr B, Bjordahl R, Zhang B, Rezner B, Rogers P, et al. GSK3 Inhibition Drives Maturation of NK Cells and Enhances Their Antitumor Activity. Cancer Res 2017; 77(20):5664–5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malladi S, Macalinao DG, Jin X, He L, Basnet H, Zou Y, et al. Metastatic Latency and Immune Evasion through Autocrine Inhibition of WNT. Cell 2016; 165(1):45–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao Q, Wu J, Wang WJ, Chen S, Zheng Y, Yu X, et al. DKK2 imparts tumor immunity evasion through beta-catenin-independent suppression of cytotoxic immune-cell activation. Nat Med 2018; 24(3):262–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 2015; 523(7559):231–235. [DOI] [PubMed] [Google Scholar]
- 12.Luke JJ, Bao R, Sweis RF, Spranger S, Gajewski TF. WNT/beta-catenin Pathway Activation Correlates with Immune Exclusion across Human Cancers. Clin Cancer Res 2019; 25(10):3074–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lau W, Barker N, Low TY, Koo BK, Li VS, Teunissen H, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 2011; 476(7360):293–297. [DOI] [PubMed] [Google Scholar]
- 14.Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A 2011; 108(28):11452–11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagano K. R-spondin signaling as a pivotal regulator of tissue development and homeostasis. Jpn Dent Sci Rev 2019; 55(1):80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Lau W, Peng WC, Gros P, Clevers H. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev 2014; 28(4):305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lebensohn AM, Rohatgi R. R-spondins can potentiate WNT signaling without LGRs. Elife 2018; 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szenker-Ravi E, Altunoglu U, Leushacke M, Bosso-Lefevre C, Khatoo M, Thi Tran H, et al. RSPO2 inhibition of RNF43 and ZNRF3 governs limb development independently of LGR4/5/6. Nature 2018; 557(7706):564–569. [DOI] [PubMed] [Google Scholar]
- 19.Snippert HJ, Haegebarth A, Kasper M, Jaks V, van Es JH, Barker N, et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 2010; 327(5971):1385–1389. [DOI] [PubMed] [Google Scholar]
- 20.Ren W, Lewandowski BC, Watson J, Aihara E, Iwatsuki K, Bachmanov AA, et al. Single Lgr5- or Lgr6-expressing taste stem/progenitor cells generate taste bud cells ex vivo. Proc Natl Acad Sci U S A 2014; 111(46):16401–16406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fullgrabe A, Joost S, Are A, Jacob T, Sivan U, Haegebarth A, et al. Dynamics of Lgr6(+) Progenitor Cells in the Hair Follicle, Sebaceous Gland, and Interfollicular Epidermis. Stem Cell Reports 2015; 5(5):843–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blaas L, Pucci F, Messal HA, Andersson AB, Josue Ruiz E, Gerling M, et al. Lgr6 labels a rare population of mammary gland progenitor cells that are able to originate luminal mammary tumours. Nat Cell Biol 2016; 18(12):1346–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marcus A, Mao AJ, Lensink-Vasan M, Wang L, Vance RE, Raulet DH. Tumor-Derived cGAMP Triggers a STING-Mediated Interferon Response in Non-tumor Cells to Activate the NK Cell Response. Immunity 2018; 49(4):754–763 e754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hegde S, Krisnawan VE, Herzog BH, Zuo C, Breden MA, Knolhoff BL, et al. Dendritic Cell Paucity Leads to Dysfunctional Immune Surveillance in Pancreatic Cancer. Cancer Cell 2020; 37(3):289–307 e289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016; 352(6282):189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov 2019; 9(8):1102–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmiedel BJ, Singh D, Madrigal A, Valdovino-Gonzalez AG, White BM, Zapardiel-Gonzalo J, et al. Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell 2018; 175(6):1701–1715 e1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mabbott NA, Baillie JK, Brown H, Freeman TC, Hume DA. An expression atlas of human primary cells: inference of gene function from coexpression networks. BMC Genomics 2013; 14:632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu C, Jin X, Tsueng G, Afrasiabi C, Su AI. BioGPS: building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res 2016; 44(D1):D313–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collins PL, Cella M, Porter SI, Li S, Gurewitz GL, Hong HS, et al. Gene Regulatory Programs Conferring Phenotypic Identities to Human NK Cells. Cell 2019; 176(1-2):348–360 e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heng TS, Painter MW, Immunological Genome Project C. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 2008; 9(10):1091–1094. [DOI] [PubMed] [Google Scholar]
- 32.Choi J, Baldwin TM, Wong M, Bolden JE, Fairfax KA, Lucas EC, et al. Haemopedia RNA-seq: a database of gene expression during haematopoiesis in mice and humans. Nucleic Acids Res 2019; 47(D1):D780–D785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Lau WB, Snel B, Clevers HC. The R-spondin protein family. Genome Biol 2012; 13(3):242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol 2013; 13(2):145–149. [DOI] [PubMed] [Google Scholar]
- 35.Niida A, Hiroko T, Kasai M, Furukawa Y, Nakamura Y, Suzuki Y, et al. DKK1, a negative regulator of Wnt signaling, is a target of the beta-catenin/TCF pathway. Oncogene 2004; 23(52):8520–8526. [DOI] [PubMed] [Google Scholar]
- 36.Whitfield JR, Beaulieu ME, Soucek L. Strategies to Inhibit Myc and Their Clinical Applicability. Front Cell Dev Biol 2017; 5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim KA, Wagle M, Tran K, Zhan X, Dixon MA, Liu S, et al. R-Spondin family members regulate the Wnt pathway by a common mechanism. Mol Biol Cell 2008; 19(6):2588–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leushacke M, Barker N. Lgr5 and Lgr6 as markers to study adult stem cell roles in self-renewal and cancer. Oncogene 2012; 31(25):3009–3022. [DOI] [PubMed] [Google Scholar]
- 39.Hao HX, Xie Y, Zhang Y, Charlat O, Oster E, Avello M, et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 2012; 485(7397):195–200. [DOI] [PubMed] [Google Scholar]
- 40.Nusse R, Clevers H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017; 169(6):985–999. [DOI] [PubMed] [Google Scholar]
- 41.Loftus RM, Assmann N, Kedia-Mehta N, O'Brien KL, Garcia A, Gillespie C, et al. Amino acid-dependent cMyc expression is essential for NK cell metabolic and functional responses in mice. Nat Commun 2018; 9(1):2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong H, Adams NM, Xu Y, Cao J, Allan DSJ, Carlyle JR, et al. The IRE1 endoplasmic reticulum stress sensor activates natural killer cell immunity in part by regulating c-Myc. Nat Immunol 2019; 20(7):865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zakiryanova GK, Kustova E, Urazalieva NT, Baimuchametov ET, Nakisbekov NN, Shurin MR. Abnormal Expression of c-Myc Oncogene in NK Cells in Patients with Cancer. Int J Mol Sci 2019; 20(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fehniger TA, Cai SF, Cao X, Bredemeyer AJ, Presti RM, French AR, et al. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity 2007; 26(6):798–811. [DOI] [PubMed] [Google Scholar]
- 45.Wagner JA, Wong P, Schappe T, Berrien-Elliott MM, Cubitt C, Jaeger N, et al. Stage-Specific Requirement for Eomes in Mature NK Cell Homeostasis and Cytotoxicity. Cell Rep 2020; 31(9):107720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sigal M, Logan CY, Kapalczynska M, Mollenkopf HJ, Berger H, Wiedenmann B, et al. Stromal R-spondin orchestrates gastric epithelial stem cells and gland homeostasis. Nature 2017; 548(7668):451–455. [DOI] [PubMed] [Google Scholar]
- 47.Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, et al. Recurrent R-spondin fusions in colon cancer. Nature 2012; 488(7413):660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Storm EE, Durinck S, de Sousa e Melo F, Tremayne J, Kljavin N, Tan C, et al. Targeting PTPRK-RSPO3 colon tumours promotes differentiation and loss of stem-cell function. Nature 2016; 529(7584):97–100. [DOI] [PubMed] [Google Scholar]
- 49.Han T, Schatoff EM, Murphy C, Zafra MP, Wilkinson JE, Elemento O, et al. R-Spondin chromosome rearrangements drive Wnt-dependent tumour initiation and maintenance in the intestine. Nat Commun 2017; 8:15945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hilkens J, Timmer NC, Boer M, Ikink GJ, Schewe M, Sacchetti A, et al. RSPO3 expands intestinal stem cell and niche compartments and drives tumorigenesis. Gut 2017; 66(6):1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019; 47(W1):W556–W560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su MJ, Melms JC, et al. A Cancer Cell Program Promotes T Cell Exclusion and Resistance to Checkpoint Blockade. Cell 2018; 175(4):984–997 e924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 2018; 46(W1):W537–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009; 25(9):1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Amezquita RA, Lun ATL, Becht E, Carey VJ, Carpp LN, Geistlinger L, et al. Orchestrating single-cell analysis with Bioconductor. Nat Methods 2020; 17(2):137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 2013; 8(9):1765–1786. [DOI] [PubMed] [Google Scholar]
- 57.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 2007; 8(9):R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Pelt GW, Kjaer-Frifeldt S, van Krieken J, Al Dieri R, Morreau H, Tollenaar R, et al. Scoring the tumor-stroma ratio in colon cancer: procedure and recommendations. Virchows Arch 2018; 473(4):405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.