Abstract
Fifty-seven Escherichia coli O26 strains isolated from patients in six countries were investigated by PCR restriction fragment length polymorphism (RFLP) analysis of the flagellin-encoding (fliC) gene (fliC RFLP analysis). The strains were determined by serotyping to belong to five different H types or were nonmotile. The fliC RFLP analysis revealed only two different patterns among the 57 strains. One fliC RFLP pattern was displayed by 54 strains and was identical to that of E. coli H11 reference strain Su4321-41. The other fliC RFLP pattern was observed for three strains and was identical to that for E. coli H32 reference strain K10. The 54 strains with the H11 fliC RFLP pattern included 22 strains of serotype O26:H11, 23 nonmotile strains, and 9 strains that were initially serotyped as H2, H8, H21, and H32 but that were confirmed to express H11 by repeat serotyping. All 54 strains with the H11 fliC RFLP pattern contained the attaching-and-effacing (eae) gene. The three strains with the H32 fliC RFLP pattern belonged to serotype O26:H32, and all were eae negative. The fliC genes of 14 selected E. coli O26:H11 strains isolated between 1964 and 1999 had identical nucleotide sequences. Our results demonstrate that E. coli O26 strains that carry the eae gene belong exclusively to the H11 clonal complex. Since there were no H11 fliC allelic variations among the O26 strains tested, E. coli O26:H11 may have emerged recently. The fliC PCR-RFLP test is a reliable, easy-to-perform, and rapid method for determination of the H types of E. coli O26 isolates.
Escherichia coli strains of serotypes O26:H11 and O26:H− (nonmotile) were recognized as causes of infantile diarrhea in 1951 (13), and since then O26 has been considered one of the most important serogroups of enteropathogenic E. coli (EPEC). In 1977, production of Shiga toxin (Stx) was identified in E. coli O26 strains isolated from infants with diarrhea (10). Since the beginning of the 1980s Stx-producing E. coli (STEC) O26 strains have been increasingly recognized as causes of diarrhea, hemorrhagic colitis, and the hemolytic-uremic syndrome (HUS) (8, 21, 22, 23). During the last few years, STEC O26 strains have been the most frequently isolated non-O157 STEC strains associated with human disease (1, 2, 9, 24). Most E. coli O26 strains isolated from patients belong to serotype O26:H11 (2, 21), but strains that possess other H antigens such as H2, H8, H21, H30, H32, H36, and H46 have also been reported (3, 6, 25, 26).
H serotyping is a reference method for determination of H antigens (4, 14). However, it is labor-intensive and time-consuming and is performed in a limited number of reference laboratories. A considerable obstacle associated with conventional H serotyping is the fact that a substantial proportion of diarrheagenic E. coli strains are nonmotile upon isolation. These isolates require several passages through semisolid media to induce their motility and to express immunoreactive H antigens so that H serotyping can be performed. This delays the H-antigen determination and results in the delay of epidemiological investigations. Another drawback of conventional H serotyping is the occurrence of cross-reactions between particular H antigens that share common epitopes (4). Moreover, some E. coli isolates do not react with the presently available H-typing sera, thus making the determination of their H antigens impossible.
Recently, a PCR-restriction fragment length polymorphism (RFLP) method (the fliC RFLP method) was developed to identify and characterize the fliC gene, which encodes flagellin, a protein subunit of H antigens (5). Using the fliC RFLP method, Fields et al. (5) showed that STEC O157:H− strains possess a fliC gene which encodes the H7 antigen. Moreover, they demonstrated that almost all of the other 52 flagellar antigens investigated in their study had distinct fliC RPLP patterns after restriction with RsaI that could thus be used for identification of the respective H antigen (5). On the basis of their findings, the authors suggested that the fliC RFLP test could potentially be useful for H typing of other clinically important E. coli strains. In the study described here we applied the fliC RFLP method to characterization of the fliC genes of 57 E. coli O26 strains that were isolated in six countries, mostly from patients with diarrhea, and that were determined by serotyping to belong to five different H types or to be nonmotile.
MATERIALS AND METHODS
Bacterial strains.
The 57 E. coli O26 strains investigated in this study are summarized in Table 1. The strains were isolated between 1964 and 1999 in six different countries including Germany (n = 32), the Czech Republic (n = 19), Denmark (n = 3), France (n = 1), Norway (n = 1), and the United States (n = 1). Three strains originated from patients with HUS, 51 strains originated from patients with diarrhea, and 3 strains were from asymptomatic carriers. Thirty-four strains belonged to five different serotypes including O26:H2, O26:H8, O26:H11, O26:H21, and O26:H32; 23 isolates were nonmotile (O26:H−) (Table 1). Two strains, 3820/99 (O26:H8, stx2+) and 3821/99 (O26:H11, stx1+, stx2+), were received from Matthias Pulz (Niedersächsisches Landesgesundheitsamt, Hannover, Germany). Strain TB 285A (O26:H2, stx1+) was kindly provided by Phillip I. Tarr (Children's Hospital and Regional Medical Center, Seattle, Wash.) and was described previously (3). E. coli H reference strains Bi7455-41 (O43:K−:H2), Ap32oc (O2:K−:H8), Su4321-41 (O13:K11:H11), U11a-44 (O8:K49:H21), and K10 (O114:K−:H32) were obtained from the collection of the International Escherichia and Klebsiella Centre (World Health Organization [WHO]), Copenhagen, Denmark.
TABLE 1.
Characteristics of E. coli O26 strains investigated in this study
| Serotypea | No. of strains | stxb,c | eaec | fliC RFLP pattern |
|---|---|---|---|---|
| O26:H2 | 1 | + | + | H11d |
| O26:H8 | 2 | + | + | H11 |
| O26:H11 | 3 | + | + | H11 |
| O26:H11 | 19 | − | + | H11 |
| O26:H21 | 4 | + | + | H11 |
| O26:H21 | 1 | − | + | H11 |
| O26:H32 | 1 | − | + | H11 |
| O26:H32 | 3 | − | − | H32e |
| O26:H−f | 2 | + | + | H11 |
| O26:H− | 21 | − | + | H11 |
Serotypes determined by initial serotyping.
stx genotypes were stx1 (six strains), stx2 (five strains), and stx1 stx2 (one strain).
+ or −, presence or absence of stx and eae genes, respectively.
The fliC RFLP pattern designated H11 was identical to that of E. coli H11 reference strain Su4321-41.
fliC-RFLP pattern designated H32 was identical to that of E. coli H32 reference strain K10.
H−, nonmotile strains.
Serotyping.
Serotyping was performed by standard procedures (4, 14) with antisera to E. coli somatic antigens O1 to O170 and flagellar antigens H1 to H56. The antisera were prepared by immunization of rabbits as recommended by Edwards and Ewing (4). Strains isolated in Germany were serotyped at the National Reference Centre for Salmonella, Hamburg; isolates from the Czech Republic were serotyped at the Institute for Medical Microbiology, Charles University, Prague; and isolates from Denmark, France, and Norway were serotyped at the International Escherichia and Klebsiella Centre (WHO).
fliC PCR-RFLP analysis.
The fliC PCR-RFLP procedure was performed as described by Fields et al. (5), with slight modifications. Primers F-FLIC1 and R-FLIC2 (5) were used to amplify the fliC gene. PCR was performed in the GeneAmp PCR System 9600 (Perkin-Elmer Applied Biosystems, Weiterstadt, Germany) in a volume of 50 μl containing 5 μl of bacterial suspension (106 bacteria), each deoxynucleoside triphosphate at a concentration of 200 μM, 30 pmol of each primer, 5 μl of 10-fold-concentrated polymerase synthesis buffer, 4 μl of MgCl2 solution, and 2.0 U of AmpliTaq DNA polymerase (Perkin-Elmer Applied Biosystems). The PCR conditions included denaturation for 30 s at 94°C, annealing for 60 s at 60°C, and extension for 120 s at 72°C for 36 cycles. Fifteen microliters of each PCR product was digested with restriction endonuclease RsaI (Gibco BRL, Eggenstein, Germany), as recommended by the manufacturer. Restriction fragments were separated on a 2% (wt/vol) agarose gel and visualized by staining with ethidium bromide.
fliC sequence analysis.
The fliC PCR amplification products were purified with a QIAquick PCR purification kit (Qiagen, Hilden, Germany). The sequencing was performed with an automated DNA sequencer (model 377; Perkin-Elmer Applied Biosystems) with primers F-FLIC1 and R-FLIC2 and with customized primers. A fluorescence procedure with the Taq Prism Ready Reaction DyeDeoxy Terminator Cycle Sequencing Kit (Perkin-Elmer Applied Biosystems) was applied according to the manufacturer's instructions. Nucleotide sequence analysis was performed with the HUSAR program package (Heidelberg Unix Sequence Analysis Resources; German Cancer Research Center, Heidelberg, Germany) and with the DNASIS program (Hitachi Software, San Bruno, Calif.).
PCR techniques.
To detect stx genes, primers KS7-KS8 (stx1B) and GK3-GK4 (stx2B, stx2cB) were used as described previously (7, 17). stx2 and stx2c were differentiated by restriction analysis of GK3-GK4 amplification products with HaeIII or FokI (16). An 863-bp fragment of the 5′ conserved region of the intimin-encoding eae gene was amplified with primers SK1 and SK2 (18).
Nucleotide sequence accession number.
The nucleotide sequence of the fliC gene of E. O26:H11 strain 6061/96 has been entered into the EMBL database library under accession number AJ243796.
RESULTS
fliC RFLP analysis of E. coli H reference strains.
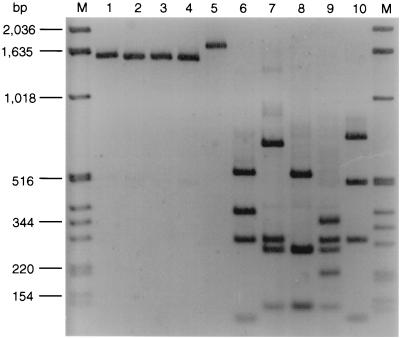
The E. coli O26 isolates chosen for analysis of their fliC genes were determined by serotyping to belong to five different H types (Table 1). To investigate the ability of the fliC RFLP method to differentiate the fliC genes that encode the respective H antigens, E. coli reference strains that express H2, H8, H11, H21, or H32 were subjected to fliC RFLP analysis. All strains produced single bands between 1.5 and 1.7 kb with primers F-FLIC1 and R-FLIC2 (Fig. 1, lanes 1 to 5). Following digestion with RsaI, the H2, H8, H11, H21, and H32 fliC amplification products displayed distinct restriction patterns (Fig. 1, lanes 6 to 10).
FIG. 1.
fliC PCR products and fliC RFLP patterns of E. coli H reference strains. Lanes M, molecular mass markers (1-kb DNA ladder; Gibco, BRL). In lanes 1 to 5 and 6 to 10, the fliC PCR products and fliC RFLP patterns, respectively, of the following strains (serotypes in parentheses) are shown: Bi7455-41 (O43:K−:H2) (lanes 1 and 6), Ap32oc (O2:K−:H8) lanes 2 and 7), Su4321-41 (O13:K11:H11) (lanes 3 and 8), U11a-44 (O8:K49:H21) (lanes 4 and 9), and K10 (O114:K−:H32) (lanes 5 and 10).
fliC RFLP patterns of E. coli O26 strains.
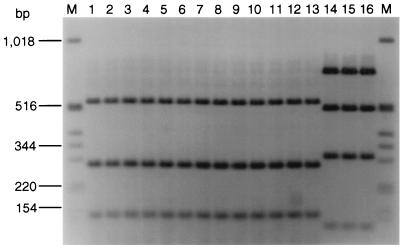
To investigate the fliC RFLP patterns of 57 E. coli O26 clinical isolates, 34 motile strains of five different H serotypes and 23 nonmotile strains (Table 1) were subjected to PCR with primers F-FLIC1 and R-FLIC2 and subsequent restriction with RsaI. All strains demonstrated a single PCR product of approximately 1.5 or 1.7 kb. After restriction with RsaI, two different fliC RFLP patterns were observed among the 57 strains (Fig. 2; Table 1). All 22 strains of serotype O26:H11 and all 23 nonmotile isolates shared a fliC RFLP pattern of three bands of 520, 280, and 150 bp (Fig. 2, lanes 1 to 4) that was identical to that of E. coli H11 reference strain Su4321-41 (Fig. 1, lane 8). Surprisingly, the same fliC RFLP pattern was displayed by all eight E. coli O26 strains whose H antigens were determined by serotyping to be H2 (Fig. 2, lane 5), H8 (Fig. 2, lanes 6 and 7), or H21 (Fig. 2, lanes 8 to 12) and by one of four strains serotyped as H32 (Fig. 2, lane 13). In contrast, the other three isolates that expressed the H32 antigen (Table 1) demonstrated a different fliC RFLP pattern (Fig. 2, lanes 14 to 16) that was identical to that of E. coli H32 reference strain K10 (Fig. 1, lane 10).
FIG. 2.
fliC RFLP patterns of E. coli O26 isolates with different H antigens determined by serotyping or determined to be nonmotile (H−). Lanes M, molecular mass marker (1-kb DNA ladder; Gibco, BRL). Lanes 1 to 16, E. coli O26 strains (H antigens in parentheses) 1616/96 (H11) (lane 1), 8590/64 (H11) (lane 2), 6422/94 (H−) (lane 3), 4393/66 (H−) (lane 4), TB 285A (H2) (lane 5), 3820/99 (H8) (lane 6), 4118/99 (H8) (lane 7), C1137-76 (H21) (lane 8), C1138-76 (H21) (lane 9), C1218-96 (H21) (lane 10), C1540-99 (H21) (lane 11), C1215-99 (H21) (lane 12), 15/77 (H32) (lane 13), 98/78 (H32) (lane 14), 1897/88 (H32) (lane 15), and 1976/88 (H32) (lane 16). On the basis of the data derived from nucleotide sequence analysis of the fliC gene in E. coli O26:H11 strain 6061/96 (accession number AJ243796), bands of 280 and 150 bp in the H11 fliC RFLP pattern (lanes 1 to 13) each consist of two fragments of similar sizes (269 and 276 bp and 141 and 142 bp, respectively) that could not be separated on the gel. Three additional fragments of 41, 31, and 14 bp could not be detected on the gel.
Analysis of discrepant results between H serotypes and fliC-RFLP pattern.
The nine strains that demonstrated discrepant results between their H serotypes and the fliC RFLP patterns are characterized in Table 2. To explain the discrepancies, the strains were subjected to repeat H serotyping. All strains agglutinated at high titers (1:6,400 to 1:25,600) with H11 antiserum but did not cross-react with the respective heterologous antisera including the H2, H8, H21, and H32 antisera. This confirmed the presence of the H11 antigen in all nine strains that displayed the H11 fliC RFLP pattern (Table 2). Moreover, the fliC genes of four selected isolates whose H serotypes were initially determined to be H2, H8, H21, and H32 were sequenced (Table 2) and shown to have nucleotide sequences identical to that of a representative E. coli O26:H11 strain, 6061/96 (accession number AJ243796).
TABLE 2.
Characteristics of E. coli O26 strains that showed discrepancies between H serotypes and fliC RFLP patterns
| Strain no. | Country of isolation | stx gene | Initial serotype | fliC RFLP patterna | H antigen by repeat serotyping | Pattern by fliC sequencing |
|---|---|---|---|---|---|---|
| TB 285A | United States | stx1 | O26:H2 | H11 | H11 | H11b |
| 3820/99 | Germany | stx2 | O26:H8 | H11 | H11 | H11 |
| 4118/99 | Germany | stx1 | O26:H8 | H11 | H11 | NPc |
| C1137-76 | Denmark | stx1 | O26:H21 | H11 | H11 | H11 |
| C1138-76 | Denmark | stx1 | O26:H21 | H11 | H11 | NP |
| C1218-96 | Denmark | stx1 | O26:H21 | H11 | H11 | NP |
| C1540-99 | Norway | −d | O26:H21 | H11 | H11 | NP |
| C1215-99 | France | stx1 | O26:H21 | H11 | H11 | NP |
| 15/77 | Czech Republic | − | O26:H32 | H11 | H11 | H11 |
The fliC RFLP patterns of all strains (shown in Fig. 2, lanes 5 to 13) were identical to that of E. coli H11 reference strain Su4321-41 (Fig. 1, lane 8).
The fliC nucleotide sequences were identical to that of E. coli O26:H11 strain 6061/96 (accession number AJ243796).
NP, not performed.
−, negative for stx gene.
Presence of stx and eae genes in E. coli O26 strains with different fliC RFLP patterns.
Only 12 of 57 E. coli O26 strains harbored stx genes (Table 1). We therefore investigated the correlation between the presence of the eae gene and fliC RFLP patterns. As shown in Table 1, all 54 strains that demonstrated the H11 fliC RFLP pattern possessed the eae gene. In contrast, eae was not detected in any of three strains that showed the H32 fliC RFLP pattern.
Sequence analysis of fliC genes of E. coli O26:H11 strains isolated from 1964 to 1999.
To determine whether the H11 fliC gene demonstrated allelic variations among strains isolated during a 35-year period, the fliC genes of 14 E. coli O26:H11 isolates from 1964 to 1999 were sequenced and their nucleotide sequences were compared. The fliC genes of all 14 strains were identical.
DISCUSSION
A number of different H antigens, including H2, H8, H11, H21, H30, H32, H36, and H46, have previously been identified by conventional H serotyping in E. coli O26 strains that cause diarrhea and/or HUS in humans (3, 6, 25, 26). Using fliC-RFLP and fliC sequence analysis, we have demonstrated that 57 E. coli O26 strains isolated from patients and determined to belong to five different H serotypes possessed only fliC genes that encode the H11 or H32 antigen. The H11 fliC gene was demonstrated in all 22 strains of serotype O26:H11 and in all 23 nonmotile strains. However, it was also found in nine strains that were originally serotyped as H2, H8, H21, and H32. All nine strains that showed discrepant results between the H serotype and the fliC RFLP pattern were confirmed to express the H11 antigen by repeat serotyping. Importantly, the eae gene, which is a determinant of the pathogenicity island LEE (locus of enterocyte effacement) and which encodes intimin, which plays an important role in the pathogenesis of EPEC- and STEC-associated diseases (12), was present in all 54 E. coli O26 strains that contained the H11-encoding fliC. In contrast, eae was absent from all three strains that harbored the fliC gene that encodes H32. These findings suggest that E. coli O26 strains that carry the eae gene and that are thus capable of causing human disease belong exclusively to the H11 clonal complex. Interestingly, while all 54 H11 fliC-containing strains possessed eae genes, only 12 (22%) of them also harbored stx genes. This parallels the situation observed among E. coli O157:H7 strains in that such strains also always possess the eae gene but stx-negative isolates have been reported to be causes of human diseases (19). However, while stx-negative variants of E. coli O157:H7 occur with a low frequency (19), stx-negative isolates accounted for 78% of eae-positive fliC H11-containing E. coli O26 strains investigated in this study. The stx-negative and eae-positive E. coli O26 strains could fall into three different categories: (i) they could be “atypical EPEC,” as defined by Nataro and Kaper (12), (ii) they could be STEC strains that lost their stx genes, or (iii) they could be progenitors of STEC O26:H11 that could, in the future, become STEC following transduction with stx-converting phages. Further investigation of the virulence characteristics of these strains is needed to establish whether they represent a distinct pathogroup or belong to different pathogroups. Although the three H32 fliC-containing E. coli O26 strains in our study were also isolated from patients with diarrhea, the lack of both the stx and the eae genes in these strains makes their etiological role in the associated disease uncertain. It is also possible that such strains possess yet unknown virulence factors.
To avoid difficulties associated with the conventional H serotyping, the fliC RFLP method was developed primarily for the detection and identification of the H7 antigen in nonmotile STEC O157 strains (5). However, the performance of the method for H typing of other E. coli strains has not yet been evaluated. We demonstrated in this study that the fliC RFLP approach is a reliable, rapid, and easy-to-perform method for determination of the H types of E. coli O26 clinical isolates, including nonmotile strains that belong to the H11 clone but that cannot be H typed by serotyping. We found the fliC RFLP method to be superior to H serotyping since it correctly identified the H types of all strains investigated, including those nine strains which were initially determined not to be H11 by serotyping. The results for these nine strains originally serotyped as H2, H8, H21, and H32 are probably the result of a laboratory failure to correctly identify the H11 antigen. This illustrates the objective difficulties inherent to conventional H serotyping (4) and strengthens the usefulness of the fliC RFLP approach for H typing of E. coli O26 isolates. The fliC RFLP method, which allows determination of the H types of isolates in 48 h, can be particularly useful in epidemiological investigations when prompt information about the H type is urgently needed.
Sequence analysis of the flagellins in pathogenic E. coli strains showed that the amino-terminal and carboxy-terminal parts of the molecules, which are responsible for secretion and polymerization, are highly conserved (15, 20). In comparison, the central region which produces the surface-exposed antigenic part of the flagellin is highly variable within the species (15, 20) and gives rise to serotype-specific epitopes (11). However, on a molecular basis, diversity within an H type exists. For example, Reid et al. (15) identified four distinct alleles within each of the fliC genes that encode H6 and H7. We have observed no allelic variation within the fliC gene that encodes H11. This could mean that E. coli O26 strains that carry H11 emerged recently. Another explanation for the identical fliC sequences within the E. coli O26:H11 strains is that this segment of the chromosome has only recently been acquired by horizontal transfer and has not had time to develop polymorphisms.
In conclusion, by using the fliC RFLP method and fliC sequence analysis, we have demonstrated that the eae-positive diarrheagenic E. coli O26 strains investigated in this study and isolated from patients in different geographical areas belong to the H11 clonal complex, even though we detected various H antigens in the strains by serotyping. On the basis of the findings presented here, we strongly suspect that E. coli O26 strains isolated from patients with diarrhea or HUS in other studies and serotyped as possessing various non-H11 flagellar antigens may also belong, in fact, to the H11 clonal complex. RFLP analysis of fliC genes appears to be a powerful method for determination of whether such strains can be reclassified as E. coli O26:H11.
ACKNOWLEDGMENTS
This study was supported by grants from the Bundesministerium für Bildung und Forschung (BMBF) Verbundprojekt, Forschungsnetzwerk “Emerging foodborne pathogens in Germany” (grants FKZ 01KI 9903 and FKZ 01KI 9902/6). Investigation of Czech isolates was partially supported by grant IGA 4563-3 from the Ministry of Health of the Czech Republic.
We thank Phillip I. Tarr for critical reading of the manuscript and helpful discussions. The excellent technical assistance of Barbara Plaschke (Würzburg) and Alena Reischelova (Prague) is highly appreciated.
REFERENCES
- 1.Bielaszewska M, Janda J, Blahova K, Feber J, Potuznik V, Souckova A. Verocytotoxin-producing Escherichia coli in children with hemolytic uremic syndrome in the Czech Republic. Clin Nephrol. 1996;46:42–44. [PubMed] [Google Scholar]
- 2.Bockemühl J, Karch H, Tschäpe H. Zur Situation der Infektionen des Menschen durch enterohämorrhagische Escherichia coli (EHEC) in Deutschland, 1997. Bundesgesundheitblatt. 1998;1998(Suppl.):2–5. doi: 10.1007/s00103-002-0458-4. [DOI] [PubMed] [Google Scholar]
- 3.Bokete T N, Whittam T S, Wilson R S, Clausen C R, O'Callahan C M, Moseley S L, Fritsche T R, Tarr P I. Genetic and phenotypic analysis of Escherichia coli with enteropathogenic characteristics isolated from Seattle children. J Infect Dis. 1997;175:1382–1389. doi: 10.1086/516470. [DOI] [PubMed] [Google Scholar]
- 4.Edwards P R, Ewing W H. Identification of Enterobacteriaceae. 3rd ed. Minneapolis, Minn: Burgess Publishing; 1972. Genus Escherichia; pp. 67–107. [Google Scholar]
- 5.Fields P I, Blom K, Hughes H J, Helsel L O, Feng P, Swaminathan B. Molecular characterization of the gene encoding H antigen in Escherichia coli and development of a PCR-restriction fragment length polymorphism test for identification of E. coli O157:H7 and O157:NM. J Clin Microbiol. 1997;35:1066–1070. doi: 10.1128/jcm.35.5.1066-1070.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giammanco A, Maggio M, Giammanco G, Morelli R, Minelli F, Scheutz F, Caprioli A. Characteristics of Escherichia coli strains belonging to enteropathogenic E. coli serogroups isolated in Italy from children with diarrhea. J Clin Microbiol. 1996;34:689–694. doi: 10.1128/jcm.34.3.689-694.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunzer F, Böhm H, Rüssmann H, Bitzan M, Aleksic S, Karch H. Molecular detection of sorbitol-fermenting Escherichia coli O157 in patients with hemolytic-uremic syndrome. J Clin Microbiol. 1992;30:1807–1810. doi: 10.1128/jcm.30.7.1807-1810.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karch H, Heesemann J, Laufs R. Phage-associated cytotoxin production by and enteroadhesiveness of enteropathogenic Escherichia coli isolated from infants with diarrhea. J Infect Dis. 1987;155:707–715. doi: 10.1093/infdis/155.4.707. [DOI] [PubMed] [Google Scholar]
- 9.Karch H, Bielaszewska M, Bitzan M, Schmidt H. Epidemiology and diagnosis of Shiga toxin-producing Escherichia coli infections. Diagn Microbiol Infect Dis. 1999;34:229–243. doi: 10.1016/s0732-8893(99)00031-0. [DOI] [PubMed] [Google Scholar]
- 10.Konowalchuk J, Speirs J I, Stavric S. Vero response to a cytotoxin of Escherichia coli. Infect Immun. 1977;18:775–779. doi: 10.1128/iai.18.3.775-779.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuwajima G. Flagellin domain that affects H antigenicity of Escherichia coli. J Bacteriol. 1988;170:485–488. doi: 10.1128/jb.170.1.485-488.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nataro J, Kaper J. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ørskov F. On the occurrence of E. coli belonging to O group 26 in cases of infantile diarrhoea and white scours. APMIS. 1951;29:373–378. [PubMed] [Google Scholar]
- 14.Ørskov F, Ørskov I. Serotyping of Escherichia coli. Methods Microbiol. 1984;1:43–112. [Google Scholar]
- 15.Reid S D, Selander R K, Whittam T S. Sequence diversity of flagellin (fliC) alleles in pathogenic Escherichia coli. J Bacteriol. 1999;181:153–160. doi: 10.1128/jb.181.1.153-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rüssmann H, Schmidt H, Heesemann J, Caprioli A, Karch H. Variants of Shiga-like toxin II constitute a major toxin component in Escherichia coli O157 strains from patients with haemolytic uraemic syndrome. J Med Microbiol. 1994;40:338–343. doi: 10.1099/00222615-40-5-338. [DOI] [PubMed] [Google Scholar]
- 17.Rüssmann H, Kothe E, Schmidt H, Franke S, Harmsen D, Caprioli A, Karch H. Genotyping of Shiga-like toxin genes in non-O157 Escherichia coli strains associated with haemolytic uraemic syndrome. J Med Microbiol. 1995;42:404–410. doi: 10.1099/00222615-42-6-404. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt H, Plaschke B, Franke S, Rüssmann H, Schwarzkopf A, Heesemann J, Karch H. Differentiation in virulence patterns of Escherichia coli possessing eae genes. Med Microbiol Immunol. 1994;183:23–31. doi: 10.1007/BF00193628. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt H, Scheef J, Huppertz H I, Frosch M, Karch H. Escherichia coli O157:H7 and O157:H− strains that do not produce Shiga toxin: phenotypic and genetic characterization of isolates associated with diarrhea and hemolytic-uremic syndrome. J Clin Microbiol. 1999;37:3491–3496. doi: 10.1128/jcm.37.11.3491-3496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoenhals G, Whitfield C. Comparative analysis of flagellin sequences from Escherichia coli strains possessing serologically distinct flagellar filaments with a shared complex surface pattern. J Bacteriol. 1993;175:5395–5402. doi: 10.1128/jb.175.17.5395-5402.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scotland S M, Day N P, Rowe B. Production of a cytotoxin affecting Vero cells by strains of Escherichia coli belonging to traditional enteropathogenic serogroups. FEMS Microbiol Lett. 1980;7:15–17. [Google Scholar]
- 22.Scotland S M, Willshaw G A, Smith H R, Rowe B. Properties of strains of Escherichia coli O26:H11 in relation to their enteropathogenic or enterohemorrhagic classification. J Infect Dis. 1990;162:1069–1074. doi: 10.1093/infdis/162.5.1069. [DOI] [PubMed] [Google Scholar]
- 23.Sramkova L, Bielaszewska M, Janda J, Blahova K, Hausner O. Verocytotoxin-producing strains of Escherichia coli in children with haemolytic uraemic syndrome and diarrhoea in Czechoslovakia. Infection. 1990;18:204–209. doi: 10.1007/BF01643386. [DOI] [PubMed] [Google Scholar]
- 24.Tarr P I, Neill M A. Perspective: the problem of non-O157 Shiga toxin (Verocytotoxin)-producing Escherichia coli. J Infect Dis. 1996;174:1136–1139. doi: 10.1093/infdis/174.5.1136. [DOI] [PubMed] [Google Scholar]
- 25.Whittam T S, Wolfe M L, Wachsmuth I K, Orskov F, Orskov I, Wilson R. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect Immun. 1993;61:1619–1629. doi: 10.1128/iai.61.5.1619-1629.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.World Health Organization. Zoonotic non-O157 Shiga toxin-producing Escherichia coli (STEC). Report of a WHO Scientific Working Group Meeting. Geneva, Switzerland: World Health Organization; 1999. [Google Scholar]