Abstract
Host innate and adaptive immune responses play a vital role in clearing infected viruses. Meanwhile, viruses also evolve a series of mechanisms to weaken the host immune responses and evade immune defense. Recently, N6-methyladenosine (m6A), the most prevalent mRNA modification, has been revealed to regulate multiple steps of RNA metabolism, such as mRNA splicing, localization, stabilization, and translation, thus participating in many biological phenomena, including viral infection. In the process of virus–host interaction, the m6A modification that presents on the virus RNA impedes capture by the pattern recognition receptors, and the m6A modification appearing on the host immune-related molecules regulate interferon response, immune cell differentiation, inflammatory cytokine production, and other immune responses induced by viral infection. This review summarizes the research advances about the regulatory role of m6A modification in the innate and adaptive immune responses during viral infections.
Keywords: N6-methyladenosine modification, viral infection, immune recognition, innate immunity, adaptive immunity
Introduction
The discovery of modifications residing in DNA and histone proteins has proposed epigenetics, which provides a new perspective on regulation of gene expression and many other important biological processes. Besides this, there are more than 170 covalent modifications in the other layer of the central dogma, RNA, predominantly in tRNA and rRNA (Boccaletto et al., 2018). Those RNA modifications, such as N6-methyladenosine (m6A), N1-methyladenosine (m1A), 5-methylcytidine (m5C), pseudouridine (Ψ), 2′O-methylation (2′OMe), 7-methylguanosine (m7G), and N6, 2′O-methyladenosine (m6Am), are critical for RNA metabolism, function, and localization, thus becoming a research hot spot (Nachtergaele and He, 2018).
Currently, emerging research indicates that m6A, as the ubiquitous modification in internal mRNA, is dynamically regulated by the functional interplay among m6A methyltransferases, demethylases, and reader proteins. It is generally believed that the “write-in” of a methyl group to the N6 position of adenosine is catalyzed by the S-adenosyl-L-methionine (SAM)-dependent multisubunit methyltransferase complex composed of METTL3, METTL14, and other accessory components. The m6A modification specifically occurs in fractional mRNA and in the consensus sequence, DRACH (D = A, G, or U; R = G or A; H = A, C or U) (Fu et al., 2014). m6A codes are interpreted through being bound by the particular m6A RNA-binding proteins, such as the YTH domain-containing proteins (YTHDC1-2, YTHDF1-3) (Wang et al., 2015). In addition, the RNA structure can be destabilized due to the weaker base pair interactions between m6A and U; thus, heterogeneous nuclear ribonucleoprotein (hnRNP) may be recruited to bind to the hidden RNA-binding sites (Liu et al., 2017). Therefore, m6A readers are used to characterize the mRNA-binding proteins whose affinity to mRNA can be influenced by the presence of m6A and/or m6A-induced RNA structure changes (Shi et al., 2019). These m6A readers execute the function of m6A in multiple processes of mRNA fate, such as splicing, nuclear export, cap-independent translation, and decay. The oxidative demethylation of m6A is proved to be carried out by the demethylases ALKBH5 and FTO, known as erasers, which confer the reversibility of m6A modification in the life cycle of mRNA (Zaccara et al., 2019). Furthermore, m6A modification heavily influences a variety of physiological and pathological events, such as embryonic development, cell differentiation, viral infection, and tumorigenesis by fine-tuning RNA biology (Bi et al., 2019).
Numerous studies show that viral infection can induce host m6A machinery rearrangement; meanwhile, m6A-associated proteins positively or negatively regulate the viral replication cycle and pathogenesis by changing the m6A modification status of viral RNA reciprocally (Yang et al., 2019). By transcriptome-wide mapping of m6A sites and manipulation of writers, erasers, or readers to perturb m6A, it is reported that the decoration of m6A in influenza A virus (IAV) genomic RNA or mRNA increased hemagglutinin expression (Courtney et al., 2017), whereas Zika virus (ZIKV) replication was inhibited by m6A modification (Lichinchi et al., 2016b). As to hepatitis B virus (HBV), m6A that distributed at the 5′ epsilon stem loop was required for efficient reverse transcription of pregenomic RNA (pgRNA), whereas m6A at the 3′ epsilon stem loop resulted in destabilization of all HBV transcripts, including mRNA and pgRNA (Imam et al., 2018). However, there are some conflicting opinions about how m6A modifications influence the replication of human immunodeficiency virus-1 (HIV-1) (Kennedy et al., 2016; Lichinchi et al., 2016a; Tirumuru et al., 2016). The appearance, location, and function of m6A modification in diverse viral RNA are summarized in detail in previous reviews (Kennedy et al., 2017; Wu et al., 2019). The outcomes of viral infections depend on not only the magnitude of virus amplification or their cytocidal effects, but also the host immune status to a large extent.
It is well characterized that innate and adaptive immune responses are invoked in succession upon a virus invading. As the host’s first line of defense against viruses, pattern recognition receptors (PRR) are critical in the recognition of conserved pathogen-associated molecular patterns (PAMPs) and launching a series of protective immune responses rapidly (Schlee and Hartmann, 2016). PRR, such as Toll-like receptors (TLR), the RIG-I-like receptor family (RLR), and the NOD-like receptor family (NLR), capture viral RNA specifically and signal through the adaptor myeloid differentiation primary response protein 88 (MyD88) or mitochondrial antiviral signaling protein (MAVS). Upon sensing viral RNA, macrophages produce a large amount of cytokines, for instance, interleukin-1β (IL-1β), IL-6, tumor necrosis factor (TNF), and interferon (IFN), eliciting inflammatory responses, building an antiviral state to block virus reproduction, and enhancing the phagocytosis or cytotoxicity effects of neutrophils and natural killer (NK) cells (Chen et al., 2017; McFadden et al., 2017). Subsequently, host cellular and humoral immune responses are often activated by antigen presenting cells (APC) to eliminate viruses. As a critical subset of CD4+ T cells, helper T lymphocytes (Th) function in orchestrating antiviral responses by producing cytokines, including IFN-γ, IL-2, IL-4, and IL-5 (Zhu, 2018). Regulatory T cells (Treg), as a group of immunosuppressive cells, participate in regulation of infection or inflammatory responses to minimize immune pathogenesis in infectious conditions (Rakebrandt et al., 2016). It is conceivable that, except for acting on viral RNA directly, the m6A modification likewise has remarkable regulatory control on the immune system and other host reactions, which gives rise to either strengthen or weaken antiviral effects. In this review, we outline the recent advances in the field about the regulation of m6A modification in the antiviral-related immune processes mentioned above, highlighting the innate immunity in response to viral infection.
m6A Modification in Non-Self RNA Recognition
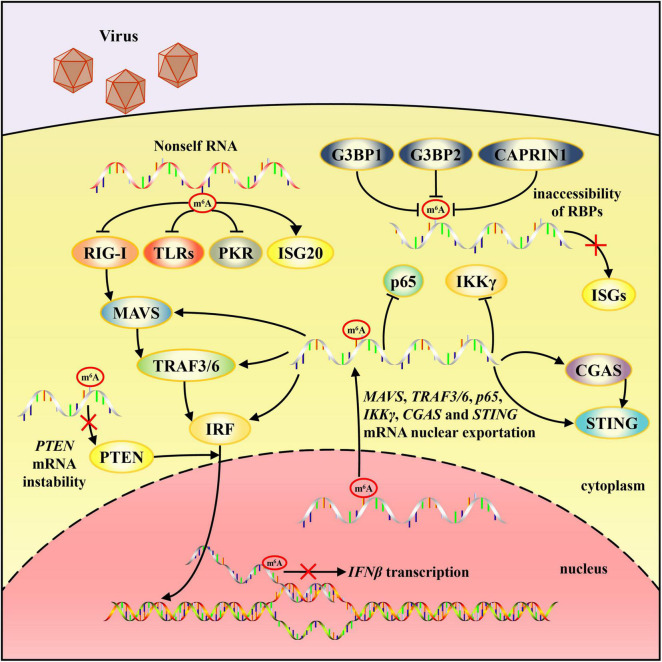
An intrinsic feature of PRR is the ability to discriminate between exogenous and host RNA, which is essential for clearance of viruses while ensuring dormancy of autoimmune responses. It is proved that RNA possessing 5′-triphosphate, double-strand, local folded, or other signatures are all recognized as non-self by PRR (Schlee and Hartmann, 2016). Given that m6A modifications are naturally found in most cellular mRNA, early views believed that, like the DNA restriction-modification system in bacteria, it served as a mark for immune sensors to distinguish self from non-self RNA (Sitaraman, 2016). However, the increasing discovery of m6A in almost all kinds of viruses demonstrates that m6A incorporation into viral RNA may be an approach whereby viruses imitate the host RNA to evade recognition by RLR and TLR, just like 2′OMe, another form of viral RNA modification (Ringeard et al., 2019).
Retinoic Acid-Induced Gene I
In the presence of K63-linked polyubiquitin, RIG-I can be activated by binding with exogenous RNA and then undergo conformational change and recruit MAVS to activate the IFN transcription factors (Malik and Zhou, 2020). However, previous research illustrates that in vitro synthesized RNA containing m6A modifications binds RIG-I poorly and could not trigger RIG-I conformational conversion or induce innate immunity (Durbin et al., 2016). Similar phenomena also occurred on circular RNA (circRNA) or short interfering RNA (siRNA), and YTHDF2 binding to the m6A modified RNA may account for the decreased immunogenicity (Chen et al., 2019; Imaeda et al., 2019). Until recently, the role of m6A modification in virus immune evasion has been deciphered. According to the result of a human metapneumovirus (HMPV) infection model, m6A-ablated HMPV was more likely to be trapped by RIG-I but not melanoma differentiation-associated gene-5 (MDA5) and facilitated RIG-I conformational change and oligomerization. The authors conclude that the m6A modification inhibits type I IFN production through protecting the viral RNA from being recognized by RIG-I both in vitro and in vivo (Lu et al., 2020). Since then, several studies have been published that show m6A modifications on different viruses, such as HBV, HCV, HIV-1, MeV, SeV, vesicular stomatitis virus (VSV), and severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), have the same effect on the process of RIG-I recognition (Kim et al., 2020b; Chen et al., 2021; Li et al., 2021; Lu et al., 2021; Qiu et al., 2021). These studies also detail that m6A-modified viral RNA recruited YTHDF2 and YTHDF3, and these reader proteins sequestered the viral RNA from RIG-I sensing (Kim et al., 2020b; Lu et al., 2021). m6A modification could reduce the local double-stranded structure of viral RNA, which is the critical signature to be recognized by RIG-I (Qiu et al., 2021).
Toll-Like Receptors and Other RNA Sensors
There are some similar findings in other RNA sensors, such as TLR, protein kinase R (PKR), and IFN-stimulated gene 20 (ISG20). One of the studies finds that substitution of A with m6A blocke the activity of RNA to activate dendritic cells (DC) in vitro through signaling of TLR3, TLR7, and TLR8 (Kariko et al., 2005). PKR can specifically detect highly structured viral RNA to restrain virus multiplication and is found to be activated by the less modified noncoding RNA in NIPBL mutated lymphoblastoid cells (Yuen et al., 2016; Bou-Nader et al., 2019). Therefore, it is very likely that m6A modification is involved in viral RNA sensing by PKR molecules. However, a completely contrary role of m6A modification was unveiled in HBV RNA. Using YTHDF2 as an intermediate, m6A-modified HBV RNA can be selectively recognized and degraded by ISG20 through its 3′-5′ exonuclease activity (Imam et al., 2020). Together, it still needs more systematic and in-depth research to elucidate the versatile roles and mechanisms of viral RNA m6A modification in innate immune recognition.
Nucleoside-modified mRNA vaccines, which not only express viral antigens stably, but also avoid being recognized and degraded by the host immune system due to the depressed immunogenicity, provide new ways for the prevention of infectious diseases. Indeed, other types of nucleoside-modified mRNA vaccines have been successfully developed against certain viruses, such as IAV, ZIKV, HIV, and SARS-CoV-2 (Pardi et al., 2017, 2018; Richner et al., 2017; Cohen, 2020), and the recently approved mRNA vaccines for emergency use authorization by FDA developed by Pfizer and Moderna are demonstrated to be very potent in stimulating strong humoral and cellular immune responses (Anderson et al., 2020; Dooling et al., 2020). Based on the findings mentioned above, it is hopeful to design mRNA vaccines by incorporating m6A modifications into virus mRNA.
m6A Modification in Innate Immune Response
The innate immune system is the host’s inherent first line of defense against viruses. Studies reveal that many aspects of the innate immune response, such as expression of IFN and ISG, inflammatory response, macrophage and DC maturation are all tightly controlled by m6A modification as a consequence to either improve the antiviral effects efficiently or weaken the immune response to prevent immunopathological damage.
Interferon Response
IFNs are a class of principal cytokines that can restrict virus amplification and spread. Binding to cell membrane receptors, IFNs activate the Janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathway, leading to the transcription of a whole repertoire of antiviral ISG. To avoid the deleterious outcomes induced by excessive IFN response, strategies that the host evolves to fine-tune IFN production are equally important. m6A modification is linked to negative regulation of IFN-β production in normal human dermal fibroblasts triggered by human cytomegalovirus (HCMV) or dsDNA (Rubio et al., 2018). Herein, the slower biogenesis and faster decay of IFNB mRNA were involved in the underlying mechanism. Alternatively, m6A might deposit onto nascent IFNB mRNA co-transcriptionally, and the initiation or elongation of transcription might be obstructed by the m6A group. This finding is verified and extended in other similar research in which HCMV infection of primary human foreskin fibroblasts and murine CMV (MCMV) infection of mice were exploited (Winkler et al., 2019). The negative regulatory role of m6A on IFNB production was directly identified by comparing the expression level of putative m6A site-mutated with wild-type IFNB constructs. Despite the discovery that the stability of IFNB mRNA was increased when the m6A sites were mutated, the role of m6A on transcription in the earlier period was not considered in this study.
Earlier studies observe that several ISG transcripts translate effectively in the presence of RNA-binding proteins, including G3BP stress granule assembly factor 1 (G3BP1), G3BP2, and cytoplasmic activation/proliferation-associated protein-1 (CAPRIN1) (Bidet et al., 2014; Li et al., 2015). Furthermore, the interacting sites in mRNA and determinants that affect the binding of these three stress granule proteins were explored with the development of proteomics. Two proteomic studies tried to explain how m6A modification impacted on mRNA-protein interactions in which m6A modification repelled binding of G3BP1, G3BP2, or CAPRIN1 to the mRNA, and these three proteins were, therefore, proposed to be m6A antireaders (Arguello et al., 2017; Edupuganti et al., 2017). Hence, it can be predicted that the expression of certain ISGs could be negatively modulated by m6A modification as well. Given that host m6A-associated machinery are induced almost immediately when viral infection takes place, m6A modification may act as a suppressive signal to downregulate the magnitude of IFN response and restrict cytotoxicity; on the other hand, this mechanism can be hijacked by viruses to facilitate their replication.
Contradictorily, some studies report that the enhancement of IFN response is also attributed to m6A modification. It was indicated that, after herpes simplex virus-1 (HSV-1) infection, m6A modification of cyclic GMP-AMP synthase (CGAS), gamma-interferon-inducible protein 16 (IFI16), and stimulator of interferon gene (STING) mRNA in RAW264.7 cells led to their cytoplasm localization and expression of these transcripts, suggesting that m6A modification was crucial to drive type I IFN production (Wang L. et al., 2019). m6A modification was also found to expedite IFN production in another study by Cao and colleagues (Zheng et al., 2017). DEAD-box (DDX) helicase family member DDX46 recruited ALKBH5 via its DEAD helicase domain to demethylate m6A modified MAVS, TNF receptor-associated factor 3 (Traf3), and Traf6 mRNA in RAW264.7 cells infected with VSV. The resultant demethylation of these three mRNAs reduced their nuclear exportation and translation into proteins responsible for IFN production, which demonstrated a positive role of m6A modification in IFN response. Contrarily, another DDX helicase family member DDX5, could enhance the formation of the METTL3–METTL14 complex, which methylates p65 and IKKγ mRNA in the nuclear. The increased methylation of these transcripts results in accelerated degradation and negatively regulates IFN-β and IL-6 production after VSV infection (Xu et al., 2021). A recent study found that WTAP maintains the protein abundance of IRF3 and IFNAR1 by improving IRF3 translation efficiency and IFNAR1 mRNA stability via m6A modification (Ge et al., 2021). Coincidentally, another study revealed that m6A modification promotes the translation of certain ISGs during the IFN response, thus augmenting the antiviral innate immunity functions (McFadden et al., 2021).
Phosphatase and tensin homolog (PTEN), as an innate immune regulator, promotes dephosphorylation of interferon regulatory factor 3 (IRF3) at the Ser97 site with a corresponding facilitation of IRF3 nuclear import and IFN production. HBV could increase m6A modification of PTEN mRNA and contribute to its instability in host cells by which HBV evaded the attack from the immune system (Kim et al., 2020a). The forkhead box protein O3 (FOXO3) is a repressive transcription factor that diminishes IFN-γ production and antiviral activity. In RAW264.7 cells infected with VSV, the m6A reader protein YTHDF3 potentiated FOXO3 translation, and the latter downregulated ISG expression (Zhang et al., 2019). It is interesting that YTHDF3 bound to the initiation region of FOXO3 mRNA independently of METTL3-installed m6A. However, the authors did not analyze the m6A sites on FOXO3 mRNA or the influences of synonymous point mutation. It is still unclear whether m6A modification is really involved in the binding of YTHDF3 to FOXO3 mRNA. Taken together, it is clear that the biological significance of m6A modification for the IFN response is complex and remains to be further investigated.
Macrophage Polarization and Dendritic Cells Activation
Classical or M1 macrophages are characterized by ingestion and digestion of cells infected with viruses and proinflammatory activity. The polarization of M1 macrophages rely on transcription factors, including STAT1 and IFN regulatory factor 5 (IRF5) although STAT6 and peroxisome proliferation-activated receptor-γ (PPAR-γ) are required for differentiation of the alternatively activated M2 macrophages that orchestrate immunoregulation, fibrous tissue repair, and restrain the duration of inflammatory response (Alisjahbana et al., 2020). It seems to be contradictory about the role of m6A modification in macrophage polarization in the following two studies. Through methylated RNA immunoprecipitation, STAT1 mRNA was identified to be m6A modified at its 3′-untranslated region (UTR) in murine bone marrow-derived macrophages (BMDMs) (Liu et al., 2019c), and the m6A methylation markedly inhibited STAT1 mRNA decay and gave rise to a constant protein translation, underlying M1 BMDMs phenotypic maturation. However, another study found that m6A modification resulted in decreased mRNA stability of STAT1 and PPAR-γ via YTHDF2, thereby impeding both M1 and M2 macrophage polarization (Gu et al., 2020). Further analysis of the role of reader proteins that bind to these m6A sites would resolve this contradiction.
During activation in response to viral infection, DCs express high levels of membrane costimulatory molecules, such as CD40, CD80, CD86, and Toll/IL-1 receptor homologous region domain-containing adaptor protein (Tirap) for initiating the adaptive immune response efficiently. Research focusing on regulation of DC maturation indicates that m6A upregulates the expression of CD40, CD80, and Tirap to prime T lymphocytes (Wang H. et al., 2019). The m6A modifications in these three mRNA were recognized by YTHDF1, and subsequently, the translation was strengthened.
Inflammatory Cytokines Production
Inflammatory responses, which are featured by local recruitment of considerable leukocytes and cytokines, are destined for suppression of infection processes. Uncontrolled inflammatory response intensity and duration, such as cytokine storm, may lead to severe immunopathological damage to the host (Cao, 2020). TLR-mediated nuclear factor kappa B (NF-κB), mitogen-activated protein kinase (MAPK), and other signaling pathways are the targets of epigenetic regulation of the inflammatory response (Yasmin et al., 2015). For instance, METTL3 facilitates activation of NF-κB and MAPK pathways in human dental pulp cells and chondrocytes (Feng et al., 2018; Liu et al., 2019b), and a completely opposite biological activity of METTL3 is found in THP-1 macrophages, in which overexpression of METTL3 significantly restrained NF-κB phosphorylation and nuclear translocation (Wang J. et al., 2019). YTHDF2 is suggested to participate in the destabilization of MAPK mRNA of RAW264.7 macrophages or IL11 mRNA of hepatocellular carcinoma cells, thus reducing IL-1β, IL-6, IL-12, and TNF-α production and relieving inflammation dramatically (Hou et al., 2019; Yu et al., 2019).
In these studies, it is not compelling to draw conclusions about the regulatory role of m6A modification only by evaluating the effects of perturbing METTL3 or YTHDF2 on the expression of inflammation-related genes, and additional mapping of the m6A distribution in the transcripts of these genes is required to clarify how gene expression or the RNA process is impacted by the m6A modifications more convincingly. Together, these divergent findings indicate the complicated regulation role of m6A modification in the inflammatory response, depending on the diverse cell lines or cellular components, and a comprehensive understanding about how inflammatory response against viruses are controlled by m6A remains to be further studied.
Other Innate Immune-Related Molecules
Right open reading frame kinase 3 (RIOK3) is a protein serine/threonine kinase that can phosphorylate MDA5 and maintain MDA5 at an inactive state (Oshiumi et al., 2016). Cold-inducible RNA binding protein (CIRBP) is induced under cellular stresses and can stabilize specific mRNA and facilitate their translation (Liao et al., 2017). In the context of infection by Flaviviridae, RIOK3 methylation, and CIRBP demethylation took place, and the changed m6A status promoted translation of RIOK3 and alternative splicing of CIRBP, respectively, all benefiting Flaviviridae infection consequently (Gokhale et al., 2020).
m6A Modification in Adaptive Immune Response
Except the regulatory role for innate immunity, m6A modification was also discovered to be correlated with adaptive immune responses, for example, T lymphocyte proliferation and differentiation, DC migration to lymph nodes, and antigen presentation.
T Lymphocyte Proliferation and Differentiation
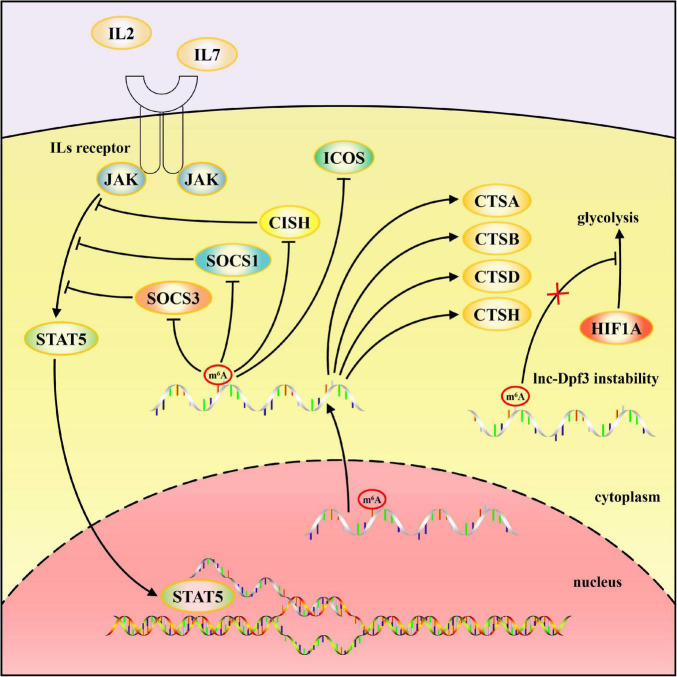
In the process of naive T cell differentiation into Th1 and Th17 cells, IL-7/STAT5 pathway activation is pivotal. Using conditional METTL3 knockout mice, it was observed that m6A deposition in the suppressor of cytokine signaling (SOCS) family SOCS1, SOCS3, and CISH mRNA accelerated their decay (Li et al., 2017). Consequently, the suppression to the IL-7/STAT5 signal pathway was removed, and this led to reprogramming of naive T lymphocytes. Subsequent research by Li and colleagues found that the immunosuppression function of Treg arose from IL-2/STAT5 pathway activation, and m6A indirectly modulated this pathway through SOCS as in Th cell differentiation (Tong et al., 2018). Another subset of T cells, follicular helper T cells (Tfh) are essential for initiating germinal center formation and activating B lymphocytes. An inducible co-stimulator (ICOS) is a signaling molecule that is indispensable for Tfh cell development. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was identified as a key target downstream of the E3 ligase VHL-hypoxia-inducible factor 1α (HIF-1α) signaling pathway to regulate the development of Tfh. It is reported that GAPDH could promote m6A modification on ICOS mRNA to reduce protein expression, thereby inhibiting the early development of Tfh (Zhu et al., 2019).
Migration and Antigen Presentation of Dendritic Cells
CCR7 chemokine receptor stimulation promotes the movement of DCs to draining lymph nodes rapidly for antigen presenting to T cells and the priming of adaptive immune responses (Bretou et al., 2017). Excessive DC migration and accumulation are related to variable inflammatory disorders; therefore, timely termination of DC migration is the key to orchestrating immune homeostasis. A long noncoding RNA lnc-Dpf3 was identified as a feedback regulator for CCR7-induced DC migration (Liu et al., 2019a). CCR7 stimulation upregulated the level of lnc-Dpf3 concurrently via m6A demethylation to prevent YTHDF2-mediated degradation, and lnc-Dpf3 directly bound to HIF-1α and abrogated transcription of the lactate dehydrogenase A (Ldha). As a result, lnc-Dpf3 inhibited glycolytic metabolism and migratory capacity of DC.
As the most powerful APC, the antigen processing and presenting of DCs can also be fine-tuned by m6A modification. To be specific, mRNA of lysosomal cathepsins, including CTSA, CTSB, CTSD, and CTSH, are m6A modified in DCs, and the expression of these proteases was reinforced by YTHDF1 (Han et al., 2019). More degradation of tumor neoantigens by these lysosomal proteases resulted in less antigen presentation, leading to the escape of tumor cells from immune surveillance. Whether the virus utilizes this immune “ignorance” caused by YTHDF1 to avoid recognition by the immune system deserves further verification. This implicates YTHDF1 as a potential therapeutic target in anticancer or antiviral immunotherapy.
m6A Modification and Antiviral-Related Components
Metabolite of Host Cells
Host cell metabolism, which encompasses metabolite availability and energy generation, can be used to shape the course of immune events and to affect the environment of viral survival. The m6A level on α-ketoglutarate dehydrogenase (OGDH) mRNA was initially increased due to the impaired enzymatic activity of ALKBH5 in RAW264.7 cells after VSV infection (Liu et al., 2019d). The m6A modification promotes OGDH transcript degradation through YTHDF2, and decreased OGDH protein expression metabolically suppresses production of itaconate, which is exploited for virus replication. This study shows the importance of m6A modification in the metabolomic response to viral infection.
Immunome of Host Cells
Recent years have witnessed technological breakthroughs, such as methylated RNA immunoprecipitation sequencing (MeRIP-seq) and m6A individual nucleotide resolution crosslinking and immunoprecipitation (miCLIP), which made it possible to profile the m6A landscape at the transcriptome level (Grozhik et al., 2017; Ovcharenko and Rentmeister, 2018).
By MeRIP-seq, 56 transcripts were identified as constitutively m6A modified in MT4 cells upon HIV-1 infection, and the most represented categories were viral gene expression and multiorganism metabolic process (Lichinchi et al., 2016a). In fact, 19 of these genes were known to be linked to HIV replication, such as EIF3M, TRAF2, and HNRNPK. However, in Jurkat and primary CD4+ T cells, the uniquely m6A modified genes upon HIV-1 infection enriched in functional clusters, such as metabolism, immune system process, multicellular organismal process, and development (Tirumuru et al., 2016). Researchers subsequently found that the binding of HIV-1 envelope glycoprotein gp120 to the CD4 receptor molecule is required for the upregulation of the m6A modification level in recipient cells (Tirumuru and Wu, 2019). There are some similar studies focusing on host m6A methylome changes upon ZIKV, respiratory syncytial virus (RSV), Kaposi’s sarcoma-associated herpesvirus (KSHV), and Flaviviridae infections (Lichinchi et al., 2016b; Hesser et al., 2018; Tan et al., 2018; Fu et al., 2019; Xue et al., 2019).
According to the results of these studies, it is certain that viral infection can rewrite the host cell methylome, and these newly gained or lost modifications often simultaneously occur at sets of genes that are enriched in confined pathways related to viral infection even if various statistical models for m6A peak calling or GO analysis algorithms were applied. Considering that genes whose expression is highly regulated often contain abundant m6A sites in their mRNA (Gokhale et al., 2020), these modular alterations might be an effective means for modulating immune related gene expression programs to promote or restrict viral infection. It is necessary to carry out deeper studies for verifying whether and how these genes or signaling pathways are regulated by m6A modification.
Who Leads the Alteration of m6A Modification?
It merits expanding research to determine how virus–host interactions drive the changed methylome or, in other words, the changed m6A machinery in the infected cells. Recently, several enlightening studies shed some light on the mechanisms. It is demonstrated that Epstein–Barr virus nuclear antigen 3C (EBNA3C) upregulates METTL14 expression depending on activation of the METTL14 promoter and stabilizes METTL14 protein (Lang et al., 2019). As a result, the increased METTL14 level facilitates EBV proliferation and self-renewal of host cells. Investigations show that the interaction between METTL3 and enterovirus 71 (EV71) nonstructural protein 2C or 3D may contribute to the cytoplasm localization of METTL3 in rhabdomyosarcoma (RD) cells (Hao et al., 2019; Yao et al., 2020). In addition, the viral protein 2A that harbors a nuclear localization signal could compete with METTL3 for nuclear importing protein karyopherin, and this partially explains the redistribution of METTL3 after EV71 infection. Siddiqui and colleagues found that HBx, an HBV-encoded regulatory protein, could interact with m6A methyltransferases and guide them to the HBV minichromosome and host PTEN chromosomal locus to achieve cotranscriptional m6A modification (Kim and Siddiqui, 2021). In HepG2 cells, Flaviviridae infection-activated innate immune and endoplasmic reticulum stress controlled the alteration of RIOK3 and CIRBP m6A conditions, respectively (Gokhale et al., 2020). The protease encoded by HIV-1 could cleave m6A reader protein YTHDF3, which incorporates into HIV-1 viral particles, antagonizing the limitation role of YTHDF3 on viral production and infectivity (Jurczyszak et al., 2020). Similarly, the 2A protease of enterovirus antagonizes the induction of ISGs in infected cells by cleaving m6A readers YTHDF1-3 (Kastan et al., 2021). These studies indeed illustrate the complex link between the viruses and m6A modification machineries.
Conclusion
The human immune system reacts to viral infection effectively by innate and adaptive immune responses. Many studies demonstrate that m6A modification regulates multiple steps of the antiviral immune response and plays an important role in the viral infection process (Figures 1, 2). In this review, we present up-to-date knowledge about m6A modification in regulating viral nucleic acid recognition, IFN production, and DC and macrophage maturation, among others. The involvement of m6A modification in antigen presentation, effector lymphocyte differentiation and other processes of adaptive immune response are also emphasized. These studies provide a basis in understanding the key role of m6A or other RNA modifications in infection and immunity in addition to providing new strategies for anti-infection immunotherapy development.
FIGURE 1.
Schematic diagram of mechanisms by which m6A modification regulates non-self RNA recognition and innate immune responses. m6A modification in exogenous RNA prevent it from being identified by RNA sensors except for ISG20. Nuclear exportation of MAVS, TRAF3/6, p65, IKKγ, CGAS, and STING mRNA can be accelerated by m6A modification. m6A decoration may be an obstacle to IFNβ and ISGs expression. PTEN, p65 and IKKγ mRNA instability can also be attributed to m6A modification.
FIGURE 2.
Schematic diagram of mechanisms by which m6A modification regulates adaptive immune responses. m6A modification of SOCS1, SOCS3, CISH mRNA, and lnc-Dpf3 accelerate their decay. Translation of lysosomal cathepsins, including CTSA, CTSB, CTSD, and CTSH, are promoted by m6A and YTHDF1, whereas the expression of ICOS is inhibited by m6A modification.
Author Contributions
XC: conceptualization and supervision. YZ, HQ, and ZG: data curation. BZ and WW: writing—original draft. BZ, WW, and XC: writing—review and editing. BZ, WW, YZ, HQ, and ZG: visualization. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Funding
This work was supported by S&T Program of Hebei (20277704D and 20372601D), the Natural Science Foundation of Hebei Province, China (H2020206352), the National Natural Science Foundation of China (81902026), and the Science and Technology Project of Hebei Education Department (QN2018150).
References
- Alisjahbana A., Mohammad I., Gao Y., Evren E., Ringqvist E., Willinger T. (2020). Human macrophages and innate lymphoid cells: tissue-resident innate immunity in humanized mice. Biochem. Pharmacol. 174:113672. 10.1016/j.bcp.2019.113672 [DOI] [PubMed] [Google Scholar]
- Anderson E. J., Rouphael N. G., Widge A. T., Jackson L. A., Roberts P. C., Makhene M., et al. (2020). Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. 383 2427–2438. 10.1056/NEJMoa2028436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello A. E., DeLiberto A. N., Kleiner R. E. (2017). RNA chemical proteomics reveals the N(6)-Methyladenosine (m(6)A)-regulated protein-RNA interactome. J. Am. Chem. Soc. 139 17249–17252. 10.1021/jacs.7b09213 [DOI] [PubMed] [Google Scholar]
- Bi Z., Liu Y., Zhao Y., Yao Y., Wu R., Liu Q., et al. (2019). A dynamic reversible RNA N(6) -methyladenosine modification: current status and perspectives. J. Cell Physiol. 234 7948–7956. 10.1002/jcp.28014 [DOI] [PubMed] [Google Scholar]
- Bidet K., Dadlani D., Garcia-Blanco M. A. (2014). G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog. 10:e1004242. 10.1371/journal.ppat.1004242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P., Machnicka M. A., Purta E., Piatkowski P., Baginski B., Wirecki T. K., et al. (2018). MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46 D303–D307. 10.1093/nar/gkx1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou-Nader C., Gordon J. M., Henderson F. E., Zhang J. (2019). The search for a PKR code-differential regulation of protein kinase R activity by diverse RNA and protein regulators. RNA 25 539–556. 10.1261/rna.070169.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretou M., Saez P. J., Sanseau D., Maurin M., Lankar D., Chabaud M., et al. (2017). Lysosome signaling controls the migration of dendritic cells. Sci. Immunol. 2:eaak9573. 10.1126/sciimmunol.aak9573 [DOI] [PubMed] [Google Scholar]
- Cao X. (2020). COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 20 269–270. 10.1038/s41577-020-0308-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Xia P., Li S., Zhang T., Wang T. T., Zhu J. (2017). RNA sensors of the innate immune system and their detection of pathogens. IUBMB Life 69 297–304. 10.1002/iub.1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Kumar S., Espada C. E., Tirumuru N., Cahill M. P., Hu L., et al. (2021). N6-methyladenosine modification of HIV-1 RNA suppresses type-I interferon induction in differentiated monocytic cells and primary macrophages. PLoS Pathog. 17:e1009421. 10.1371/journal.ppat.1009421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. G., Chen R., Ahmad S., Verma R., Kasturi S. P., Amaya L., et al. (2019). N6-Methyladenosine modification controls circular RNA immunity. Mol. Cell 76 96–109e109. 10.1016/j.molcel.2019.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. (2020). Vaccine designers take first shots at COVID-19. Science 368 14–16. 10.1126/science.368.6486.14 [DOI] [PubMed] [Google Scholar]
- Courtney D. G., Kennedy E. M., Dumm R. E., Bogerd H. P., Tsai K., Heaton N. S., et al. (2017). Epitranscriptomic enhancement of influenza a virus gene expression and replication. Cell Host Microbe 22 377–386e375. 10.1016/j.chom.2017.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooling K., McClung N., Chamberland M., Marin M., Wallace M., Bell B. P., et al. (2020). The advisory committee on immunization practices’ interim recommendation for allocating initial supplies of COVID-19 vaccine - United States, 2020. MMWR Morb. Mortal. Wkly. Rep. 69 1857–1859. 10.15585/mmwr.mm6949e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin A. F., Wang C., Marcotrigiano J., Gehrke L. (2016). RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. mBio 7 e833–e816. 10.1128/mBio.00833-16 * e00833-16, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edupuganti R. R., Geiger S., Lindeboom R. G. H., Shi H., Hsu P. J., Lu Z., et al. (2017). N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24 870–878. 10.1038/nsmb.3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Li Q., Meng R., Yi B., Xu Q. (2018). METTL3 regulates alternative splicing of MyD88 upon the lipopolysaccharide-induced inflammatory response in human dental pulp cells. J. Cell Mol. Med. 22 2558–2568. 10.1111/jcmm.13491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Dominissini D., Rechavi G., He C. (2014). Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 15 293–306. 10.1038/nrg3724 [DOI] [PubMed] [Google Scholar]
- Fu Y., Zorman B., Sumazin P., Sanna P. P., Repunte-Canonigo V. (2019). Epitranscriptomics: correlation of N6-methyladenosine RNA methylation and pathway dysregulation in the hippocampus of HIV transgenic rats. PLoS One 14:e0203566. 10.1371/journal.pone.0203566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Ling T., Wang Y., Jia X., Xie X., Chen R., et al. (2021). Degradation of WTAP blocks antiviral responses by reducing the m(6) a levels of IRF3 and IFNAR1 mRNA. EMBO Rep. 22:e52101. 10.15252/embr.202052101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale N. S., McIntyre A. B. R., Mattocks M. D., Holley C. L., Lazear H. M., Mason C. E., et al. (2020). Altered m(6)a modification of specific cellular transcripts affects flaviviridae infection. Mol Cell 77 542–555e548. 10.1016/j.molcel.2019.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozhik A. V., Linder B., Olarerin-George A. O., Jaffrey S. R. (2017). Mapping m(6)A at individual-nucleotide resolution using crosslinking and immunoprecipitation (miCLIP). Methods Mol. Biol. 1562 55–78. 10.1007/978-1-4939-6807-7_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Zhang Y., Li D., Cai H., Cai L., Xu Q. (2020). N6-methyladenosine demethylase FTO promotes M1 and M2 macrophage activation. Cell Signal 69:109553. 10.1016/j.cellsig.2020.109553 [DOI] [PubMed] [Google Scholar]
- Han D., Liu J., Chen C., Dong L., Liu Y., Chang R., et al. (2019). Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature 566 270–274. 10.1038/s41586-019-0916-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H., Hao S., Chen H., Chen Z., Zhang Y., Wang J., et al. (2019). N6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Res. 47 362–374. 10.1093/nar/gky1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesser C. R., Karijolich J., Dominissini D., He C., Glaunsinger B. A. (2018). N6-methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi’s sarcoma-associated herpesvirus infection. PLoS Pathog. 14:e1006995. 10.1371/journal.ppat.1006995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Zhang H., Liu J., Zhao Z., Wang J., Lu Z., et al. (2019). YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol. Cancer 18:163. 10.1186/s12943-019-1082-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaeda A., Tomoike F., Hayakawa M., Nakamoto K., Kimura Y., Abe N., et al. (2019). N(6)-methyl adenosine in siRNA evades immune response without reducing RNAi activity. Nucleosides Nucleotides Nucleic Acids 38 972–979. 10.1080/15257770.2019.1641205 [DOI] [PubMed] [Google Scholar]
- Imam H., Khan M., Gokhale N. S., McIntyre A. B. R., Kim G. W., Jang J. Y., et al. (2018). N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc. Natl. Acad. Sci. U.S.A. 115 8829–8834. 10.1073/pnas.1808319115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam H., Kim G. W., Mir S. A., Khan M., Siddiqui A. (2020). Interferon-stimulated gene 20 (ISG20) selectively degrades N6-methyladenosine modified Hepatitis B Virus transcripts. PLoS Pathog. 16:e1008338. 10.1371/journal.ppat.1008338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurczyszak D., Zhang W., Terry S. N., Kehrer T., Bermudez Gonzalez M. C., McGregor E., et al. (2020). HIV protease cleaves the antiviral m6A reader protein YTHDF3 in the viral particle. PLoS Pathog. 16:e1008305. 10.1371/journal.ppat.1008305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariko K., Buckstein M., Ni H., Weissman D. (2005). Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23 165–175. 10.1016/j.immuni.2005.06.008 [DOI] [PubMed] [Google Scholar]
- Kastan J. P., Tremblay M. W., Brown M. C., Trimarco J. D., Dobrikova E. Y., Dobrikov M. I., et al. (2021). Enterovirus 2A(pro) cleavage of the YTHDF m(6)a readers implicates YTHDF3 as a mediator of Type I interferon-driven JAK/STAT signaling. mBio 12 e116–e121. 10.1128/mBio.00116-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy E. M., Bogerd H. P., Kornepati A. V., Kang D., Ghoshal D., Marshall J. B., et al. (2016). Posttranscriptional m(6)a editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe 19 675–685. 10.1016/j.chom.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy E. M., Courtney D. G., Tsai K., Cullen B. R. (2017). Viral epitranscriptomics. J. Virol. 91 e2263–e2216. 10.1128/JVI.02263-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G. W., Imam H., Khan M., Mir S. A., Kim S. J., Yoon S. K., et al. (2020a). HBV-induced increased N6 methyladenosine modification of PTEN RNA affects innate immunity and contributes to HCC. Hepatology 73 533–547. 10.1002/hep.31313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G. W., Imam H., Khan M., Siddiqui A. (2020b). N (6)-Methyladenosine modification of hepatitis B and C viral RNAs attenuates host innate immunity via RIG-I signaling. J. Biol. Chem. 295 13123–13133. 10.1074/jbc.RA120.014260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G. W., Siddiqui A. (2021). Hepatitis B virus X protein recruits methyltransferases to affect cotranscriptional N6-methyladenosine modification of viral/host RNAs. Proc. Natl. Acad. Sci. U.S.A. 118 e2019455118. 10.1073/pnas.2019455118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang F., Singh R. K., Pei Y., Zhang S., Sun K., Robertson E. S. (2019). EBV epitranscriptome reprogramming by METTL14 is critical for viral-associated tumorigenesis. PLoS Pathog. 15:e1007796. 10.1371/journal.ppat.1007796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. B., Tong J., Zhu S., Batista P. J., Duffy E. E., Zhao J., et al. (2017). m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 548 338–342. 10.1038/nature23450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. M., MacDonald M. R., Rice C. M. (2015). To translate, or not to translate: viral and host mRNA regulation by interferon-stimulated genes. Trends Cell. Biol. 25 320–329. 10.1016/j.tcb.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Hui H., Bray B., Gonzalez G. M., Zeller M., Anderson K. G., et al. (2021). METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS-CoV-2 infection. Cell Rep. 35:109091. 10.1016/j.celrep.2021.109091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Tong L., Tang L., Wu S. (2017). The role of cold-inducible RNA binding protein in cell stress response. Int. J. Cancer 141 2164–2173. 10.1002/ijc.30833 [DOI] [PubMed] [Google Scholar]
- Lichinchi G., Gao S., Saletore Y., Gonzalez G. M., Bansal V., Wang Y., et al. (2016a). Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 1:16011. 10.1038/nmicrobiol.2016.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi G., Zhao B. S., Wu Y., Lu Z., Qin Y., He C., et al. (2016b). Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe 20 666–673. 10.1016/j.chom.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Zhou K. I., Parisien M., Dai Q., Diatchenko L., Pan T. (2017). N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45 6051–6063. 10.1093/nar/gkx141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zhang X., Chen K., Cheng Y., Liu S., Xia M., et al. (2019a). CCR7 chemokine receptor-inducible lnc-Dpf3 restrains dendritic cell migration by inhibiting HIF-1alpha-mediated glycolysis. Immunity 50 600–615 e615. 10.1016/j.immuni.2019.01.021 [DOI] [PubMed] [Google Scholar]
- Liu Q., Li M., Jiang L., Jiang R., Fu B. (2019b). METTL3 promotes experimental osteoarthritis development by regulating inflammatory response and apoptosis in chondrocyte. Biochem. Biophys. Res. Commun. 516 22–27. 10.1016/j.bbrc.2019.05.168 [DOI] [PubMed] [Google Scholar]
- Liu Y., Liu Z., Tang H., Shen Y., Gong Z., Xie N., et al. (2019c). The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 facilitates M1 macrophage polarization through the methylation of STAT1 mRNA. Am. J. Physiol. Cell Physiol. 317 C762–C775. 10.1152/ajpcell.00212.2019 [DOI] [PubMed] [Google Scholar]
- Liu Y., You Y., Lu Z., Yang J., Li P., Liu L., et al. (2019d). N (6)-methyladenosine RNA modification-mediated cellular metabolism rewiring inhibits viral replication. Science 365 1171–1176. 10.1126/science.aax4468 [DOI] [PubMed] [Google Scholar]
- Lu M., Xue M., Wang H. T., Kairis E. L., Ahmad S., Wei J., et al. (2021). Nonsegmented negative-sense RNA viruses utilize N (6)-Methyladenosine (m(6)A) as a common strategy to evade host innate immunity. J. Virol. 95 e1939–e1920. 10.1128/JVI.01939-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Zhang Z., Xue M., Zhao B. S., Harder O., Li A., et al. (2020). N(6)-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat. Microbiol. 5 584–598. 10.1038/s41564-019-0653-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik G., Zhou Y. (2020). Innate immune sensing of influenza a virus. Viruses 12:755. 10.3390/v12070755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden M. J., Gokhale N. S., Horner S. M. (2017). Protect this house: cytosolic sensing of viruses. Curr. Opin. Virol. 22 36–43. 10.1016/j.coviro.2016.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden M. J., McIntyre A. B. R., Mourelatos H., Abell N. S., Gokhale N. S., Ipas H., et al. (2021). Post-transcriptional regulation of antiviral gene expression by N6-methyladenosine. Cell Rep. 34:108798. 10.1016/j.celrep.2021.108798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtergaele S., He C. (2018). Chemical modifications in the life of an mRNA transcript. Annu. Rev. Genet. 52 349–372. 10.1146/annurev-genet-120417-031522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi H., Kouwaki T., Seya T. (2016). Accessory factors of cytoplasmic viral RNA sensors required for antiviral innate immune response. Front. Immunol. 7:200. 10.3389/fimmu.2016.00200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko A., Rentmeister A. (2018). Emerging approaches for detection of methylation sites in RNA. Open Biol. 8:180121. 10.1098/rsob.180121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Parkhouse K., Kirkpatrick E., McMahon M., Zost S. J., Mui B. L., et al. (2018). Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 9:3361. 10.1038/s41467-018-05482-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Secreto A. J., Shan X., Debonera F., Glover J., Yi Y., et al. (2017). Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat. Commun. 8:14630. 10.1038/ncomms14630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W., Zhang Q., Zhang R., Lu Y., Wang X., Tian H., et al. (2021). N(6)-methyladenosine RNA modification suppresses antiviral innate sensing pathways via reshaping double-stranded RNA. Nat. Commun. 12:1582. 10.1038/s41467-021-21904-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakebrandt N., Littringer K., Joller N. (2016). Regulatory T cells: balancing protection versus pathology. Swiss Med. Wkly. 146:w14343. 10.4414/smw.2016.14343 [DOI] [PubMed] [Google Scholar]
- Richner J. M., Himansu S., Dowd K. A., Butler S. L., Salazar V., Fox J. M., et al. (2017). Modified mRNA vaccines protect against zika virus infection. Cell 168 1114–1125 e1110. 10.1016/j.cell.2017.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringeard M., Marchand V., Decroly E., Motorin Y., Bennasser Y. (2019). FTSJ3 is an RNA 2’-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature 565 500–504. 10.1038/s41586-018-0841-4 [DOI] [PubMed] [Google Scholar]
- Rubio R. M., Depledge D. P., Bianco C., Thompson L., Mohr I. (2018). RNA m(6) a modification enzymes shape innate responses to DNA by regulating interferon beta. Genes Dev. 32 1472–1484. 10.1101/gad.319475.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M., Hartmann G. (2016). Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 16 566–580. 10.1038/nri.2016.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Wei J., He C. (2019). Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74 640–650. 10.1016/j.molcel.2019.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaraman R. (2016). The role of DNA restriction-modification systems in the biology of Bacillus anthracis. Front. Microbiol. 7:11. 10.3389/fmicb.2016.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B., Liu H., Zhang S., da Silva S. R., Zhang L., Meng J., et al. (2018). Viral and cellular N(6)-methyladenosine and N(6),2’-O-dimethyladenosine epitranscriptomes in the KSHV life cycle. Nat. Microbiol. 3 108–120. 10.1038/s41564-017-0056-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirumuru N., Wu L. (2019). HIV-1 envelope proteins up-regulate N (6)-methyladenosine levels of cellular RNA independently of viral replication. J. Biol. Chem. 294 3249–3260. 10.1074/jbc.RA118.005608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirumuru N., Zhao B. S., Lu W., Lu Z., He C., Wu L. (2016). N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. Elife 5:e15528. 10.7554/eLife.15528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J., Cao G., Zhang T., Sefik E., Amezcua Vesely M. C., Broughton J. P., et al. (2018). m(6)A mRNA methylation sustains Treg suppressive functions. Cell Res. 28 253–256. 10.1038/cr.2018.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Hu X., Huang M., Liu J., Gu Y., Ma L., et al. (2019). Mettl3-mediated mRNA m(6)A methylation promotes dendritic cell activation. Nat. Commun. 10:1898. 10.1038/s41467-019-09903-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Yan S., Lu H., Wang S., Xu D. (2019). METTL3 attenuates LPS-induced inflammatory response in macrophages via NF-kappaB signaling pathway. Mediators Inflamm. 2019:3120391. 10.1155/2019/3120391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wen M., Cao X. (2019). Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science 365:eaav0758. 10.1126/science.aav0758 [DOI] [PubMed] [Google Scholar]
- Wang X., Zhao B. S., Roundtree I. A., Lu Z., Han D., Ma H., et al. (2015). N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161 1388–1399. 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler R., Gillis E., Lasman L., Safra M., Geula S., Soyris C., et al. (2019). m(6)A modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 20 173–182. 10.1038/s41590-018-0275-z [DOI] [PubMed] [Google Scholar]
- Wu F., Cheng W., Zhao F., Tang M., Diao Y., Xu R. (2019). Association of N6-methyladenosine with viruses and related diseases. Virol. J. 16:133. 10.1186/s12985-019-1236-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Cai Y., Ma Z., Jiang B., Liu W., Cheng J., et al. (2021). The RNA helicase DDX5 promotes viral infection via regulating N6-methyladenosine levels on the DHX58 and NFkappaB transcripts to dampen antiviral innate immunity. PLoS Pathog. 17:e1009530. 10.1371/journal.ppat.1009530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M., Zhao B. S., Zhang Z., Lu M., Harder O., Chen P., et al. (2019). Viral N(6)-methyladenosine upregulates replication and pathogenesis of human respiratory syncytial virus. Nat. Commun. 10:4595. 10.1038/s41467-019-12504-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Wang H., Zhang W. (2019). Regulation of virus replication and T cell homeostasis by N(6)-Methyladenosine. Virol. Sin. 34 22–29. 10.1007/s12250-018-0075-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M., Dong Y., Wang Y., Liu H., Ma H., Zhang H., et al. (2020). N(6)-methyladenosine modifications enhance enterovirus 71 ORF translation through METTL3 cytoplasmic distribution. Biochem. Biophys. Res. Commun. 527 297–304. 10.1016/j.bbrc.2020.04.088 [DOI] [PubMed] [Google Scholar]
- Yasmin R., Siraj S., Hassan A., Khan A. R., Abbasi R., Ahmad N. (2015). Epigenetic regulation of inflammatory cytokines and associated genes in human malignancies. Mediators Inflamm. 2015:201703. 10.1155/2015/201703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R., Li Q., Feng Z., Cai L., Xu Q. (2019). m6A Reader YTHDF2 Regulates LPS-induced inflammatory response. Int. J. Mol. Sci. 20:1323. 10.3390/ijms20061323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen K. C., Xu B., Krantz I. D., Gerton J. L. (2016). NIPBL controls RNA biogenesis to prevent activation of the stress kinase PKR. Cell Rep. 14 93–102. 10.1016/j.celrep.2015.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara S., Ries R. J., Jaffrey S. R. (2019). Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20 608–624. 10.1038/s41580-019-0168-5 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang X., Zhang X., Wang J., Ma Y., Zhang L., et al. (2019). RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc. Natl. Acad. Sci. U.S.A. 116 976–981. 10.1073/pnas.1812536116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., Hou J., Zhou Y., Li Z., Cao X. (2017). The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nat. Immunol. 18 1094–1103. 10.1038/ni.3830 [DOI] [PubMed] [Google Scholar]
- Zhu J. (2018). T helper cell differentiation, heterogeneity, and plasticity. Cold Spring Harb. Perspect. Biol. 10 a030338. 10.1101/cshperspect.a030338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Zhao Y., Zou L., Zhang D., Aki D., Liu Y. C. (2019). The E3 ligase VHL promotes follicular helper T cell differentiation via glycolytic-epigenetic control. J. Exp. Med. 216 1664–1681. 10.1084/jem.20190337 [DOI] [PMC free article] [PubMed] [Google Scholar]