Abstract
Background
Type 2 Diabetes mellitus (T2DM) is a chronic metabolic disorder. It is a major non-communicable disease affecting 463 million people globally in 2019 and is expected to be double to about 700 million by 2045. The majority are Asians with Indian ethnicity in Malaysia reported as the highest prevalence of T2DM. Cardiovascular disease, renal failure, blindness and neuropathy, as well as premature death are the known morbidity and mortality resulted from T2DM. T2DM is characterized by the dysfunctional insulin physiology that causes reduction of glucose transport into the cells which lead to hyperglycaemia. Hence, one of the important treatments is an oral antidiabetic drug that lowers the serum glucose level in patients with T2DM. This drug will be transported across cell membranes by organic cation transporters (OCT). Therefore, it is important to identify the OCT candidate gene polymorphisms related to T2DM especially among the Indian ethnicity in Malaysia.
Methods
Blood samples were collected from 132 T2DM patients and 133 controls. Genotyping of OCT1 (rs628031), OCT2 (rs145450955), OCT3 (rs3088442 and rs2292334) was performed using (PCR-RFLP).
Results
No association was observed for genotypic and allelic distributions in all the gene polymorphisms of OCT genes (P > 0.05). However, a logistic regression analysis stratified by gender in a dominant model showed a significant difference for OCT3 among males with T2DM (P = 0.006). Significant association was also observed for OCT3 when stratified to subjects aged > 45 years old (P = 0.009).
Conclusion
Based on these findings, the association of OCT3 (rs2292334) could be considered as a possible genetic risk factor for the development of T2DM among Indian males alone.
Keywords: Type 2 diabetes mellitus, Organic cation transporter, OCT polymorphisms, Indian ethnicity
1. Introduction
Diabetes mellitus is defined as “a chronic metabolic disease characterized by hyperglycemia and abnormal carbohydrate metabolism caused by the body’s impaired ability to produce or respond to insulin” (World Health Organization, 2019). Diabetes mellitus can be classified into two classes, Type 1 diabetes (T1DM), which is caused by deficit in total insulin secretion and Type 2 diabetes mellitus (T2DM), a combination of inadequate insulin secretion and resistance to insulin (Tuomi et al., 2014, Zolk, 2012).
Diabetes is developing rapidly around the world as the standard of living rises (Wild et al., 2004). Around 415 million people worldwide are believed to have diabetes, which is expected to reach 642 million by 2040 (Ogurtsova et al., 2017). Malaysia, a country comprising a multi-ethnic population, has one of the highest prevalence of T2DM compared to the other Asian countries, and the trend is projected to continue to rise (Abdullah et al., 2017). The National Health and Morbidity Surveys (NHMS) 2015, reported the prevalence of T2DM in Malaysia to be highest among Indians (22.1%), followed by Malays (14.6%), Chinese (12.0%), and indigenous Malaysian ethnicities (10.7%) (Institute for Public Health NHMS, 2015).
T2DM is a chronic, progressive disorder that develops as a result of the interaction between genetic and environmental factors (overweight, obesity, high carbohydrate intake, and sedentary lifestyle) (Chiasson and Rabasa-Lhoret, 2004). Treatments for diabetes mellitus are aimed toward reducing microvascular and macrovascular complications through optimal control (van Leeuwen et al., 2013). Oral antidiabetic drugs are crucial as first-line treatments for T2DM patients due to their mild hydrophobic property and requirement of transporters to cross the plasma membrane into the cells (Umamaheswaran et al., 2011).
A family of transporters known as “organic cation transporters” (OCT1, OCT2, and OCT3) is involved in the transfer of endogenous physiological amino compounds and is encoded by the Solute carrier family 22 member 1 SLC22A1, Solute carrier family 22 member 2 SLC22A2, and Solute carrier family 22 member 3 SLC22A3 genes, respectively (Ghaffari-Cherati et al., 2016). The OCT (SLC22A) gene family encodes>30 proteins and are mapped onto chromosome 6q25.3. The gene contains 11 exons and is involved in the excretion of a wide variety of drugs, toxins, and metabolites in the kidney, liver, and other tissues. Therefore, human reaction to anti-diabetics medication is possibly influenced by genetic variants in the OCT (SLC22A) genes (Hakooz et al., 2017, Kashi et al., 2015, Kang et al., 2007).
The OCT family in humans consists of three organic cation transporters (OCT1, OCT2, and OCT3), with OCT3 less studied compared to OCT1 and OCT2 (Chen et al., 2015, Koepsell, 2004). It is well established that OCT3 interferes with substrate specificity of two other OCT family transporters and that it is an efficient transporter for drugs (Hosseyni-Talei et al., 2017, Chen et al., 2015, Zhu et al., 2012). OCT3, being a major transporter, also contributes to the overall regulation of neurotransmission and the maintenance of homeostasis within the central nervous system (Massmann et al., 2014). OCT1 and OCT2 are expressed at different levels in the liver, small intestine, kidney and brain (Koepsell, 2011). OCT1 is mainly expressed in the basolateral membrane of hepatocytes that initiate uptake from the blood, whereas OCT2 is primarily expressed in the basolateral membrane of kidney epithelial cells (König et al., 2011, Meyer zu Schwabedissen et al., 2010, Müller et al., 2013, Sato et al., 2008). Several factors including the nature of the disease and the target organ, influence drug response; however, estimates indicate that genetic variation accounts for 20% to 95% of the variability in the response to similar anti-diabetic drugs (Ghaffari-Cherati et al., 2016).
Several single nucleotide polymorphisms (SNPs) in the three genes have been identified through population genetic analyses studies (Du Plessis et al., 2015). These polymorphisms may contribute to the inter-individual variability in the pharmacokinetics disposition, efficacy, and toxicity of therapeutic drugs. Recent researches have demonstrated inter-patient differences in response to pharmacological therapy may be related to a variety of genetic variants in the OCT genes (Du Plessis et al., 2015).
Early detection of high-risk individuals is critical for preventing or delaying the onset of diabetes. Individuals develop diabetes as a result of the interaction of genetic and environmental factors. Thus, a susceptibility gene may exhibit a variable phenotype across populations or regions as numerous studies have also shown inconsistent findings regarding the association between OCT1, OCT2, and OCT3 gene variants and diabetes in different ethnic groups (Phate et al., 2020). There is a paucity of knowledge in Malaysia about the OCT gene polymorphisms associated with T2DM. In Malaysia, the Indian ethnic group has the greatest prevalence of T2DM when compared to other ethnic groups (Institute for Public Health NHMS, 2015). Thus, this study will ascertain the genetic variation of the OCT1, OCT2, and OCT3 genes and their association with T2DM in Malaysia's Indian ethnic group. Additionally, this study enhances our understanding of additional factors, such as phenotypic characteristics, that might contribute to T2DM and may have a significant impact on future individual medical treatment for T2DM.
2. Materials and methods
2.1. Ethics statement
The present case-control study was registered with the Malaysian National Medical Research Register (NMRR) (reference number NMRR-18–3028-44490) and the study protocol was approved by the Medical Research and Ethics Committee (MREC), Ministry of Health (MoH), Malaysia.
2.2. Study population
All subjects were recruited from Hospital Serdang, Malaysia, and were between the age of 18 and 65 years old and of Indian ethnicity. All participants were required to sign a written informed consent form prior to participation in the study, and socio-demographic profiles were obtained. Participants diagnosed with T2DM by the medical practitioner were recruited as case subjects. The T2DM diagnosis was based on a fasting plasma glucose (FPG) of ≥ 7.0 mmol/L and HbA1c ≥ 6.3% following the T2DM clinical practice guidelines from the Ministry of Health, Malaysia (Ministry of Health, 2020). Healthy participants without T2DM and any history of chronic or acute diseases (coronary artery, myocardial infarction, stroke, T1DM, prior history of renal failure, autoimmune disease, liver disease) were recruited as control subjects. An amount of 2 to 3 ml of blood was collected into EDTA tubes (Becton Dickinson, New Jersey, USA) from all subjects and stored at −20 °C for further analysis.
2.3. Gene and SNP selection
Based on the obtained literature reviews, OCT1 (rs628031), OCT2 (rs145450955), and OCT3 (rs3088442 and rs2292334) gene polymorphisms were among the significant SNPs studied in T2DM (Mahrooz et al., 2017), and thus, were selected as a candidate SNP in the present study. Furthermore, the SNP was selected based on functional significance and location. All the SNPs were not in linkage disequilibrium, which were performed using the Haploview software (http://www.broad.mit.edu/mpg/haploview). The RegulomeDB (http://www.regulomedb.org/) was used to determine the effect of the OCT SNPs on allele-specific transcription factor binding and link on the expression of the gene target. All SNPs scored a RegulomeDB of 4 and 5 which indicate the variants had minimal transcription factor binding evidence.
2.4. Genotyping
Genomic DNA was extracted using the QIAamp DNA blood mini kit (Qiagen, Valencia, CA, USA) following the manufacturer's protocol. Genotyping was then performed using PCR-RFLP method for the four gene polymorphisms; OCT1 (rs628031), OCT2 (rs145450955), and OCT3 (rs3088442 and rs2292334). The PCR was carried out in a 25 µL volume of a reaction mixture comprising 1 unit of Taq DNA polymerase, 400 to 500 ng genomic DNA, 200 mM of each dNTP, 1.5 mM MgCL2, 280 nM of the following primers: forward: 5′-CTAAACCCAGTGATTCATGCTCTTT-3 ' and reverse: 5′-TTTGTTCTCATT-CCAGAGGCTTATC-3′ for OCT1 (rs628031); forward: 5′-CTGTGCCTCCTGGATTCATA-3′ and reverse: 5′-AGGTTGCTTTTGTTCTACAGT-3′ for OCT2 (rs145450955); forward: 5′-AGATTGCATGGAGGATGAAC-3′ and reverse: 5′-TGT TCCAGAGGAGGTGGACG-3′ for OCT3 (rs3088442); forward: 5′-GTGGTGGAACTGCCAGGA-3′ and reverse: 5′-CTACAGAACCAATCTCTTACTTCG-3′ for OCT3 (rs2292334). The PCR cycling conditions were carried out for 35 cycles with denaturation at 94 °C for 30 s (rs628031), 93 °C for 35 s (rs145450955), 93 °C for 50 s (rs3088442 and rs2292334), annealing at 56 °C for 60 s (rs628031), 54 °C for 35 s (rs145450955), 54 °C for 40 s (rs3088442), 55 °C for 40 s (rs2292334), and extended at 72 °C for 60 s (rs628031), 35 s (rs145450955) and 40 s (rs3088442 and rs2292334) respectively. The amplification products were digested with MscI (rs628031), BsaAI (rs145450955) (Fermentas, Lithuania), and AciI (rs3088442 and rs2292334) restriction enzymes (Thermo Scientific, Lithuania) and visualized under UV light with the Alpha Imager (Alpha Innotech, San Leandro, CA).
2.5. Statistical analysis
All statistical data were analysed using IBM SPSS software (version 24.0, SPSS Inc., Chicago, USA). The Hardy-Weinberg equilibrium and linkage disequilibrium of alleles in each SNPs were confirmed using the Haploview software (http://www.broad.mit.edu/mpg/haploview). Categorical variables were compared using the chi-square test, while normal continuous variables using the Student’s t-test and one-way analysis of variance (ANOVA). Skewed continuous variables are compared using Mann-Whitney U test. Logistic regression analysis was performed to determine the association between OCT1 (rs628031), OCT2 (rs145450955), and OCT3 (rs3088442 and rs2292334) gene polymorphisms with T2DM under the assumption of three genetic models (additive, dominant and recessive) and also stratified by gender. Bonferroni adjustment for multiple statistical testing was applied to correct the threshold of P-value. Any subjects with missing data were excluded from the study and a P-value of p < 0.05 was considered statistically significant.
3. Results
3.1. Clinical and biochemical parameters of study subjects
A total of 132 T2DM patients and 133 controls were recruited based on the inclusion and exclusion criteria. Table 1 illustrates the clinical and parameters between T2DM and control subjects. T2DM subjects have a mean age of 49.83 ± 13.68 years, whereas the control subjects with a mean of 46.65 ± 14.20 years. There is a significant difference in HbA1c levels between T2DM and controls (P < 0.001), with higher level of HbA1c among the control subjects (8.26 ± 1.89).
Table 1.
Clinical and biochemical parameters of study subjects.
| Parameters | T2DM (n = 132) | Controls (n = 133) | P-value† | |
|---|---|---|---|---|
| Age (Years) | Total | 49.83 ± 13.68 | 46.65 ± 14.20 | 0.069 |
| ≤45 years | 47 (35.6) | 61 (45.9) | ||
| >45 years | 85 (64.4) | 72 (54.1) | 0.089§ | |
| Gender | Male | 84 (63.6) | 71 (53.4) | |
| Female | 48 (36.4) | 62 (46.6) | 0.09§ | |
| BMI (Kg/m2) | 27.83 ± 4.72 | 27.70 ± 4.96 | 0.625‡ | |
| Blood glucose | FBS(mmol/L) | 7.84 ± 3.00 | 7.47 ± 2.64 | 0.669 |
| HbA1C | 7.84 ± 6.61 | 8.26 ± 1.89 | <0.001* |
Data are presented as mean ± SD and data reported as number (percentage) of subjects for gender; BMI = body mass index, FBS = fasting blood sugar, HbA1C = glycated haemoglobin
*Significant P-value, P < 0.05
Mann-Whitney U test
Student’s t-test
Pearson chi-square test
3.2. Genotypic and allelic distributions
Genotyping was performed for all the four OCT gene polymorphisms as presented in Fig. 1. No significant association was recorded in both genotype and allele frequency in the four OCT gene polymorphisms when compared between T2DM and control subjects (P > 0.05). A significant association (P = 0.009) was observed only for OCT3 (rs2292334) gene polymorphism when stratified to subjects aged > 45 years old (Table 2). Likewise, all gene variants when compared based on an additive, dominant and recessive genetic model were not significantly different (P > 0.05) (Table 3). However, a significant difference was observed in OCT3 (rs2292334) gene polymorphism in a dominant genetic model when stratified by male gender and for all ages (P = 0.006). No significant difference was recorded for the other gene polymorphisms after stratifying by gender (P > 0.05) (Table 4).
Fig. 1.
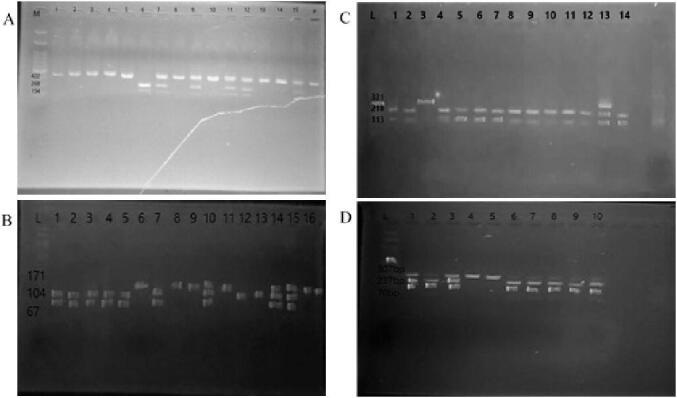
A: PCR-RFLP product for OCT1 (rs628031) gene polymorphism (Lane M: 100 bp ladder; Lane 1-lane 5, 8, 10, 13 and 14: WT, 422 bp; Lane 7,9,11,12, and 13: HT, 422 bp, 268 bp and 154 bp; Lane 6: MT, 268 bp and 154 bp). B: PCR-RFLP product for OCT3 (rs3088442) gene polymorphism (Lane L: 100 bp ladder; Lane 1–5 and 7: WT, 104 bp and 57 bp; Lane 10, 14 and 15: HT, 171 bp, 104 bp and 67 bp; Lane 6, 8, 9, 11 and 16: MT, 171 bp). C: PCR-RFLP product for OCT2 (rs145450955) gene polymorphism (Lane L: 100 bp ladder, Lane 3: WT, 331 bp; Lane 13: HT, 331 bp, 218 bp and 113 bp; Lane 1,2,4–12, and 14: MT, 218 bp and 113 bp). D: PCR-RFLP product for OCT3 (rs2292334) gene polymorphism (Lane L: 100 bp ladder; Lane 2, 6–10: WT, 237 bp and 70 bp; Lane 1, and 3, HT, 307 bp, 237 bp and 70 bp; Lane 4 and 5: MT, 307 bp).
Table 2.
Genotypic and allelic distributions of SLC22A1 rs628031, SLC22A2 rs145450955, SLC22A3 rs3088442 and SLC22A3 rs2292334 gene polymorphisms between T2DM and control subjects.
| SNPs | Genotypes (%) | P-value† | Alleles (%) | P-value† | Odd ratio (95% CI) |
|||
|---|---|---|---|---|---|---|---|---|
| All age | ||||||||
| rs628031 T2DM Controls |
AA 70(53.4) 83(62.4) |
AG 51(38.9) 42(31.6) |
GG 10(7.7) 8 (6.0) |
0.336 |
A 191(72.9) 208(78.2) |
G 71(27.1) 58(21.8) |
0.157 |
1.333 (0.895–1.986) |
| rs145450955 T2DM Controls |
CC 71(53.8) 82(61.7) |
CT 17(12.9) 14(10.5) |
TT 44(33.3) 37(27.8) |
0.431 |
C 159(60.2) 178(66.9) |
T 105(39.8) 88(33.1) |
0.11 |
1.336 (0.937–1.905) |
| rs3088442 T2DM Controls |
GG 83(62.9) 87(65.4) |
GA 16(12.1) 16(12.0) |
AA 33(25.0) 30(22.6) |
0.89 |
G 182(68.9) 190(31.1) |
A 82(31.1) 76(28.6) |
0.531 |
0.888 (0.612–1.288) |
| rs2292334 T2DM Controls |
GG 82(62.1) 94(71.2) |
GA 19(14.4) 15(11.4) |
AA 31(23.5) 23(17.4) |
0.29 |
G 183(69.3) 203(76.9) |
A 81(30.7) 61(23.1) |
0.05 |
1.473 (1.000–2.171) |
| Age > 45 years | ||||||||
| rs628031 T2DM Controls |
AA 44(50.6) 43(49.4) |
AG 34(56.7) 26(43.3) |
GG 7(70.0) 3(30.0) |
0.446 |
A 122(71.8) 112(78.9) |
G 48(28.2) 32(22.2) |
0.223 |
1.377 (0.822–2.306) |
| rs145450955 T2DM Controls |
CC 43(51.2) 41(48.8) |
CT 12(60.0) 8(40.0) |
TT 30(56.6) 23(43.4) |
0.705 |
C 98(57.6) 90(62.5) |
T 72(42.4) 54(37.5) |
0.382 |
1.224 (0.777–1.929) |
| rs3088442 T2DM Controls |
GG 49(50.0) 49(50.0) |
GA 12(60.0) 8(40.0) |
AA 24(61.5) 15(38.5) |
0.404 |
G 110(64.7) 106(73.6) |
A 60(35.3) 38(26.4) |
0.090 |
1.522 (0.936–2.474) |
| rs2292334 T2DM Controls |
GG 52(49.1) 54(50.9) |
GA 10(62.5) 6(37.5) |
AA 23(67.6) 11(32.4) |
0.132 |
G 114(67.1) 114(79.2) |
A 56(32.9) 28(19.4) |
0.009* |
2.000 (1.186–3.373) |
SNPs = single nucleotide polymorphisms, T2DM = type 2 diabetes mellitus, CI = confidence interval.
*Significant P-value, P < 0.05.
Pearson Chi-square test.
Table 3.
Genotypes association based on genetic models.
| OR (95% CI) for diabetics |
||||||
|---|---|---|---|---|---|---|
| Gene variant | Additive | Dominant | Recessive | p† | p‡ | p§ |
| All age | ||||||
| rs628031 | 1.49 (0.55–4.06) | 1.37 (0.84–2.26) | 1.34 (0.50–3.56) | 0.428 | 0.212 | 0.56 |
| rs145450955 | 1.29 (0.75–2.24) | 1.28 (0.78–2.11) | 1.25 (0.73–2.12) | 0.359 | 0.335 | 0.419 |
| rs3088442 | 1.09 (0.61–1.96) | 1.11 (0.67–1.85) | 1.07 (0.60–1.89) | 0.772 | 0.685 | 0.824 |
| rs2292334 | 1.56 (0.84–2.91) | 1.61 (0.95–2.73) | 1.43 (0.78–2.64) | 0.163 | 0.075 | 0.249 |
| Age > 45 years | ||||||
| rs628031 | 2.26 (0.55–9.34) | 1.39 (0.73–2.62) | 2.05 (0.51–8.23) | 0.258 | 0.313 | 0.314 |
| rs145450955 | 1.27 (0.63–2.53) | 1.31 (0.69–2.47) | 1.18 (0.61–2.31) | 0.506 | 0.404 | 0.625 |
| rs3088442 | 1.56 (0.72–3.35) | 1.53 (0.79–2.97) | 1.45 (0.69–3.07) | 0.259 | 0.207 | 0.328 |
| rs2292334 | 2.17 (0.96–4.90) | 2.04 (1.01–4.10) | 2.02 (0.90–4.49) | 0.062 | 0.046 | 0.087 |
OR = odds ratio, CI = confidence interval.
p†, for the additive model; p‡, for the dominant model; and p§, for the recessive model.
†‡§ Logistic regression analysis
Table 4.
Genotypes associations based on genetic models and genders.
| OR (95% CI) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Age | Gender | Gene Variant | Additive | Dominant | Rececssive | p† | p‡ | p§ |
| All | Male | rs628031 | 1.97 (0.50–7.71) | 1.77 (0.90–3.48) | 1.37 (0.37–5.06) | 0.332 | 0.756 | 0.633 |
| rs145450955 | 1.03 (0.49–2.18) | 1.05 (0.54–2.04) | 1.12 (0.56–2.22) | 0.937 | 0.885 | 0.755 | ||
| rs3088442 | 0.81 (0.37–1.77) | 0.81 (0.40–1.66) | 0.94 (0.45–1.96) | 0.6 | 0.569 | 0.866 | ||
| rs2292334 | 3.11 (1.25–7.73) | 2.91 (1.37–6.19) | 2.92 (1.21–7.08) | 0.014 | 0.006* | 0.018 | ||
| Female | rs628031 | 0.69 (0.11–4.11) | 0.83 (0.36–1.89) | 1.09 (0.21–5.67) | 0.679 | 0.658 | 0.917 | |
| rs145450955 | 1.78 (0.70–4.51) | 1.99 (0.86–4.63) | 1.56 (0.63–3.84) | 0.224 | 0.107 | 0.338 | ||
| rs3088442 | 1.57 (0.54–4.55) | 1.43 (0.63–3.22) | 1.22 (0.45–3.31) | 0.407 | 0.392 | 0.703 | ||
| rs2292334 | 0.53 (0.18–1.52) | 0.70 (0.31–1.61) | 0.55 (0.19–1.50) | 0.237 | 0.401 | 0.241 | ||
| >45 years | Male | rs628031 | 1.45 (0.29–7.13) | 1.91 (0.84–4.31) | 1.07 (0.23–5.07) | 0.651 | 0.121 | 0.930 |
| rs145450955 | 1.13 (0.45–2.83) | 1.09 (0.49–2.48) | 1.11 (0.47–2.63) | 0.799 | 0.826 | 0.808 | ||
| rs3088442 | 1.69 (0.66–4.29) | 1.82 (0.76–4.33) | 1.55 (0.62–3.88) | 0.273 | 0.178 | 0.353 | ||
| rs2292334 | 3.30 (1.09–10.1) | 2.69 (1.04–6.93) | 3.14 (1.04–9.47) | 0.035 | 0.040 | 0.042 | ||
| Female | rs628031 | NA | 0.73 (0.25–2.12) | NA | NA | 0.565 | NA | |
| rs145450955 | 1.28 (0.43–3.86) | 1.49 (0.51–4.41) | 1.15 (0.39–3.45) | 0.659 | 0.462 | 0.797 | ||
| rs3088442 | 1.25 (0.32–4.91) | 1.22 (0.42–3.50) | 1.19 (0.32–4.45) | 0.748 | 0.713 | 0.795 | ||
| rs2292334 | 1.23 (0.34–4.37) | 1.43 (0.48–4.19) | 1.11 (0.32–3.84) | 0.752 | 0.520 | 0.872 | ||
OR = odds ratio, CI = confidence interval, NA = not available.
p†, for the additive model; p‡, for the dominant model; and p§, for the recessive model.
*Significant P-value, P < 0.01 after Bonferroni correction (0.05/4 for 4 variants).
†‡§ Logistic regression analysis, adjusted for age.
4. Discussion
A total of 265 subjects were recruited for this study, including 132 subjects with T2DM and 133 control subjects. To our knowledge, this is the first study reporting the frequency of OCT1, OCT2 and OCT3 gene polymorphisms among Malaysia’s Indian ethnic group. From our findings, there was no significant association in both genotypic and allelic distribution observed in OCT1 (rs628031), OCT2 (rs145450955), OCT3 (rs3088442 and rs2292334) gene polymorphisms between T2DM and control subjects (P > 0.05). The allelic distributions of all OCT gene polymorphisms studied on T2DM in different populations are shown in Table 5 with contradictory findings. These inconsistent results could be explained by various factors including ethnic differences, environmental factors, and different study methodology.
Table 5.
Allele frequency distribution of OCT gene polymorphisms.
| SNPs | Population | Case/control | Alleles in cases |
Alleles in controls |
P-value | Reference | ||
|---|---|---|---|---|---|---|---|---|
| G | A | G | A | |||||
|
OCT1 rs628031 |
China | 153/124 | 0.72 | 0.28 | 0.74 | 0.26 | * | Zhou et al., 2015 |
| Indonesia | 86/NA | 0.61 | 0.39 | NA | NA | NS | Vitarani et al., 2017 | |
| Saudi Arabia | 201/89 | 0.69 | 0.31 | 0.71 | 0.29 | NS | Altall et al., 2019 | |
| Latvia | 53/193 | 0.27 | 0.73 | 0.24 | 0.76 | * | Tarasova et al., 2012 | |
| Iran | 63/77 | 0.32 | 0.68 | 0.33 | 0.66 | NS | Shokri et al., 2016 | |
| Present study | 132/133 | 0.27 | 0.73 | 0.22 | 0.78 | NS | ||
| C | T | C | T | |||||
|
OCT2 rs145450955 |
Iran | 40/NA | 0.8 | 0.2 | NA | NA | * | Kashi et al., 2015 |
| Present study | 132/133 | 0.6 | 0.4 | 0.67 | 0.33 | NS | ||
| G | A | G | A | |||||
|
OCT3 rs3088442 |
Iran | 150/152 | NR | NR | NR | NR | ** | Mahrooz et al., 2017 |
| Pakistan | 600/300 | 0.67 | 0.33 | 0.54 | 0.46 | ** | Moeez et al., 2019 | |
| Present study | 132/133 | 0.69 | 0.31 | 0.31 | 0.29 | NS | ||
| G | A | G | A | |||||
|
OCT3 rs2292334 |
Iran | 150/152 | 0.65 | 0.35 | 0.75 | 0.25 | * | Mahrooz et al., 2017 |
| Present study | 132/133 | 0.69 | 0.31 | 0.77 | 0.23 | NS | ||
NA, not available; NR, not reported; NS, not significant; *(p < 0.05); **(P < 0.001).
Apart from the genetic factors, clinical and biochemical parameters such as abdominal obesity, family history of diabetes and hypertension, older age and poor diet are associated with diabetes. Recently, the Malaysian Ministry of Health projected that the prevalence of T2DM could rise to as much as 20.1% in 2020 if the major associated factors (overweight/obesity, poor diet and inadequate physical activity) are not comprehensively addressed (Ministry of Health., 2013, Institute for Public Health NHMS, 2015). The average age of T2DM onset is 45 years old and a 10-year prospective cohort study conducted in India reported that subjects older than 45 years had a1.6 times higher risk of developing T2DM (Vijayakumar et al., 2019). Hence, the present study included an additional analysis excluding those<45 years old as there might be a possibility of the young subjects underpowering the study since they might be future cases carrying the genetic variants.
In this study, a significant association was recorded for the rs2292334 variant of the OCT3 gene when analyzed among subjects aged>45 years old (P = 0.009). A similar variant also showed a significant difference when stratified by the male gender (P < 0.05). The mutant variant was also seen to be at the highest risk compared to the other variants.
A study by Chen et al. reported that rs2292334 is involved in the therapeutic action of metformin and with an A allele frequency of 0.45 could be considered as a polymorphic variant in the population even with different ethnic groups (Chen et al., 2010). This is consistent with the present study and also a study among the Iranian population that reported a frequency of 0.65 major G alleles and 0.35 of minor A alleles of the rs2292334 variant in T2DM patients (Mahrooz et al., 2017).
According to Aoyama et al., 2006, Nies et al., 2009, the rs2292334 variant plays a critical role in the gene expression of OCT3 and influences the final production of mRNA. The minor A allele of rs2292334 was also linked to a higher risk of T2DM among obese patients in comparison with non-obese patients (Mahrooz et al., 2017). A combination of obesity and parental history of diabetes among males has also shown a significant interaction that increases the risk of T2DM (OR = 2.4) (Wikner et al., 2013). This further proves the findings from the present study that showed a significant association of OCT3 rs2292334 gene polymorphism with Indian ethnicity male subjects with T2DM. In the Pakistani population, the A allele of OCT3 rs3088442 was a protective factor and associated with the clinical efficacy of metformin in patients who have type 2 diabetes (Moeez et al., 2019).
Despite the findings, the present study has several limitations. This study lacked access to data on gene expression, which could be used to assess the association between corresponding polymorphisms. Additionally, this study’s findings are confined among Malaysian Indians. Future research should include Malays and Chinese as well, to better understand the genetic risk factor for OCT gene variants to develop in T2DM patients in a general Malaysian population. While advanced age is a risk factor for T2DM, this study enrolled patients of adults aged above 18 years old. However, the current study includes a stratified analysis of participants aged over 45 years to address this limitation. The contradictory findings from this study in contrast to other populations could be attributed to a variety of reasons, including ethnic diversity which can result in distinctly different genetic predispositions. Certain countries may have heterogeneous ethnicities; this may result in different genetic predispositions in comparison to countries with homogenous ethnicity. There is also the matter of varying sample sizes in comparison with other epidemiological studies. In this study, the relatively small number of samples and low frequency of the risk allele could contribute to the lack of association. Although GWAS studies have suggested that OCT gene polymorphisms were associated with T2DM in various populations, there is a lack of information on the relationship between OCT gene polymorphisms and Malaysian Indians Therefore, the candidate gene approach used in this study focuses on a few SNPs that were selected based on their low minor allele frequency and potential significance. To our best knowledge, this study is the first to evaluate the association of OCT gene polymorphisms with T2DM among the Indian ethnicity in Malaysia.
5. Conclusion
In conclusion, the association of OCT3 rs2292334 gene polymorphism could be considered as a genetic susceptibility to the development of T2DM among Malaysian Indian ethnicity male subjects. However, further study with a larger sample size is needed to confirm the association. This study could be used as a platform to further understanding genetic factors that cause T2DM and, in the future, may have a significant impact on individualizing medical treatment for T2DM particularly for Indian ethnicity.
CRediT authorship contribution statement
Sabah Ghasan Abood Al-Ashoor: Formal analysis, Investigation, Writing – original draft, Writing – review & editing. Vasudevan Ramachandran: Conceptualization, Methodology, Validation, Resources, Writing – review & editing, Supervision, Project administration, Funding acquisition. Liyana Najwa Inche Mat: Validation, Resources, Writing – review & editing, Supervision. Nur Afiqah Mohamad: Validation, Formal analysis, Data curation, Writing – review & editing. Mohd Hazmi Mohamed: Validation, Resources, Writing – review & editing, Supervision. Wan Aliaa Wan Sulaiman: Conceptualization, Methodology, Resources, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
The authors would also like to put on record their appreciation to the participants of the study for their cooperation and involvement in this study.
Funding
This work was supported by the Putra Grant (Grant no: GP-IPS/2018/9648800) from Universiti Putra Malaysia.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Sabah Ghasan Abood Al-Ashoor, Email: asuphd@gmail.com.
Vasudevan Ramachandran, Email: vasudevanramachandran@bharathuniv.ac.in.
Wan Aliaa Wan Sulaiman, Email: wanaliaa@upm.edu.my.
References
- Abdullah, N., Abdul Murad, N. A., Mohd Haniff, E. A., Syafruddin, S. E., Attia, J., Oldmeadow, C., . . . Holliday, E. G., 2017. Predicting type 2 diabetes using genetic and environmental risk factors in a multi-ethnic Malaysian cohort. Public Health. 149, 31-38. https://doi: 10.1016/j.puhe.2017.04.003. [DOI] [PubMed]
- Altall, R. M., Qusti, S. Y., Filimban, N., Alhozali, A. M., Alotaibi, N. A., Dallol, A., . . . Bakhashab, S., 2019. SLC22A1 And ATM Genes Polymorphisms Are Associated With The Risk Of Type 2 Diabetes Mellitus In Western Saudi Arabia: A Case-Control Study. Appl Clin Genet. 12, 213-219. https://doi: 10.2147/TACG.S229952. [DOI] [PMC free article] [PubMed]
- Aoyama, N., Takahashi, N., Kitaichi, K., Ishihara, R., Saito, S., Maeno, N., . . . Ozaki, N., 2006. Association between gene polymorphisms of SLC22A3 and methamphetamine use disorder. [Research Support, Non-U.S. Gov't]. Alcohol Clin Exp Res. 30, 1644-1649. https://doi: 10.1111/j.1530-0277.2006.00215.x. [DOI] [PubMed]
- Chen, E. C., Liang, X., Yee, S. W., Geier, E. G., Stocker, S. L., Chen, L., & Giacomini, K. M., 2015. Targeted disruption of organic cation transporter 3 attenuates the pharmacologic response to metformin. Mol Pharmacol. 88, 75-83. https://doi: 10.1124/mol.114.096776. [DOI] [PMC free article] [PubMed]
- Chen, L., Pawlikowski, B., Schlessinger, A., More, S. S., Stryke, D., Johns, S. J. Giacomini, K. M., 2010. Role of organic cation transporter 3 (SLC22A3) and its missense variants in the pharmacologic action of metformin. Pharmacogenet Genomics. 20, 687-699. https://doi:10.1097/FPC.0b013e32833fe789. [DOI] [PMC free article] [PubMed]
- Chiasson, J. L., & Rabasa-Lhoret, R., 2004. Prevention of type 2 diabetes: insulin resistance and beta-cell function. Diabetes. 53 Suppl 3, S34-38. https://doi: 10.2337/diabetes.53.suppl_3.s34. [DOI] [PubMed]
- Du Plessis, M., Pearce, B., Jacobs, C., Hoosain, N., & Benjeddou, M., 2015. Genetic polymorphisms of the organic cation transporter 1 gene (SLC22A1) within the Cape Admixed population of South Africa. Mol Biol Rep. 42, 665-672. https://doi: 10.1007/s11033-014-3813-2. [DOI] [PubMed]
- Ghaffari-Cherati, M., Mahrooz, A., Hashemi-Soteh, M. B., Hosseyni-Talei, S. R., Alizadeh, A., & Nakhaei, S. M., 2016. Allele frequency and genotype distribution of a common variant in the 3 -untranslated region of the SLC22A3 gene in patients with type 2 diabetes: Association with response to metformin. J Res Med Sci. 21, 92. https://doi: 10.4103/1735-1995.192508. [DOI] [PMC free article] [PubMed]
- Hakooz, N., Jarrar, Y. B., Zihlif, M., Imraish, A., Hamed, S., & Arafat, T., 2017. Effects of the genetic variants of organic cation transporters 1 and 3 on the pharmacokinetics of metformin in Jordanians. Drug Metab Pers Ther. 32, 157-162. https://doi: 10.1515/dmpt-2017-0019. [DOI] [PubMed]
- Hosseyni-Talei, S. R., Mahrooz, A., Hashemi-Soteh, M. B., Ghaffari-Cherati, M., & Alizadeh, A., 2017. Association between the synonymous variant organic cation transporter 3 (OCT3)-1233G>A and the glycemic response following metformin therapy in patients with type 2 diabetes. Iran J Basic Med Sci. 20, 250-255. https://doi: 10.22038/IJBMS.2017.8351. [DOI] [PMC free article] [PubMed]
- Institute for Public Health. National Health and Morbidity Survey 2015 (NHMS 2015). Vol. II: Non-Communicable Diseases, Risk Factors & Other Health Problems [database online]. http://www.moh.gov.my/moh/resources/nhmsreport2015vol2.pdf. 2015. Accessed December 15, 2019.
- Kang, H. J., Song, I. S., Shin, H. J., Kim, W. Y., Lee, C. H., Shim, J. C., Shin, J. G., 2007. Identification and functional characterization of genetic variants of human organic cation transporters in a Korean population. Drug Metab Dispos. 35, 667-675. https://doi: 10.1124/dmd.106.013581. [DOI] [PubMed]
- Kashi, Z., Masoumi, P., Mahrooz, A., Hashemi-Soteh, M. B., Bahar, A., & Alizadeh, A., 2015. The variant organic cation transporter 2 (OCT2)-T201M contribute to changes in insulin resistance in patients with type 2 diabetes treated with metformin. Diabetes Res Clin Pract. 108, 78-83. https://doi:10.1016/j.diabres.2015.01.024. [DOI] [PubMed]
- Koepsell, H., 2004. Polyspecific organic cation transporters: their functions and interactions with drugs. Trends Pharmacol Sci. 25, 375-381. https://doi: 10.1016/j.tips.2004.05.005. [DOI] [PubMed]
- Koepsell, H., 2011. Substrate recognition and translocation by polyspecific organic cation transporters. Biol Chem. 392, 95-101. https://doi: 10.1515/BC.2011.009. [DOI] [PubMed]
- Konig, J., Zolk, O., Singer, K., Hoffmann, C., & Fromm, M. F., 2011. Double-transfected MDCK cells expressing human OCT1/MATE1 or OCT2/MATE1: determinants of uptake and transcellular translocation of organic cations. Br J Pharmacol. 163, 546-555. https://doi: 10.1111/j.1476-5381.2010.01052.x. [DOI] [PMC free article] [PubMed]
- Mahrooz, A., Alizadeh, A., Hashemi-Soteh, M. B., Ghaffari-Cherati, M., & Hosseyni-Talei, S. R., 2017. Polymorphic Variants rs3088442 and rs2292334 in the Organic Cation Transporter 3 (OCT3) Gene and Susceptibility Against Type 2 Diabetes: Role of their Interaction. Arch Med Res. 48, 162-168. https://doi: 10.1016/j.arcmed.2017.03.010. [DOI] [PubMed]
- Massmann, V., Edemir, B., Schlatter, E., Al-Monajjed, R., Harrach, S., Klassen, P., Ciarimboli, G., 2014. The organic cation transporter 3 (OCT3) as molecular target of psychotropic drugs: transport characteristics and acute regulation of cloned murine OCT3. Pflugers Arch. 466, 517-527. https://doi: 10.1007/s00424-013-1335-8. [DOI] [PubMed]
- Meyer zu Schwabedissen, H. E., Verstuyft, C., Kroemer, H. K., Becquemont, L., & Kim, R. B., 2010. Human multidrug and toxin extrusion 1 (MATE1/SLC47A1) transporter: functional characterization, interaction with OCT2 (SLC22A2), and single nucleotide polymorphisms. Am J Physiol Renal Physiol. 298, F997-F1005. https://doi: 10.1152/ajprenal.00431.2009. [DOI] [PubMed]
- Ministry of Health. 2013. National Diabetes Registry Report Volume 1, 2009-2012. Kuala Lumpur, Ministry of Health, Malaysia. [database online]. https://www.moh.gov.my/moh/resources/Penerbitan/Rujukan/NCD/Diabetes/National_Diabetes_Registry_Report_Vol_1_2009_2012.pdf. Accessed December, 2019.
- Ministry of Health. 2020. Practice Guidelines: Management of Type 2 Diabetes Mellitus (6th Edition). Kuala Lumpur, Ministry of Health, Malaysia. [database online]. https://www.moh.gov.my/moh/resources/Penerbitan/CPG/Endocrine/CPG_T2DM_6th_Edition_2020_13042021.pdf. Accessed August, 2021.
- Moeez, S., Riaz, S., Masood, N., Kanwal, N., Arif, M. A., Niazi, R., & Khalid, S., 2019. Evaluation of the rs3088442 G>A SLC22A3 Gene Polymorphism and the Role of microRNA 147 in Groups of Adult Pakistani Populations With Type 2 Diabetes in Response to Metformin. Can J Diabetes. 43, 128-135 e123. https://doi: 10.1016/j.jcjd.2018.07.001. [DOI] [PubMed]
- Muller, F., Konig, J., Hoier, E., Mandery, K., & Fromm, M. F., 2013. Role of organic cation transporter OCT2 and multidrug and toxin extrusion proteins MATE1 and MATE2-K for transport and drug interactions of the antiviral lamivudine. Biochem Pharmacol. 86, 808-815. https://doi: 10.1016/j.bcp.2013.07.008. [DOI] [PubMed]
- Nies, A. T., Koepsell, H., Winter, S., Burk, O., Klein, K., Kerb, R., Schaeffeler, E., 2009. Expression of organic cation transporters OCT1 (SLC22A1) and OCT3 (SLC22A3) is affected by genetic factors and cholestasis in human liver. Hepatology. 50, 1227-1240. https://doi: 10.1002/hep.23103. [DOI] [PubMed]
- Ogurtsova K., da Rocha Fernandes J.D., Huang Y., Linnenkamp U., Guariguata L., Cho N.H., Cavan D., Shaw J.E., Makaroff L.E. IDF Diabetes Atlas: Global estimates for the prevalence of diabetes for 2015 and 2040. Diabetes Res. Clin. Pract. 2017;128:40–50. doi: 10.1016/j.diabres.2017.03.024. [DOI] [PubMed] [Google Scholar]
- Phate S.D., Daswani B.R., Mishra D.N., Joshi K.S. Genetic analysis of SLC47A1, SLC22A1, SLC22A2, ATM gene polymorphisms among diabetics in an Indian population. Int. J. Basic Clin. Pharmacol. 2020;9(6):891. doi: 10.18203/2319-2003.ijbcp20202189. [DOI] [Google Scholar]
- Sato T., Masuda S., Yonezawa A., Tanihara Y., Katsura T., Inui K. Transcellular transport of organic cations in double-transfected MDCK cells expressing human organic cation transporters hOCT1/hMATE1 and hOCT2/hMATE1. Biochem. Pharmacol. 2008;76:894–903. doi: 10.1016/j.bcp.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Shokri F., Ghaedi H., Ghafouri Fard S., Movafagh A., Abediankenari S., Mahrooz A., Omrani M.D. Impact of ATM and SLC22A1 Polymorphisms on Therapeutic Response to Metformin in Iranian Diabetic Patients. Int. J. Mol. Cell Med. 2016;5(1):1–7. [PMC free article] [PubMed] [Google Scholar]
- Tarasova, L., Kalnina, I., Geldnere, K., Bumbure, A., Ritenberga, R., Nikitina-Zake, L., Klovins, J., 2012. Association of genetic variation in the organic cation transporters OCT1, OCT2 and multidrug and toxin extrusion 1 transporter protein genes with the gastrointestinal side effects and lower BMI in metformin-treated type 2 diabetes patients. Pharmacogenet Genomics. 22, 659-666. https://doi: 10.1097/FPC.0b013e3283561666. [DOI] [PubMed]
- Tuomi, T., Santoro, N., Caprio, S., Cai, M., Weng, J., & Groop, L., 2014. The many faces of diabetes: a disease with increasing heterogeneity. Lancet. 383, 1084-1094. https://doi: 10.1016/S0140-6736(13)62219-9. [DOI] [PubMed]
- Umamaheswaran, G., Praveen, R. G., Arunkumar, A. S., Das, A. K., Shewade, D. G., & Adithan, C., 2011. Genetic analysis of OCT1 gene polymorphisms in an Indian population. Indian J Hum Genet. 17, 164-168. https://doi: 10.4103/0971-6866.92094. [DOI] [PMC free article] [PubMed]
- van Leeuwen, N., Swen, J. J., Guchelaar, H. J., & t Hart, L. M., 2013. The role of pharmacogenetics in drug disposition and response of oral glucose-lowering drugs. Clin Pharmacokinet. 52, 833-854. https://doi: 10.1007/s40262-013-0076-3. [DOI] [PubMed]
- Vijayakumar, G., Manghat, S., Vijayakumar, R., Simon, L., Scaria, L. M., Vijayakumar, A., Jaleel, A., 2019. Incidence of type 2 diabetes mellitus and prediabetes in Kerala, India: results from a 10-year prospective cohort. BMC Public Health. 19, 140. https://doi: 10.1186/s12889-019-6445-6. [DOI] [PMC free article] [PubMed]
- Vitarani, D. A. N., Zullies, I., Ahmad, H. S., & Mohammad, R. I., 2017. Allele Frequencies of Two Main Metformin Transporter Genes: SLC22A1 rs628031 A>G and SLC47A1 rs2289669 G>A among the Javanese Population in Indonesia. Current Pharmacogenomics and Personalized Medicine. 15, 121-128. https://doi: http://dx.doi.org/10.2174/1875692115666170706113120.
- WHO. Diabetes. World Health Organization. https://www.who.int/news-room/fact-sheets/detail/diabetes. Published October, 2018. Accessed December 15, 2019.
- Wikner, C., Gigante, B., Hellenius, M. L., de Faire, U., & Leander, K., 2013. The risk of type 2 diabetes in men is synergistically affected by parental history of diabetes and overweight. PLoS One. 8, e61763. https://doi: 10.1371/journal.pone.0061763. [DOI] [PMC free article] [PubMed]
- Wild, S., Roglic, G., Green, A., Sicree, R., & King, H., 2004. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 27, 1047-1053. https://doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed]
- Zhou Y., Ye W., Wang Y., Jiang Z., Meng X., Xiao Q., Yan J. Genetic variants of OCT1 influence glycemic response to metformin in Han Chinese patients with type-2 diabetes mellitus in Shanghai. Int. J. Clin. Exp. Pathol. 2015;8:9533–9542. [PMC free article] [PubMed] [Google Scholar]
- Zhu, H. J., Appel, D. I., Grundemann, D., Richelson, E., Markowitz, J.S., Evaluation of organic cation transporter 3 (SLC22A3) inhibition as a potential mechanism of antidepressant action. Pharmacol Res., 2012, 65, 491-496. https://doi: 10.1016/j.phrs.2012.01.008. [DOI] [PubMed]
- Zolk, O., Disposition of metformin: variability due to polymorphisms of organic cation transporters, Ann Med., 2012, 44, 119-129. https://doi: 10.3109/07853890.2010.549144. [DOI] [PubMed]


