Abstract
Introduction
Predicting the postoperative neurological function of cervical spondylotic myelopathy (CSM) patients is generally based on conventional magnetic resonance imaging (MRI) patterns, but this approach is not completely satisfactory. This study utilized radiomics, which produced advanced objective and quantitative indicators, and machine learning to develop, validate, test, and compare models for predicting the postoperative prognosis of CSM.
Materials and methods
In total, 151 CSM patients undergoing surgical treatment and preoperative MRI was retrospectively collected and divided into good/poor outcome groups based on postoperative modified Japanese Orthopedic Association (mJOA) scores. The datasets obtained from several scanners (an independent scanner) for the training (testing) cohort were used for cross‐validation (CV). Radiological models based on the intramedullary hyperintensity and compression ratio were constructed with 14 binary classifiers. Radiomic models based on 237 robust radiomic features were constructed with the same 14 binary classifiers in combination with 7 feature reduction methods, resulting in 98 models. The main outcome measures were the area under the receiver operating characteristic curve (AUROC) and accuracy.
Results
Forty‐one (11) radiomic models were superior to random guessing during CV (testing), with significant increased AUROC and/or accuracy (P AUROC < .05 and/or P accuracy < .05). One radiological model performed better than random guessing during CV (P accuracy < .05). In the testing cohort, the linear SVM preprocessor + SVM, the best radiomic model (AUROC: 0.74 ± 0.08, accuracy: 0.73 ± 0.07), overperformed the best radiological model (P AUROC = .048).
Conclusion
Radiomic features can predict postoperative spinal cord function in CSM patients. The linear SVM preprocessor + SVM has great application potential in building radiomic models.
Keywords: cervical spondylotic myelopathy, machine learning, radiomics
This study builds and analysis 98 radiomic machine learning models to predict postoperative spinal cord function in CSM, and finds out that the optimal radiomic model is better than the optimal radiological model.
1. INTRODUCTION
Cervical spondylotic myelopathy (CSM), an age‐related degenerative disease that is common worldwide, 1 is mainly caused by the compression of the spinal cord and may possibly lead to disability. 2 , 3 , 4 Surgery to reduce direct compression of the spinal cord might alleviate disease progression 1 ; however, due to individual differences, some patients do not benefit from surgery. 5 Prognostic prediction is important because it affects subsequent treatment decision making. Currently, prognosis is generally based on magnetic resonance imaging (MRI) with a detailed macrostructural evaluation of the spinal cord. 6 Unfortunately, the use of conventional MRI indicators (eg, increased intensity signal [ISI]) to predict CSM outcomes has been controversial 5 , 7 because of their subjectivity or the insufficient information contained therein.
Radiomics, which makes full use of medical images with objective measurements, has contributed greatly to the study of predictive models. 8 , 9 Machine learning (ML) effectively utilities radiomic features and has the potential to build effective and reliable models. Recent studies revealed that radiomics and corresponding models demonstrated advantages in multiple tumor diseases, 10 , 11 , 12 however, their application in nontumor diseases are still in the initial stage. To date, radiomic studies in CSM are still lacking.
In this article, we constructed radiological and radiomic models based on classifiers with/without feature reduction methods and validated, tested, and compared these models. We aimed to utilize preoperative MRI and identify the optimal model to predictive postsurgical spinal cord function in CSM patients.
2. MATERIALS AND METHODS
2.1. Patients
The study design was approved by the appropriate ethics review board, which waived the requirement of informed consent due to the retrospective nature of the study. A total of 151 patients (99 men and 52 women) who underwent surgical treatment in our hospital from January 2017 to June 2017 were included in our study. The inclusion criteria were (a) diagnosed with CSM and operated on by a specific team which was led by one senior orthopedist; (b) available preoperative MRI results; (c) high‐quality image data with no motion artifacts; and (d) preoperative and long‐term follow‐up (≥3 years) modified Japanese Orthopedic Association (mJOA) score. The exclusion criteria were as follows: (a) prior head or neck surgery; and (b) a history of notable additional diseases (spinal cord tumor, multiple sclerosis, syringomyelia, spinal cord injury, or motor neuron disease). Clinical data collected included age, sex, symptom duration, and surgery. Neurological impairment was measured using mJOA. Participants were classified into a poor outcome group (postoperative mJOA score < 16) and a good outcome group (postoperative mJOA score ≥ 16), as patients with a postoperative mJOA score less than 16 still have severe residual deficits. 5 , 13
2.2. MRI methods
MR scans were performed with 3 T MR (GE Healthcare, Waukesha Wisconsin and Siemens Medical Solutions, Erlangen, Germany) and 1.5 T MR (GE Healthcare, Waukesha, Wisconsin) scanners with patients in the head‐first supine position. The parameters are shown in Table S1.
The dataset obtained from one scanner (n = 41) was regarded as the testing cohort, and the dataset obtained from the rest scanners (n = 110) was regarded as the training cohort.
2.3. Radiologic evaluation
ISI in T2 *WI was classified into four types 14 : type 0, no ISI; type 1, ISI with a diffuse boundary (≥2/3 spinal cord); type 2: ISI with a diffuse boundary (<2/3 spinal cord); and type 3: ISI with a distinct boundary (<2/3 spinal cord). Two radiologists classified ISI on axial images independently under the supervision of a senior radiologist without knowledge of preoperative and postoperative neurological function, and disagreements were discussed until a consensus was reached. The compression ratio (CR) was computed automatically with the help of spinal cord toolbox (SCT) to describe the severity of the compression of the spinal cord, 15 , 16 , 17 defined as follows:
The details of the automatic computation are described in the following section. The slice with the lowest CR, which indicated the most severe compression, 15 , 16 , 17 over the whole spine was chosen as the maximum compressed level (MCL).
2.4. Image preprocessing
To limit the differences among images, we applied a standardized MRI preprocessing pipeline. 18 First, resample the images to ensure the same resolution. Second, pre‐crop images around the centerline of the spinal cord were taken to ensure the same size. Third, the intensity of images was normalized to ensure that the intensity of the same tissue was consistent. Fourth, the 2D image at the MCL was selected, and the Z‐score was used to standardize the image. Steps 1 to 3 were achieved by SCT (Version 4.0.0; https://github.com/neuropoly/spinalcordtoolbox) 19 (Figure 1). Z‐score standardization was achieved by setting parameters in PyRadiomics (Version 3.0, git://github.com/Radiomics/pyradiomics) 20 when extracting radiomic features.
FIGURE 1.
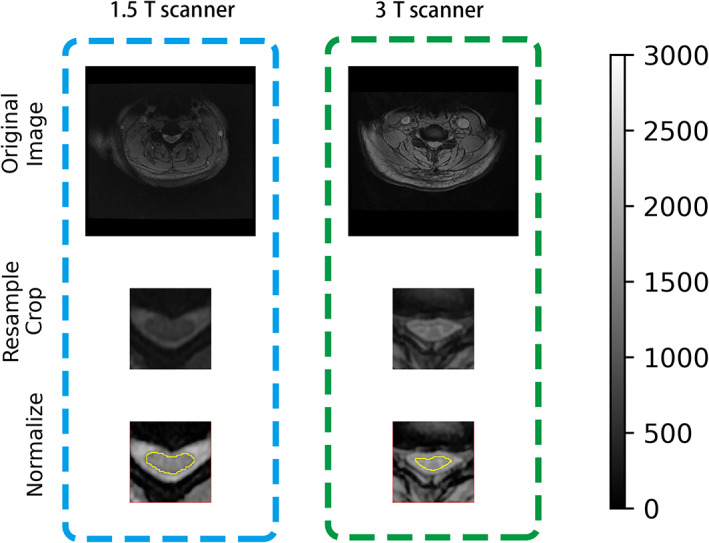
Image preprocessing pipeline. The left (right) column represents images collected from a 1.5 T scanner (3 T scanner). After resampling, cropping, and intensity normalization, images were comparable across scanners. Corresponding automatic segmentation of the spinal cord (yellow line) is shown
2.5. Radiomics: Segmentation
The spinal cord in T2 *WI was segmented automatically by SCT and then manually corrected by two independent radiologists supervised by a senior radiologist. Based on two radiologists' segmentations, the dice coefficient score (DCS, a measurement of similarity) was (median [IQR, interquartile range]) 0.93 (0.90‐0.95), as were the intercorrelation coefficients (ICCs) of CR. For each patient, the slice with the minimum average CR was referred to as the MCL. Under the supervision of a senior radiologist, two independent radiologists checked the selection of MCL and finally confirmed the location of MCL.
2.6. Radiomics: Region of interest
The region of interest (ROI), whose border is widely accepted for radiomic analysis, should include sufficient pixels, refined and effective information, and repeated segmentation; this is the case for the area covered by the spinal cord at the MCL on axial T2 *WI. 21 , 22 , 23 Therefore, we referred to this area as the ROI. In contrast, it is difficult to find an accurate, repeatable, and widely acceptable region on sagittal images, which is why we focused our study on T2 *WI.
2.7. Radiomics: Feature extraction
For the preprocessed T2 *WI at the MCL, three class features (shape, first‐order statistics, and textures [e.g., gray level co‐occurrence matrix (GLCM), gray level size zone matrix (GLSZM), gray level run length matrix (GLRLM), neighboring gray‐tone difference matrix (NGTDM), and gray level dependence matrix (GLDM)]) were extracted from the ROI with/without seven built‐in suitable filters (wavelet, square, square root, logarithm, exponential, gradient, and local binary pattern) for a small ROI, resulting in 1032 features. Related details are available online (https://pyradiomics.readthedocs.io/en/latest/index.html). Excellent robust features (ICC ≥0.90 between the two radiologists' segmentations) were enrolled for subsequent analysis.
2.8. Machine learning
Machine learning (ML) is defined as programming computers to optimize a performance criterion based on previous experience or example dataset. 24 ML is recommended to handle the high‐dimensional data provided by radiomics. 1 To apply ML in radiomic studies, a general pipeline is widely accepted, including feature reduction, modeling methodology, and evaluation. 12 Essential information is kept after feature reduction and serves as input to build a model; finally, the application value of the model is assessed. 12 For excellent robust radiomic features, 14 widely applied built‐in binary classifiers combined with 7 compatible feature reduction methods, totaling 98 radiomic models, were constructed by auto‐sklearn (a Python package that can preprocess the input dataset, reduce the number of features, construct and validate the ML model automatically, Version 0.12.0; https://github.com/automl/autosklearn). The same classifiers were applied for the radiological features, including CR and the types of ISI, resulting in 14 radiological models. A list of the methods and their abbreviations are presented in Table 1. Five‐fold cross‐validation (CV) is a method to effectively use the small sample data. It randomizes the training set into 5‐folds and uses 4‐folds for training and the rest for validation, then repeated five times. CV was applied to train, and validate models. The testing dataset was kept unseen during the whole procedure. The pipeline of radiomic analysis is shown in Figure 2.
TABLE 1.
Abbreviations for the feature reduction methods and classifiers
| Feature Preprocessor | Classifiers | ||
|---|---|---|---|
| Select percentile | DT | Decision tree | |
| Select rates | RF | Random forest | |
| Linear SVM preprocessor | Linear support vector machine preprocessor | ET | Extra trees |
| ET preprocessor | Extra trees preprocessor | Adaboost | Adaptive boosting |
| Fast ICA | Fast independent component analysis | GBDT | Gradient boosting decision trees |
| FA | Feature agglomeration | BNB | Bernoulli naïve Bayes |
| PCA | Principal component analysis | GNB | Gaussian naïve Bayes |
| PA | Passive aggressive | ||
| QDA | Quadratic discriminant analysis | ||
| LDA | Linear discriminant analysis | ||
| Linear SVM | Linear support vector machine | ||
| SVM | Support vector machine | ||
| KNN | K‐nearest neighbors | ||
| SGD | Stochastic gradient descent | ||
FIGURE 2.
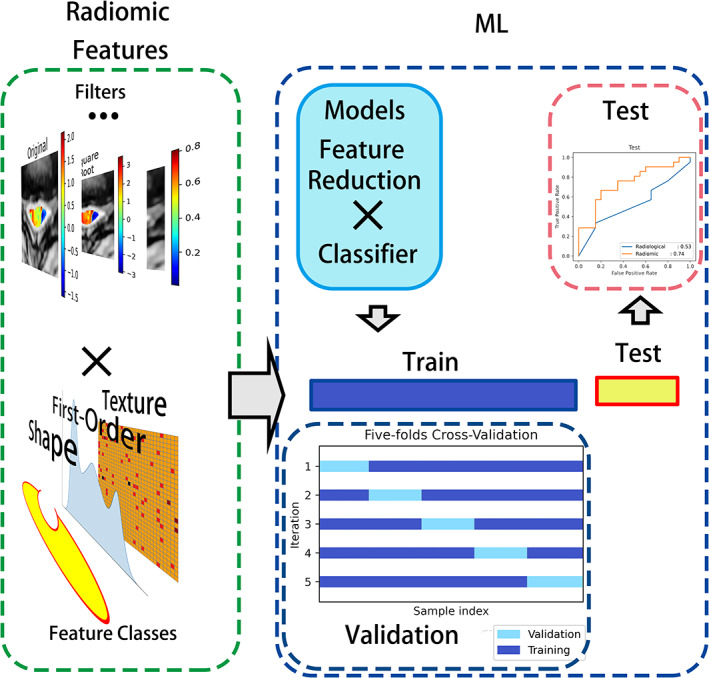
Radiomics analysis pipeline. Radiomic features were extracted from the spinal cord at the MCL of preprocessed images with or without filters. Feature reduction methods combined with binary classifiers resulted in ML models. Models were trained and cross validated on the training dataset and tested on the testing dataset. ML, machine learning; MCL, maximum compression level
2.9. Model evaluation
The area under the receiver operating characteristic (AUROC) and accuracy, widely used overall indicators in medicine and computer science, were regarded as the main evaluation index in our study. A random guessing model whose AUROC (accuracy) equals 0.5 (no information rate [NIR], denoting the best guessing given no information beyond the overall distribution of binary classes), was referred to as the baseline. We arbitrarily subdivided model performance into three groups based on their AUROC and accuracy compared with the random guessing model: (1) high potential clinical application value with significantly increased AUROC and accuracy (P AUROC < .05, P accuracy < .05); (2) low potential clinical application value with significantly increased AUROC or accuracy ([P AUROC < .05, P accuracy > .05] or [P AUROC > .05, P accuracy < .05]); and (3) no potential clinical application value with comparable or decreased AUROC and accuracy. If the grading of performance during CV and testing showed disagreement, the performance on the testing cohort was considered, as the performance on this cohort is more meaningful.
2.10. Model selection and comparison
For overall radiomic or radiological models, the Technique for Order Preference by Similarity to Ideal Solution (TOPSIS), a multiple criteria decision‐making method, was used by the package PyTOPS in Python (Version 0.1; http://home.iitb.ac.in/~skarmakar/index.html). This method determines the best solution from a set of alternatives with certain attributes. The best alternative is chosen based on its Euclidean distance from the ideal solution. We used PyTOPS to select the best radiomic or radiological model based on the AUROC and accuracy. The AUROC, accuracy, corresponding standard deviation (SD), and relative SD (RSD, the ratio of SD to the mean value), as well as the sensitivity, specificity, precision and F1 score, were measured and compared. The SDs were measured by a proportion test based on the binomial distribution or bootstrapping 1000 times.
2.11. Statistical analysis
2.11.1. Clinical factors and radiological evaluation
For normally (nonnormally) distributed continuous variables, we applied Student's t‐test (Mann‐Whitney U test). To compare categorical variables, we used the chi‐square test.
2.11.2. Model comparison
AUROCs (accuracy, sensitivity, specificity, precision) were compared by the DeLong test (paired or unpaired proportion test).
2.11.3. Analysis software
Overall, the above statistical analyses were performed using Python modules (SciPy, Version 1.15.0, https://www.scipy.org; Statsmodels, Version 0.13.0, https://www.statsmodels.org; Mlxtend, Version 0.18.0, https://rasbt.github.io/mlxtend/) and R language (pROC, Version 1.17.0.1, https://cran.r-project.org/web/packages/pROC/index.html; caret, Version 6.0.88, https://cran.r-project.org/web/packages/caret/vignettes/caret.html). RSDAUROC and RSDaccuracy were compared by the Forkman J method (MedCalc, Version 0.20.3, MedCalc Software Ltd, Belgium).
3. RESULTS
3.1. Patient characteristics and conventional MRI features
Images from 80 patients with good outcomes and 71 patients with poor outcomes, comprising a total of 151 subjects, were divided into the training and testing cohorts. No significant differences in clinical factors or radiological factors were observed between the training and testing groups (P > .05) (Table 2).
TABLE 2.
Clinical and radiological factors of 151 subjects
| Train (n = 110) | Test (n = 41) | P | |
|---|---|---|---|
| Clinical factors | |||
| Age (years) a | 54.1 ± 10.6 | 56.5 ± 8.1 | .194 |
| Sex (F/M) | 37/73 | 15/26 | .883 |
| Symptom duration (months) b | 12.0 (3.3‐37.2) | 12.0 (6.0‐48.0) | .359 |
| Preoperative mJOAa | 13.5 ± 2.0 | 13.2 ± 2.1 | .435 |
| Operation (anterior/posterior) | 65/45 | 24/17 | .901 |
| Outcome (good/poor) | 60/50 | 20/21 | .654 |
| Radiological factors | |||
| CRa | 0.37 ± 0.08 | 0.38 ± 0.10 | .859 |
| ISI | .054 | ||
| Type 0 | 23 | 6 | |
| Type 1 | 20 | 8 | |
| Type 2 | 58 | 17 | |
| Type 3 | 9 | 10 |
Abbreviations: CR, compression ratio; IQR, interquartile range; ISI, increased signal intensity; mJOA, modified Japanese Orthopedic Association score.
Normally distributed continuous variables (mean ± SD) were statistically analyzed by Student's t‐test.
Nonnormally distributed continuous variables (median [IQR]) were statistically analyzed by the Mann‐Whitney U test.
3.2. Model application value: Models vs random guessing
3.2.1. Radiological model
Radiological models yielded AUROCs and accuracy ranges of 0.51 to 0.61 and 0.51 to 0.58 (0.36‐0.53 and 0.49‐0.59) during CV (testing). The SVM revealed potential clinical application value during CV (P AUROC = .049, P accuracy = .255) (Figure 3 and Figure S1). However, no radiological models showed potential clinical application value in the testing cohort (Figure 4 and Figure S2).
FIGURE 3.
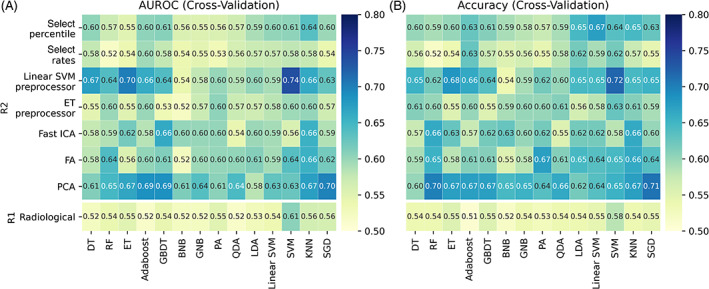
Heatmaps of AUROC and accuracy through 5‐fold CV. R1 (R2) referred radiological (radiomic) models. (A) AUROC; (B) accuracy. CV, cross‐validation; AUROC, area under the receiver operating characteristic curve
FIGURE 4.
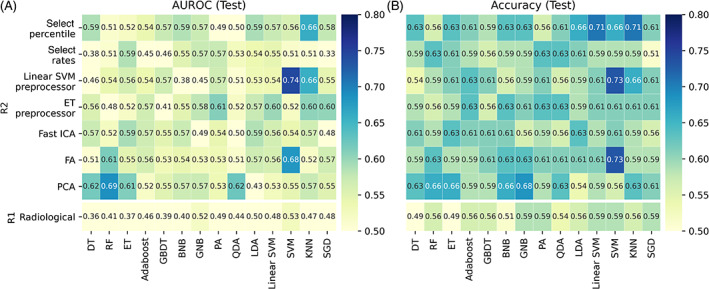
Heatmaps of AUROC and accuracy on the testing cohort. R1 (R2) referred radiological (radiomic) models. (A) ROC‐AUC; (B) accuracy. AUROC, area under the receiver operating characteristic curve
3.2.2. Radiomic model
In total, 237 excellent robust features (108 first‐order features, 9 shape features, and 120 texture features) were retained from 1032 features with ICC ≥ 0.9. During CV, 25 radiomic models demonstrated high potential clinical application value, with AUROC and accuracy ranged 0.61 to 0.74 and 0.64 to 0.72, respectively. Meanwhile, 16 radiomic models demonstrated low potential clinical application value on CV. The remaining 57 models had no potential clinical application value (Figure 3 and Figure S1). In the testing cohort, three radiomic models (linear SVM preprocessor + SVM, FA + SVM, and PCA + RF) were observed to have high potential clinical application value, with AUROCs and accuracies ranging from 0.68 to 0.74 and 0.66 to 0.73. A total of eight radiomic models had low potential clinical application value, while the remaining models had no potential clinical application value (Figure 4 and Figure S2).
3.3. Model comparison: The best radiological model vs the best radiomic model
With TOPSIS, the linear SVM preprocessor + SVM (SVM) was selected as the best radiomic (radiological) model, with an F1 score of 0.72 ± 0.08 (0.45 ± 0.11). The performance of the models is summarized in Table 3. The best radiomic model, based on 13 radiomic features ([filters] feature names), including five shape features ([no filters] pixel surface, minor axis length, maximum diameter, elongation), seven first‐order features ([wavelet‐LL, HL, and gradient] range, [wavelet‐LL] 10 percentiles, [wavelet‐HL] energy, [local binary pattern] mean, and [exponential] variance), and one texture feature ([wavelet‐LL] GLCM cluster prominence), overperformed the best radiological model, showing significantly higher AUROC, stability, and sensitivity (P AUROC = .048, P RSDAUROC = .008, P RSDAccuracy = .024, P sensitivity = .039).
TABLE 3.
Comparison between the best radiological and radiomic models in the testing cohort
| The best radiological model | The best radiomic model | P | |
|---|---|---|---|
| AUROC a | 0.53 ± 0.09 | 0.74 ± 0.08 | .048 |
| RSDAUROC b | 0.17 | 0.11 | .008 |
| Accuracy c | 0.59 ± 0.08 | 0.73 ± 0.07 | .181 |
| RSDAccuracy b | 0.13 | 0.09 | .024 |
| Sensitivity c | 0.33 ± 0.10 | 0.67 ± 0.10 | .039 |
| Specificity c | 0.85 ± 0.08 | 0.80 ± 0.09 | 1.000 |
| Precision d | 0.70 ± 0.14 | 0.78 ± 0.10 | .645 |
Abbreviations: AUROC, area under the receiver operating characteristic curve; RSD, relative SD.
AUROCs (mean ± SD) were compared by DeLong test.
RSDs were compared by Forkman J methods.
Proportion indicators (mean ± SD) were compared by paired proportion test (i.e., McNemar's test).
Proportion indicators (mean ± SD) were compared by nonpaired proportion test.
4. DISCUSSION
In our study, we utilized radiomics and advanced ML to predict the postoperative prognosis of patients with CSM, compared radiomic models to radiological models, reported the advantages of the radiomic models, and demonstrated the potential clinical application value of the linear SVM preprocessor + SVM, which was identified as the best algorithm for radiomic models.
The preprocessing of images and the choice of ROI are the cornerstones of this research. The repeatability of radiomics is an essential problem, as the signal intensity of MRI varies among scanners and scanning protocols. 25 Normalization is a common solution to increase the reproducibility of images and has been applied in a multi‐center study in the spinal cord. 18 After normalization, some radiomic models performed well cross multi‐scanners in our study, which might due to the reduced variation of signal intensity. The choice of ROI is also crucial; it is a feasible and practical way to predict the prognosis of CSM by extracting the imaging features from the spinal cord at MCL of T2 *WI rather than ISI. Although ISI is regarded as the lesion of CSM, its size is generally small. Due to the partial volume effect, the boundary is unreliable, and the radiomic features are unstable, 26 indicating that ISI is not suitable for radiomic analysis. The cross‐section of the spinal cord is an alternative, as the reproducibility and repeatability of ROI and radiomic features is increased along with the increasing size of the ROI. 26 , 27 The information contained in the ROI includes information on not only small lesions but also the influence and changes around the ISI and over the whole spinal cord section, which are meaningful and useful. 21 , 22 , 23 We have proven that the morphology, first‐order, and some texture features extracted from ROI are stable and can be used for the effective prediction of CSM.
Model selection is another important part to consider. 28 Multiple algorithms are recommended and applied in spinal diseases, 28 however, the best ML for radiomics in CSM remained unknown. In our study, we propose a protocol to select the best radiomic model. As the reproducibility of radiomic models determines their value for extensive application, the performance with the testing dataset is more meaningful. 29 In addition, various indicators, with their specific advantages and disadvantages, can be used to measure the performance of models from different aspects, but no one alone can synthesize all metrics and comprehensively measure the performance of the models. 30 , 31 , 32 Our study also confirmed that the changes in the AUROC and accuracy are not exactly consistent. The TOPSIS method, which has been applied in engineering, marketing management and so on, 33 provides an alternative method to select models based on various criteria. 34 In TOPSIS analysis, various dimensional criteria are converted into nondimensional criteria, the positive ideal solution with maximum benefits and minimum costs and the negative ideal solution with minimum benefits and maximum costs are formed, and an alternative is evaluated and selected based on its distance to these solutions. 33 , 35 Therefore, we applied a model selection method based on the AUROC and accuracy of the testing cohort and comprehensively evaluated the performance of the model. The selected model reported the highest AUROC and accuracy, consistent with our expectation.
Our work recommends linear SVM preprocessor + SVM as the best algorithms for radiomics in CSM. ML makes it possible to handle complex and numerous data, however, the optimal ML for radiomics is under debated 36 , 37 and the selection of ML depends on researchers' preference. 11 , 12 Although tree‐based models (eg, RF and Adaboost) combined with feature reduction were reported to be more preferable, 8 , 38 the SVM combined with the linear SVM preprocessor was observed to be the best in our study. The core reason is the nonlinear nature of medical problems; along with nonlinear kernels, which could transform linear input into nonlinear input, SVM has been reported to have the ability to solve nonlinear problems, 8 , 39 , 40 similar to tree‐based models. As suggested by Gu et al, radiomic models based on SVM with nonlinear kernels performed better than the one with a linear kernel. 41 Additionally, SVM has special advantage for the small‐size samples. 42
In conclusion, we utilized radiomics to predict CSM prognosis using numerous ML methods, validated and tested the models, and identified the optimal model, namely, the linear SVM preprocessor + SVM, which was superior to radiological models. We acknowledge that there are still some limitations to this study. Our radiomic models were trained on limited sample from a single center. Normalization and an independent testing dataset were applied to enhance models' applicability. The method can be used in future multi‐center data collection when standardization is needed. Meanwhile, we conservatively used the spinal cord on axial T2 *WI but not sagittal T2WI as the ROI in radiomic analysis. Compared with the sagittal spinal cord, the axial spinal cord at the MCL is a widely recognized ROI with better gray matter contrast, including a high intramedullary signal and the potential to provide even more information. 21 , 22 , 23 Referring to it as the ROI can enhance the repeatability and reliability of radiomics. Our models based on multiple scanners suggested the credibility of this ROI and the ability of the models to be extensively used, thereby providing a foundation for further prospective multi‐center studies. Additionally, the comparisons performed in this study offer a potential reference for the development of new models that may be useful for other radiomics studies.
5. CONCLUSION
Radiomics has high potential application value for the preoperative prediction of CSM outcomes. The optimal model, the linear SVM preprocessor + SVM, provides an alternative approach for physicians to use in their clinical practice.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Figure S1 Comparison of performance between constructed models and the random guessing model through 5‐fold CV. R1 (R2) referred radiological (radiomic) models. (a) P values of the difference in AUROCs between the models and the random guessing model; (b) P values of the difference in accuracy between the models and the random guessing model. CV, cross‐validation; AUROC, area under the receiver operating characteristic curve
Figure S2. Comparison of performance between constructed models and the random guessing model on the testing cohort. R1 (R2) referred radiological (radiomic) models. (a) P values of the difference in AUROCs between the models and the random guessing model; (b) P values of the difference in accuracy between the models and the random guessing model. AUROC, area under the receiver operating characteristic curve
Table S1 Parameters of the scanners
ACKNOWLEDGMENTS
We appreciate the support from Jing‐Jing Cui and Guang‐Ming Zhang and their helpful discussions on machine learning models. Nan Li also provided useful suggestions on the analytical methods. The National Multidisciplinary Cooperative Diagnosis and Treatment Capacity Building Project for Major Diseases, Peking University Third Hospital's Research, Innovation and Transformation Fund (BYSYZHKC2020116), Key Clinical Projects of Peking University Third Hospital (BYSY2018003), Beijing Natural Science Foundation (7204327, Z190020), Capital's Funds for Health Improvement and Research (2020‐4‐40916), Clinical Medicine Plus X‐Young Scholars Project, Peking University, the Fundamental Research Funds for the Central Universities (PKU2021LCXQ005), and National Natural Science Foundation of China (82102638) supported this study.
Zhang, M.‐Z. , Ou‐Yang, H.‐Q. , Jiang, L. , Wang, C.‐J. , Liu, J.‐F. , Jin, D. , Ni, M. , Liu, X.‐G. , Lang, N. , & Yuan, H.‐S. (2021). Optimal machine learning methods for radiomic prediction models: Clinical application for preoperative T2 *‐weighted images of cervical spondylotic myelopathy. JOR Spine, 4(4), e1178. 10.1002/jsp2.1178
Meng‐Ze Zhang and Han‐Qiang Ou‐Yang contributed equally to this work.
Funding information The National Multidisciplinary Cooperative Diagnosis and Treatment Capacity Building Project for Major Diseases; Key Clinical Projects of Peking University Third Hospital, Grant/Award Number: BYSY2018003; Peking University Third Hospital's Research, Innovation and Transformation Fund, Grant/Award Number: BYSYZHKC2020116; Beijing Natural Science Foundation, Grant/Award Numbers: 7204327, Z190020; Capital's Funds for Health Improvement and Research, Grant/Award Number: 2020‐4‐40916; Clinical Medicine Plus X‐Young Scholars Project, Peking University, The Fundamental Research Funds for the Central Universities, Grant/Award Number: PKU2021LCXQ005; National Natural Science Foundation of China, Grant/Award Number: 82102638
Contributor Information
Liang Jiang, Email: jiangliang@bjmu.edu.cn.
Hui‐Shu Yuan, Email: huishuy@bjmu.edu.cn.
REFERENCES
- 1. Toledano M, Bartleson JD. Cervical spondylotic myelopathy. Neurol Clin. 2013;31:287‐305. [DOI] [PubMed] [Google Scholar]
- 2. Tetreault L, Goldstein CL, Arnold P, et al. Degenerative cervical myelopathy: a spectrum of related disorders affecting the aging spine. Neurosurgery. 2015;77:S51‐S67. [DOI] [PubMed] [Google Scholar]
- 3. Iyer A, Azad TD, Tharin S. Cervical spondylotic myelopathy. Clin Spine Surg. 2016;29:408‐414. [DOI] [PubMed] [Google Scholar]
- 4. Bakhsheshian J, Mehta VA, Liu JC. Current diagnosis and management of cervical spondylotic myelopathy. Global Spine J. 2017;7:572‐586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nouri A, Tetreault L, Cote P, Zamorano JJ, Dalzell K, Fehlings MG. Does magnetic resonance imaging improve the predictive performance of a validated clinical prediction rule developed to evaluate surgical outcome in patients with degenerative cervical myelopathy? Spine. 2015;40:1092‐1100. [DOI] [PubMed] [Google Scholar]
- 6. Chatley A, Kumar R, Jain VK, Behari S, Sahu RN. Effect of spinal cord signal intensity changes on clinical outcome after surgery for cervical spondylotic myelopathy. J Neurosurg Spine. 2009;11:562‐567. [DOI] [PubMed] [Google Scholar]
- 7. Karpova A, Arun R, Davis AM, et al. Predictors of surgical outcome in cervical spondylotic myelopathy. Spine. 2013;38:392‐400. [DOI] [PubMed] [Google Scholar]
- 8. Du D, Feng H, Lv W, et al. Machine learning methods for optimal radiomics‐based differentiation between recurrence and inflammation: application to nasopharyngeal carcinoma post‐therapy PET/CT images. Mol Imaging Biol. 2020;22:730‐738. [DOI] [PubMed] [Google Scholar]
- 9. Yin P, Mao N, Zhao C, et al. Comparison of radiomics machine‐learning classifiers and feature selection for differentiation of sacral chordoma and sacral giant cell tumour based on 3D computed tomography features. Eur Radiol. 2019;29:1841‐1847. [DOI] [PubMed] [Google Scholar]
- 10. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563‐577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kumar V, Gu Y, Basu S, et al. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30:1234‐1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749‐762. [DOI] [PubMed] [Google Scholar]
- 13. Tetreault LA, Kopjar B, Vaccaro A, et al. A clinical prediction model to determine outcomes in patients with cervical spondylotic myelopathy undergoing surgical treatment: data from the prospective, multi‐center AOSpine North America study. J Bone Joint Surg Am. 2013;95:1659‐1666. [DOI] [PubMed] [Google Scholar]
- 14. You JY, Lee JW, Lee E, Lee GY, Yeom JS, Kang HS. MR classification system based on axial images for cervical compressive myelopathy. Radiology. 2015;276:553‐561. [DOI] [PubMed] [Google Scholar]
- 15. Ogino H, Tada K, Okada K, et al. Canal diameter, anteroposterior compression ratio, and spondylotic myelopathy of the cervical spine. Spine (Phila Pa 1976). 1983;8:1‐15. [DOI] [PubMed] [Google Scholar]
- 16. Bucciero A, Vizioli L, Tedeschi G. Cord diameters and their significance in prognostication and decisions about management of cervical spondylotic myelopathy. J Neurosurg Sci. 1993;37:223‐228. [PubMed] [Google Scholar]
- 17. Tsurumi T, Goto N, Shibata M, Goto J, Kamiyama A. A morphological comparison of cervical spondylotic myelopathy: MRI and dissection findings. Okajimas Folia Anat Jpn. 2005;81:119‐122. [DOI] [PubMed] [Google Scholar]
- 18. Gros C, De Leener B, Badji A, et al. Automatic segmentation of the spinal cord and intramedullary multiple sclerosis lesions with convolutional neural networks. Neuroimage. 2019;184:901‐915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De Leener B, Levy S, Dupont SM, et al. SCT: spinal cord toolbox, an open‐source software for processing spinal cord MRI data. Neuroimage. 2017;145:24‐43. [DOI] [PubMed] [Google Scholar]
- 20. van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational Radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104‐e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wen CY, Cui JL, Liu HS, et al. Is diffusion anisotropy a biomarker for disease severity and surgical prognosis of cervical spondylotic myelopathy? Radiology. 2014;270:197‐204. [DOI] [PubMed] [Google Scholar]
- 22. Ellingson BM, Salamon N, Grinstead JW, Holly LT. Diffusion tensor imaging predicts functional impairment in mild‐to‐moderate cervical spondylotic myelopathy. Spine J. 2014;14:2589‐2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang W, Han X, Guo H, et al. Usefulness of conventional magnetic resonance imaging, diffusion tensor imaging and neurite orientation dispersion and density imaging in evaluating postoperative function in patients with cervical spondylotic myelopathy. J Orthop Translat. 2018;15:59‐69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. El Naqa I, Murphy MJ. What is machine learning? In: El Naqa I, Li R, Murphy MJ, eds. Machine Learning in Radiation Oncology. Cham: Springer; 2015:3‐11. [Google Scholar]
- 25. Scalco E, Belfatto A, Mastropietro A, et al. T2w‐MRI signal normalization affects radiomics features reproducibility. Med Phys. 2020;47:1680‐1691. [DOI] [PubMed] [Google Scholar]
- 26. Yip SS, Aerts HJ. Applications and limitations of radiomics. Phys Med Biol. 2016;61:R150‐R166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li Q, Bai H, Chen Y, et al. A fully‐automatic multiparametric Radiomics model: towards reproducible and prognostic imaging signature for prediction of overall survival in Glioblastoma Multiforme. Sci Rep. 2017;7:14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Azimi P, Yazdanian T, Benzel EC, et al. A review on the use of artificial intelligence in spinal diseases. Asian Spine J. 2020;14:543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kocak B, Durmaz ES, Erdim C, Ates E, Kaya OK, Kilickesmez O. Radiomics of renal masses: systematic review of reproducibility and validation strategies. AJR Am J Roentgenol. 2020;214:129‐136. [DOI] [PubMed] [Google Scholar]
- 30. Lobo JM, Jiménez‐Valverde A, Real R. AUC: a misleading measure of the performance of predictive distribution models. Glob Ecol Biogeogr. 2008;17:145‐151. [Google Scholar]
- 31. Byrne S. A note on the use of empirical AUC for evaluating probabilistic forecasts. Electron J Statist. 2016;10:380‐393. [Google Scholar]
- 32. Cook NR. Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation. 2007;115:928‐935. [DOI] [PubMed] [Google Scholar]
- 33. Behzadian M, Otaghsara SK, Yazdani M, Ignatius J. Review: a state‐of the‐art survey of TOPSIS applications. Expert Syst Appl. 2012;39:13051‐13069. [Google Scholar]
- 34. Vazquezl MYL, Peñafiel LAB, Muñoz SXS, Martinez MAQ. A framework for selecting machine learning models using TOPSIS. In: Ahram T, ed. Advances in Artificial Intelligence, Software and Systems Engineering. Cham: Springer International Publishing; 2021:119‐126. [Google Scholar]
- 35. Aruldoss M, Lakshmi TM, Venkatesan VP. A survey on multi criteria decision making methods and its applications. Am J Inf Syst. 2013;1:31‐43. [Google Scholar]
- 36. Zhang B, He X, Ouyang F, et al. Radiomic machine‐learning classifiers for prognostic biomarkers of advanced nasopharyngeal carcinoma. Cancer Lett. 2017;403:21‐27. [DOI] [PubMed] [Google Scholar]
- 37. Duan C, Liu F, Gao S, et al. Comparison of Radiomic models based on different machine learning methods for predicting Intracerebral hemorrhage expansion. Clin Neuroradiol. 2021. [DOI] [PubMed] [Google Scholar]
- 38. Zhang Y, Zhang B, Liang F, et al. Radiomics features on non‐contrast‐enhanced CT scan can precisely classify AVM‐related hematomas from other spontaneous intraparenchymal hematoma types. Eur Radiol. 2019;29:2157‐2165. [DOI] [PubMed] [Google Scholar]
- 39. Fan RE, Chang KW, Hsieh CJ, Wang XR, Lin CJ. LIBLINEAR: a library for large linear classification. J Mach Learn Res. 2008;9:1871‐1874. [Google Scholar]
- 40. Chang CC, Lin CJ. LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol. 2011;2:1‐27. [Google Scholar]
- 41. Gu J, Zhu J, Qiu Q, Wang Y, Bai T, Yin Y. Prediction of immunohistochemistry of suspected thyroid nodules by use of machine learning‐based Radiomics. AJR Am J Roentgenol. 2019;213:1348‐1357. [DOI] [PubMed] [Google Scholar]
- 42. Chen J, Yin Y, Han L, Zhao F. Optimization approaches for parameters of SVM. Proceedings of the 11th International Conference on Modelling, Identification and Control (ICMIC2019). Cham: Springer; 2020:575‐583. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Comparison of performance between constructed models and the random guessing model through 5‐fold CV. R1 (R2) referred radiological (radiomic) models. (a) P values of the difference in AUROCs between the models and the random guessing model; (b) P values of the difference in accuracy between the models and the random guessing model. CV, cross‐validation; AUROC, area under the receiver operating characteristic curve
Figure S2. Comparison of performance between constructed models and the random guessing model on the testing cohort. R1 (R2) referred radiological (radiomic) models. (a) P values of the difference in AUROCs between the models and the random guessing model; (b) P values of the difference in accuracy between the models and the random guessing model. AUROC, area under the receiver operating characteristic curve
Table S1 Parameters of the scanners


