Abstract
Trichomonas vaginalis, the causative agent for human trichomoniasis, is a problematic sexually transmitted disease mainly in women, where it may be asymptomatic or cause severe vaginitis and cervicitis. Despite its high prevalence, the genetic variability and drug resistance characteristics of this organism are poorly understood. To address these issues, genetic analyses were performed on 109 clinical isolates using three approaches. First, two internal transcribed spacer (ITS) regions flanking the 5.8S subunit of the ribosomal DNA gene were sequenced. The only variation was a point mutation at nucleotide position 66 of the ITS1 region found in 16 isolates (14.7%). Second, the presence of a 5.5-kb double-stranded RNA T. vaginalis virus (TVV) was assessed. TVV was detected in 55 isolates (50%). Finally, a phylogenetic analysis was performed based on random amplified polymorphic DNA data. The resulting phylogeny indicated at least two distinct lineages that correlate with the presence of TVV. A band-sharing index indicating relatedness was created for different groups of isolates. It demonstrated that isolates harboring the virus are significantly more closely related to each other than to the rest of the population, and it indicated a high level of relatedness among isolates with in vitro metronidazole resistance. This finding is consistent with the hypothesis that drug resistance to T. vaginalis resulted from a single or very few mutational events. Permutation tests and nonparametric analyses showed associations between metronidazole resistance and phylogeny, the ITS mutation, and TVV presence. These results suggest the existence of genetic markers with clinical implications for T. vaginalis infections.
Trichomonas vaginalis is a protozoan parasite that infects the human urogenital tract and is the causative agent of trichomoniasis, the most common nonviral sexually transmitted disease (19). Infected individuals manifest a wide range of symptoms, from an asymptomatic presentation to severe sequelae. T. vaginalis infections have also been related to complications during pregnancy, including premature rupture of membranes (16), low birth weight, and preterm labor (2). Another serious aspect of trichomoniasis is the association between T. vaginalis infection and an increased risk of transmission of other sexually transmitted diseases, including human immunodeficiency virus (10).
Despite estimates of more than 170 million T. vaginalis infections per year worldwide (27), very little is known about the degree of T. vaginalis strain variation in patient isolates and how these characteristics impact the clinical manifestation of T. vaginalis infections. Most analyses of isolate differences in T. vaginalis have been limited to phenotypic observations, such as the level of drug resistance in vitro. The few studies of genotypic variations have been limited to small numbers of isolates, and most of the studies do not consider clinical phenotypes. Broad studies of genetic polymorphisms can lead to the distinction between strains with different phenotypes and could be useful to link problematic infections to specific treatment strategies.
In one of the few studies involving genetic polymorphisms and clinical phenotypes, Vanacova et al. (22) compared random amplified polymorphic DNA (RAPD) data from 18 isolates of T. vaginalis to determine if this technique would be useful for strain typing of clinical isolates. They found the RAPD data correlated with in vivo and in vitro metronidazole resistance, geographic origin, and clinical presentation. Their results suggest that RAPDs are powerful markers to analyze genetic diversity in T. vaginalis and that study of the T. vaginalis genome could help identify strains that are associated with clinically significant data.
Ribosomal gene sequencing is another method commonly used to find genetic polymorphisms between organisms. These genes are useful primarily because of their conserved, ubiquitous nature. However, internal transcribed spacer (ITS) regions are often more useful in studies that involve closely related species. These regions exist between the ribosomal genes and are found in all eukaryotic organisms, but because they form no part of the functional ribosomal structure, they are much less conserved than the actual genes. Even closely related species can have ITS regions that vary widely in both composition and length; therefore, ITS regions are ideal candidates for intraspecies and intragenus comparisons (6).
An additional characteristic of T. vaginalis that may be helpful in determining strain types or genetic relatedness is the presence of the Trichomonas vaginalis virus (TVV). TVV was first identified in 1985 by Wang and Wang (23). It has no known effect on the parasite but is found in approximately 50% of clinical isolates (25). The genome of TVV consists of a 5.5-kb double-stranded RNA. Many other early diverging eukaryotes, such as Giardia lamblia (15), Babesia spp. (7), Leishmania spp. (26), and Phytomonas spp. (13), also harbor similar double-stranded RNA viruses. TVV has no known lytic cycle, and attempts to infect isolates that are uninfected have been unsuccessful (24). Therefore, it is probable that the virus is acquired exclusively through vertical transmission, making its presence or absence a useful genetic marker.
One of the most significant clinical traits of T. vaginalis is resistance to metronidazole, the only drug currently licensed for its treatment in the United States. Clinical resistance to metronidazole is associated with a patient's failure to clear infection following treatment with more than one standard course of metronidazole. In the majority of cases, isolates from these patients will also demonstrate metronidazole resistance in vitro under aerobic conditions (9, 17; E. M. Narcisi, unpublished data).
This study examined the existence of metronidazole resistance as it relates to the different genotypes found in a large population of T. vaginalis isolates. The in vitro metronidazole resistance level was determined for each isolate. Differences in molecular characteristics were evaluated by assaying each isolate for TVV, sequencing the rRNA ITS region, and performing RAPD analysis. Statistical and phylogenetic analyses were performed to determine the relationship of these characteristics in this large population of isolates.
MATERIALS AND METHODS
Isolates.
Isolates used in this study were obtained from three different sources. The first group of isolates was obtained from a single large urban teaching hospital (ATL isolates; n = 67). A vaginal swab from every woman who was admitted to the women's urgent care center from March to May 1997 was cultured in TYM media (3) with 10% heat-inactivated fetal bovine serum (pH 6.0). Cultures were tested for T. vaginalis by examination of a wet mount preparation. Patients were informed of the study, and clinical data were collected on consenting patients whose cultures were wet mount positive. The second group of isolates came from cultures sent to the Centers for Disease Control and Prevention for the metronidazole susceptibility testing service (MSA isolates; n = 34). These isolates were collected between 1995 and 1998 by U.S. gynecologists from patients who did not respond to at least two courses of standard metronidazole treatment. The isolates were cryopreserved after no more than 30 days in culture. An additional eight isolates that are commonly used in T. vaginalis studies and have well-defined characteristics were also included in the study as reference isolates. Personal identifiers were removed and the source of isolates was kept anonymous to protect patient confidentiality.
Isolation of DNA.
Isolates of T. vaginalis were grown in 50 ml of TYM media for 2 to 4 days until the culture populations were in the log phase of growth. Cells for DNA extraction were obtained by centrifugation of the 50-ml cultures at 430 × g for 20 min at 4°C. The resulting pellets were washed once with phosphate-buffered saline (pH 7.4) and centrifuged again at 1,730 × g for 20 min at 4°C. Total nucleic acid was extracted with the Isoquick nucleic acid extraction kit (ORCA Research, Inc., Bothell, Wash.) according to the manufacturer's specifications.
The extracted nucleic acids were dissolved in RNase-free water. DNA concentrations for each isolate were based on optical density readings at a wavelength of 260 nm.
Trichomonas vaginalis virus.
All isolates used in the study were screened for the presence of TVV by electrophoresis of total nucleic acid extracts on an agarose gel stained with ethidium bromide (EtBr) as described previously (23, 25). TVV migrates at approximately 5.5 kb.
Metronidazole susceptibility assay.
An in vitro metronidazole assay was performed for each isolate as originally described by Meingässner et al. (14) and modified by Narcisi and Secor (18). Briefly, cells were incubated in increasing levels of metronidazole (0.2 to 400 μg/ml) for 48 h under aerobic conditions. The minimum lethal concentration (MLC) was determined to be the lowest concentration of metronidazole at which no motile cells were found. Each assay was performed twice in triplicate for each isolate tested, and dimethyl sulfoxide was used as a drug carrier control.
PCR amplification of the ITS fragments.
The ITS region of the ribosomal DNA (rDNA) was amplified through PCR using primers TVITSF and TVITSR (ACCGCCCGTCGCTCCTACCGA and CTCCGCTTAATGAGATGCTTC, respectively), which were designed from the conserved 3′ end of the small ribosomal subunit gene and the 5′ end of the large ribosomal subunit gene (1). Reactions were performed in a total volume of 50 μl, each reaction mixture containing 200 μmol of each deoxynucleoside triphosphate, 20 pmol of each primer, 3 mM MgCl2, and 1.25 U of Taq polymerase. Then, 200 mM of DNA template was added to each reaction mixture. The reaction mixtures were overlaid with mineral oil, and amplification was carried out in a Hybaid Omnigene thermocycler (Vanguard International, Inc., Neptune, N.J.). The temperature profile consisted of a 1-min denaturation step at 94°C followed by 40 cycles at a melting temperature of 94°C for 1 min, an annealing temperature of 54°C for 1 min, and an extension temperature of 72°C for 1 min. After amplification, the PCR products were removed from mineral oil and stored at −20°C. The presence of product was confirmed after EtBr staining of a 1.5% agarose gel. The expected fragment migrated at 450 bp. Amplified products consisting of single, intense bands were then purified from PCR primers using the Wizard DNA purification system (Promega Corporation, Madison, Wis.).
Sequence of ITS fragments.
Automated sequencing was used to determine the sequence of the ITS region for each isolate. The reaction mixture consisted of 10 pmol of one primer, either TVITSF or TVITSR for forward- and reverse-strand sequencing, respectively; 8 μl of the ready reaction mixture (PE Applied Biosystems, Foster City, Calif.); and 13 μl of template. Reactions were carried out in a total reaction mixture volume of 20 μl on either a GeneAmp 9600 or a GeneAmp 2400 thermocycler (PE Applied Biosystems). Amplification consisted of 25 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. Dye terminators were then removed from the sequencing PCR products using Centri-sep columns (Princeton Separations, Inc., Adelphia, N.J.). The final sequencing was performed on the ABI 377 automated sequencer (PE Applied Biosystems) using a 4% acrylamide gel.
Random amplified polymorphic DNA.
Each isolate was analyzed with three different RAPD reactions. The three RAPD PCR mixtures contained either 10 pmol of primer OPD3 (GTCGCCGTCA), 20 pmol of primer OPD5 (TGAGCGGACA), or 10 pmol each of primer OPD1 (ACCGCGAAGG) and primer OPD2 (GGACCCAACC). The primers used were designed from Operon D kit primers (Operon Technologies, Alameda, Calif.). Each reaction mixture also contained 2.5 mM MgCl2, 250 μM of each deoxynucleoside triphosphate, 1.25 U of Taq polymerase, and 25 ng of template DNA. The samples were then overlaid with mineral oil and amplified. Amplification consisted of 5 min at 94°C followed by 40 cycles of 94°C for 1 min, 36°C for 1 min, and 72°C for 2 min. A final 15-min extension at 72°C was performed. Bands were separated on a 0.75% agarose gel for 1.5 to 2 h at 108 V. The gel was then stained in 300 ml of 1× Tris acetate-EDTA (TAE)–30 μl of EtBr for 15 min and destained in 1× TAE for 5 min. The banding pattern was determined from observation of the gel on a UV transilluminator. The presence or absence of each band was scored visually.
Statistical and epidemiological analyses.
A database consisting of the molecular and phenotypic data for each isolate was created in Epi Info 6 (Epidemiology Program Office, Centers for Disease Control and Prevention, Atlanta, Ga.). Any significant associations between characteristics, except the MLC values, were determined using the Fisher exact test. A Mann-Whitney nonparametric test was used to determine significant associations between the MLC values and other characteristics.
Phylogenetic analyses.
Phylogenetic analyses were performed using RAPD data. A database was created consisting of every isolate and its corresponding RAPD band pattern. To prevent the source of isolates from confounding the results and to allow for population size limitations of the analysis programs used, it was also necessary to create smaller databases that included a portion of the total isolates sampled. The three smaller databases used in these analyses consisted of the following groupings: the 42 non-ATL isolates (34 MSA isolates and 8 reference isolates), all 67 ATL isolates, and a subpopulation of 29 ATL isolates for which patient clinical data were collected and whose phenotypes represented those of all ATL isolates. The RAPDistance program (version 1.04; Research School of Biological Sciences, Australian National University, Canberra, Australia [http://life.anu.edu.au/molecular/software/rapd.html]) was then used to determine pairwise distances between isolates for each smaller grouping and for the entire set of isolates. For this calculation, Pearson's phi coefficient (20), an algorithm that determines genetic distance based on shared traits (RAPD bands), was used. Trees were designed using the neighbor-joining method based on pairwise distances and are unrooted.
Treept, a program created by Flegr and Zaboj (4), was used to evaluate the neighbor-joining trees and their concordance to phenotypic traits based on a permutation tail probability (PTP) test. The program was used to create a total of 5,000 trees by random permutation. For each permutated tree, the average distance between the isolates sharing the same traits was calculated. The distance between isolates with the same traits in the neighbor-joining tree was then compared to the permutated trees' average distances. The percentage of random trees with a higher concordance to the phenotype being analyzed was determined. Phenotypes were considered to be significantly concordant with a neighbor-joining tree when there was less than a 5% chance of a random tree with a higher concordance. Concordance with metronidazole resistance was tested in one of two ways. The 42 non-ATL isolates were tested by assigning each isolate with an MLC of less than 50 μg/ml a value of 1, those with borderline resistance (MLC of 50 μg/ml) a value of 2, and the resistant isolates (MLC > 50 μg/ml) a value of 3. Due to there being no highly resistant ATL strains and only a few borderline strains, the exact MLC values were used to determine the concordance of the ATL isolates. Concordance with TVV was tested by assigning a “1” to isolates with the virus and a “0” to isolates without it. The ITS C66T mutation was tested by assigning a “1” to the isolates with the mutation and a “0” to those without it.
The final analysis examined the band data to determine if isolates that share phenotypes have significantly more similar band patterns than the total set of isolates. A band-sharing index was created for the group of isolates being examined by determining a band-shared value, S, that is defined in the following formula: Sxy = 2nxy/(nx + ny) (11), where nxy is the number of bands shared by isolates x and y, and nx and ny represent the number of bands present for isolates x and y, respectively. The band-shared value is an arbitrary number assigned to each group and has no significance by itself. The significance of the band-shared index was evaluated by a PTP test that chooses a random sample, equal in size to the group being examined, from all 109 isolates. The test was performed 2,000 times. If the original band-shared value fell in the upper 95th percentile of the permutated values, then the group members were considered to be significantly more closely related to each other than to the population as a whole.
RESULTS
Prevalence of phenotypes.
Isolates with an in vitro aerobic MLC of greater than or equal to 50 μg/ml are considered resistant to metronidazole (12). Of the 67 ATL isolates, 6 (8.9%) showed at least borderline aerobic resistance in vitro. The most resistant ATL isolates had an MLC of 100 μg/ml. The MSA isolates showed a resistance prevalence of 91% (31 of 34), but six of these isolates had only borderline resistant MLCs (MLC = 50 μg/ml). The other 25 resistant MSA isolates had higher MLC values (100 μg/ml to >400 μg/ml). Of the eight non-ATL, non-MSA isolates, five had high levels of metronidazole resistance (>400 μg/ml) and the other three were susceptible.
TVV was found in 50% of the total isolates (55 of 109). There was very little difference between the frequency of positive isolates in any of the three groups. Table 1 summarizes the distribution of TVV among the three groups.
TABLE 1.
Prevalence of metronidazole resistance and T. vaginalis virus in 109 cultures of T. vaginalis
| Isolate group | Total no. of cultures | No. (%) of cultures
|
|
|---|---|---|---|
| Metronidazole resistanta | TVV present | ||
| ATL | 67 | 6 (9) | 36 (54) |
| MSA | 34 | 31 (91) | 15 (44) |
| Other | 8 | 5 (63) | 4 (50) |
| Total | 109 | 42 (39) | 55 (50) |
MLC ≥ 50 μg/ml under aerobic conditions.
Clinical data were collected on 29 patients who did not show any signs of concomitant vaginal infections. The patients were scored on a seven-point scale based on the number of symptoms present. There were no significant correlations between the virulence score and TVV, metronidazole resistance, rRNA sequence data, or genetic relatedness based on RAPD data (data not shown).
Genetic polymorphisms.
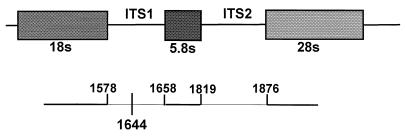
Figure 1 is a diagram of the rDNA gene of T. vaginalis. The majority of the isolates sequenced were identical to the consensus sequence previously published by Katiyar et al. (8). However, 16 of the 109 isolates (15%) had a point mutation at nucleotide position 66 of the ITS1 region in which a thymidine replaces a cytosine. The lower portion of Fig. 1 shows the location of the C66T mutation. Individually, 9% (6 of 67) of the ATL isolates, 26% (9 of 34) of the MSA isolates, and 13% (1 of 8) of the other isolates were positive for the C66T sequence. The 5.8S subunit and ITS2 region of all 109 isolates were identical to each other and to the previously published sequences (8).
FIG. 1.
Schematic representation of the rDNA gene of T. vaginalis. The rDNA gene of eukaryotic organisms is arranged in tandem repeats. The ITS regions are transcribed with the gene and then are removed before the ribosome is assembled. In the rDNA gene of T. vaginalis, ITS1 begins at nucleotide position 1578 and ends at nucleotide position 1658. A point mutation at the 66th position of the ITS1 region (nucleotide position 1644 of the rDNA gene), in which a cytosine is replaced by a thymidine, occurs in less than 15% of the isolates.
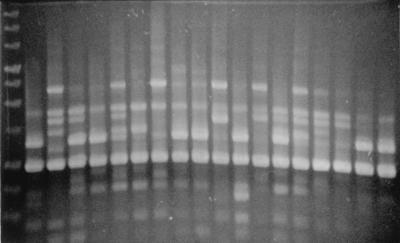
To obtain RAPD data, five different primers were tested but only two, OPD3 and OPD5, produced patterns consistent with detectable isolate differences on EtBr-stained agarose gels. The primers that did not produce patterns were combined to form multiplex primer sets. One such primer set was chosen using OPD1 and OPD2, referred to as the OPD1-2 primer set. RAPD analysis was performed on all isolates using the two primers and the primer set. For OPD3, bands at eight definitive molecular weights were scored as either present or absent for each isolate. For the primers OPD5 and OPD1-2, we used 9 and 12 bands, respectively. The total banding pattern of 29 bands was used as character data in phylogenetic analysis. Figure 2 is an example of a gel on which OPD3 PCR products were run.
FIG. 2.
Representative agarose gel with OPD3 RAPD products. At least two separate PCRs were performed for every isolate using each primer. The gel shows the banding pattern of the PCR using OPD3 for 18 isolates. The lane on the far left was loaded with a 1-kb DNA ladder (Promega).
Associations of TVV, ITS C66T mutation, and metronidazole resistance.
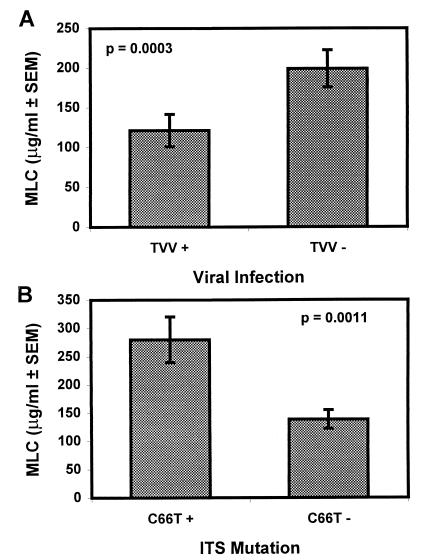
Nonparametric analysis was used to examine the total data set for relationships among phenotypes and molecular characteristics. This was done to better understand the mechanisms responsible for clinically significant phenotypes and to identify areas where future research may help to uncover better treatment or diagnostic methods. For example, as shown in Table 2, when the absence of TVV is compared with the existence of resistance to metronidazole, considered among the total population as an MLC of ≥50 μg/ml, there is a significant inverse relationship (P = 0.0145). Figure 3A shows this relationship by comparing the average MLC value among TVV-positive and TVV-negative isolates. These results indicate that isolates not harboring TVV are more likely to be resistant to metronidazole. There is also a significant inverse relationship between the absence of TVV and the presence of the rDNA ITS C66T mutation (P < 0.0001). In fact, all isolates that harbored this point mutation were negative for the virus.
TABLE 2.
Statistical relationships between phenotypes and genetic polymorphisms of 109 T. vaginalis isolates
| Characteristic | Metronidazole resistance | Absence of TVV | Presence of ITS C66T mutation |
|---|---|---|---|
| Absence of TVV | 0.0145a | ||
| Presence of ITS C66T mutation | 0.0018a | <0.0001a | |
| MLC | <0.0001b | 0.0348b | 0.0012b |
Fisher exact test.
Mann-Whitney nonparametric test.
FIG. 3.
Average MLC for isolates with different characteristics. (A) The average MLCs of isolates grouped based on the presence or absence of TVV. (B) The average MLCs of isolates grouped based on the presence or absence of the rDNA ITS C66T mutation. Isolates with MLC values of >400 μg/ml were treated as 400 μg/ml.
A Mann-Whitney nonparametric analysis indicated that isolates with the rDNA C66T ITS mutation had significantly higher MLC values than those isolates without the mutation (P = 0.0012). The difference is clearly shown in Fig. 3B, which compares the average MLC value for isolates with and without the mutation. As expected, a Fisher exact test confirmed a significant relationship between the rDNA ITS C66T mutation and the presence of metronidazole resistance (P = 0.0018).
Grouping of isolates by RAPD data.
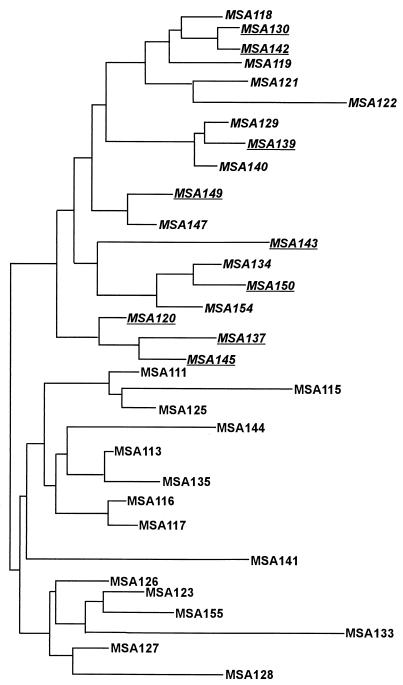
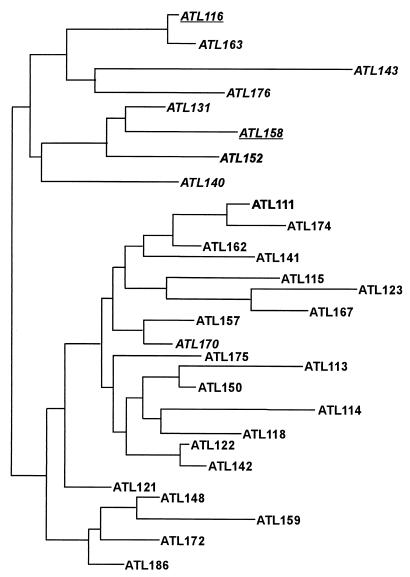
The RAPD band data were then used to characterize the isolates through phylogenetic analysis. Phylogenetic trees were built using the RAPDistance program (see above). The 34 MSA isolates fell into two major groups (Fig. 4). The upper half of the tree consists of 18 isolates, all of which are negative for TVV. Also, all of the isolates with the ITS C66T mutation are included in the upper half. The lower branches consist of 14 isolates, all of which are positive for TVV. The dendrogram shown in Fig. 5 demonstrates that the 29 ATL isolates for which we had clinical data also fell into two major groups that correlated with the presence or absence of TVV, with one exception. Isolate ATL 170 was TVV negative but grouped with the TVV-positive isolates. Dendrograms constructed from RAPD data for all 67 ATL isolates as well as the entire population of 109 isolates demonstrated the same major groupings (results not shown). These trees suggest that isolates with TVV are more closely related to other isolates with TVV than to isolates without the virus. This may be the result of a few, or possibly only one, genetic event that occurred during the evolution of T. vaginalis in which the virus was either lost or gained.
FIG. 4.
Dendrogram for MSA isolates based on RAPD data. The tree was built by the neighbor-joining method. The distance matrix was calculated with Pearson's phi coefficient based on RAPD data from three primers (29 bands). Isolates negative for TVV (italicized) all aggregated in the same (upper) branch, and TVV-positive isolates grouped together in a separate branch. All isolates with the rDNA ITS C66T mutation (underlined) also sorted into the main upper branch. Branch lengths are drawn to scale based on genetic distances between isolates.
FIG. 5.
Dendrogram for ATL isolates based on RAPD data. The tree was built by the neighbor-joining method. The distance matrix was calculated with Pearson's phi coefficient based on RAPD data from three primers (29 bands). All isolates negative for TVV (italicized), except ATL170, aggregated in one branch. All TVV-positive isolates grouped together in the main lower branch. Isolates with the rDNA ITS C66T mutation (underlined) were again associated with the TVV-negative isolates in the main upper branch. Branch lengths are drawn to scale based on genetic distances between isolates.
Concordance of traits.
The significant associations of individual traits discussed previously were further supported by additional analyses based on RAPD data using the Treept program. This program tested for concordance between the trees shown in Fig. 4 and 5 and each phenotype. The trees consisting of 67 ATL isolates and 109 total isolates were not analyzed because of sample size limitations of the Treept program. A PTP test was performed with Treept in which the order of the isolates in each tree was compared to a phenotype of each isolate. A significant association indicated that there is a relationship between the phenotype and the evolutionary assumptions that the tree makes. In other words, this test was designed to determine if a phenotype is significantly more likely to occur in groups of isolates that are more closely related. The concordance of such phenotypes with a described phylogeny can lend support to the phylogeny and can help to describe strains or lineages. The trees drawn based on RAPD data showed a highly significant association with the presence of TVV in both sets analyzed (P < 0.001). Similarly, the dendrogram shown in Fig. 4 was significantly correlated with the presence of the ITS C66T mutation (P = 0.0095). A comparison of the ATL dendrogram (Fig. 5) with the ITS C66T mutation was not applicable because of the low incidence of the mutation in this set of isolates. However, a significant association was found between this tree and the MLC values of the isolates (P = 0.0004).
Shared-band analysis.
A shared-band index that evaluated the relatedness of isolates within a group confirmed many of the previously described relationships. The RAPD band data were used to compute this index. Table 3 shows the shared-band index (S value) and its corresponding P value determined for several groups of isolates which share a particular trait. For example, isolates that harbor TVV have a significantly high level of relatedness (P = 0.005). In other words, these isolates share more bands than would be expected from a random grouping of 55 isolates in this total population. Isolates without TVV also appear to be closely related although the relatedness is not significant (P = 0.137). The only other group with a significant level of relatedness is the group containing isolates with metronidazole resistance (P = 0.003). This result indicates that drug-resistant isolates have genomes that are more similar to each other than they are to the group as a whole, suggesting that they probably derived from a common ancestor. There also appears to be a high level of relatedness among the ATL isolates and among the isolates with the ITS C66T mutation, although the P values do not achieve significance (P = 0.095 and 0.065, respectively).
TABLE 3.
Results from index of shared-band analysis
| Isolate group | No. of isolates | Shared-band indexa | P valueb |
|---|---|---|---|
| TVV+ | 55 | 0.2618 | 0.005 |
| TVV− | 54 | 0.2539 | 0.137 |
| ATL | 67 | 0.2436 | 0.095 |
| MSA | 42 | 0.2568 | 0.730 |
| C66T+ | 15 | 0.2680 | 0.065 |
| C66T− | 94 | 0.2472 | 0.661 |
| MetR+ | 42 | 0.2638 | 0.003 |
| MetR− | 67 | 0.2395 | 0.980 |
For members of the indicated group.
P values were determined from a permutation test performed on the RAPD band data collected from all 109 isolates in order to determine if groups of isolates which share characteristics have a higher level of relatedness to each other than the level of relatedness among the entire group of isolates. P values of ≤0.05 indicate groups of isolates that are significantly more closely related to each other and are shown in bold.
DISCUSSION
RAPD trees were useful in the detection of evolutionary relationships among T. vaginalis isolates with similar molecular and phenotypic traits. This was demonstrated by the correlation of all trees with the presence of TVV. Since there is no known lytic cycle for TVV, it is unlikely that isolates acquire the virus through horizontal transmission (24). Rather, the inheritance of the virus from a parent strain to its progeny is the most probable, and possibly the only, mode of transmission. The virus can thus be a useful molecular marker for T. vaginalis. It has been postulated that molecular markers correlating with phylogenetic trees can be useful in lending support to the predicted evolutionary relationships (21). Therefore, the nonrandom association of TVV and the RAPD band patterns, two unrelated genotypic characteristics, lends further support to the accuracy of the phylogenetic trees produced in this study. In Fig. 5, the virus-negative isolate, ATL170, is located among the TVV-positive group. It is possible that this isolate lost the virus relatively recently. It would therefore remain closely related to isolates harboring the virus. This theory is supported by the observation of isolates that have lost the virus after prolonged cultivation in vitro (25).
The division of the trees into two distinct lineages corresponding with the presence or absence of TVV (Fig. 4 and 5) is further supported by the shared-band analysis for the TVV-positive population, indicating that the isolates harboring the virus are more closely related to each other than to TVV-negative isolates. This may suggest that a single predecessor of the TVV-positive isolates acquired the virus or that the virus was lost multiple times from the predecessors of the TVV-negative isolates. We also investigated the possibility that this correlation could have resulted from the amplification of the viral genome by the RAPD primers. However, there was no evidence to suggest that viral genome amplification occurred (data not shown).
The most interesting finding of this study is the association between relatedness of metronidazole-resistant isolates based on RAPD data. These results suggest that the development of resistant isolates has occurred in only a few rare instances. Our data are consistent with the findings of Vanacova et al. (22), where in vitro and in vivo metronidazole resistance correlated with phylogenies based on RAPD patterns. Unlike the previous study, the RAPD data presented here did not correlate with the geographic origin of the isolates (data not shown). However, the isolates in this study were from different cities in the same country, as opposed to those of Vanacova et al., who studied isolates that originated on different continents (22). Phylogenies from the study by Vanacova and colleagues did not correlate with TVV, but it is possible that the small sample size used prevented the finding of a significant association.
As with all studies using RAPD as a typing technique, care must be taken in the analysis of the results. Because of the arbitrary nature of the PCR priming used in this technique, there is the possibility that amplification from different loci may comigrate on a gel or that variations in template concentration may result in sporadic fragments. To guard against this possibility, we evaluated the primers over a range of template concentrations to ensure that the banding patterns were consistent (data not shown). Also, the correlation between our RAPD results and heritable genotypic and phenotypic traits (TVV and metronidazole resistance, respectively) suggests that the relationships between the various isolates based on the RAPD data are reliable.
The ITS region of rDNA is often used in intraspecies studies because variation is often found there. However, the lack of ITS sequence variation among the T. vaginalis isolates shown here suggests a high level of relatedness among the isolates and a relatively short period of time since the isolates have diverged. This lack of variability in the rDNA ITS region is consistent with findings by Gunderson et al., which suggested that trichomonad species diverged from each other relatively recently (5).
This study was designed to investigate the genetic diversity of T. vaginalis in relation to clinical phenotypes of a large collection of isolates. A number of correlations between molecular and phenotypic traits have been identified. Isolates with TVV are significantly more likely to be susceptible to metronidazole, whereas isolates with the ITS C66T mutation are significantly less likely to be susceptible. Perhaps the most interesting finding is the evidence that metronidazole resistance exists in a closely related group of isolates, indicating that only one or a very few mutations have occurred which result in resistance. This suggests that it may be possible to identify a marker for resistance that could lead to improved treatment strategies.
ACKNOWLEDGMENTS
We acknowledge Hugo Moreno and the Women's Health Clinic at Grady Memorial Hospital, Atlanta, Ga. We also thank Jarsov Flegr from Charles University for his cooperation in using the Treept program. Epidemiologic support was provided by George Schmid and Debbie Mosure, and Virginia Secor provided editorial assistance.
This study was funded in part by a grant from the Office of Women's Health, CDC, Atlanta, Ga.
REFERENCES
- 1.Chakrabarti D, Dame J B, Gutell R R, Yowell C A. Characterization of the rDNA unit and sequence analysis of the small subunit rRNA and 5.8S rRNA genes from Tritrichomonas foetus. Mol Biochem Parasitol. 1992;52:75–84. doi: 10.1016/0166-6851(92)90037-k. [DOI] [PubMed] [Google Scholar]
- 2.Cotch M F, Pastorek J G, Nugent R P, Hillier S L, Gibbs R S, Martin D H, Exchenbach D A, Edelman R, Carey C, Regan J A, Drohn M A, Kebanoff M A, Vijaya A, Rhoads G G the Vaginal Infections and Prematurity Study Group. Trichomonas vaginalis associated with low birth weight and preterm delivery. Sex Transm Dis. 1997;24:1–8. doi: 10.1097/00007435-199707000-00008. [DOI] [PubMed] [Google Scholar]
- 3.Diamond L S. The establishment of various trichomonads of animals and man in axenic cultures. J Parasitol. 1957;43:488–490. [PubMed] [Google Scholar]
- 4.Flegr J, Zaboj P. PTPT, the freeware program for permutation testing concordance between phylogeny and the distribution of phenetic traits. Acta Soc Zool Bohem. 1998;61:91–95. [Google Scholar]
- 5.Gunderson J, Hinkle G, Leipe D, Morrison H G, Stickel S K, Odelson D A, Breznak J A, Nerad T A, Muller M, Sogin M L. Phylogeny of trichomonads inferred from small-subunit rRNA sequences. J Eukaryot Microbiol. 1995;42:411–415. doi: 10.1111/j.1550-7408.1995.tb01604.x. [DOI] [PubMed] [Google Scholar]
- 6.Hillis D M, Dixon M T. Ribosomal DNA: molecular evolution and phylogenetic inference. Q Rev Biol. 1991;66:411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- 7.Hotzel I, Kabakoff R, Ozaki L S. Small extrachromosomal nucleic acid segments in protozoan parasites. Vet Parasitol. 1995;57:57–60. doi: 10.1016/0304-4017(94)03110-i. [DOI] [PubMed] [Google Scholar]
- 8.Katiyar S K, Visvesvara G S, Edlind T D. Comparisons of ribosomal RNA sequences from a mitochondrial protozoa: implications for processing, mRNA binding and paromomycin susceptibility. Gene. 1995;152:27–33. doi: 10.1016/0378-1119(94)00677-k. [DOI] [PubMed] [Google Scholar]
- 9.Kulda J. Trichomonads, hydrogenosomes and drug resistance. Int J Parasitol. 1999;29:199–212. doi: 10.1016/s0020-7519(98)00155-6. [DOI] [PubMed] [Google Scholar]
- 10.Laga M, Manoka A, Kivuvu M, et al. Non-ulcerative sexually transmitted diseases as risk factors for HIV-1 transmission in women: results from a cohort study. AIDS. 1993;7:95–102. doi: 10.1097/00002030-199301000-00015. [DOI] [PubMed] [Google Scholar]
- 11.Lehman T, Besansky N J, Hawley W A, Fahey T G, Kamua L, Collins F H. Microgeographic structure of Anopheles gambiae in western Kenya based on mtDNA and microsatellite loci. Mol Ecol. 1997;6:243–253. doi: 10.1046/j.1365-294x.1997.00177.x. [DOI] [PubMed] [Google Scholar]
- 12.Lossick J G. Therapy of urogenital trichomoniasis. In: Honigberg B M, editor. Trichomonads parasitic in man. New York, N.Y: Springer-Verlag; 1989. pp. 324–341. [Google Scholar]
- 13.Marche S, Roth C, Manohar S K, Dollet M, Baltz T. RNA virus-like particles in pathogenic plant trypanosomatids. Mol Biochem Parasitol. 1993;57:261–268. doi: 10.1016/0166-6851(93)90202-9. [DOI] [PubMed] [Google Scholar]
- 14.Meingässner J G, Havelec L, Mieth H. Studies on strain sensitivity of Trichomonas vaginalis to metronidazole. Br J Vener Dis. 1978;54:72–76. doi: 10.1136/sti.54.2.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller R L, Wang A L, Wang C C. Purification and characterization of the Giardia lamblia double-stranded RNA virus. Mol Biochem Parasitol. 1988;28:189–196. doi: 10.1016/0166-6851(88)90003-5. [DOI] [PubMed] [Google Scholar]
- 16.Minkoff H, Grunebaum A N, Schwarz R H, Feldman J, Cummings M, Crumbleholme W, Clark L, Pringle G, McCormack W M. Risk factors for prematurity and premature rupture of membranes: a prospective study of the vaginal flora in pregnancy. Am J Obstet Gynecol. 1984;150:965–972. doi: 10.1016/0002-9378(84)90392-2. [DOI] [PubMed] [Google Scholar]
- 17.Muller M, Lossick J G, Gorrell T E. In vitro susceptibility of Trichomonas vaginalis to metronidazole and treatment outcome in vaginal trichomoniasis. Sex Transm Dis. 1988;15:17–24. doi: 10.1097/00007435-198801000-00004. [DOI] [PubMed] [Google Scholar]
- 18.Narcisi E M, Secor W E. In vitro effect of tinidazole and furazolidone on metronidazole-resistant Trichomonas vaginalis. Antimicrob Agents Chemother. 1996;40:1121–1125. doi: 10.1128/aac.40.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrin D, Delgaty K, Bhatt R, Garber G. Clinical and microbiological aspects of Trichomonas vaginalis. Clin Microbiol Rev. 1998;11:300–317. doi: 10.1128/cmr.11.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sokal R R, Sneath P H A. Principles in numerical taxonomy. New York, N.Y: W. H. Freeman & Co.; 1963. p. 134. [Google Scholar]
- 21.Tibayrenc M. Beyond strain typing and molecular epidemiology: integrated genetic epidemiology of infectious diseases. Parasitol Today. 1998;14:323–329. doi: 10.1016/s0169-4758(98)01286-1. [DOI] [PubMed] [Google Scholar]
- 22.Vanacova S, Tachezy J, Kulda J, Flegr J. Characterization of trichomonad species and strains by PCR fingerprinting. J Eukaryot Microbiol. 1997;44:545–552. doi: 10.1111/j.1550-7408.1997.tb05960.x. [DOI] [PubMed] [Google Scholar]
- 23.Wang A L, Wang C C. A linear double-stranded RNA in Trichomonas vaginalis. J Biol Chem. 1985;260:3697–3702. [PubMed] [Google Scholar]
- 24.Wang A L, Wang C C. The double-stranded RNA in Trichomonas vaginalis may originate from virus-like particles. Proc Natl Acad Sci USA. 1986;83:7956–7960. doi: 10.1073/pnas.83.20.7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang A L, Wang C C, Alderete J F. Trichomonas vaginalis phenotypic variation occurs only among trichomonads infected with the double-stranded RNA virus. J Exp Med. 1987;166:142–150. doi: 10.1084/jem.166.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Widmer G, Dooley S. Phylogenetic analysis of Leishmania RNA virus and Leishmania suggests ancient virus-parasite association. Nucleic Acids Res. 1995;23:2300–2304. doi: 10.1093/nar/23.12.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.World Health Organization; World Health Organization, editors. Global program on AIDS. Geneva, Switzerland: World Health Organization; 1995. An overview of selected curable sexually transmitted diseases; pp. 2–27. [Google Scholar]