Abstract
The efficacy of cisplatin (CDDP) has been demonstrated in the treatment of various cancers as monotherapy and combination therapy with immunotherapy. However, acquired CDDP resistance is a major obstacle to successful treatment. In the present study, the mechanisms underlying acquired CDDP resistance were examined using ACR20 cells, which are CDDP-resistant cells derived from A549 lung cancer cells. CDDP induces cytotoxicity by binding nuclear DNA and generating reactive oxygen species (ROS). Contrary to our expectation, ROS levels were elevated in ACR20 cells not treated with CDDP. Pretreatment with an ROS inhibitor enhanced the sensitivity of ACR20 cells to CDDP and prevented the activation of nuclear factor (NF)-кB signaling and upregulation of inhibitor of apoptosis proteins (IAPs). Notably, evaluation of the mitochondrial oxygen consumption rate and mitochondrial superoxide levels revealed a deterioration of mitochondrial function in ACR20 cells. Mitochondrial DNA PCR-RFLP analysis revealed four mutations with varying percentage levels in ACR20 cells. In addition, in cytoplasmic hybrids with mitochondria from ACR20 cells, intrinsic ROS levels were elevated, expression of IAPs was increased, and complex I activity and sensitivity to CDDP were decreased. Analysis of three-dimensional structure data indicated that a mutation (ND2 F40L) may impact the proton translocation pathway, thereby affecting mitochondrial complex I activity. Together, these findings suggest that intrinsic ROS levels were elevated by mitochondrial DNA mutations, which decreased the sensitivity to CDDP via activation of NF-κB signaling and induction of IAP expression in ACR20 cells. These findings indicate that newly identified mutations in mitochondrial DNA may lead to acquired cisplatin resistance in cancer.
Keywords: cisplatin, acquired resistance, mitochondria, complex I, reactive oxygen species
Introduction
For approximately 40 years, cisplatin (CDDP) has been approved for medical use in the treatment of a number of cancers, including testicular, ovarian, bladder, and lung cancer, among others (1). Recently, CDDP has been used for the treatment of patients with acquired resistance to molecular-targeted drugs. It has also been used in combination with immune checkpoint inhibitors, such as pembrolizumab (2,3). However, acquisition of CDDP resistance leads to the failure of cancer therapy; therefore, overcoming CDDP resistance remains an important obstacle to successful cancer treatment. Although many researchers have investigated the mechanism of acquired CDDP resistance, including reduced drug accumulation (4), increased detoxification by glutathione (5), and increased repair of CDDP-DNA adducts (6), to date, there are still no approved therapies for the treatment of patients with acquired CDDP resistance.
We previously established high-grade CDDP-resistant A549 cells (A549 cell-derived CDDP-resistant at 20 µM, ACR20 cells) by culturing cells with 20 µM CDDP. The resistance of ACR20 cells was 18.5-fold higher than that of A549 cells (7). We reported that CD44v overexpression increased the expression of cystine-glutamate transporter xCT in the plasma membrane, which led to the acquisition of CDDP resistance (7). However, in ACR20 cells treated with an xCT inhibitor or siRNA, the sensitivity to CDDP was not recovered to the same level as that in A549 cells (7). Therefore, other mechanisms for the acquisition of CDDP resistance may exist. (7). CDDP is imported into cells and induces reactive oxygen species (ROS) generation, which causes cytotoxicity (8). One report showed that cytoplasmic ROS production was reduced in CDDP-resistant cells derived from gastric cancer cells (9). Glutathione, a primary cellular antioxidant, is composed of glutamate, cysteine, and glycine. Therefore, we predicted that the level of glutathione was decreased and that the levels of ROS were reduced in ACR20 cells.
Mitochondria are cellular organelles that generate most of the energy required for biochemical reactions in cells. Mitochondria have an oxidative phosphorylation system, which comprises five complexes (I–V). Mitochondria possess their own genome, mitochondrial DNA (mtDNA), which encodes mitochondrial respiratory chain components, including seven subunits of complex I [NADH dehydrogenase (ND)1, ND2, ND3, ND4L, ND4, ND5, and ND6], one subunit of complex III [cytochrome b (CYTB)] and three subunits of complex IV [cytochrome c oxidase (COX) I, II, and III], 22 tRNAs, and 2 rRNAs. Mutations in nuclear genes or mitochondrial genes that encode mitochondrial respiratory chain proteins result in mitochondrial dysfunction. Numerous mtDNA mutations have been identified in the development and progression of various pathologies and mitochondrial dysfunction is implicated in various human diseases, including diabetes, Alzheimer's disease, and cancer (10). mtDNA is a critical target of CDDP (11) and the levels of CDDP adducts are increased in mtDNA compared with nuclear DNA (12). This suggests that the binding of CDDP to mtDNA may cause mtDNA damage and subsequent mitochondrial dysfunction. In addition, in ρ° cells depleted of mtDNA, which lose mitochondrial function, sensitivity to doxorubicin or CDDP was found to decrease (13). Thus, mitochondrial dysfunction may lead to the acquisition of CDDP resistance. However, how CDDP induces mitochondrial dysfunction and how mitochondrial dysfunction reduces cell sensitivity to CDDP is still unclear.
In the present study, the mechanism underlying CDDP resistance was investigated in ACR20 cells. It was demonstrated that mtDNA mutations in CDDP-resistant ACR20 cells caused loss of mitochondrial function, upregulation of intrinsic ROS levels (IRLs), and expression of inhibitor of apoptosis proteins (IAPs), which contributed to the decreased sensitivity to CDDP. These results suggest that mtDNA mutations play a role in the acquisition of CDDP resistance.
Materials and methods
Cell culture
Human lung carcinoma cell line A549 (cat. # RCB0098, RRID: CVCL_0023) were provided by RIKEN BioResource Research Center through the National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science and Technology (Japan). ACR20 cells, CDDP-resistant cells, were established and cultured as previously described (7). Both cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM, Nacalai Tesque, Japan, cat. 08459-64) with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 µg/ml streptomycin in the presence or absence of 20 µM CDDP (Wako Pure Chemical Industries, Ltd., Osaka, Japan, cat. 033-20091) at 37°C with 5% CO2 and 95% air. Human cervical cancer HeLa cells (cat. no. CCL-2, RRID: CVCL_0030) were obtained from American Type Culture Collection. ρ° HeLa cells, which are deficient in mtDNA and resistant to 6-thioguanine, were isolated as previously described (14).
Flow cytometry analysis
Cells (1.2×105 cells) were seeded on a 60-mm dish and incubated in culture media at 37°C in 5% CO2 and 95% air. After a 24-h incubation, the cells were treated with 80 µM CDDP for 24 h. Annexin V/PI staining (Thermo Fisher Scientific, Inc., cat. no. V13245) was performed following the manufacturer's protocol. The stained cells were analyzed using FACSCalibur (BD Biosciences) and FlowJo software 10.5.3 (Tree Star, Inc.). Apoptotic cells were identified as Annexin V+/PI− (early apoptosis) and Annexin V+/PI+ (late apoptosis) cells.
Western blotting
Cells were seeded at 7×105 cells on a 60-mm dish and incubated in culture media at 37°C in 5% CO2 and 95% air. In experiments examining cleaved caspase-3 levels, after a 24-h incubation, the cells were treated with 80 µM CDDP for 24 h. In experiments examining IAPs (namely, c-IAP1, c-IAP2, and XIAP) and phosphorylation of IκB-α and NF-кB, after a 1-h incubation, the cells were washed with PBS and treated with 40 mM N-acetyl-L-cysteine (NAC; Sigma-Aldrich; Merck KGaA, cat. no. A7250), an ROS inhibitor, for 24 h. Western blot assays were performed as previously described (15). The antibodies are listed in Table SI. After probing of anti-phospho-IκB-α and anti-phospho-NF-кB antibodies, the blots were stripped and reprobed with anti-IκB-α or anti-NF-кB antibodies. After probing of anti-XIAP, anti-c-IAP1 antibody, antic-IAP2, or anti-NDUFB8 antibodies, the blots were stripped and reprobed with anti-GAPDH or anti-β-actin antibodies.
Cytotoxic activity assay
To evaluate whether ROS affected the sensitivity of cells to CDDP, cells were seeded at 4.8×104 cells/well on a 24-well plate and incubated in culture media at 37°C in 5% CO2 and 95% air. After a 1-h incubation, the cells were treated with 40 mM NAC for 24 h. The treated cells were washed with PBS and cultured in culture media containing 80 µM CDDP for 48 h. The half maximal inhibitory concentration (IC50) of CDDP was determined following the previous study protocol (7). Cells were fixed with 4% paraformaldehyde, stained with 0.5% crystal violet and lysed with 10% acetic acid. The optical density (O.D.) was measured at 600 nm. The percentage of cell viability and 50% growth inhibitory concentration was calculated as previously described (7).
ROS measurement
Cells were incubated for 48 h, washed with HBSS (Nacalai Tesque, cat. no. 09735-75), stained with 10 µM CM-H2DCFDA (Thermo Fisher Scientific, Inc., cat. no. C6827) or 5 µM MitoSox (Thermo Fisher Scientific, Inc., cat. no. M36008), and incubated for 30 min at 37°C in 5% CO2 and 95% air. Cells were washed with HBSS and observed with a fluorescence microscope using FITC filters. The number of cells with positive fluorescence was counted and the average number of cells in three fields was taken as the number of ROS-producing cells per field.
ρ A549 cell establishment
Respiration-deficient ρ A549 cells were established by treatment of A549 cells with 50 ng/ml EtBr as previously described (16) for 14 days (Fig. S1A and B).
Cytoplasmic hybrid (Cybrid) establishment
Cybrids were established as previously described (17). In brief, A549 cells or ACR20 cells were seeded at 1.5×106 cells or 3.0×106 cells, respectively, on a 100-mm dish for 48 h. To prepare enucleated A549 cells or ACR20 cells (the mtDNA donor), cells were pretreated with 10 µg/ml cytochalasin B (Sigma-Aldrich; Merck KGaA, cat. no. 14930-96-2) for 15 min and centrifuged at 17,300 × g for 20 min. The resultant cytoplasts were fused with ρ° HeLa cells, using polyethylene glycol. After 2 days, selective isolation of the cybrids was performed by culture in selection medium with 6-thioguanine, to exclude unfused A549 cells or ACR20 cells, and without uridine and pyruvate, to exclude parental ρ° HeLa cells. The resultant cybrids contained nuclear DNA from ρ° HeLa cells and mtDNA from enucleated A549 cells or ACR20 cells.
mtDNA extraction and mtDNA sequencing
mtDNA extraction (Wako Pure Chemical Industries, Ltd., cat. no. 291-55301), polymerase chain reaction (PCR) (Takara Shuzo, Otsu, Japan, cat. no. RR001C), and purification of the PCR products (Takara Shuzo, cat. no. 740609) were performed following the manufacturers' protocols. Primer sequences are shown in Table SII. DNA sequencing was conducted by Eurofins Genomics (Tokyo, Japan).
PCR-restriction fragment length polymorphism (PCR-RFLP)
The percentage levels of mtDNA mutations in A549 cells and ACR20 cells were examined by PCR-RFLP. In addition, to confirm that the mtDNA derived from A549 and ACR20 cells was incorporated into A549cyb and ACR20cyb cells, respectively, we performed PCR-RFLP. In brief, mtDNA was extracted from the A549 and ACR20 cells. Total DNA was extracted from A549cyb and ACR20cyb cells. PCR amplification was performed according to the manufacturer's protocol. Purified PCR products were digested with the restriction enzyme. The restriction fragments were analyzed by gel electrophoresis. PCR primer sequences, restriction enzyme, and annealing temperature are shown in Table SIII.
Mitochondrial activity assay
A XF24 Extracellular Flux Analyzer (Seahorse Biosciences, USA) was used to determine the intracellular bioenergetic profiles. A549 or ACR20 cells were seeded at 4×104 cells/well on the XF24 V7 Cell Culture microplate. Extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) were measured as previously described (18). Mitochondria from cultured cells were isolated using the Mitochondria Isolation Kit (BioVision, Inc., cat. no. K288-50). Protein concentrations of isolated mitochondrial samples were estimated using Bradford assays. Isolated mitochondria were stored at −80°C until used. Mitochondrial complex activities were measured using MitoCheck Complex I Activity Assay Kit (Cayman Chemical Company, cat. no. 700930). Complex I activity was determined as activity without inhibitor of complex I subtracted from the activity with complex I inhibitor.
Quantitative reverse transcription real-time PCR (RT-qPCR)
RT-qPCR was established as previously described (7). RT-qPCR was performed using PrimeScript™ RT Master Mix (Takara Shuzo, cat. no. RR036) and TB Green® Premix Ex Taq™ II (Takara Shuzo, cat. no. RR820) according to the manufacturer's protocol. Primer sequences are shown in Table SIV. Relative expression was calculated using the ΔΔCt method (19) with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as the reference gene.
Bioinformatic analysis of NADH-ubiquinone oxidoreductase chain (ND) proteins
The structures of the mutant ND proteins were analyzed at Altif Laboratories Inc. (Tokyo, Japan).
Statistical analysis
Data are expressed as the mean ± standard error (SEM) of independent determinations for each experiment. Normality was assessed by Kolmogorov-Smirnov normality test. Unpaired t-test was used to evaluate the differences between two groups. Comparisons of more than two groups were evaluated by one-way analysis of variance (ANOVA), followed by the Student-Newman-Keuls post hoc test. P-value <0.05 is indicative of a statistically significant difference.
Results
Impairment of CDDP-induced apoptosis in ACR20 cells
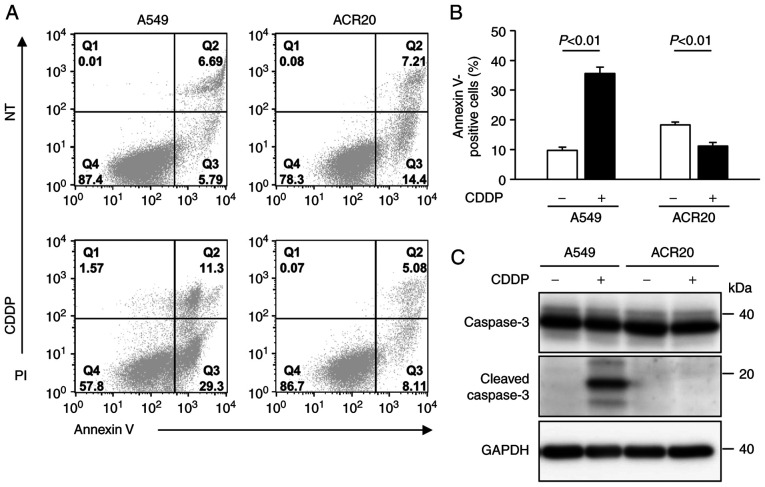
We previously established CDDP-resistant cells derived from A549 cells, which we named ACR20 cells (7). To determine whether ACR20 cells are resistant to CDDP-induced apoptosis, we evaluated CDDP-induced apoptosis using Annexin V/PI staining assays. In the CDDP-treated A549 cells, a high percentage of cells were positively stained for Annexin V and cleaved caspase-3 expression was detected by western blotting, thus indicating that CDDP induced apoptosis in the A549 cells (Fig. 1A-C). In contrast, the percentage of Annexin V-stained cells was low (11%) and cleaved caspase-3 was not detected in the CDDP-treated ACR20 cells. These results indicate that CDDP-induced apoptosis was impaired in the ACR20 cells.
Figure 1.
ACR20 cells show resistance to CDDP-induced apoptosis. (A) Cells were treated with or without 80 µM cisplatin (CDDP) for 24 h followed by Annexin V/PI staining. Representative flow cytometry profiles of A549 and ACR20 cells untreated (NT) or treated with 80 µM CDDP for 24 h. (B) Quantification of apoptosis by Annexin V binding to A549 and ACR20 cells. Bars represent mean ± SE (n=6). (C) Western blot analysis of cleaved and total caspase-3 expression levels in A549 and ACR20 cells treated as indicated.
Increase in intrinsic ROS in ACR20 cells is involved in the impairment of CDDP-induced apoptosis
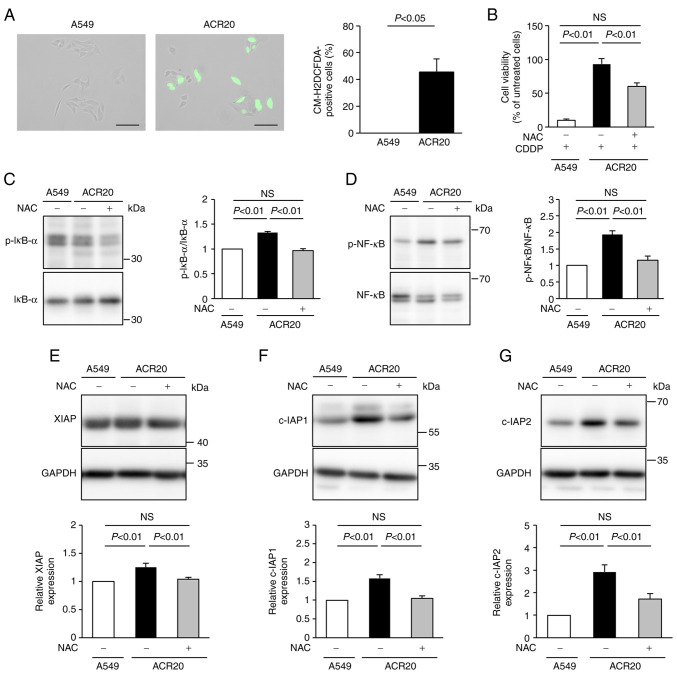
To determine the mechanism underlying the impairment of CDDP-induced apoptosis in ACR20 cells, we analyzed the production of ROS using the ROS indicator CM-H2DCFDA. Green fluorescence was not detected in A549 cells not treated with CDDP but was observed after CDDP treatment (Figs. S2 and 2A). This result indicates that CDDP induced ROS generation in the cytoplasm of the A549 cells. Unexpectedly, green fluorescence was detected in ACR20 cells not treated with CDDP, and the number of green fluorescence-positive cells was increased compared to that of the A549 cells (Fig. 2A). These results indicate that IRLs were elevated in ACR20 cells. NAC, an inhibitor of ROS, pretreatment improved the sensitivity of ACR20 cells to CDDP (Fig. 2B). Together, these findings suggest that the elevation of IRLs was involved in the acquisition of CDDP resistance.
Figure 2.
Elevated levels of intrinsic ROS are involved in the mechanism of acquisition of CDDP resistance. (A) Cells were incubated with CM-H2DCFDA and observed by fluorescence microscopy. Representative images under the fluorescence microscope are shown on the left. The numbers of CM-H2DCFDA-stained cells were counted. On the right, bars represent mean ± SE (n=3). Scale bar, 100 µm. (B) Cells were treated with or without NAC for 24 h. Cells were washed with PBS and incubated with 80 µM CDDP for 72 h. Cell viability was measured using crystal violet assay. (C and D) Cells were treated with or without NAC for 24 h and then lysed. Phosphorylation levels of IκB-α and NF-κB were analyzed by western blotting, and p-IκB-α/IκB-α (C) and p-NF-κB/NF-κB (D) ratios were determined. Bars represent mean ± SE (n=3). (E-G) Expression levels of XIAP (E), c-IAP1 (F), and c-IAP2 (G) were analyzed by western blotting. Bars represent mean ± SE (n=3). ROS, reactive oxygen species; CDDP, cisplatin; NAC, N-acetyl-L-cysteine; IAP, inhibitors of apoptosis protein.
NF-κB regulates the gene expression of IAPs (20). ROS-stimulated IκB kinase leads to phosphorylation of IκB and NF-κB, which promotes IκB degradation and modulates NF-κB activation (21). Thus, we hypothesized that the elevation of IRLs in ACR20 cells would induce the expression of IAPs and impair CDDP-induced apoptosis. The expression levels of phosphorylated IκB-α and NF-κB in ACR20 cells were significantly higher than those in A549 cells (P<0.01), and these levels were suppressed in ACR20 cells by NAC pretreatment (Fig. 2C and D). Similarly, the levels of IAPs in ACR20 cells were significantly elevated compared to those in A549 cells (P<0.01), and the expression of IAPs in ACR20 cells was suppressed by NAC pretreatment (Fig. 2E-G). The expression of phosphorylated IκB-α, phosphorylated NF-κB, and IAPs in A549 cells was not affected by NAC pretreatment (data not shown). There results suggest that elevated IRLs decreased the sensitivity of ACR20 cells to CDDP by activating NF-κB signaling and inducing IAP expression.
Loss of mitochondrial function in ACR20 cells is associated with CDDP resistance
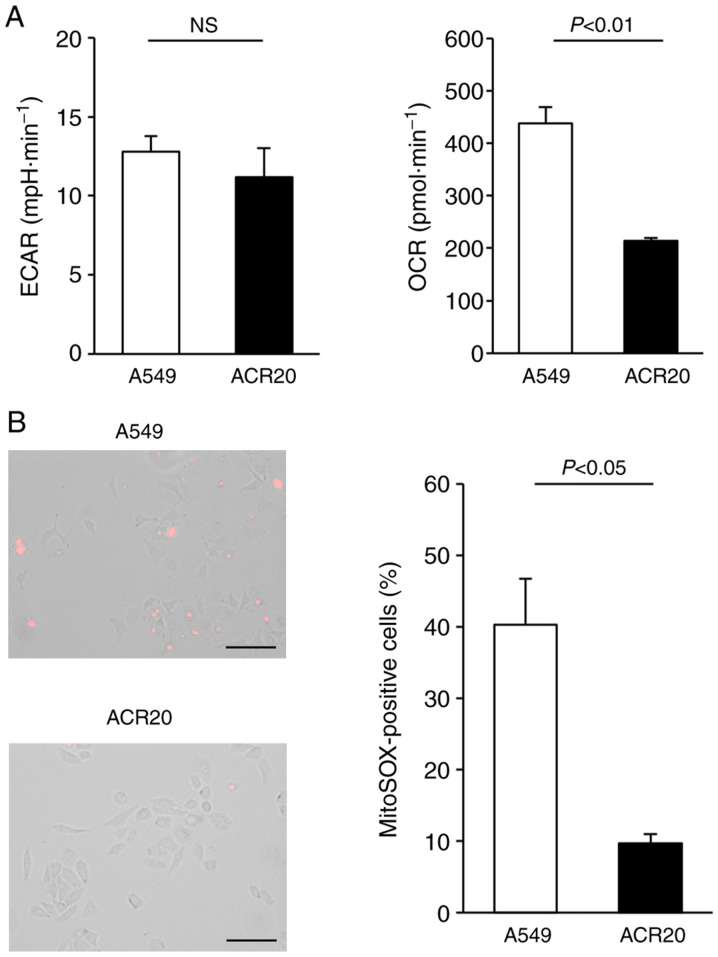
Intracellular levels of ROS are dependent on the balance between ROS generation and ROS elimination. Increased IRLs in cancer cells may be caused by malfunction of the mitochondrial respiratory chain (22,23). Extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) values are important indicators of mitochondrial respiration and glycolysis. We found that ECAR was not changed and that OCR was decreased in the ACR20 cells when compared to the A549 cells (Fig. 3A). Mitochondrial superoxide is generated as a byproduct of ATP production (24,25). In A549 cells loaded with MitoSox, an indicator of mitochondrial superoxide, red fluorescence was observed; however, in contrast, red fluorescence was not detected in the ACR20 cells (Fig. 3B). Together, these results indicate a loss of mitochondrial function in ACR20 cells. To investigate whether mitochondrial dysfunction causes CDDP resistance in A549 cells, we analyzed CDDP sensitivity in mtDNA-depleted ρ A549 cells, which showed loss of mitochondrial function. The IC50 values of CDDP in A549 and ρ A549 cells were 4.64±1.04 and 11.56±2.26 µM, respectively (Fig. S1C). Together, these results indicate that mitochondrial dysfunction causes CDDP-resistance in the A549 cells.
Figure 3.
ACR20 cells show loss of mitochondrial function. (A) Extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) were measured using the XF24 Analyzer. Bars represent mean ± SE (n=6). (B) Cells were incubated with MitoSox and observed by fluorescence microscopy. Representative images under the fluorescence microscope are shown. The number of MitoSox-stained cells was counted. Bars represent mean ± SE (n=3). Scale bar, 100 µm.
mtDNA mutations causes the decrease in mitochondrial complex I activity in ACR20 cells
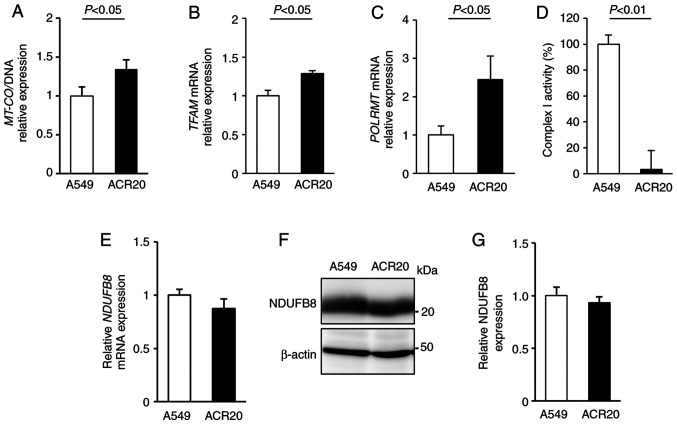
The mitochondrial genome encodes mitochondrial respiratory chain components. Thus, decreased quality and quantity of mtDNA lead to mitochondrial dysfunction (26–28). To examine whether a change in the number of mtDNA copies in ACR20 cells is involved in the loss of mitochondrial function, we evaluated the expression of mitochondria-encoded COX I (MT-COI) levels normalized to α-tubulin levels to determine the number of mtDNA copies (29,30). Contrary to our expectations, MT-COI expression was increased in the ACR20 cells when compared to the expression level in the A549 cells (Fig. 4A). The number of mtDNA copies is regulated by transcription factor A, mitochondrial (TFAM) and RNA polymerase mitochondrial (POLRMT) (31,32). We found that the expression of TFAM and POLRMT mRNAs was increased in the ACR20 cells when compared to the A549 cells (Fig. 4B and C). These results indicate that the number of mtDNA copies was increased in ACR20 cells, and this increase might cause the upregulation of TFAM and POLRMT mRNA expression.
Figure 4.
ACR20 cells show an increase in the number of mtDNA copies and decrease in Complex I activity. (A) Number of mtDNA copies was determined by qPCR of MT-COI relative to the nuclear α-tubulin in A549 and ACR20 cells. (B and C) Expression of TFAM (B) and POLRMT (C) mRNA was analyzed by RT-qPCR. Bars represent mean ± SE (n=4). (D) Complex I activity of extracted mitochondria was measured using MitoCheck® Complex I Activity Assay kit. Bars represent mean ± SE (n=4). (E) Expression of NDFUB8 (E) mRNA was analyzed by RT-qPCR. Bars represent mean ± SE (n=4). (F and G) NDUFB8 expression was analyzed by western blotting. Bars represent mean ± SE (n=3). mtDNA, mitochondrial DNA.
To determine the quality of mtDNA, mtDNA was extracted from the A549 and ACR20 cells for sequencing and identification of mtDNA mutations. Moreover, the percentage levels of mtDNA mutations were analyzed by PCR-RFLP. 4587 T>C and 11384 C>A were included in 100% of mtDNAs in the ACR20 cells (Fig. S3A and B and Table I). 13148 C>A and 14162 G>T were specifically detected in ACR20 cells and were included in 44.9 and 53.3% of the mtDNAs, respectively (Fig. S3C and D and Table I). Thus, four mtDNA mutations with varying percentage levels were identified in ACR20 cells and these mutations were located in genes encoding mitochondrial complex I. We further found that the activity of mitochondrial complex I was decreased in ACR20 cells compared to the A549 cells (Fig. 4D). NDUFB8 is a component of the supernumerary subunits in mammalian complex I, which are central to the structure, stability, and assembly (33). mRNA and proteins levels of NDUFB8 were comparable between the A549 and ACR20 cells (Fig. 4E-G). Thus, these results suggest that the identified mtDNA mutations caused the decrease of mitochondrial complex I activity in ACR20 cells.
Table I.
Identified mtDNA mutations with varying percentage levels in ACR20 cells.
| Percentage levels of mutation | ||||
|---|---|---|---|---|
|
|
||||
| Mutations | Amino acid change | Gene | A549 cells (%) | ACR20 cells (%) |
| 4587T>C | Phe (40)>Leu | MT-ND2 | 44.00 | 100 |
| 11384C>A | Leu (209)>Ile | MT-ND4 | 4.28 | 100 |
| 13148C>A | Pro (271)>Gln | MT-ND5 | 0 | 44.90 |
| 14612G>T | Ser (21)>Tyr | MT-ND6 | 0 | 53.30 |
mtDNA, mitochondrial DNA.
mtDNA mutations in ACR20 cells are involved in CDDP resistance
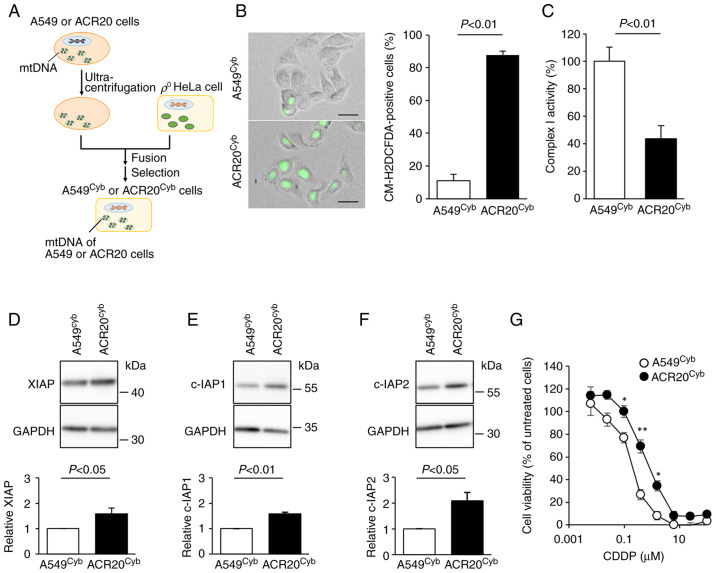
To clarify whether the identified mtDNA mutations in ACR20 cells contributed to the elevation of IRLs and the decreased activity of complex I, cybrids (cells with the same physiological functions such as nuclear DNA, but different only in mtDNA) were established (17) (Fig. 5A). mtDNA derived from A549 and ACR20 cells was incorporated into A549cyb and ACR20cyb cells, respectively (Fig. S4). The number of ROS-producing cells was significantly increased and the activity of complex I was decreased in the ACR20cyb cells compared to the A549cyb cells (P<0.01) (Fig. 5B and C). The expression of IAPs was significantly increased in the ACR20cyb cells compared with the A549cyb cells (XIAP and c-IAP1, P<0.05 and P<0.01; c-IAP2; P<0.05) (Fig. 5D-F). We further examined whether the mtDNA mutations impacted the sensitivity to CDDP. The IC50 value of CDDP in the A549cyb and ACR20cyb cells were 0.27±0.02 and 0.69±0.02 nM, respectively (Fig. 5G). Together, these results indicate that the four identified mtDNA mutations with varying percentage levels in ACR20 cells caused CDDP resistance through the increased expression of IAPs via elevation of IRLs and inactivation of complex I.
Figure 5.
Mitochondrial dysfunction leads to the decreased sensitivity of ACR20 cells to CDDP. (A) A scheme showing the methods for generation of A549cyb and ACR20cyb cells. (B) Cells were incubated with CM-H2DCFDA and observed by fluorescence microscopy (EVOS® FL). Representative images under the fluorescence microscope are shown. The number of CM-H2DCFDA-stained cells was counted. Bars represent mean ± SE (n=3). Scale bar, 20 µm. (C) Complex I activity of extracted mitochondria was measured using MitoCheck® Complex I Activity Assay kit. Bars represent mean ± SE (n=4). (D-F) Expression levels of XIAP (D), c-IAP1 (E) and c-IAP2 (F) were analyzed by western blotting. Bars represent mean ± SE (n=4). (G) Cells were incubated in culture medium containing various concentrations of cisplatin (CDDP) for 72 h. Cell viability was measured using crystal violet assay. Each value represents mean ± SE (n=4). Unpaired t-test; *P<0.05, **P<0.01. IAP, inhibitors of apoptosis protein; mtDNA, mitochondrial DNA.
Structural analysis of proteins with the identified mtDNA mutations
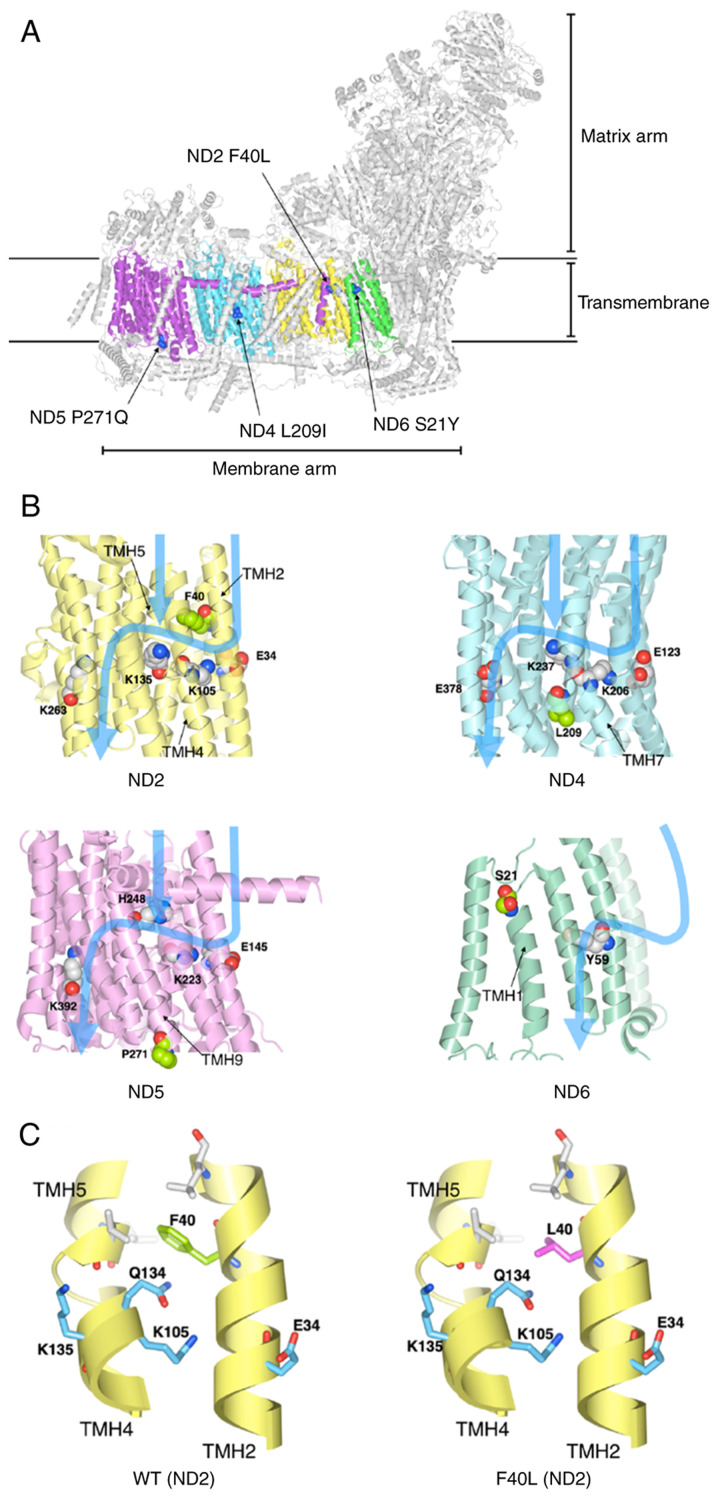
We next determined which of the four identified mtDNA mutations may be associated with complex I inactivation using the three-dimensional structure data of human mitochondrial complex I. The four identified mutations (ND2 F40L, ND4 L209I, ND5 P271Q, and ND6 S21Y) in the mitochondrial complex I were marked on the structure of human respiratory complex I (PDB ID: 5XTB) (Fig. 6A). The mutations were located in the transmembrane. The electric charge and polar amino acid, which exist in proton channels affect proton translocation (34,35). Therefore, the protein structure of the four mutations was modeled and the effect of the mutations on polar amino acids in proton translocation was investigated. Four mutations and key residues were marked to three-dimensional protein structures of complex I and the pathway of proton translocation was marked with reference to Fiedorczuk et al (34) and Fiedorczuk and Sazanov (35) (Fig. 6B). ND2 F40L is located near the charged side chains (Glu34, Lys105, and Lys135) and hydrophilic side chains (Gln134) (Fig. 6B and C). The side chain of Phe residue contacts TMH5 and is packed by hydrophobic interactions with the surrounding hydrophobic residues (Fig. 6C). Although Leu is hydrophobic, the side chain of Leu is smaller than that of Phe. Thus, we speculated that the F40L mutation eliminated these hydrophobic interactions and the packing between TMH2 and TMH5 (Fig. 6C). Furthermore, the conformation of Glu134 may be changed by displacement from Phe40 to Ile40, which may cause the structure change of the proton translocation pathway. In contrast, the other mutations were present at different positions of the proton translocation pathway (Fig. 6B). These results show the possibility that ND2 F40L affects the proton pathway, leading to the decrease of complex I activity.
Figure 6.
Structural analysis of proteins with the identified mtDNA mutations. (A) Locations of mutations in the three-dimensional structure of mitochondrial complex I. ND2, ND4, ND5, and ND6 are depicted in yellow, cyan, purple and green, respectively. The mutated amino acid residues are shown in blue. (B) Blue arrows indicate the path of proton translocation. Space-filling model indicates the charged amino acid residues (key residues) that are thought to be involved in proton translocation. The amino acid residues of four identified mutations are depicted in light green. (C) The F40L mutation may alter ND2 protein structure. Phe40 (left) and Leu40 (right) are depicted in green and magenta, respectively. Important amino acids for proton translocation are depicted in light blue. mtDNA, mitochondrial DNA; ND, NADH-ubiquinone oxidoreductase chain.
Discussion
In the present study, it was demonstrated that elevated intrinsic ROS levels (IRLs) in ACR20 cells led to increased phosphorylation of IκB-α and NF-κB activity, increased inhibitors of apoptosis protein (IAP) expression and decreased cell sensitivity to cisplatin (CDDP). NF-κB signaling is activated by reactive oxygen species (ROS) (21) and NF-κB regulates the expression of IAPs (20). c-IAP1, cIAP2 and XIAP bind to caspase-3 and inhibit the activity of caspase-3 (36–40). This suggests that ROS-induced activation of NF-κB signaling increases the expression of IAPs and suppresses CDDP-induced apoptosis, leading to the acquisition of CDDP resistance. N-acetyl-L-cysteine (NAC) pretreatment improved the sensitivity of ACR20 cells. The percentage of Annexin V/PI-positive cells in the ACR20 cells untreated with CDDP was higher than that in the ACR20 cells treated with 80 µM CDDP. It was considered possible that non-apoptotic cells were stained with Annexin V in ACR20 cells under normal conditions. Annexin V forms a shield around negatively charged phospholipid molecules. Moreover, the lipid contents in the cytomembrane were found to be altered in cisplatin- and doxorubicin-resistant MCF-7 cells (41). Therefore it is possible that the lipid content of the cytomembrane in ACR20 cells may be changed and affected by Annexin V staining; however, the detailed reason for this is still unclear. These findings suggest that the IRLs elevation is partly involved in the mechanism of acquisition of CDDP resistance and that NAC administration before treatment with CDDP may be effective for cancer patients who show resistance to CDDP.
The elevation of IRLs in cancer cells are thought to be caused by oncogenic signals (42,43), oncogenic transformation (44), or malfunction of the mitochondrial respiratory chain (22,23). Depletion of mitrochondrial DNA (mtDNA) in hepatocarcinoma cells, which showed a loss of mitochondrial function, was found to result in chemoresistance to CDDP (13). Thus, mitochondrial function may affect cell sensitivity to CDDP. In the present study, we demonstrated that mitochondrial function was decreased in ACR20 cells and that sensitivity to CDDP in ρ A549 cells was decreased compared to A549 cells. These findings indicate that mitochondrial dysfunction caused the decreased sensitivity to CDDP in A549 cells.
A previous study showed that HeLa cybrids containing mutant mtDNA derived from the pancreatic cancer cell lines CFPAC-1 (10970 T>C in ND4; 8696 T>C and 9070 T>G in ATPase; 2905 A>G in the 16S rRNA gene) or CAPAN-2 (6267 G>A in COI; 10176 G>A in ND3) were resistant to 5-fluororacil and CDDP (45). In this study, we identified four mutations in mtDNA (4587 T>C, 11384 C>A, 13148 C>A, and 14162 G>T) with varying percentage levels in ACR20 cells. These mutations caused the decreased activity of complex I, elevation of IRLs, and induction of IAP expression, which may have led to the CDDP resistance. Furthermore, the three-dimensional structure data suggest that the 11384 C>A mutation may be associated with the decreased activity of complex I. Future studies will be needed to confirm that the 1384 C>A regulates the proton translocation pathway that affects the activity of complex I. Taken together, 11384 C>A may be a new biomarker of sensitivity to CDDP. Studies using clinical samples may provide a clue to this possibility.
Mitochondrial function is associated with the number of mtDNA copies (27). Although mitochondrial function was reduced in ACR20 cells, the number of mtDNA copies was increased. In addition, the expression levels of TFAM and POLRMT mRNA, which encode proteins that regulate the number of mtDNA copies, were increased in ACR20 cells. These results suggest that upregulation of TFAM and POLRMT mRNA caused the increased number of mtDNA copies. Our results were consistent with a previous report, in which TFAM was upregulated in CDDP-resistant cells (46). TFAM is not only required for both transcription and maintenance of mtDNA but also the regulation of anti-apoptotic genes, such as BIRC5 (47) and BCL2 (48). Expression of BIRC5 mRNA was increased in ACR20 cells, while expression of BCL2 mRNA was comparable between A549 and ACR20 cells (data not shown). Thus, TFAM upregulation in ACR20 cells may be, in part, associated with the acquisition of CDDP resistance.
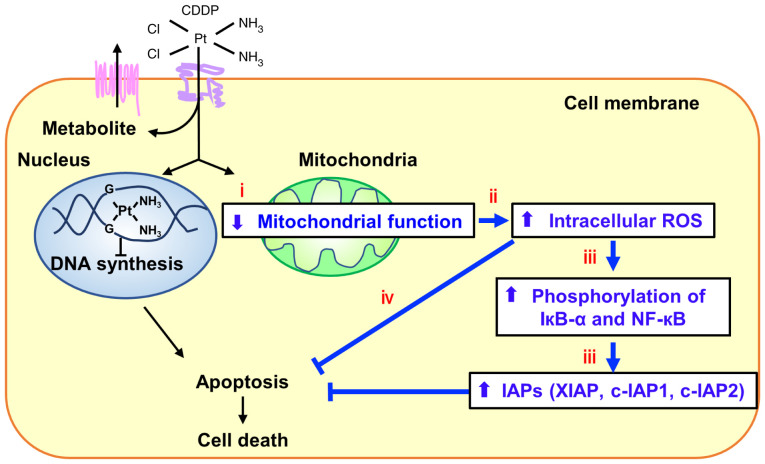
In summary, we demonstrated that several newly identified mtDNA mutations caused the elevation of IRLs, which promoted CDDP resistance through induction of NF-кB signaling and increased expressions of IAPs (Fig. 7). NAC treatment prior to CDDP administration may be an effective strategy against CDDP resistance, and the newly identified mutations in mtDNA may be potential biomarkers for sensitivity to CDDP.
Figure 7.
mtDNA mutations with varying percentage levels may lead to acquired CDDP resistance in cancer. The identified mtDNA mutations were found to cause the decrease of mitochondrial complex I activity and the elevation of intracellular ROS levels. ROS upregulate the expression of IAPs and phosphorylation of IкB-α and NF-кB, which led to the decreased sensitivity to CDDP. CDDP, cisplatin; mtDNA, mitochondrial DNA; ROS, reactive oxygen species; IAPs, inhibitors of apoptosis proteins.
Supplementary Material
Acknowledgements
We thank Gabrielle White Wolf, PhD for editing the draft of this manuscript.
Glossary
Abbreviations
- CDDP
cisplatin
- DMEM
Dulbecco's modified Eagle's medium
- IAPs
inhibitors of apoptosis proteins
- IRLs
intrinsic ROS levels
- NAC
N-acetyl-L-cysteine
- ND
NADH-ubiquinone oxidoreductase chain
- mtDNA
mitochondrial DNA
- ROS
reactive oxygen species
- RT-qPCR
reverse transcription quantitative real-time PCR
- TMH
transmembrane helix
- TG
6-thioguanine
Funding Statement
The present study was supported by a Grant-in-Aid for Scientific Research (no. 20K07169) to SH from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Availability of data and materials
The datasets used and analyzed during the current study are registered in the DDBJ under the accession number LC655995 and LC657585 and are available from the corresponding authors upon reasonable request.
Authors' contributions
SH designed the experiments. SH analyzed the data. KI and KN established the cybrids. MW and NT performed and analyzed the mitochondrial activity. SH wrote the paper. SH, SK, NS, TT and YR evaluated and confirmed the authenticity of all the raw data. All authors read and approved the final manuscript for publication.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 2.Gandhi L, Rodriguez-Abreu D, Gadgeel S, Esteban E, Felip E, Angelis FD, Domine M, Clingan P, Hochmair MJ, Powell SF, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med. 2018;378:2078–2092. doi: 10.1056/NEJMoa1801005. [DOI] [PubMed] [Google Scholar]
- 3.Paz-Ares LG, Luft A, Tafreshi A, Gumus M, Mazieres J, Hermes B, Senler FC, Fülöp A, Rodriguez-Cid J, Sugawara S, et al. Phase 3 study of carboplatin-paclitaxel/nab-paclitaxel (Chemo) with or without pembrolizumab (Pembro) for patients (Pts) with metastatic squamous (Sq) non-small cell lung cancer (NSCLC) J Clin Oncol. 2018;36((15_Suppl)):S105–S105. doi: 10.1200/JCO.2018.36.15_suppl.105. [DOI] [Google Scholar]
- 4.Komatsu M, Sumizawa TS, Mutoh M, Chen ZS, Terada K, Furukawa T, Yang XL, Gao H, Miura N, Sugiyama T, Akiyama S. Copper-transporting P-type adenosine triphosphatase (ATP7B) is associated with cisplatin resistance. Cancer Res. 2000;60:1312–1316. [PubMed] [Google Scholar]
- 5.Lai GM, Ozols RF, Young RC, Hamilton TC. Effect of glutathione on DNA repair in cisplatin-resistant human ovarian cancer cell lines. J Natl Cancer Inst. 1989;7:535–539. doi: 10.1093/jnci/81.7.535. [DOI] [PubMed] [Google Scholar]
- 6.Zeng-Rong N, Paterson J, Alpert L, Tsao MS, Viallet J, Alaoui-Jamali MA. Elevated DNA repair capacity is associated with intrinsic resistance of lung cancer to chemotherapy. Cancer Res. 1995;55:4760–4764. [PubMed] [Google Scholar]
- 7.Horibe S, Kawauchi S, Tanahashi T, Sasaki N, Mizuno S, Rikitake Y. CD44v-dependent upregulation of xCT is involved in the acquisition of cisplatin-resistance in human lung cancer A549cells. Biochem Biophys Res Commun. 2018;507:426–432. doi: 10.1016/j.bbrc.2018.11.055. [DOI] [PubMed] [Google Scholar]
- 8.Choi YM, Kim HK, Shim W, Anwar MA, Kwon JW, Kwon HK, Kim HJ, Jeong H, Kim HM, Hwang D, et al. Mechanism of cisplatin-induced cytotoxicity is correlated to impaired metabolism due to mitochondrial ROS generation. PLoS One. 2015;10:e0135083. doi: 10.1371/journal.pone.0135083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang SF, Chen MS, Chou YC, Ueng YF, Yin PH, Yeh TS, Lee HC. Mitochondrial dysfunction enhances cisplatin resistance in human gastric cancer cells via the ROS-activated GCN2-eIF2α-ATF4-xCT pathway. Oncotarget. 2016;7:74132–74151. doi: 10.18632/oncotarget.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. Mitochondrial DNA mutations and human disease. Biochim Biophys Acta. 2010;1797:113–128. doi: 10.1016/j.bbabio.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Yang Z, Schumaker LM, Egorin MJ, Zuhowski EG, Guo Z, Cullen KJ. Cisplatin preferentially binds mitochondrial DNA and voltage-dependent anion channel protein in the mitochondrial membrane of head and neck squamous cell carcinoma: Possible role in apoptosis. Clin Cancer Res. 2006;12:5817–5825. doi: 10.1158/1078-0432.CCR-06-1037. [DOI] [PubMed] [Google Scholar]
- 12.Olivero OA, Semino C, Kassim A, Lopez-Larraza DM, Poirier MC. Preferential binding of cisplatin to mitochondrial DNA of Chinese hamster ovary cells. Mutat Res. 1995;346:221–230. doi: 10.1016/0165-7992(95)90039-X. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez-Sanchez E, Marin JJ, Perez MJ. The expression of genes involved in hepatocellular carcinoma chemoresistance is affected by mitochondrial genome depletion. Mol Pharm. 2014;11:1856–1868. doi: 10.1021/mp400732p. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi J, Ohta S, Kikuchi A, Takemitsu M, Goto Y, Nonaka I. Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proc Natl Acad Sci USA. 1991;88:10614–10618. doi: 10.1073/pnas.88.23.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horibe S, Matsuda A, Tanahashi T, Inoue J, Kawauchi S, Mizuno S, Ueno M, Takahashi K, Maeda Y, Maegouchi T, et al. Cisplatin resistance in human lung cancer cells is linked with dysregulation of cell cycle associated proteins. Life Sci. 2015;124:31–40. doi: 10.1016/j.lfs.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 16.King MP, Attardi G. Human cells lacking mtDNA: Repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 17.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi JI. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 18.Semba H, Takeda N, Isagawa T, Sugiura Y, Honda K, Wake M, Miyazawa H, Yamaguchi Y, Miura M, Jenkins DM, et al. HIF-1α-PDK1 axis-induced active glycolysis plays an essential role in macrophage migratory capacity. Nat Commun. 2016;7:11635. doi: 10.1038/ncomms11635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Cai X, Lu W, Yang Y, Yang J, Ye J, Gu Z, Hu C, Wang X, Cao P. Digitoflavone inhibits IκBα kinase and enhances apoptosis induced by TNFα through downregulation of expression of nuclear factor κB-regulated gene products in human pancreatic cancer cells. PLoS One. 2013;8:e77126. doi: 10.1371/journal.pone.0077126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karin M. The beginning of the end: IkappaB kinase (IKK) and NF-kappaB activation. J Biol Chem. 1999;274:27339–27342. doi: 10.1074/jbc.274.39.27339. [DOI] [PubMed] [Google Scholar]
- 22.Murphy MP. Mitochondrial dysfunction indirectly elevates ROS production by the endoplasmic reticulum. Cell Metab. 2013;18:145–146. doi: 10.1016/j.cmet.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Tafani M, Sansone L, Limana F, Arcangeli T, Santis ED, Polese M, Fini M, Russo MA. The interplay of reactive oxygen species, hypoxia, inflammation, and sirtuins in cancer initiation and progression. Oxid Med Cell Longev. 2016;2016:3907147. doi: 10.1155/2016/3907147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saybaşili H, Yüksel M, Haklar G, Yalçin AS. Effect of mitochondrial electron transport chain inhibitors on superoxide radical generation in rat hippocampal and striatal slices. Antioxid Redox Signal. 2001;3:1099–1104. doi: 10.1089/152308601317203602. [DOI] [PubMed] [Google Scholar]
- 25.Staniek K, Gille L, Kozlov AV, Nohl H. Mitochondrial superoxide radical formation is controlled by electron bifurcation to the high and low potential pathways. Free Radic Res. 2002;36:381–387. doi: 10.1080/10715760290021225. [DOI] [PubMed] [Google Scholar]
- 26.Wu J, Jin Z, Zheng H, Yan LJ. Sources and implications of NADH/NAD(+) redox imbalance in diabetes and its complications. Diabetes Metab Syndr Obes. 2016;9:145–153. doi: 10.2147/DMSO.S106087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiedemann FR, Manfredi G, Mawrin C, Beal MF, Schon EA. Mitochondrial DNA and respiratory chain function in spinal cords of ALS patients. J Neurochem. 2002;80:616–625. doi: 10.1046/j.0022-3042.2001.00731.x. [DOI] [PubMed] [Google Scholar]
- 28.Rusecka J, Kaliszewska M, Bartnik E, Tońska K. Nuclear genes involved in mitochondrial diseases caused by instability of mitochondrial DNA. J Appl Genet. 2018;59:43–57. doi: 10.1007/s13353-017-0424-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cote HC, Brumme ZL, Craib KJ, Alexander CS, Wynhoven B, Ting L, Wong H, Harris M, Harrigan PR, O'Shaughnessy MV, Montaner JS. Changes in mitochondrial DNA as a marker of nucleoside toxicity in HIV-infected patients. N Engl J Med. 2002;346:811–820. doi: 10.1056/NEJMoa012035. [DOI] [PubMed] [Google Scholar]
- 30.Mambo E, Gao X, Cohen Y, Guo Z, Talalay P, Sidransky D. Electrophile and oxidant damage of mitochondrial DNA leading to rapid evolution of homoplasmic mutations. Proc Natl Acad Sci USA. 2003;100:1838–1843. doi: 10.1073/pnas.0437910100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang D, Kim SH, Hamasaki N. Mitochondrial transcription factor A (TFAM): Roles in maintenance of mtDNA and cellular functions. Mitochondrion. 2007;7:39–44. doi: 10.1016/j.mito.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 32.Kuhl I, Miranda M, Posse V, Milenkovic D, Mourier A, Siira SJ, Bonekamp NA, Neumann U, Filipovska A, Polosa PL, et al. POLRMT regulates the switch between replication primer formation and gene expression of mammalian mtDNA. Sci Adv. 2016;2:e1600963. doi: 10.1126/sciadv.1600963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu J, Vinothkumar KR, Hirst J. Structure of mammalian respiratory complex I. Nature. 2016;536:354–358. doi: 10.1038/nature19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fiedorczuk K, Letts JA, Degliesposti G, Kaszuba K, Skehel M, Sazanov LA. Atomic structure of the entire mammalian mitochondrial complex I. Nature. 2016;538:406–410. doi: 10.1038/nature19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fiedorczuk K, Sazanov LA. Mammalian mitochondrial complex I structure and disease-causing mutations. Trends Cell Biol. 2018;28:835–867. doi: 10.1016/j.tcb.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 36.Deveraux QL, Takahashi R, Salvesen GS, Reed JC. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi R, Deveraux Q, Tamm I, Welsh K, Assa-Munt N, Salvesen GS, Reed JC. A single BIR domain of XIAP sufficient for inhibiting caspases. J Biol Chem. 1998;273:7787–7790. doi: 10.1074/jbc.273.14.7787. [DOI] [PubMed] [Google Scholar]
- 38.Roy N, Deveraux QL, Takahashi R, Salvesen GS, Reed JC. The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J. 1997;16:6914–6925. doi: 10.1093/emboj/16.23.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maier JKX, Lahoua Z, Gendron NH, Fetni R, Johnston A, Davoodi J, Rasper D, Roy S, Slack RS, Nicholson DW, MacKenzie AE. The neuronal apoptosis inhibitory protein is a direct inhibitor of caspases 3 and 7. J Neurosci. 2002;22:2035–2043. doi: 10.1523/JNEUROSCI.22-06-02035.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davoodi J, Lin L, Kelly J, Liston P, MacKenzie AE. Neuronal apoptosis-inhibitory protein does not interact with Smac and requires ATP to bind caspase-9. J Biol Chem. 2004;279:40622–40628. doi: 10.1074/jbc.M405963200. [DOI] [PubMed] [Google Scholar]
- 41.Todor IN, Lukyanova NY, Chekhun VF. The lipid content of cisplatin- and doxorubicin-resistant MCF-7 human breast cancer cells. Exp Oncol. 2012;34:97–100. [PubMed] [Google Scholar]
- 42.Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function. Mol Cell. 2002;9:1031–1044. doi: 10.1016/S1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 43.Hlavata L, Aguilaniu H, Pichová A, Nyström T. The oncogenic RAS2(val19) mutation locks respiration, independently of PKA, in a mode prone to generate ROS. EMBO J. 2003;22:3337–3345. doi: 10.1093/emboj/cdg314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim JKM, Leprivier G. The impact of oncogenic RAS on redox balance and implications for cancer development. Cell Death Dis. 2019;10:955. doi: 10.1038/s41419-019-2192-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizutani S, Miyato Y, Shidara Y, Asoh S, Tokunaga A, Tajiri T, Ohta S. Mutations in the mitochondrial genome confer resistance of cancer cells to anticancer drugs. Cancer Sci. 2009;100:1680–1687. doi: 10.1111/j.1349-7006.2009.01238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kidani A, Izumi H, Yoshida Y, Kashiwagi E, Ohmori H, Tanaka T, Kuwano M, Kohno K. Thioredoxin2 enhances the damaged DNA binding activity of mtTFA through direct interaction. Int J Oncol. 2009;35:1435–1440. doi: 10.3892/ijo_00000462. [DOI] [PubMed] [Google Scholar]
- 47.Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- 48.Cory S, Adams JM. The Bcl2 family: Regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the current study are registered in the DDBJ under the accession number LC655995 and LC657585 and are available from the corresponding authors upon reasonable request.