Abstract
The rise of antibiotic resistance has increased the need for alternative ways of preventing and treating enteropathogenic bacterial infection. Various probiotic bacteria have been used in animal and human. However, Saccharomyces boulardii is the only yeast currently used in humans as probiotic. There is scarce research conducted on yeast species commonly found in kefir despite its claimed potential preventative and curative effects. This work focused on adhesion properties, and antibacterial metabolites produced by Kluyveromyces lactis and Saccharomyces unisporus isolated from traditional kefir grains compared to Saccharomyces boulardii strains. Adhesion and sedimentation assay, slide agglutination, microscopy and turbidimetry assay were used to analyze adhesion of Salmonella Arizonae and Salmonella Typhimurium onto yeast cells. Salmonella growth inhibition due to the antimicrobial metabolites produced by yeasts in killer toxin medium was analyzed by slab on the lawn, turbidimetry, tube dilution and solid agar plating assays. Alcohol and antimicrobial proteins production by yeasts in killer toxin medium were analyzed using gas chromatography and shotgun proteomics, respectively. Salmonella adhered onto viable and non-viable yeast isolates cell wall. Adhesion was visualized using scanning electron microscope. Yeasts-fermented killer toxin medium showed Salmonella growth inhibition. The highest alcohol concentration detected was 1.55%, and proteins with known antimicrobial properties including cathelicidin, xanthine dehydrogenase, mucin-1, lactadherin, lactoperoxidase, serum amyloid A protein and lactotransferrin were detected in yeasts fermented killer medium. These proteins are suggested to be responsible for the observed growth inhibition effect of yeasts-fermented killer toxin medium. Kluyveromyces lactis and Saccharomyces unisporus have anti-salmonella effect comparable to Saccharomyces boulardii strains, and therefore have potential to control Salmonella infection.
Keywords: Kluyveromyces lactis, Probiotics, Kefir, Saccharomyces unisporus, Saccharomyces boulardii, Salmonella, Yeasts, Shotgun proteomics
Abbreviations: WHO, World Health Organization; FAO, Food Agriculture Organization; ATCC, American type Culture Collection; YEPDA, Yeast Extract Peptone Dextrose Agar; YEPDB, Yeast Extract Peptone Dextrose Broth; PBS, Phosphate buffered saline; CFU, Colony Forming Unit; DSR, Desk Sputter Coater ; HCL, Hydrochloric Acid; NaOH, Sodium hydroxide; KTM, Killer Toxin Cedium; CFS, Cell Free Supernatant; DTT, Dithiothreitol ; LC-MS/MS, Liquid Chromatography with tandem mass spectrometry/Liquid Chromatography with tandem mass spectrometry; HPLC, High-performance liquid chromatography; mL, Milliliter; Min, Minute; h, Hour; RSLC, Rapid Separation Liquid Chromatography; AGC, Automatic Gain Control; SPSS, Statistical Package for the Social Sciences; IBM, International Business Machines ; SD, Standard Deviation; LFQ, Label Free Quantitation; GIT, The gastrointestinal tract; DNA, Deoxyribonucleic Acid; RNA, Ribonucleic Acid; ATP, Adenosine triphosphate
1. Introduction
Salmonella resistance to current antibiotic drugs is rising at an alarming rate worldwide especially in Africa and Asia. For example in Malawi, the percentage of multi-drug resistant Salmonella enterica serovar Typhi increased from 7% in 2010 to 97% in 2014 (Feasey et al., 2015). This has raised the need for alternative prophylaxis and therapies. One of the ways of preventing and treating infectious diseases in human and animal is the use of probiotic microorganisms (Feye et al., 2019, Gut et al., 2018, Kogan and Kocher, 2007). Probiotics are defined by World Health Organization (WHO) as ‘live microorganisms which when administered in adequate amounts confer a health benefit on the host’ (FAO/WHO, 2002). Lactobacillus, Bifidobacterium, Streptococcus thermophilus, Enterococcus, Leuconostoc species, Escherichia coli Nissle 1917, Bacillus species and Saccharomyces boulardii are currently used as probiotics in humans against bacterial pathogens infection (Bakken, 2014, Bekar et al., 2011, Khodadad et al., 2013, Nami et al., 2015). Probiotics are commonly found in popular fermented functional foods such as yoghurt, milk, kefir, cheese, soybean, fruits, sourdough and vegetable products (Plessas et al., 2016, Prado et al., 2015, Priyodip et al., 2017, Saarela et al., 2000) or formulated into either lyophilized forms as capsules, powders or in aqueous solutions (Martins et al., 2009). These microorganisms exert antagonism against susceptible bacteria by production of antibacterial molecules, prevention of biofilm formation and pathogen invasiveness; adhesion of bacteria cells onto cell walls, and degradation of bacterial toxins among others (Gut et al., 2018).
Kefir drink is a probiotic product made from a kefir grain, a consortium of many microorganisms and exopolysaccharide, consumed in many parts of the world including the Caucasus Mountains of Russia, Europe, Asia, South and North America due to its health benefits conferred by the probiotic components (Garrote et al., 1997). Bacterial cultures of kefir include Lactobacillus, Lactococcus, Leuconostoc and Streptococcus genera whereas the yeast cultures include Kluyveromyces, Candida, Saccharomyces and Pichia (Plessas et al., 2016).
The aim of this study was to screen kefir yeast isolates for potential application in Salmonella infection control. The study hypothesized that yeasts isolated from kefir and Saccharomyces boulardii may show Salmonella binding capability, as well as growth inhibition due to production of antimicrobial metabolites. Specifically, Salmonella enterica serovar Arizonae (S. Arizonae) and Salmonella enterica serovar Typhimurium (S. Typhimurium) adhesion onto Saccharomyces unisporus ATCC 10612 (S. unisporus) and Kluyveromyces lactis var. lactis ATCC 56498 (K. lactis) as well as growth inhibition due to antibacterial metabolites were analyzed in an in vitro experiments in comparison to Saccharomyces boulardii strains {Saccharomyces var boulardii MYA-796 (SB48) and Saccharomyces var boulardii MYA-797 (SB49)}.
2. Materials and methods
2.1. Materials
Microbiological cultures including kefir yeast isolates (S. unisporusd and K. lactis), S. boulardii strains (Victoria University culture collection). S. Arizonae ATCC 13314 and S. Typhimurium ATCC 14028 (In vitro Technologies, Melbourne, Australia). Microbiological media including yeast extract peptone dextrose agar (YEPDA), YEPD broth (YEPDB), nutrient agar, chloramphenicol drug (100 mg/L), and nutrient broth (Basingstoke, United Kingdom). Phosphate buffered saline (PBS), cycloheximide (0.1%), bile extract porcine (containing glycine & taurine conjugate of hyodeoxycholic acid), glycerol, citrate-phosphate buffer, 0.22 μm sterile filters (Sigma Aldrich, St. Louis, USA) and absolute ethanol (Rowe Scientific, Melbourne, Australia).
2.2. Preparation of Salmonella and yeast cultures
Salmonella and yeast cultures were prepared as described in a previous study (Tiago et al., 2012) with some modifications. Well-isolated S. Arizonae and S. Typhimurium colonies initially grown on nutrient agar and incubated at 37 °C for 24 h were inoculated into 10 mL nutrient broth and incubated at 37 °C for 24 h. One milliliter of each culture was serially diluted to approximately 103 CFU/mL. Another set of Salmonella cultures with approximately 109 CFU/mL (undiluted culture) was set aside. Well-isolated yeast colonies on YEPDA were picked and inoculated into 10 mL YEPDB and incubated at 25 °C for 24 h in a shaking incubator at 100 horizontal strokes/min (Innova 4230 New Brunswick Scientific, Edison, NJ, USA). The yeast cultures with approximately 108 CFU/mL were labelled accordingly. All microbial cultures were kept in a refrigerator at 4 °C prior to use when necessary.
2.3. Adhesion of Salmonella onto yeast cell wall
2.3.1. Salmonella adhesion onto yeast cell wall
Adhesion of Salmonella onto yeast cell wall was performed as previously described (Tiago et al., 2012) with some modifications. One milliliter of viable yeast culture broth (approximately 108 CFU/mL) was added to a 15 mL centrifuge tube containing 0.5 mL of Salmonella culture (approximately 109 CFU/mL). The bacteria-yeast mixture was vortexed for 30 s and incubated at 37 °C for 4 h. One hundred microliter was removed from the top and was serially diluted in 0.1% peptone water to 10−7. One milliliter of each dilution was added to a Petri dish and then 20 mL molten nutrient agar at 45 °C (supplemented with 0.1% cycloheximide to suppress yeasts growth) was added and mixed gently. Plates were incubated at 37 °C for 24 h. S. Arizonae and S. Typhimurium colonies were counted and expressed as log10 CFU/mL. For the controls, 1 mL sterile YEPDB (containing no yeast cells) was added to 0.5 mL of each Salmonella serovars and treated as above.
To assess adhesion of Salmonella onto non-viable yeast cells, 10 mL of yeast cultures were heated at 100 °C for 5 min to obtain inactivated cells. Adhesion and sedimentation were performed as per the protocol for viable yeast cells described above.
Slide agglutination was performed as described in the literature (Perez-Sotelo et al., 2005) by inoculating 0.02 mL viable and non-viable yeast suspension with 0.01 mL Salmonella on microscopic slides. The two cultures were mixed using sterile inoculating loop before gentle rocking the slide. Agglutination was observed under illumination (Perez-Sotelo et al., 2005).
Salmonella (0.01 mL, 109 CFU/mL) and yeast (108 CFU/mL 0.02 mL) were added to a clean microscope slide and mixed using sterile inoculating loop. The smear was allowed to air dry before fixing with 0.1% glutaraldehyde, washed three times with 1X PBS and allowed to air dry again. Smear on slides were dehydrated using classical dehydration process (Kiekens et al., 2019, Piroeva et al., 2013) with modification. Briefly, slides were soaked stepwise in 50%, 75% and absolute ethanol consequentially for 30 min at room temperature before drying at 37 °C for 1 h. The slides were then glued onto scanning electron microscope stubs and gold-coated by sputtering for 5 min using a desk sputter coater DSR1 (Markham, Canada). The adhesion was qualitatively observed using a Teneo scanning electron microscope (Thermo Fisher Scientific, Hillsboro, USA). Adhesion of Salmonella onto both viable and heat-inactivated yeast cell wall was analyzed at magnification of 10,000×.
2.3.2. Salmonella growth in presence of inactivated yeast cells
Growth behaviour of Salmonella in presence of inactivated yeast cells was analyzed using turbidimetry assay. Six milliliters of yeast (109 CFU/mL) and 3 mL of Salmonella (109 CFU/mL) cultures were centrifuged (Eppendorf 5810 R, Hamburg, Germany) at 4000g for 15 min at 4 °C and washed twice with 1X PBS respectively. The yeast pellets were re-suspended in 6 mL of 1X PBS whereas the Salmonella pellets were re-suspended in 3 mL 1X PBS and 3 mL nutrient broth (as source of nutrients for growth). One milliliter of the yeast suspension (102 CFU/mL) and 0.5 mL of the Salmonella suspension (103 CFU/mL) were added to a 15 mL centrifuge tube and vortexed for 20 s to mix. Salmonella cultures in 1X PBS and nutrient broth were used as control. All the tubes containing cultures including controls were supplemented with 0.1% cycloheximide (4 mL/L) to inactivate yeasts. Prepared Salmonella-yeasts mixtures (0.2 mL) and controls were dispensed into flat-bottom 96 well microtiter plate (Biorad, California, USA). The optical densities were read at 0, 2, 4, 6, 12 and 24 h using microplate reader at 595 nm (Biorad, California, USA).
2.3.3. Factors affecting Salmonella adhesion onto yeast cell wall
The pH of yeast and Salmonella culture broths were adjusted separately to 2.0, 4.0, 5.0, 7.0, and 8.0 with either 1 M HCl or 1 M NaOH. Salmonella (20 µL, 109 CFU/mL) and yeast (10 µL, 108 CFU/mL) of same pH were added onto a clean microscope slide, mixed using a sterile inoculating loop, rocked gently and observed under illumination for agglutination as described in the literature (Perez-Sotelo et al., 2005). Both viable and non-viable yeasts were tested.
Effect of bile salt on Salmonella adhesion onto yeast cell wall was performed as described in the literature (Tiago et al., 2012) with some modifications. Briefly, 6 mL each of yeast (108 CFU/mL) and Salmonella (109 CFU/mL) broths were centrifuged (Eppendorf 5810 R, Hamburg, Germany) separately at 4000g for 15 min at 4 °C and washed twice with 1X PBS. The pellets were each re-suspended in 6 mL 1X PBS containing 3 g/L bile (supplemented with 0.1 % cycloheximide 4 mL/L). For the control, S. Arizonae and S. Typhimurium prepared separately without yeast cells, were similarly treated as above. Qualitative and quantitative analysis were performed as described under adhesion and sedimentation assays. However, experimental conditions including time, temperature, initial culturing of yeast and Salmonella in bile-free media were maintained to reflect standardized simulated intestinal digestive system (Minekus et al., 2014).
2.4. Salmonella growth inhibition by yeast metabolites
2.4.1. Salmonella growth inhibition assay
Salmonella growth inhibition by yeast colonies was performed as described in the literature (Gut et al, 2019). Yeast colonies on YEPDA grown for 72 h at 25 °C were cut and placed onto Muller-Hinton agar (supplemented with 0.1% cycloheximide) initially inoculated with 104 CFU/mL of S. Arizonae and S. Typhimurium using spread plate technique. The Muller-Hinton agar plates were incubated at 37 °C for 24 h. Plates were checked for growth inhibition indicated by complete or partial clearing around the yeast slabs.
Further experimentation to investigate Salmonella growth inhibition (Bajaj et al., 2013) was carried out with some modifications. Yeasts were grown in a killer toxin medium (KTM) composed of YEPDB plus glycerol (50 g/L), buffered at pH 5 using 50 mM citrate-phosphate buffer, and incubated at 25 °C under shaking (180 rpm) for 24–48 h. KTM has been reported to enhance production of killer toxin by yeast species (Bajaj et al., 2013). Sampling for KTM cell free supernatant (KTM-CFS) was performed at 24 and 48 h. The yeast fermented KTM was centrifuged (Eppendorf 5810 R, Hamburg, Germany) at 4000g for 30 min at 4 °C and filter sterilized using 0.22 μm membrane filter.
KTM-CFS was analyzed for antibacterial potential using established turbidimetry assay (broth dilution) with some modifications (Balouiri et al., 2016). Briefly, 0.1 mL of 104 CFU/mL of Salmonella culture in fresh nutrient broth was dispensed into flat-bottom 96 well microtiter plate (Bio-Rad, California, USA) containing 0.1 mL of KTM-CFS. The optical density was read using microplate reader at 595 nm (Biorad, California, USA) at 0, 2, 4, 6, 12 and 24 h. Growth curves were constructed using a Microsoft Excel.
To determine if the effect of KTM-CFS on Salmonella demonstrated by turbidimetry assay is bacteriostatic or bactericidal, 5 mL of the KTM-CFS was inoculated with 0.5 mL of Salmonella cultures (approximately 103 CFU/mL). Sampling was performed at < 1, 30, 60, 90, 120 and 240 min by plating 0.1 mL onto nutrient agar, incubated in 37 °C for 24 h before counting colonies. For control, unfermented KTM was inoculated with Salmonella and tested along with the samples.
2.4.2. Determination of alcohol content in KTM and its effect on Salmonella species
Alcohol content of KTM-CFS was analyzed quantitatively by gas chromatography as described in a previous study (Nikolaou et al., 2017) with some modifications. Gas chromatography (Shimazdzu, Kyoto, Japan) with SGE BP20 GC capillary column (12.0 m length, 0.22 mm inner diameter, 0.25 μm film thickness, Fisher Scientific, Hampton, USA) and flame ionization detector at 200 °C was used. Samples were filter sterilized through a 0.22 μm membrane filter (Sigma Aldrich, St. Louis, USA) and 5 μL was injected into the column. The oven temperature was set as 35 °C for 5 min, then increased to 200 °C at a rate of 10 °C/min. The injector temperature was maintained at 200 °C, with a split ratio of 50:1 and flow rate of 1.1/min. Analysis of the results were performed using a Lab Solution software (Shimazdzu, Kyoto, Japan). Alcohol concentration was calculated using 6 point standards (0, 125, 250, 500, 750 and 1000 mM/L; standard curve (R2 > 0.99) created with absolute ethanol.
To determine if the concentration of ethanol in yeast fermented KTM is responsible for Salmonella growth inhibition demonstrated by the turbidimetry assay, ethanol control (1, 1.5 and 2% in sterile distilled water) effect on Salmonella was investigated. Five milliliters of each control concentration was inoculated with 0.5 mL of Salmonella cultures (approximately 103 CFU/mL). Sampling time, plating, incubation and colony counting were performed as described under 2.4.1.
To determine if the growth inhibition was due to other volatile metabolites other than ethanol, KTM-CFS was heated in loosely capped centrifuge tubes at 60 °C for 10 min in a shaking incubator at 120 horizontal strokes per min. Five milliliters of the heat treated KTM-CFS was inoculated with 0.5 mL of Salmonella cultures (approximately 103 CFU/mL) prepared above. Sampling time, plating, incubation and colony counting were performed as described under 2.4.1.
2.4.3. Shotgun proteomics
Yeast-fermented KTM and unfermented KTM (control) samples were analyzed for proteins and peptides identification using shotgun proteomics. Samples were diluted to between 1 and 5 mg/mL total protein concentration, and approximately 10 µg/L total protein of each was buffer exchanged into 50 mM ammonium bicarbonate and the protein was reduced in 2.5 mM DTT at 95 °C for 5 min followed by alkylation with 10 mM chloroacetamide for 30 min at ambient temperature. Trypsin was then added at the rate of 0.5 µg per 10 µg of protein and incubated at 37 °C overnight. All enzyme digests were analyzed by LC-MS/MS using the QExactive Plus mass spectrometer (Thermo Scientific, Bremen, Germany) coupled online with a RSLC nano HPLC (Ultimate 3000, Thermo Scientific, Bremen, Germany). Two hundred nanograms of sample was injected and concentrated on a 100 µm, 2 cm nanoviper pepmap100 trap column with 97.5% buffer A (0.1% Trifluoroacetic acid) at a flow rate of 15 min−1. The peptides were then eluted and separated with a Thermo RSLC pepmap100, 75 µm × 50 cm, 100 Ǻ pore size, reversed phase nano column with a 30 min gradient of 92.5% buffer A (0.1% formic acid) to 42.5% B (80% acetonitrile 0.1% formic acid), at a flow rate of 250 nL min−1. The eluent was nebulised and ionised using the Thermo nano electrospray source with a distal coated fused silica emitter (New Objective, Woburn, MA, USA) with a capillary voltage of 1900 V. Peptides were selected for MS/MS analysis in full MS/dd-MS2 (TopN) mode with the following parameter settings: TopN 10, resolution 70000, MS/MS AGC target 5e5, 118 ms Max IT, NCE 27, 1.8 m/z isolation window, dynamic exclusion was set to 10 s. Results were analyzed using MaxQuant to obtain protein identifications and their respective label-free quantification values using in-house standard parameters. Data were normalized based on the assumption that the majority of proteins do not change between the different conditions. Protein identification numbers were also used to verify protein names using Uniprot database.
2.5. Statistical analysis
Experiments were performed in triplicates and subsampled. Results were expressed as the mean ± standard deviation (SD). Means differences were statistically analyzed using Student’s t-test. Mean differences were considered significant at p < 0.05. Statistical analysis was performed using SPSS version 26 Statistical software (IBM, New York, USA). Growth curves were constructed using Microsoft Excel 2016 (Microsoft, Washington, United States). The means for all the experiment followed a normal distribution. For shotgun proteomics, statistical analysis was performed using Perseus. The LFQ data was converted to log2 scale, samples were grouped by conditions and missing values were imputed based on normal distributions after all proteins were eliminated that had 2 or less valid values. Protein fold-changes were calculated and their significance was determined using a two-sided T-test with error-corrected P- values.
3. Results
3.1. Adhesion of Salmonella onto yeast cells
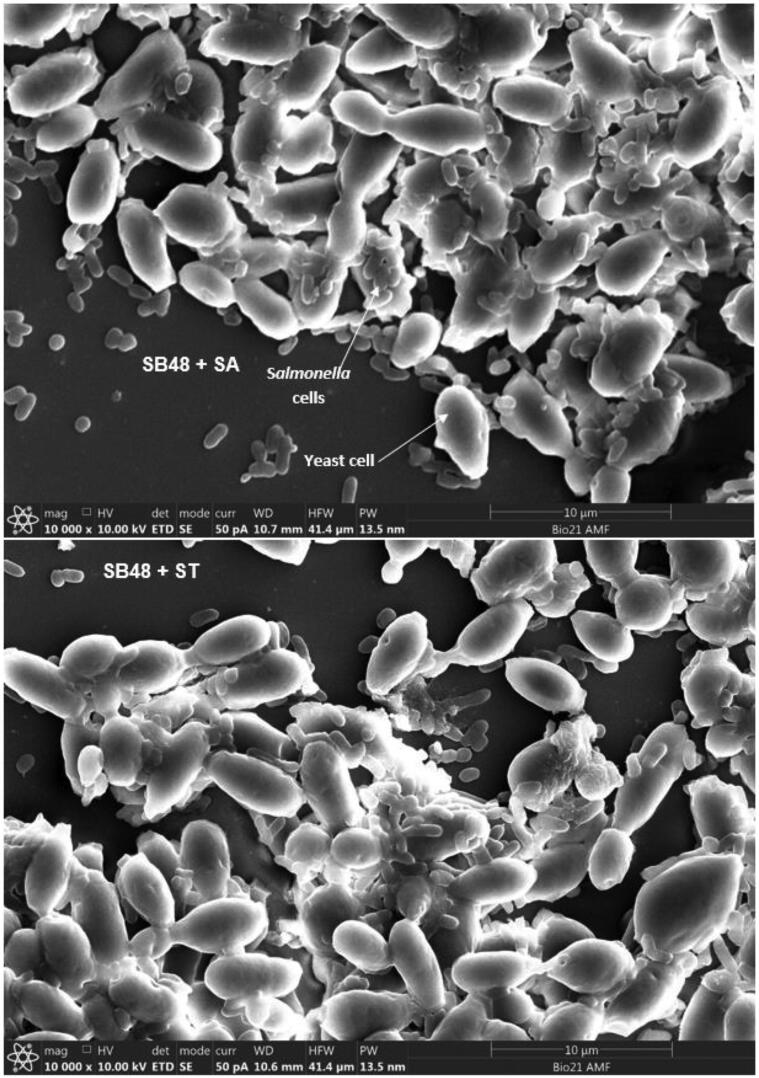
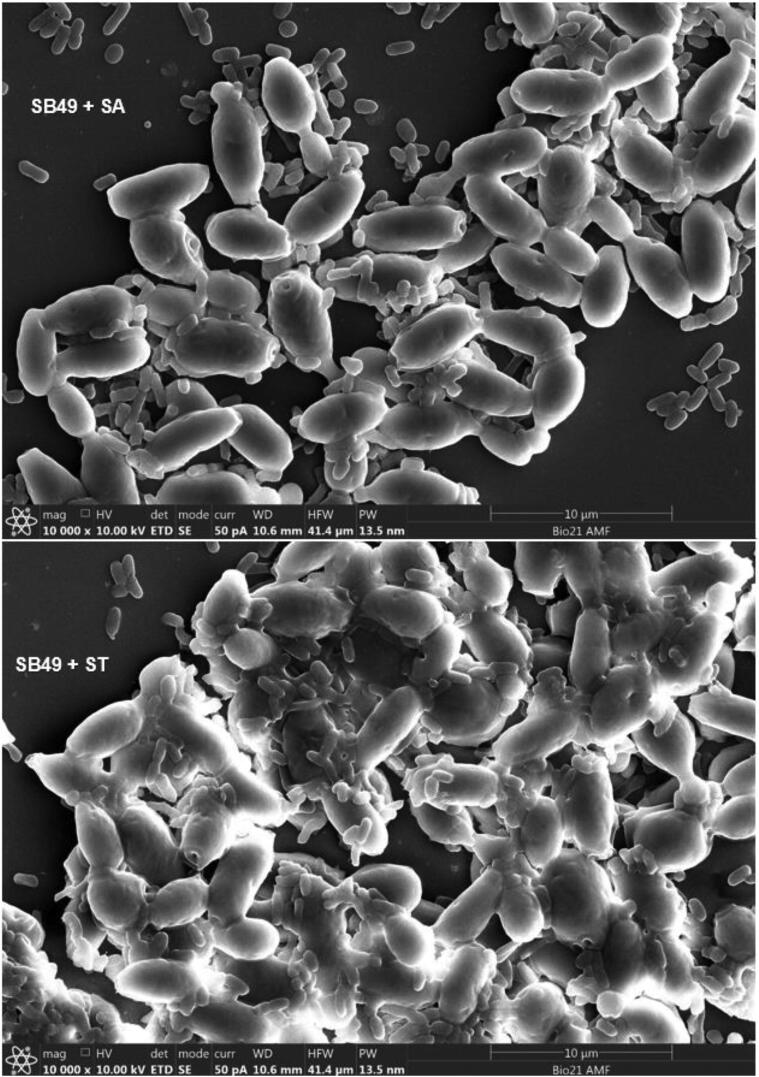
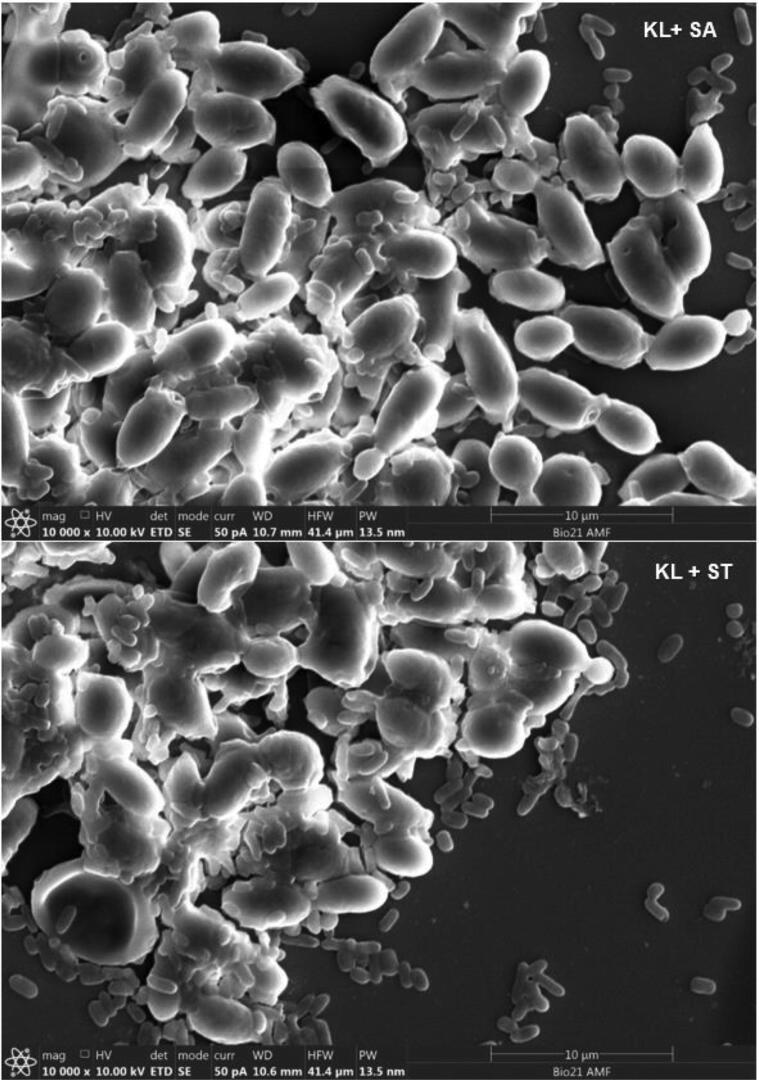
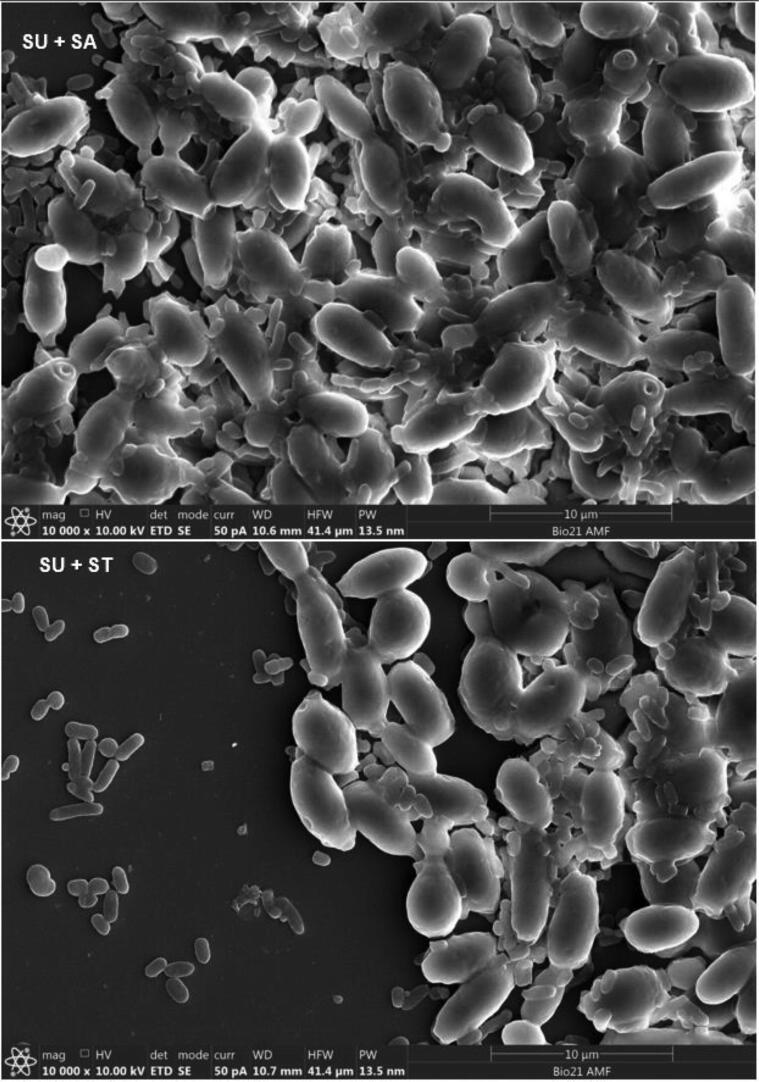
Adherence of S. Arizonae and S. Typhimurium onto viable and non-viable yeast cells was qualitatively and quantitatively analyzed (Table 1). Colony count of the two Salmonella serovars after sedimentation with K. lactis, S. unisporus and S. boulardii strains were lower (p < 0.05) compared to the controls for both viable and non-viable yeast cells. Agglutination on the slides which is an indication of aggregation (between Salmonella and yeast cells) was also observed in all cases (Table 1). Fig. 1A, Fig. 1B, Fig. 1C, Fig. 1D show clear adherence of Salmonella onto viable yeast strains observed using a scanning electron microscope. Observed adhesions of Salmonella onto non-viable kefir yeast isolates and S. boulardii strain (data not shown) appeared similar to that onto viable yeasts. Scanning electron microscope results provide clear visual evidence that Salmonella count reduced after treatment with yeast, likely caused by attachment to yeast cells and subsequent settling at the bottom of the tubes.
Table 1.
Adhesion of Salmonella onto yeast cell wall: quantitative and qualitative analysis results.
| Yeast strains/control | S. Arizonae Count (log10 CFU/ mL) | S. Typhimurium Count (log10 CFU/ mL) | |
|---|---|---|---|
| Live yeast | Control (YEPD) | 9.14 ± 0.05b | 9.20 ± 0.05b |
| SB48 | 8.69 ± 0.40 ++a | 8.66 ± +++a | |
| SB49 | 8.63 ± ++a | 8.82 ± +++a | |
| KL | 8.88 ± ++a | 8.81 ± 0.11 +++a | |
| SU | 8.97 ± ++a | 8.84 ± 0.20 +++a | |
| Heat-killed yeast | Control (YEPD) | 8.92 ± 0.04b | 8.93 ± 0.06b |
| SB48 | 8.67 ± ++ a | 8.32 ± 0.17 ++ a | |
| SB49 | 8.68 ± ++ a | 8.28 ± 0.19 ++ a | |
| KL | 8.68 ± ++ a | 8.69 ± 0.11 ++ a | |
| SU | 8.69 ± ++ a | 8.67 ± 0.09 ++ a | |
| Bile | Control (bilee) | 9.04 ± 0.05b | 8.96 ± 0.07b |
| SB48 | 8.74 ± 0.10 ++a | 8.68 ± 0.09 ++a | |
| SB49 | 8.72 ± ++a | 8.86 ± 0.03 ++a | |
| KL | 7.72 ± ++a | 8.88 ± ++a | |
| SU | 8.78 ± 0.06 ++a | 8.67 ± 0.13 ++a | |
| pH 2.0, 4.0, 5.0 and 7.0, and 8.0 (live yeast) | SB48 | ++ | +++ |
| SB49 | ++ | +++ | |
| KL | ++ | +++ | |
| SU | ++ | +++ | |
The experiments was performed twice and in triplicate and the values are reported as the mean plus standard deviation; + = very weak agglutination seen after 5 s of gentle rocking of slide; ++ medium level agglutination seen after 5 s of gentle rocking of slide; +++ = very strong instant agglutination seen after rocking of slide. Means with different superscript are significantly different (p < 0.05
Fig. 1A.
Adhesion of Salmonella servers onto SB48 cell walls observed using scanning electron microscope. SA = S. Arizonae; ST = S. Typhimurium.
Fig. 1B.
Adhesion of Salmonella servers onto SB49 cell walls observed using scanning electron microscope. SA = S. Arizonae; ST = S. Typhimurium.
Fig. 1C.
Adhesion of Salmonella servers onto KL cell walls observed using scanning electron microscope. SA = S. Arizonae; ST = S. Typhimurium, KL = K. lactis.
Fig. 1D.
Adhesion of Salmonella servers onto SU cell walls observed using scanning electron microscope. SA = S. Arizonae; ST = S. Typhimurium, SU = S. unisporus.
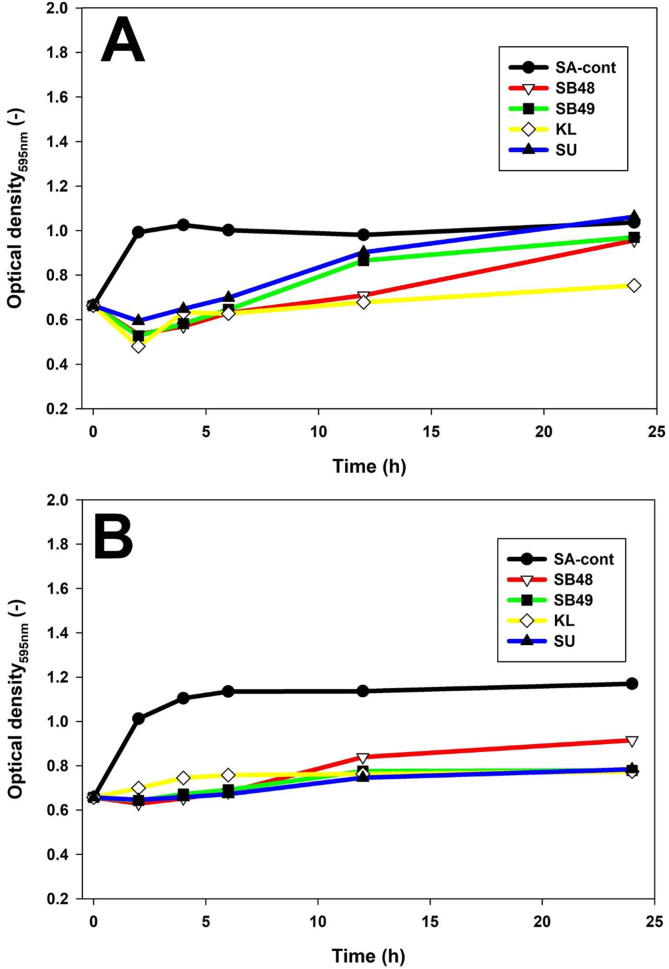
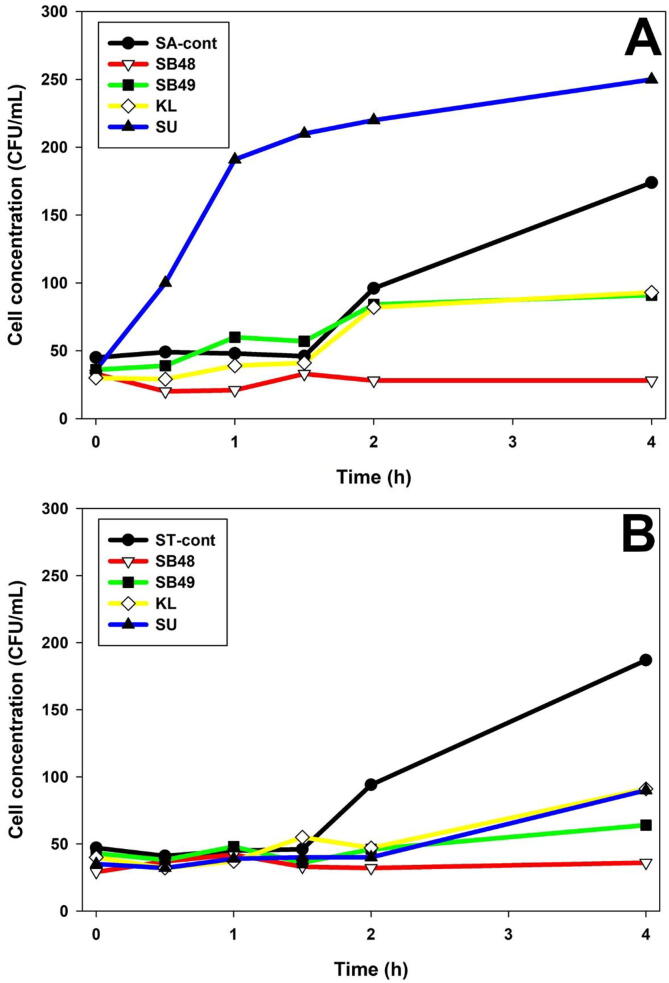
The growth behavior of Salmonella in presence of inactivated yeast cells was determined at different time points using optical density (Fig. 2). Growth rate of S. Arizonae was lower in all yeast cells compared to the control (p < 0.05) until 12th h (Fig. 2). On the other hand, S. Typhimurium had its growth significantly suppressed by all yeasts compared to the control (P < 0.05) until the experiment was stopped at 24 h (Fig. 2).
Fig. 2.
Salmonella real time growth analysis in presence of cyloheximide inactivated yeast cells. A, S. Arizonae; B, S. Typhimurium.
3.2. Factors affecting Salmonella adhesion onto yeast cell walls
The effect of pH on adhesion was investigated qualitatively at pH 2, 4, 5, 7 and 8 (Table 1). The gastrointestinal tract (GIT) pH ranged from 3 to 7, with the stomach being more acidic (Minekus et al., 2014). There was no difference in observable agglutination with all the pH levels, which indicated that pH did not interfere with adhesion.
Bile salt (0.3%) had no effect on adhesion of Salmonella onto K. lactis, S. unisporus, and S. boulardii strains (Table 1). Slide agglutination in presence of bile did not interfere with adhesion of Salmonella onto yeast cell walls with exception of K. lactis, which showed weak clumping on slide agglutination.
3.3. Salmonella growth inhibition by yeast antimicrobial metabolites
3.3.1. Salmonella growth inhibition by yeasts metabolites
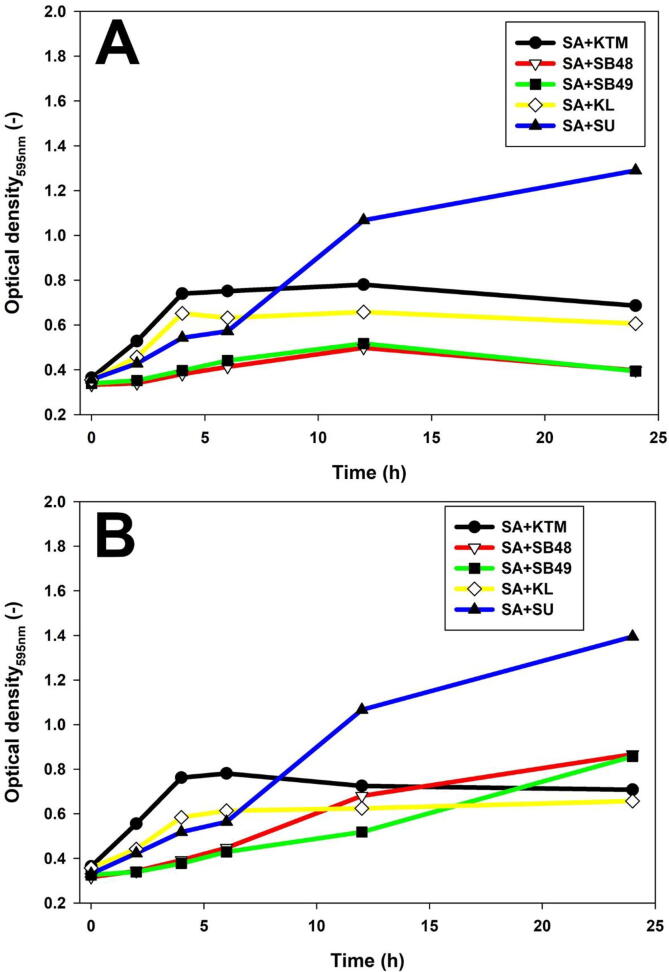
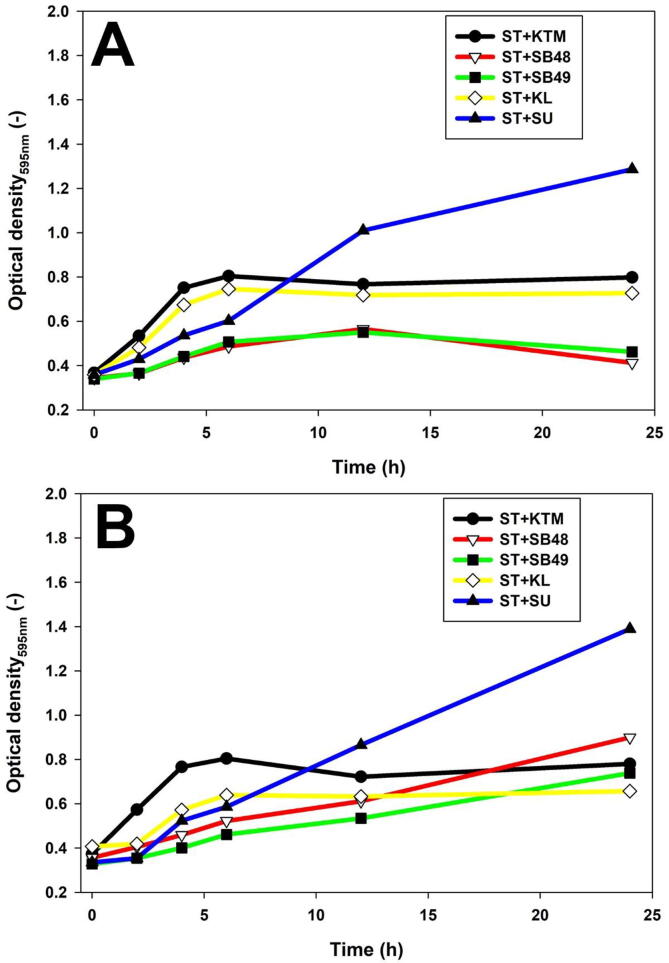
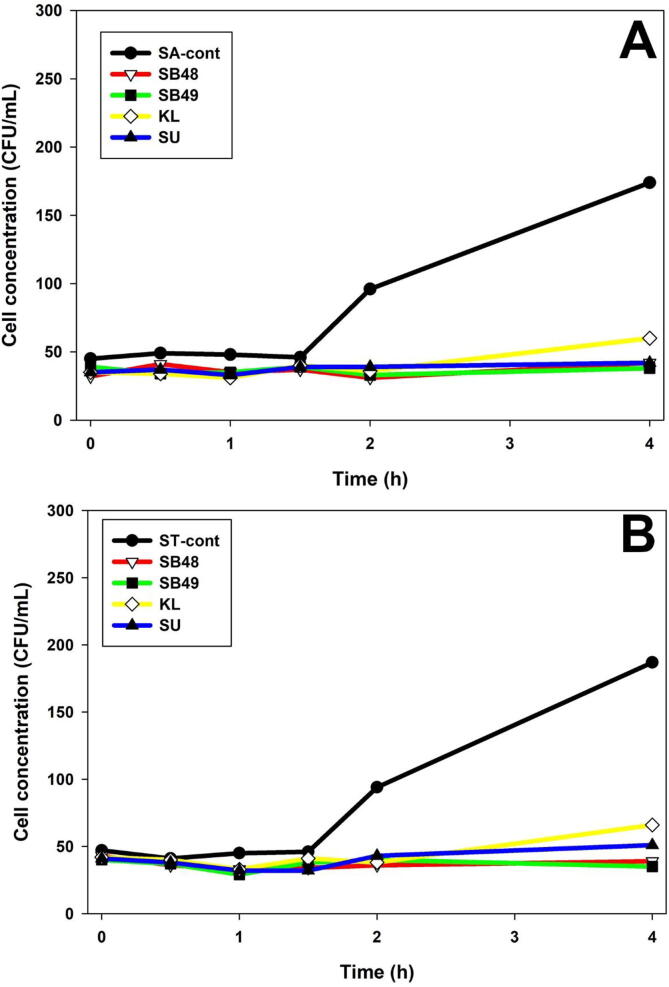
Potential presence of anti-Salmonella molecules produced by yeast was investigated using slab on the lawn assay, and growth inhibition was not observed (data not shown). The effect of potentially antimicrobial metabolites in KTM-CFS was assessed (Fig. 3, Fig. 4) using a turbidimetry assay. Salmonella growth was suppressed by S. unisporus KTM-CFS in the first 5 h, but started to grow rapidly thereafter, compared to the control. However, K. lactis exhibited growth rate suppression of S. Typhimurium at all-time points analyzed. Forty-eight hour fermentation of KTM with K. lactis showed stronger growth inhibition of Salmonella compared to 24 h (Fig. 3, Fig. 4). S. boulardii strains showed significant growth rate reduction of Salmonella compared to kefir yeast isolates (Fig. 3, Fig. 4). Fig. 5 showed that Salmonella count remained constant without significant decrease or increase in yeast-fermented KTM, which indicated growth inhibition effect.
Fig. 3.
S. Arizonae growth inhibition by yeast fermented KTM using optical turbidimetry assay. A, KTM fermented for 24 h; B, KTM fermented for 48 h.
Fig. 4.
S. Typhimurium growth inhibition by yeast fermented KTM using optical turbidimetry assay. A, KTM fermented for 24 h; B, KTM fermented for 48 h.
Fig. 5.
Determination of bacteriostatic and bactericidal effect of KTM on Salmonella using colony forming unit counting essay. A; S. Arizonae; B S. Typhimurium.
3.3.2. Assessment of alcohol content in KTM
To confirm the presence of alcohol in KTM-CFS, GC analysis was performed with data shown in Table 2. Fermentation of KTM with S. boulardii showed significantly higher concentration of alcohol after 24 h compared to that at 48 h. The concentration of alcohol produced by kefir yeast isolates in KTM-CFS from 48 h fermentation was also lower than that at 24 h (Table 2). Experimentation to determine if alcohol in yeasts-fermented KTM is responsible for growth inhibition above showed no effect on Salmonella, possibly due to low concentration (data not shown). Heat treatment of the fermented KTM-CFS to remove potential volatile antimicrobial compounds did not have effect on Salmonella growth for K. lactis, and S. boulardii strains. However, S. unisporus fermented KTM-CFS growth enhancing effect on S. arizonae was lost (Fig. 6).
Table 2.
Analysis of alcohol content in fermented KTM using GC.
| Yeast strain | Alcohol concentration (% V/V) |
|
|---|---|---|
| 24 h KTM fermentation | 48 h KTM fermentation | |
| SB48 | 1.55 ± 0.16a | 0.64 ± 0.01a |
| SB49 | 1.42 ± 0.01a | 0.43 ± 0.04a |
| KL | 1.30 ± 0.06a | 1.21 ± 0.17a |
| SU | 1.46 ± 0.01a | 1.32 ± 0.07a |
| KTM blank | 0.08 ± 0.05b | 0.08 ± 0.05b |
Means with different superscript are significantly different (p < 0.05)
Fig. 6.
Determination of bacteriostatic and bactericidal effect of heat treated KTM on Salmonella using colony forming unit counting essay. A; S. Arizonae; B S. Typhimurium.
3.3.3. Assessment of antimicrobial proteins in KTM
Production of proteins with potential antimicrobial properties in yeasts–fermented KTM was assessed using shotgun proteomics. Proteins shown in Table 3 were produced in substantial quantities in yeasts-fermented KTM, but present in trace amount or absent in KTM blank.
Table 3.
Shotgun proteomic analysis of yeast fermented KTM.
| SB48 | SB49 | KL | SU |
|---|---|---|---|
| 40S ribosomal protein S14-A 40S ribosomal protein S14-B 40S ribosomal protein S29 60 kDa chaperonin 60S ribosomal protein L13 60S ribosomal protein L20-B 60S ribosomal protein L20-A 60S ribosomal protein L28 60S ribosomal protein L34 60S ribosomal protein L35a, 60S ribosomal protein L35a 60S ribosomal protein L36 60S ribosomal protein L37a 60S ribosomal protein L8 Actin, cytoplasmic 1 Acyl-CoA-binding protein Adenosylhomocysteinase Alcohol dehydrogenase 2 Alcohol dehydrogenase 1 ATP synthase subunit alpha mitochondrial ATP-dependent molecular chaperone HSC82 ATP-dependent molecular chaperone HSP82 Broad substrate specificity ATP-binding cassette transporter ABCG ATP-binding cassette sub-family G member 2 Butyrophilin subfamily 1 member A1 Calmodulin Cathelicidin-2* cathelicidin-4* cathelicidin-6* cathelicidin-7* CD59 glycoprotein CD9 antigen Cystatin E/M Cysteine-rich secretory protein 2 Elongation factor 2 Endoplasmic reticulum chaperone BiP Enolase 1 Enolase 2 Enoyl-CoA hydratase, mitochondrial Eukaryotic initiation factor 4A-I Eukaryotic initiation factor 4A-II Eukaryotic translation initiation factor 2 subunit gamma Fatty acid-binding protein heart FGG protein Fibrinogen gamma-B chain Folate receptor alpha Fructose-bisphosphate aldolase Glucan 1,3-beta-glucosidase I/II Glyceraldehyde-3-phosphate dehydrogenase 2 Glyceraldehyde-3-phosphate dehydrogenase 3 Glycoprotein 2 Heat shock protein SSB2;Heat shock protein SSB1 Heterogeneous nuclear ribonucleoproteins A2/B1 Histatherin Ig-like domain-containing protein Immunoglobulin J chain Inositol polyphosphate-5-phosphatase E Keratin 24 Lactadherin* Lactoperoxidase* Lactotransferrin* Lipoprotein lipase Monocyte differentiation antigen CD14 Mucin-1* NPC intracellular cholesterol transporter 2 Nucleobindin 2 Nucleobindin-1 Parathyroid hormone-related protein Peptidyl-prolyl cis-trans isomerase Peptidyl-prolyl cis-trans isomerase A Perilipin-2 Platelet glycoprotein 4 Polymeric immunoglobulin receptor, Prostaglandin-H2 D-isomerase Protein BMH1 Protein BMH2 Ribosomal protein L21e 60S ribosomal protein L21 Secretoglobin family 1D member Selenoprotein M Serum amyloid A protein* SET nuclear oncogene snRNA-associated Sm-like protein LSm4 Sodium-dependent phosphate transport protein 2B Solute carrier family 38 member 10 Sulfhydryl oxidase TGOLN2 protein Translationally-controlled tumor protein homolog Transthyretin Triosephosphate isomerase Ubiquitin-40S ribosomal protein S27a Ubiquitin-60S ribosomal protein L40 Polyubiquitin-B, Polyubiquitin-C Ubiquitin-like protein SMT3 Uncharacterized protein Uncharacterized protein Uncharacterized protein Vacuolar protein sorting-associated protein 53 homolog WAP four-disulfide core domain 2 Xanthine dehydrogenase/oxidase* Zinc-alpha-2-glycoprotein |
40S ribosomal protein S14-A 40S ribosomal protein S14-B 60S ribosomal protein L20-B 60S ribosomal protein L20-A 60S ribosomal protein L36 Actin cytoplasmic 1 Adenosylhomocysteinase Alcohol dehydrogenase 2 Alcohol dehydrogenase 1 ATP-dependent molecular chaperone HSC82;ATP-dependent molecular chaperone HSP82 Cathelicidin-2* Cathelicidin-6* Cathelicidin-7* Cysteine-rich secretory protein 2 Elongation factor 2 Enolase 1 Enolase 2 Fructose-bisphosphate aldolase Glucan 1,3-beta-glucosidase I/II Glyceraldehyde-3-phosphate dehydrogenase 2 Glyceraldehyde-3-phosphate dehydrogenase 3 Heat shock protein SSB2 Heat shock protein SSB1 Histatherin Immunoglobulin J chain Inositol polyphosphate-5-phosphatase E Keratin 24 NPC intracellular cholesterol transporter 2 Nucleobindin 2 Peptidyl-prolyl cis-trans isomerase Pyruvate kinase 1 Secretoglobin family 1D member Selenoprotein M Translationally-controlled tumor protein homolog Transthyretin Triosephosphate isomerase Ubiquitin-like protein SMT3 Uncharacterized protein Uncharacterized protein Vacuolar protein sorting-associated protein 53 homolog |
Fructose-bisphosphate aldolase Lactadherin* 40S ribosomal protein S14-A 40S ribosomal protein S14-B 40S ribosomal protein S24 40S ribosomal protein S29 60 kDa chaperonin 60S ribosomal protein L13 60S ribosomal protein L14 60S ribosomal protein L20-B 60S ribosomal protein L20-A 60S ribosomal protein L21 60S ribosomal protein L28 60S ribosomal protein L34 60S ribosomal protein L34-A 60S ribosomal protein L34-B 60S ribosomal protein L35a 60S ribosomal protein L36 60S ribosomal protein L37a 60S ribosomal protein L8 Actin, cytoplasmic 1 Acyl-CoA-binding protein Adenosylhomocysteinase Alcohol dehydrogenase 2 Alcohol dehydrogenase 1 ATP synthase subunit alpha mitochondrial ATP synthase subunit beta mitochondrial ATP-dependent molecular chaperone HSC82; ATP-dependent molecular chaperone HSP82 Beta-1,4-galactosyltransferase 1 Beta-2-microglobulin Broad substrate specificity ATP-binding cassette transporter ABCG2 Butyrophilin subfamily 1 member A1 Calmodulin Cathelicidin-1* Cathelicidin-2* Cathelicidin-4* Cathelicidin-6* Cathelicidin-7* CD59 glycoprotein CD9 antigen Cellular repressor of E1A stimulated genes 1 Collagen alpha-2(I) chain Cystatin domain Cystatin E/M Cystatin-C Cysteine-rich secretory protein 2 Elongation factor 2 Endoplasmic reticulum chaperone BiP Enolase 1 Enolase 2 Enoyl-CoA hydratase, mitochondrial Eukaryotic initiation factor 4A-I Eukaryotic initiation factor 4A-II Eukaryotic translation initiation factor 2 subunit gamma Fatty acid-binding protein heart FGG protein Fibroblast growth factor-binding protein 1 Folate receptor alpha Glucan 1,3-beta-glucosidase I/II Glyceraldehyde-3-phosphate dehydrogenase Glyceraldehyde-3-phosphate dehydrogenase 2 Glyceraldehyde-3-phosphate dehydrogenase 3 Glycoprotein 2 Glycosylation-dependent cell adhesion molecule 1 Granulin precursor Heat shock 70 kDa protein 1A Heat shock protein SSB2 Heat shock protein SSB1 Helix-destabilizing protein Heterogeneous nuclear ribonucleoprotein K Heterogeneous nuclear ribonucleoproteins A2/B1 HHIP like 2 Histatherin Ig-like domain-containing protein Immunoglobulin J chain Inorganic pyrophosphatase Inositol polyphosphate-5-phosphatase E Keratin 24 Lactoperoxidase* Lactotransferrin* Lipoprotein lipase Monocyte differentiation antigen CD14 Mucin-1* Myosin heavy chain 9 NPC intracellular cholesterol transporter 2 Nucleobindin 2 Parathyroid hormone-related protein Peptidyl-prolyl cis-trans isomerase Peptidyl-prolyl cis-trans isomerase A Perilipin-2 Platelet glycoprotein 4 Polymeric immunoglobulin receptor Prostaglandin-H2 D-isomerase Pyruvate kinase 1 Ribonuclease pancreatic Ribosomal protein L37 Secretoglobin family 1D member Selenoprotein M SET nuclear oncogene Sodium-dependent phosphate transport protein 2B Solute carrier family 38 member 10 Sulfhydryl oxidase TGOLN2 protein Translationally-controlled tumor protein homolog Transthyretin Triosephosphate isomerase U6 snRNA-associated Sm-like protein LSm4 Ubiquitin thioesterase Ubiquitin-60S ribosomal protein L40, Polyubiquitin-C, Polyubiquitin-B Ubiquitin-60S ribosomal protein L40 Ubiquitin-40S ribosomal protein S31 Polyubiquitin Ubiquitin Ubiquitin-like protein SMT3 Uncharacterized protein Uncharacterized protein Uncharacterized protein Uncharacterized protein Vacuolar protein sorting-associated protein 53 homolog WAP four-disulfide core domain 2 Xanthine dehydrogenase/oxidase* Zinc-alpha-2-glycoprotein |
60 kDa chaperonin 60S ribosomal protein L20-B 60S ribosomal protein L20-A 60S ribosomal protein L21 60S ribosomal protein L28 60S ribosomal protein L34 60S ribosomal protein L36 60S ribosomal protein L8 Actin, cytoplasmic 1 Alcohol dehydrogenase 2 Alcohol dehydrogenase 1 ATP-dependent molecular chaperone HSC82; ATP-dependent molecular chaperone HSP82 Broad substrate specificity ATP-binding cassette transporter ABCG2 ATP-binding cassette sub-family G member 2 Calmodulin Cathelicidin-2* Cathelicidin-6* Cathelicidin-7* CD59 glycoprotein Cystatin E/M Cystatin-C Cysteine-rich secretory protein 2 Endoplasmic reticulum chaperone BiP Enolase 1 Enolase 2 Eukaryotic initiation factor 4A-I Fatty acid-binding protein, heart Folate receptor alpha, Folate receptor 2Fructose-bisphosphate aldolase Glucan 1,3-beta-glucosidase I/II Glycoprotein 2 Heat shock protein SSB2 Heat shock protein SSB1 Heterogeneous nuclear ribonucleoproteins A2/B1 Histatherin Immunoglobulin J chain Inositol polyphosphate-5-phosphatase E Keratin 24 Lactadherin Lactoperoxidase Lipoprotein lipase Monocyte differentiation antigen CD14 Mucin-1* NPC intracellular cholesterol transporter 2 Nucleobindin 1 Nucleobindin 2 Parathyroid hormone-related protein Peptidyl-prolyl cis-trans isomerase A Polymeric immunoglobulin receptor Prostaglandin-H2 D-isomerase Pyruvate kinase 1 Secretoglobin family 1D member Selenoprotein M SET nuclear oncogene Solute carrier family 38 member 10 TGOLN2 protein Translationally-controlled tumor protein homolog Transthyretin U6 snRNA-associated Sm-like protein LSm4 Ubiquitin-40S ribosomal protein S27a Ubiquitin-60S ribosomal protein L40 Polyubiquitin-B Polyubiquitin-C Ubiquitin-like protein SMT3 Uncharacterized protein Uncharacterized protein Uncharacterized protein Vacuolar protein sorting-associated protein 53 homolog WAP four-disulfide core domain 2 Xanthine dehydrogenase/oxidase |
Key: * = protein with known antimicrobial properties.
4. Discussion
Adhesion is defined as a process whereby cells attach to surfaces of each other with the aid of adhesins (Brückner & Mösch, 2012). The adherence of Salmonella and other enteric bacterial pathogens onto yeast cell wall is postulated to be due to specific binding of type 1 fimbriae on bacteria cell with mannan oligosaccharides on yeast cell wall (Gut et al., 2018). Moreover, non-specific adhesion mechanisms including electrostatic and hydrophobic attachment between bacteria and yeasts have been reported (Adegbola and Old, 1985, Pérez-Sotelo et al., 2005, Tiago et al., 2012).
Adhesion of the two Salmonella serovars onto kefir yeast isolates and S. boulardii strains (Table 1, Fig. 1A-D) are consistent with a previous study in which Gram negative enteric bacteria (Escherichia coli and Enterobacter aerogenes) were bound to kefir yeast isolates and S. boulardii strains (Gut et al., 2019). Furthermore, similar results showed that enteropathogenic bacteria including S. Typhimurium (ATCC 14028) adhered onto viable and non-viable yeast cells (França et al., 2015, Martins et al., 2010, Posadas et al., 2017). The growth trend of Salmonella in the presence of inactivated yeasts (Fig. 2) was likely due to Salmonella binding to yeast cells and subsequent sedimentation. Bacteria cells in tight auto-aggregation have been reported to show no cell division (Hassani et al., 2009). This therefore resulted in reduced Salmonella population, as was indicated in a less turbid medium compared to the control. Both kefir yeast isolates and S. boulardii strains were found to survive well in simulated GIT conditions (Gut et al., 2019). Yeasts survival and proliferation may lead to higher numbers in GIT due to potential growth, increasing the capacity to bind Salmonella and subsequent shedding in feces. Furthermore, the advantage of yeasts is the fact that they are not affected by drugs targeting infectious bacteria such as Salmonella, making them suitable candidates for complementary therapy with antibiotics. For example chloramphenicol is an antibiotic typically used in Salmonella treatment (Gut et al., 2018) and works against bacteria by binding to ribosomes and blocking protein synthesis but does not affect yeasts (Das and Patra, 2017, Gut et al., 2019). The current study also established that non-viable yeast cells were as effective in Salmonella attachment as live yeast cells. This may have critical therapeutic or prophylactic application advantage as viable yeast cells have been associated with at least 100 fungemia cases (Gut et al., 2018). For example, the use of viable yeast cells in immunocompromised people or those with gastrointestinal disease has been reported to pose serious threat of fungemia (Kelesidis & Pothoulakis, 2012). Use of non-viable kefir yeast either prophylactically or as a complementary therapy for Salmonella infection may be a better choice for people with GIT diseases or compromised immunity.
Effect of pH on adhesion of Salmonella onto yeast cell walls (Table 1) was in line with a previous research (Tiago et al., 2012), which reported that pH between 4 and 8 had no effect on bacteria attachment onto yeast cell wall. Adhesion of Salmonella onto yeast cell wall at GIT pH is important because it is where invasion of pathogen and gastroenteritis occur. Therefore, if Salmonella could attach to yeast cells in the GIT under acidic condition, invasion may be prevented. Furthermore, bile salt has high surface activity (Attili et al., 1986) and reduces surface hydrophobicity (Tiago et al., 2012), therefore may prevent adhesion. However, 0.3% bile salt did not prevent adhesion in this study (Table 1) which correlated with previous reports (Gomez Zavaglia et al., 2002, Guglielmetti et al., 2009, Tiago et al., 2012). Adherence in presence of bile is important since it will not interfere with potential prophylactic or therapeutic application in GIT.
Growth inhibitory properties of yeast against bacteria have been previously proposed to include production of ethanol and other antibacterial metabolites such as killer toxins (Bajaj et al., 2013, Muccilli and Restuccia, 2015). Many mechanisms of probiotics including yeasts against susceptible microbial cells have been proposed and involve destabilization of the cell membrane, cell lysis, degradation of nucleic acid, inhibition of protein synthesis and binding onto yeasts (Gut et al., 2021).
Lack of Salmonella growth inhibition by yeast colonies correlated with a previous study on S. boulardii effect on enteric bacterial growth (Rajkowska et al., 2012). However, a study reported that killer toxin produced by bakery Saccharomyces showed growth inhibition of Escherichia coli and S. Typhimurium as a result of cell membrane destruction (Alsoufi & Aziz, 2017). Therefore further experimentation involving use of KTM was required. Stronger growth inhibition exhibited by KTM fermented with K. lactis for 48 h (Fig. 3, Fig. 4) may be due to accumulation of antibacterial metabolites produced by K. lactis when fermentation time was prolonged, consistent with a previous report in which K. lactis and Kluyveromyces marxinus showed antagonistic effect on Salmonella Paratyphi B, S. Typhimurium ATCC 14028 and Salmonella Enteriditis when fermentation time was extended by downregulating chromosomal sopD gene (Ceugniez et al., 2017). Salmonella growth promotion by S. unisporus KTM-CFS observed in this study after the initial 5 h (Fig. 3, Fig. 4) was likely due to the loss of potential antibacterial molecules coupled with possible presence of growth factors such as amino acids and vitamins released by yeasts (Bechtner et al., 2019, Gut et al., 2021, Stadie et al., 2013). This growth promoting effect of S. unisporus on S. Arizonae was lost likely due to heat inactivation or evaporation of some volatiles (Fig. 6). However, Salmonella population generally remained stable in the presence of yeast-fermented KTM due to its bacteriostatic effect compared to the control which showed significant increase (Fig. 5).
Studies have confirmed production of alcohol by Kluyveromyces and Saccharomyces species commonly isolated from kefir (Ho et al., 2012, Magalhães et al., 2010). Concentration of alcohol produced by kefir yeast isolates in Table 2, was consistent with reported alcohol concentrations in the literature (Magalhães et al., 2010, Nuñez, 2016). Furthermore, production of alcohol in KTM fermented with S. boulardii is consistent with a previous study in which this yeast strain was used in a beer production (Mulero-Cerezo et al, 2019). The differences in concentration between 24 and 48 h fermentation could be due to the attainment of stationary growth phase resulting in constant metabolic activities at 24 h (Mulero-Cerezo et al., 2019), and the likelihood of loss due to evaporation during fermentation. Lack of bacteriostatic effect by 2% ethanol control showed that ethanol in the KTM may not be responsible for bacteriostatic effect observed, an indication of presence of other potential antimicrobial metabolites. Suppression of Salmonella growth by KTM fermented with SB48, SB49, K. lactis and S. unisporus after evaporation suggested presence of non-volatile molecules, possibly antimicrobial proteins which showed anti-salmonella activities. Antimicrobial proteins produced by yeasts (Saccharomyces and Kluyveromyces species) have been documented to be effective against bacteria (Branco et al., 2017, Rima et al., 2012, Al-Sahlany et al., 2020, Hasan et al., 2019, Liu et al., 2018). The shotgun proteomics analysis of KTM-CFS confirmed the presence of such proteins which have been shown to have antimicrobial properties. Previous studies have reported antimicrobial activity of cathelicidin (Xia et al., 2015), xanthine dehydrogenase (Okamura et al., 2018), lactotransferrin and mucin-1 (Gut et al., 2018), lactadherin (Sabha et al., 2018), lactoperoxidase (Bafort et al., 2014) and serum amyloid A protein (Kagan et al., 2012). The mechanisms of these proteins against susceptible bacteria have been reported to involve DNA, RNA, ATP synthesis, or protein synthesis inhibition, as well as disruption in membrane and ionic potential of the cell membrane (Biadała et al., 2020). The proteins marked with asterisk in Table 3 may have inhibited Salmonella growth in the current study however, further studies are needed.
5. Conclusion
Kefir yeast isolates obtained from traditional kefir grains showed comparable anti-salmonella effect to that of S. boulardii with respect to adhesion as well as growth inhibition due to antimicrobial metabolites production. Shot-gun proteomics analysis showed presence of cathelicidin, xanthine dehydrogenase, mucin-1, lactadherin, lactoperoxidase, serum amyloid A protein and lactotransferrin in yeast fermented killer toxin medium which have anti-bacterial properties. These proteins in KTM may be responsible for bacteriostatic effect observed in this study. K. lactis and S. unisporus have potential to be used prophylactically and therapeutically in control of Salmonella infection. However, further studies involving cell lines, animals as well as human trials are needed to prove these kefir isolates efficacy in prevention and treatment of Salmonella infection.
Credit authorship contribution statement
Abraham Majak Gut: Writing – original draft, Writing – review & editing. Todor Vasiljevic: Supervision, Writing – review & editing. Thomas Yeager: Supervision, Writing – review & editing. Osaana N. Donkor: Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors acknowledge Victoria University for funding this work, Bio21 Institute of The University of Melbourne for the opportunity to use their scanning electron microscope and Monash University Proteomics and Metabolic Facility for Shotgun proteomics analysis.
References
- Adegbola R.A., Old D.C. Fimbrial and non-fimbrial haemagglutinins in Enterobacter aerogenes. J. Med. Microbiol. 1985;19(1):35–43. doi: 10.1099/00222615-19-1-35. [DOI] [PubMed] [Google Scholar]
- Al-Sahlany S.T.G., Altemimi A.B., Abd Al-Manhel A.J., Niamah A.K., Lakhssassi N., Ibrahim S.A. Purification of bioactive peptide with antimicrobial properties produced by Saccharomyces cerevisiae. Foods. 2020;9(3):324. doi: 10.3390/foods9030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsoufi M.A., Aziz R.A. Use killer toxin extracted from bakery yeast for extending shelf life of fruits. Pak. J. Biotechnol. 2017;14(1):23–27. [Google Scholar]
- Attili A.F., Angelico M., Cantafora A., Alvaro D., Capocaccia L. Bile acid-induced liver toxicity: relation to the hydrophobic-hydrophilic balance of bile acids. Med. Hypotheses. 1986;19(1):57–69. doi: 10.1016/0306-9877(86)90137-4. [DOI] [PubMed] [Google Scholar]
- Bafort F., Parisi O., Perraudin J.-P., Jijakli M. Mode of action of lactoperoxidase as related to its antimicrobial activity: a review. Enzyme Res. 2014 doi: 10.1155/2014/517164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj B.K., Raina S., Singh S. Killer toxin from a novel killer yeast Pichia kudriavzevii RY55 with idiosyncratic antibacterial activity. J. Basic Microbiol. 2013;53(8):645–656. doi: 10.1002/jobm.201200187. [DOI] [PubMed] [Google Scholar]
- Bakken J.S. Staggered and tapered antibiotic withdrawal with administration of kefir for recurrent Clostridium difficile infection. Clin. Infect. Dis. 2014;59(6):858–861. doi: 10.1093/cid/ciu429. [DOI] [PubMed] [Google Scholar]
- Balouiri M., Sadiki M., Ibnsouda S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016;6(2):71–79. doi: 10.1016/j.jpha.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtner J., Xu D., Behr J., Ludwig C., Vogel R.F. Proteomic Analysis of Lactobacillus nagelii in the Presence of Saccharomyces cerevisiae Isolated From Water Kefir and Comparison With Lactobacillus hordei. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekar O., Yilmaz Y., Gulten M. Kefir improves the efficacy and tolerability of triple therapy in eradicating Helicobacter pylori. J. Med. Food. 2011;14(4):344–347. doi: 10.1089/jmf.2010.0099. [DOI] [PubMed] [Google Scholar]
- Biadała A., Szablewski T., Lasik-Kurdyś M., Cegielska-Radziejewska R. Antimicrobial activity of goat’s milk fermented by single strain of kefir grain microflora. Eur. Food Res. Technol. 2020;246(6):1231–1239. doi: 10.1007/s00217-020-03483-2. [DOI] [Google Scholar]
- Branco P., Francisco D., Monteiro M., Almeida M.G., Caldeira J., Arneborg N., Prista C., Albergaria H. Antimicrobial properties and death-inducing mechanisms of saccharomycin, a biocide secreted by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2017;101(1):159–171. doi: 10.1007/s00253-016-7755-6. [DOI] [PubMed] [Google Scholar]
- Brückner S., Mösch H.-U. Choosing the right lifestyle: adhesion and development in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2012;36(1):25–58. doi: 10.1111/j.1574-6976.2011.00275.x. [DOI] [PubMed] [Google Scholar]
- Ceugniez A., Coucheney F., Jacques P., Daube G., Delcenserie V., Drider D. Anti-Salmonella activity and probiotic trends of Kluyveromyces marxianus S-2-05 and Kluyveromyces lactis S-3-05 isolated from a French cheese, Tomme d'Orchies. Res. Microbiol. 2017;168(6):575–582. doi: 10.1016/j.resmic.2017.03.004. [DOI] [PubMed] [Google Scholar]
- Das B., Patra S. In: Nanostructures for Antimicrobial Therapy. Ficai A., Grumezescu A.M., editors. Elsevier; 2017. Chapter 1 - Antimicrobials: Meeting the Challenges of Antibiotic Resistance Through Nanotechnology; pp. 1–22. [Google Scholar]
- FAO/WHO, 2002. Probiotics in food Health and nutritional properties and guidelines for evaluation: Report of a Joint FAO/WHO Expert Consultation on Evaluation of health and Nutritional Properties of Probiotics in Food including powder Milk with Live lactic Acid bacteria. Cordoba, Argentina 2002; Report of a Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food London, Ontario, Canada 2002.
- Feasey, N.A., Gaskell, K., Wong, V., Msefula, C., Selemani, G., Kumwenda, S., et al., 2015. Rapid emergence of multidrug resistant, H58-lineage Salmonella typhi in Blantyre, Malawi. PLoS Neglected Tropical Diseases 9(4). [DOI] [PMC free article] [PubMed]
- Feye, K.M., Carroll, J.P., Anderson, K.L., Whittaker, J.H., Schmidt-McCormack, G.R., McIntyre, D.R., et al., 2019. Saccharomyces cerevisiae Fermentation Products That Mitigate Foodborne Salmonella in Cattle and Poultry. Front. Veterinary Sci., 6. [DOI] [PMC free article] [PubMed]
- França R.C., Conceição F.R., Mendonça M., Haubert L., Sabadin G., de Oliveira P.D., et al. Pichia pastoris X-33 has probiotic properties with remarkable antibacterial activity against Salmonella Typhimurium. Appl. Microbiol. Biotechnol. 2015;99(19):7953–7961. doi: 10.1007/s00253-015-6696-9. [DOI] [PubMed] [Google Scholar]
- Garrote G.L., Abraham A.G., De Antoni G.L. Preservation of kefir grains, a comparative study. LWT-Food Sci. Technol. 1997;30(1):77–84. [Google Scholar]
- Gomez Zavaglia A., Kociubinski G., Perez P., Disalvo E., De Antoni G. Effect of bile on the lipid composition and surface properties of bifidobacteria. J. Appl. Microbiol. 2002;93(5):794–799. doi: 10.1046/j.1365-2672.2002.01747.x. [DOI] [PubMed] [Google Scholar]
- Guglielmetti S., Tamagnini I., Minuzzo M., Arioli S., Parini C., Comelli E., Mora D. Study of the adhesion of Bifidobacterium bifidum MIMBb75 to human intestinal cell lines. Curr. Microbiol. 2009;59(2):167–172. doi: 10.1007/s00284-009-9415-x. [DOI] [PubMed] [Google Scholar]
- Gut A.M., Vasiljevic T., Yeager T., Donkor O. Salmonella infection-prevention and treatment by antibiotics and probiotic yeasts: a review. Microbiology. 2018;164(11):1327–1344. doi: 10.1099/mic.0.000709. [DOI] [PubMed] [Google Scholar]
- Gut A.M., Vasiljevic T., Yeager T., Donkor O.N. Characterization of yeasts isolated from traditional kefir grains for potential probiotic properties. J. Funct. Foods. 2019;58:56–66. [Google Scholar]
- Gut A.M., Vasiljevic T., Yeager T., Donkor O.N. Kefir characteristics and antibacterial properties-potential applications in control of enteric bacterial infection. Int. Dairy J. 2021;118:105021. doi: 10.1016/j.idairyj.2021.105021. [DOI] [Google Scholar]
- Hasan M., Moghal M.M.R., Saha S.K., Yamazaki M. The role of membrane tension in the action of antimicrobial peptides and cell-penetrating peptides in biomembranes. Biophys. Rev. 2019;11(3):431–448. doi: 10.1007/s12551-019-00542-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassani A.S., Malekzadeh F., Amirmozafari N., Hamdi K., Ordouzadeh N., Ghaemi A. Phage shock protein G, a novel ethanol-induced stress protein in Salmonella typhimurium. Current Microbiol. 2009;58(3):239–244. doi: 10.1007/s00284-008-9314-6. [DOI] [PubMed] [Google Scholar]
- Ho C.-Y., Chang J.-J., Lee S.-C., Chin T.-Y., Shih M.-C., Li W.-H., Huang C.-C. Development of cellulosic ethanol production process via co-culturing of artificial cellulosomal Bacillus and kefir yeast. Appl. Energy. 2012;100:27–32. doi: 10.1016/j.apenergy.2012.03.016. [DOI] [Google Scholar]
- Kagan B.L., Jang H., Capone R., Teran Arce F., Ramachandran S., Lal R., Nussinov R. Antimicrobial properties of amyloid peptides. Mol. Pharm. 2012;9(4):708–717. doi: 10.1021/mp200419b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelesidis T., Pothoulakis C. Efficacy and safety of the probiotic Saccharomyces boulardii for the prevention and therapy of gastrointestinal disorders. Therap. Adv. Gastroenterol. 2012;5(2):111–125. doi: 10.1177/1756283X11428502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodadad, A., Farahmand, F., Najafi, M., Shoaran, M., 2013. Probiotics for the treatment of pediatric helicobacter pylori infection: a randomized double blind clinical trial. [PMC free article] [PubMed]
- Kiekens S., Vandenheuvel D., Broeckx G., Claes I., Allonsius C., De Boeck I., et al. Impact of spray-drying on the pili of Lactobacillus rhamnosus GG. Microb. Biotechnol. 2019 doi: 10.1111/1751-7915.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan G., Kocher A. Role of yeast cell wall polysaccharides in pig nutrition and health protection. Livestock Sci. 2007;109(1):161–165. doi: 10.1016/j.livsci.2007.01.134. [DOI] [Google Scholar]
- Liu Z., Zhu M., Chen X., Yang G., Yang T., Yu L., Hui L., Wang X. Expression and antibacterial activity of hybrid antimicrobial peptide cecropinA-thanatin in Pichia pastoris. Front. Lab. Med. 2018;2(1):23–29. [Google Scholar]
- Magalhães K.T., Pereira M.A., Nicolau A., Dragone G., Domingues L., Teixeira J.A., de Almeida Silva J.B., Schwan R.F. Production of fermented cheese whey-based beverage using kefir grains as starter culture: Evaluation of morphological and microbial variations. Bioresour. Technol. 2010;101(22):8843–8850. doi: 10.1016/j.biortech.2010.06.083. [DOI] [PubMed] [Google Scholar]
- Martins F.S., Dalmasso G., Arantes R.M., Doye A., Lemichez E., Lagadec P., et al. Interaction of Saccharomyces boulardii with Salmonella enterica serovar Typhimurium protects mice and modifies T84 cell response to the infection. PLoS One. 2010;5(1):e8925. doi: 10.1371/journal.pone.0008925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins F.S., Veloso L.C., Arantes R.M., Nicoli J.R. Effects of yeast probiotic formulation on viability, revival and protection against infection with Salmonella enterica ssp. enterica serovar Typhimurium in mice. Lett. Appl. Microbiol. 2009;49(6):738–744. doi: 10.1111/j.1472-765X.2009.02732.x. [DOI] [PubMed] [Google Scholar]
- Minekus, Alminger M., Alvito P., Ballance S., Bohn T., Bourlieu C., et al. A standardised static in vitro digestion method suitable for food–an international consensus. Food Funct. 2014;5(6):1113–1124. doi: 10.1039/c3fo60702j. [DOI] [PubMed] [Google Scholar]
- Muccilli S., Restuccia C. Bioprotective Role of Yeasts. Microorganisms. 2015;3(4):588. doi: 10.3390/microorganisms3040588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero-Cerezo J., Briz-Redón Á., Serrano-Aroca Á. Saccharomyces Cerevisiae Var. Boulardii: Valuable Probiotic Starter for Craft Beer Production. Appl. Sci. 2019;9(16):3250. [Google Scholar]
- Nami Y., Haghshenas B., Abdullah N., Barzegari A., Radiah D., Rosli R., Khosroushahi A.Y. Probiotics or antibiotics: future challenges in medicine. J. Med. Microbiol. 2015;64(Pt 2):137–146. doi: 10.1099/jmm.0.078923-0. [DOI] [PubMed] [Google Scholar]
- Nikolaou A., Galanis A., Kanellaki M., Tassou C., Akrida-Demertzi K., Kourkoutas Y. Assessment of free and immobilized kefir culture in simultaneous alcoholic and malolactic cider fermentations. LWT - Food Sci. Technol. 2017;76:67–78. doi: 10.1016/j.lwt.2016.10.034. [DOI] [Google Scholar]
- Nuñez M. In: Non-Bovine Milk and Milk Products. Tsakalidou E., Papadimitriou K., editors. Academic Press; San Diego: 2016. Chapter 7 - Existing Technologies in Non-cow Milk Processing and Traditional Non-cow Milk Products; pp. 161–185. [Google Scholar]
- Okamura Y., Inada M., Elshopakey G.E., Itami T. Characterization of xanthine dehydrogenase and aldehyde oxidase of Marsupenaeus japonicus and their response to microbial pathogen. Mol. Biol. Rep. 2018;45(4):419–432. doi: 10.1007/s11033-018-4177-9. [DOI] [PubMed] [Google Scholar]
- Pérez-Sotelo L.S., Talavera-Rojas M., Monroy-Salazar H.G., Lagunas-Bernabé S., Cuarón-Ibargüengoytia J.A., Jimenez R., Vázquez-Chagoyán J.C. In vitro evaluation of the binding capacity of Saccharomyces cerevisiae Sc47 to adhere to the wall of Salmonella spp. Rev. Latinoam. Microbiol. 2005;47(3–4):70–75. [PubMed] [Google Scholar]
- Perez-Sotelo L.S., Talavera-Rojas M., Monroy-Salazar H.G., Lagunas-Bernabe S., Cuaron-Ibarguengoytia J.A., Jimenez R.M., Vazquez-Chagoyan J.C. In vitro evaluation of the binding capacity of Saccharomyces cerevisiae Sc47 to adhere to the wall of Salmonella spp. Rev. Latinoam. Microbiol. 2005;47(3–4):70–75. [PubMed] [Google Scholar]
- Piroeva I., Atanassova-Vladimirova S., Dimowa L., Sbirkova H., Radoslavov G., Hristov P., Shivachev B. A simple and rapid scanning electron microscope preparative technique for observation of biological samples: application on bacteria and DNA samples. Bulg. Chem. Commun. 2013;45(4):510–515. [Google Scholar]
- Plessas S., Nouska C., Mantzourani I., Kourkoutas Y., Alexopoulos A., Bezirtzoglou E. Microbiological Exploration of Different Types of Kefir Grains. Fermentation. 2016;3(1):1. [Google Scholar]
- Posadas G.A., Broadway P.R., Thornton J.A., Carroll J.A., Lawrence A., Corley J.R., et al. Yeast pro-and paraprobiotics have the capability to bind pathogenic bacteria associated with animal disease. Trans. Animal Sci. 2017;1(1):60–68. doi: 10.2527/tas2016.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado M.R., Blandón L.M., Vandenberghe L.P.S., Rodrigues C., Castro G.R., Thomaz-Soccol V., Soccol C.R. Milk kefir: composition, microbial cultures, biological activities, and related products. Front. Microbiol. 2015;6:1177. doi: 10.3389/fmicb.2015.01177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyodip P., Prakash P.Y., Balaji S. Phytases of Probiotic Bacteria: Characteristics and Beneficial Aspects. Indian J. Microbiol. 2017;57(2):148–154. doi: 10.1007/s12088-017-0647-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowska K., Kunicka-Styczyńska A., Rygala A. Probiotic activity of Saccharomyces cerevisiae var. Boulardii against human pathogens. Food Technol. Biotechnol. 2012;50(2):230–236. [Google Scholar]
- Rima H., Steve L., Ismail F. Antimicrobial and probiotic properties of yeasts: from fundamental to novel applications. Front. Microbiol. 2012;3:421. doi: 10.3389/fmicb.2012.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarela M., Mogensen G., Fondén R., Mättö J., Mattila-Sandholm T. Probiotic bacteria: safety, functional and technological properties. J. Biotechnol. 2000;84(3):197–215. doi: 10.1016/S0168-1656(00)00375-8. [DOI] [PubMed] [Google Scholar]
- Sabha B.H., Alzahrani F., Almehdar H.A., Uversky V.N., Redwan E.M. Disorder in milk proteins: lactadherin multifunctionality and structure. Curr. Protein Pept. Sci. 2018;19(10):983–997. doi: 10.2174/1389203719666180608091849. [DOI] [PubMed] [Google Scholar]
- Stadie J., Gulitz A., Ehrmann M.A., Vogel R.F. Metabolic activity and symbiotic interactions of lactic acid bacteria and yeasts isolated from water kefir. Food Microbiol. 2013;35(2):92–98. doi: 10.1016/j.fm.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Tiago, Martins F.D.S., Souza E., Pimenta P.F.P., Araújo H.R.C., Castro I.D.M., et al. Adhesion to the yeast cell surface as a mechanism for trapping pathogenic bacteria by Saccharomyces probiotics. J. Med. Microbiol. 2012;61(9):1194–1207. doi: 10.1099/jmm.0.042283-0. [DOI] [PubMed] [Google Scholar]
- Xia X., Zhang L., Wang Y. The antimicrobial peptide cathelicidin-BF could be a potential therapeutic for Salmonella typhimurium infection. Microbiol. Res. 2015;171:45–51. doi: 10.1016/j.micres.2014.12.009. [DOI] [PubMed] [Google Scholar]