Abstract
Infrequent restriction site PCR (IRS-PCR) is a recently described DNA fingerprinting technique based on selective amplification of restriction endonuclease-cleaved fragments. Bartonella isolates associated with human disease and related nonhuman isolates were analyzed by IRS-PCR genomic fingerprinting. Preparation of DNA templates began with double digestion using three different restriction endonuclease combinations. Combinations included the frequently cutting endonuclease HhaI in conjunction with an infrequently cutting endonuclease, EagI, SmaI, or XbaI. Digestion was followed by ligation of oligonucleotide adapters designed with ends complementary to the restriction endonuclease sites. Amplification of fragments flanked with an EagI, SmaI, or XbaI site in combination with an HhaI site produced a series of different-sized amplicons resolvable into patterns by polyacrylamide gel electrophoresis (PAGE). The pattern complexity was varied by the addition of selective nucleotides to the 3′ ends of the EagI-, SmaI-, or XbaI-specific primers. Amplicons were also generated with fluorescently labeled primers and were subsequently resolved and detected by capillary electrophoresis. Analysis by traditional slab PAGE and capillary electrophoresis provided suitable resolution of patterns produced with the enzyme combinations EagI-HhaI and SmaI-HhaI. However, the combination of XbaI-HhaI produced too many fragments for sufficient resolution by traditional PAGE, thus requiring the better resolving properties of capillary electrophoresis. Due to the flexibility in modulating the pattern complexity and electrophoresis methods, these techniques allow for a high level of experimental optimization. The results provide evidence of the discriminatory power, ease of use, and flexibility of the IRS-PCR method as it applies to the identification of human-pathogenic Bartonella species.
The recent increased recognition of Bartonella-associated human diseases has been made possible, in part, by the advent of novel methods for the genotypic identification of human pathogenic Bartonella species (4, 5). For example, Bartonella quintana, the etiologic agent of trench fever, was, until recently, widely considered a relic human pathogen associated with past World Wars (39); however, B. quintana has now been isolated and identified as a sporadic, modern-era disease-causing agent in the United States, France, Russia, and Peru (7, 8, 13, 16, 27, 35). Furthermore, epidemic trench fever has now been documented in central Africa (28). Genotypic methods were central to the demonstration that Bartonella henselae is the causative agent of cat scratch disease and of related disease among immune-system-impaired persons (12, 29, 30), and B. henselae continues to be implicated in various other complicated-disease manifestations, such as neuroretinitis (21) and encephalopathy (2, 10). Bartonella elizabethae was first isolated from a patient with endocarditis (11). Recent studies that employed genotypic methods for the characterization of Bartonella species isolates have now demonstrated the widespread distribution of B. elizabethae in urban rats (genus Rattus) (14). Bartonella bacilliformis, the etiologic agent of Carrion's disease, has historically been recognized as endemic in specific regions of the foothills of the Peruvian Andes (9). However, genotypic characterization of isolates from bacteremic persons without classic signs of Carrion's disease, found in regions not previously recognized as regions of B. bacilliformis endemicity, have begun to shed new light on a more complex and incompletely understood natural history and epidemiology of this important disease (15). Additional Bartonella species have been tentatively associated with a wide variety of sporadic, presumably zoonotic, human diseases. These include Bartonella grahamii, Bartonella clarridgeiae, Bartonella vinsonii subsp. arupensis, and Bartonella vinsonii subsp. berkhoffii (18, 19, 33, 40).
Several DNA-based molecular typing techniques have been used for the identification and differentiation of Bartonella spp. (29, 32, 34, 36, 38). These techniques include pulsed-field gel electrophoresis (PFGE), enterobacterial repetitive intergenic consensus PCR (ERIC-PCR), repetitive extragenic palindromic PCR (REP-PCR), arbitrarily primed PCR (AP-PCR), and PCR-restriction fragment length polymorphism (RFLP) analysis. All of these methods have inherent experimental difficulties. PFGE is a relatively time-consuming technique that can be difficult to perform and requires a large amount of high-quality DNA, and the large fragments are difficult to resolve and to size accurately. Techniques such as ERIC-PCR, REP-PCR, and AP-PCR are sensitive to experimental variation, making reproducibility and standardization difficult. PCR-RFLP analysis is relatively simple, quick and reproducible, but it provides a limited amount of experimental data from a small region of the genome. Sequence analysis of various genes (16S rRNA, gltA, ftsZ, ribC, htrA, the intergenic spacer region [ITS], and groEL) has also been used to identify and differentiate Bartonella isolates (6, 17, 20, 22, 34, 37). DNA sequence analysis is highly reproducible, is information rich, and is often considered to be a “gold standard” for microbial typing. However, DNA gene sequence analysis, like PCR-RFLP analysis, typically considers only a small segment of the genome. Additionally, an ideal gene segment must have a significantly variable sequence flanked by conserved PCR primer regions and should not be susceptible to lateral gene transfer, and multiple genes representing different regions of the genome should be analyzed (24). Extensive sequence analysis may not be practical for some laboratories because of the expense and time required. Thus, there remains a need for simple, high-throughput, high-resolution, and reproducible analytical tools for the routine analysis of Bartonella genotypes, as well as of other bacteria.
Recently, a new bacterial typing method known as infrequent-restriction-site PCR (IRS-PCR) has been described (23). Three subsequent studies have shown that IRS-PCR results are at least as discriminatory as those of PFGE and field-inversion gel electrophoresis (FIGE) but did not require the amount of time and large quantities of high-quality genomic DNA required by these techniques (25, 31, 41).
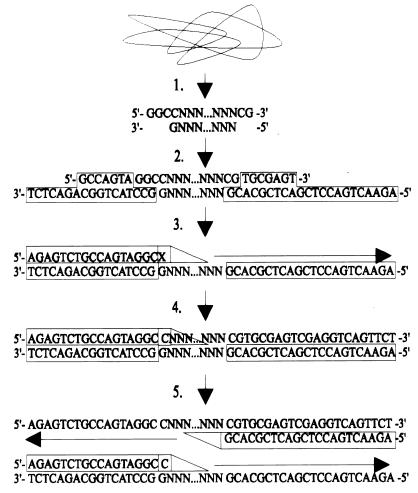
IRS-PCR, as originally described, begins with double digestion of genomic DNA, using a combination of an infrequently and a frequently cutting restriction endonuclease (Fig. 1) (23). This combination produces three types of restriction fragments grouped by their flanking cleaved ends: (i) fragments flanked on both sides by the infrequent restriction site (which occur rarely, if ever), (ii) fragments flanked by the infrequent and the frequent restriction site (a majority of the analyzed fragments), and (iii) fragments flanked on both sides by the frequent restriction site (most abundant, but will not amplify because elongation from the infrequent restriction site is required for primer binding at the frequent restriction site [see step 5, Fig. 1]). Following digestion, oligonucleotide adapters with specificity for the cleaved DNA ends are ligated. These adapters are subsequently used as primer binding sites for PCR fragment amplification. Primers are designed as complements to adapter sequences with the addition of a 3′ nucleotide extension. This nucleotide extends past the adapter sequence into the unknown fragment sequence. If this nucleotide extension is complementary to the unknown fragment nucleotide, Taq polymerase will read through and extension will occur. If it is noncomplementary, elongation will be terminated and amplification will not occur. Successful amplification produces a series of fragments that can be separated and visualized by gel electrophoresis.
FIG. 1.
Schematic of IRS-PCR analysis. (Step 1) Double digestion using EagI and HhaI. (Step 2) Adapter ligation. (Step 3) Primary denaturation followed by EagI primer binding. (Step 4) If the selective primer extension is a cytosine, then Taq polymerase reads through and initial extension occurs. If X ≠ C, extension does not occur. (Step 5) Initial extension produces complementary binding sequence for oligonucleotide AH1 (from HhaI adapter).
Automated DNA sequencers permit the separation and detection of fluorescently labeled PCR products. Labeling one of the IRS-PCR fragments with a fluorescently labeled primer produces a set of amplicons suitable for analysis with automated DNA sequencers. Although this method requires specialized equipment, it produces highly resolved genomic fingerprints suitable for further analysis, which are stored for use in future studies.
The IRS-PCR genomic fingerprinting method has been applied to only a few bacterial species; however, it has shown potential for becoming a universal tool for the differentiation of bacteria. Our application of IRS-PCR genomic fingerprinting to a set of well-recognized Bartonella spp. produced patterns easily used for systematic differentiation. The technique was reproducible, could be used by any laboratory capable of performing the PCR, and can be greatly elaborated on with automated fluorescence analysis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bartonella species used in this study included B. henselae Houston-1 (ATCC 49882), B. elizabethae F9251 (ATCC 49927), B. quintana Fuller (ATCC VR-358), B. bacilliformis KC583 (ATCC 35685), B. vinsonii subsp. berkhoffii (ATCC 51672), B. clarridgeiae (ATCC 51734), B. grahamii (ATCC 700132), B. vinsonii subsp. arupensis (ATCC 700727), and four strains from a Centers for Disease Control and Prevention collection, B. quintana strain OK90-268, B. henselae strain Marseilles, B. henselae strain Tiger-2, and a previously unpublished cat isolate, B. weissii species nova (R. L. Regnery, submitted for publication). All strains were cultivated directly on commercially available rabbit blood heart infusion agar (Becton Dickinson Microbiology Systems, Cockeysville, Md.) and incubated at 32°C in a humidified 5% CO2 environment or at 28°C with no CO2 (29). The cultures were grown for varying lengths of time (e.g., 5 to 10 days), depending on the growth characteristics of the individual species.
Preparation of template DNA.
Bacterial cultures were washed from plates with 10 mM Tris buffer (pH 8.0) containing 1 mM EDTA. Cells were vortexed and subsequently pelleted by centrifugation. Genomic DNA was extracted from the bacterial pellets with the Easy-DNA kit (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. DNA quantity and purity were assessed by UV spectrophotometry. The final DNA concentration was adjusted to 50 ng/μl. All enzymes were acquired from New England Biolabs (Beverly, Mass.). One microliter of genomic DNA was digested with 10 U of HhaI and 10 U of either EagI, SmaI, or XbaI in the appropriate digestion buffer (final volume, 50 μl) for 2 h at 37°C. A ligation master mix was prepared by combining T4 DNA ligase (400 U), ATP (12.6 pmol), 10× ligation buffer (0.75 μl), the HhaI adapter (20 pmol), either the EagI, the SmaI, or the XbaI adapter (20 pmol), and water for a total volume of 7.5 μl per reaction. An aliquot of 12.5 μl of the genomic DNA double-digestion reaction mixture was mixed with 7.5 μl of the ligation master mix. This solution was incubated at 16°C for 2 h for ligation, at 37°C for 30 min to ensure cleavage of any religated fragments (for SmaI reactions, 25°C for 15 min and then 37°C for 15 min), and at 65°C for 20 min to inactivate the remaining enzymes. The solution could then be stored at 4°C until further use.
Adapters and primers.
Adapters were constructed as described by Mazurek et al. (23). All oligonucleotides were supplied by the Biotechnology Core Facility, Centers for Disease Control and Prevention. Complete sequences for each adapter and corresponding primers are shown in Table 1. Adapter pairs were designed to ligate to corresponding cleaved ends produced by restriction endonuclease digestion. The longer oligonucleotide of the EagI, SmaI, and XbaI adapters is phosphorylated at the 5′ end to allow ligation to the 3′ end of the restriction fragment. To prepare the adapters, equimolar amounts of individual oligonucleotide adapter pairs were combined in 1× PCR buffer [10× PCR buffer contains Tris-Cl, KCl, (NH4)2SO4 15 mM MgCl2 (pH 8.7) at 20°C]. The oligonucleotides were allowed to anneal by heating at 90°C for 5 min and were slowly cooled to 4°C for 30 min in a thermocycler. Stock adapter was stored at −20°C at a concentration of 20 μM. Due to the short length of AH2, the HhaI adapter was never allowed to reach room temperature, where it would be expected to dissociate.
TABLE 1.
Adapter and primer oligonucleotides
| Oligonucleotide | Sequence |
|---|---|
| HhaI adapter | |
| AH1 | 5′-AGA ACT GAC CTC GAC TCG CAC G-3′a |
| AH2 | 3′-TG AGC GT-5′ |
| EagI adapter | |
| EagI-ad1 | 5′-PO4-GGC CTA CTG GCA GAC TCT-3′ |
| AX2 | 3′-AT GAC CG-5′b |
| EagI primers, PE-Nc | 5′-AGA GTC TGC CAG TAG GCC GN-3′ |
| SmaI adapter | |
| SmaI-ad1 | 5′-PO4-CGA ACG ATC TCC AGG AGG-3′ |
| SmaI-ad2 | 3′-GCT TGC TAG AGG TCC T-5′ |
| SmaI primers, PSmaI-Nc | 5′-CCT CCT GGA GAT CGT TCG GGG N-3′ |
| XbaI adapter | |
| AX1 | 5′-PO4-CTA GTA CTG GCA GAC TCT-3′ |
| AX2 | 3′-AT GAC CG-5′ |
| XbaI primers, PX-Nc | 5′-AGA GTC TGC CAG TAC TAG AN-3′ |
AH1 serves as part of the adapter and as the HhaI primer.
EagI and XbaI use the same sequence for this portion of the adapter.
N denotes any nucleotide A, T, G, or C. Four primers are actually synthesized.
Primers were constructed to complement SmaI-ad1, EagI-ad1, and AX1 (see Table 1). Four primers, differing by a single nucleotide extension at the 3′ terminus, were designed for each individual adapter. These primers are designated the “forward” reaction primers. Primer AH1 is the same as the longer oligonucleotide of the HhaI adapter pair and is designated the “reverse” primer. Forward primers were synthesized with and without a 5′ 6-carboxyfluorescein (FAM) (Glen Research, Sterling, Va.). Primers were diluted to 20 μM and stored at −20°C in the dark until needed.
Amplification.
Each PCR mixture contained 1 μl of restricted-ligated genomic DNA, 0.5 U of HotStarTaq DNA Polymerase (Qiagen, Valencia, Calif.), deoxynucleoside triphosphates (200 μM each), and appropriate primers (1.0 μM each) in 1× PCR buffer. Four different reactions were performed for each restriction endonuclease combination. For example, if the enzymes SmaI and HhaI were used, then the primer PSmaI-A (or PSmaI-T, -G, or -C) would be combined in individual reaction tubes with primer AH1. This primer combination would allow the selective amplification of fragments flanked by SmaI and HhaI sites only when the nucleotide directly downstream from the SmaI site is a thymine. All PCR assays were performed using a PTC200 DNA-Engine (MJ Research, Inc., Waltham, Mass.). Thermocycler conditions varied depending on the primer combination used. For EagI-HhaI, the program consisted of initial denaturing at 95°C for 15 min; 25 cycles of denaturing at 94°C for 1 min, primer annealing at 67°C for 30 s, and extension at 72°C for 2 min; and a final extension at 72°C for 10 min. For SmaI-HhaI, the program was initial denaturing at 95°C for 15 min; 25 cycles of denaturing at 94°C for 1 min and annealing and extension at 72°C for 2 min; and a final extension at 72°C for 10 min. For XbaI-HhaI, initial denaturing took place at 95°C for 15 min, followed by 25 cycles of denaturing at 94°C for 1 min, primer annealing at 61°C for 30 s, and extension at 72°C for 2 min, with a final extension at 72°C for 10 min. All reaction conditions remained the same if FAM-labeled primers were used. All PCR assays included negative controls consisting of all reactants except digested and ligated genomic DNA.
Pattern visualization.
For slab gel polyacrylamide gel electrophoresis (PAGE), 10 μl of each amplified reaction mixture was electrophoresed on precast 10% polyacrylamide gels (NOVEX, San Diego, Calif.) in 1× Tris-borate-EDTA (TBE) buffer (0.045 M Tris-borate–0.001 M EDTA). Fragments were electrophoresed at 180 V for 1 to 1 1/2 h and subsequently stained with 1× SYBR Gold (Molecular Probes, Eugene, Oreg.) for 30 min. Gels were visualized by UV transillumination, photographed with a charge-coupled device (CCD) camera, and analyzed with the Advanced Quantifier software package (Genomic Solutions, Ann Arbor, Mich.).
Resolution and detection of FAM-labeled fragments was achieved on the ABI 310 Genetic Analyzer capillary electrophoresis system (Perkin-Elmer, Norwalk, Conn.). Analyzed product mixtures consisted of 1 μl of amplified product combined with 24 μl of deionized formamide and 1 μl of internal-lane size standard labeled with ROX dye (GeneScan 1000 ROX; Applied Biosystems, Foster City, Calif.). The mixtures were heated at 95°C for 5 min before being cooled on ice for 10 min. Each sample was electrophoresed on performance-optimized polymer POP-4 (Applied Biosystems) for 35 min. Data were collected and analyzed using the ABI 310 collection package, v. 1.0.4 and GeneScan, v. 3.1 (Applied Biosystems). Fragments were sized in the range of 50 to 700 nucleotides.
Further numerical analysis of the patterns was done using the BIONUMERICS software package (Applied Maths, Kortrijk, Belgium). The similarity between pairs of separate genomic fingerprints was calculated after normalization and subtraction of background by using both the product moment correlation coefficient to examine whole densitometric curves and the Dice and/or Jaccard band matching coefficients (26).
RESULTS
Pattern variation.
This study used IRS-PCR patterns to differentiate clinically relevant isolates of Bartonella. Under the conditions used, the resulting patterns provided sufficient information to readily differentiate all species tested. Both slab and capillary PAGE were used to separate fragments. Slab gel PAGE allowed for sufficient fragment resolution with the enzyme combinations EagI-HhaI and SmaI-HhaI (Fig. 2 and 3). Patterns were also resolved and detected with the ABI 310 capillary electrophoresis Genetic Analyzer. Capillary electrophoresis successfully resolved a large number of labeled DNA fragments produced after XbaI-HhaI digestion and subsequent amplification (Fig. 4). Fragments between 50 and 700 bp were found to be the most reproducible. Fragments above and below this range were ignored in further analysis. Pattern variation between species was high (pairwise similarity as measured by the Dice coefficient, 23.0 to 32.4%), while pattern variation among isolates of a single species was relatively low (79.4 to 100% similarity).
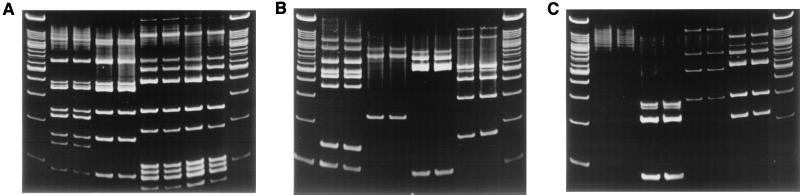
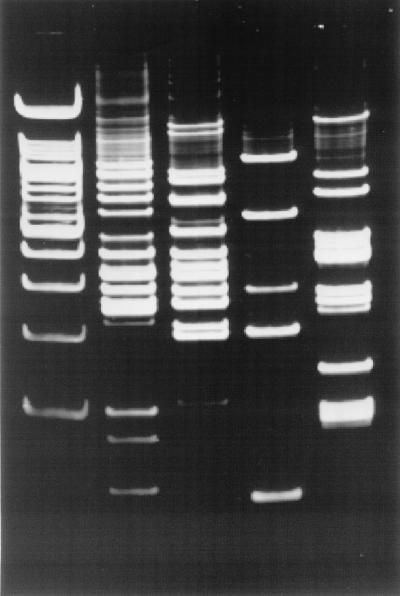
FIG. 2.
Duplicate IRS-PCR patterns produced by amplification of restricted-ligated SmaI-HhaI DNAs using primer PSmaI-C. The first and last lanes in each gel are 100-bp ladders. (A) Second and third lanes, B. henselae Houston-1; fourth and fifth lanes, B. henselae Marseilles; sixth and seventh lanes, B. quintana Fuller; eighth and ninth lanes, B. quintana OK90-268. (B) Second and third lanes, B. bacilliformis KC583; fourth and fifth lanes, B. elizabethae; sixth and seventh lanes, B. clarridgeiae; eighth and ninth lanes, B. grahamii. (C) Second and third lanes, B. vinsonii subsp. vinsonii; fourth and fifth lanes, B. vinsonii subsp. arupensis; sixth and seventh lanes, B. vinsonii subsp. berkhoffii; eighth and ninth lanes, B. weissii.
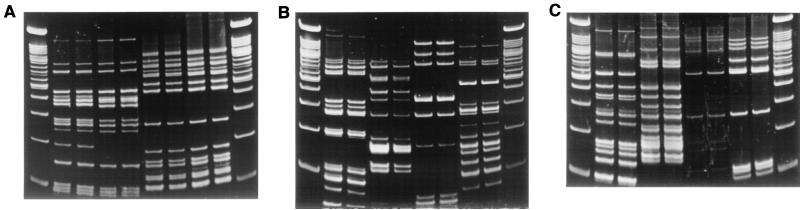
FIG. 3.
Duplicate IRS-PCR patterns produced by amplification of restricted-ligated EagI-HhaI DNAs using primer PEagI-C. Lane assignments are the same as for Fig. 2.
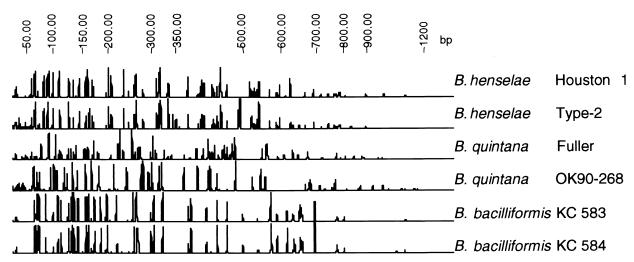
FIG. 4.
Examples of IRS-PCR electropherograms generated by analysis of fluorescent IRS-PCR products with capillary electrophoresis. Patterns were generated using fluorescent primer PX-C. Patterns were subsequently normalized and sized against a common internal standard.
Pattern complexity.
Addition of selective nucleotides to the primer sequence permitted the production of multiple patterns for each restricted-ligated DNA (Fig. 5). Pattern complexity was modified based on the nucleotide in the position immediately downstream from the ligated adapter sequence. With four different selective primers and three enzyme combinations, it was possible to generate 12 unique patterns for each DNA. Enzyme combination EagI-HhaI produced anywhere from 10 to 22 bands, and SmaI-HhaI produced 0 to 20 bands. By using capillary electrophoresis and the combination XbaI-HhaI, it was possible to generate anywhere from 75 to 125 bands with PX-A and 25 to 60 bands with PX-C. In general, primers with the A or T extension produced more fragments than primers with the G or C extension, reflecting the high AT content of Bartonella genomes (34).
FIG. 5.
Effects of selective nucleotide addition on IRS-PCR patterns of B. henselae Houston-1. Patterns were generated by amplification of EagI-HhaI restricted-ligated fragments by using various forward primers in combination with primer AH-1. First lane, 100-bp ladder; second lane, primer EagI-A; third lane, PEagI-T; fourth lane, PEagI-G; fifth lane, PEagI-C.
Pattern reproducibility.
All IRS-PCR genomic fingerprints resolved by slab gel electrophoresis were run in duplicate (reactions were completed separately from start to finish, i.e., DNA extraction, restriction endonuclease digestion, adapter ligation, PCR amplification, and electrophoresis). There was little to no pattern variability between duplicate reactions, only minor variations in band intensity. Patterns generated with the enzyme combination SmaI-HhaI tended to have greater amounts of high-molecular-weight background than the EagI-HhaI patterns. This was perhaps due to incomplete extension of large PCR fragments.
Pattern reproducibility was also examined using a fluorescence-detecting capillary electrophoresis system. Ten different colonies from a plate of B. henselae strain Tiger-2 were picked and replated for growth before DNA extraction. The entirety of the remaining protocol was completed individually for each separate isolated DNA (data not shown). We found that the run-to-run correlation varied from 94.3 to 98.3% based on band matching using the Dice coefficient, and 75.7 to 98% based on whole densitometric profiles using the Pearson correlation.
DISCUSSION
IRS-PCR has been shown to be a robust method for the molecular characterization of bacteria (23, 25, 31, 41). The technique allows for a high level of flexibility and can produce several different genomic fingerprints of varying complexity for each sample analyzed, depending on enzyme combination and primer modification. Restriction endonuclease digestion is sequence specific, primers used in the PCR are specific to the previously ligated sequence (not arbitrary as with ERIC-PCR and related techniques), and only limited information about the target DNA is needed. Relative to typing techniques such as PFGE, the technique is more efficient and minimal amounts of genomic DNA are required. Due to the small size of the fragment amplified by the IRS-PCR procedure, genomic DNA may be extracted from the organism in a variety of ways with only limited concern for DNA integrity (23).
Optimal pattern generation is initially controlled by the number of DNA fragments generated by the selected restriction endonuclease combination. Selection of restriction endonuclease pairs can be simplified if the basic genomic organization is known (G+C content, complete genome sequence, size, and DNA modification). For example, if more fragments are required for the analysis, a restriction endonuclease combination with an increased predicted cutting frequency could be used, and vice versa. The essential experimental design remains the same regardless of the restriction endonuclease pairs used. Several restriction endonuclease combinations can be utilized to generate optimized, unique, and easy-to-interpret patterns.
In this study, three different restriction endonuclease combinations were used. Each combination used one rare-cutting enzyme (GC-rich 6-base recognition sequence), either EagI, SmaI, or XbaI, and a relatively infrequently cutting restriction endonuclease, HhaI (GC-rich 4-base recognition sequence). EagI and SmaI were chosen based on previously published PFGE data for Bartonella (34). Both of these enzymes, in conjunction with HhaI, produced patterns easily resolved by traditional slab PAGE. XbaI was chosen based on the original IRS-PCR experiment by Mazurek et al. (23). In the original publication, the use of XbaI-HhaI produced informative patterns for Mycobacterium avium and Mycobacterium intracellulare isolates, Pseudomonas aeruginosa isolates, and Staphylococcus isolates. All amplified DNA fragment patterns were resolved by traditional electrophoretic methods. However, when the same conditions were used for Bartonella isolates, the banding patterns were much more complex and difficult to resolve using traditional PAGE.
The use of fluorescently labeled primers combined with capillary electrophoresis has been described for the resolution of amplified fragment length polymorphism (AFLP) analysis (1, 3). Analogous techniques were used in this study for the resolution and detection of IRS-PCR patterns. Fluorescently labeled forward primers were used to amplify fragments under the same conditions as the nonlabeled primers. Products were separated and detected using the ABI 310 Genetic Analyzer. Resulting patterns were highly resolved, complex, and accurately sized with internally labeled DNA fragment size standards. Complicated DNA fragment patterns were challenging to analyze by hand; however, reproducible pattern analysis was greatly facilitated by the BioNumerics and GeneScan software.
A major advantage with fluorescent detection systems such as the ABI 310 Genetic Analyzer is that they are highly automated and provide uniform data collection and analysis. It is practical to analyze EagI-HhaI and SmaI-HhaI patterns with PAGE; however, analysis with the fluorescent detection system accurately resolves band sizes within 1 to 2 bp and automatically collects, stores, and analyzes these patterns for further studies.
Selective nucleotide addition to the forward primer allowed a further degree of pattern selectivity. This step facilitated the production of four unique patterns usable for comparison between isolates. Patterns produced using selective nucleotides and the enzyme combination SmaI-HhaI were different for all Bartonella species. However, the number of bands produced was generally very small. EagI-HhaI patterns produced more fragments than SmaI-HhaI, and XbaI-HhaI generated the most-complex patterns. With all three systems as tools for the analysis of Bartonella, experimental design can undergo significant optimization as required by specific laboratory capabilities and according to the needs of the individual experiment. One difficulty with the use of SmaI-HhaI for the analysis of Bartonella isolates is the potential lack of SmaI sites. Bartonella vinsonii subsp. vinsonii was shown to be devoid of SmaI cutting sites by PFGE (34). This is reflected in Fig. 2 by the absence of any defined IRS-PCR fragments. To avoid this complication, analysis of related Bartonella species should not be dependent on SmaI-HhaI IRS-PCR.
Patterns produced with the restriction endonuclease combination XbaI-HhaI and fluorescently labeled primers were highly resolved using the ABI 310 Genetic Analyzer. When multiple selective primers were used, patterns containing hundreds of fragments were obtained. These data may be suited for cluster analysis using distance and/or parsimony methods. A quantitative summary of a subset of data is shown in Table 2.
TABLE 2.
IRS-PCR data using PX-A and PX-T selective primersb
| Species or isolate | No. of comigrating bands/no. of fragments present for the indicated pair of species and/or isolates
|
|||
|---|---|---|---|---|
| B. henselae Houston-1 (PX-A = 119)a | B. quintana Fuller (PX-A = 101) | B. elizabethae (PX-A = 106) | B. bacilliformis KC583 (PX-A = 75) | |
| B. henselae Houston-1 (PX-T = 73) | 82/220 | 86/225 | 55/194 | |
| B. quintana Fuller (PX-T = 46) | 20/119 | 75/207 | 50/176 | |
| B. elizabethae (PX-T = 80) | 25/153 | 25/126 | 48/181 | |
| B. bacilliformis KC583 (PX-T = 48) | 24/121 | 16/94 | 15/128 | |
Values in parentheses indicate the total number of bands generated by the indicated selective primer set.
Comigrating fragments generated with PX-A selective primers are displayed in the upper right quadrant, whereas comigrating fragments generated with PX-T selective primers are indicated in the lower left quadrant.
In conclusion, the described IRS-PCR fingerprinting techniques are useful, pangenomic tools that are not limited to analysis of individual genes. These methods can be used for the identification and differentiation of known strains of pathogenic Bartonella when comprehensive isolate identification is required, and the techniques are relatively simple and rapid and require minimal amounts of genomic DNA. Experimental flexibility allows for the optimization of data production through variation either in the restriction endonuclease combination used or in selective primers during amplification. Fluorescent detection systems allow for high resolution of DNA fragment patterns along with convenient computerized data acquisition, analysis, storage, and retrieval. The highly reproducible quality of the DNA fingerprints produced by IRS-PCR makes the method suitable for archiving and sharing information for future isolate identification and possible phylogenetic analysis.
REFERENCES
- 1.Aarts H J, Hakemulder L E, Van Hoef A M. Genomic typing of Listeria monocytogenes strains by automated laser fluorescence analysis of amplified fragment length polymorphism fingerprint patterns. Int J Food Microbiol. 1999;49:95–102. doi: 10.1016/s0168-1605(99)00057-4. [DOI] [PubMed] [Google Scholar]
- 2.Armengol C E, Hendley J O. Cat-scratch disease encephalopathy: a cause of status epilepticus in school-aged children. J Pediatr. 1999;134:635–638. doi: 10.1016/s0022-3476(99)70252-0. [DOI] [PubMed] [Google Scholar]
- 3.Arnold C, Metherell L, Clewley J P, Stanley J. Predictive modelling of fluorescent AFLP: a new approach to the molecular epidemiology of E. coli. Res Microbiol. 1999;150:33–44. doi: 10.1016/s0923-2508(99)80044-8. [DOI] [PubMed] [Google Scholar]
- 4.Bass J W, Vincent J M, Person D A. The expanding spectrum of Bartonella infections. I. Bartonellosis and trench fever. Pediatr Infect Dis J. 1997;16:2–10. doi: 10.1097/00006454-199701000-00003. [DOI] [PubMed] [Google Scholar]
- 5.Bass J W, Vincent J M, Person D A. The expanding spectrum of Bartonella infections. II. Cat-scratch disease. Pediatr Infect Dis J. 1997;16:163–179. doi: 10.1097/00006454-199702000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Birtles R J, Raoult D. Comparison of partial citrate synthase gene (gltA) sequences for phylogenetic analysis of Bartonella species. Int J Syst Bacteriol. 1996;46:891–897. doi: 10.1099/00207713-46-4-891. [DOI] [PubMed] [Google Scholar]
- 7.Brouqui P, Houpikian P, Dupont H T, Toubiana P, Obadia Y, Lafay V, Raoult D. Survey of the seroprevalence of Bartonella quintana in homeless people. Clin Infect Dis. 1996;23:756–759. doi: 10.1093/clinids/23.4.756. [DOI] [PubMed] [Google Scholar]
- 8.Brouqui P, Lascola B, Roux V, Raoult D. Chronic Bartonella quintana bacteremia in homeless patients. N Engl J Med. 1999;340:184–189. doi: 10.1056/NEJM199901213400303. [DOI] [PubMed] [Google Scholar]
- 9.Caceres-Rios H, Rodriguez-Tafur J, Bravo-Puccio F, Maguina-Vargas C, Diaz C S, Ramos D C, Patarca R. Verruga peruana: an infectious endemic angiomatosis. Crit Rev Oncog. 1995;6:47–56. doi: 10.1615/critrevoncog.v6.i1.40. [DOI] [PubMed] [Google Scholar]
- 10.Carithers H A, Margileth A M. Cat-scratch disease. Acute encephalopathy and other neurologic manifestations. Am J Dis Child. 1991;145:98–101. doi: 10.1001/archpedi.1991.02160010104026. [DOI] [PubMed] [Google Scholar]
- 11.Daly J S, Worthington M G, Brenner D J, Moss C W, Hollis D G, Weyant R S, Steigerwalt A G, Weaver R E, Daneshvar M I, O'Connor S P. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993;31:872–881. doi: 10.1128/jcm.31.4.872-881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolan M J, Wong M T, Regnery R L, Jorgensen J H, Garcia M, Peters J, Drehner D. Syndrome of Rochalimaea henselae adenitis suggesting cat scratch disease. Ann Intern Med. 1993;118:331–336. doi: 10.7326/0003-4819-118-5-199303010-00002. [DOI] [PubMed] [Google Scholar]
- 13.Drancourt M, Mainardi J L, Brouqui P, Vandenesch F, Carta A, Lehnert F, Etienne J, Goldstein F, Acar J, Raoult D. Bartonella (Rochalimaea) quintana endocarditis in three homeless men. N Engl J Med. 1995;332:419–423. doi: 10.1056/NEJM199502163320702. [DOI] [PubMed] [Google Scholar]
- 14.Ellis B A, Regnery R L, Beati L, Bacellar F, Rood M, Glass G G, Marston E, Ksiazek T G, Jones D, Childs J E. Rats of the genus Rattus are reservoir hosts for pathogenic Bartonella species: an Old World origin for a New World disease? J Infect Dis. 1999;180:220–224. doi: 10.1086/314824. [DOI] [PubMed] [Google Scholar]
- 15.Ellis B A, Rotz L D, Leake J A, Samalvides F, Bernable J, Ventura G, Padilla C, Villaseca P, Beati L, Regnery R, Childs J E, Olson J G, Carrillo C P. An outbreak of acute bartonellosis (Oroya fever) in the Urubamba region of Peru, 1998. Am J Trop Med Hyg. 1999;61:344–349. doi: 10.4269/ajtmh.1999.61.344. [DOI] [PubMed] [Google Scholar]
- 16.Jackson L A, Spach D H, Kippen D A, Sugg N K, Regnery R L, Sayers M H, Stamm W E. Seroprevalence to Bartonella quintana among patients at a community clinic in downtown Seattle. J Infect Dis. 1996;173:1023–1026. doi: 10.1093/infdis/173.4.1023. [DOI] [PubMed] [Google Scholar]
- 17.Kelly T M, Padmalayam I, Baumstark B R. Use of the cell division protein FtsZ as a means of differentiating among Bartonella species. Clin Diagn Lab Immunol. 1998;5:766–772. doi: 10.1128/cdli.5.6.766-772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerkhoff F T, Bergmans A M, van Der Zee A, Rothova A. Demonstration of Bartonella grahamii DNA in ocular fluids of a patient with neuroretinitis. J Clin Microbiol. 1999;37:4034–4038. doi: 10.1128/jcm.37.12.4034-4038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kordick D L, Hilyard E J, Hadfield T L, Wilson K H, Steigerwalt A G, Brenner D J, Breitschwerdt E B. Bartonella clarridgeiae, a newly recognized zoonotic pathogen causing inoculation papules, fever, and lymphadenopathy (cat scratch disease) J Clin Microbiol. 1997;35:1813–1818. doi: 10.1128/jcm.35.7.1813-1818.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosoy M Y, Regnery R L, Tzianabos T, Marston E L, Jones D C, Green D, Maupin G O, Olson J G, Childs J E. Distribution, diversity, and host specificity of Bartonella in rodents from the Southeastern United States. Am J Trop Med Hyg. 1997;57:578–588. doi: 10.4269/ajtmh.1997.57.578. [DOI] [PubMed] [Google Scholar]
- 21.Lombardo J. Cat-scratch neuroretinitis. J Am Optom Assoc. 1999;70:525–530. [PubMed] [Google Scholar]
- 22.Marston E L, Sumner J W, Regnery R L. Evaluation of intraspecies genetic variation within the 60-kDa heat shock protein gene (groEL) of Bartonella species. Int J Syst Bacteriol. 1999;49:1015–1023. doi: 10.1099/00207713-49-3-1015. [DOI] [PubMed] [Google Scholar]
- 23.Mazurek G H, Reddy V, Marston B J, Haas W H, Crawford J T. DNA fingerprinting by infrequent-restriction-site amplification. J Clin Microbiol. 1996;34:2386–2390. doi: 10.1128/jcm.34.10.2386-2390.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olive D M, Bean P. Principles and applications of methods for DNA-based typing of microbial organisms. J Clin Microbiol. 1999;37:1661–1669. doi: 10.1128/jcm.37.6.1661-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park Y H, Yoo J H, Huh D H, Cho Y K, Choi J H, Shin W S. Molecular analysis of fluoroquinolone-resistance in Escherichia coli on the aspect of gyrase and multiple antibiotic resistance (mar) genes. Yonsei Med J. 1998;39:534–540. doi: 10.3349/ymj.1998.39.6.534. [DOI] [PubMed] [Google Scholar]
- 26.Rademaker J L, Hoste B, Louws F J, Kersters K, Swings J, Vauterin L, Vauterin P, de Bruijn F J. Comparison of AFLP and rep-PCR genomic fingerprinting with DNA-DNA homology studies: Xanthomonas as a model system. Int J Syst Evol Microbiol. 2000;50:665–677. doi: 10.1099/00207713-50-2-665. [DOI] [PubMed] [Google Scholar]
- 27.Raoult D, Birtles R J, Montoya M, Perez E, Tissot-Dupont H, Roux V, Guerra H. Survey of three bacterial louse-associated diseases among rural Andean communities in Peru: prevalence of epidemic typhus, trench fever, and relapsing fever. Clin Infect Dis. 1999;29:434–436. doi: 10.1086/520229. [DOI] [PubMed] [Google Scholar]
- 28.Raoult D, Ndihokubwayo J B, Tissot-Dupont H, Roux V, Faugere B, Abegbinni R, Birtles R J. Outbreak of epidemic typhus associated with trench fever in Burundi. Lancet. 1998;352:353–358. doi: 10.1016/s0140-6736(97)12433-3. [DOI] [PubMed] [Google Scholar]
- 29.Regnery R L, Anderson B E, Clarridge J E, Rodriguez-Barradas M C, Jones D C, Carr J H. Characterization of a novel Rochalimaea species, R. henselae sp. nov., isolated from blood of a febrile, human immunodeficiency virus-positive patient. J Clin Microbiol. 1992;30:265–274. doi: 10.1128/jcm.30.2.265-274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Relman D A, Loutit J S, Schmidt T M, Falkow S, Tompkins L S. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N Engl J Med. 1990;323:1573–1580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 31.Riffard S, Lo Presti F, Vandenesch F, Forey F, Reyrolle M, Etienne J. Comparative analysis of infrequent-restriction-site PCR and pulsed-field gel electrophoresis for epidemiological typing of Legionella pneumophila serogroup 1 strains. J Clin Microbiol. 1998;36:161–167. doi: 10.1128/jcm.36.1.161-167.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez-Barradas M C, Hamill R J, Houston E D, Georghiou P R, Clarridge J E, Regnery R L, Koehler J E. Genomic fingerprinting of Bartonella species by repetitive element PCR for distinguishing species and isolates. J Clin Microbiol. 1995;33:1089–1093. doi: 10.1128/jcm.33.5.1089-1093.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roux V, Eykyn S J, Wyllie S, Raoult D. Bartonella vinsonii subsp. berkhoffii as an agent of afebrile blood culture-negative endocarditis in a human. J Clin Microbiol. 2000;38:1698–1700. doi: 10.1128/jcm.38.4.1698-1700.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roux V, Raoult D. Inter- and intraspecies identification of Bartonella (Rochalimaea) species. J Clin Microbiol. 1995;33:1573–1579. doi: 10.1128/jcm.33.6.1573-1579.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rydkina E B, Roux V, Gagua E M, Predtechenski A B, Tarasevich I V, Raoult D. Bartonella quintana in body lice collected from homeless persons in Russia. Emerg Infect Dis. 1999;5:176–178. doi: 10.3201/eid0501.990126. . (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sander A, Buhler C, Pelz K, von Cramm E, Bredt W. Detection and identification of two Bartonella henselae variants in domestic cats in Germany. J Clin Microbiol. 1997;35:584–587. doi: 10.1128/jcm.35.3.584-587.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sander A, Posselt M, Bohm N, Ruess M, Altwegg M. Detection of Bartonella henselae DNA by two different PCR assays and determination of the genotypes of strains involved in histologically defined cat scratch disease. J Clin Microbiol. 1999;37:993–997. doi: 10.1128/jcm.37.4.993-997.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sander A, Ruess M, Bereswill S, Schuppler M, Steinbrueckner B. Comparison of different DNA fingerprinting techniques for molecular typing of Bartonella henselae isolates. J Clin Microbiol. 1998;36:2973–2981. doi: 10.1128/jcm.36.10.2973-2981.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varela G, Vinson J W, Molina-Pasquel C. Trench fever. II. Propagation of Rickettsia quintana on cell-free medium from the blood of two patients. Am J Trop Med Hyg. 1969;18:708–712. [PubMed] [Google Scholar]
- 40.Welch D F, Carroll K C, Hofmeister E K, Persing D H, Robison D A, Steigerwalt A G, Brenner D J. Isolation of a new subspecies, Bartonella vinsonii subsp. arupensis, from a cattle rancher: identity with isolates found in conjunction with Borrelia burgdorferi and Babesia microti among naturally infected mice. J Clin Microbiol. 1999;37:2598–2601. doi: 10.1128/jcm.37.8.2598-2601.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoo J H, Choi J H, Shin W S, Huh D H, Cho Y K, Kim K M, Kim M Y, Kang M W. Application of infrequent-restriction-site PCR to clinical isolates of Acinetobacter baumannii and Serratia marcescens. J Clin Microbiol. 1999;37:3108–3112. doi: 10.1128/jcm.37.10.3108-3112.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]