Abstract
The ability to manipulate the electrophysiology of electrically active cells and tissues has enabled a deeper understanding of healthy and diseased tissue states. This has primarily been achieved via input/output (I/O) bioelectronics that interface engineered materials with biological entities. Stable long-term application of conventional I/O bioelectronics advances as materials and processing techniques develop. Recent advancements have facilitated the development of graphene-based I/O bioelectronics with a wide variety of functional characteristics. Engineering the structural, physical, and chemical properties of graphene nanostructures and integration with modern microelectronics have enabled breakthrough high-density electrophysiological investigations. Here, we review recent advancements in 2D and 3D graphene-based I/O bioelectronics and highlight electrophysiological studies facilitated by these emerging platforms. Challenges and present potential breakthroughs that can be addressed via graphene bioelectronics are discussed. We emphasize the need for a multidisciplinary approach across materials science, micro-fabrication, and bioengineering to develop the next generation of I/O bioelectronics.
I. INTRODUCTION
The first intracellular recording of action potential was made by Hodgkin and Huxley.1 This seminal breakthrough was recorded from squid axon (ca. 0.5 mm in diameter) due to the scale of the available bioelectrical tools at the time. Today, we know that tightly controlled specific ion movement is found in and regulates all levels of cellular and subcellular activities. The tremendous progress in all scientific fields, including materials science, has provided the tools and level of knowledge that allow us to study electrophysiology at micro-scales relevant to a wide variety of cell types. Electrophysiology and related biochemical processes have been studied in vivo, ex vivo, and in vitro at the cellular and protein level, and we have detailed descriptions of those. The relationship between the physiology and electrical activity of cells and tissues has facilitated an in-depth understanding of intracellular processes crucial to higher-order functions.2–6 However, some remain enigmatic, and many are still left for us to discover. In addition to gaining knowledge, understanding of those processes enables us to affect them by both electrical and chemical signals.7,8 These capabilities are utilized to treat pathologies such as Parkinson's disease9 and heart arrhythmias.10 This is implemented using medical technologies to measure and modulate electrical activity of the affected biological systems.
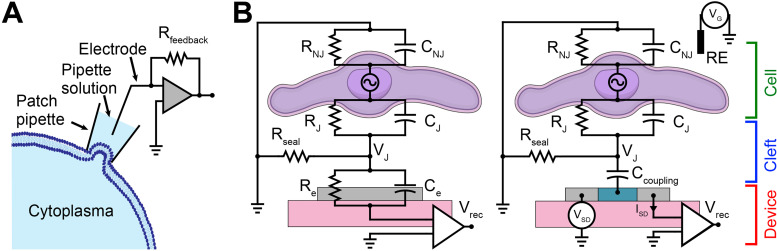
Input/output (I/O) bioelectronic platforms have been developed to facilitate electrophysiology studies and clinical applications.8,11–13 I/O platforms are designed to both produce a signal (input) and record a signal (output). Such capabilities can be integrated into platforms capable of chemical and optical I/O in addition to electrical I/O.14 These platforms can be broadly categorized as intracellular or extracellular bioelectronics (Fig. 1). Intracellular bioelectronics examine the electrical potentials inside and across the cell membrane of single cells.2,15,16 Traditionally, these tools have relied on the patch-clamp approach, which provides direct intracellular access allowing accurate recording of action potentials and corresponding cellular communication paradigms [Fig. 1(a)].15,16 Extracellular bioelectronics, primarily classified as active (such as field-effect transistors, FETs) or passive (microelectrode arrays, MEAs), allow investigations of local field potentials in the extracellular space [Fig. 1(b)].4,11 The generated extracellular field potentials lead to corresponding changes in the junction potential (VJ) when interfaced with either MEAs or FETs.11 These changes in VJ are recorded as corresponding changes in potential (in interfaced MEAs) and channel current (in interfaced FETs).11 Both MEAs and FETs have facilitated simultaneous multi-scale recording and stimulation of cells and tissues in vitro and in vivo over extended periods.17–20
FIG. 1.
I/O bioelectronic interfaces. (a) Schematic of the whole-cell patch-clamp technique for recording cellular electrophysiology. (b) Electrical equivalent circuit of the (I) cell–MEA and (II) cell–FET interfaces. RJ, RNJ, Rseal, and Re represent junctional, non-junctional, seal, and electrode resistances, respectively. CJ, CNJ, Ccoupling, and Ce represent junctional, non-junctional, coupling, and electrode capacitances, respectively. VJ, VSD, VG, Vrec, and ISD represent junctional voltage across the cleft, source–drain voltage, gate voltage, recorded voltage, and source–drain current, respectively. RE represents reference electrode.
Recent advances in materials, electrical, and biomedical engineering have enabled the development of breakthrough I/O bioelectronics for real-time high-density electrophysiological mapping.13,18,21,22 This has been coupled with the emergence of nanomaterials with tunable physical and chemical properties.4,12,13,23 In this review, we provide a brief overview of conventional bioelectronics and then focus our attention on emerging trends in materials engineering for the development of next-generation of I/O bioelectronics.
II. EVOLUTION OF BIOELECTRONICS
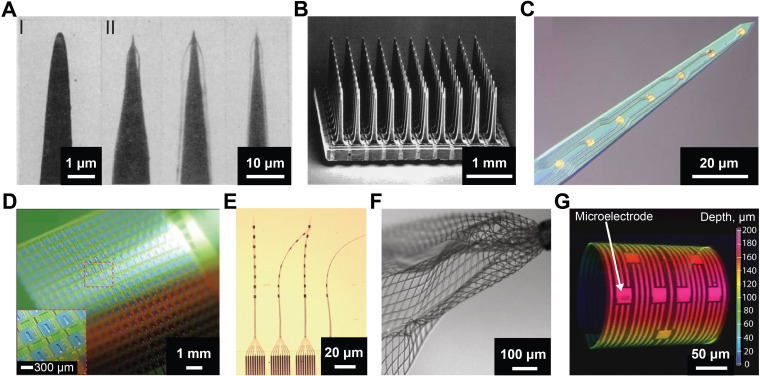
Bioelectronics development relies on existing materials and processing techniques.23,24 The dimensions of the available tools enforce a physical limitation on the scale of cells and tissues that can be investigated. Early-stage research was focused on macro-scale nerves and axons.1 Although early microelectrode iterations contributed extensively to neuroscience research, they were limited to a single channel composed of sharpened W, Pt, or Pt-Ag/AgCl wires [Fig. 2(a)].8,25,26 Technological breakthroughs in micro-fabrication enabled the development of multiplexed micro-scale bioelectronics, such as Utah arrays and Michigan probes,27,28 for precise investigations of cells and tissues both in vitro and in vivo. Utah arrays consist of high-density 80–100 μm diameter silicon tips [Fig. 2(b)] and have been implanted into the cortex for in vivo electrophysiology recording and mapping with little pathological complications [Fig. 2(b)].27 They have preliminary been used for in vivo electrical recordings in animals, especially non-human primates, and have served as a promising toolset for clinical use.29 Michigan probes also leverage on silicon as a substrate and are composed of multiple channels integrated on a single probe [Fig. 2(c)].28 Further modification of the exposed electrode surface with materials such as conductive polymers and hydrogels has resulted in dramatic enhancements of probe sensitivity, robustness, and biocompatibility.28 Although these platforms facilitated the next wave of seminal breakthroughs in neuroscience and therapeutic interventions, their rigid nature results in chronic inflammatory response leading to the generation of capsules and scars.8,13
FIG. 2.
Evolution of microelectrodes for bioelectronics. (a) Electron micrograph of (I) an uncoated, sharpened tungsten wire; and (II) optical images of coated electrodes immersed in water to show the coating. Reproduced with permission from Hebul, Science 125, 3247 (1957). Copyright 2002 Science.25 (b) Scanning electron micrograph of Utah electrode array with 100 microelectrodes. Reproduced with permission from Normann et al., Vision Res. 39(15), 2577–2587 (1999). Copyright 1999 Elsevier Science Ltd.27 (c) Optical micrograph of Michigan probe with eight-channel recording gold sites. Reproduced with permission from Abidian et al., Adv. Funct. Mater. 19(4), 573–585 (2009). Copyright 2009 Wiley-VCH.28 (d) Photograph of a flexible 360-channel high density active electrode array. Reproduced with permission from Viventi et al., Nat. Neurosci. 14, 1599–1605 (2011). Copyright 2011 Nature Publishing Group.30 (e) Photograph of four threads on a NET-e device panel. Reproduced with permission from Wei et al., Adv. Sci. 5(6), 1700625 (2018). Copyright 2018 Author(s), licensed under a Creative Commons Attribution 4.0 License.31 (f) Optical image of mesh electronics emerging from the tip (upper right) of a 95 μm diameter needle into 1× phosphate-buffered saline (PBS) solution. Reproduced with permission from Liu et al., Nat. Nanotechnol. 10, 629–636 (2015). Copyright 2015 Springer Nature.32 (g) 3D confocal microscopy image of 3D-SR-BA with microelectrodes. Color bar represents the depth in micrometers. Reproduced with permission from Kalmykov et al., Sci. Adv. 5(8), eaax0729 (2020). Copyright 2019 Author(s), licensed under a Creative Commons Attribution-NonCommercial License 4.0.33
Numerous efforts have been made to address mechanical mismatch of bioelectronics and further minimize platform dimensions.4,24 Viventi et al. demonstrated a novel approach toward silicon-based flexible bioelectronics by fabricating ultrathin silicon nanomembrane transistors arrays and successfully recording in vivo brain activity of a cat [Fig. 2(d)].30 Wei et al. developed ultra-flexible nanoelectronic threads (NET) with more than eight electrodes housed on a single thread within a cross-sectional area of 10 μm2 [Fig. 2(e)].31 This platform enabled neural mapping in a mouse model with high signal-to-noise ratio (SNR) and without chronic neuronal degeneration.31 To achieve seamless integration of bioelectronics with in vivo systems in a minimally invasive manner, Liu et al. fabricated syringe injectable microporous mesh electronics [Fig. 2(f)].32 Injection of mesh electronics into biological cavities was achieved through small injections sites by using syringes with a diameter less than 100 μm.32 Once injected into the hippocampus of live rodent brains, mesh electronics facilitated multichannel recordings of the electrical activity, such as the δ-wave local field potentials.32 On the other hand, to study cellular communication in in vivo like three-dimensional (3D) cell cultures, Kalmykov et al. leveraged pre-stressed thin films to fabricate a 3D self-rolling biosensor array (3D-SR-BA) [Fig. 2(g)] and recorded electrical activity of cardiac and neural spheroids.33,34 Park et al. integrated electrical, optical, chemical, and thermal biointerfaces onto a single platform to study the coordinated electrical activity across the surface of cortical spheroids.35
Although the evolution of bioelectronic platforms has been multifaceted, there is still an imminent requirement for technological innovations of the biointerfaces themselves to enable multimodal platforms. Engineering nanomaterials for bioelectronic interfaces by tailoring material structures and properties provides an opportunity to address the limitations of existing bioelectronics platforms and overcome the current technological challenges.12,36 Carbon-based nanomaterials, especially graphene-based systems, are promising candidates for bioelectronic interfaces due to their high mechanical flexibility, structural stability, and high tunability of physical and chemical properties.4,12,37
III. GRAPHENE NANOSTRUCTURES
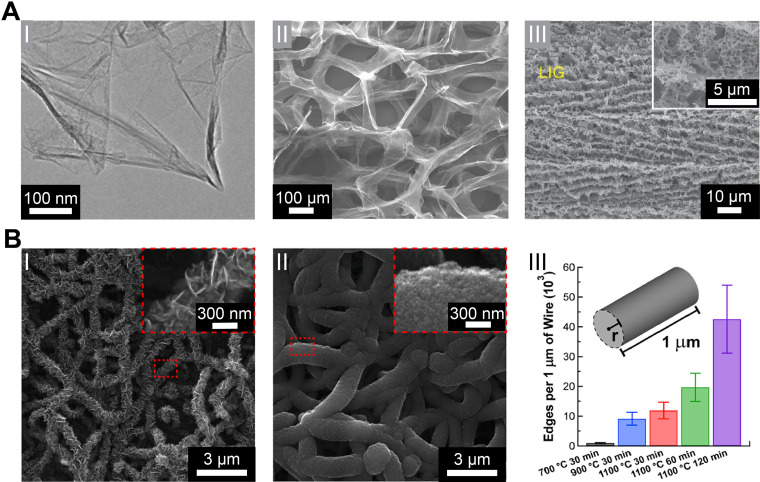
Graphene consists of a honeycomb sp2 hybridized two-dimensional (2D) carbon lattice [Fig. 3(a-I)] with many favorable properties for bioelectronics such as its high electrical conductivity (charge carrier mobility up-to 200 000 cm2 V−1 s−1),38 mechanical strength (Young's modulus of ∼1 TPa),39 high surface-to-volume ratio (theoretical value of ∼2630 m2 g−1),40 and chemical stability.41 Due to its excellent processability, multiple approaches have been developed for implementing graphene-based nanostructures in platforms for electrophysiology. Bottom–up synthesis of graphene, such as through chemical vapor deposition (CVD),42 leverages the self-assembling properties of aromatic carbon structures, while top–down approaches such as chemical exfoliation of graphite40 and laser-induced graphene (LIG) synthesis allow high yield rates.43 Graphene can readily be implemented as a building block for other carbon allotropes or hierarchical structures.12 For example, Yavari et al. demonstrated a 3D graphene foam (GF) network, which represented a significant stride in creating higher dimensionality graphene structures [Fig. 3(a-II)].44 Lin et al. demonstrated patterned LIG-based devices [Fig. 3(a-III)], which allowed for quick fabrication of graphene nanostructures on flexible platforms.43 LIG has since been further developed for specific bioelectronic applications.45
FIG. 3.
Graphene nanostructures. (a) (I) A typical low-magnification transmission electron microscopy (TEM) image of synthesized graphene sheets. Reproduced with permission from Dato et al., Chem. Commun. 40, 6095–6097 (2009). Copyright 2009 The Royal Society of Chemistry.51 (II) Scanning electron micrograph of the microporous graphene foam (GF) structure showing a continuous network of 3D interconnected graphene sheets that comprise the walls of the foam-like structure. Reproduced with permission from Yavari et al., Sci. Rep. 1, 166 (2011). Copyright 2011 Author(s), licensed under a Creative Comments CC-BY-NC-ND License.44 (III) Scanning electron microscopy (SEM) image of the 3D porous LIG film patterned on polyimide (PI) substrates. Reproduced with permission from Lin et al., Nat. Commun. 5, 5714 (2014). Copyright 2014 Nature Publishing Group.43 (b) Truly 3D topology of graphene. (I–II) Representative SEM images of NT-3DFG synthesized for 30 min at 700 and 1100 °C, respectively. The insets represented by red-dashed boxes show out-of-plane graphene on SiNWs. (III) Number of flake edges along a 1 μm length of the nanowire of radius, r, for all synthesis conditions. Results are presented as mean ± SD (n = 3). Reproduced with permission from San Roman et al., ACS Catal. 10(3), 1993–2008 (2020). Copyright 2020 American Chemical Society.49
A barrier to the implementation of graphene nanomaterials is the lack of scalable assembly methods that allow tunable 3D topological arrangements. Graphene nanostructures that can be precisely synthesized for flake size, flake density, and placement hold promise as an approach for next-generation electrode interfaces. Recently, graphene has been shown to grow out-of-plane from growth substrates through plasma-enhanced chemical vapor deposition (PECVD) synthesis processes.46 Garg et al. and San Roman et al. demonstrated the tunability of the graphene flake size and density of 3D fuzzy graphene (3DFG) on Si nanowire templates (NT-3DFG) through varying PECVD conditions such as CH4 partial pressure, synthesis temperature, and synthesis time [Fig. 3(b)].47–50 These unique graphene structures allow complex device interfaces and closer contact with biological systems.
IV. GRAPHENE BIOINTERFACES
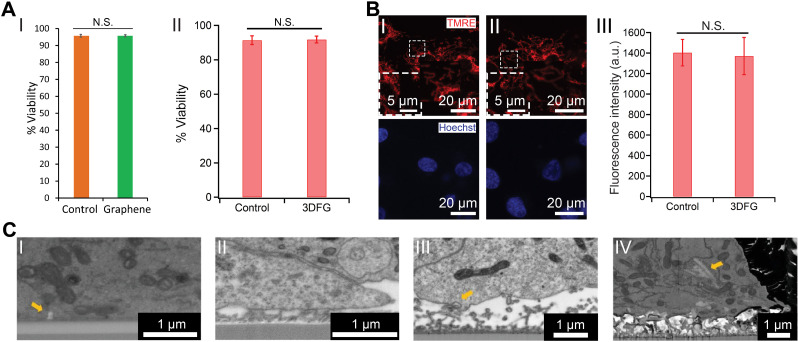
Prior to building interfaces between bioelectronics and biological entities, the biocompatibility of the interfacing materials must be evaluated. Extensive efforts have been focused on using different assays to evaluate biocompatibility from various perspectives.52,53 Rastogi et al. investigated the viability of human embryonic stem cell-derived cardiomyocytes (hESC-CMs) after 10 days in culture on 2D graphene and observed no statistically significant difference between the control and the graphene substrates [Fig. 4(a-I)].54 Beyond 2D planar graphene, Dipalo et al. also investigated the biocompatibility of hESC-CMs on 3D graphene structures, specifically 3DFG, and did not observe any significant decrease in cellular viability after 6 days in culture [Fig. 4(a-II)].55 Furthermore, both 2D and 3D graphene nanostructures did not induce cellular stress in interfaced cells as evaluated through the tetramethylrhodamine ethyl ester (TMRE) assay.55,56 For example, no significant change in the mitochondrial membrane potential (MMP) of cardiomyocytes cultured on 3DFG was detected through the TMRE assay, indicating an absence of cellular stress [Fig. 4(b)].55 However, incubating dispersed graphene flakes with murine microphages (RAW 267.4) for 24 h resulted in detectable changes in the MMP, which indicates that varying the size of the graphene flakes has an impact on cellular health.57 Therefore, graphene-based nanostructure properties (such as geometry, surface charge, and chemical reactivity), the biological platforms (such as cell type), and experimental conditions (such as input dose, incubation time, and incubation temperature) should be evaluated for each system.52,53
FIG. 4.
Graphene biointerfaces. (a) The effect of graphene nanostructures on the viability of cardiomyocytes. (I) Quantification of %Viability of hESC-CMs cultured on glass (orange) and graphene (green) substrates. N.S. denotes no statistically significant difference. Results are presented as mean ± SD (n = 3). Reproduced with permission from Rastogi et al., Cel. Mol. Bioeng. 11, 407–418 (2018). Copyright 2018 Biomedical Engineering Society.54 (II) %Viability of hESC-CMs cultured on Si/SiO2 control and 3DFG substrates. N.S. denotes no statistically significant difference. Results are presented as mean ± SD (n = 4). Reproduced with permission from Dipalo et al., Sci. Adv. 7(15), eabd5175 (2021). Copyright 2021 Author(s), licensed under Creative Commons Attribution-NonCommercial License 4.0.55 (b) TMRE assay performed on hESC-CMs at 6 days in vitro (DIV), cultured on (I) Si/SiO2 control and (II) 3DFG substrates. Red (TMRE) and blue (Hoechst) denote mitochondria and cell nuclei, respectively. (III) Relative fluorescence readout of the TMRE-labeled hESC-CMs cultured on Si/SiO2 control and 3DFG substrates. Results are presented as mean ± SD (n = 4). N.S. denotes no statistically significant difference. Reproduced with permission from Dipalo et al., Sci. Adv. 7(15), eabd5175 (2021). Copyright 2021 Author(s), licensed under Creative Commons Attribution-NonCommercial License 4.0.55 (c) Coupling between graphene nanostructures and electrogenic cells. SEM images (tilt 52°, backscattered electrons) of (I) 2D graphene-cell cross section, arrow identifies membrane invagination; (II) 3DFG-cell cross section; (III) NT-3DFG mesh-cell cross section, arrow identifies membrane wrapping at single wire. (IV) NT-3DFG vertically standing wires-cell cross section, arrow identifies a wire spontaneous penetration. Reproduced with permission from Mation et al., Adv. Mater. Interfaces 7(18), 2000699 (2020). Copyright 2020 Wiley-VCH GmbH.58
Matino et al. utilized graphene, 3DFG, and NT-3DFG platforms for characterizing the cytoskeletal arrangement as well as membrane wrapping processes that promote tight device coupling and spontaneous intracellular penetration [Fig. 4(c)].58 SEM imaging of the cell-nanomaterial interface revealed that the cell membranes had sufficient flexibility to tightly adhere on all out-of-plane graphene materials [Fig. 4(c)], and the authors noted that out-of-plane graphene did not noticeably alter cell viability.58 Further reviews for graphene-based nanomaterials' specific interactions with mammalian cells and tissues have also been completed.59,60
V. GRAPHENE-BASED OUTPUT BIOELECTRONICS
Numerous graphene bioelectronics have been reported for in vitro and in vivo electrophysiological investigations of individual cells and tissues.12 As described earlier, graphene bioelectronics can also be classified either as active or passive devices that leverage the material's unique structural, physical, and chemical properties.37 While active graphene bioelectronics provide high sensitivity and enable n- and p-type recordings from the same device, passive devices allow both electrophysiological investigation and manipulation.4,61
Using single graphene flake FETs, Cohen-Karni et al. recorded extracellular field potentials from spontaneously beating embryonic chicken cardiomyocytes with SNR > 4 [Fig. 5(a)].62 Extracellular field potentials generated during electrical activity induced a change in the conductance of the FET channel, which enabled electrophysiology recordings when measured as a function of time. Hess et al. fabricated graphene-FET (gFET) arrays for simultaneous multiplexed extracellular field potential recordings from electrogenic cells from up to eight gFETs [Fig. 5(b)].61 In order to record extracellular field potentials from organoids and spheroids, Kalmykov et al. demonstrated the fabrication of functional gFET arrays onto a 3D self-rolling platform.33
FIG. 5.
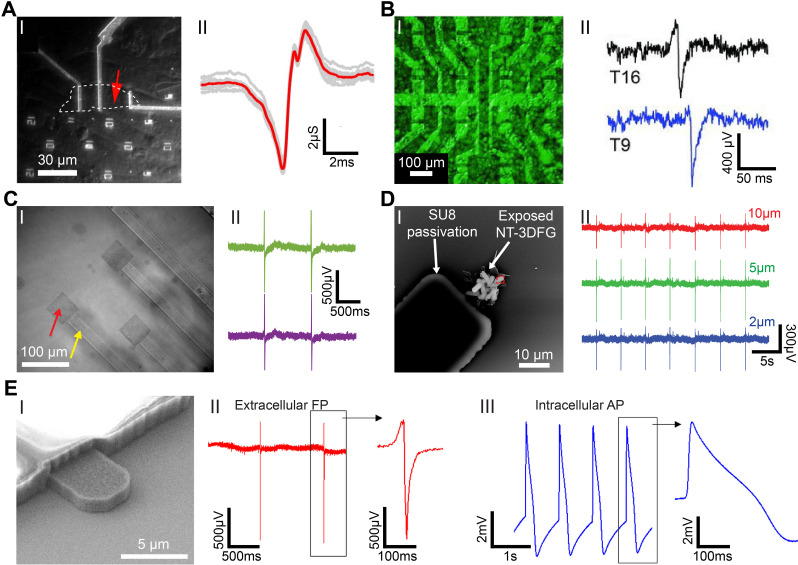
Graphene nanostructures for electrophysiology recordings. (a) Graphene FET (gFET) for electrical recording. (I) Optical microscope image of polydimethylsiloxane (PDMS)/cells interfaced with large flake gFET. Graphene flake outline is marked by white-dashed line; the measured device is marked by red arrow. (II) Recorded averaged peak (red) and raw data (gray traces) for the gFET and cell in (I). Reproduced with permission from Cohen-Karni et al., Nano Lett. 10(3), 1098–1102 (2010). Copyright 2010 American Chemical Society.62 (b) Graphene transistor arrays for recording action potentials from electrogenic cells. (I) Combination of an optical microscopy image of a transistor array and a fluorescence image of the calcein-stained cell layer on the same array. (II) Exemplary single spikes. The current response has been converted to an extracellular voltage signal. The upper spike resembles a capacitive coupling followed by the opening of voltage-gated sodium channels, whereas in the bottom one, the ion channels dominate over the capacitive coupling. Reproduced with permission from Hess et al., Adv. Mater. 23(43), 5045–5049 (2011). Copyright 2011 Wiley-VCH Verlag GmbH.61 (c) Graphene MEAs for electrical and optical measurements of human stem cell-derived cardiomyocytes. (I) Differential interference contrast (DIC) image of graphene MEAs fabricated on glass coverslip. (II) Representative recorded field potential traces using graphene MEAs. Reproduced with permission from Rastogi et al., Cel. Mol. Bioeng. 11, 407–418 (2018). Copyright 2018 Biomedical Engineering Society.54 (d) 3DFG ultra-MEAs for subcellular electrical recordings. (I) SEM image of 10 μm NT-3DFG-MEAs. (II) Representative recorded field potential traces using 10, 5, and 2 μm NT-3DFG ultra-microelectrodes. Reproduced with permission from Rastogi et al., Nano Res. 13, 1444–1452 (2020). Copyright 2020 Springer.67 (e) Intracellular action potential recordings from cardiomyocytes by ultrafast pulsed laser irradiation of fuzzy graphene MEAs. (I) SEM image of 5 μm 3DFG electrodes. (I) Representative extracellular field potential recording of hiPSC-CMs using 3DFG-MEA with 50 μm electrodes (n = 80 electrodes). (III) Representative intracellular action potential recording on 3DFG-MEA with 50-μm electrodes after optoporation (n = 70 electrodes). Reproduced with permission from Dipalo et al., Sci. Adv. 7(15), eabd5175 (2021). Copyright 2021 Author(s), licensed under Creative Commons Attribution-NonCommercial License 4.0.55
Unlike conventional thin-films and other 2D materials, 2D graphene has unique optical properties and exhibits high transparency (up to 97.7%).63 This allows 2D graphene-based electrodes to simultaneously record electrophysiology as well as optically monitor (via fluorescent dyes) intracellular Ca2+ transients; Ca2+ is an important secondary messenger for signal transduction in excitable cells.64,65 Rastogi et al. fabricated transparent graphene MEAs for simultaneous in vitro recordings of extracellular field potentials and corresponding Ca2+ transients in cardiomyocytes [Fig. 5(c)].54 The impedance of the fabricated graphene microelectrodes was determined to be similar to that of Au microelectrodes of similar dimensions.54 Surface treatment of graphene microelectrodes via HNO3 increased charge carriers and led to a reduction in electrode impedance.54 The developed graphene MEAs platform allowed real-time recording of the electrophysiological signals with an SNR of ca. 14 as well as the corresponding Ca2+ activity.54 Minimizing the overall dimensions of the recording electrodes to the ultra-micro scale has enabled recording electrophysiology with subcellular precision.66,67 Although various ultra-microelectrode (UME) platforms have been developed, they show limited long-term stability and relatively high impedances.67 3D graphene nanostructures are a promising alternative as they exhibit high surface-to-volume ratio leading to lower impedance and exhibit long-term stability.48,49,68,69 Rastogi et al. leveraged hierarchical nanomaterials with standard lithography techniques to pattern NT-3DFG UMEs for recording extracellular field potentials from hESC-CMs with electrodes as small as 2 × 2 μm2 [Fig. 5(d)].67 It was demonstrated that the impedance at 1 kHz of 2 × 2 μm2 NT-3DFG UMEs was comparable to 50 × 50 μm2 Au electrodes, facilitating a 625-fold reduction in the footprint of the recording electrodes while maintaining SNR > 6.67
Monitoring intracellular action potentials is critical for in-depth electrophysiological and toxicological investigations. Interfacing nanostructures, such as gold-mushrooms70 and nanowires,71,72 with electrically active cells allows intracellular recordings with high SNR; however, these approaches have limited throughput. Coupling microelectrode platforms with efficient cell-poration techniques provides an alternative approach for high-throughput intracellular recordings. Illumination of porous Pt microelectrodes in water with ultrafast pulsed laser has been shown to lead to the generation of hot carriers that enable localized cellular membrane poration.73 Dipalo et al. leveraged a similar mechanism to record intracellular action potentials using 3DFG microelectrodes [Fig. 5(e)].55 The high surface area of 3DFG allowed recording of extracellular field potentials from the interfaced cardiomyocytes with an SNR of ca. 27 dB [Fig. 5(e-II)].55 Illumination of the cell-3DFG interface with a 1 mW 1064 nm ultrafast pulsed laser leads to successful optoporation and facilitated recordings of intracellular action potentials [Fig. 5(e-II)].55 This technique is minimally invasive and was repeated on the same cell without adverse effects.55
Irrespective of the electrode geometry and recording mechanism, large-scale multiplexing of electrode arrays and downstream electronics is an active challenge.74,75 Innovations in material engineering and processing, fabrication schemes, and hardware designing have allowed the development of high-density electrode arrays in the form of microwires,76 multi-shank silicon probes,21 multiplexed neural threads,77 and intravascular mesh electrodes.78 In the case of conventional gFET platforms, each sensing node requires its independent signal amplifier.79 By integrating gFETs with custom-built front-end amplifiers, Garcia-Cortadella et al. demonstrated frequency-division multiplexing of neural signals.79 Increasing the density of electrodes leads to a direct increase in the data throughput. Advances in data science and machine learning are being coupled with high-density electrode arrays to streamline neural signal processing.75 This includes approaches such as neural data compression and decoding.80,81
Combining graphene's exceptional physical properties and ease of processibility with recent advances in multiplexing via hardware designing and signal processing will allow investigation of both extra- and intracellular electrophysiology with high spatiotemporal resolution.
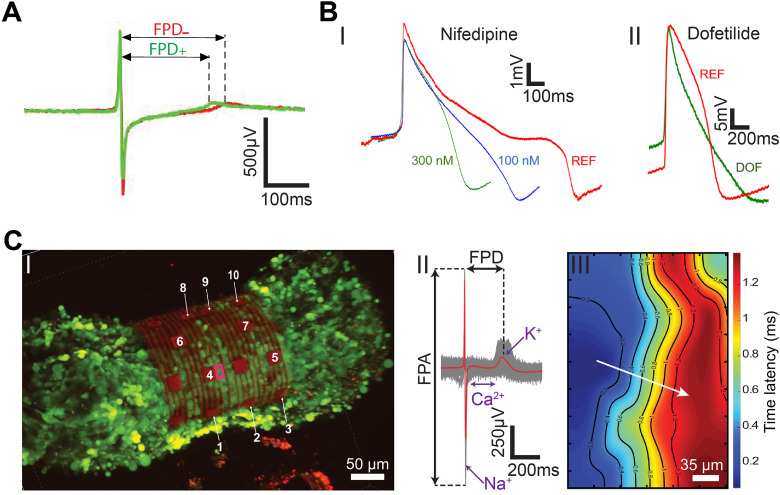
The effect of therapeutics on the physiology and the electrophysiology of cells and tissues is of great interest to pharmacological studies and drug development.82,83 Single graphene microelectrodes can be used to study the effect of therapeutics on 2D cell cultures.54,55 Using isoproterenol as a proof-of-concept drug, Rastogi et al. demonstrated that single-layer graphene microelectrodes can successfully record an increase in the beat frequency and decrease in the field potential duration (FPD) of cardiomyocytes [Fig. 6(a)].54 Concurrent Ca2+ imaging corroborated the electrophysiological recordings.54 Dipalo et al. investigated the effect of Nifedipine and Dofetilide (DOF) on the intracellular electrical activity of cardiomyocytes [Fig. 6(b)].55 Nifedipine, a Ca2+ blocker, reduced the duration of cardiac action potentials (APD90, action potential duration at 90% amplitude) from 915 ± 90 ms to 535 ± 97 ms and 332 ± 75 ms for 100 and 300 nM Nifedipine, respectively [Fig. 6(b-I)].55 On the other hand, Dofetilide, a class III antiarrhythmic agent, was observed to prolong the repolarization phase leading to an increase in APD90 from 586 ± 67 ms to 741 ± 101 ms [Fig. 6(b-II)].55 In addition to electrical recordings, graphene-based microelectrodes have also been employed for neurotransmitter sensing. Neurotransmitters such as dopamine play a crucial role in neurological disorders such as schizophrenia84 and Parkinson's disease.85,86 Castagnola et al. leveraged the high surface-area of 3DFG to demonstrate fast scan cyclic voltammetry-based sensing of dopamine.68 3DFG microelectrodes exhibited exceptional sensitivity (2.12 ± 0.05 nA/nM) and selectivity to dopamine with a 50 × 50 μm2 geometric footprint. Successful dopamine detection with high sensitivity and selectivity was retained after further miniaturization of the 3DFG microelectrodes to 2 × 2 μm2.68 Graphene-based bioelectronics, therefore, present a high-performance platform for an in-depth electrophysiological investigation that can be utilized for pharmacological drug testing and neurotransmitter sensing.
FIG. 6.
Electrophysiological investigations and mapping. (a) The effect of β-adrenergic receptor agonist on the extracellular electrophysiology using graphene MEAs. Averaged trace (70 peaks) before (red, −) and after (green, +) the application of β-adrenergic receptor agonist, isoproterenol. Reproduced with permission from Rastogi et al., Cel. Mol. Bioeng. 11, 407–418 (2018). Copyright 2018 Biomedical Engineering Society.54 (b) Intracellular electrophysiology investigations of the effect drugs on human-derived cardiomyocytes. (I) Representative cardiac action potentials before and after the administration of nifedipine at various concentrations. (II) Representative cardiac action potentials before and after the administration of 100 nM dofetilide (DOF). REF, reference signal in physiological conditions. Reproduced with permission from Dipalo et al., Sci. Adv. 7(15), eabd5175 (2021). Copyright 2021 Author(s), licensed under Creative Commons Attribution-NonCommercial License 4.0.55 (c) Mapping electrical signal propagation in 3D using the 3D-SR-BA. (I) A 3D confocal microscopy image of 3D cardiac spheroid labeled with Ca2+ indicator dye (Fluo-4, green fluorescence) encapsulated by the 3D-SR-BA. (II) Averaged field potential peak (red trace) and raw data (gray traces, n = 100 peaks recorded by channel 4). (III) 2D representation of the isochronal map of time latencies. White arrow represents average conduction velocity direction. Reproduced with permission from Kalmykov et al., Sci. Adv. 5(8), eaax0729 (2020). Copyright 2019 Author(s), licensed under a Creative Commons Attribution-NonCommercial License 4.0.33
3D cell cultures closely recapitulate in vivo systems.34,87,88 Their electrophysiological mapping is used for disease modeling and therapeutic designing.34 Kalmykov et al. fabricated a polymeric 3D-SR-BA for the electrophysiological investigation of human embryonic stem cell-derived cardiomyocyte spheroids [Fig. 6(c)].33 The 3D-SR-BA platform conformally wraps an interfaced cardiac spheroid for continuous and stable multiplexed electrophysiology recordings coupled with concurrent Ca2+ imaging.33 The high spatiotemporal resolution of 3D-SR-BA allows individual ionic currents across the cell membrane, field potential duration (FPD), and the field potential amplitude (FPA) to be detected at a single sensor level [Fig. 6(c-II)].33 Furthermore, the conduction velocity of 3D spheroids can be calculated from the 3D isochronal maps of the electrical signal propagation across the surface of the interfaced spheroid [Fig. 6(c-III)].33 3D-SR-BA has also been used to study the firings rates of individual units in 3D cortical spheroids and investigate the effect of drugs such as glutamate on neuronal action potentials.34 Park et al. designed multifunctional 3D frameworks for measuring the burst activity across the surface of cortical spheroids and assembloids.35 Engineering graphene nanostructures onto these 3D platforms will allow extra- and intracellular electrical activity recordings with high SNR.
VI. GRAPHENE-BASED INPUT BIOELECTRONICS
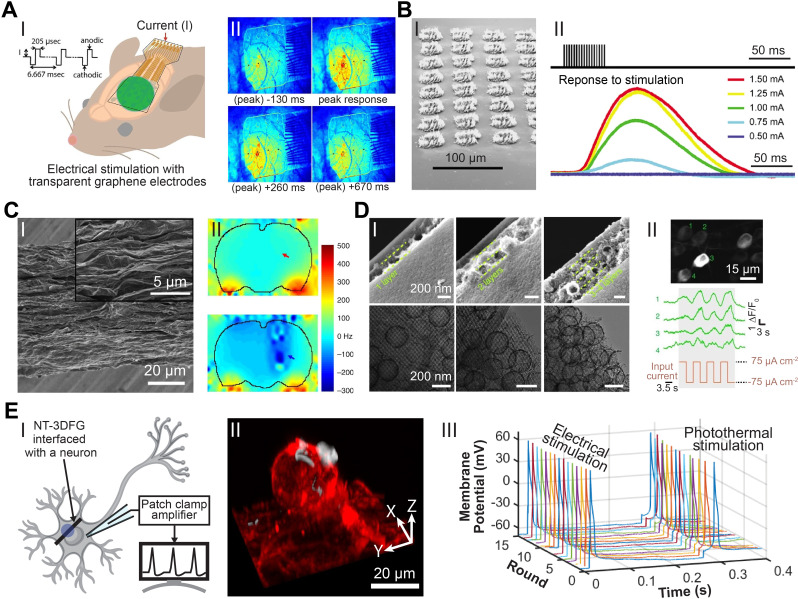
Modulating cellular electrophysiology and behavior has broadened the understanding of brain activity as well as contributed tools and therapeutic interventions for neurological disorders.4,12,89 Specifically, electrical stimulation is a promising approach for the treatment of pathologies, such as spinal cord injury and muscle atrophy.12,89 The charge storage capacitance and optical transparency of 2D graphene bioelectronics allow simultaneous electrical stimulation and imaging as demonstrated by Park et al. using graphene micro-electrocorticography (micro-ECoG) electrodes in transgenic mice [Fig. 7(a-I)].90 The graphene micro-ECoG electrodes enabled temporal mapping of fluorescence indicators, demonstrating the spatiotemporal propagation of action potentials in response to stimulating current pulses [Fig. 7(a-II)].90 Modulation of myogenesis and cell morphology in SHSY5Y human neuroblastoma cells have also been achieved through electrical stimulation via graphene electrodes.91
FIG. 7.
Graphene and carbon nanostructures for stimulation. (a) Electrical neural stimulation transparent graphene electrode arrays implanted in GCaMP6f mice. (I) Demonstration of micro-electrocorticography (micro-ECoG) implantation over sensorimotor cortex and electrical stimulation in GCaMP6f mice. (II) Visualization of the intensity of neural response to 100 μA electrical stimulation at times −130 to +670 ms of peak response with a graphene electrode array. Reproduced with permission from Park et al., ACS Nano 12(1), 148–157 (2018). Copyright 2017 American Chemical Society.90 (b) Flexible neural electrodes array based-on porous graphene for cortical stimulation. (I) Tilt SEM image of a 64-spot porous graphene array. (II) Stimulus evoking current (representing movement) response of the flex sensor in arbitrary units. Reproduced with permission from Lu et al., Sci. Rep. 6, 33526 (2016). Copyright 2016 Author(s), licensed under Creative Commons Attribution 4.0 License.69 (c) Simultaneous deep brain stimulation and fMRI with graphene fiber electrodes. (I) A representative SEM image of the axial external surface of a GF fiber. Inset, magnified image of the region in the dashed box. (II) B0 distortion maps observed in rats implanted with a GF (upper) and PtIr (lower) bipolar electrodes (electrodes are pointed by arrows). Reproduced with permission from Zhao et al., Nat. Commun. 11, 1788 (2020). Copyright 2020 Author(s), licensed under Creative Commons Attribution 4.0 License.92 (d) Micelle-enabled self-assembly of porous and monolithic carbon membranes for bioelectronic interfaces. (I) Cross-sectional view (upper panels) and associated top view (lower panels) of the hierarchical porous material. The hierarchical structures display two components: a bottom layer constructed from an ordered mesoporous structure and layers of porous vesicles assembled into multiple layers (as separated by the dashed lines). (II) Top: a retinal calcium image showing activated retinal ganglion cells (RGCs) upon the stimulation. Middle: representative calcium traces from individual RGCs (numbered in the upper image). Bottom: the input current density during the stimulation. Reproduced with permission from Fang et al., Nat. Nanotechnol. 16, 206–213 (2021). Copyright 2020 Author(s), licensed under Springer Nature Ltd.93 (e) Remote nongenetic optical modulation of neuronal activity using NT-3DFG. (I) Schematic illustrating an NT-3DFG interfaced with a neuron for photothermal stimulation. Purple spot indicates laser illumination area. (II) A 3D reconstruction of a fluorescence images of a representative DRG neuron labeled with plasma membrane stain (red, CellMask plasma membrane stain) and interfaced with NT-3DFG (white). (III) Representative recorded membrane potential of a repetitively stimulated DRG neuron (DRG neuron was patch-clamped in whole-cell configuration and current clamp mode). Electrical stimulation was performed by injecting a pulse of 100 nA for 1 ms. Photothermal stimulation was performed with 405-nm laser with a pulse of 2.28 mW power and 0.6-ms pulse duration (1.37 μJ). Reproduced with permission from Rastogi et al., Proc. Natl. Acad. Sci. U. S. A. 117(24), 13339–13349 (2020). Copyright 2020 Author(s), licensed under a creative Commons Attribution 4.0 License.94
However, materials with 2D topographies typically exhibit limited charge storage and injection capacities.12 Engineered graphene structures with 3D topography and high surface area can overcome these limitations and display enhanced electrical stimulation efficiencies.69 Lu et al. designed flexible electrode arrays based on laser-induced porous graphene with a high charge injection capacity (CIC) of 3.1 mC/cm2 [Fig. 7(b-I)].69 Electrical stimulation of the motor cortex in mice using these arrays evoked transient ankle and knee flexion in the contralateral leg [Fig. 7(b-II)].69 Zhao et al. showed that graphene fibers microelectrodes exhibit stable CIC up to 10.1 mC/cm2 for 19 days [Fig. 7(c-I)].92 Deep brain stimulation (DBS) with these electrodes alleviated symptoms of Parkinsonian motor deficits in a rat. Moreover, the presence of these electrodes did not result in detectable field distortions during functional magnetic resonance imaging (fMRI) [Fig. 7(c-II)].92 Macroscopic input bioelectronics for modulating electrical activity of tissues and organs have been realized via 3D carbon structures. Fang et al. fabricated monolithic carbon membranes with hierarchical porous surface topology by carbonizing self-assembled Resol and Pluronic F127 micelles [Fig. 7(d-I)].93 These structures were used to modulate electrical activity across multiple biological scales ranging from individual cells (primary cardiomyocytes) to tissue (mice retinal tissue) and organ (isolated rat heart) [Fig. 7(d-II)].
Optical modulation offers a minimally invasive alternative for conventional electrical stimulation.94,95 Rastogi et al. leveraged NT-3DFG's efficient photothermal energy conversion for optical stimulation of dorsal root ganglion (DRG) neurons with subcellular precision [Fig. 7(e-I)].94 Instead of being internalized, NT-3DFG adhere to the cell plasma membrane after co-incubation with DRG neurons [Fig. 7(e-II)].94 The rapid rise in local temperature induced by laser pulses incident at the NT-3DFG and neuron interface generates depolarizing currents across the cell membrane resulting in action potentials.94,96 Photothermal stimulation of neurons with sub-millisecond laser pulses is highly reproducible and does not cause damage to the cell membrane [Fig. 7(e-III)].94
VII. CHALLENGES AND PROSPECTS
Graphene-based bioelectronics is promising technology for clinical applications; however, there are certain challenges that still need to be overcome. Chemical instability, which affects device performance and longevity, is a common concern for active components of bioelectronic platforms. While graphene-based electrodes exhibit a wide water stability window,68,97 reactive sp2 defects, edges, and grain boundaries alter chemical stability.98 For biological applications, biofouling of electrode interfaces is a major concern that may be overcome through the addition of antifouling coatings.99,100
Due to graphene's high tensile strength and modulus, the mechanical integrity of graphene-based platforms often hinges on the quality of adhesion to metal interconnects and the integration of the insulating components. Robust bonds between devices and substrates can help prevent delamination or loss of graphene interfaces. Nanostructures directly produced or patterned onto source substrates can translate into industry-scale micro-fabrication techniques.67,93,101 The varied synthesis strategies for graphene-based nanomaterials allow for numerous processing pathways involving suspensions,102 composites,103 or thin films,104 which will continue to yield novel bioelectronic device architectures.
The ease of chemical functionalization of graphene-based nanostructures through covalent-bonded organic functionalities, π–π stacked functional groups, and dispersed metal/metal oxide nanoparticles provides routes for a highly flexible application-driven platform.105 Nanoparticles such as Pt-Fe3S4 have been utilized for in situ electrochemical NO generation for neuronal modulation;106 laser patterned SiC nanostructures have been employed for in situ H2O2 generation to regulate muscle contraction.107 Combining electrocatalytic and energy conversion principles with appropriate functionalization of graphene biointerfaces will lead to the development of multimodal systems for electrophysiology- and biomolecule-based sensing and actuation. Integration of sensing and actuation into a unified high-density multifunctional platform will allow closed-loop control over physiology and electrophysiology.
VIII. CONCLUSION
Here, we highlight recent advances in graphene nanostructures for electrophysiology manipulation of cells and tissues. Novel I/O bioelectronics coupling engineered materials and biology are aimed at overcoming current challenges in areas such as device miniaturization, long-term stability, and increased complexity in device architecture.8,12 Several approaches demonstrate the ability to engineer physiochemical properties of graphene and integrate graphene nanostructures with advanced backend microelectronics.55,67,68 These microelectronic platforms achieve high-density arrays for extra- and intracellular electrophysiological studies with high spatiotemporal resolution and multiplexed recording capabilities.54,55,67 While some challenges remain, graphene nanostructures promise to propel electrophysiological research capabilities and clinical translation. Multidisciplinary strategies across materials science, micro-fabrication, catalysis, and bioengineering are pivotal for incremental enhancements in multifunctional I/O bioelectronics. These improvements will yield greater scientific knowledge in human electrophysiology and benefit patient health and society for generations to come.
ACKNOWLEDGMENTS
T.C.-K. acknowledges funding support from the National Science Foundation (Award No. CBET1552833), the Defense Advanced Research Projects Agency through Cooperative Agreement D20AC00002 awarded by the U.S. Department of the Interior (DOI), Interior Business Center, and the National Institute of Health (Award No. R21EB029164). The content of the information does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred.
AUTHOR DECLARATIONS
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
R.G., D.S.R., and Y.W. contributed equally to this work.
DATA AVAILABILITY
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- 1. Hodgkin A. L. and Huxley A. F., Nature 144(3651), 710–711 (1939). 10.1038/144710a0 [DOI] [Google Scholar]
- 2. Hodgkin A. L. and Huxley A. F., J. Physiol. 117(4), 500–544 (1952). 10.1113/jphysiol.1952.sp004764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cole K. S. and Curtis H. J., J. Gen. Physiol. 22(5), 649–670 (1939). 10.1085/jgp.22.5.649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rastogi S. K., Kalmykov A., Johnson N., and Cohen-Karni T., J. Mater. Chem. B 6(44), 7159–7178 (2018). 10.1039/C8TB01600C [DOI] [PubMed] [Google Scholar]
- 5. Calabresi P., Mercuri N. B., Sancesario G., and Bernardi G., Brain 116, 433–452 (1993). [PubMed] [Google Scholar]
- 6. Josephson M. E., Clinical Cardiac Electrophysiology: Techniques and Interpretations ( Lippincott Williams & Wilkins, 2008). [Google Scholar]
- 7. Galvani L., De Viribus Electricitatis in Motu Musculari Commentarius ( Tip. Istituto delle Scienze, Bologna, 1791). [Google Scholar]
- 8. Hong G. and Lieber C. M., Nat. Rev. Neurosci. 20(6), 330–345 (2019). 10.1038/s41583-019-0140-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krack P., Batir A., Van Blercom N., Chabardes S., Fraix V., Ardouin C., Koudsie A., Limousin P. D., Benazzouz A., and LeBas J. F., N. Engl. J. Med. 349(20), 1925–1934 (2003). 10.1056/NEJMoa035275 [DOI] [PubMed] [Google Scholar]
- 10. Zimetbaum P. and Goldman A., Circulation 122(16), 1629–1636 (2010). 10.1161/CIRCULATIONAHA.109.925610 [DOI] [PubMed] [Google Scholar]
- 11. Spira M. E. and Hai A., Nat. Nanotechnol. 8(2), 83 (2013). 10.1038/nnano.2012.265 [DOI] [PubMed] [Google Scholar]
- 12. San Roman D., Garg R., and Cohen-Karni T., APL Mater. 8(10), 100906 (2020). 10.1063/5.0020455 [DOI] [Google Scholar]
- 13. Rivnay J., Wang H., Fenno L., Deisseroth K., and Malliaras G. G., Sci. Adv. 3(6), e1601649 (2017). 10.1126/sciadv.1601649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Canales A., Jia X., Froriep U. P., Koppes R. A., Tringides C. M., Selvidge J., Lu C., Hou C., Wei L., Fink Y., and Anikeeva P., Nat. Biotechnol. 33(3), 277–284 (2015). 10.1038/nbt.3093 [DOI] [PubMed] [Google Scholar]
- 15. Sakmann B. and Neher E., Annu. Rev. Physiol. 46(1), 455–472 (1984). 10.1146/annurev.ph.46.030184.002323 [DOI] [PubMed] [Google Scholar]
- 16. Verkhratsky A., Krishtal O. A., and Petersen O. H., Pflügers Arch. 453(3), 233–247 (2006). 10.1007/s00424-006-0169-z [DOI] [PubMed] [Google Scholar]
- 17. Hutzler M., Lambacher A., Eversmann B., Jenkner M., Thewes R., and Fromherz P., J. Neurophysiol. 96(3), 1638–1645 (2006). 10.1152/jn.00347.2006 [DOI] [PubMed] [Google Scholar]
- 18. Miccoli B., Lopez C. M., Goikoetxea E., Putzeys J., Sekeri M., Krylychkina O., Chang S.-W., Firrincieli A., Andrei A., and Reumers V., Front. Neurosci. 13, 641 (2019). 10.3389/fnins.2019.00641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hochberg L. R., Serruya M. D., Friehs G. M., Mukand J. A., Saleh M., Caplan A. H., Branner A., Chen D., Penn R. D., and Donoghue J. P., Nature 442(7099), 164–171 (2006). 10.1038/nature04970 [DOI] [PubMed] [Google Scholar]
- 20. Huys R., Braeken D., Jans D., Stassen A., Collaert N., Wouters J., Loo J., Severi S., Vleugels F., and Callewaert G., Lab Chip 12(7), 1274–1280 (2012). 10.1039/c2lc21037a [DOI] [PubMed] [Google Scholar]
- 21. Steinmetz N. A., Aydin C., Lebedeva A., Okun M., Pachitariu M., Bauza M., Beau M., Bhagat J., Böhm C., and Broux M., Science 372(6539), eabf4588 (2021). 10.1126/science.abf4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sahasrabuddhe K., Khan A. A., Singh A. P., Stern T. M., Ng Y., Tadić A., Orel P., LaReau C., Pouzzner D., Nishimura K., Boergens K. M., Shivakumar S., Hopper M. S., Kerr B., Hanna M.-E. S., Edgington R. J., McNamara I., Fell D., Gao P., Babaie-Fishani A., Veijalainen S., Klekachev A. V., Stuckey A. M., Luyssaert B., Kozai T. D. Y., Xie C., Gilja V., Dierickx B., Kong Y., Straka M., Sohal H. S., and Angle M. R., J. Neural Eng. 18(1), 015002 (2021). 10.1088/1741-2552/abd0ce [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang A. and Lieber C. M., Chem. Rev. 116(1), 215–257 (2016). 10.1021/acs.chemrev.5b00608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Song E., Li J., Won S. M., Bai W., and Rogers J. A., Nat. Mater. 19(6), 590–603 (2020). 10.1038/s41563-020-0679-7 [DOI] [PubMed] [Google Scholar]
- 25. Hubel D. H., Science 125(3247), 549–550 (1957). 10.1126/science.125.3247.549 [DOI] [PubMed] [Google Scholar]
- 26. Frank K. and Fuortes M. G., J. Physiol. 130(3), 625–654 (1955). 10.1113/jphysiol.1955.sp005432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Normann R. A., Maynard E. M., Rousche P. J., and Warren D. J., Vision Res. 39(15), 2577–2587 (1999). 10.1016/S0042-6989(99)00040-1 [DOI] [PubMed] [Google Scholar]
- 28. Abidian M. R. and Martin D. C., Adv. Funct. Mater. 19(4), 573–585 (2009). 10.1002/adfm.200801473 [DOI] [Google Scholar]
- 29. Choi J.-r., Kim S.-M., Ryu R.-H., Kim S.-P., and Sohn J.-w., Exp. Neurobiol. 27(6), 453 (2018). 10.5607/en.2018.27.6.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Viventi J., Kim D.-H., Vigeland L., Frechette E. S., Blanco J. A., Kim Y.-S., Avrin A. E., Tiruvadi V. R., Hwang S.-W., Vanleer A. C., Wulsin D. F., Kathryn D., Gelber C. E., Palmer L., Van der Spiegel J., Wu J., Xiao J., Huang Y., Contreras D., Rogers J. A., and Litt B., Nat. Neurosci. 14(12), 1599–1605 (2011). 10.1038/nn.2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wei X., Luan L., Zhao Z., Li X., Zhu H., Potnis O., and Xie C., Adv. Sci. 5(6), 1700625 (2018). 10.1002/advs.201700625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu J., Fu T.-M., Cheng Z., Hong G., Zhou T., Jin L., Duvvuri M., Jiang Z., Kruskal P., Xie C., Suo Z., Fang Y., and Lieber C. M., Nat. Nanotechnol. 10, 629–636 (2015). 10.1038/nnano.2015.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kalmykov A., Huang C., Bliley J., Shiwarski D., Tashman J., Abdullah A., Rastogi S. K., Shukla S., Mataev E., and Feinberg A. W., Sci. Adv. 5(8), eaax0729 (2019). 10.1126/sciadv.aax0729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kalmykov A., Reddy J. W., Bedoyan E., Wang Y., Garg R., Rastogi S. K., Cohen-Karni D., Chamanzar M., and Cohen-Karni T., J. Neural Eng. 18(5), 055005 (2021). 10.1088/1741-2552/abf290 [DOI] [PubMed] [Google Scholar]
- 35. Park Y., Franz C. K., Ryu H., Luan H., Cotton K. Y., Kim J. U., Chung T. S., Zhao S., Vazquez-Guardado A., Li K., Avila R., Phillips J. K., Quezada M. J., Jang H., Soo K. S., Min W. S., Kyeongha K., Jeong H., Bandokar A. Y., Memgdi H., Zhao H., Osher G. R., Wang H., Lee K., Zhang Y., Huang Y., Finan J. D., and Rogers J. A., Sci. Adv. 7(12), eabf9153 (2021). 10.1126/sciadv.abf9153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pennacchio F., Garma L., Matino L., and Santoro F., J. Mater. Chem. B 6(44), 7096–7101 (2018). 10.1039/C8TB01737A [DOI] [PubMed] [Google Scholar]
- 37. Tiwari A., Graphene Bioelectronics ( Elsevier, 2017). [Google Scholar]
- 38. Novoselov K. S., Geim A. K., Morozov S. V., Jiang D., Zhang Y., Dubonos S. V., Grigorieva I. V., and Firsov A. A., Science 306(5696), 666–669 (2004). 10.1126/science.1102896 [DOI] [PubMed] [Google Scholar]
- 39. Lee C., Wei X., Kysar J. W., and Hone J., Science 321(5887), 385–388 (2008). 10.1126/science.1157996 [DOI] [PubMed] [Google Scholar]
- 40. Stoller M. D., Park S., Zhu Y., An J., and Ruoff R. S., Nano Lett. 8(10), 3498–3502 (2008). 10.1021/nl802558y [DOI] [PubMed] [Google Scholar]
- 41. Geim A. K., Science 324(5934), 1530–1534 (2009). 10.1126/science.1158877 [DOI] [PubMed] [Google Scholar]
- 42. Zhang Y., Zhang L., and Zhou C., Acc. Chem. Res. 46(10), 2329–2339 (2013). 10.1021/ar300203n [DOI] [PubMed] [Google Scholar]
- 43. Lin J., Peng Z., Liu Y., Ruiz-Zepeda F., Ye R., Samuel E. L., Yacaman M. J., Yakobson B. I., and Tour J. M., Nat. Commun. 5(1), 5714 (2014). 10.1038/ncomms6714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yavari F., Chen Z., Thomas A. V., Ren W., Cheng H.-M., and Koratkar N., Sci. Rep. 1(1), 166 (2011). 10.1038/srep00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wan Z., Nguyen N.-T., Gao Y., and Li Q., Sustainable Mater. Technol. 25, e00205 (2020). 10.1016/j.susmat.2020.e00205 [DOI] [Google Scholar]
- 46. Tsounis C., Lu X., Bedford N. M., Subhash B., Thomsen L., Zhang Q., Ma Z., Ostrikov K., Bendavid A., and Scott J. A., ACS Nano 14(9), 11327–11340 (2020). 10.1021/acsnano.0c03380 [DOI] [PubMed] [Google Scholar]
- 47. Garg R., Gopalan D. P., de la Barrera S. C., Hafiz H., Nuhfer N. T., Viswanathan V., Hunt B. M., and Cohen-Karni T., Nano Lett. 19(8), 5335–5339 (2019). 10.1021/acs.nanolett.9b01790 [DOI] [PubMed] [Google Scholar]
- 48. Garg R., Rastogi S. K., Lamparski M., de la Barrera S. C., Pace G. T., Nuhfer N. T., Hunt B. M., Meunier V., and Cohen-Karni T., ACS Nano 11(6), 6301–6311 (2017). 10.1021/acsnano.7b02612 [DOI] [PubMed] [Google Scholar]
- 49. San Roman D., Krishnamurthy D., Garg R., Hafiz H., Lamparski M., Nuhfer N. T., Meunier V., Viswanathan V., and Cohen-Karni T., ACS Catal. 10(3), 1993–2008 (2020). 10.1021/acscatal.9b03919 [DOI] [Google Scholar]
- 50. Garg R., San Roman D., and Cohen-Karni T., Pure Appl. Chem. 92(12), 1929–1936 (2020). 10.1515/pac-2020-0801 [DOI] [Google Scholar]
- 51. Dato A., Lee Z., Jeon K.-J., Erni R., Radmilovic V., Richardson T. J., and Frenklach M., Chem. Commun. 2009, 6095–6097. 10.1039/b911395a [DOI] [PubMed] [Google Scholar]
- 52. Rahmati M. and Mozafari M., Front. Bioeng. Biotechnol. 7, 4 (2019). 10.3389/fbioe.2019.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Syama S. and Mohanan P., Int. J. Biol. Macromol. 86, 546–555 (2016). 10.1016/j.ijbiomac.2016.01.116 [DOI] [PubMed] [Google Scholar]
- 54. Rastogi S. K., Bliley J., Shiwarski D. J., Raghavan G., Feinberg A. W., and Cohen-Karni T., Cell. Mol. Bioeng. 11(5), 407–418 (2018). 10.1007/s12195-018-0525-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dipalo M., Rastogi S. K., Matino L., Garg R., Bliley J., Iachetta G., Melle G., Shrestha R., Shen S., and Santoro F., Sci. Adv. 7(15), eabd5175 (2021). 10.1126/sciadv.abd5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rastogi S. K., Raghavan G., Yang G., and Cohen-Karni T., Nano Lett. 17(5), 3297–3301 (2017). 10.1021/acs.nanolett.7b01215 [DOI] [PubMed] [Google Scholar]
- 57. Li Y., Liu Y., Fu Y., Wei T., Guyader L. L., Gao G., Liu R.-S., Chang Y.-Z., and Chen C., Biomaterials 33(2), 402–411 (2012). 10.1016/j.biomaterials.2011.09.091 [DOI] [PubMed] [Google Scholar]
- 58. Matino L., Rastogi S. K., Garma L. D., Cohen-Karni T., and Santoro F., Adv. Mater. Interfaces 7, 2000699 (2020). 10.1002/admi.202000699 [DOI] [Google Scholar]
- 59. Tu Z., Guday G., Adeli M., and Haag R., Adv. Mater. 30(33), 1706709 (2018). 10.1002/adma.201706709 [DOI] [PubMed] [Google Scholar]
- 60. Henriques P. C., Pereira A. T., Pires A. L., Pereira A. M., Magalhães F. D., and Gonçalves I. C., ACS Appl. Mater. Interfaces 12(18), 21020–21035 (2020). 10.1021/acsami.9b21841 [DOI] [PubMed] [Google Scholar]
- 61. Hess L. H., Jansen M., Maybeck V., Hauf M. V., Seifert M., Stutzmann M., Sharp I. D., Offenhäusser A., and Garrido J. A., Adv. Mater. 23(43), 5045–5049 (2011). 10.1002/adma.201102990 [DOI] [PubMed] [Google Scholar]
- 62. Cohen-Karni T., Qing Q., Li Q., Fang Y., and Lieber C. M., Nano Lett. 10(3), 1098–1102 (2010). 10.1021/nl1002608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nair R. R., Blake P., Grigorenko A. N., Novoselov K. S., Booth T. J., Stauber T., Peres N. M., and Geim A. K., Science 320(5881), 1308–1308 (2008). 10.1126/science.1156965 [DOI] [PubMed] [Google Scholar]
- 64. Grienberger C. and Konnerth A., Neuron 73(5), 862–885 (2012). 10.1016/j.neuron.2012.02.011 [DOI] [PubMed] [Google Scholar]
- 65. Wang Y., Garg R., Hartung J. E., Goad A., Patel D. A., Vitale F., Gold M. S., Gogotsi Y., and Cohen-Karni T., ACS Nano 15(9), 14662–14671 (2021). 10.1021/acsnano.1c04431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kozai T. D. Y., Langhals N. B., Patel P. R., Deng X., Zhang H., Smith K. L., Lahann J., Kotov N. A., and Kipke D. R., Nat. Mater. 11(12), 1065–1073 (2012). 10.1038/nmat3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rastogi S. K., Bliley J., Matino L., Garg R., Santoro F., Feinberg A. W., and Cohen-Karni T., Nano Res. 13, 1444–1452 (2020). 10.1007/s12274-020-2695-y [DOI] [Google Scholar]
- 68. Castagnola E., Garg R., Rastogi S. K., Cohen-Karni T., and Cui X. T., Biosens. Bioelectron. 191, 113440 (2021). 10.1016/j.bios.2021.113440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lu Y., Lyu H., Richardson A. G., Lucas T. H., and Kuzum D., Sci. Rep. 6(1), 33526 (2016). 10.1038/srep33526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fendyur A., Mazurski N., Shappir J., and Spira M. E., Front. Neuroeng. 4, 14 (2011). 10.3389/fneng.2011.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tian B., Cohen-Karni T., Qing Q., Duan X., Xie P., and Lieber C. M., Science 329(5993), 830–834 (2010). 10.1126/science.1192033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Robinson J. T., Jorgolli M., Shalek A. K., Yoon M.-H., Gertner R. S., and Park H., Nat. Nanotechnol. 7(3), 180–184 (2012). 10.1038/nnano.2011.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dipalo M., Melle G., Lovato L., Jacassi A., Santoro F., Caprettini V., Schirato A., Alabastri A., Garoli D., and Bruno G., Nat. Nanotechnol. 13(10), 965–971 (2018). 10.1038/s41565-018-0222-z [DOI] [PubMed] [Google Scholar]
- 74. Nurmikko A., Neuron 108(2), 259–269 (2020). 10.1016/j.neuron.2020.10.015 [DOI] [PubMed] [Google Scholar]
- 75. Rapeaux A. B. and Constandinou T. G., Curr. Opin. Biotechnol. 72, 102–111 (2021). 10.1016/j.copbio.2021.10.001 [DOI] [PubMed] [Google Scholar]
- 76. Obaid A., Hanna M.-E., Wu Y.-W., Kollo M., Racz R., Angle M. R., Müller J., Brackbill N., Wray W., Franke F., Chichilnisky E. J., Hierlemann A., Ding J. B., Schaefer A. T., and Melosh N. A., Sci. Adv. 6(12), eaay2789 (2020). 10.1126/sciadv.aay2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Musk E., J. Med. Internet Res. 21(10), e16194 (2019). 10.2196/16194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Oxley T. J., Yoo P. E., Rind G. S., Ronayne S. M., Lee C. S., Bird C., Hampshire V., Sharma R. P., Morokoff A., and Williams D. L., J. Neurointerventional Surg. 13(2), 102–108 (2021). 10.1136/neurintsurg-2020-016862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Garcia-Cortadella R., Schafer N., Cisneros-Fernandez J., Ré L., Illa X., Schwesig G., Moya A., Santiago S., Guirado G., and Villa R., Nano Lett. 20(5), 3528–3537 (2020). 10.1021/acs.nanolett.0c00467 [DOI] [PubMed] [Google Scholar]
- 80. Wu T., Zhao W., Keefer E., and Yang Z., J. Neural Eng. 15(6), 066019 (2018). 10.1088/1741-2552/aae18d [DOI] [PubMed] [Google Scholar]
- 81. Tam W.-K., Wu T., Zhao Q., Keefer E., and Yang Z., BMC Biomed. Eng. 1(1), 22 (2019). 10.1186/s42490-019-0022-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yu F., Hunziker W., and Choudhury D., Micromachines 10(3), 165 (2019). 10.3390/mi10030165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhang B., Korolj A., Lai B. F. L., and Radisic M., Nat. Rev. Mater. 3(8), 257–278 (2018). 10.1038/s41578-018-0034-7 [DOI] [Google Scholar]
- 84. Howes O. D. and Kapur S., Schizophr. Bull. 35(3), 549–562 (2009). 10.1093/schbul/sbp006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lotharius J. and Brundin P., Nat. Rev. Neurosci. 3(12), 932–942 (2002). 10.1038/nrn983 [DOI] [PubMed] [Google Scholar]
- 86. Höglinger G. U., Rizk P., Muriel M. P., Duyckaerts C., Oertel W. H., Caille I., and Hirsch E. C., Nat. Neurosci. 7(7), 726–735 (2004). 10.1038/nn1265 [DOI] [PubMed] [Google Scholar]
- 87. Takahashi T., Annu. Rev. Pharmacol. Toxicol. 59, 447–462 (2019). 10.1146/annurev-pharmtox-010818-021108 [DOI] [PubMed] [Google Scholar]
- 88. Dutta D., Heo I., and Clevers H., Trends Mol. Med. 23(5), 393–410 (2017). 10.1016/j.molmed.2017.02.007 [DOI] [PubMed] [Google Scholar]
- 89. Hejazi M., Tong W., Ibbotson M. R., Prawer S., and Garrett D. J., Front. Neurosci. 15, 403 (2021). 10.3389/fnins.2021.658703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Park D.-W., Ness J. P., Brodnick S. K., Esquibel C., Novello J., Atry F., Baek D.-H., Kim H., Bong J., and Swanson K. I., ACS Nano 12(1), 148–157 (2018). 10.1021/acsnano.7b04321 [DOI] [PubMed] [Google Scholar]
- 91. Heo C., Yoo J., Lee S., Jo A., Jung S., Yoo H., Lee Y. H., and Suh M., Biomaterials 32(1), 19–27 (2011). 10.1016/j.biomaterials.2010.08.095 [DOI] [PubMed] [Google Scholar]
- 92. Zhao S., Li G., Tong C., Chen W., Wang P., Dai J., Fu X., Xu Z., Liu X., and Lu L., Nat. Commun. 11(1), 1788 (2020). 10.1038/s41467-020-15570-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Fang Y., Prominski A., Rotenberg M. Y., Meng L., Acarón Ledesma H., Lv Y., Yue J., Schaumann E., Jeong J., Yamamoto N., Jiang Y., Elbaz B., Wei W., and Tian B., Nat. Nanotechnol. 16(2), 206–213 (2021). 10.1038/s41565-020-00805-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Rastogi S. K., Garg R., Scopelliti M. G., Pinto B. I., Hartung J. E., Kim S., Murphey C. G., Johnson N., San Roman D., Bezanilla F., Cahoon J. F., Gold M. S., Chamanzar M., and Cohen-Karni T., Proc. Natl. Acad. Sci. 117(24), 13339–13349 (2020). 10.1073/pnas.1919921117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Jiang Y., Parameswaran R., Li X., Carvalho-de-Souza J. L., Gao X., Meng L., Bezanilla F., Shepherd G. M., and Tian B., Nat. Protoc. 14(5), 1339–1376 (2019). 10.1038/s41596-019-0135-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Carvalho-de-Souza J. L., Pinto B. I., Pepperberg D. R., and Bezanilla F., Biophys. J. 114(2), 283–288 (2018). 10.1016/j.bpj.2017.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Murastov G., Bogatova E., Brazovskiy K., Amin I., Lipovka A., Dogadina E., Cherepnyov A., Ananyeva A., Plotnikov E., and Ryabov V., Biosens. Bioelectron. 166, 112426 (2020). 10.1016/j.bios.2020.112426 [DOI] [PubMed] [Google Scholar]
- 98. Bellunato A., Arjmandi-Tash H., Cesa Y., and Schneider G. F., Chem. Phys. Chem. 17(6), 785–801 (2016). 10.1002/cphc.201500926 [DOI] [PubMed] [Google Scholar]
- 99. Sabaté del Río J., Henry O. Y. F., Jolly P., and Ingber D. E., Nat. Nanotechnol. 14(12), 1143–1149 (2019). 10.1038/s41565-019-0566-z [DOI] [PubMed] [Google Scholar]
- 100. Zhang T., Xu H., Xu Z., Gu Y., Yan X., Liu H., Lu N., Zhang S., Zhang Z., and Yang M., Microchim. Acta 186(4), 240 (2019). 10.1007/s00604-019-3343-7 [DOI] [PubMed] [Google Scholar]
- 101. Gao L., Ni G.-X., Liu Y., Liu B., Castro Neto A. H., and Loh K. P., Nature 505(7482), 190–194 (2014). 10.1038/nature12763 [DOI] [PubMed] [Google Scholar]
- 102. Johnson D. W., Dobson B. P., and Coleman K. S., Curr. Opin. Colloid Interface Sci. 20(5), 367–382 (2015). 10.1016/j.cocis.2015.11.004 [DOI] [Google Scholar]
- 103. Hossain M. F., Heo J. S., Nelson J., and Kim I., Information 10(10), 325 (2019). 10.3390/info10100325 [DOI] [Google Scholar]
- 104. Bae S., Kim H., Lee Y., Xu X., Park J.-S., Zheng Y., Balakrishnan J., Lei T., Ri Kim H., Song Y. I., Kim Y.-J., Kim K. S., Özyilmaz B., Ahn J.-H., Hong B. H., and Iijima S., Nat. Nanotechnol. 5(8), 574–578 (2010). 10.1038/nnano.2010.132 [DOI] [PubMed] [Google Scholar]
- 105. Georgakilas V., Otyepka M., Bourlinos A. B., Chandra V., Kim N., Kemp K. C., Hobza P., Zboril R., and Kim K. S., Chem. Rev. 112(11), 6156–6214 (2012). 10.1021/cr3000412 [DOI] [PubMed] [Google Scholar]
- 106. Park J., Jin K., Sahasrabudhe A., Chiang P.-H., Maalouf J. H., Koehler F., Rosenfeld D., Rao S., Tanaka T., Khudiyev T., Schiffer Z. J., Fink Y., Yizhar O., Manthiram K., and Anikeeva P., Nat. Nanotechnol. 15(8), 690–697 (2020). 10.1038/s41565-020-0701-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Nair V., Yi J., Isheim D., Rotenberg M., Meng L., Shi F., Chen X., Gao X., Prominski A., Jiang Y., Yue J., Gallagher C. T., Seidman D. N., and Tian B., Sci. Adv. 6(34), eaaz2743 (2020). 10.1126/sciadv.aaz2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.