Abstract
Systemic sclerosis (SSc) is a complex multisystem disease with the highest case-specific mortality among all autoimmune rheumatic diseases, yet without any available curative therapy. Therefore, the development of novel therapeutic antifibrotic strategies that effectively decrease skin and organ fibrosis is needed. Existing animal models are cost-intensive, laborious and do not recapitulate the full spectrum of the disease and thus commonly fail to predict human efficacy. Advanced in vitro models, which closely mimic critical aspects of the pathology, have emerged as valuable platforms to investigate novel pharmaceutical therapies for the treatment of SSc. This review focuses on recent advancements in the development of SSc in vitro models, sheds light onto biological (e.g., growth factors, cytokines, coculture systems), biochemical (e.g., hypoxia, reactive oxygen species) and biophysical (e.g., stiffness, topography, dimensionality) cues that have been utilized for the in vitro recapitulation of the SSc microenvironment, and highlights future perspectives for effective drug discovery and validation.
Keywords: 3D in vitro models, animal models, fibrosis, in vitro microenvironment, scleroderma, tissue engineering
1. Introduction
Systemic sclerosis (SSc; also termed scleroderma) is an autoimmune disease that is characterized by the distinctive pathogenetic triad of microvascular dysfunction, dysregulation of innate and adaptive immunity, and generalized fibrosis in multiple organs.[1] SSc has the highest case-specific mortality of any of the autoimmune rheumatic diseases, with more than half of patients dying as a direct consequence of the pathology.[2] For this reason, SSc is a disease with a high unmet clinical need. Although intensive research in the last years has improved the understanding of the disease, only one drug, nintedanib, has thus far been approved by the Food and Drug Administration (FDA) for the treatment of SSc-associated interstitial lung disease.[3] Furthermore, there are no generally accepted therapies for skin and organ fibrosis, which are known to be key manifestations of SSc, leaving the need for novel antifibrotic therapeutic strategies in SSc.[4] Although significant strides have been achieved using various animal models, these systems are expensive for the purposes of routine drug development/screening studies and have limited replicability due to different physiology and genetics in comparison to the human disease.[5] These shortcomings of animal models impose the need for standardize protocols to increase reproducibility[6] and development of more reliable, clinically relevant in silico and in vitro models.[7]
In vitro cell-based studies have been proven to be valuable tools in drug discovery programs, especially due to their low cost and high speed of testing compounds. Cell cultures represents an immense value in the investigation of cellular and functional aspects of disease processes for improved therapeutic interventions.[8] The most commonly utilized model systems are based on conventional 2D monolayer cultures exposed to exogenous profibrotic stimuli, commonly transforming growth factor-β (TGF-β).[9] However, customarily used cell-based models frequently fail to give predictable and reliable data for in vivo responses. A critical component of this failure results from the lack of recapitulating the native in vivo microenvironment. In the SSc scenario, the histopathological and physicochemical cues of the disease microenvironment are critical for the stimulation of biological functions mediated by cell signaling.[10] Multiple cytokines, chemokines, and signal transduction pathways are implicated in the progression of SSc as well as structural features of the extracellular matrix (ECM) such as stiffness, viscoelasticity, and topography. Moreover, although 2D culture systems are simple and economical, they do not consider the spatial organization of cells within the 3D architecture of organs and do not replicate native cell–cell and cell–ECM interactions and signaling pathways.[11] Recent advancements in 3D cell culture technologies and tissue engineering strategies have made it possible to engineer advanced physiologically relevant 3D in vitro models not only to study disease mechanism and progression, but also to use as a platform to design new therapeutic compounds and to screen for drug efficacy and safety.[12]
This review summarizes the utility and limitations of various animal models of SSc and focuses on the most recent advances in in vitro SSc models, highlighting the crucial role of biological, biochemical, and biophysical cues in mimicking SSc microenvironment. The potential of bioengineered tissues as in vitro models to investigate molecular and cellular mechanisms involved in the onset and progression of systemic sclerosis and/or to serve as screening platforms to test novel pharmaceutical therapies for the treatment of the disease, will be discussed. In addition, we will shed light to the next challenges and future directions that must be addressed toward an effective 3D in vitro model for SSc.
2. Mechanisms and Pathophysiology of Systemic Sclerosis (SSc)
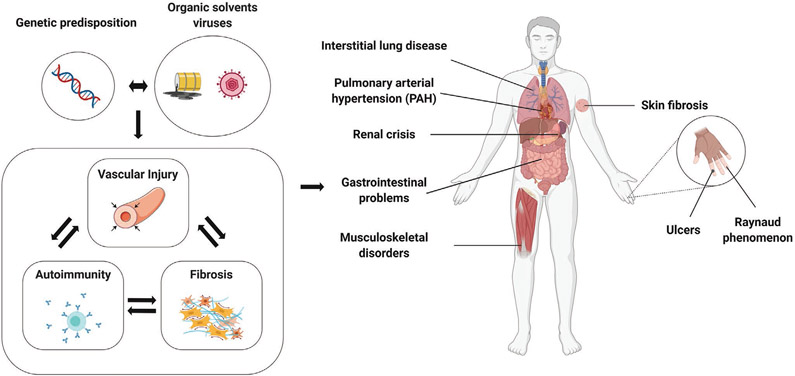
Systemic sclerosis is a complex chronic and often progressive disease characterized by vascular alterations, inflammation and autoimmunity, and multisystemic excessive fibrosis (Figure 1). Although skin fibrosis is the distinguishing hallmark, the pathological involvement of the viscera including the lungs, gastrointestinal tract, kidneys and heart determines the clinical outcome.[13] Patients are characterized by subtypes based on the extent of skin involvement, with limited cutaneous systemic sclerosis (lcSSc) and diffuse cutaneous systemic sclerosis (dcSSc) subsets delineated on the basis of distal or proximal skin involvement.[14] The pathogenesis of systemic sclerosis is complex and remains elusive. An interplay between genetic factors and environmental events, such as job-related exposures to silica dust, vinyl chloride and organic solvents or viruses and other infectious agents, is likely to play a part in the origin of the disease.[15] The onset of vascular injury in SSc includes endothelial activation and vascular damage, thickening of the vessel wall due to intimal and smooth muscle cell proliferation, and finally vessel narrowing and obliteration, which lead to tissue ischemia, oxidative stress and ultimately organ dysfunction.[16] Infiltration of inflammatory cell within the lesions is common in patients with early-stage disease, and inflammatory and immune cells are an important source of profibrotic mediators such as TGF-β, platelet-derived growth factor (PDGF), interleukin 1 (IL-1) and interleukin 6 (IL-6). Figure 2 depicts these multiple pathologic processes.
Figure 1.
Pathophysiology of SSc. Genetic and environmental factors trigger the onset of SSc. SSc is characterized by vascular alterations, inflammation and autoimmunity, and multisystemic excessive fibrosis, which ultimately lead to severe and life-threatening organ complications. Created with BioRender.com.
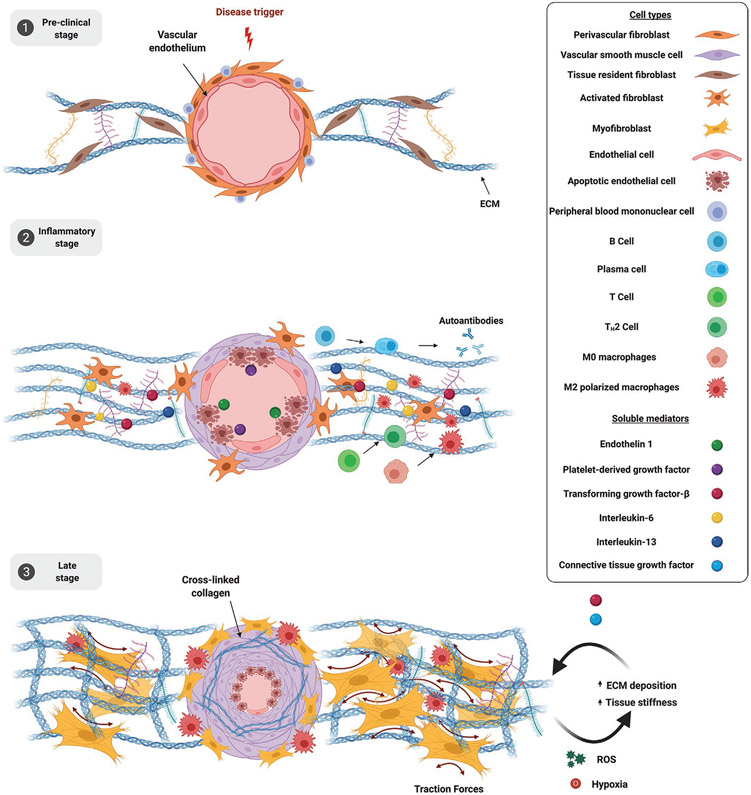
Figure 2.
Molecular mechanisms of SSc. 1) Preclinical stage. Vascular injury is the earliest event in SSc which leads to endothelial cell activation and entrapment of peripheral blood mononuclear cells. 2) Inflammatory stage. Progressive vascular damage causes endothelial cell apoptosis, which in turn secrete ET-1 and PDGF that stimulate smooth muscle cell proliferation, leading to luminal narrowing, and inflammatory cells recruitment. Plasma cells secrete autoantibodies (anti-Scl-70, anticentromere, anti-RNA-polymerase III) and IL-6. Type 2 T helper (TH2) cells secrete TGF-β and IL-13. Polarized M2 macrophages secrete TGF-β. These soluble mediators contribute to fibroblasts activation and increase ECM deposition. 3) Late stage. Progressive endothelial cells apoptosis, smooth muscle cells proliferation and vessel narrowing lead to tissue hypoxia and oxidative stress which contribute to the maintenance of fibrosis. Fibroblasts undergo complete myofibroblasts differentiation and increase ECM deposition leading to mechanical stress and perpetuating the fibrotic process. M2 polarized macrophages infiltration further increases TGF-β secretion. Created with BioRender.com.
Dysregulation of both innate and adaptive immunity is also a prominent factor that contributes to systemic sclerosis pathogenesis. Antinuclear antibodies are present in up to 95% of SSc patients and specific autoantibodies, such as antitopoisomerase 1, anticentromere, and anti-RNA polymerase III antibodies, directed against intracellular nuclear components, are present in over 75% of patients.[17] Besides the presence of autoantibodies, evidence of dysregulated immune responses are represented by inflammatory cells and inflammatory molecules in target tissues such as the skin and lungs and a prominent type I interferon (IFN) signature in circulating and tissue-infiltrating immune cells.[18]
The pathologic hallmark of SSc is extensive fibrosis involving multiple organs, which can lead to significant organ failure.[19] Fibrosis is characterized by replacement of normal tissue architecture with rigid and mechanically stressed connective tissue rich in collagen and other ECM macromolecules, such as elastin, glycosaminoglycan, and fibronectin.[20] The abnormal accumulation of ECM results from increased synthesis by activated fibroblasts, enhanced assembly and deposition catalyzed by prolyl and lysyl-oxidase and transglutaminase 2 and aberrant ECM degradation.[21]
The differentiation of fibroblasts into myofibroblasts is a critical step in the onset of fibrosis. Myofibroblasts are specialized fibroblasts that acquire characteristics of smooth muscle cells, including the expression of α-SMA. In contrast to physiological wound healing, where myofibroblasts are present only transiently within granulation tissue before undergoing apoptosis, myofibroblasts in SSc are persistent. These contractile cells secrete not only matrix proteins, but also TGF-β and other profibrotic components, and thus further promote ECM deposition and remodeling.[22] In addition to activation and proliferation of resident fibroblasts, other sources of activated fibroblasts include recruitment of circulating fibrocytes and the differentiation from epithelial cells.[23] Epithelial cells have been demonstrated to trans differentiate into fibroblasts and myofibroblasts, undergoing an epithelial to mesenchymal transition (EMT) in response to TGF-β and other growth factors and/or cytokines during the development of fibrosis.[24] In addition to epithelial cells, endothelial cells have also been shown to transdifferentiate into fibroblasts through endothelial to mesenchymal transition (EndMT).[25] Other sources of fibroblasts include trans differentiation of pericytes and adipocytes.[26] Scientific advances have considerably augmented the understanding of the pathophysiological mechanism of SSc. The antifibrotic drugs nintedanib and pirfenidone have been approved for the treatment of patients with idiopathic pulmonary fibrosis, and nintedanib recently received approval for SSc-ILD, but there are still a dearth of effective anti-fibrotic agents for the full array of SSc manifestations.[27] There is therefore an urgent unmet need to develop new anti-fibrotic therapies for use in SSc.
3. Animal Models of SSc
Animals models have been extensively used to study the complex mechanisms involved in the pathogenesis of SSc and ultimately to bring new insight for the development of therapeutic strategies.
Recent years have seen a plethora of genetic, transgenics and induced animal models that have contributed to our knowledge of the initiating events of systemic sclerosis (Table 1).[28]
Table 1.
Representative animal models of SSc.
| Classification | Model | Pathological features | Major limitations | Refs. |
|---|---|---|---|---|
| Genetic | Tight skin 1 (TSK-1) mouse model | Skin fibrosis, antitopoisomerase-1 autoantibodies | Dermal sclerosis is lacking, no sign of vasculopathy, absence of inflammation | [214] |
| Tight skin 2 (TSK-2) mouse model | Skin fibrosis, mononuclear cell infiltration in the dermis and adipose tissues, antinuclear antibody | No sign of vasculopathy | [215] | |
| University of California at Davis line 200 (UCD-200) chickens | Skin and organ fibrosis, perivascular lymphocytic infiltration, endothelial injury, antinuclear antibodies | Avian background limits molecular studies, high cost | [216] | |
| Transgenic | Fos-related antigen-2 (Fra-2) mouse model | Skin fibrosis, microangiopathy, disease course similar to human SSc | Poor characterization, absence of autoimmunity | [217] |
| Endothelin-1 mouse model | Glomerulosclerosis and interstitial fibrosis | Absence of autoimmunity | [218] | |
| TGF-β receptor I transgenic mouse model | Collagen accumulation in dermis and pulmonary vessels, epidermal thinning, loss of adipose tissues in subcutis | Absence of autoimmune and inflammatory aspects | [219] | |
| Friend leukemia integration factor-1 (Fli-1+/−) / Krüppel-like factor 5 (KLF5+/−) | Fibrosis and vasculopathy of the skin and lung, B-cell activation and autoantibody production | Poor characterization, mild inflammation | [220] | |
| Knockout | Caveolin-1 KO mouse model | Skin and lung fibrosis, vascular disease | Mild inflammation and absence of autoimmunity | [221] |
| Fli-1 KO mouse | Vascular disease, skin fibrosis | Poor characterization, absence of autoimmunity | [222] | |
| Induced | Bleomycin mouse model of fibrosis (intradermal injection) | Dermal or pulmonary fibrosis, antinuclear antibodies, inflammatory response | Fibrosis is limited to the injection site and not systemic, vascular phenomena are usually absent | [36] |
| Bleomycin rat model of fibrosis (intratracheal injection) | Strong pulmonary fibrosis, antinuclear antibodies, inflammatory response | Fibrosis is limited to the lung site and not systemic, vascular phenomena are usually absent | [37] | |
| Hypochlorous mouse model (HOCl) | Dermal and pulmonary fibrosis, antitopoisomerase antibodies, | Poor characterization | [223] | |
| Sclerodermatous graft-versus-host disease (GVHD) mouse model | Dermal fibrosis and pulmonary, presence of inflammatory infiltrate | Sophisticated technical skills required | [224] |
Genetic animal models spontaneously develop mutations to the genome with manifestations similar to those of SSc. One of the best-characterized genetic animal models of SSc are tight skin-1 (Tsk-1) mice, in which a tandem duplication in the gene for fibrillin 1 (Fbn1), a mediator of elastic fibers assembly, is responsible for the pathogenic phenotype. In heterozygous mice, this mutation leads to thickening of the subcutaneous tissue (hypodermis) and endothelial cell apoptosis. Fibrosis in these mice develops from excessive production of ECM by activated fibroblasts upon activation of the TGF-β pathway.[29] A related genetic model of SSc is tight skin 2 (Tsk-2) mouse, which presents mutations in the gene for type III collagen alpha.[30] Tsk-2 mice demonstrate increased type I and III collagen, which lead to abnormal ECM deposition, and an inflammatory dermal mononuclear cell infiltrate.[31]
Transgenic mouse models with the pathological cascade of SSc have been established to further understand the process of the disease. The transgenic mouse model overexpressing the Fos-related antigen-2 (FRA-2) gene showed many of the important factors resulting in the vascular damage and progressive skin and lung fibrosis of systemic sclerosis as well as pulmonary hypertension.[32] A TNF-transgenic model was recently shown to develop spontaneous and severe pulmonary hypertension and have genomic overlap with SSc-PAH but to lack systemic fibrosis.[33] Another model is represented by urokinase-type plasminogen activator receptor (uPAR)-deficient mice. Urokinase-type plasminogen activator receptor is a glycosylphosphatidylinositol-anchored cell surface receptor which concentrates its ligand, urokinase-type plasminogen activator (uPA), at the cell–matrix interface. The uPA/uPAR complex promotes the fibrinolysis and the degradation of other ECM, serving as a key regulator of ECM homeostasis and angiogenesis. uPAR deficient mice reproduce the fibrotic and vascular features of SSc, such as increase collagen content and perivascular inflammatory cells infiltration in skin and lungs.[34]
Inducible animal models are quicker and easier to evaluate than genetic models and offer valuable clues to study the role of selected target molecules in the developmental process of SSc. The bleomycin model of fibrosis is probably the most utilized model of SSc. Bleomycin was originally isolated from the fungus streptomyces verticillus, and is often used as an anti-tumor medication for the treatment of various kinds of malignancy. Bleomycin hydrolase inactivates bleomycin by hydrolyzing the amide bond in the β-aminoalanineamide moiety. Bleomycin-induced toxicity occurs predominantly in the lungs and the skin, due to the deficiency of the enzyme in these organs.[35] For this reason, the bleomycin mouse model of fibrosis has been frequently used to replicate common features of SSc such as dermal or pulmonary fibrosis. Local dermal injections of bleomycin in mice induced collagen synthesis at the injection site over 4 weeks. The overall effects were found to be systemic because the lung similarly showed increased collagen synthesis.[36] Bleomycin can also be delivered via the intratracheal route, resulting in severe pulmonary fibrosis.[37] Although the bleomycin model replicates critical aspects observed in SSc, this model lacks the typical autoantibody patterns present in the pathology and bleomycin induced fibrosis was found to be strain specific.[38] Another inducible animal models of SSc is represented by the hypochlorous mouse model (HOCl). Repeated intradermal injections of hypochlorous acid generates hydroxyl radicals, which lead to enhanced synthesis of collagen in the lung and skin tissues. In addition, this model mimics the pathological damages observed in the systemic sclerosis kidneys and induces antitopoisomerase antibodies.[39] The mechanism of action of hypochlorous acid-induced fibrosis is not fully understood, thus restricting its commonality of use. Another model is represented by the angiotensin II-inducible model of fibrosis. Angiotensin II (Ang II) is a vasoactive peptide that induces vascular constriction, water and salt retention, and high blood pressure.[40] Subcutaneous injection of Ang II
induced both inflammation and fibrosis in the skin by accumulating activated fibroblasts and promoting EndMT of circulating blood cells. However, it is not known whether these animal models developed autoantibodies specific for systemic sclerosis.[41]
These animal models offer essential clues for the improved knowledge of the molecular mechanisms of SSc pathology and the identification of potential therapeutic targets for the treatment of this disease.
As explained above, none of the currently available models encompasses all aspects of SSc in humans. Therefore, multiple models should be utilized when studying drug efficacy to account for deficiencies and limitations of single models, resulting however in high costs while not guaranteeing clinical translatability. In addition, although animal models predict biological relevant pharmacokinetic responses to drug administration, their different physiology and genetics from humans hamper the exact recapitulation of the human diseases.[28a] There have been numerous drugs which have been successful in animal models which have not performed well in clinical trials given the complexity of SSc and imperfection of each of the models.[42] The high number of animals required during preclinical studies remain an ethical issue, besides being cost-intensive and laborious.[43] Thus, it is becoming imperative the need to develop more predictable in vitro models that can mimic aspects of human in vivo cellular behavior.
4. SSc: the Need for Advanced In Vitro Models
In the last few years, progress in the understanding of the pathogenesis of SSc energized the design of numerous promising clinical studies. Several recent reviews have summarized therapies for SSc that are currently in clinical trials and shed light on novel potential therapeutic targets for the management of the disease.[44a,44b,4,44c,44d] For example, tocilizumab, a humanized monoclonal antibody against the human IL-6 receptor-α, has shown encouraging results by improving both skin and lung fibrosis[45] and has reached phase III clinical trials. Another novel promising therapy is seen in trials of the endocannabinoid receptor type 2 agonist lenabasum. This synthetic molecule has emerged as a potent modulator both of skin and lung inflammation have antifibrotic potential as well.[46] Nevertheless, despite the positive signs of clinical response in subsets of patients, these two clinical trials have been unsuccessful in meeting their endpoints and failed to gain regulatory approval.[47,46] Furthermore, considering the emerging of new potential therapeutics, along with the repurposing of existing drugs, clinical trials in SSc are more active than ever. However, the limited numbers of patients available for trials poses the need to refine pre-clinical research in order to select the optimal drug candidates with the best chance of clinical success.
In light of the limitations associated with animal models, cell systems and in vitro tissue equivalents represents precious tools to investigate the disease’s molecular pathways and to generate a platform for drug screening for early-phase studies.
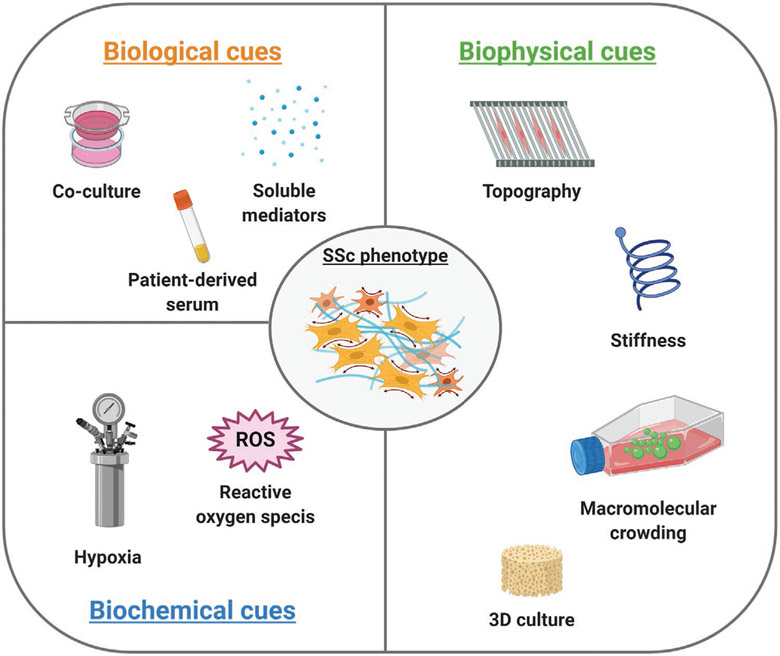
In vitro cell-based models are an important element of the drug discovery process. In contrast to cost-intensive animal models, assays using cultured cells are simple, fast, and cost-effective as well as versatile and easily reproducible.[48] The efficacy of an in vitro model is determined by its capacity to closely replicate relevant characteristics of the in vivo microenvironment.[49] Different approaches can be utilized to develop in vitro models that recapitulate the SSc phenotype. One approach comprises the use of cells isolated from healthy donors, which are converted into a disease-specific phenotype by the addition of profibrotic modulators during the culture time to induce the expression and secretion of fibrotic markers and increase the deposition of ECM. Nevertheless, the use of exogenous stimuli does not result in a disease-activating mechanism. Due to short culture periods that do not model disease progression, cells do not acquire the full disease-specific patterns of gene expression and are fundamentally limited in representing the complexity of the disease.[50] For this reason, cells derived from SSc patients have become one of the most important materials in the study of the pathology. For example, fibroblasts derived from SSc skin lesions have been demonstrated to secrete an abnormal amount of ECM proteins (collagens, fibronectin) and fibrogenic modulators (TGF-β, CTGF) and fibrotic markers (α-SMA) in vitro.[51] Despite the tremendous utility of patient-derived cells, in vitro studies are limited by challenges including availability of patient donor cells, particularly in a rare disease such as SSc. In vitro expansion of scleroderma fibroblasts has been associated with loss of the SSc phenotype over time in culture, showing a marked decrease in collagen production in fibroblasts cultured for up to ten passages,[52] and a reduction of the disease transcriptional signature after four passages.[53] Moreover, patient-derived cells showed high heterogeneity with regard to inflammatory as well as fibrotic signatures, which can be lost during cell proliferation into any daughter lineage, leading to cell pools that do not recapitulate the variety of cells in vivo.[54] Therefore, especially given the complexity of SSc, it is imperative to achieve a system that allows for spatiotemporal control over the biological, biochemical, and biophysical cues of the in vitro extracellular microenvironment to properly mimic the pathological condition of SSc (Figure 3).
Figure 3.
Overview of biological, biochemical, and biophysical cues used in vitro to recapitulate the SSc microenvironment. Created with BioRender.com.
5. Biological Cues
5.1. Growth Factors Supplementation
A multitude of soluble growth factors are implicated in systemic sclerosis (Table 2) and TGF-β is commonly recognized as the master regulator of fibrosis.[55] TGF-β belongs to a superfamily of proteins that includes bone morphogenetic proteins (BMPs), growth differentiation factors (GDFs) activins, inhibins, myostatin, nodal and anti-Mullerian hormone (AMH) proteins.[56] There are three isoforms of TGF-β (TGF-β1, TGF-β2, and TGF-β3), which contain highly conserved regions but diverge in several amino acid regions. The three TGF-β isoforms function through the same receptor heterodimers, TGF-β receptor type 1(TGFR-1) and TGF-β receptor type 2 (TGFR2) and activate the same canonical mothers against decapentaplegic homologue (SMAD)-2–SMAD3 signaling pathway.[57] In this review we refer solely to the TGF-β family, and in particular to the TGF-β1 isoform, unless otherwise stated.
Table 2.
Influence of growth factors on SSc phenotype and drug testing (α-SMA, α-smooth muscle actin; CTGF, connective tissue growth factor; ECM, extracellular matrix; EMT, epithelial-mesenchymal transition; EndMT, endothelial-mesenchymal transition; ET-1, endothelin 1; FGF, fibroblast growth factor; FGFR, fibroblast growth factor receptor; IGF, insulin-like growth factor 1; IGF1R, insulin-like growth factor 1 receptor; JNK, c-Jun N-terminal kinase; MCP-1, monocyte chemoattractant protein-1; MMP-1, matrix metalloproteinase-1; PDGF, platelet-derived growth factor; PI3K, phosphoinositide 3-kinases; TGF-β, transforming growth factor-β; TKI, tyrosine kinase; VE, vascular endothelial; VEGF, vascular endothelial growth factor; ZO-1, zonula occludens-1 protein.
| Soluble mediator | Cell type | Experimental output | Therapeutics | Refs. |
|---|---|---|---|---|
| TGF-βa) | Primary human dermal fibroblasts | TGF-β2 increased collagen I deposition | Histone deacetylase inhibitor trichostatin A (TSA) inhibited the TGF-β-induced collagen I synthesis by suppressing Sp1 activity in skin fibroblasts | [225] |
| Human dermal fibroblast cell line (BJ) | TGF-β1 (25 ng mL−1) increased the expression of on collagen I, MMP-1 and CTGF and induced expression of α-SMA protein | S-adenosyl-l-homocysteine hydrolase (SAHH) inhibitor (DZ2002) reversed the stimulatory effect of TGF-β on collagen I, MMP-1 and CTGF expression | [226] | |
| Human epidermal keratinocytes | TGF-β (10 ng mL−1) induced EMT down-regulating E-cadherin and ZO-1 and upregulating vimentin and fibronectin | Smad inhibitor (SB431542) reversed established EMT | [227] | |
| Microvascular endothelial cells (MVECs) | TGF-β (10 ng mL−1) induced EndMT synergistically with SSc-derived fibroblasts, reducing tube formation ability, CD31 and VEGF-A expression, and upregulating mesenchymal markers such as α-SMA, collagen I, and TGF-β | ET-1 dual receptor antagonists bosentan (BOS) and macitentan (MAC) reduced the expression of mesenchymal markers and restored CD31 expression and tube formation ability | [228] | |
| SSc-derived human dermal microvascular endothelial cells | TGF-β (10 ng mL−1) induced EndMT upregulating pro-collagen I and a-SMA and downregulating CD31 and VE-cadherin | – | [99] | |
| CTGF | Human WI-38 lung fibroblasts cell | CTGF (10 ng mL−1) showed augmented α-SMA levels | Inhibitors of Src-family kinases (SFK) and PI3K, blocked CTGF-dependent α-SMA upregulation | [229] |
| SSc-derived human dermal and lung fibroblasts | CTGF (40 ng mL−1) stimulated fibroblastic cell proliferation and increased collagen I and fibronectin deposition | – | [230] | |
| PDGF | SSc-derived human dermal fibroblasts | PDGF (40 ng mL−1) stimulated the synthesis of collagen I | Dasatinib and nilotib (inhibitors of abl kinases and PDGF receptors) reduced the mRNA and protein levels of ECM proteins | [231] |
| SSc-derived human dermal fibroblasts | PDGF (10 ng mL−1 or 40 ng mL−1) stimulate the expression of MCP-1, which is associated with the presence of inflammatory infiltrates in SSc skin lesions | Antibodies blocking MCP-1 decreased the chemotactic activity of SSc fibroblasts | [232] | |
| SSc-derived human dermal fibroblasts | PDGF-AA (10 ng mL−1) or PDGF-BB (10 ng mL−1) increased cell proliferation and ECM deposition | Crenolanib (inhibitor of PDGF receptor signaling) inhibited cell proliferation and migration | [233] | |
| SSc-derived human dermal fibroblasts | PDGF-BB (40 ng mL−1) stimulated the synthesis of collagen I | Imatinib (tyrosine kinase inhibitor) blocked the stimulatory effects of PDGF in a dose-dependent manner | [234] | |
| IGF | SSc-derived lung fibroblasts | IGF-II (100 ng mL−1 or 200 ng mL−1) induced collagen I and fibronectin deposition, without affecting fibroblasts proliferation | Inhibitors of PI3 kinase and JNK were able to block IGF-II-induced collagen and fibronectin production | [235] |
| Healthy and SSc-derived lung fibroblasts | IGF-II (200 ng mL−1) stimulated gene expression of fibronectin, collagen and α-SMA and decreased the expression of IGF-receptors and insulin receptor in SSc fibroblasts | Tyrphostin AG 538, a specific inhibitor of the IGF1R TKI, reduced intracellular collagen and fibronectin | [236] | |
| FGF9 | Human dermal fibroblasts | FGF9 (10 ng mL−1) promoted the expression of collagen type I and α-SMA and the formation of stress fibers, as markers of fibroblast-to myofibroblast differentiation, by activating FGFR3 signaling | Treatment with the selective FGFR3 inhibitor PD173074 or knockdown of FGFR3 by siRNA abrogated the stimulatory effects of FGF9 on FGFR3 profibrotic target genes, and blocked FGF9-induced fibroblast activation | [237] |
| VEGF | SSc-derived human dermal fibroblasts | VEGF (10 or 20 ng mL−1) showed direct profibrotic effects by upregulating procollagen I and III expression | – | [238] |
References serve as representative examples.
The bioavailability of TGF-β is regulated by its secretion from macrophages and other cells as an inactive precursor, which is then converted to its biologically active matrix-bound latent form via integrin-mediated processes.[58] TGF-β is a master regulator of fibroblast phenotype and function. Upon TGF-β stimulation, fibroblasts are become activated and undergo phenotypic transition into myofibroblasts, which leads to excessive matrix deposition and unbalance between matrix synthesis/degradation signals.[59] Furthermore TGF-β plays an important role in the EMT of epithelial cells to myofibroblasts.[24,60] In addition, TGF-β can play a role in the vasculopathy observed in SSc. TGF-β stimulates the expression of vascular endothelial growth factor (VEGF) and endothelin-1 (ET-1) in endothelial cells,[61] thereby mediating the vasoconstriction seen in patients with SSc.[62] Thus, the complex effects of TGF-β on proangiogenic and antiangiogenic factors partially explain the complex vascular phenotype seen in patients with SSc. The delivery of TGF-β to in vitro fibrosis platforms is a critical element in studying fibrotic mechanisms. The delivery in vitro is generally performed by simple addition of soluble TGF-β to the culture medium. For instance, studies proved that the supplementation of TGF-β to human skin fibroblasts increased the deposition of collagen type I.[63] In response to the need for an effective therapeutic for dermal fibrosis, a plethora of TGF-β stimulated in vitro models have been used as screening platform for antifibrotic molecules. As one example of many, human dermal fibroblasts stimulated with TGF-β1 to induce differentiation into profibrotic myofibroblast cells were used to assess the antifibrotic potential of Pirfenidone (PFD), a synthetic molecule already FDA-approved for the treatment of idiopathic pulmonary fibrosis. It was shown that PFD inhibited fibrogenic signals of TGF-β by abrogating p38-mediated MAPK activation, downregulating the transcription of profibrotic genes, such as type I and type III collagen, and blocking myofibroblast differentiation.[64] One of the limitations of this approach consists in the fact that the bioavailability of TGF-β in vivo is controlled by ECM-mediated integrins and mechanosensing mechanisms that inhibit or activate its binding to the corresponding receptors. Therefore, the development of culture conditions with tailored (patho-) physiological substrate stiffness could better mimic TGF-β in vivo bioavailability and may advance the relevance of its use in vitro culture models.[65]
Another critical growth factor involved in the pathogenesis of SSc is connective tissue growth factor (CTGF). CTGF is a cysteine-rich matricellular protein that functions in combination with TGF-β to enhance fibrotic responses. CTGF is not normally expressed in dermal fibroblasts, but is constitutively overexpressed by fibroblasts present in skin and pulmonary fibrotic lesions of scleroderma patients. The overexpression of CTGF promotes fibroblast proliferation, myofibroblast differentiation, and matrix deposition.[66] Moreover, CTFG plays a role in leading endothelial cells to transdifferentiate toward myofibroblasts.[67] Supplementation of CTGF in the culture media was showed to stimulate fibroblastic cell proliferation and ECM synthesis.[68] A recent study demonstrated that knockdown of CTGF using a mesoporous silica nanoparticle-based small interfering RNA (siRNA) delivery system prevented collagen deposition, activation and differentiation of fibroblast.[69]
Platelet-derived growth factors (PDGFs) have been demonstrated to have a critical role in fibrosis. PDGF is secreted by platelets, endothelial cells, macrophages, and fibroblasts that function as potent mitogens and chemoattractants for mesenchymal progenitor cells.[66a] Elevated expression of PDGF and its receptors has been found in scleroderma skin and lung tissues, and contributes to persistent fibrosis by the generation of reactive oxygen species and consequent fibroblast activation.[70] The supplementation of PDGF to normal human dermal fibroblasts in vitro has been shown to increase the mRNA and protein levels of matrix metalloproteinase 1 (MMP-1) and tissue inhibitor of metalloproteinase (TIMP)-1, but not type I collagen, fibronectin, or TIMP-2. Additionally, PDGF induced the mitogenic and migratory activity of human dermal fibroblasts in a dose-dependent manner.[71]
Another important mediator involved in the pathogenesis of SSc is endothelin 1 (ET-1). Endothelins are potent vasomodulatory peptides produced by endothelial cells, macrophages, fibroblasts and other cell types and can function as downstream mediators of TGF-β responses.[72] ET-1 signaling via endothelin receptors A (ETRA) and B (ETRB) on fibroblasts induces fibroblast migration and myofibroblast differentiation. Primary cultured dermal fibroblasts from SSc patients and healthy controls treated with ET-1 upregulated collagen type I, CTGF, type I plasminogen activator inhibitor (PAI-1) and pAkt in a time-dependent manner within 72 h.[73] In addition, the synergistic treatment of endothelial cells isolated from patients with SSc with ET-1 and TGF-β induced activation of the endothelial to mesenchymal transition (EndMT) process. Treatment with macitentan (MAC), an ET-1 receptor antagonist which is clinically used in pulmonary hypertension, prevented EndMT and fibroblast accumulation.[74]
It is worth to note that, due to the multifactorial nature of SSc, a multitude of interconnected growth factor-activated molecular pathways occur across multiple tissues, leading to aberrant signaling crosstalk (which includes also cytokines, chemokines, adipokines, neurotrophins, and metabolites) and ultimately to organ alterations. For this reason, results obtained in vitro from individual cell populations exposed to single growth factors need to be treated with caution, representing only a minimal fraction of SSc complexity.
5.2. Cytokines Supplementation
A wide range of cytokines have been found to be potent regulators of tissue fibrosis and endothelial damage.[75] The interleukin (IL)-1 family is a group of 11 proinflammatory and anti-inflammatory cytokines that have been reported to be involved in the pathogenesis in SSc. For example endogenous IL-1α expression by SSc fibroblasts has been demonstrated to increase the expression of IL-6 and PDGF, determining the abnormal function of SSc fibroblasts.[76] IL-6 is a pleiotropic and pro-inflammatory cytokine that is produced by activated immune cells and stromal cells, including T cells, macrophages, endothelial cells and fibroblasts, and is associated with a wide range of biological functions.[77] In particular, IL-6 is a potent inducer of matrix production in fibroblasts by increasing TGF-β expression, TIMP-1 synthesis and myofibroblast differentiation, resulting in collagen accumulation.[78] Treatment with anti-IL-6 therapy (tocilizumab) modified the biological characteristics of dermal fibroblasts derived from SSc patients, restoring functional properties, and reversing TGF-β-activated molecular pathways which were present prior to treatment.[79] IL-4 is a type 2 cytokine activated by CD4+ and CD8+ T cells and mast cells.[80] IL-4 has been demonstrated to be a profibrotic cytokine participating in cutaneous, cardiac fibrosis, pulmonary fibrosis, and hepatic fibrosis. IL-4 supplementation was shown to induce fibroblasts proliferation, myofibroblasts differentiation and collagen production in vitro.[81] IL-13 is another type 2 cytokine that is increased in the serum and lesional tissue of patient affect by SSc.[82] Supplementation of primary dermal fibroblasts with IL-13 stimulated cell proliferation and ECM synthesis.[83] IL-17 has been reported to be increased in the peripheral blood and target organs, including skin. It amplifies inflammatory responses by inducing the production of IL-6, CCL2 and CXCL8 (IL-8), MMPs and the expression of adhesion molecules in stromal cells including fibroblasts and endothelial cells.[84] Supplementation of IL-17 enhanced the proliferation of dermal fibroblasts and induced the expression of adhesion molecules and IL-1 production in endothelial cells in vitro.[85] TNF-α is a proinflammatory cytokine which has been reported to be elevated in patients with SSc and favors the development of pulmonary fibrosis and pulmonary arterial hypertension.[86] TNF-α supplementation induced high levels of IL-6 in SSc-derived fibroblasts, participating in the self-perpetuation of inflammation during SSc.[87]
In summary, a myriad of soluble mediators is involved in the pathogenesis and progression of SSc, which are secreted by several cell populations according to the stage of the disease. In order to properly supplement these molecules in vitro, continued efforts to understand native pathophysiological signaling pathways will be necessary. This requirement will help to recapitulate the concentrations and spatial and temporal distributions of bioactive factors during these processes. To date, biomaterial-based GF delivery systems have been optimized to provide differential immobilization efficiency and release kinetics.[88] While these systems have been extensively used in regenerative medicine, we also foresee their utility for the design of in vitro SSc models.
5.3. Serum Supplementation
The beneficial effect of animal sera has long been recognized as means to promote in vitro cell attachment, expansion, maintenance, and proliferation by providing essentials nutrients and growth factors.[89] However, animal derived sera, such as fetal bovine serum (FBS) or fetal calf serum (FCS), have several technical disadvantages associated with to batch-to-batch variation, xenoimmunization, and possible contamination with mycoplasma, viruses, endotoxins, and prions.[90] These limitations can affect the phenotype and the behavior of cells expanded in culture, preventing a quality-by-design approach.[91] Human-derived sera can replace FBS and FCS supplemented media and can create an in vitro microenvironment that more accurately resembles the human environment.[92] It has been demonstrated that human dermal fibroblasts viability cultured in human serum (HS) supplementation was much higher compared to FBS supplementation. Furthermore, gene expression analysis showed that fibroblasts cultured with HS supplementation maintained expression of collagen type I, increased expression of collagen type III and fibronectin, and reduced expression of a-smooth muscle actin (a-SMA) compared to FBS.[93] Serum derived from patients affected by SSc has been demonstrated to contain characteristic serum autoantibodies, profibrotic chemokines and growth factors such as IL-4, IL-17, and CTGF, and sonic hedgehog (SHH), which stimulate fibroblast-to-myofibroblast transition and promote dermal fibrosis.[94] In addition, a recent study identified a discriminant metabolic profile between the serum derived from patients affected by SSc and healthy patients, suggesting the importance of SSc serum not only as a diagnostic tool for the diagnosis and classification of the disease.[95] Very few studies have thus far investigated the effect of SSc sera in vitro. In particular, immune complexes containing scleroderma-specific autoantibodies derived from patients’ serum, have been showed to elicit proinflammatory and profibrotic effects in skin fibroblasts.[96] Another study demonstrated that autoantibodies purified from SSc-patient sera directed to platelet-derived growth factor receptor (PDGFR) were able to induce growth and a pro-fibrotic state in vascular smooth muscle cells through the epidermal growth factor receptor (EGFR).[97] Another study showed that H2O2 production by endothelial cells and fibroblasts was higher after incubation with SSc sera than with healthy sera. Moreover, this model allowed to test the efficacy of bosentan and N-acetylcystein potentiated 5-fluorouracil (5FU) on the inhibition of oxidative stress.[98] Treatment with SSc sera has been reported to induce EndoMT of dermal microvascular endothelial cells by reducing the expression of endothelial markers such as CD31 and VE-cadherin, and upregulating of mesenchymal markers, including α-SMA and collagen type I.[99]
Despite these interesting results on the use of patient-derived serum in cell culture, a limited number of investigations have assessed the potential of SSc serum as a tool to recreate the pathological microenvironment in vitro.
5.4. Cocultures
In the native tissue milieu, various cell populations interact between each other, stimulating different signaling pathways, and thus influencing numerous aspects of cell function. In vitro coculture models have been developed to recapitulate the in vivo physical contact and paracrine signaling between cell types. Coculture systems can be carried out either by directly seeding different cell types together in the same culture dish, or indirectly, using transwell inserts, whereby cells are located in the same media, without being in contact.[100] Multiple cell types, including epithelial cells, fibroblasts, endothelial cells, pericytes and leukocytes respond to noxious stimuli in the pathogenesis of SSc. Several in vitro models have been established to investigate the effect of complex intracellular interactions during pathogenic events. For example, in order to elucidate the influence of SSc epidermal keratinocytes on dermal fibroblasts, dispase-dissociated epidermal layers were directly cocultured within a collagen-embedded fibroblast gel. Results showed that SSc epidermal sheets strongly stimulated fibroblast activation, causing gel contraction and induction of CTGF via IL-1, ET-1, and TGF-β. The addition of exogenous IL-1 receptor antagonist (IL-1ra) blocked gel contraction by SSc epidermis, suggesting a potential therapeutic implication.[101]
Another study investigated the effect of dermal fibroblasts in the impairment of angiogenesis in SSc. By oversecreting pigment epithelium-derived factor (PEDF), a major antiangiogenic factor, SSc-derived fibroblast suppressed tube formation when cocultured with human dermal microvascular endothelial cells (MVECs) in an angiogenesis in vitro assay. PEDF knockdown in SSc fibroblasts reversed this process and rescued the number of tubule formed by MVECs. This pathway may present a promising target for new therapeutic interventions in SSc.[102]
6. Biochemical Cues
6.1. Hypoxia
Tissue hypoxia is a characteristic feature of SSc and contributes directly to the progression of the disease.[103] It was demonstrated that fibrotic lesions in the skin of SSc patients exhibit significantly decreased oxygen levels in comparison to SSc nonfibrotic skin and the skin of healthy individuals.[104] Molecular responses to hypoxia are regulated by the transcription factor hypoxia-inducible factor 1 (HIF-1). While HIF-1 is rapidly degraded after translation under normoxic conditions, its activity increases exponentially after exposure to low oxygen levels. The activation of HIF-1 thus plays a critical role in the transcriptional activation of downstream signaling involved in cell proliferation, angiogenesis and fibrogenesis.[105] Tissue oxygenation in SSc is impaired by microvascular alterations and reduced capillary density, which result in a decrease of the blood flow and poor oxygen supply.[104,106] Oxygen supply is further reduced by accumulation of ECM, which impairs diffusion from blood vessels to cells.[107] Chronic tissue hypoxia causes a vicious cycle by overexpression of VEGF, which in turn leads to aberrant vessel formation and TGF-β activation, thereby increasing tissue fibrosis.[108] Hypoxia induces multiple ECM proteins in dermal fibroblasts in vitro, such as thrombospondin 1, collagens, fibronectins, insulin-like growth factor binding proteins, and transforming growth factor β-induced protein, in a time-dependent manner.[109]
Numerous studies have contributed to the understanding of the role of hypoxia on the molecular mechanism of SSc. For instance, it has been showed that dermal fibroblasts stimulated with hypoxia (1% oxygen tension) showed increased CTGF expression via activation of HIF-1α, contributing to the progression of fibrosis.[110] Hypoxia can also drive epithelial-mesenchymal transition (EMT). It was demonstrated that severe hypoxia (1.5% oxygen tension) as well as moderate hypoxia (3% oxygen tension) induced the expression of α-smooth muscle actin (α-SMA) and vimentin and decreased the expression of E-cadherin in alveolar epithelial cells (AEC). In addition, hypoxia increased the levels of TGF-β, and preincubation of cells with an inhibitor of the TGF-type I receptor kinase prevented the hypoxia-induced EMT, suggesting that the process was TGF-β dependent.[111] Therapeutic strategies targeting hypoxia have been tested in vitro. 2-Methoxyestradiol (2-ME), a potent inhibitor of HIF-1α, inhibited the fibrotic effect of hypoxia on SSc fibroblasts by down-regulating CTGF and collagen I through the PI3K/Akt/mTOR/HIF-1α signaling pathway. In addition, 2-ME induced apoptosis and inhibited proliferation of SSc fibroblasts.[112] Despite the importance of hypoxia in SSc, conventional in vitro conditions expose the cells to a non-physiological hyperoxic environment (20% oxygen tension) that is far from recapitulating the pathological microenvironment.
6.2. Reactive Oxygen Species
Oxidative stress, as defined by an imbalance between oxidants (reactive oxygen and nitrogen species (ROS/RNS) and antioxidants, is consistently observed in patients with SSc.[113] Unpaired electrons make free radicals highly reactive and among them, the superoxide radical (·O2−), hydrogen peroxide (H2O2), hydroxyl radical (·OH−), hypochlorous acid (HOCl) and peroxynitrite (ONOO−) are key oxidative molecules within the ROS family.[114] Several oxidative stress biomarkers, such as nitric oxide, malondialdehyde (MDA-a marker of lipid peroxydation), asymmetric dimethylarginine (ADMA) and hydroperoxides are elevated in the blood of SSc patients compared to healthy controls.[115] Oxidative stress has been demonstrated to cause the activation and damage of ECs, leading to vascular hyperreactivity, apoptosis and impaired angiogenesis.[116] Increased ROS generation has been reported to mediate TGF-β-induced EndMT.[117] ROS have been shown to support chronic inflammation and autoimmunity through the genesis of neoepitopes and the activation of T and B lymphocytes and macrophages.[118] Permanent overproduction of ROS stimulates the proliferation and activation of fibroblasts and their synthesis of ECM.[119] Also, fibroblasts from SSc have been shown to maintain an overproduction of ROS in SSc through the upregulation of nicotinamide adenine dinucleotide phosphate (NADPH) oxidase (NOX)-2 and NOX-4 proteins.[120] NOX is a family of enzymes involved in the generation of ROS, acting via the transfer of a single electron from NADPH to oxygen.[121] The activation of NOX has been demonstrated to trigger fibroblast proliferation and expression of type I collagen genes in SSc cells, thereby maintaining an autocrine feedback mechanism of ROS generation.[119]
As oxidative stress impacts many aspects of the pathophysiology of SSc, several in vitro studies assessed the potential of natural and synthetic antioxidants in the supportive treatment of SSc. The antioxidant epigallocatechin-3-gallate (EGCG), a polyphenol present in green tea extracts, has been shown to reduce oxidative stress and the fibrotic effects on activated dermal fibroblasts from SSc patients.[122] The antioxidant effect of kaempferol, a natural flavonoid, was investigated on H2O2-induced intracellular ROS accumulation in SSc fibroblasts and suppressed the intracellular accumulation of ROS and reduced H2O2-induced apoptosis.[123] N-acetylcysteine (NAC), a scavenger of free radicals and a precursor of the major antioxidant glutathione, inhibits fibroblast proliferation and collagen synthesis[119] and reduces peroxynitrite (ONOO−) synthesis by activated lung macrophages from SSc patients in vitro.[124]
Although ROS seem to have an important role in fibrosis, a therapeutic strategy utilizing antioxidants is not yet clear and further investigations are needed to further elucidate the mechanisms linking ROS dynamics and SSc pathogenesis.
7. Biophysical Cues
7.1. Substrate Stiffness
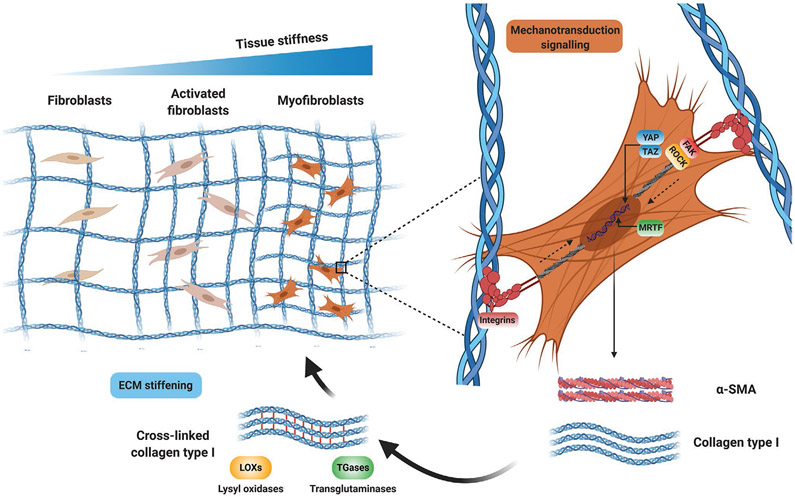
In addition to biological and biochemical signals, the dysregulation of biophysical properties of the tissue microenvironment in skin, lung, and other organs have been associated with the progression of fibrosis in SSc.[125] Excessive deposition of ECM increases tissue stiffness and reduces the elasticity of affected organs, leading to mechanical stress (Table 3).[126] Tissue stiffness can be measured as the elastic modulus, defined as the resistance to deformation, and expressed as the magnitude of a stress (compression, elongation, or shear force, normalized to area) divided by the strain (deformation) induced by the stress.[127] Increased matrix stiffness anticipates the development of fibrosis, which suggests that tissue stiffening may induce the early activation of myofibroblasts.[4] Matrix stiffness orchestrates fibrosis by controlling fundamental profibrotic mechanisms including mechano-activation of myofibroblasts via mechano-transduction pathways. Mechano-transduction involves cell surface integrins and changes in cytoskeletal tension mediated by focal adhesion kinase (FAK) and RHO-associated kinase (ROCK). These signals activate the downstream effectors YAP (Yes-associated protein), TAZ (transcriptional coactivator with PDZ-binding motif) and myocardin-related transcription factor (MRTF) which increase fibroblast activation and further perpetuate the fibrotic process (Figure 4).[128] Lung fibroblasts cultured on stiff substrates showed an increase in proliferation and differentiation into myofibroblasts in comparison to soft substrates.[129] Moreover, high substrate stiffness increased the synthesis of ECM and the expression of α-SMA and decreased the expression of matrix proteolytic genes and prostaglandin E2 (PGE-2).[129] The increase in myofibroblasts’ contractility leads to further matrix remodeling and stiffening and activation of TGF-β, amplifying the signal and leading to a positive feedback loop.[130] TGF-β1 is secreted and stored in the ECM in a latent multiprotein complex with latency associated peptide (LAP) and latent TGF-β1 binding proteins (LTBPs).[131] The activation of TGF-β1 first requires the binding of αv integrin to arginine-glycine-aspartic acid (RGD) sequences in LAP.[132] Mechanical forces exerted by cells on LAP via integrin-based adhesions then lead to changes in the conformational state of this complex, which ultimately release and activate TGF-β1 for receptor binding.[133]
Table 3.
Influence of substrate stiffness on the fibrotic phenotype in SSc and drug testing (α-SMA, α-smooth muscle actin; DFs, dermal fibroblasts; EKs, epidermal keratinocytes; IPF, idiopathic pulmonary fibrosis; MRTF-A, myocardin-related transcription factor A; PAAm, polyacrylamide; PDMS, polydimethylsiloxane; PI3K, phosphoinositide 3-kinases; TAZ, transcriptional coactivator with PDZ-binding motif; TCP, tissue culture plastic; TGF-β, transforming growth factor-β; YAP, yes-associated protein).
| Model | Cell type | Experimental output | Therapeutics | Refs. |
|---|---|---|---|---|
| PAAm gels with moduli of 0.6 (healthy tissue) or 6 kPa (sclerotic tissue) | Healthy and SSc-derived human DFs | High substrate stiffness led to a pronounced increase in TAZ/YAP nuclear localization and increase in the number and size of actin stress fibers indicative of a fibrotic transition | Dimethyl fumarate (DMF), via inhibition of the PI3K/Akt pathway, reduced nuclear localization of both YAP and TAZ in fibroblasts on stiff gels. In addition, it reduced the cell spread area and the number and size of stress fibers | [139] |
| Soft (5 kPa) and stiff (50 kPa) collagen coated PAAm gels | Healthy and SSc-derived human DFs | High substrate stiffness enhanced nuclear translocation of MRTF-A in dermal fibroblasts. The SSc cells had more nuclear MRTF-A on soft and stiff matrices than healthy control cells. Collagen expression and synthesis increases on stiffer matrices | Inhibitor of nuclear translocation of MRTF-A (CCG-1423) or knockdown of MRTF-A reduces contractility and suppresses fibrotic targets in SSc fibroblasts | [239] |
| PDMS substrates with varying stiffness to mimic healthy (1–10 kPa) and fibrotic (15–50 kPa) dermis | Primary adult human DFs | Cells cultured on skin-soft silicones were α-SMA-negative and displayed low mRNA levels of fibrosis-associated genes. Cells grown on 50 kPa and on TCP had α-SMA-positive stress fibers | – | [145] |
| Collagen-coated PAAm hydrogels with compliant (0.5 kPa) or stiff (12 kPa) matrices | Normal human primary EKs | NHEKs grown on soft matrix expressed more E-cadherin and less α-SMA than cells grown on stiff matrix. Stiff matrix augmented the TAZ protein expression level compared to soft matrix | Transient receptor potential vanilloid 4 (TRPV4) antagonist (GSK219) blocked both matrix stiffness-induced and TGF-β-induced expression of YAP and TAZ proteins | [240] |
| Collagen I-coated PAAm gels of elastic moduli 0.4 kPa (healthy lung tissue) and 25 kPa (fibrotic lung tissue) | Normal IPF- pulmonary derived fibroblasts | Cells exhibited predominant nuclear localization of YAP/TAZ on stiff matrix and far fewer cells exhibiting distinct nuclear localization on soft matrix | YAP and TAZ siRNA knockdown attenuates profibrotic matrix synthesis, contraction, and proliferation, preferentially on pathologically stiffened matrices | [128a] |
| Engineered microtissue based on micropillar device | Normal human lung fibroblasts | Stiffness of TGF-β1 treated microtissues was higher (24.0 ± 8.0 kPa) than untreated microtissues (5.5 ± 2.8 kPa) and matched those of fibrotic lungs | Nintedanib and pirfenidone strongly inhibited TGF-β1-induced tissue stiffening and maintained tissue stiffness and compliance comparable to untreated samples | [241] |
| Silicone culture substrates with tunable Young’s modulus (5–47 kPa) | Rat lung myofibroblasts (MFs) | Induction of MF contraction with thrombin enhanced latent TGF-β1 activation on 9–47-kPa substrates but had no effect on 5-kPa soft substrates. Increased α-SMA expression on stiff (47 kPa) substrates | – | [130] |
| Collagen I-coated PAAm gels of various stiffnesses (1, 2, 8, 25, or 50 kPa) | Normal human lung fibroblasts | Expression of α-SMA proteins was higher on the stiffer substrates (25 kPa gel and TCP) than on the soft 2 kPa gel. Migration of fibroblasts on stiff substrates was higher than cells on 2 kPa gel | Short interfering RNA for α-SMA inhibited cell migration | [242] |
| Gelatin-coated PAAm gels with a stiffness of 13 kPa | Mesenchymal-like cells generated from iPSCs | Cells were highly proliferative, showing increased gene and protein expression of collagen I, a-SMA, and TGF-β | Antifibrotic small molecule AA-5 downregulated gene and protein expression of a-SMA and collagen I and decrease in cell stiffness | [243] |
Figure 4.
Matrix stiffness and mechanotransduction in fibrosis. Mechanotransduction pathways mediate matrix stiffness-induced myofibroblast activation. Stiffness-mediated traction forces are transmitted across integrins, which induce actomyosin cell contractility mediated by focal adhesion kinase (FAK) and RHO-associated kinase (ROCK). These signals activate the downstream effectors YAP (Yes-associated protein), TAZ (transcriptional coactivator with PDZ-binding motif) and myocardin-related transcription factor (MRTF), which increase the expression of profibrotic markers such as α-SMA and collagen type I. Increased collagen deposition and crosslinking further increases ECM stiffening, creating a profibrotic positive feedback loop between matrix stiffness and myofibroblast activation. Created with BioRender.com.
Culture systems with tailored mechanical properties have been engineered to mimic aspects of diseased tissues and investigate the mechanism of myofibroblast activation. For instance, stiff collagen hydrogels promoted stress fibers formation, smooth muscle actin (SMA) expression and TGF-β1-induced response in human fibroblasts.[134] Another study identified that a stiff polyacrylamide substrate induced lung myofibroblast differentiation through actin cytoskeletal remodeling-mediated activation of megakaryoblastic leukemia factor-1 (MKL1), which transduces mechanical stimuli from the ECM, leading to the induction of a fibrogenic program.[135] A recent study showed that the activity of the transient receptor potential vanilloid 4 (TRPV4) channel was increased when cells were plated on stiff collagen-coated polyacrylamide gel matrices within the pathophysiological range seen in diseased/fibrotic dermal tissue. Genetic ablation or pharmacological antagonism of the TRPV4 channel abrogated Ca2+ influx and matrix stiffness-induced myofibroblast differentiation, evidencing that therapeutic inhibition of TRPV4 activity may provide a targeted approach to the treatment of scleroderma.[136]
Given the increasing recognition that ECM stiffness is a major factor contributing to the progression of SSc, it has become evident that the identification of optimal substrate stiffness which replicates pathological fibrotic conditions will enable the development of more precise mechano-therapeutic interventions for tissue stiffening. Therapeutic strategies targeting mechanotransduction signaling mediated by integrins,[132b] FAK,[137] ROCK,[138] or YAP/TAZ[139] have been promisingly tested in preclinical and clinical studies.
7.2. Substrate Topography
The topographical organization of the ECM significantly influences cell morphology and behavior, including growth, adhesion, and migration.[140] Cells sense their underlying topography via focal adhesion interactions, pushing and pulling against the matrix and activating a cascade of cellular and molecular events, which ultimately influence gene and protein expression.[141] Morphological changes to the dermal collagen organization and focal adhesion complexes have been reported in skin biopsies from SSc patients. These changes are characterized by the presence of highly aligned collagen bundles, and a loss of normal “basket-weave” collagen organization that is characteristic of the healthy dermis.[142] The alteration of the stiffness of the ECM significantly contributes to the alignment of the collagen fibrils, and further amplifies the fibrotic process.[143] The ability of cells to mechanically sense these changes may be due to the deposition and realignment of new collagen fibrils in which cells generate myosin-generated tensile forces applied through cell-matrix adhesions.[144] In vitro studies have demonstrated that increasing stress fiber formation and ECM alignment increase the elastic modulus of the fibroblast-populated collagen gels over a culture period of 21 d.[145] Moreover, several studies have reported that mechanical strength of anisotropic nanofibers is significantly higher than that of the disordered nanofibers.[146] The anisotropic ECM ultrastructure within the fibrotic microenvironment is a critical cue in maintaining the myofibroblasts phenotypes in SSc. This was demonstrated by using a 3D model of either randomly oriented or aligned electrospun poly-caprolactone (PCL) nanofibers with adsorbed type I collagen. Guidance cues from aligned collagen fibers enhanced the fibrogenic potential of dermal fibroblasts by increasing cell migration, adhesion, and guidance signaling pathways. Arhgdib (Rho GDP-dissociation inhibitor 2) was one of the most upregulated genes following fibroblast culture on aligned fiber substrates, and siRNA knockdown of Arhgdib significantly reduced directed cell migration under aligned fiber culture conditions.[147] Another study utilized glass slides coated with aligned fibers to investigate alignment and migration of lung fibroblasts isolated from SSc patients. The results indicated that migration took place when lung fibroblasts were cultured on aligned collagen following stimulation with PDGF, but was not induced on the woven, randomly orientated collagen substrates.[148] In addition, heparin, which binds ligands including PDGF and stem cell factor (SCF), and imatinib, which blocks downstream tyrosine kinase receptors, both inhibited lung fibroblast migration individually. Importantly, the two drugs showed synergistic effect in SSc cells, supporting a possible pilot evaluation of combination therapy.[148] A recent study investigated the effect of collagen microarchitecture on myofibroblast differentiation and fibrosis. Adipose stromal cells (ASCs) were cultured on collagen gels consisting of networks with thin fibers and low porosity, or scaffolds with thicker collagen fibers with larger pores. Interestingly, ASCs contractility on collagen matrices with thicker fibers and larger pores resulted in collagen fiber densification and alignment in the direction of cell polarization and migration, increasing stiffness in a physiologically relevant regime of strain. The stiffening of local matrix, in turn, stimulated a contractile phenotype via a positive feedback loop, thereby modulating myofibroblast differentiation and fibrosis.[149] Due to the interaction between increased ECM alignment and the formation of fibrotic lesions, it is of considerable interest to further investigate the influence of patterned cell culture substrates on tissue mechanics and collagen alignment.
7.3. Macromolecular Crowding
The recapitulation of collagen matrix formation in vitro has proven challenging, partly due to the omission of important cofactors in the culture media and partially because of sub-optimal cell culture conditions. The omission of ascorbate in cell culture results in minimal production and deposition of collagen within the cell layer. Ascorbic acid is a crucial cosubstrate for the enzymes prolyl hydroxylase and lysyl hydroxylase, which are responsible for the posttranslational hydroxylation of prolyl and lysyl residues on collagen fibers. Hydroxylation of the prolyl residues renders the collagen triple helix thermostable and hydroxylation of the lysyl residues is responsible for the extracellular cross-linking of collagen fibers.[150] However, even with ascorbate supplementation, cells deposit only subphysiological amounts of secreted collagen I into their matrices. Indeed, in standard cell culture settings, the conversion of water-soluble procollagen to insoluble collagen is relatively slow, since the proteinases required for the enzymatic cleavage of procollagen are dispersed in dilute culture media.[151] To overcome this limitation, macromolecular crowding (MMC) has been introduced as a means to accelerate ECM deposition in vitro. The addition of macromolecules into the culture media emulates the naturally crowded in vivo milieu, and thus amplifies deposition of cell-secreted ECM.[152] The addition of macromolecules to the culture media also results in a more efficient volume occupancy, preventing the dispersion of active molecules, consequently accelerating the conversion of procollagen to collagen and ultimately deposition of the latter.[153] Moreover, use of MMC has been demonstrated to drive the molecular assembly of collagen fibrils in vitro and to stabilize the formed matrix through enzymatic crosslinking.[154] One application of MMC to produce the full deposition of collagen in fibroblast cultures has been the development of a valuable screening tool, the so-called “Scar in a Jar”, for antifibrotic compound screening.[155] The original scar in a jar model consisted of human fibroblasts cultured in the presence either of 500 kDa dextran sulphate (DxS) or a mixture of neutral 70 and 400 kDa Ficoll (Fc). Crowding of culture medium with dextran sulphate served as the rapid deposition mode as it yielded maximum granular deposition of collagen I by 48 h, whereas neutral mixed Ficoll served as the accelerated mode (Fc), which resulted in the deposition of a fibrillar collagen meshwork after 6 d of culture in comparison with non-crowded cultures.[156] This system has been used to evaluate the potential of antifibrotic compounds effective at the epigenetic, post-transcriptional/translational level. Another study utilized the principle of macromolecular crowding to create an ECM-rich in vitro hypertrophic scar model that more closely recapitulates “in vivo-like” conditions than customarily used monolayer culture systems.[157] This model was used to test the antifibrotic effect of a series of naphthalene derivatives derived from medicinal herbs on human dermal fibroblasts. Interestingly, shikonin and naphthazarin were shown to inhibit the transcription and translation of collagen in human scar-derived fibroblasts and induced apoptosis via the mitochondrial apoptosis pathways, suggesting their potential therapeutic value for the treatment of dermal fibrosis and scarring.[157] A similar study, combined macromolecular crowding with TGF-β to develop a robust, high throughput, phenotypic screening assay using pulmonary fibroblasts derived from patients affected by idiopathic pulmonary fibrosis.[158] The Scar-in-a-Jar offers a novel pathophysiologically relevant screening and evaluation tool for antifibrotic compounds interfering with different key steps in the collagen biosynthesis pathway.
Overall, despite all these progresses in the modeling of fibrosis using MMC in vitro, the importance of the crowded extracellular niche is still underestimated,[159] likely due to the fact that optimal crowding agents remain still elusive and further studies are required to reveal and unravel their diverse effects on cell behavior and phenotype. Optimization of MMC protocols will contribute to the generation of models with high levels of biomimicry which can be used as an instrument to recreate SSc conditions.
7.4. Dimensionality and 3D Architecture
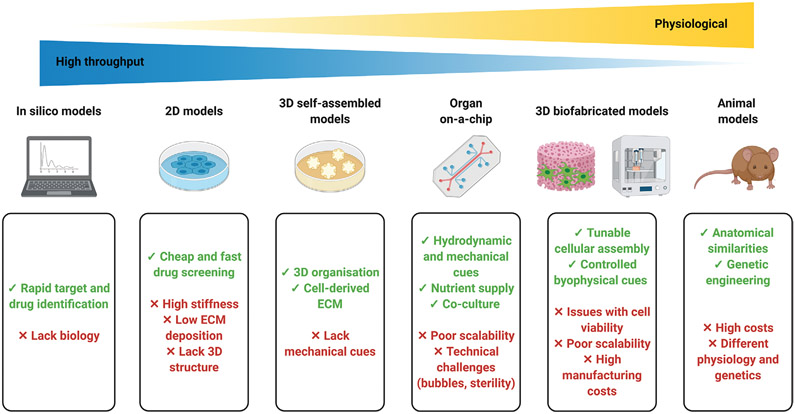
Cell-based assays have been crucial in drug discovery and provide a simple, rapid, and cost-effective tool to support screening of therapeutics before large-scale and cost-intensive animal testing. To date, the majority of cell-based assays are based on traditional 2D monolayer cultures grown on flat dishes optimized for cell attachment and growth.[160] However, 2D culture systems have multiple limitations, including loss of tissue-specific architecture, non-physiological mechanical and biochemical signals, and non-physiologic cell-to-cell and cell-to-matrix interactions.[161] The standard 2D environment may therefore provide misleading results regarding the predicted responses of cells to drug treatments (Figure 5).[162] To overcome these important limitations, there has been tremendous progress in tissue engineering and regenerative medicine over the past few decades that has led to the development of a wide range of 3D cell culture systems. Indeed, multicellular organisms reside within a complex 3D environment, rich with multiple ECM components, several cell populations and soluble factors, which not only provide structural support to tissues and organs, but also physiological conditions that allow for optimal functionality and delineate specific microenvironments.[163] Table 4 summarizes several 3D scaffold-free and scaffold-based SSc models, including the therapeutic approaches used to evaluate each model’s feasibility for drug development and screening.
Figure 5.
Advantages and limitations of in silico, in vitro, and in vivo models utilized to replicate complex pathophysiologies. Created with BioRender.com.
Table 4.
3D in vitro scaffold-free and scaffold-based models of SSc and their application in drug screening (α-SMA, α-smooth muscle actin; DAMPs, damage-associated molecular patterns; DFs, dermal fibroblasts; EKs, epidermal keratinocytes; ECM, extracellular matrix; iPSC, induced pluripotent stem cell; LOXL-4, lysyl oxidase-like-4; MD-2, myeloid differentiation-2; SAS, self-assembled stromal tissues; TGF-β, transforming growth factor-β; TLR4, toll-like receptor-4).
| Model | Scaffold | Cells | Experimental output | Therapeutic | Refs. |
|---|---|---|---|---|---|
| Scaffold-free models | |||||
| Self-assembled dermal equivalents | – | Early-stage SSc or late-stage SSc-derived human DFs | TGFβ-1 induced a significant increase in dermal thickness early-stage SSc equivalents | – | [173] |
| Self-assembled skin equivalents | – | Normal human EKs and DFs |
Differentiated keratinocytes increased collagen and TGF-β gene and protein expression, but not undifferentiated keratinocytes | – | [244] |
| 3D skin organoid | – | DFs and EKs generated from iPSCs derived from cord blood mononuclear cells (CBMCs) | TGF-β treatment increased the thickness and collagen gene and protein expression deposition of the 3D iPSC-fibroblasts layer | Pirfenidone attenuated the increase in the thickness and collagen expression |
[182] |
| Tissue spheroids | – | Human primary fibroblasts and human macrophages | Fibroblasts in the 3D spheroids had significantly higher expression levels of α SMA and collagen I compared to the 2D culture. Addition of macrophages to the spheroids enhanced collagen I maturation | – | [167] |
| Scaffold-based models | |||||
| 3D organotypic human skin equivalents | Collagen type I (rat/bovine) hydrogel | Human DFs, human dermal microvascular endothelial cells, foreskin keratinocyte human mast cell line (HMC-1) | HMC-1 cells induced α-SMA expression by fibroblasts and stimulated fibroblast contraction of collagen gels | Tryptase inhibitors eliminated the ability of HMC-1 cells to stimulate fibroblast contraction | [245] |
| Human DFs and EKs | Exogenous fibronectin extra domain A (FnEDA, 10 μg mL−1) had potent effects on collagen gene expression, myofibroblast differentiation, increased matrix stiffness and collagen cross-linking in human skin equivalents | RNA interference and inhibitor of TLR-4 prevented cutaneous fibrosis, collagen deposition, and myofibroblast accumulation | [246] | ||
| Healthy human dermal DFs and EKs | TGF-β induced the expression of collagen I, fibronectin and α-SMA in skin equivalents | Agonist peptides targeting adiponectin receptor abrogated the stimulation of fibroblast migration, and attenuated fibrotic expression in unstimulated SSc fibroblasts | [247] | ||
| Healthy or SSc-derived DFs | SSc fibroblasts progressively remodel their dermal microenvironment, with time-dependent increases in collagen deposition, matrix reorganization, accumulation of DAMPs and substrate rigidity | Inhibitors of MD-2/TLR-4 complex formation significantly reduced rigidity and collagen content of the tissue equivalents | [248] | ||
| Healthy or SSc-derived DFs and human EKs | SSc-fibroblasts demonstrated enhanced stromal rigidity with increased collagen crosslinking, upregulation of LOXL-4 expression and innate immune signaling genes | Knockdown of LOXL-4 suppressed rigidity, contraction and α-SMA expression in SSc-fibroblasts skin equivalents and TGF-β induced ECM aggregation and collagen crosslinking in SAS | [249] | ||
| Vascularized human skin equivalents | Decellularized segment of porcine jejunum supplied by a single artery-vein pair with intact outer vascular system | Human EKs, DFs and microvascular endothelial cells | Skin equivalents perfused at a physiological pressure formed a functional vessel system. Exposure to TGF-β induced fibroblast to myofibroblast transition, increased release of collagen and excessive deposition of extracellular matrix | Nintedanib attenuated TGF-β signaling, reduced fibroblast to myofibroblast transition and decreased ECM deposition | [199] |
| Decellularized scaffold | Decellularized scaffolds prepared from healthy and fibrotic scleroderma lung explants | Healthy and SSC-derived peripheral blood mononuclear cells (PBMCs) | Scleroderma scaffolds increased procollagen type I production by PBMCs, which was stimulated by enhanced stiffness and abnormal ECM composition. Enhanced Netrin-1 expression was seen on SSc-derived cells | Antibody mediated netrin-1 neutralization attenuated procollagen I detection | [250] |
7.4.1. Self-Assembled Models
Tissues are formed by building blocks that self-assemble into highly organized structures that enable and regulate their functions.[164] Tissue engineering by self-assembly (TESA) relies on the inherent capacity of cells to self-assemble into highly organized 3D tissue-like constructs and to produce their own ECM, without the need of an external scaffold. These assemblies can be created through self-assembled aggregation, cell sheets, tissue strands or direct bioprinting.[165]
3D cell aggregates allow for the fabrication of organotypic microtissues due to their multidimensional cell–cell interactions and communication. Multicellular spheroids are scaffold-free cellular models based on the spontaneous aggregation of cells into spherical compact clusters on nonadherent substrates. The complex cell interactions recapitulate spatial and functional characteristics of the native tissue modulating cell activities and signaling.[166] For this reasons, 3D multicellular spheroids have been utilized as in vitro models of fibrosis. Recently, 3D human fibroblasts spheroids have been created for the development of an in vitro fibrogenesis model. Fibroblast-based spheroids showed significantly higher expression levels of fibrotic genes (αSMA and collagen I) compared to 2D monolayer culture. Furthermore, since the absence of immune cell mediators was recognized as a likely limit to physiologic model behavior, hybrid spheroids were fabricated with fibroblasts and macrophages. The addition of macrophages to the fibroblasts spheroids at a ratio of 1:16 (macrophages-fibroblasts) resulted in an increase of fibroblast activation and myofibroblast differentiation. Similarly, more macrophages were polarized toward an inflammatory type M1 in this group with greater CCR7 and pSTAT1 expression. In addition, hybrid spheroids demonstrated high expression of fibrosis-related genes (collagen I, collagen III and TGF-β) and inflammatory genes (TNF, IL-1β and IL-6). This system thus represents a valuable in vitro fibrogenesis model for high-throughput antifibrosis therapy screening.[167] Building on the success of spheroids, researchers have also focused on organoids, which generally better replicate tissue morphology and organization, and embed multiple cell populations that are distributed physiologically.[168] Organoids are in vitro self-assembling 3D organ-like architectures grown from tissue-specific adult stem cells or pluripotent stem cells, such as embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs).[169] Organoids have been engineered to model pathogenetic mechanisms that affect lung homeostasis and involve complex interactions between different cell types, such as those occurring in interstitial lung diseases, which is a frequent complication of systemic sclerosis.[170] For instance, lung organoids composed of iPSC-derived mesenchymal cells, treated with exogenous TGF-β1, demonstrated increased contraction and the development of fibroblastic foci by expressing of collagen 1 and α-SMA.[171] Organoids represent valuable tools for the screening of compounds with pro-regenerative, antifibrotic or tissue protective capabilities for precision medicine. However future applications of organoids may be limited by the fact that they lack mechanical cues, vasculature, and immune components, and may be prone to tumorigenicity in case of IPSCs.
Another strategy to fabricate cellular 3D constructs consists of stacking of multiple cell sheets, which takes advantage of the cell-deposited ECM network that intertwines the different cell sheets.[172] Self-assembled reconstructed dermis equivalents have been created to study the fibrotic phenotype of SSc fibroblasts. Fibroblasts isolated from lesional or nonlesional skin of early- and late-stage SSc patients were grown for 35 d to form sheets and two fibroblast sheets were layered and maturated to generate a thick dermal-like layer.[173] The study showed that in this system, development of skin fibrosis resulted in progressive changes at the fibroblast level, from a normal phenotype to a sustained and autonomous fibrotic phenotype. Based on these results, the authors suggested that antifibrotic treatments of SSc could gain efficacy if they were tailored to disease duration and severity. Another study developed a tissue-engineered model of self-assembled reconstructed skin to mimic interactions between dermal and epidermal cells.[174] Four dermal fibroblasts layers were superimposed to form a reconstructed dermis and undifferentiated, differentiated and unsorted keratinocytes were seeded onto the dermal sheets. Results showed that undifferentiated keratinocytes inhibited dermal fibrosis through downregulation of TGF-β, induction of FGF-β (fibroblasts growth factor β) and initiation of desmosome formation, indicating that undifferentiated keratinocytes may be a promising option for fibrotic scar prevention. Despite the utility of self-assembled skin models for disease modeling, these systems lack the presence of critical components such as hair follicles, glands, tactile corpuscles, and subcutaneous adipose tissue, which play a critical role in physiological functioning. However, progress in biofabrication techniques will likely address the incorporation of such appendages.[175]
7.4.2. Scaffold-Based Models
Scaffold-based models of 3D tissues consist of cells grown in the presence of support scaffolds consisting of either hydrogel-based or polymeric fiber-based materials. These scaffolds can be composed of natural, synthetic, or combinations of different polymers, and represent a 3D construct that is structurally, mechanically and functionally similar to the biological tissue.[176,175]
For the fabrication of human skin equivalents, dermal fibroblasts are generally cultured in collagen matrices to recreate a dermal-like layer, whereas keratinocytes are cultured on top of these dermal equivalents, followed by air–liquid interface culture to promote full epidermal maturation.[177] For example, bioengineered human skin equivalents were fabricated using both SSc-derived dermal fibroblasts and normal dermal fibroblasts.[178] Stromal equivalents were assembled by embedding dermal fibroblasts into collagen type I hydrogels. Full skin equivalents were fabricated by adding keratinocytes onto the collagen gel, after the maturation of the dermal layer. Results evidenced that SSc fibroblasts altered collagen architecture, as seen by a more mature and aligned fibrillar structure, enhanced stromal rigidity with increased collagen crosslinking, and upregulated LOXL-4 expression and innate immune signaling genes. Interestingly, knockdown of LOXL-4 suppressed rigidity, contraction and α-SMA expression in the SSc skin equivalents and TGF-β-induced ECM aggregation and collagen crosslinking in the stromal compartment. This skin-like tissue platform provided a suitable tool to test mechanisms that mediate skin fibrosis. Nevertheless, the lack of vascular and immune cells into the 3D skin-like tissues limits its potential.
Bioengineered skin equivalents have also been utilized to develop skin-humanized mouse models to test the progression of SSc and to monitor the response to antifibrotic drugs in vivo. For instance, 3D bioengineered skin, based on plasma-derived hydrogels and containing human SSc-derived keratinocytes and fibroblasts, were grafted onto immunodeficient (SCID) mice. Results showed that the human skin-SCID mouse models closely replicate the SSc fibrotic phenotype in vivo up to 16 weeks.[179]
Despite the relevance of cell populations derived from SSc patients, the fabrication of 3D tissue equivalents is limited by the availability of donor cells. In this regard, human iPSCs offer great potential to generate relevant cell types and can provide an alternative source of cells for tissue engineering purposes.[180] Currently, patient-specific iPSCs are generated by reprogramming of adult somatic cells by ectopic expression of pluripotency-associated transcription factors including OCT4, SOX2, KLF4, and c-MYC.[181]
Cord blood mononuclear cells (CBMCs)-derived iPSCs have been used to generate dermal fibroblasts, which have been used to create 3D dermal equivalents. Treatment with TGF-ß1 activated iPSC-fibroblasts and increased their proliferation rate and ECM production. In addition, TGF-β1 treatment increased the thickness of the 3D iPSC-fibroblasts construct. Treatment with pirfenidone, a drug used to treat idiopathic pulmonary fibrosis, elicited an antifibrotic effect, attenuating the increase in dermal thickness and expression of fibrosis genes. These results suggest that the use of iPSC-derived fibroblasts in skin equivalents could be utilized for drug screening purposes.[182]
A simple and versatile technique used for the fabrication of ECM-mimicking fibrous scaffolds is electrospinning. Electrospun fibrous scaffolds create nano- to microscale fibrillar network with interconnecting pores, resembling natural ECM in tissues, and facilitating the formation of artificial tissues in vitro.[183] Poly-caprolactone (PCL) electrospun scaffolds coated with bleomycin treated lung extracts, obtained from solubilization with a glass homogenizer, have been used as a model of idiopathic pulmonary fibrosis (IPF) to induce bone marrow-derived cells to differentiate into myofibroblast-like cells.[184] This approach combines biological extracts isolated from fibrotic lungs with synthetic nanofibers that serve as an ECM-like substrate to determine the effect of biochemical signals present in the fibrotic microenvironment. This model has the potential to help identify compounds that either mitigate or reverse the fibrotic differentiation of bone marrow -derived cells. Moreover, bone marrow-derived cells cultured on electrospun fibers with higher elastic modulus displayed increased fibrotic gene expression, demonstrating the importance of matrix modulus in cell differentiation.[184]
The design of electrospun scaffolds with appropriate topographical features is critical to determine cell function and to foster desired cell differentiation. For example, a recent study showed that nanometer scale electrospun fibers upregulated the expression of α-SMA, TGF-β, and vimentin filaments in comparison to micrometer scale fibers. The size change of the electrospun fibers (from micrometers to nanometers) altered fibroblast differentiation and led to higher α-SMA expression and more contractile myofibroblasts.[185] Another study demonstrated that electrospun fiber diameter can modulate epithelial to mesenchymal transition (EMT). Indeed, epithelial cells grown on fibers with an average diameter of 5μm exhibited a downregulation of epithelial markers such E-cadherin and upregulation of mesenchymal markers such as vimentin. However, cells grown on fibers with an average diameter of 0.5 μm grew as compact colonies with a stable epithelial phenotype.[186]
Despite considerable advances in biomaterials design and development for the engineering of physiological-relevant tissue models, several challenges hinder the applicability of such models as part of daily pharmaceutical research. One such limitation is the scalability and manufacturing of complex, bio-mimetic and reproducible scaffold-based models in compliance with current good manufacturing practices (cGMPs), which result in cost-intensive processes, not comparable to 2D and self-assembled models.
7.4.3. Decellularized Matrix Bioscaffolds
Given the complexity of ECM composition and structure, designing and fabricating a biomaterial-based scaffold that fully mimics the biochemical and structural properties of native tissue ECM is currently challenging. However, decellularization of whole tissues and organs by removing cellular components provides a useful method for harvesting an ECM which retains tissue-specific 3D morphology, biochemical, and biomechanical cues.[187,163a] A 3D model of the fibrotic lung microenvironment created from decellularized lung explants was used to determine whether the lung ECM of patients with scleroderma leads to the development of fibrocytes from peripheral blood mononuclear cells. Fibrocytes are collagen-producing leukocytes abundantly present in patients with SSc-related interstitial lung disease (ILD) via unknown mechanisms that have been associated with altered expression of neuroimmune proteins. Culture of control cells and patient-derived cells in lung scaffolds from patients with SSc-related ILD increased production of procollagen I, which was stimulated by enhanced stiffness and abnormal ECM composition. Moreover, enhanced detection of netrin-1 (a laminin-like protein that regulates cell–matrix interactions) expression in cells from patients with SSc-related ILD was observed, and antibody-mediated netrin-1 neutralization attenuated detection of collagen-producing leukocytes in all settings.[188] This study demonstrated the utility of decellularized platforms for disease modeling and potential drug discovery. Other studies provided insights into ECM-mediated positive-feedback loops using decellularized lung scaffold from patients with IPF.[189] In the absence of exogenous factors, IPF ECM alone can promote normal lung fibroblasts to become activated myofibroblasts, and once fibroblasts are activated, IPF ECM sets up a positive feedback loop capable of sustaining progressive fibrosis. Although successful decellularization has been achieved for many organs, standardized decellularization protocols still need to be defined with the final goal of advancing the creation of in vitro models.[190] Moreover, the development of decellularized skin matrices will allow further exploration in the pathogenesis of SSc dermal fibrosis.
7.4.4. Organ on Chip
Recent advances in microfabrication and microfluidics have enabled the development of microengineered models of human organs—known as organs-on-chips—that have the potential to provide platforms to model organ level responses for drug discovery in an in vitro setting.[191] Important cues involved in the pathogenesis of fibrosis, such as mechanical strain, fluid flow, and hydrostatic pressures, can be integrated in such models. Recently, a 3D-bioengineered pulmonary fibrotic (Eng-PF) tissue was developed utilizing silk collagen-I hydrogels seeded with pulmonary fibroblasts, airway epithelial cells, and microvascular endothelial cells. Eng-PF tissue was cultured under tension, tethered along the longitudinal axis of a bioreactor plate, and had the capacity to be cyclically strained with perfusion ability. Eng-PF tissues were able to model myofibroblast differentiation and permit evaluation of antifibrotic drugs, such as pirfenidone and nintedanib. Further, Eng-PF tissues were used to model epithelial injury with the addition of bleomycin and cellular recruitment by perfusion of cells through a hydrogel microchannel.[192] Unlike other 3D tissue systems, lung-on-chip platforms are standalone devices, therefore scaling these devices for high throughput antifibrotic drug screening could be challenging. Organ-on-chip technologies have also been advanced in the production of skin equivalents. The main advantages supported by microfluidics are the presence of fluid flow and fine control of the microenvironment, which yield improved epidermal morphogenesis and differentiation, and enhanced barrier function.[193] Although a skin-on-a-chip model consisting of epidermal, dermal and endothelial cells has been used to stimulate skin inflammation and edema,[194] a skin chip-model that replicate dermal fibrosis still needs to be developed.
8. Missing Players: Vasculature and Hematopoietic Immune Cells
Recent research has enabled the development of in vitro models which recapitulate different aspects of SSc, which could significantly contribute to early-stage drug discovery. However, current bioengineered systems often lack the presence of perfusable (micro-) vasculature and organ-specific immune responses, which need to be incorporated into the tissue models to achieve patho-physiologically relevant levels of function.
It is known that patients affected by SSc present a series of vascular abnormalities. Among them are direct damage of vascular and perivascular cells, abnormal vasoreactivity, hypoxia, impaired angiogenesis, and platelet dysfunction. These vascular changes result in decreased capillary blood flow, and subsequently in clinically overt symptoms such as Raynaud’s syndrome and fingertip ulcers.[195] In addition, vascular damage contributes to the production of a cascade of soluble mediators that ultimately influence the onset and progression of tissue fibrosis.[196]
Given the importance of the vasculature in SSc, the integration of vascular structures into in vitro cultured tissues is of paramount importance to provide realistic models of complex interactions, which modulates tissue remodeling and fibrosis in the tissue of interest.
One common strategy relies on the spontaneous organization of endothelial cells to form vascular networks within biomaterials scaffold or in multicellular assemblies.[197] Initially, endothelial cells form a primitive network within an avascular tissue, which is similar to vasculogenesis. In order to maturate into a functional vascular network with sufficient vascular cell viability and function, perfusion bioreactors are required to mimic in vivo like flow rate.[198] For example, a recent study focused on the development of vascularized skin equivalents as an advanced model of human skin with a fully polarized epidermal layer, a stratified dermis and a functional vascular system with physiological perfusion pressures.[199] These models were induced to undergo fibrotic transformation and resembled key features of SSc skin, with accumulation of ECM, fibroblast to myofibroblast transition and aberrant activation of TGF-β signaling. In addition, treatment with nintedanib in a pharmacologically relevant dose exerted antifibrotic effects in vascularized human skin equivalents by attenuating TGF-β signaling, reducing fibroblast to myofibroblast transition and decreasing ECM deposition.[199] By incorporating a mature vascular network, this platform provided a pathophysiologically relevant human setting for the evaluation of antifibrotic drugs, potentially improving predictive value.
Considering that the immune system plays a pivotal role in this disease,[200] another key element which is generally missing during the development of in vitro tissue analogues is the presence of relevant immune cells. Activation of both innate and adaptive immune responses leads to activation of fibroblasts, differentiation into myofibroblasts, ECM deposition and finally fibrosis. Monocytes, macrophages and dendritic cells all release soluble mediators that can directly affect fibroblast activation and tissue remodeling, or indirectly affect it by inducing the release of profibrotic factors by other cell types, including T cells and B cells.[18]
Using cocultures of immune cells with fibroblasts, a number of valuable in vitro models have tried to approximate the role of innate/adaptive immunity activation in fibroblasts differentiation and organ fibrosis. For example, it was demonstrated that coculture of SSc fibroblasts and SSc plasma-differentiated macrophages (expressing high levels of CCL2, IL-6, and TGF-β) using transwell plates resulted in activation of signaling pathways involved in the regulation of inflammation and fibrosis, suggesting that therapeutic targeting of these cells may be beneficial in ameliorating SSc progression.[201] Another study demonstrated that B cells cocultured with human dermal fibroblasts (HDFs) derived from SSc patients are potent inducers of IL-6, CCL2, and TGF-β1, which enhance collagen production by fibroblasts.[202] The integration of key immune cell subsets is likely a needed strategy to improve the relevance of SSc in vitro models. However, the complex mechanisms by which the immune system orchestrates organs and tissues are challenging to replicate, especially in prolonged culture conditions. New approaches such as “on-a-chip” platforms can incorporate multiple cell types under controlled biochemical and biophysical conditions contributing to the formation and progression of SSc.[203]
9. Challenges and Future Directions
Among autoimmune diseases, SSc is one of the most devastating pathologies, and its heterogeneity and complexity pose unique challenges for the discovery and development of effective therapeutic strategies. Although tremendous scientific progress has significantly increased the knowledge of the biological and molecular mechanism at the basis of SSc, numerous unanswered questions remain in the field of SSc therapeutics. Animal models represent indispensable for preclinical drug testing, even though none of these models faithfully recapitulates the full spectrum of SSc. For this reason, the development of in vitro models that replicate the complex and dynamic SSc milieu are essential to overcome some of the in vivo disease models’ shortcomings. As herein reviewed, a variety of biological, biochemical, and biophysical cues have integrated into in vitro platforms to mimic the pathophysiological signals typical of SSc, thereby improving their suitability for the identification and screening of effective drugs. However, in vitro models have not yet fully recovered the SSc phenotype to a level comparable with the human disease. Due to the complexity of SSc, it is likely unrealistic to create models that fully embrace all aspect of the pathology, including vascular component, immune system and organ fibrosis. Multifactorial approaches combining synergistic microenvironmental cues are one way forward to develop more complex in vitro systems. Advancements in biomaterial science will contribute to the development of new platforms that not only give structural dimensionality to the models, but will also modulate cell behavior by providing heterogeneous spatial organization and spatiotemporal controlled biological and mechanical signals.[204] Progress in iPSCs technologies have the potential to provide disease-relevant cells in a personalized manner and could facilitate the development of patient-specific SSc models for personalized medicine without requiring multiple tissue collections.[205]
Emerging technologies that go beyond well-established ex vivo assays for the characterization of fibrotic hallmarks (SDS-PAGE,[206] quantification of hydroxyproline content,[206] Sircol collagen assay,[207] histological and immunohistological analysis), not only help to better understand the biology of the disease, but also lead to improved assessment of the therapeutic effects of potential drug candidates. Examples include assessment of ECM structure and stiffness by second harmonic generation (SHG) microscopy[208] and atomic force microscopy,[145] as well as noninvasive imaging of tissue and organ damage[209] by magnetic resonance imaging (MRI),[210] computed tomography (CT),[211] ultrasound (US),[212] and positron emission tomography (PET).[213] These imaging techniques can be readily utilized to measure treatment responses over time without the need to sacrifice experimental animals, thereby facilitating the clinical translation of antifibrotic therapies.
Overall, despite the wide recognition of the utility and potential of 3D models in the drug development pipeline, there is not substantial evidence that such systems outweigh the data obtained from 2D models or the cost of testing in animals. Indeed, drug discovery programs are still far from being driven by primary screens in 3D and it is still premature to claim that 3D models improve the clinical success rates of drug candidates. In addition, the majority of models are conventionally not designed for automated analyses, being incompatible with high-content screening platforms, and thus are held back by routine applications into industrial settings. Therefore, future efforts should be directed not only to the standardization and validation of such models, but also to their miniaturization in order to enhance experimental efficiency with automation.
While further advances are needed, in vitro models of SSc represent promising platforms for disease modeling and for drug discovery, increasing our understanding of the mechanisms underlying this devastating disease.
Acknowledgements
This work is part of the SKINTEGRITY project under the umbrella of University Medicine Zurich/Hochschulmedizin Zürich. Financial support was received from Skintegrity, the Novartis Foundation (18B082) and the Balgrist Foundation. Dr. Korman is supported by NIH/NIAMS award K08 AR070285.
Biography

Andrea De Pieri received his B.Sc. in Health Biotechnologies and M.Sc. in Medical Biotechnologies from Universita degli Studi di Padova (Italy) in 2013 and 2015, respectively. He completed his Ph.D. in Biomedical Engineering at National University of Ireland Galway (Ireland) in 2020. He is currently a postdoctoral researcher in the Department of Biomedical Engineering at the Rochester Institute of Technology (RIT), USA. His current research interests include the development of electrospun-based 3D skin models for in vitro disease modeling.

Karin Wuertz-Kozak is a pharmacist (2003) with a Ph.D. in Human Biology (2006) and an MBA in Leadership and Sustainability (2015). She holds the position of Kate Gleason Endowed Full Professor of Biomedical Engineering at the Rochester Institute of Technology and leads the Tissue Regeneration and Mechanobiology Laboratory. Her research aims to understand the cellular mechanisms underlying musculoskeletal and skin pathologies for subsequent development of novel treatment options that allow for tissue regeneration by use of cells, biomaterials, biologics, genome engineering and/or mechanical cues.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Andrea De Pieri, Department of Biomedical Engineering, Rochester Institute of Technology (RIT), 106 Lomb Memorial Rd., Rochester, NY 14623, USA.
Benjamin D. Korman, Department of Medicine, Division of Allergy, Immunology and Rheumatology, University of Rochester Medical Center, Rochester, NY 14623, USA
Astrid Jüngel, Center of Experimental Rheumatology, University Clinic of Rheumatology, Balgrist University Hospital, University Hospital Zurich, Zurich 8008, Switzerland; Department of Physical Medicine and Rheumatology, Balgrist University Hospital, University of Zurich, Zurich 8008, Switzerland.
Karin Wuertz-Kozak, Department of Biomedical Engineering, Rochester Institute of Technology (RIT), 106 Lomb Memorial Rd., Rochester, nY 14623, USA; Schon Clinic Munich Harlaching, Spine Center, Academic Teaching Hospital and Spine Research Institute of the Paracelsus Medical University Salzburg (Austria), Munich 81547, Germany.
References
- [1].Gabrielli A, Avvedimento EV, Krieg T, N. Engl. J. Med 2009, 360, 1989. [DOI] [PubMed] [Google Scholar]
- [2].Denton CP, Ong VH, Nat. Rev. Rheumatol 2013, 9, 451. [DOI] [PubMed] [Google Scholar]
- [3].a) Distler O, Highland KB, Gahlemann M, Azuma A, Fischer A, Mayes MD, Raghu G, Sauter W, Girard M, Alves M, Clerisme-Beaty E, Stowasser S, Tetzlaff K, Kuwana M, Maher TM, N. Engl. J. Med 2019, 380, 2518; [DOI] [PubMed] [Google Scholar]; b) Kuwana M, Azuma A, Mod. Rheumatol 2020, 30, 225. [DOI] [PubMed] [Google Scholar]
- [4].Volkmann ER, Varga J, Nat. Rev. Rheumatol 2019, 15, 208. [DOI] [PubMed] [Google Scholar]
- [5].Gaspar D, Zeugolis DI, Drug Discovery Today 2016, 21, 1341. [DOI] [PubMed] [Google Scholar]
- [6].a) Green S, Nature 2008, 455, 460; [DOI] [PubMed] [Google Scholar]; b) Pusztai L, Hatzis C, Andre F, Nat. Rev. Clin. Oncol 2013, 10, 720; [DOI] [PubMed] [Google Scholar]; c) Frye SV, Arkin MR, Arrowsmith CH, Conn PJ, Glicksman MA, Hull-Ryde EA, Slusher BS, Nat. Rev. Drug Discovery 2015, 14, 733. [DOI] [PubMed] [Google Scholar]
- [7].a) Bergström CAS, Charman WN, Porter CJH, Adv. Drug Delivery Rev 2016, 101, 6; [DOI] [PubMed] [Google Scholar]; b) Nagy AL, Catoi C, Socaciu C, Pintea A, Oros NA, Coman C, Rugina D, Matea CT, Mocan T, Coccini T, De Simone U, De Angelis I, Bertero A, Sambuy Y, Caloni F, ALTEX 2018, 35, 420; [DOI] [PubMed] [Google Scholar]; c) Piñero J, Furlong LI, Sanz F, Curr. Opin. Pharmacol 2018, 42, 111; [DOI] [PubMed] [Google Scholar]; d) Jeong CG, Dal Negro G, Getsios S, Ekert JE, in Microfluidic Cell Culture Systems, 2nd ed., (Eds: Borenstein JT, Tandon V, Tao SL, Charest JL), Elsevier, Newbury Park, California: 2019, pp. 121–158. [Google Scholar]
- [8].Shingatgeri V, in In Vitro Toxicology (Eds: Dhawan A, Kwon S), Academic Press, San Diego, CA: 2018, pp. 187–207. [Google Scholar]
- [9].Clark RAF, McCoy GA, Folkvord JM, McPherson JM, J. Cell. Physiol 1997, 170, 69. [DOI] [PubMed] [Google Scholar]
- [10].Abraham DJ, Varga J, Trends Immunol. 2005, 26, 587. [DOI] [PubMed] [Google Scholar]
- [11].Justice BA, Badr NA, Felder RA, Drug Discovery Today 2009, 14, 102. [DOI] [PubMed] [Google Scholar]
- [12].Nam K-H, Smith AST, Lone S, Kwon S, Kim D-H, J. Lab. Autom 2014, 20, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].a) Denton CP, Khanna D, Lancet 2017, 390, 1685; [DOI] [PubMed] [Google Scholar]; b) Korman B, Transl. Res 2019, 209, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].a) Sinnathurai P, Schrieber L, Intern. Med. J 2013, 43, 476; [DOI] [PubMed] [Google Scholar]; b) Varga J, Hinchcliff M, Nat. Rev. Rheumatol 2014, 10, 200; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Moinzadeh P, Aberer E, Ahmadi-Simab K, Blank N, Distler JHW, Fierlbeck G, Genth E, Guenther C, Hein R, Henes J, Herich L, Herrgott I, Koetter I, Kreuter A, Krieg T, Kuhr K, Lorenz H-M, Meier F, Melchers I, Mensing H, Mueller-Ladner U, Pfeiffer C, Riemekasten G, Sárdy M, Schmalzing M, Sunderkoetter C, Susok L, Tarner IH, Vaith P, Worm M, Wozel G, Zeidler G, Hunzelmann N, Ann. Rheum. Dis 2015, 74, 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].a) Tyndall AJ, Bannert B, Vonk M, Airò P, Cozzi F, Carreira PE, Bancel DF, Allanore Y, Müller-Ladner U, Distler O, Iannone F, Pellerito R, Pileckyte M, Miniati I, Ananieva L, Gurman AB, Damjanov N, Mueller A, Valentini G, Riemekasten G, Tikly M, Hummers L, Henriques MJ, Caramaschi P, Scheja A, Rozman B, Ton E, Kumánovics G, Coleiro B, Feierl E, Szucs G, Von Mühlen CA, Riccieri V, Novak S, Chizzolini C, Kotulska A, Denton C, Coelho PC, Kötter I, Simsek I, de la Pena Lefebvre PG, Hachulla E, Seibold JR, Rednic S, Štork J, Morovic-Vergles J, Walker UA, Ann. Rheum. Dis 2010, 69, 1809; [DOI] [PubMed] [Google Scholar]; b) Katsumoto TR, Whitfield ML, Connolly MK, Annu. Rev. Pathol.: Mech. Dis 2011, 6, 509. [DOI] [PubMed] [Google Scholar]
- [16].Trojanowska M, Nat. Rev. Rheumatol 2010, 6, 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Solomon DH, Kavanaugh AJ, Schur PH, Arnett FC, Curr. Opin. Rheumatol 2006, 18, 579. [DOI] [PubMed] [Google Scholar]
- [18].Kania G, Rudnik M, Distler O, Nat. Rev. Rheumatol 2019, 15, 288. [DOI] [PubMed] [Google Scholar]
- [19].Bhattacharyya S, Wei J, Varga J, Nat. Rev. Rheumatol 2012, 8, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Varga J, Trojanowska M, Kuwana M, J. Scleroderma Relat. Disord 2017, 2, 137. [Google Scholar]
- [21].Allanore Y, Simms R, Distler O, Trojanowska M, Pope J, Denton CP, Varga J, Nat. Rev. Dis. Primers 2015, 1, 15002. [DOI] [PubMed] [Google Scholar]
- [22].Ho YY, Lagares D, Tager AM, Kapoor M, Nat. Rev. Rheumatol 2014, 10, 390. [DOI] [PubMed] [Google Scholar]
- [23].a) Hoyles RK, Derrett-Smith EC, Khan K, Shiwen X, Howat SL, Wells AU, Abraham DJ, Denton CP, Am J. Respir. Crit. Care Med 2011, 183, 249; [DOI] [PubMed] [Google Scholar]; b) Griffiths K, Habiel DM, Jaffar J, Binder U, Darby WG, Hosking CG, Skerra A, Westall GP, Hogaboam CM, Foley M, Sci. Rep 2018, 8, 3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Stone RC, Pastar I, Ojeh N, Chen V, Liu S, Garzon KI, Tomic-Canic M, Cell Tissue Res. 2016, 365, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Piera-Velazquez S, Li Z, Jimenez SA, Am. J. Pathol 2011, 179, 1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].a) Rajkumar VS, Sundberg C, Abraham DJ, Rubin K, Black CM, Arthritis Rheum. 1999, 42, 930; [DOI] [PubMed] [Google Scholar]; b) Rajkumar VS, Howell K, Csiszar K, Denton CP, Black CM, Abraham DJ, Arthritis Res. Ther 2005, 7, R1113; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ohgo S, Hasegawa S, Hasebe Y, Mizutani H, Nakata S, Akamatsu H, Exp. Dermatol 2013, 22, 769; [DOI] [PubMed] [Google Scholar]; d) Marangoni RG, Korman BD, Wei J, Wood TA, Graham LV, Whitfield ML, Scherer PE, Tourtellotte WG, Varga J, Arthritis Rheumatol. 2015, 67, 1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Maher TM, Strek ME, Respir. Res 2019, 20, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].a) Artlett CM, Open Access Rheumatol.: Res. Rev 2014, 6, 65; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Asano Y, J. Dermatol 2016, 43, 19. [DOI] [PubMed] [Google Scholar]
- [29].Baxter RM, Crowell TP, McCrann ME, Frew EM, Gardner H, Lab. Invest 2005, 85, 1199. [DOI] [PubMed] [Google Scholar]
- [30].Long KB, Li Z, Burgwin CM, Choe SG, Martyanov V, Sassi-Gaha S, Earl JP, Eutsey RA, Ahmed A, Ehrlich GD, Artlett CM, Whitfield ML, Blankenhorn EP, J. Invest. Dermatol 2015, 135, 718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].a) Christner PJ, Hitraya EG, Peters J, McGrath R, Jiménez SA, Arthritis Rheum. 1998, 41, 2132; [DOI] [PubMed] [Google Scholar]; b) Jimenez SA, Christner PJ, Curr. Opin. Rheumatol 2002, 14, 671. [DOI] [PubMed] [Google Scholar]
- [32].a) Maurer B, Busch N, Jüngel A, Pileckyte M, Gay RE, Michel BA, Schett G, Gay S, Distler J, Distler O, Circulation 2009, 120, 2367; [DOI] [PubMed] [Google Scholar]; b) Maurer B, Reich N, Juengel A, Kriegsmann J, Gay RE, Schett G, Michel BA, Gay S, Distler JHW, Distler O, Ann. Rheum. Dis 2012, 71, 1382; [DOI] [PubMed] [Google Scholar]; c) Birnhuber A, Biasin V, Schnoegl D, Marsh LM, Kwapiszewska G, Cell Signalling 2019, 64, 109408. [DOI] [PubMed] [Google Scholar]
- [33].Bell RD, White RJ, Garcia-Hernandez ML, Wu E, Rahimi H, Marangoni RG, Slattery P, Duemmel S, Nuzzo M, Huertas N, Yee M, O’Reilly MA, Morrell C, Ritchlin CT, Schwarz EM, Korman BD, Arthritis Rheumatol 2020, 72, 1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].a) Manetti M, Rosa I, Milia AF, Guiducci S, Carmeliet P, Ibba-Manneschi L, Matucci-Cerinic M, Ann. Rheum. Dis 2014, 73, 1700; [DOI] [PubMed] [Google Scholar]; b) Manetti M, Rosa I, Fazi M, Guiducci S, Carmeliet P, Ibba-Manneschi L, Matucci-Cerinic M, Ann. Rheum. Dis 2016, 75, 474. [DOI] [PubMed] [Google Scholar]
- [35].Moeller A, Ask K, Warburton D, Gauldie J, Kolb M, Int. J. Biochem. Cell Biol 2008, 40, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yamamoto T, Takagawa S, Katayama I, Yamazaki K, Hamazaki Y, Shinkai H, Nishioka K, J. Invest. Dermatol 1999, 112, 456. [DOI] [PubMed] [Google Scholar]
- [37].Phan SH, Thrall RS, Ward PA, Am. Rev. Respir. Dis 1980, 121, 501. [DOI] [PubMed] [Google Scholar]
- [38].Yamamoto T, Kuroda M, Nishioka K, Arch. Dermatol. Res 2000, 292, 535. [DOI] [PubMed] [Google Scholar]
- [39].a) Servettaz A, Goulvestre C, Kavian N, Nicco C, Guilpain P, Chéreau C, Vuiblet V, Guillevin L, Mouthon L, Weill B, Batteux F, J. Immunol 2009, 182, 5855; [DOI] [PubMed] [Google Scholar]; b) Batteux F, Kavian N, Servettaz A, Curr. Opin. Rheumatol 2011, 23, 511. [DOI] [PubMed] [Google Scholar]
- [40].Zaman MA, Oparil S, Calhoun DA, Nat. Rev. Drug Discovery 2002, 1, 621. [DOI] [PubMed] [Google Scholar]
- [41].Stawski L, Han R, Bujor AM, Trojanowska M, Arthritis Res. Ther 2012, 14, R194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Allanore Y, Matucci-Cerinic M, Distler O, RMD Open 2016, 2, e000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Graham ML, Prescott MJ, Eur. J. Pharmacol 2015, 759, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].a) Aringer M, Denton CP, J. Scleroderma Relat. Disord 2018. 3, 193; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sierra-Sepülveda A, Esquinca-González A, Benavides-Suárez SA, Sordo-Lima DE, Caballero-Islas AE, Cabral-Castañeda AR, Rodríguez-Reyna TS, Biomed. Res. Int 2019, 2019, 4569826; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chung MP, Chung L, Expert Opin. Invest. Drugs 2020, 29, 349; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Denton CP, Yee P, Ong VH, Eur. J. Rheumatol 2020, 7, S242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Khanna D, Denton CP, Lin CJF, van Laar JM, Frech TM, Anderson ME, Baron M, Chung L, Fierlbeck G, Lakshminarayanan S, Allanore Y, Pope JE, Riemekasten G, Steen V, Müller-Ladner U, Spotswood H, Burke L, Siegel J, Jahreis A, Furst DE, Ann. Rheum. Dis 2018, 77, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Spiera R, Hummers L, Chung L, Frech TM, Domsic R, Hsu V, Furst DE, Gordon J, Mayes M, Simms R, Lafyatis R, Martyanov V, Wood T, Whitfield ML, Constantine S, Lee E, Dgetluck N, White B, Arthritis Rheumatol. 2020, 72, 1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Khanna D, Lin CJF, Furst DE, Goldin J, Kim G, Kuwana M, Allanore Y, Matucci-Cerinic M, Distler O, Shima Y, van Laar JM, Spotswood H, Wagner B, Siegel J, Jahreis A, Denton CP, Lancet Respir. Med 2020, 8, 963.32866440 [Google Scholar]
- [48].Kelm JM, Lal-Nag M, Sittampalam GS, Ferrer M, Drug Discovery Today 2019, 24, 26. [DOI] [PubMed] [Google Scholar]
- [49].Bhadriraju K, Chen CS, Drug Discovery Today 2002, 7, 612. [DOI] [PubMed] [Google Scholar]
- [50].Kawakami T, Ihn H, Xu W, Smith E, LeRoy C, Trojanowska M, J. Invest. Dermatol 1998, 110, 47. [DOI] [PubMed] [Google Scholar]
- [51].a) Ihn H, Yamane K, Kubo M, Tamaki K, Arthritis Rheum. 2001, 44, 474; [DOI] [PubMed] [Google Scholar]; b) Qi Q, Guo Q, Tan G, Mao Y, Tang H, Zhou C, Zeng F, J. Eur. Acad. Dermatol. Venereol 2009, 23, 160; [DOI] [PubMed] [Google Scholar]; c) Usategui A, del Rey MJ, Pablos JL, Expert Rev. Clin. Immunol 2011, 7, 491. [DOI] [PubMed] [Google Scholar]
- [52].Vuorio T, Mäkelä JK, Vuorio E, J. Cell. Biochem 1985, 28, 105. [DOI] [PubMed] [Google Scholar]
- [53].Chadli L, Sotthewes B, Li K, Andersen SN, Cahir-McFarland E, Cheung M, Cullen P, Dorjée A, de Vries-Bouwstra JK, Huizinga TWJ, Fischer DF, DeGroot J, Viney JL, Zheng TS, Aarbiou J, Gardet A, Sci. Rep 2019, 9, 4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Gardner H, Shearstone JR, Bandaru R, Crowell T, Lynes M, Trojanowska M, Pannu J, Smith E, Jablonska S, Blaszczyk M, Tan FK, Mayes MD, Arthritis Rheum. 2006, 54, 1961. [DOI] [PubMed] [Google Scholar]
- [55].Varga J, Pasche B, Nat. Rev. Rheumatol 2009, 5, 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].a) Rahimi RA, Leof EB, J. Cell. Biochem 2007, 102, 593; [DOI] [PubMed] [Google Scholar]; b) Hanna A, Frangogiannis NG, Front. Cardiovasc. Med 2019, 6, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lafyatis R, Nat. Rev. Rheumatol 2014, 10, 706. [DOI] [PubMed] [Google Scholar]
- [58].Biernacka A, Dobaczewski M, Frangogiannis NG, Groundwater 2011, 29, 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].a) Leask A, Abraham DJ, FASEB J. 2004, 18, 816; [DOI] [PubMed] [Google Scholar]; b) Schiller M, Javelaud D, Mauviel A, J. Dermatol. Sci 2004, 35, 83; [DOI] [PubMed] [Google Scholar]; c) Meng X-M, Nikolic-Paterson DJ, Lan HY, Nat. Rev. Nephrol 2016, 12, 325. [DOI] [PubMed] [Google Scholar]
- [60].Salton F, Volpe MC, Confalonieri M, Medicina 2019, 55, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hummers LK, Hall A, Wigley FM, Simons M, J. Rheumatol 2009, 36, 576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Vancheeswaran R, Magoulas T, Efrat G, Wheeler-Jones C, Olsen I, Penny R, Black CM, J. Rheumatol 1994, 21, 1838. [PubMed] [Google Scholar]
- [63].a) Roberts AB, Sporn MB, Assoian RK, Smith JM, Roche NS, Wakefield LM, Heine UI, Liotta LA, Falanga V, Kehrl JH, Proc. Natl. Acad. Sci. USA 1986, 83, 4167; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Appling WD, O’Brien WR, Johnston DA, Duvic M, FEBS Lett. 1989, 250, 541. [DOI] [PubMed] [Google Scholar]
- [64].Hall CL, Wells AR, Leung KP, Lab Invest. 2018, 98, 640. [DOI] [PubMed] [Google Scholar]
- [65].a) Porras AM, Hutson HN, Berger AJ, Masters KS, Curr. Opin. Biotechnol 2016, 40, 24; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Robertson IB, Rifkin DB, Cold Spring Harb Perspect Biol. 2016, 8, a021907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].a) Denton CP, Abraham DJ, Curr. Opin. Rheumatol 2001, 13, 505; [DOI] [PubMed] [Google Scholar]; b) Leask A, Denton CP, Abraham DJ, J. Invest. Dermatol 2004, 122, 1; [DOI] [PubMed] [Google Scholar]; c) Ramazani Y, Knops N, Elmonem MA, Nguyen TQ, Arcolino FO, van den Heuvel L, Levtchenko E, Kuypers D, R. Goldschmeding, Matrix Biol 2018, 68-69, 44. [DOI] [PubMed] [Google Scholar]
- [67].Serratì S, Chillà A, Laurenzana A, Margheri F, Giannoni E, Magnelli L, Chiarugi P, Dotor J, Feijoo E, Bazzichi L, Bombardieri S, Kahaleh B, Fibbi G, Del Rosso M, Arthritis Rheum. 2013, 65, 258. [DOI] [PubMed] [Google Scholar]
- [68].Frazier K, Williams S, Kothapalli D, Klapper H, Grotendorst GR, J. Invest. Dermatol 1996, 107, 404. [DOI] [PubMed] [Google Scholar]
- [69].Kang S, Kim J, Ahn M, Kim J, Heo M-G, Min D-H, Won C, Nanoscale 2020, 12, 6385. [DOI] [PubMed] [Google Scholar]
- [70].a) Svegliati Baroni S, Santillo M, Bevilacqua F, Luchetti M, Spadoni T, Mancini M, Fraticelli P, Sambo P, Funaro A, Kazlauskas A, Avvedimento EV, Gabrielli A, N. Engl. J. Med 2006, 354, 2667; [DOI] [PubMed] [Google Scholar]; b) Dragun D, Distler JHW, Riemekasten G, Distler O, Arthritis Rheum. 2009, 60, 907; [DOI] [PubMed] [Google Scholar]; c) Iwayama T, Olson LE, Curr. Rheumatol. Rep 2013, 15, 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Jinnin M, Ihn H, Mimura Y, Asano Y, Yamane K, Tamaki K, J. Cell. Physiol 2005, 202, 510. [DOI] [PubMed] [Google Scholar]
- [72].Shi-wen X, Kennedy L, Renzoni EA, Bou-Gharios G, du Bois RM, Black CM, Denton CP, Abraham DJ, Leask A, Arthritis Rheum. 2007, 56, 4189. [DOI] [PubMed] [Google Scholar]
- [73].Jing J, Dou TT, Yang JQ, Chen XB, Cao HL, Min M, Cai SQ, Zheng M, Man XY, Eur. Cytokine Network 2015, 26, 10. [DOI] [PubMed] [Google Scholar]
- [74].Cipriani P, Di Benedetto P, Ruscitti P, Capece D, Zazzeroni F, Liakouli V, Pantano I, Berardicurti O, Carubbi F, Pecetti G, Turricchia S, Alesse E, Iglarz M, Giacomelli R, J. Rheumatol 2015, 42, 1808. [DOI] [PubMed] [Google Scholar]
- [75].Dantas AT, Almeida A. R. d., Sampaio MCPD, Cordeiro MF, Oliveira P. S. S. d., Mariz H. d. A., Pereira MC, Rego M. J. B. d. M., Pitta I. d. R., Duarte ALBP, Pitta M. G. d. R., Immunol. Lett 2018, 198, 12. [DOI] [PubMed] [Google Scholar]
- [76].Kawaguchi Y, Hara M, Wright TM, J. Clin. Invest 1999, 103, 1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Choy E, Rose-John S, J. Scleroderma Relat. Disord 2017, 2, S1. [Google Scholar]
- [78].a) Richards CD, Kerr C, Tanaka M, Hara T, Miyajima A, Pennica D, Botelho F, Langdon CM, J. Immunol 1997, 159, 2431; [PubMed] [Google Scholar]; b) Gallucci RM, Lee EG, Tomasek JJ, J. Invest. Dermatol 2006, 126, 561; [DOI] [PubMed] [Google Scholar]; c) Kawaguchi Y, J. Scleroderma Relat. Disord 2017, 2, S6. [Google Scholar]
- [79].Denton CP, Ong VH, Xu S, Chen-Harris H, Modrusan Z, Lafyatis R, Khanna D, Jahreis A, Siegel J, Sornasse T, Ann. Rheum. Dis 2018, 77, 1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Atamas SP, White B, Clin. Diagn. Lab. Immunol 1999, 6, 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Salmon-Ehr V, Serpier H, Nawrocki B, Gillery P, Clavel C, Kalis B, Birembaut P, Maquart F-X, Arch. Dermatol 1996, 132, 802. [PubMed] [Google Scholar]
- [82].Gasparini G, Cozzani E, Parodi A, Cytokine 2020, 125, 154799. [DOI] [PubMed] [Google Scholar]
- [83].Fuschiotti P, Larregina AT, Ho J, Feghali-Bostwick C, Medsger TA Jr, Arthritis Rheum. 2013, 65, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Chizzolini C, Dufour AM, Brembilla NC, Immunol. Lett 2018, 195, 61. [DOI] [PubMed] [Google Scholar]
- [85].Kurasawa K, Hirose K, Sano H, Endo H, Shinkai H, Nawata Y, Takabayashi K, Iwamoto I, Arthritis Rheum. 2000, 43, 2455. [DOI] [PubMed] [Google Scholar]
- [86].Hasegawa M, Fujimoto M, Kikuchi K, Takehara K, J. Rheumatol 1997, 24, 663. [PubMed] [Google Scholar]
- [87].Antonelli A, Ferri C, Ferrari SM, Mancusi C, Colaci M, Sebastiani M, Fallahi P, Scand. J. Rheumatol 2011, 40, 453. [DOI] [PubMed] [Google Scholar]
- [88].Wang Z, Wang Z, Lu WW, Zhen W, Yang D, Peng S, NPG Asia Mater. 2017, 9, e435. [Google Scholar]
- [89].a) Dessels C, Potgieter M, Pepper MS, Front. Cell Dev. Biol 2016, 4, 115; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) van der Valk J, Bieback K, Buta C, Cochrane B, Dirks WG, Fu J, Hickman JJ, Hohensee C, Kolar R, Liebsch M, Pistollato F, Schulz M, Thieme D, Weber T, Wiest J, Winkler S, Gstraunthaler G, ALTEX 2018, 35, 99. [DOI] [PubMed] [Google Scholar]
- [90].Gstraunthaler G, Lindl T, van der Valk J, Cytotechnology 2013, 65, 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Kim MM, Audet J, Commun. Biol 2019, 2, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].a) Kocaoemer A, Kern S, Klüter H, Bieback K, Stem Cells 2007, 25, 1270; [DOI] [PubMed] [Google Scholar]; b) Savelli S, Trombi L, D’Alessandro D, Moscato S, Pacini S, Giannotti S, Lapi S, Scatena F, Petrini M, Cytotherapy 2018, 20, 556. [DOI] [PubMed] [Google Scholar]
- [93].Mazlyzam AL, Aminuddin BS, Saim L, Ruszymah BHI, Arch. Med. Res 2008, 39, 743. [DOI] [PubMed] [Google Scholar]
- [94].a) Hasegawa M, Fujimoto M, Kikuchi K, Takehara K, J. Rheumatol 1997, 24, 328; [PubMed] [Google Scholar]; b) Sato S, Nagaoka T, Hasegawa M, Tamatani T, Nakanishi T, Takigawa M, Takehara K, J. Rheumatol 2000, 27, 149; [PubMed] [Google Scholar]; c) Fineschi S, Goffin L, Rezzonico R, Cozzi F, Dayer J-M, Meroni PL, Chizzolini C, Arthritis Rheum. 2008, 58, 3913; [DOI] [PubMed] [Google Scholar]; d) Horn A, Palumbo K, Cordazzo C, Dees C, Akhmetshina A, Tomcik M, Zerr P, Avouac J, Gusinde J, Zwerina J, Roudaut H, Traiffort E, Ruat M, Distler O, Schett G, Distler JHW, Arthritis Rheum. 2012, 64, 2724; [DOI] [PubMed] [Google Scholar]; e) O’Reilly S, Hügle T, van Laar JM, Rheumatology 2012, 51, 1540; [DOI] [PubMed] [Google Scholar]; f) Beyer C, Huscher D, Ramming A, Bergmann C, Avouac J, Guiducci S, Meier F, Vettori S, Siegert E, Jaeger VK, Maurer B, Riemekasten G, Walker U, Müller-Ladner U, Valentini G, Matucci-Cerinic M, Allanore Y, Distler O, Schett G, Distler JHW, Ann. Rheum. Dis 2018, 77, 626; [DOI] [PubMed] [Google Scholar]; g) Burbelo PD, Gordon SM, Waldman M, Edison JD, Little DJ, Stitt RS, Bailey WT, Hughes JB, Olson SW, PLoS One 2019, 14, e0214202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Murgia F, Svegliati S, Poddighe S, Lussu M, Manzin A, Spadoni T, Fischetti C, Gabrielli A, Atzori L, Sci. Rep 2018, 8, 7626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Raschi E, Chighizola CB, Cesana L, Privitera D, Ingegnoli F, Mastaglio C, Meroni PL, Borghi MO, Arthritis Res. Ther 2018, 20, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Arts MR, Baron M, Chokr N, Fritzler MJ, G. the Canadian Scleroderma Research, Servant MJ, PLoS One 2014, 9, e100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Servettaz A, Guilpain P, Goulvestre C, Chéreau C, Hercend C, Nicco C, Guillevin L, Weill B, Mouthon L, Batteux F, Ann. Rheum. Dis 2007, 66, 1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Manetti M, Romano E, Rosa I, Guiducci S, Bellando-Randone S, De Paulis A, Ibba-Manneschi L, Matucci-Cerinic M, Ann. Rheum. Dis 2017, 76, 924. [DOI] [PubMed] [Google Scholar]
- [100].a) Bogdanowicz DR, Lu HH, Biotechnol. J 2013, 8, 395; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Paschos NK, Brown WE, Eswaramoorthy R, Hu JC, Athanasiou KA, J. Tissue Eng. Regener. Med 2015, 9, 488. [DOI] [PubMed] [Google Scholar]
- [101].Aden N, Nuttall A, Shiwen X, de Winter P, Leask A, Black CM, Denton CP, Abraham DJ, Stratton RJ, J. Invest. Dermatol 2010, 130, 2191. [DOI] [PubMed] [Google Scholar]
- [102].Liakouli V, Elies J, El-Sherbiny YM, Scarcia M, Grant G, Abignano G, Derrett-Smith EC, Esteves F, Cipriani P, Emery P, Denton CP, Giacomelli R, Mavria G, Del Galdo F, Ann. Rheum. Dis 2018, 77, 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].a) Silverstein JL, Steen VD, Medsger TA, Falanga V Jr, Arch. Dermatol 1988, 124, 1379; [PubMed] [Google Scholar]; b) Beyer C, Schett G, Gay S, Distler O, Distler JHW, Arthritis Res. Ther 2009, 11, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Distler O, Distler JHW, Scheid A, Acker T, Hirth A, Rethage J, Michel BA, Gay RE, Müller-Ladner U, Matucci-Cerinic M, Plate KH, Gassmann M, Gay S, Circ. Res 2004, 95, 109. [DOI] [PubMed] [Google Scholar]
- [105].a) Koyasu S, Kobayashi M, Goto Y, Hiraoka M, Harada H, Cancer Sci. 2018, 109, 560; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lee JW, Ko J, Ju C, Eltzschig HK, Exp. Mol. Med 2019, 51, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Distler JHW, Gay S, Distler O, Rheumatology 2006, 45, iii26. [DOI] [PubMed] [Google Scholar]
- [107].a) Lokmic Z, Musyoka J, Hewitson TD, Darby IA, Int. Rev. Cell Mol. Biol 2012, 296, 139; [DOI] [PubMed] [Google Scholar]; b) Darby IA, Hewitson TD, Cell Tissue Res. 2016, 365, 553. [DOI] [PubMed] [Google Scholar]
- [108].a) Drake CJ, Little CD, Proc. Natl. Acad. Sci. USA 1995, 92, 7657; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Corpechot C, Barbu V, Wendum D, Kinnman N, Rey C, Poupon R, Housset C, Rosmorduc O, Hepatology 2002, 35, 1010; [DOI] [PubMed] [Google Scholar]; c) Xiong A, Liu Y, Front. Pharmacol 2017, 8, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Distler JHW, Jüngel A, Pileckyte M, Zwerina J, Michel BA, Gay RE, Kowal-Bielecka O, Matucci-Cerinic M, Schett G, Marti HH, Gay S, Distler O, Arthritis Rheum. 2007, 56, 4203. [DOI] [PubMed] [Google Scholar]
- [110].Hong K-H, Yoo S-A, Kang S-S, Choi J-J, Kim W-U, Cho C-S, Clin. Exp. Immunol 2006, 146, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Zhou G, Dada LA, Wu M, Kelly A, Trejo H, Zhou Q, Varga J, Sznajder JI, Am. J. Physiol.: Lung Cell. Mol. Physiol 2009, 297, L1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Zhou X, Liu C, Lu J, Zhu L, Li M, Rheumatology 2018, 57, 1675. [DOI] [PubMed] [Google Scholar]
- [113].Yamamoto T, in Systems Biology of Free Radicals and Antioxidants (Ed: Laher I), Springer, Berlin: 2014; pp. 3737–3752. [Google Scholar]
- [114].Thuan DTB, Zayed H, Eid AH, Abou-Saleh H, Nasrallah GK, Mangoni AA, Pintus G, Front. Immunol 2018, 9, 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Luo J-Y, Liu X, Jiang M, Zhao H-P, Zhao J-J, Mod. Rheumatol 2017, 27, 306. [DOI] [PubMed] [Google Scholar]
- [116].a) Grygiel-Górniak B, Puszczewicz M, Mediators Inflammation 2014, 2014, 389582; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Abdulle AE, Diercks GFH, Feelisch M, Mulder DJ, Goor H. v., Front. Physiol 2018, 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Thuan DTB, Zayed H, Eid AH, Abou-Saleh H, Nasrallah GK, Mangoni AA, Pintus G, Front. Immunol 2018, 9, 1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Doridot L, Jeljeli M, Chêne C, Batteux F, Redox Biol. 2019, 25, 101122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Sambo P, Baroni SS, Luchetti M, Paroncini P, Dusi S, Orlandini G, Gabrielli A, Arthritis Rheum. 2001, 44, 2653. [DOI] [PubMed] [Google Scholar]
- [120].Spadoni T, Svegliati Baroni S, Amico D, Albani L, Moroncini G, Avvedimento EV, Gabrielli A, Arthritis Rheumatol. 2015, 67, 1611. [DOI] [PubMed] [Google Scholar]
- [121].Bedard K, Krause K-H, Physiol. Rev 2007, 87, 245. [DOI] [PubMed] [Google Scholar]
- [122].Dooley A, Shi-Wen X, Aden N, Tranah T, Desai N, Denton CP, Abraham DJ, Bruckdorfer R, Rheumatology 2010, 49, 2024. [DOI] [PubMed] [Google Scholar]
- [123].Sekiguchi A, Motegi S-I, Fujiwara C, Yamazaki S, Inoue Y, Uchiyama A, Akai R, Iwawaki T, Ishikawa O, J. Dermatol. Sci 2019, 96, 8. [DOI] [PubMed] [Google Scholar]
- [124].Ammendola R, Ruocchio MR, Chirico G, Russo L, De Felice C, Esposito F, Russo T, Cimino F, Arch. Biochem. Biophys 2002, 397, 253. [DOI] [PubMed] [Google Scholar]
- [125].Eckes B, Wang F, Moinzadeh P, Hunzelmann N, Krieg T, Front. Med. (Lausanne) 2017, 4, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].a) Wells RG, Discher DE, Sci. Signaling 2008, 1, p e13; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Santos A, Lagares D, Curr. Rheumatol. Rep 2018, 20, 2. [DOI] [PubMed] [Google Scholar]
- [127].a) Wells RG, Biochim. Biophys. Acta, Mol. Basis Dis 2013, 1832, 884; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Guimarães CF, Gasperini L, Marques AP, Reis RL, Nat. Rev. Mater 2020, 5, 351. [Google Scholar]
- [128].a) Liu F, Lagares D, Choi KM, Stopfer L, Marinković A, Vrbanac V, Probst CK, Hiemer SE, Sisson TH, Horowitz JC, Rosas IO, Fredenburgh LE, Feghali-Bostwick C, Varelas X, Tager AM, Tschumperlin DJ, Am. J. Physiol.: Lung Cell. Mol. Physiol 2015, 308, L344; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Herrera J, Henke CA, Bitterman PB, J. Clin. Invest 2018, 128, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Liu F, Mih JD, Shea BS, Kho AT, Sharif AS, Tager AM, Tschumperlin DJ, J. Cell Biol 2010, 190, 693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Wipff P-J, Rifkin DB, Meister J-J, Hinz B, J. Cell Biol 2007, 179, 1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Hinz B, Matrix Biol. 2015, 47, 54. [DOI] [PubMed] [Google Scholar]
- [132].a) Munger JS, Sheppard D, Cold Spring Harbor Perspect. Biol 2011, 3, a005017; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Henderson NC, Arnold TD, Katamura Y, Giacomini MM, Rodriguez JD, McCarty JH, Pellicoro A, Raschperger E, Betsholtz C, Ruminski PG, Griggs DW, Prinsen MJ, Maher JJ, Iredale JP, Lacy-Hulbert A, Adams RH, Sheppard D, Nat. Med 2013, 19, 1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Hinz B, Curr. Rheumatol. Rep 2009, 11, 120. [DOI] [PubMed] [Google Scholar]
- [134].Arora PD, Narani N, McCulloch CA, Am. J. Pathol 1999, 154, 871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Huang X, Yang N, Fiore VF, Barker TH, Sun Y, Morris SW, Ding Q, Thannickal VJ, Zhou Y, Am. J. Respir. Cell Mol. Biol 2012, 47, 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Sharma S, Goswami R, Merth M, Cohen J, Lei KY, Zhang DX, Rahaman SO, Am. J. Physiol. Cell Physiol 2017, 312, C562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Wong VW, Rustad KC, Akaishi S, Sorkin M, Glotzbach JP, Januszyk M, Nelson ER, Levi K, Paterno J, Vial IN, Kuang AA, Longaker MT, Gurtner GC, Nat. Med 2012, 18, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Jiang C, Huang H, Liu J, Wang Y, Lu Z, Xu Z, Int. J. Mol. Sci 2012, 13, 8293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Toyama T, Looney AP, Baker BM, Stawski L, Haines P, Simms R, Szymaniak AD, Varelas X, Trojanowska M, J. Invest. Dermatol 2018, 138, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].a) Curtis A, Wilkinson C, Biomaterials 1997, 18, 1573; [DOI] [PubMed] [Google Scholar]; b) Park J, Kim D-H, Levchenko A, Biophys. J 2018, 114, 1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].a) Ross AM, Jiang Z, Bastmeyer M, Lahann J, Small 2012, 8, 336; [DOI] [PubMed] [Google Scholar]; b) Sousa MP, Caridade SG, Mano JF, Adv. Healthcare Mater 2017, 6, 1601462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].a) de Vries HJC, Enomoto DNH, van Marle J, van Zuijlen PPM, Mekkes JR, Bos JD, Lab. Invest 2000, 80, 1281; [DOI] [PubMed] [Google Scholar]; b) Strupler M, Pena AM, Hernest M, Tharaux PL, Martin JL, Beaurepaire E, Schanne-Klein MC, Opt. Express 2007, 15, 4054. [DOI] [PubMed] [Google Scholar]
- [143].Chen G, Xia B, Fu Q, Huang X, Wang F, Chen Z, Lv Y, Int. J. Biol. Sci 2019, 15, 2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Coelho NM, Wang A, McCulloch CA, Biochim. Biophys. Acta, Mol. Basis Dis 2019, 1866, 118510. [DOI] [PubMed] [Google Scholar]
- [145].Achterberg VF, Buscemi L, Diekmann H, Smith-Clerc J, Schwengler H, Meister J-J, Wenck H, Gallinat S, Hinz B, J. Invest. Dermatol 2014, 134, 1862. [DOI] [PubMed] [Google Scholar]
- [146].a) Kamada A, Levin A, Toprakcioglu Z, Shen Y, Lutz-Bueno V, Baumann KN, Mohammadi P, Linder MB, Mezzenga R, Knowles TPJ, Small 2020, 16, 1904190; [DOI] [PubMed] [Google Scholar]; b) Liu Z, Zhang Z, Ritchie RO, Adv. Funct. Mater 2020, 30, 1908121. [Google Scholar]
- [147].Cao L, Lafyatis R, Burkly LC, PLoS One 2017, 12, e0180751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Ahmed Abdi B, Lopez H, Karrar S, Renzoni E, Wells A, Tam A, Etomi O, Hsuan JJ, Martin GR, Shiwen X, Denton CP, Abraham D, Stratton R, Sci. Rep 2017, 7, 2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Seo BR, Chen X, Ling L, Song YH, Shimpi AA, Choi S, Gonzalez J, Sapudom J, Wang K, Andresen Eguiluz RC, Gourdon D, Shenoy VB, Fischbach C, Proc. Natl. Acad. Sci. USA 2020, 117, 11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].a) Murad S, Grove D, Lindberg KA, Reynolds G, Sivarajah A, Pinnell SR, Proc. Natl. Acad. Sci. USA 1981, 78, 2879; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sorushanova A, Delgado LM, Wu Z, Shologu N, Kshirsagar A, Raghunath R, Mullen AM, Bayon Y, Pandit A, Raghunath M, Zeugolis DI, Adv. Mater 2019, 31, 1801651. [DOI] [PubMed] [Google Scholar]
- [151].Kumar P, Satyam A, Fan X, Collin E, Rochev Y, Rodriguez BJ, Gorelov A, Dillon S, Joshi L, Raghunath M, Pandit A, Zeugolis DI, Sci. Rep 2015, 5, 8729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Satyam A, Kumar P, Fan X, Gorelov A, Rochev Y, Joshi L, Peinado H, Lyden D, Thomas B, Rodriguez B, Raghunath M, Pandit A, Zeugolis D, Adv. Mater 2014, 26, 3024. [DOI] [PubMed] [Google Scholar]
- [153].a) Cigognini D, Gaspar D, Kumar P, Satyam A, Alagesan S, Sanz-Nogués C, Griffin M, O’Brien T, Pandit A, Zeugolis DI, Sci. Rep 2016, 6, 30746; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gaspar D, Fuller KP, Zeugolis DI, Acta Biomater. 2019, 88, 197; [DOI] [PubMed] [Google Scholar]; c) De Pieri A, Rana S, Korntner S, Zeugolis DI, Int. J. Biol. Macromol 2020, 164, 434. [DOI] [PubMed] [Google Scholar]
- [154].a) Lareu RR, Subramhanya KH, Peng Y, Benny P, Chen C, Wang Z, Rajagopalan R, Raghunath M, FEBS Lett. 2007, 581, 2709; [DOI] [PubMed] [Google Scholar]; b) Chen C, Loe F, Blocki A, Peng Y, Raghunath M, Adv. Drug Delivery Rev 2011, 63, 277. [DOI] [PubMed] [Google Scholar]
- [155].Badowski C, Iskander A, Gaspar D, Zeugolis DI, Raghunath M, in Cell Engineering and Regeneration (Eds: Gimble JM, Marolt D, Oreffo R, Redl H, Wolbank S), Springer International Publishing, Cham, Switzerland: 2018, pp. 1–27. [Google Scholar]
- [156].Chen CZC, Peng YX, Wang ZB, Fish PV, Kaar JL, Koepsel RR, Russell AJ, Lareu RR, Raghunath M, Br. J. Pharmacol 2009, 158, 1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Fan C, Lim LKP, Loh SQ, Ying Lim KY, Upton Z, Leavesley D, Chem.-Biol. Interact 2019, 310, 108747. [DOI] [PubMed] [Google Scholar]
- [158].Good RB, Eley JD, Gower E, Butt G, Blanchard AD, Fisher AJ, Nanthakumar CB, BMC Biomed. Eng 2019, 1, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Ellis RJ, Trends Biochem. Sci 2001, 26, 597. [DOI] [PubMed] [Google Scholar]
- [160].a) Fang Y, Eglen RM, SLAS Discovery 2017, 22, 456; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Langhans SA, Front. Pharmacol 2018, 9, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].a) Knight E, Przyborski S, J. Anat 2015, 227, 746; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ravi M, Paramesh V, Kaviya SR, Anuradha E, Solomon FDP, J. Cell. Physiol 2015, 230, 16. [DOI] [PubMed] [Google Scholar]
- [162].Chaicharoenaudomrung N, Kunhorm P, Noisa P, World J Stem Cells 2019, 11, 1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].a) Hussey GS, Dziki JL, Badylak SF, Nat. Rev. Mater 2018, 3, 159; [Google Scholar]; b) Hippler M, Lemma ED, Bertels S, Blasco E, Barner-Kowollik C, Wegener M, Bastmeyer M, Adv. Mater 2019, 31, 1808110. [DOI] [PubMed] [Google Scholar]
- [164].Gaspar VM, Lavrador P, Borges J, Oliveira MB, Mano JF, Adv. Mater 2020, 32, 1903975. [DOI] [PubMed] [Google Scholar]
- [165].Ouyang L, Armstrong JPK, Salmeron-Sanchez M, Stevens MM, Adv. Funct. Mater 2020, 30, 1909009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].a) Fennema E, Rivron N, Rouwkema J, van Blitterswijk C, de Boer J, Trends Biotechnol. 2013, 31, 108; [DOI] [PubMed] [Google Scholar]; b) Laschke MW, Menger MD, Trends Biotechnol. 2017, 35, 133. [DOI] [PubMed] [Google Scholar]
- [167].Tan Y, Suarez A, Garza M, Khan AA, Elisseeff J, Coon D, Biomater. Sci 2020, 8, 1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].a) Lancaster MA, Knoblich JA, Science 2014, 345, 1247125; [DOI] [PubMed] [Google Scholar]; b) Dutta D, Heo I, Clevers H, Trends Mol. Med 2017, 23, 393. [DOI] [PubMed] [Google Scholar]
- [169].Kim S, Cho A-N, Min S, Kim S, Cho S-W, Adv. Ther 2019, 2, 1800087. [Google Scholar]
- [170].Perelas A, Silver RM, Arrossi AV, Highland KB, Lancet Respir. Med 2020, 8, 304. [DOI] [PubMed] [Google Scholar]
- [171].Wilkinson DC, Alva-Ornelas JA, Sucre JMS, Vijayaraj P, Durra A, Richardson W, Jonas SJ, Paul MK, Karumbayaram S, Dunn B, Gomperts BN, Curr. Protoc. Stem Cell Biol 2017, 6, 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Kikuchi T, Shimizu T, Wada M, Yamato M, Okano T, Biomaterials 2014, 35, 2428. [DOI] [PubMed] [Google Scholar]
- [173].Corriveau M-P, Boufaied I, Lessard J, Chabaud S, Senécal J-L, Grodzicky T, Chartier S, Raymond Y, Moulin VJ, J. Pathol 2009, 217, 534. [DOI] [PubMed] [Google Scholar]
- [174].Wang X, Liu Y, Deng Z, Dong R, Liu Y, Hu S, Li Y, Jin Y, J. Dermatol. Sci 2009, 53, 103. [DOI] [PubMed] [Google Scholar]
- [175].Randall MJ, Jüngel A, Rimann M, Wuertz-Kozak K, Front. Bioeng. Biotechnol 2018, 6, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Freed LE, EngelmayrJr GC, Borenstein JT, Moutos FT, Guilak F, Adv. Mater 2009, 21, 3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Pereira RF, Barrias CC, Granja PL, Bartolo PJ, Nanomedicine 2013, 8, 603. [DOI] [PubMed] [Google Scholar]
- [178].Huang M, Cai G, Baugh LM, Liu Z, Smith A, Watson M, Popovich D, Zhang T, Stawski LS, Trojanowska M, Georgakoudi I, BlackIII LD, Pioli PA, Whitfield ML, Garlick J, Arthritis Rheumatol. 2020, 72, 791. [DOI] [PubMed] [Google Scholar]
- [179].Luchetti MM, Moroncini G, Jose Escamez M, Svegliati Baroni S, Spadoni T, Grieco A, Paolini C, Funaro A, Avvedimento EV, Larcher F, Del Rio M, Gabrielli A, Arthritis Rheumatol. 2016, 68, 2263. [DOI] [PubMed] [Google Scholar]
- [180].a) Murry CE, Keller G, Cell 2008, 132, 661; [DOI] [PubMed] [Google Scholar]; b) Pessôa L. V. d. F., Bressan FF, Freude KK, World J. Stem Cells 2019, 11, 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S, Cell 2007, 131, 861. [DOI] [PubMed] [Google Scholar]
- [182].Kim Y, Park N, Rim YA, Nam Y, Jung H, Lee K, Ju JH, Stem Cell Res. Ther 2018, 9, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].a) Fuller K, Pandit A, Zeugolis DI, Pharm. Nanotechnol 2014, 2, 23; [Google Scholar]; b) Ding Y, Li W, Zhang F, Liu Z, Ezazi N. Zanjanizadeh, Liu D, Santos HA, Adv. Funct. Mater 2019, 29, 1802852. [Google Scholar]
- [184].Fischer SN, Johnson JK, Baran CP, Newland CA, Marsh CB, Lannutti JJ, Biomaterials 2011, 32, 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Li Y, Xiao Z, Zhou Y, Zheng S, An Y, Huang W, He H, Yang Y, Li S, Chen Y, Xiao J, Wu J, ACS Appl. Mater. Interfaces 2019, 11, 28377. [DOI] [PubMed] [Google Scholar]
- [186].Ravikrishnan A, Ozdemir T, Bah M, Baskerville KA, Shah SI, Rajasekaran AK, Jia X, ACS Appl. Mater. Interfaces 2016, 8, 17915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Rajab TK, O’Malley TJ, Tchantchaleishvili V, Artif. Organs 2020, 44, 1031. [DOI] [PubMed] [Google Scholar]
- [188].Sun H, Zhu Y, Pan H, Chen X, Balestrini JL, Lam TT, Kanyo JE, Eichmann A, Gulati M, Fares WH, Bai H, Feghali-Bostwick CA, Gan Y, Peng X, Moore MW, White ES, Sava P, Gonzalez AL, Cheng Y, Niklason LE, Herzog EL, Arthritis Rheumatol. 2016, 68, 1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].a) Booth AJ, Hadley R, Cornett AM, Dreffs AA, Matthes SA, Tsui JL, Weiss K, Horowitz JC, Fiore VF, Barker TH, Moore BB, Martinez FJ, Niklason LE, White ES, Am J. Respir. Crit. Care Med 2012, 186, 866; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Parker MW, Rossi D, Peterson M, Smith K, Sikström K, White ES, Connett JE, Henke CA, Larsson O, Bitterman PB, J. Clin. Invest 2014, 124, 1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Garreta E, Oria R, Tarantino C, Pla-Roca M, Prado P, Fernández-Avilés F, Campistol JM, Samitier J, Montserrat N, Mater. Today 2017, 20, 166. [Google Scholar]
- [191].Esch EW, Bahinski A, Huh D, Nat. Rev. Drug Discovery 2015, 14, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Sundarakrishnan A, Zukas H, Coburn J, Bertini BT, Liu Z, Georgakoudi I, Baugh L, Dasgupta Q, Black LD, Kaplan DL, ACS Biomater. Sci. Eng 2019, 5, 2417. [DOI] [PubMed] [Google Scholar]
- [193].Sriram G, Alberti M, Dancik Y, Wu B, Wu R, Feng Z, Ramasamy S, Bigliardi PL, Bigliardi-Qi M, Wang Z, Mater. Today 2018, 21, 326. [Google Scholar]
- [194].Wufuer M, Lee G, Hur W, Jeon B, Kim BJ, Choi TH, Lee S, Sci. Rep 2016, 6, 37471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Pattanaik D, Brown M, Postlethwaite AE, J. Inflammation Res 2011, 4, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Matucci-Cerinic M, Kahaleh B, Wigley FM, Arthritis Rheum. 2013, 65, 1953. [DOI] [PubMed] [Google Scholar]
- [197].a) Chen Y-C, Lin R-Z, Qi H, Yang Y, Bae H, Melero-Martin JM, Khademhosseini A, Adv. Funct. Mater 2012, 22, 2027; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Grebenyuk S, Ranga A, Front. Bioeng. Biotechnol 2019, 7, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Lovett M, Lee K, Edwards A, Kaplan DL, Tissue Eng., Part B 2009, 15, 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Matei A-E, Chen C-W, Kiesewetter L, Györfi A-H, Li Y-N, Trinh-Minh T, Xu X, Tran Manh C, van Kuppevelt T, Hansmann J, Jüngel A, Schett G, Groeber-Becker F, Distler JHW, Ann. Rheum. Dis 2019, 78, 1686. [DOI] [PubMed] [Google Scholar]
- [200].Brown M, O’Reilly S, Clin. Exp. Immunol 2019, 195, 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Bhandari R, Ball MS, Martyanov V, Popovich D, Schaafsma E, Han S, ElTanbouly M, Orzechowski NM, Carns M, Arroyo E, Aren K, Hinchcliff M, Whitfield ML, Pioli PA, Arthritis Rheumatol. 2020, 72, 1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].François A, Chatelus E, Wachsmann D, Sibilia J, Bahram S, Alsaleh G, Gottenberg J-E, Arthritis Res. Ther 2013, 15, R168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Miller CP, Shin W, Ahn EH, Kim HJ, Kim D-H, Trends Biotechnol. 2020, 38, 857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Davidson MD, Burdick JA, Wells RG, Adv. Healthcare Mater 2020, 9, 1901682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].a) Liu C, Oikonomopoulos A, Sayed N, Wu JC, Development 2018, 145, dev156166; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Doss MX, Sachinidis A, Cells 2019, 8, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Capella-Monsonís H, Coentro JQ, Graceffa V, Wu Z, Zeugolis DI, Nat. Protoc 2018, 13, 507. [DOI] [PubMed] [Google Scholar]
- [207].Coentro JQ, Capella-Monsonís H, Graceffa V, Wu Z, Mullen AM, Raghunath M, Zeugolis DI, Methods Mol. Biol 2017, 1627, 341. [DOI] [PubMed] [Google Scholar]
- [208].Rouède D, Schaub E, Bellanger JJ, Ezan F, Scimeca JC, Baffet G, Tiaho F, Sci. Rep 2017, 7, 12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Baues M, Dasgupta A, Ehling J, Prakash J, Boor P, Tacke F, Kiessling F, Lammers T, Adv. Drug Delivery Rev 2017, 121, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Low AH, Lax M, Johnson SR, Lee P, J. Rheumatol 2009, 36, 961. [DOI] [PubMed] [Google Scholar]
- [211].Bocchino M, Bruzzese D, D’Alto M, Argiento P, Borgia A, Capaccio A, Romeo E, Russo B, Sanduzzi A, Valente T, Sverzellati N, Rea G, Vettori S, Sci. Rep 2019, 9, 9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Li H, Furst DE, Jin H, Sun C, Wang X, Yang L, He J, Wang Y, Liu A, Arthritis Res. Ther 2018, 20, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Peelen DM, Zwezerijnen B, Nossent EJ, Meijboom LJ, Hoekstra OS, Van der Laken CJ, Voskuyl AE, Rheumatology 2020, 59, 1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Green MC, Sweet HO, Bunker LE, Am. J. Pathol 1976, 82, 493. [PMC free article] [PubMed] [Google Scholar]
- [215].Christner PJ, Peters J, Hawkins D, Siracusa LD, Jiménez SA, Arthritis Rheum. 1995, 38, 1791. [DOI] [PubMed] [Google Scholar]
- [216].Nguyen VA, Sgonc R, Dietrich H, Wick G, J. Autoimmun 2000, 14, 143. [DOI] [PubMed] [Google Scholar]
- [217].Maurer B, Distler JHW, Distler O, Vasc. Pharmacol 2013, 58, 194. [DOI] [PubMed] [Google Scholar]
- [218].a) Hocher B, Thöne-Reineke C, Rohmeiss P, Schmager F, Slowinski T, Burst V, Siegmund F, Quertermous T, Bauer C, Neumayer HH, Schleuning WD, Theuring F, J. Clin. Invest 1997, 99, 1380; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hocher B, Schwarz A, Fagan KA, Thöne-Reineke C, El-Hag K, Kusserow H, Elitok S, Bauer C, Neumayer H-H, Rodman DM, Theuring F, Am. J. Respir. Cell Mol. Biol 2000, 23, 19. [DOI] [PubMed] [Google Scholar]
- [219].Sonnylal S, Denton CP, Zheng B, Keene DR, He R, Adams HP, VanPelt CS, Geng YJ, Deng JM, Behringer RR, de Crombrugghe B, Arthritis Rheum. 2007, 56, 334. [DOI] [PubMed] [Google Scholar]
- [220].Noda S, Asano Y, Nishimura S, Taniguchi T, Fujiu K, Manabe I, Nakamura K, Yamashita T, Saigusa R, Akamata K, Takahashi T, Ichimura Y, Toyama T, Tsuruta D, Trojanowska M, Nagai R, Sato S, Nat. Commun 2014, 5, 5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Castello-Cros R, Whitaker-Menezes D, Molchansky A, Purkins G, Soslowsky LJ, Beason DP, Sotgia F, lozzo RV, Lisanti MP, Cell Cycle 2011, 10, 2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Asano Y, Stawski L, Hant F, Highland K, Silver R, Szalai G, Watson DK, Trojanowska M, Am. J. Pathol 2010, 176, 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Meng M, Tan J, Chen W, Du Q, Xie B, Wang N, Zhu H, Wang K, Front. Immunol 2019, 10, 1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Zhang Y, McCormick LL, Desai SR, Wu C, Gilliam AC, J. Immunol 2002, 168, 3088. [DOI] [PubMed] [Google Scholar]
- [225].Ghosh AK, Mori Y, Dowling E, Varga J, Biochem. Biophys. Res. Commun 2007, 354, 420. [DOI] [PubMed] [Google Scholar]
- [226].Zhang Z, Wu Y, Wu B, Qi Q, Li H, Lu H, Fan C, Feng C, Zuo J, Niu L, Tang W, Arthritis Res. Ther 2019, 21, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].O’Kane D, Jackson MV, Kissenpfennig A, Spence S, Damkat-Thomas L, Tolland JP, Smyth AE, Denton CP, Stuart Elborn J, McAuley DF, O’Kane CM, Exp. Dermatol 2014, 23, 497. [DOI] [PubMed] [Google Scholar]
- [228].Corallo C, Cutolo M, Kahaleh B, Pecetti G, Montella A, Chirico C, Soldano S, Nuti R, Giordano N, Arthritis Res. Ther 2016, 18, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Laplante P, Sirois I, Raymond MA, Kokta V, Béliveau A, Prat A, Pshezhetsky AV, Hébert MJ, Cell Death Differ. 2010, 17, 291. [DOI] [PubMed] [Google Scholar]
- [230].Shi-wen X, Pennington D, Holmes A, Leask A, Bradham D, Beauchamp JR, Fonseca C, du Bois RM, Martin GR, Black CM, Abraham DJ, Exp. Cell Res 2000, 259, 213. [DOI] [PubMed] [Google Scholar]
- [231].Akhmetshina A, Dees C, Pileckyte M, Maurer B, Axmann R, Jüngel A, Zwerina J, Gay S, Schett G, Distler O, Distler JHW, FASEB J. 2008, 22, 2214. [DOI] [PubMed] [Google Scholar]
- [232].Distler O, Pap T, Kowal-Bielecka O, Meyringer R, Guiducci S, Landthaler M, Schölmerich J, Michel BA, Gay RE, Matucci-Cerinic M, Gay S, Müller-Ladner U, Arthritis Rheum. 2001, 44, 2665. [DOI] [PubMed] [Google Scholar]
- [233].Makino K, Makino T, Stawski L, Mantero JC, Lafyatis R, Simms R, Trojanowska M, J. Invest. Dermatol 2017, 137, 1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Distler JHW, Jüngel A, Huber LC, Schulze-Horsel U, Zwerina J, Gay RE, Michel BA, Hauser T, Schett G, Gay S, Distler O, Arthritis Rheum. 2007, 56, 311. [DOI] [PubMed] [Google Scholar]
- [235].Hsu E, Feghali-Bostwick CA, Am. J. Pathol 2008, 172, 1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Garrett SM, Hsu E, Thomas JM, Pilewski JM, Feghali-Bostwick C, PLoS One 2019, 14, e0225422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Chakraborty D, Zhu H, Jüngel A, Summa L, Li YN, Matei AE, Zhou X, Huang J, Trinh-Minh T, Chen CW, Lafyatis R, Dees C, Bergmann C, Soare A, Luo H, Ramming A, Schett G, Distler O, Distler JHW, Sci. Transl. Med 2020, 12, eaaz5506. [DOI] [PubMed] [Google Scholar]
- [238].Maurer B, Distler A, Suliman YA, Gay RE, Michel BA, Gay S, Distler JHW, Distler O, Ann. Rheum. Dis 2014, 73, 1880. [DOI] [PubMed] [Google Scholar]
- [239].Shiwen X, Stratton R, Nikitorowicz-Buniak J, Ahmed-Abdi B, Ponticos M, Denton C, Abraham D, Takahashi A, Suki B, Layne MD, Lafyatis R, Smith BD, PLoS One 2015, 10, e0126015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Sharma S, Goswami R, Zhang DX, Rahaman SO, J. Cell. Mol. Med 2019, 23, 761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Asmani M, Velumani S, Li Y, Wawrzyniak N, Hsia I, Chen Z, Hinz B, Zhao R, Nat. Commun 2018, 9, 2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Asano S, Ito S, Takahashi K, Furuya K, Kondo M, Sokabe M, Hasegawa Y, Physiol. Rep 2017, 5, e13281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Vijayaraj P, Minasyan A, Durra A, Karumbayaram S, Mehrabi M, Aros CJ, Ahadome SD, Shia DW, Chung K, Sandlin JM, Darmawan KF, Bhatt KV, Manze CC, Paul MK, Wilkinson DC, Yan W, Clark AT, Rickabaugh TM, Wallace WD, Graeber TG, Damoiseaux R, Gomperts BN, Cell Rep. 2019, 29, P3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Wang X, Liu Y, Deng Z, Dong R, Liu Y, Hu S, Li Y, Jin Y, J. Dermatol. Sci 2009, 53, 103. [DOI] [PubMed] [Google Scholar]
- [245].Gailit J, Marchese MJ, Kew RR, Gruber BL, J. Invest. Dermatol 2001, 117, 1113. [DOI] [PubMed] [Google Scholar]
- [246].Bhattacharyya S, Tamaki Z, Wang W, Hinchcliff M, Hoover P, Getsios S, White ES, Varga J, Sci. Transl. Med 2014, 6, 232ra250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Marangoni RG, Masui Y, Fang F, Korman B, Lord G, Lee J, Lakota K, Wei J, Scherer PE, Otvos L, Yamauchi T, Kubota N, Kadowaki T, Asano Y, Sato S, Tourtellotte WG, Varga J, Sci. Rep 2017, 7, 4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Bhattacharyya S, Wang W, Qin W, Cheng K, Coulup S, Chavez S, Jiang S, Raparia K, De Almeida LMV, Stehlik C, Tamaki Z, Yin H, Varga J, JCI Insight 2018, 3, e98850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Huang M, Cai G, Baugh LM, Liu Z, Smith A, Watson M, Popovich D, Zhang T, Stawski LS, Trojanowska M, Georgakoudi I, BlackIII LD, Pioli PA, Whitfield ML, Garlick J, Arthritis Rheumatol. 2020, 72, 791. [DOI] [PubMed] [Google Scholar]
- [250].Sun H, Zhu Y, Pan H, Chen X, Balestrini JL, Lam TT, Kanyo JE, Eichmann A, Gulati M, Fares WH, Bai H, Feghali-Bostwick CA, Gan Y, Peng X, Moore MW, White ES, Sava P, Gonzalez AL, Cheng Y, Niklason LE, Herzog EL, Arthritis Rheumatol. 2016, 68, 1251. [DOI] [PMC free article] [PubMed] [Google Scholar]