Figure 1.
Barcode Cycle Sequencing (BCS): A strategy for converting amino acids into DNA barcodes directly on a next-generation sequencing chip
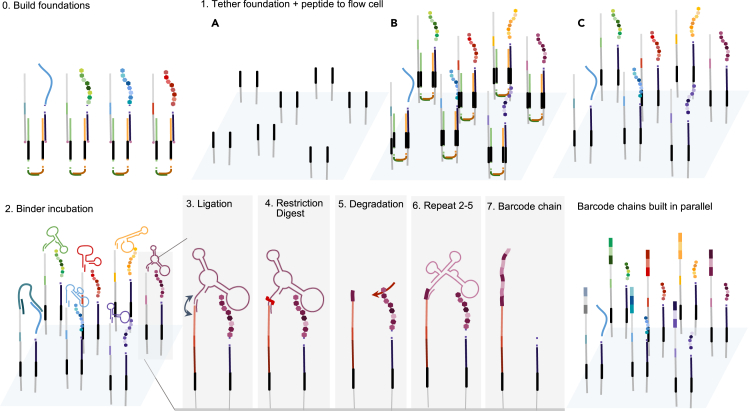
This schematic depicts the seeding of foundations and subsequent per round barcode capture. Step 0 depicts the off-chip construction of a target-foundation complex to ensure colocalization between the foundation and target, as described in Figure 2. Step 1 includes the tethering of the peptide-foundation complex onto solid substrate on the flow cell. Step 2 includes incubating the bound proteins or peptides with a barcoded binder library under conditions that allow the appropriate aptamer to bind specifically to the appropriate N-terminal amino acid. Step 3 includes ligating the aptamer tail to a second oligonucleotide bound to the substrate. Step 4 includes cleaving off the binder, leaving the DNA barcode associated with that particular amino acid bound to the second oligonucleotide. For a full-fledged ProtSeq technology, after or at the same time with binder removal, Step 5 would consist of a degradation step in which the terminal amino acid is cleaved. After a washing cycle, Steps 2–5 are repeated, generating a chain of DNA barcodes that reflect binding events to the colocalized target. Refer to Figure S8 for alternative methods.