Figure 8.
Isolation of N-terminal amino acid-binding aptamers using a semi-automated and parallel replicates-and-switch selection strategy
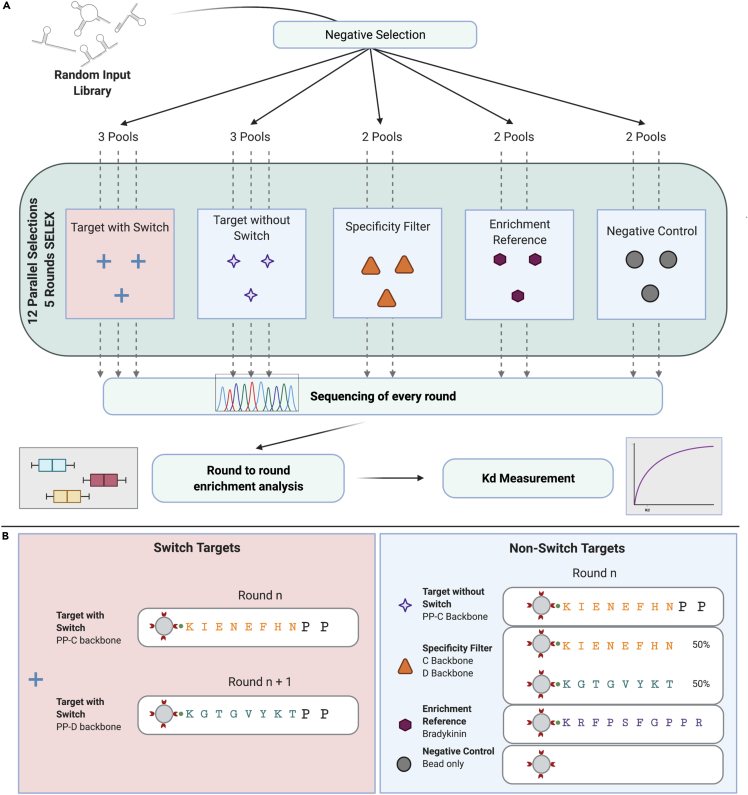
(A) Schematic diagram of replicates-and-switch selection strategy. Twelve selections comprising replicates of each target mixtures were run for five rounds in parallel. The workflow begins with a negative selection against streptavidin beads on an initial pool of ssDNA and split across 12 random pools. Two parallel selections were performed on each control reference target and three parallel selections on the target (Proline-Proline) with and without the switching of backbones (C and D backbones) in alternating rounds. A representative pool of ssDNA from every round of every selection was sequenced and analyzed for round-to-round enrichment of sequences. Refer to Figures S5, S6, and S7 for digestion quality control assay, enrichment profile of top 10 aptamers, and contamination analysis, respectively.
(B) Target compositions and amino acid sequences in Non-Switch and Switch SELEX.