Highlights
-
•
Environmental pollutants dyes, pesticides, pharmaceuticals, explosive waste and polyaromatic hydrocarbons.
-
•
Environmental pollutants toxicity.
-
•
Possible microbial biodegradation pathways of environmental pollutants.
Keywords: Biodegradation, Microbes, Pesticides, Pharmaceuticals, Dyes, Explosive waste, PAHs
Abstract
Industrialization and human activities have led to serious effects on environment. With the progress taking place in the biodegradation field, it is important to summarize the latest advancement. In this review, we intend to provide insights on the recent progress on the biodegradation of environmental contaminants such as dyes, pesticides, pharmaceuticals, explosive waste and polyaromatic hydrocarbons by microorganisms. Along with the biodegradation of environmental contaminants, toxicity effects have also been discussed.
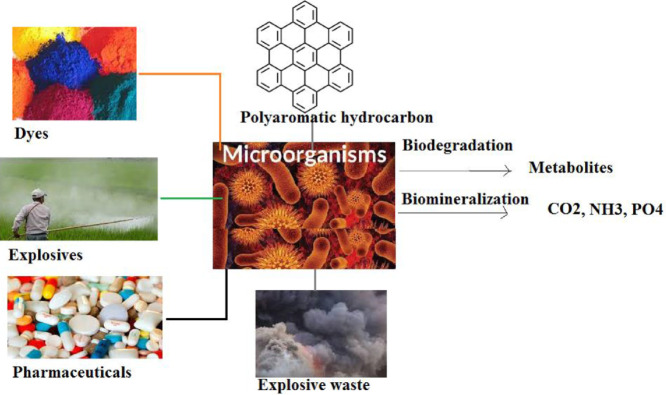
Graphical abstract
1. Introduction
One of the most problematic aspects of continuous anthropogenic activities and industrialization is the release of toxic waste into the environment. The release of such contaminants leads to disturbance in the nature which ultimately reflects in ecological processes. Direct effects of such contaminants may also lead to toxicity in various organisms including humans. The various kinds of environmental contaminants discussed in this review are dyes, pesticides, pharmaceuticals, explosive waste and persistent organic pollutants (POP). Dyes have been found to have carcinogenicity and allergenic effects (Chung, 2016). Immuno-toxic effects of pesticides can also be found in literature (Corsini et al., 2013). The increased understanding of the dangerous effects of environmental pollutants has directed to a striking increase in research on various strategies that may be applied to clean up the environment. The physical and chemical treatment technologies presently used for the remediation of pollutants are expensive and cannot sufficiently mitigate or remediate these contaminants. Biodegradation is the natural process where substances are degraded biologically. Biodegradation is defined as the biologically catalyzed reduction in complexity of chemical compounds (Andeer et al., 2012). Whereas bioremediation is an engineered process of employing microorganisms to clean the environmental contaminants. It is the utilization of the biodegradation ability of microorganisms in such a manner that increases the speed of the process and provides a practical aspect of the property to clean the environment. In this review, the potential of microorganisms mediated remediation is looked upon. Various taxonomic groups such as fungi, archaebacteria and eubacteria are abundant in members which can perform biodegradation of environmental contaminants. For performing the process of biodegradation, the microorganism first must be able to survive in the presence of particular contaminant. This review covers the research carried out in the field of bioremediation of dyes, PAHs, pharmaceuticals, explosive waste, and pesticides in the last decade and the toxicity effects of the respective pollutants in the last couple of decades.
2. Dyes
Dyes are soluble chemicals which provide color to the materials. They diffuse through the materials. Dyes have presence of at least one chromophore group in them. Dyes can be natural or synthetic. Dyes are widely used in food, pharmaceutical, leather, cosmetic and textile industries. These dyes when discharged into water bodies without any check or regulation pose a great threat to the aquatic life and consequently to the environment (B. Singh and Singh, 2016a). Among the different kinds of dyes used, the most common are azo, anthraquinone and deoxidizing dyes. An example of toxic effects of dyes is of Malachite Green (MG) a member of cationic triphenylmethane dye which has its use mostly as a fungicide or as a disinfectant agent. It is toxic to the mammalian cells in concentration as low as 0.1 mg/ml (Cleinmensen et al., 1984). MG has been banned in many countries but due to its low cost and efficacy it is still used in some countries. Azo dyes account for the majority of the synthetic dyes used in commercial applications. Azo dyes are aromatic compounds with one or more -N = N- groups. Therefore, proper degradation or removal of these dyes from the environment is of high priority.
2.1. Toxicity effects of dyes
The textile effluent and their products have been found to be of toxic nature in the environment. When the textile effluent containing azo dyes is released into the environment it leads to many problems because of their teratogenic, mutagenic and carcinogenic effects (Tan et al., 2016). Various reports have observed and described the ecotoxicological effects of dyes on aquatic life (Bae et al., 2006; Meriç et al., 2005). Daphnia and Danio are the most common organisms on which the evaluation of both acute and chronic toxicity is routinely done (Y. Verma, 2008). However, the results obtained from the toxicity studies on single organism cannot be extrapolated to other taxa levels because of different responses of each organism to the contaminant. The link between carcinogenic and mutagenic effects of some azo dyes has been well established as of now (Chequer et al., 2011). An indirect link was established between hypoactivity of zebrafish larvae and energy consumed. It was found that zebrafish larvae exposed with dye Basic Red 51 were less active (Abe et al., 2018). Moreover, certain metabolites produced by breakdown of dyes were found to be even more toxic than parent compounds. For example, the toxicity of Acid Violet 7 increases after biodegradation by Pseudomonas putida due to the formation of metabolites 4′-aminoacetanilide and 5-acetamido-2-amino-1- hydroxy‑3,6-naphtalene disulphonic acid (Mansour et al., 2010). Natural dyes have been implemented in various uses such as cosmetics, drugs, textiles, these are more biodegradable than synthetic dyes and have less harmful effects at same concentrations as that of synthetic dyes. However, more research needs to be done to evaluate toxicity effects of natural dyes as there is less evidence available in literature on natural dyes compared to synthetic dyes (Abe et al., 2018).
2.2. Microbe mediated remediation of dyes
Many physical (adsorption, coagulation, flocculation, membrane filtration etc.) and chemical methods (oxidation process, Fenton's reagent, ozonation etc.) are available for the removal of dyes. But these methods have their own disadvantages which are: low efficiency, selective over types of dyes, sludge production, generation of toxic by-products, and sometime high cost. Biodegradation using microorganisms provides a cost effective, feasible and environmental-friendly alternative to the physiochemical methods. Generally, microorganisms are isolated and characterized from anthropogenic polluted environment because they have adapted and are competent to remediate such sites (Asad et al., 2007). Examples of the microorganisms which have been isolated from such sites and were able to grow on certain optimal conditions are mentioned in the Table 1: (based on literature).
Table 1.
Microorganisms capable of biodegrading dyes.
| Microorganism/Co-Culture/Consortium | Dyes | Isolation from or Source | Degradation Pathway/Enzymes Involved | Degradation Product (Metabolite) | Percentage Transformation | Techniques Used | Reference |
|---|---|---|---|---|---|---|---|
| ShewanellaputrefaciensCN32, Bacillus circulansBWL1061 | Sudan I | – | Synergistic effect of Azoreductase enzyme and Non-specific reductive decolorization | – | 90.23% in 108 h | Drop plate method | (Liu et al., 2018) |
| Bacillus pseudomycoidesMH229766 | Acid Black 24 (AB24) |
Wastewater treatment plant in Noida, India | activities of oxido-reductive enzymes lignin peroxidase, laccase and azoreductase were induced |
– | 96.79% at 40 mg/L initial dye concentration | HPLC, FTIR, UV Spectroscopy | (Kumar et al., 2019) |
| Fungal Strain VITAF-1 |
Reactive Green Dye (RGD) | dye contaminated sites of Tirupur district, T.N, India | oxidoreductive enzymes like laccase, LiP and DCIP reductase | benzoic acid, 2(−1-oxopropyl) | 97.9% within 48 h | UV–Vis spectrophotometer, HPLC,FTIR, GC–MS |
(Sinha et al., 2016) |
| Phanerochaete chrysosporium | Reactive Black 5 (RB5) | – | secretion of the extracellular enzyme MnP |
– | 90.3% in 72 h for initial dye concentration 100 mg/L |
TLC, UV–vis spectroscopy, FTIR | (Enayatizamir et al., 2011) |
|
Pseudomonas sp. strain DY1 |
Malachite green (MG) | – | Mn-peroxidase, NADH–DCIP and MG reductase were involved |
malachite green carbinol, (dimethyl amino-phenyl)-phenyl-methanone, N,N-dimethylaniline, (methyl amino-phenyl)-phenyl-methanone, (amino phenyl)-phenyl methanone and di-benzyl methane |
90.3–97.2% at concentrations of MG 100–1000 mg/l under shaking condition within 24 h. |
UV–vis GC–MS LC-MS |
(Du et al., 2011) |
|
Bacillussp. V1DMK, Lysinibacillussp. V3DMK, Bacillussp. V5DMK, Bacillussp. V7DMK, Ochrobacteriumsp. V10DMK, Bacillussp. V12DMK. SB4 |
Reactive Violet 5R (RV5) | Soil samples collected from Kharicutcanal, Gujarat, India | asymmetrical cleavage of azo linkage | 1-diazo-2-naphthol, 4-hydroxybenzenesulphonic acid, 2-naphthol and benzenesulphonic acid | decolorized 200 mg/L of RV5 within 18 h under static condition |
FTIR, NMR GC–MS |
(Jain et al., 2012) |
|
Micrococcus luteus strain SSN2 |
Direct Orange 16 (DO-16) | textile industry near Ranipet, Tamil Nadu, India |
reduction of the azo bond | – | 96% efficiency at 3% NaCl in 6 h under static conditions |
UV–vis TLC FTIR HPLC |
(R.L. Singh et al., 2015) |
| GalactomycesgeotrichumMTCC 1360 | Reactive Yellow-84A | Microbial Type Culture Collection, Chandigarh, India |
azoreductase, laccase and tyrosinase enzyme activities |
4(5‑hydroxy, 4-amino cyclopentane) sulfobenzene and 4(5‑hydroxy cyclopentane) sulfobenzene |
86% decolorization of azo dye | HPLC, FTIR, GC–MS and HPTLC |
(Govindwar et al., 2014) |
|
Pleurotus eryngii F032 |
Reactive Black 5 (RB5) | recreational forest, UniversitiTeknologi Malaysia (UTM) |
– | – | 93.56% decolorization of 10 mg/L RB5 within 72 h of incubation in dark condition with agitation |
UV–vis spectroscopy | (Hadibarata et al., 2013) |
| Enterobacter asburiaestrain XJUHX-4TM | Malachite green (MG) | dye-contaminated wastewater of a smallscale dyeing industry situated at Habra, West Bengal, India |
significant increase in the activities of enzymes laccase, dichlorophenolindopnenol reductase and malachite green reductase were observed |
leucomalachite green, desmethylleucomalachite green, didesmethylleucomalachite green, (dimethyl amino phenyl)-phenyl methanone, (methyl amino phenyl)-phenyl methanone, (amino phenyl)-phenyl methanone and aniline |
>95% decolorizaton | UV–vis spectroscopy, TLC, GC–MS | (Mukherjee and Das, 2014) |
| Enterobacter sp. SXCR | Congo red | from petroleum contaminated soil from Ranchi, Jharkhand, India |
cleavage of azo bonds by azoreductase | – | 98% dye removal observed at0.1–0.3 g/L of dye | UV–visible spectral analysis, HPLC, and FTIR |
(Prasad and Aikat, 2014) |
| Acinetobacter baumannii | Reactive red 198 | Kovalam sea shore in Tamil Nadu, India | biotransformation by various oxidative and reductive enzymes |
– | 96.20% decolorization was observed in 500 mg/L of reactive red 198 after 72 h. |
UV-visible spectroscopy and Fourier-transform infrared (FTIR) | (Unnikrishnan et al., 2018) |
| Arthrobacter soli BS5 | Reactive black 5 (RB5) | effluent from textile industries located at Industrial area, Panki site 5, Kanpur, India |
– | – | 98% after 120 h of incubation | Atomic absorption spectrophotometer and GC–MS |
(Khan et al., 2018) |
|
Staphylococcus sp. K2204 |
Remazol Brilliant Blue R (RBBR) |
textile wastewater | enzymes like laccase, manganese and lignin peroxidase facilitated the catalysis of the asymmetric cleavage |
– | complete decolorization of RBBR within 12 h. | FTIR,HPLC | (Velayutham et al., 2018) |
| Pichiasp. Strain TCL | Acid Red B | sea mud collected in Heishijiao Beach Park (Dalian, China) |
reductive cleavage of azo groups |
4-amino-naphthalene-1-sulfonic acid;3-amino-4‑hydroxy-naphthalene-1-sulfonic acid; 3,4-dihydroxy-naphthalene-1-sulfonic acid;naphthalene-1,2,3,4-tetraol; catechol; 3–7-dihydroxy-octahydro-naphthalene-2,6‑dione. |
90% of dye (100 mg/L)decolorized within 10 h | UV–vis, HPLC analysis | (Qu et al., 2012) |
| Providenciasp. SRS82 | Acid Black 210 (AB210) | Soil and wastewater samples from the vicinity of textile dyeing industries located in Indore, India |
Induction of intracellular and extracellular lignin peroxidase, intracellular laccase and tyrosinase, azoreductase, and DCIP reductase |
Benzene, naphthalene and 4-aminophenyl-N-(4-aminophenyl) benzene sulphonamide. | degrade 100 mg/L dye within 90 min under optimum conditions | FTIR, HPTLC, HPLC, GC/MS and LCMS |
(Agrawal et al., 2014) |
| BrevibacilluslaterosporusandGalactomycesgeotrichum | Reactive Red 198 (RR 198) | Microbial Type Culture Collection, Chandigarh, India | veratryl alcohol oxidase, laccase, NADH-DCIP reductase and azo-reductase (Biomineralization) |
(ethylsulfonyl)benzene and 1,3,5-triazine | 92% | FTIR,HPTLC,GC–MS | (Kurade et al., 2015) |
| Bacillus vietnamensis sp. MSB17 | Malachite Green (MG) | continental slope of the eastern Arabian Sea | activities of enzymes such as tyrosinase, laccase, and manganese peroxidase were observed | methanone, [4-(dimethylamino) phenyl] phenyl- and 2, 6-bis (1, 1- dimethylethyl) phenol |
complete decolorization of dye (50 mg/L) was attained within 4 h of incubation |
UV–VIS, FT-IR, and GC–MS analysis |
(Kabeer et al., 2019) |
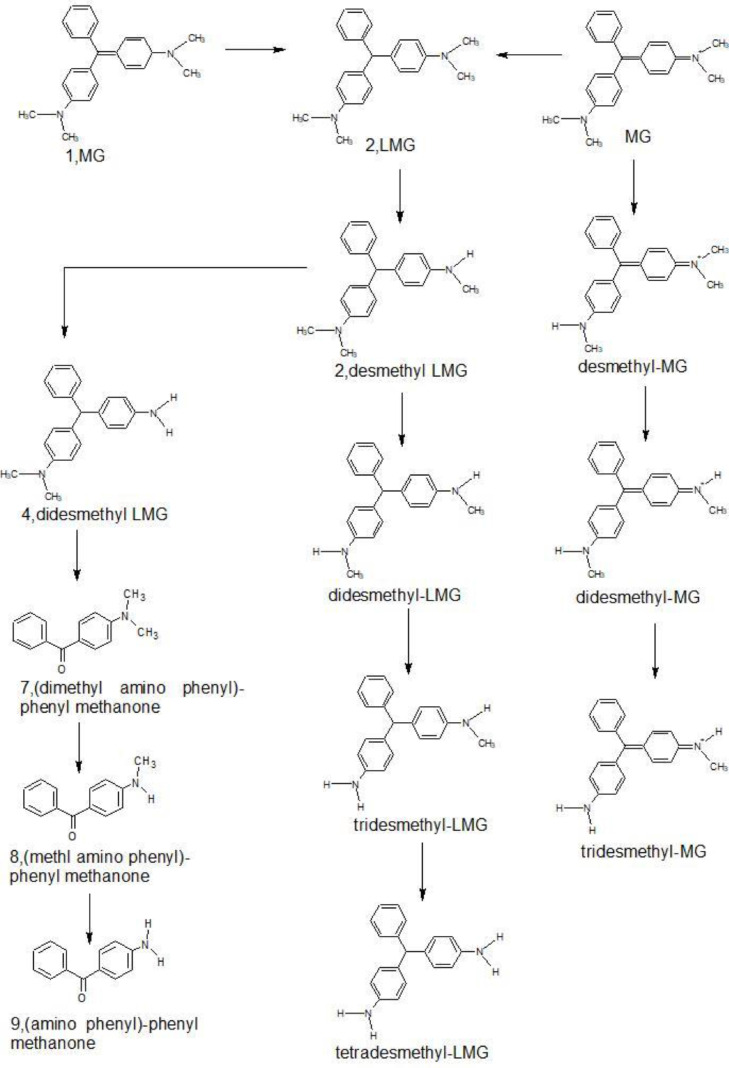
As it is evident by now, Malachite green has multiple toxicity effects on body and is a recalcitrant dye. A species of Enterobacter genus was found to degrade malachite green with more than 98% efficiency when it was provided with sucrose and beef extract (as carbon and nitrogen sources respectively) in a ratio of 5:1. Biodegradation pathway of Malachite green has been well studied, it involves enzymes such as laccase, reductase and cytochrome P450 enzymes and involves production of metabolites such as leucomalachite green, desmethyl leucomalachite green, didesmethyl leucomalachite green and aniline (Ref. to Fig. 1) (Mukherjee and Das, 2014; J. a. Wang et al., 2012)
Fig. 1.
Microbial degradation pathway of Malachite green. (Mukherjee and Das, 2014; J. a. Wang et al., 2012). LMG - leucomalachite green.
Optimization of parameters such as pH, temperature, salt concentration is done using various statistical approaches such as response surface methodology (RSM) to improve the biodegradation efficiency (Kumar et al., 2019). The genes from these tolerant strains can be characterized and can be utilized by genetic engineering to form new recombinant microorganisms which will exhibit the biodegradation capability of the donor gene microorganism. Various microorganisms such as bacteria, fungi and yeast etc. are responsible for biodegradation of dyes. However bacteria are preferred over other microorganisms due to their fast replication, easy manipulation and ability to tolerate harsh conditions (Rathour et al., 2018). Microorganisms have been utilized in various forms - pure, mixed, living and dead for biodegradation. Isolated bacteria are more often immobilized (on alginate beads) for biodegradation as it provides certain advantages such as higher degradation efficiency, capability of reuse and higher biomass loading. A great deal of advancement has been done in the development of bioreactors. Various bioreactors such as stirred tank bioreactors, airlift bioreactors, fluidized bed bioreactors, wave bioreactors, combined or sequential bioreactors have been employed (Vikrant et al., 2018). Various enzymes have been reported in literature which are linked with the process of biodegradation of dyes such as - azoreductases, laccases, tyrosinases, lignin peroxidases, Mn peroxidases and DCIP–NADH (R. L. R.L. Singh et al., 2015). The concept of simultaneous degradation of dye and production bioelectricity has been utilized in the form of Microbial Fuel Cell (MFC) (Fernando et al., 2014; Ilamathi and Jayapriya, 2018). Multiple dyes like Reactive Black 5, Reactive Orange 16, Disperse Red 78 and Direct Red 81 were degraded by a single consortium consisting of bacteria Providencia rettgeri strain HSL1 and Pseudomonas sp. SUK1 (Lade et al., 2015).
3. Pesticides
The growing population demands its need of food to be met unanimously. Population growth kinetics suggests that the overall population of humans by 2050 is going cross the nine billion mark. To provide the need of food to live to such an enormous number of individuals would require increase in production of food by 70 percent (According to Food and Agriculture Organization; FAO report Rome 12–13 october,2009). The first and most important thing in achieving this feat would be to minimize the losses to crops by pests. Pesticides are chemicals that are used to control the population of pest to the level at which they would cause minimalistic harm to the crops. Pesticides cover a wide range of target pests including mites, snails, insects, rodents, fungi, birds and even viruses (Velázquez-Fernández et al., 2012). They can be classified on the basis of their persistence in the environment. They can be classified as non-persistent (readily degradable) or persistent pesticides. Non-persistent pesticides include – methoxychlor, malathion, paraquat etc. Whereas persistent pesticides include – DDT, aldrin, tordon, turbacil, etc. (J. P. Verma et al., 2014)
3.1. Toxicity effects of pesticides
Pesticide residues pose a great threat to the soil quality and health of living organisms. There are various effects of pesticides on aquatic life such as delayed metamorphosis, disruption of steroid metabolism, low rate of opercular movement, erratic swimming etc. (Sidhu et al., 2019). There is enough evidence of toxic effects of pesticides on both aquatic as well as terrestrial life on both plants as well as animals. Atrazine is one of the pesticides that has toxic effects on wide range of organisms including humans. It can affect central nervous system, reproductive system, cardiovascular system and immune system. There has been tremendous development in the field of statistics and artificial intelligence. To reduce the time and effort, assessment of toxicity of pesticides in rats has been done by one such example of artificial intelligence i.e. QSAR model (Quantitative Structure–Activity Relationship) (Hamadache et al., 2016). Triamidefon and its metabolite triadimenol affected endocrine machinery of Xenopus laevis (African frog). Triadimefon was found to be causing more toxic effects than triamidenol. Moreover, the frogs exhibited sex-linked differences liver histology, antioxidant enzyme activities and thyroid hormone levels (W. Zhang et al., 2020). At sublethal concentrations, flumethrin has been found to have high acute toxicity in honey bees. The toxic effects were found to be a result of increased oxidative stress and damage to the midgut by apoptosis (Qi et al., 2020). Acetamiprid is a member of neonicotinoids, it was thought to be safe in prospective of mammals but recent reports suggest that it also exhibits toxic effects towards mammals. It works by binding to nicotinic acetylcholine receptors in insects. Exposure to acetamiprid was found to be associated with decreased neurogenesis in mice and abnormal neuronal distribution in newborn mice (Kagawa and Nagao, 2018). Fenvalerate is a widely used pesticide and is known to cause impairment of male reproductive system but the mechanism is not clear. Recently, it has come to light that fenvalerate may impair male reproductive system through changes in circadian rhythm gene levels. It was found that fenvalerate inhibited testosterone synthesis, altered the expression of circadian rhythm mRNA and increased intracellular calcium ion levels in mouse Leydig cells (Guo et al., 2017). Organophosphate pesticides have been shown to hinder various metabolic processes in plants such as photosynthesis, carbon metabolism, chlorophyll biosynthesis and nitrogen metabolism (Sidhu et al., 2019).
3.2. Microbe mediated remediation of pesticides
Earlier studies of microbial remediation of pesticide residues can be traced back to 1940. In case of DDT it was observed that if co-substrate starch (slow releasing carbon source) was added for co-metabolism, it led to complete mineralization and detoxification of DDT by a developed microbial consortium under nitrogen-fixing conditions (Khan et al., 2015). Various studies of isolation, enrichment, characterization and subsequent degradation of a particular pesticide have been published throughout decades (Ref. to Table 2 for some of the latest examples of pesticide degrading microorganisms).
Table 2.
Biodegradation of pesticides by microbes.
| Microorganism/Consortium | Pesticide | Isolated from/Source | Degradation Pathway | Degradation Intermediates (Metabolites) | Efficiency | Technique used | Reference |
|---|---|---|---|---|---|---|---|
| Fusarium solani | Lindane | Soil samples from the premises of IndiaPesticide Limited, Uttar Pradesh, India |
release of chloridewhenLindane was used as sole carbon source |
– | 59.4% | Gas Chromatography | (Sagar et al., 2011) |
| Streptomyces sp. Strain AC5 and Streptomyces sp. Strain AC7 | Chlorpyrifos (CP) | Soil sampleswere collected from a blueberry field that was located in the city of Gorbea in southern Chile |
Phosphomonoesterase act in hydrolyzing O-P bonds leaving phosphorus available for uptake as asourceof phosphorus and to release ethanol as a carbonsource |
3,5,6- trichloro-2-pyridinol (TCP) |
90% degradation after 24 h of incubation |
HPLC Gas Chromatography |
(Briceno et al., 2012) |
| Fusarium verticillioides | Lindane | from Agave tequilana leaves by enrichment techniques | aerobic carboxylation is suggested | gamma-pentachlorocyclohexene and benzoic acid derivatives |
with Agave leaves treatment, the degradation efficiency was found to be 86% |
GC-ECD SEM |
(Guillén-Jiménez et al., 2012) |
| Aspergillus terreusStrain JAS1 | Chlorpyrifos | Paddy field soil sample was collected from the top layer 0–20 cm in Vellore district, Tamil Nadu, India | chlorpyrifos employed as a sole carbon and energy source | 3,5,6-trichloro-2-pyridinol | complete removal or Chlorpyrifos in 24 h | HPLC, FTIR |
(Silambarasan et al., 2013) |
| Alcaligenes faecalis StrainJBW4 | endosulfan | activated sludge samples were collected from an endosulfan company |
non-oxidative pathway | Endosulfan diol and endosulfan lactone | 87.5% of alpha endosulfanand 83.9% of beta endosulfan degraded within 5 days | GC–MS | (Kong et al., 2013) |
| Pseudomonas aeruginosa Strain Is 6 | acephate | Composite surface soil samples were collected fromagricultural sites ofTanjore, Tamilnadu, India |
The oxidative degradation of acephate was found to be due to hydrolysis of carboxyl group by carboxylesterase enzyme and releasing acetic acid residue |
No accumulative products were detected ; the metabolites might have formed and been immediately degraded |
the strain Is-6 showed 92% degradation of acephate (1000 mg/ L) within 7 days of incubation |
HPLC ESI-MS |
(Ramu et al., 2014) |
| Streptomyces sp. Strain A14 | Methoxychlor (MTX) | surface soil samples were taken from an experimental site northwest of San Miguel deTucuman, Argentina |
dominantly degraded by dechlorination, dehydrogenation and CN-replacement |
1,1-dichloro-2,2-bis(4-methoxyphenyl)ethane, 1,1-dichloro- 2,2-bis(4-methoxyphenyl)ethylene, 1‑chloro-2,2-bis(4-methoxyphenyl)ethane, and 2,2-bis(4- hidroxypheny)acetonitrile |
For conc of pesticide 8.33 and 16.60 mg/kg, bacterium reached its maximum removal percentages (40% and 76%) after 28 days of incubation |
GC–MS | (Cai et al., 2014) |
| Ochrobactrum sp. strain HZM | quinalphos (QP) | pesticide-contaminated soil samples |
hydrolysis of organophosphate compounds | 2-Hydroxyquinoxaline and diethyl phosphate | 84.61% | HPLC, GC–MS | (Talwar et al., 2014) |
|
Pseudomonas aeruginosa Strain RRA, Bacillus megateriumStrain RRB, SphingobacteriumsiyangensisStrain RSA, StenotrophomonaspavaniiStrain RSB and Curtobacterium plantarumStrain RSC |
chlorpyrifos (CP) | bacteria were isolated from chlorpyrifos (CP) treated rice plants |
enzyme catalysis | – | five isolates degraded more than 90% of CP in 24 h when the initial concentration was lower than 5 mg/L. |
CLSM GC-ECD |
(Feng et al., 2017) |
| Chryseobacteriumindologenes Strain SSJ1 | flubendiamide | samples were collected from groundnut cultivating soil of Dharwad district, Karnataka, India, |
isolate utilized the flubendiamide as a sole carbon and nitrogen source |
– | 89.06% initial pesticide was removed by the isolate with 5 days incubation period |
UV–vis Spectroscopy, HPLC |
(Jadhav and David, 2016) |
| Bacillus sp. Strain SG2 | cypermethrin | Pesticide-contaminated soils were collected from a rice field of Udham Singh Nagar, Uttarakhand, India |
ester hydrolysis of pyrethroid takes place by carboxylesterases, results in acid and alcohol production |
4-propylbenzoate, 4-propylbenzaldehyde, phenol M-tertbutyl and 1-dodecanol |
bacteria degraded the compound up to 81.6% within 15 days |
GC–MS FTIR |
(Sharma et al., 2016) |
| Bacillus tequilensis | trichlorfon (TCF) | Soil samples were collected from the surface layer of a pesticide-polluted field in Hubei Province, China |
deoxidation and dehydration (including the cleavage of the P–C phosphonate bond and the C–O bond) |
DDCV, dimethyl phosphite, Trichloroethanal, dimethyl hydrogen phosphite and chloral hydrate |
degradation of 71.1% at an initial TCF concentration of 200 mg/L within 5 days |
HPLC GC–MS |
(Tian et al., 2016) |
|
Bacillus subtilis and Fomitopsispinicola |
DDT | Culture collection | (1) dechlorination to DDD, (2) dehydrochlorination to DDE, and (3) formation of DDMU |
DDD (1,1-dichloro- 2,2-bis(4-chlorophenyl) ethane), DDE (1,1-dichloro-2,2-bis(4- chlorophenyl) ethylene), and DDMU (1‑chloro-2,2-bis(4-chlorophenyl) ethylene) |
addition of 10 mL of B. subtilis into F. pinicola culture showed the highest DDT degradation of 86% during the 7days incubation period |
HPLC GC–MS |
(Sariwati et al., 2017) |
| Citricoccus sp. TT3 | Atrazine | Soil samples were collected from the wastewater outfall of the Tianjin Huayu Pesticide Factory in China |
proposed: atrazine–hydroxyatrazine–N-isopropylammelide– cyanuric acid. These steps are catalyzed by the enzymesencoded by trzN, atzB, and atzC, respectively |
– | the strain removed 50 mg/L atrazine in 66 h with 1% inoculum | PCR, SEM and Agarose gel electrophoresis |
(Yang et al., 2018) |
| Paenibacilluspolymyxa | fluazinam, BHC, PCNB, chlorpyrifos and DDT | General Microbiology Center of the China Microbial Culture Collection Management Committee |
– | the main degradation products were alkanes, which are nontoxic |
the degradation rates of fluazinam, BHC, PCNB, chlorpyrifos, and DDT in the medium were 94.77%, 70.34%,77.92%,78.30%,66.70%, |
GC–MS HPLC |
(Zhang et al., 2019) |
|
Rhodococcusrhodochrous sp. AQ1, Bacillus tequilensis sp. AQ2, Bacillus aryabhattai sp. AQ3 and Bacillus safensis sp. AQ4 |
Metribuzin (MB) | soil samples were collected from potato vegetated field at Arifwala, Pakistan |
Complete biomineralization into water and carbon di-oxide | desamino-metribuzin (DA), diketo-metribuzin (DK) and desamino-diketometribuzin (DADK) |
98.63% MB degradation was observed | GC–MS HPLC |
(Wahla et al., 2019) |
| Cupriavidussp. ISTL7 | Carbofuran | waste sampling performed at Ghazipur landfill Delhi, India | hydrolysis pathway starting from carbofuran todegrade and form carbofuran-7-phenol and methylamine Carbofuran-7-phenol further degrades to form 3-(2‑hydroxy-2-methylpropyl) benzene-1,2-diol whilemethylamineentersthe glyphosate pathway |
carbofuran- 7-phenol, methylamine, 2‑hydroxy-3-(3-methylpropan-2-ol)benzene-N-methyl-carbamate etc. |
strain ISTL7 efficiently degraded approximately 98% of carbofuran (400 ppm) within 96 h |
FTIR GC–MS |
(Gupta et al., 2019) |
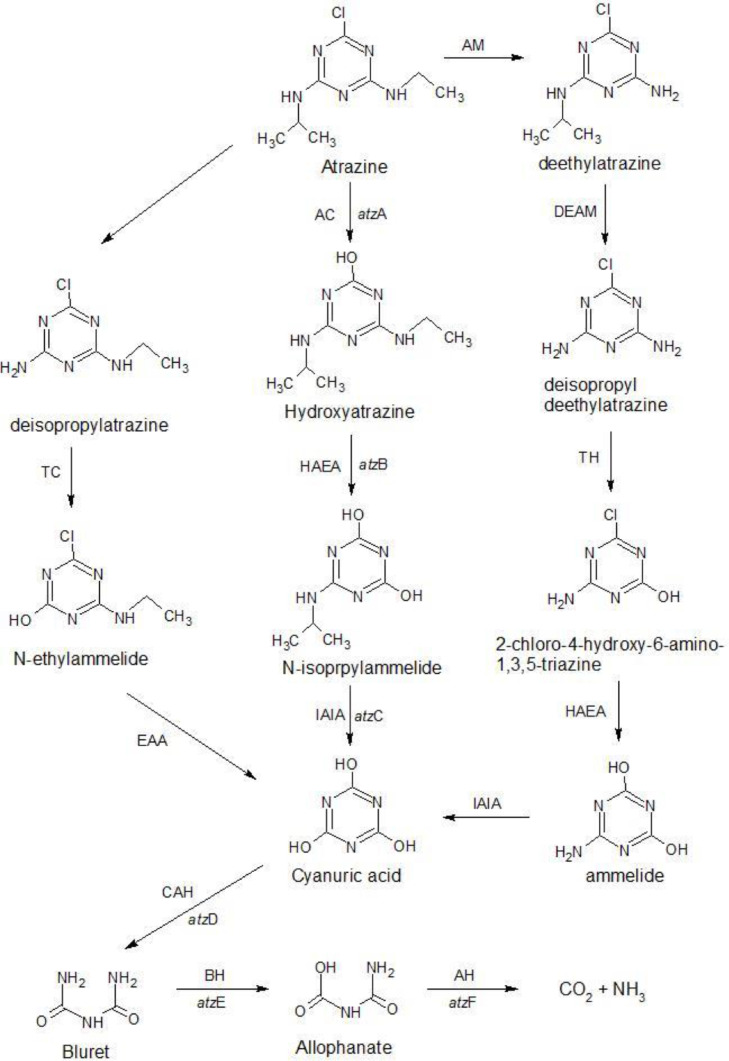
Biodegradation of Atrazine is well studied and the genes involved are well characterized. The major genes that take part in its biodegradation process are atzA, atzB, atzC, atzD, atzE, atzF (Ref. to Fig. 2) found in different atrazine degrading bacteria (B. Singh and Singh, 2016b)
Fig. 2.
Example of biodegradation pathway - Atrazine (with genes and enzymes) (De Souza et al., 1998; Mandelbaum et al., 1993; Martinez et al., 2001). AM, atrazine Monooxygenase; AC, atrazine chlorohydrolase; AH, allophanate hydrolase; BH, biuret hydrolase; CAH, cyanuric acid hydrolase; DIHA, deisopropyhidroxylatrazine amidohydrolase; DEAM, deethylatrazine monooxygenase; EAA, N-ethylammelide amidohydrolase; HAEA, hidroxyatrazine ethylaminohydrolase; IAIA, N-isopropylammelide isopropylamidohydrolase; TC, s-triazine chlorohydrolase; TH, s-triazine hydrolase.
Several enzymes responsible for pesticide degradation have been identified and isolated that include transferases, isomerases, hydrolases, ligases, esterases, peroxidases and oxidases. These enzymes perform reactions such as hydrolysis, oxidation, reduction of nitro group to amino group, ring cleavage etc. (J. P. Verma et al., 2014). An increase in the efficiency of lindane removal was observed when pure culture Streptomyces Sp. were immobilized on different matrices (Saez et al., 2012). Several environmental factors have been found to affect the biotransformation efficiency including pH, temperature, salinity, carbon dioxide and oxygen concentration (Yichen Y. Huang et al., 2018). Optimization of these factors were done by Taguchi design of experiment (DOE) method to achieve 98.63% degradation of metribuzin under pH 7, temperature 30 °C and pesticide concentration 45 mg/L by a microbial consortium consisting of microbial species Rhodococcus rhodochrous sp. AQ1, Bacillus tequilensis sp. AQ2, Bacillus aryabhattai sp. AQ3 and Bacillus safensis sp. AQ4 (Wahla et al., 2019). The taguchi DOE is used to improve the optimization of reducing the effect of noise factors. It utilizes orthogonal arrays to optimize various physio-chemical parameters (Basak et al., 2013). The effect of surfactant in improving the solubilization of pesticide aiding in the process of biodegradation by the microorganism. It was found that rhamnolipid and sophorolipid enhanced the solubilization of chlorinated pesticide hexachlorocyclohexane by 3–9 folds resulting in increased efficiency of biodegradation of the pesticide by Sphingomonas sp. In both liquid medium and soil slurry (Manickam et al., 2012). Biofilms are already known to be important biogeochemical cycling and removal of pollutants from the ecosystem. The ability of natural river biofilm to degrade carbofuran and carbaryl was studied and effect of different seasons on the biodegradation efficiency by biofilms was also checked. It was observed that the ability of river biofilms to degrade carbofuran in four different seasons were similar (54.1–59.5%) but the biofilms showed low efficiency in degrading carbaryl (0–27.5%) (Tien et al., 2013). Genetic engineering has enormous potential in improving the process of biodegradation by making recombinant strains. Lindane is a highly persistent toxic pesticide which impairs photosynthesis, respiration and nitrogen-fixation in Anabaena. To solve this problem linA2 gene encoding dehydrochlorinase (obtained from Sphingomonas paucimobilis B90) was knocked-in and overexpressed in Anabaena genome. The resulting recombinant Anabaena was able to degrade over 98% of 10 ppm lindane within 10 days (Chaurasia et al., 2013). Microbial fuel cells (MFCs) are also being constructed to degrade organic waste and to simultaneously generate electricity. An experiment was conducted in which soil MFC could remove 71.15% of the hexachlorobenzene provided (Cao et al., 2015). Apart from soil and water sources from environmentally contaminated area, the biodegrading microbes can also be isolated from higher living organisms present there. Five different bacterial strains capable of degrading endosulfan were isolated from microflora of Blatta orientalis (cockroach). The isolated bacteria were identified as Pseudomonas aeruginosa G1, Stenotrophomonas maltophilia G2, Bacillus atrophaeus G3, Citrobacter amolonaticus G4 and Acinetobacter lwoffii G5 based on morphological, biochemical and fatty acid profile analysis (FAME). They were capable of degrading endosulfan and had efficiency range between 56 and 89%. Similarly five different strains of endophytic bacteria were isolated from rice plants and were found to be capable of degrading chlorpyrifos both invivo as well as invitro (Feng et al., 2017)
4. Pharmaceuticals
Pharmaceuticals comprise a class of relatively emerging contaminants compared to others. Pharmaceuticals are utilized all over the world to treat diseases or to maintain the health humans or animals. Various kinds of pharmaceuticals contaminating the environment include antibiotics, analgesics, antacids, tranquillizers, stimulants, antipyretic, lipid regulators, anti-depressants, and other various prescription and non-prescription drugs (Rana et al., 2017). Although pharmaceuticals have been present in water bodies for decades the paradigm of considering them environment contaminants has shifted towards the start of 21st century. Pharmaceuticals are excreted out of the human system after being transformed into a metabolite or without transformation. The excreta from humans as sewage carries these pharmaceuticals towards wastewater treatment plants (WWTP). If the wastewater is not treated properly, the effluent from WWTP becomes a cause of concern for the aquatic ecosystem after being release into the water bodies (Rivera-Utrilla et al., 2013). Aquaculture, hospital wastewater and illegal drug disposal can be the other sources of contamination (Caracciolo et al., 2015). Pharmaceuticals as emerging contaminants are unique in the sense that they are designed to be active even at low concentrations. Moreover, their target includes enzymes or receptors which can be conserved among evolutionary distant organisms. Impact of these contaminants in the range of ng/L to μg/L have shown to cause sub-lethal effects in non-target organisms in literature (Hampel et al., 2010; Mimeault et al., 2005).
4.1. Toxicity effects of pharmaceuticals
It is quite obvious how pharmaceuticals play a major role in increasing the life span of humans by decreasing the potential risk of diseases and ultimately treating them. The possible effects on other organisms and environment are becoming clearer with the help of research focusing on ecotoxicology effects of pharmaceuticals. NSAID (non-steroidal anti-inflammatory drugs) is the most important class of drugs – Ibuprofen, ketoprofen and aspirin are its most important members. These drugs are incompletely degraded and their discharge into sewage or release into surface water ultimately leads to their accumulation which poses a threat to aquatic life (Gómez-Oliván et al., 2014). Ibuprofen is linked with nephrotoxic effects in Rhamdia quelen (Mathias et al., 2018). NSAIDs are also linked with decreased photosynthetic and respiratory rates in green algae Scenedesmus obliquus (H. Wang et al., 2020). Salicylic acid, primary metabolite of acetylsalicylic acid (aspirin), was found to be responsible for oxidative stress and neurotoxicity in Mytilus galloprovincialis (mussel). It was observed that the acetylcholinesterase activity was decreased when treated with ketoconazole and erythromycin both singly as well as in combination, providing indication of potential neurotoxicity to the animal (Liu et al., 2017). When wistar rats were treated with pharmaceutical wastewater, necrosis of renal epithelial cells in kidney, inflammation in endocardium and cellular swelling in liver were observed (Sharif et al., 2016). Pharmaceutical wastewater constitutes mixture of pharmaceuticals in low concentrations rather than isolated drugs. Therefore, studies focusing on treatment with mixture of pharmaceuticals might provide a more suitable approach to find potential toxicity in organisms present in environment. In environment, drugs may interact with each other and interfere with the mode of action or work independently. This may lead to increased or decreased effect on the non-target organisms (Geiger et al., 2016). For example, the toxic effect on Lissodelphis peronii in the form of loss of tactile response was relatively higher on exposure to mixture of Naproxen, Carbamazepine and Sulfamethoxazole as compared to when individual compounds were tested, however the concentrations of drugs used were much higher than those found in the environment (Melvin et al., 2014). To predict potential targets of drugs on evolutionary close or distant species, databases such as ECOdrug (www.ecodrug.org) can also be utilized.
4.2. Microbe mediated remediation of pharmaceuticals
As it is evident by now that microbes play a major role in biodegradation of xenobiotics, pharmaceuticals have also been found to be degraded by the microbes. In fact, some microbes utilize these contaminants as source of their energy by complete mineralization. Biodegradation provides a feasible method of removing contaminants because the physical methods, advanced oxidation process, activated carbon are limited by high energy requirement and production of toxic by-products (Homem and Santos, 2011; Schwarzenbach et al., 2006). Some of the examples of biodegradation by microbes are shown in table below:
An important pharmaceutical contaminant is Paracetamol (PAM). It is a common over-the-counter drug used most commonly as antipyretic. Microbial fuel cell was coupled with Fenton oxidation process to provide a method by which PAM could be degraded without external power supply (L. Zhang et al., 2015). MFC generally consists of anode and cathode. Microorganisms are grown on anode and are known as electricigens. They promote the electron transfer to cathode and the oxidized pollutants on cathode are reduced (Logan, 2009). Nootropic drugs as environmental contaminants and their ecotoxicological effects have sparked little interest. These drugs are not readily metabolized in the system and as much as 90% of the administered drug is reported to be excreted out through urine (Mache et al., 2012). Piracetam (2-oxo-1-pyrrolidine acetamide) is an example of nootropic drugs which was found to be completely mineralized by two species of Ochrobactrum. Its biodegradation occurs through cleavage of the heterocyclic ring at the C–N bond (Woźniak-Karczewska et al., 2018). It was the first report on complete biodegradation of piracetam. However, more insight is needed to elucidate its complete biodegradation pathway. A strain of thermophilic microorganism, Thermus thermophilus C419 was found to be capable of biodegrading members of fluoroquinolones such as ciprofloxacin, ofloxacin, norfloxacin, enrofloxacin. This suggests that microbes can be also utilized to treat harsh environments which are also contaminated (Pan et al., 2018). Acrylonitrile degrading Corynebacterum sp. D5 utilized nitrile hydratase and amidase in a two-step reaction to generate acrylamide and acrylic acid, although it couldn't completely mineralize it (Sunarko and Sulistinah, 2019). Over 90% of Iopromide and 70% of Carbamazepine was found to be degraded by fungi Gymnopilus luteofolius and Stropharia rugosoannulata respectively when self-immobilized in pellet morphology (Castellet-Rovira et al., 2018)
5. Explosive waste
It wouldn't be surprising for anyone to consider the residues from explosives as environmental contaminants. Explosives are used worldwide most commonly for the use as military ammunition. Explosives are also used in underground mining, demolition work and in construction industry for new roads. Explosives have high content of nitrogen and oxygen which on explosion leave toxic waste in the environment.
Various classes of explosives include nitrate esters, nitroaromatics, and (B. Singh et al., 2012). Conventional methods of treating explosive contaminated site (incineration and composting) suffer limitations such as high expenditure of energy and cost as well as exposure of workers to toxins (Esteve-Núñez et al., 2001)
5.1. Toxicity effects of explosive waste
Most of the data concerning toxic effects of explosive waste comes from TNT and RDX. Dissolved oxygen (DO), chemical oxygen demand (COD), total dissolved solids (TDS) and conductivity are some of the most important parameters that are studied in context to understand and evaluate the toxicity due to the effluents from production of explosives.
TNT wastewater has been shown to have poor biodegradability because of high COD (Ye et al., 2011). Effect of TNT (provided in oral form) was studied in wild cotton rats (Sigmodon hispidus), histopathological studies revealed that TNT caused splenic congestion, lymphoid hyperplasia and increase in liver weight in both males and females. The number of erythrocytes and level of hemoglobin were decreased in both sexes, whereas increase in level of methemoglobin and enhanced activity of glutathione S-transferases (GST) were observed in males (Reddy et al., 2000). Recent advancement in technology provided direct manner for studying toxicity effects of environmental contaminants. Single Plane Illumination Microscopy (SPIM) provided a 3D approach to visualize toxicity effects in zebrafish in developmental stages due to exposure to TNT. The resultant toxic effects observed were – high level of apoptosis in actively developing tissues, cardiac looping defects and hypoplastic heart chamber formation (Eum et al., 2016).
5.2. Microbe mediated remediation of explosive waste
Microbes have been found to successfully metabolize the residual from explosive waste as the source of their growth. Biodegradation of nitrate esters occurs through denitration reaction (Christodoulatos et al., 1997). PETN reductase enzyme was isolated from Enterobacter species (Ref. to Table 4) which was able to transform PETN (pentaerythritol tetranitrate) into nitrites and nitrates (Binks et al., 1996). TNT (2,4,6-trinitrotoluene) is member of the class nitroaromatic compounds. The main pathway by which TNT is biodegraded starts with initial hydrogenation reaction to yield hydride-Meisenheimer complex of TNT (H-TNT) (Vorbeck et al., 1998). Environmental factors also contribute to the degree of biodegradation. For example, it was found that using initial neutral or slightly acidic medium (pH 6) favored the biodegradation of TNT by Yarrowia lipolytica AN-L15 which grew and added to the acidity of the medium by release of organic acids (Ziganshin et al., 2010). It was observed that if co-substrate starch (slow releasing carbon source) was added for co-metabolism by a developed microbial consortium led to complete mineralization and detoxification of DDT under nitrogen-fixing conditions (Khan et al., 2015).
Table 3.
Microbial biodegradation of pharmaceuticals.
| Microorganism/Consortium | Pharmaceutical | Isolated from/Source | Degradation Pathway | Degradation Intermediates (Metabolites) | Efficiency | Technique used | Reference |
|---|---|---|---|---|---|---|---|
| Trametes versicolor | clofibric acid (CLOFI, lipid regulator) and carbamazepine (CARBA, antiepileptic/analgetic) | American Type Culture Collection. | cytochrome P450 system may be involved in the first step of CLOFI and CARBA oxidation by T. versicolor |
– | CLOFI (91%) and CARBA (58%) | (GC–CIRMS) NMR |
(Marco-Urrea et al., 2009) |
| Pseudomonas sp. SA01 | Phenol | samples were taken from pharmaceutical plant wastewater effluent located west of Tehran |
meta-cleavage pathway. | – | isolated strain started to degrade 0.7 g/l of phenol after an initial very short lag phase, and phenol decomposition was then rapidly completed within 30 h |
UV–vis SEM |
(Shourian et al., 2009) |
| Mixed culture of Heterotrophic Bacteria | clofibric acid | mixture of soil contaminated with several herbicides, including propanil, and soil from organic rice agriculture supplemented with (NH4)2SO4 and propanil |
biocatalysis and biodegradation database (BBD) software developed by the University of Minnesota (UM) was used to simulate and predict the biodegradation pathway of CLF |
_-hydroxyisobutyric acid, lactic acid and 4-chlorophenol. | 51% biodegradation (initial CLF concentration = 2 mg/L) | HPLC–DAD GC–MS |
(Salgado et al., 2012) |
| Pseudomonas sp. Strain CE21 and Strain CE 22 | cefalexin | Wastewater samples containing activated sludge were collected from sewage treatment plant in Hong Kong | – | 2‑hydroxy-3-phenyl pyrazine | Strain CE22 was able to degrade over 90% of cefalexin, while CE21 was able to remove46.7% of cefalexin after incubation for 24 h | HPLC MS-MS |
(Lin et al., 2015) |
| Achromobacterdenitrificans PR1 | Sul-famethoxazole (SMX) | Samples of activated sludge and treated domestic wastewater collected from a wastewater treatment plant in the North of Portugal | sulfonamide was used as sole source of carbon, nitrogen andenergy | 3-amino-5-methylisoxazole | Strain PR1 was able to removeSMX at a rate of 73.6 μmolSMX/gcell dry weight | DGGE HPLC–UV–vis |
(Reis et al., 2014) |
| Labrys portucalensis strain F11 | Fluoxetine (FLX) | Sediment sample collected from an industrially contaminated site in Northern Portugal |
fluorobenzene (FB) was used as sole carbon and energy source | stoichiometric liberation of fluoride | 2 μM of racemic FLX was completely removed of both enantiomers in 30 d | HPLC analysis | (Moreira et al., 2014) |
| Streptomyces MIUG 4.89 | carbamazepine | Microbial Cultures Collection of the Bioaliment Research Center,‘Dunarea de Jos’ University of Galati, Romania. |
extracellular laccase production thought to play role in degradation |
– | 35% degradation at an initial concentration of 0.2 mg/L of carbamazepine | HPLC | (Popa et al., 2014) |
| Ustilago sp. SMN03 | Cefdinir | Wastewater was collected from a pharmaceutical industry located in Ranipet, Vellore Dist., India |
Cefdinir was utilized as a sole carbon source |
six novel intermediates formed |
isolate was found to degrade 81% of cefdinir within 6 days and an initial cefdinir concentration of 200 mg/L |
UV–vis LC-MS FTIR |
(Selvi et al., 2014) |
| Ochrobactrumsp. Strain WX-J1 | Erythromycin A(EA) | Soil contaminated by EA was collected at a site near a pharmaceutical factory, Henan, China |
Strain WX-J1 can utilized EA as a sole source of carbon and energy |
3-depyranosyloxy erythromycin A, 7,12-dyhydroxy-6-deoxyerythronolide B, 6-deoxyerythronolide B and propionaldehyde |
when the initial Erythromycin A concentration was 100 mg/L, 97% of Erythromycin A was degraded |
HPLC–(UV)-MS | (C. Zhang et al., 2017) |
|
Citrobacter amalonaticusRashtia |
Paclitaxel | samples were collected from wastewater chamber of the Sobhan oncology pharmaceutical company |
The isolate utilized Paclitaxel as the sole carbon source. Aerobic degradation pathway is suggested by authors |
– | 87–93% efficacy under aerobic condition | HPLC | (Zamani et al., 2016) |
Table 4.
Biodegradation of explosive waste by microbes.
| Microorganism | Explosive waste | Isolated From | Degradation pathway | Degradation product | Efficiency/Specific degradation rate | Technique used | Ref. |
|---|---|---|---|---|---|---|---|
| Phanerochaetechrysosporium | TNT | Forest Products Laboratory | Degradation occurs by reduction of nitro groups | 2amDNT 4amDNT |
The initial concentration of TNT was 30 mg/L. This concentration of TNT was reduced to less than 60 μg/L at the end of the 96 h incubation |
HPLC NMR GC–MS |
(Bumpus and Tatarko, 1994) |
| Stenotrophomonas maltophiliaPB1 | RDX | Soil and water samples were collected from a site that had been heavily contaminated with RDX and HMX |
isolate from the culture used RDX as a sole source of nitrogen for growth | methylene-N-(hydroxymethyl)-hydroxylamine-N’-(hydroxymethyl) nitroamine |
specific degradation rate was a value of 0.22 mmol of N per s/kg of protein |
HPLC NMR Mass Spectrometry |
(Binks et al., 1995) |
|
Enterobacter cloacaePB2 |
PETN | Soil and water samples were collected from a site that had been heavily contaminated with munition compounds |
Isolate was found to use PETN as a sole source of nitrogen for growth |
pentaerythritol dinitrate, 3‑hydroxy-2,2-bis-[(nitrooxy) methyl] propanal, and 2,2-bis [(nitrooxy)methyl]-propanedial | specific degradation rate gave a value of 1.03 mmol of PETN/g of protein per hour |
Mass Spectrometry NMR HPLC |
(Binks et al., 1996) |
| Pseudomonas putida strain TP1 and Pseudomonas aeruginosa strain TP6 | TNT | Soil samples collected from a TNT-contaminated site located in southern Taiwan |
Both strains demonstrated the ability to grow on the medium containing TNT as a carbon, energy, and nitrogen source |
– | More than 90% of the TNT in the growth medium was degraded by both strains after 22 days incubation |
HPLC | (Chien et al., 2014) |
| Mixed culture | NTO | Soil Samples | degradation occurred via reduction of nitro-groups | 3- amino-1,2,4-triazol-5-one (ATO) and 3-hydroxyamino-1,2,4- triazol-5-one (HTO) |
– | HPLC-DAD QToF-MS |
(Krzmarzick et al., 2015) |
| Mixed Culture | TET and PETN | textile wastewater treatment plant activated sludge | PETNdegradation in the aerobic condition follows a successive reductive degradation pathway with the release of NO2- in each denitration step. TNT biodegradation involved reduction of one nitro group to form a hydroxylamino group and subsequent reduction of the other nitro group to an amino group | pentaerythritoldinitrate,3‑hydroxy-2,2-bis [(nitrooxy)methyl]propanal,and2,2-bis-[(nitrooxy)methyl]-propanedial for PETN and amino-4, 6-dinitrotoluene and 4-amino-2, 6-dinitrotoluene for TNT |
Addition of rhamnolipid surfactant (60 mg/l) increased the removal efficiencies of TNT and PETN from 53% and 57% to 98% and 91%, respectively | HPLC LC-MS |
(Karami et al., 2017) |
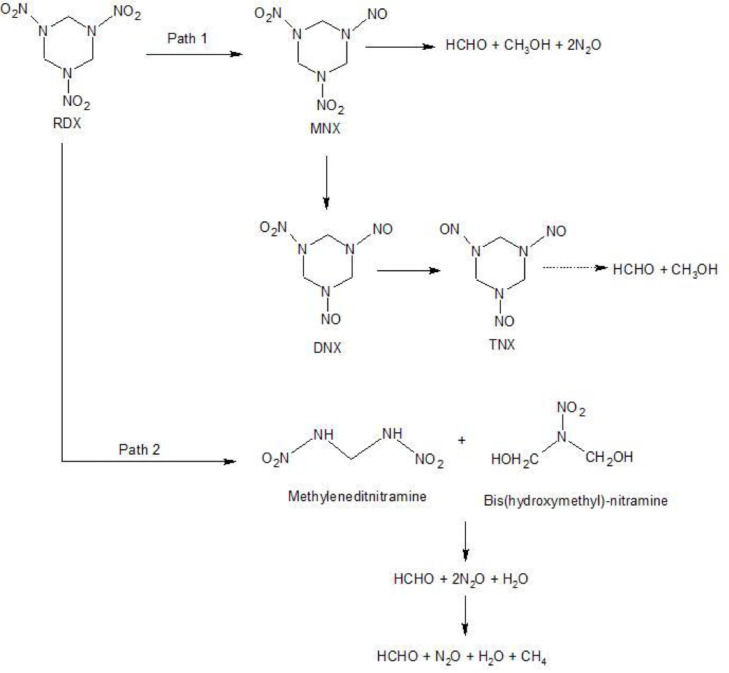
RDX (Hexahydro-1,3,5-trinitro-1,3,5-triazine), a member of the nitramines class of explosives has been widely used in military munition leading to soil and groundwater contamination. It has been found to be degraded via sequential reduction of the N—NO2 groups leading to the generation of mono-, di- and tri-nitroso derivatives (Ref. to Fig. 3). Alternatively, RDX can also proceed to degradation via direct ring cleavage pathway (Halasz et al., 2002). Another study pointed towards the role of electron transport machinery in degradation of RDX. cymA gene was disrupted by transposon sequence in RDX-defective strain of Shewanella oneidensis. This isolated defective strain degraded RDX at a minimal rate (10% of the wild type) compared to the wild strain providing evidence that cymA (c-type cytochrome) has a major role to play in anaerobic reduction of RDX (Perreault et al., 2012). Several species capable of biodegradation have been isolated, enriched and identified from contaminated area as shown in Table 4. Whereas some species have also been identified by the use of stable isotope probing which is a culture independent method that targets only active organisms. The method involves uptake of labelled substrate and subsequent incorporation of labelled atoms into nucleic acids (Andeer et al., 2012). Members of the classes Spirochaetes, Bacteroidia and a-Proteobacteria which were not previously observed were found to be capable of RDX degradation using this method.
Fig. 3.
RDX microbial biodegradation pathways. Path 1 (via nitroso derivatives) and Path 2 (direct ring cleavage pathway) are illustrated (Hawari et al., 2000). MNX, DNX and TNX are mono, di- and tri- nitroso derivates of RDX respectively.
6. PAHs
Polycyclic aromatic hydrocarbons (PAHs) are a class of organic compounds composed of multiple fused benzene rings released from incomplete combustion or pyrolysis of materials containing carbon and hydrogen. PAHs are derived from various natural and anthropogenic activities. The natural sources include volcano eruptions and forest fires. While the primary source of PAHs are anthropogenic sources such as coal combustion, vehicle exhaust, petroleum volatilization, biomass burning, coke plants, steel plants and many other industries. The US EPA and the EU have categorized sixteen PAH compounds as priority pollutants because of their potential toxicity. They include naphthalene, acenaphthylene, acenaphthene, fluorene, phenanthrene, anthracene, fluoranthene, pyrene, benzo[a]anthracene, chrysene, benzo[b]fluoranthene, benzo[k]fluoranthene, benzo[a]pyrene, benzo[g,h,i]perylene, indeno[1,2,3-c,d]pyrene, and dibenz[a,h]anthracene (Hussar et al., 2012). PAHs are present in both gas phase and particulate phase, they are widely dispersed in air, fresh and sea water, sediment, soil and biota. Light molecular weight (LMW) PAHs consisting of 2 to 3 benzene rings have higher vapor pressures, thereby being dominant in gas phase whereas the high molecular weight (HMW) PAHs with 5–6 benzene rings contribute to particle phase. The medium molecular weight (MMW) PAHs with 4 rings are present in both gas and particulate phases (Dat and Chang, 2017). Properties of PAHs like hydrophobicity, low water solubility and strong tendency to absorb to the soil matrix make them persistent in the environment. These factors contribute to low PAH bioavailability and consequently low biodegradation rate. (Abdel-Shafy and Mansour, 2016)
6.1. Toxicity effects of PAHs
Various PAH pollutants pose severe threat to the environment and human health because of their potential toxicity. Most important sources of exposure to PAHs include food containing PAHs, smoke from open fireplaces, and cigarettes (Abdel-Shafy and Mansour, 2016). Some PAHs are well known to have properties of carcinogenicity, mutagenicity, and teratogenicity. Toxicity arises due to metabolism of PAHs leading to formation of reactive metabolites such as diol-epoxides, radical cations and o-quinones (active carcinogens). These reactive metabolites result in DNA adducts, which lead to DNA mutations, ultimately resulting in alteration of gene expression profiles (Moorthy et al., 2015). Increased exposure to PAHs affects human fertility and has been associated with incidences of male sterility, loss of ovarian functions (Bidgoli et al., 2011). and early onset of natural menopause (Yun Y. Huang et al., 2018). Epidemiological studies indicate that the ability of PAHs to cross the placental barrier can lead to developmental toxicity. Prenatal or early postnatal exposure to PAHs can lead to various complications like intrauterine growth retardation, low IQ, problems with behavior, allergies or asthma (Drwal et al., 2019). Exposure to PAHs is also linked with diabetes (Yang et al., 2017), oxidative stress (Wang et al., 2015), hepatotoxicity (F. Li et al., 2020b), cutaneous toxicity (Prieux et al., 2019) and many short-term health effects including nausea, vomiting, and inflammation (Gao et al., 2018).
6.2. Microbe mediated remediation of PAHs
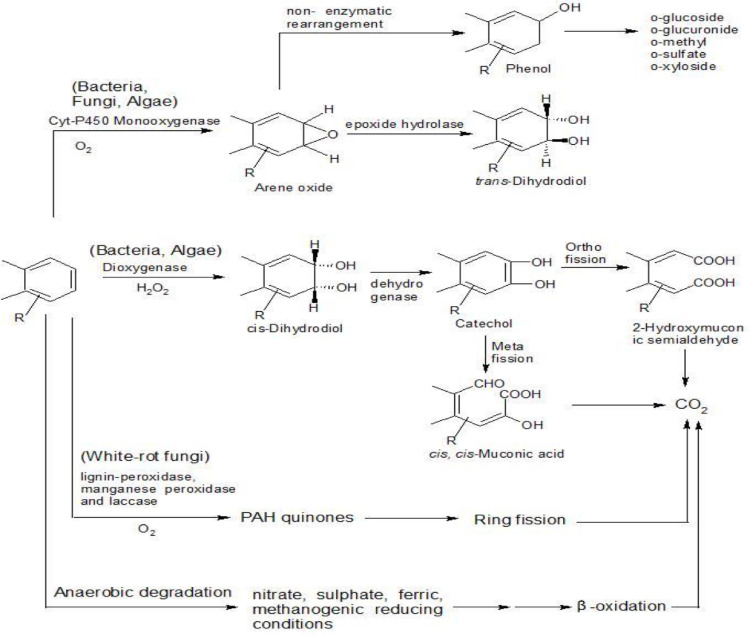
A large variety of bacteria, fungi and algae have been isolated that are capable of degrading PAHs using varying metabolic pathways (Ref to Table 5). Aerobic bacteria as well as algae use mono and di-oxygenases for the activation and cleavage of benzene rings, whereas anaerobic bacteria follow PAH catabolism by entirely different pathways and enzymes with metal- and/or flavin-containing cofactors. Various lignolytic and non-lignolytic fungi are able to oxidize PAH and the enzymes involved in the initial attack are mainly Cytochrome P-450 mono oxygenase and lignin degrading enzymes such as manganese peroxidase, lignin peroxidase, phenoloxidases (laccases, tyrosinases), and H2O2-producing enzymes (Kadri et al., 2019) (Ref to Fig. 4).
Table 5.
Biodegradation of different PAHs by various microbes.
| Microorganism/Co-Culture/Consortium | PAHs | Isolation from or Source | Degradation Pathway/Enzymes Involved | Degradation Product (Metabolite) | Percentage Transformation | Techniques Used | Reference |
|---|---|---|---|---|---|---|---|
| Halomonas sp. | Phenanthrene (Phe), pyrene (Pyr), naphthalene (NaP), and benzo [a]pyrene (BaP) | Brackish water sample from Pichavaram mangrove, Tamil Nadu, India, | – | – | Phe (67%), Pyr (63%), NaP (60%), BaP (58%) |
– | (Govarthanan et al., 2020) |
| Ganoderma sp. | Naphthalene, phenanthrene and fluorene | – | Extracellular ligninolytic enzymes (laccase and non-specific peroxidases) | variable | naphthalene 34—73%, phenanthrene 9—67%, fluorene 11—64% | GC–MS | (Torres-Farradá et al., 2019) |
| Pleurotus ostreatus | napthalene | Pharmaceutical Microbiology Laboratory (NCRRT -Egypt) |
Naphthalene dioxygenase and ligninolytic enzymes |
α, β-naphthol, salicylic and benzoic acid |
86.47% | HPLC and Thin layer chromatography (TLC) | (Elhusseiny et al., 2019) |
| Aspergillus terricola var americanus | Benz (a) Anthracene, Dibenz (a, h) Anthracene and Indeno [1, 2, 3-cd] Pyrene | Microbial Type Culture Collection and Gene Bank (MTCC), Chandigarh | both extracellular (Laccase enzyme) and intracellular (cytochrome P450 monooxygenase oxidation) pathways | variable | 94.80, 90.16, and 93.80%, respectively, after 10 days | GC–MS | (Guntupalli et al., 2019) |
| Pseudomonas sp. JPN2 | pyrene | crude oil was collected from Dagang Oilfield, Tianjin Province, Northern China | Aerobic degradation through dioxygenase enzyme system | 4,5-dihydroxy-4,5-dihydropyrene, 4-phenanthrol, 1‑hydroxy-2-naphthoic acid and phthalate | 82.88% after 25 d | GC–MS | (Jin et al., 2016) |
| Pseudomonas sp. JP1 | benzo[a]pyrene (BaP), fluoranthene, and phenanthrene | Shantou Bay, Shantou, China | Anaerobic biodegradation with nitrate as the electron acceptor | variable | 30, 47, and 5%, respectively | GC/MS | (Liang et al., 2014) |
| Ulva prolifera | Phenanthrene | coastal water (Rushan City, China) | – | – | 91.3% | – | (C. Zhang et al., 2017) |
| Chlorella vulgaris | fluorene | Culture Collection of Algae of Bushehr Shrimp Research Institute, Iran | dioxygenase enzyme system based degradation | N-Hydroxymethylcarbazol, Dibutyl phthalate, Hexadecanoic acid, ethyl ester, 1,2-Benzenedicarboxylic acid, dioctyl ester |
– | GC–MS | (Asghari et al., 2019) |
| Anabaena fertilissima | anthracene (ant) and pyrene (pyr) | center for conservation and utilization of blue green algae, iari, new delhi, india | – | degraded product for ANT was 2, 4-Dimethyl-1-heptene and for PYR it was 2, 3, 4-Trimethylhexane | degradation of ANT by 46% and PYR by 33%, at 5.0 mg/L and 3.0 mg/L | GC/MS | (Patel et al., 2016) |
| Cellulosimicrobium cellulans CWS2 | benzo(a)pyrene | PAH contaminated soil | Anaerobic degradation under nitrate-reducing conditions | pyrene, 1-aminopyrene, phenanthrene, 1-methylphenanthrene, 1,7-dimethylnaphthalene, 1-(2-hydroxypropyl)naphthalene, 1-methylnaphthalene, 2‑hydroxy-3-(3-methyl-2-butenyl)−1,4-naphthalenedione, diethyl phthalate, and 2-acetyl-3-methoxybenzoc acid | 78.8%) was observed in 13 days | GC–MS | (Qin et al., 2018) |
| Achromobacter xylosoxidans Strain DN002 | Fluoranthene | petroleum-contaminated soil | Aerobic degradation through dioxygenases (catechol 1,2 dioxygenase and catechol 2,3 dioxygenase) | – | 92.8% after 14 days | – | (Ma et al., 2015) |
| Hydrogenophaga sp. PYR1 | pyrene and benzo[a]pyrene | river sediments in the east area of Taihu Lake (a large shallow lake in China) | Anaerobic degradation under ferric iron reduction conditions | benzoic acid, 2‑hydroxy-phenyl ester and naphthalene,1,2,3-trimethyl-4-propenyl | 94% pyrene within 15 d | GC–MS | (Yan et al., 2017) |
| Mycobacterium gilvum | pyrene | activated sludge from a coking wastewater treatment plant of SGIS Songshan Co., Ltd., China | Aerobic degradation through dioxygenases | Phthalic acid, 1-Naphthol, 4-Phenanthrenol, 4-Phenanthrenecarboxylic acid | 95% of pyrene (50 mg L − 1) in 7 days | GC–M– | (Wu et al., 2019) |
Fig. 4.
Different pathways for biodegradation of PAHs by microbes (Bogan et al., 1996; Cerniglia, 1992; Eaton and Chapman, 1992; Gibson and Parales, 2000; Mueller et al., 1995).
Most hydrocarbon-contaminated sites such as soil or groundwater system are anoxic, but anaerobic hydrocarbon biodegradation rates are extremely low.
Activated carbon (AC) has been shown to act as conductive material facilitating direct interspecies electron transfer (DIET) between bacteria attached to the AC particles (Lovley, 2017). Under anaerobic conditions the biodegradation potential of naphthalene was strongly stimulated (96%) by the AC addition and the diversity of microbial communities increased and was structurally changed with increase in abundance of Geobacter, Thiobacillus, Sulfuricurvum, and methanogenic archaea (Bonaglia et al., 2020). Microbial co-metabolism has also been found to increase degradation rate of PAHs. Microbacterium sp. strain mediated degradation of BaP (benzo[a]pyrene) increased notably with the addition of PHE (phenanthrene), and was not accelerated by PYR (pyrene) under denitrifying conditions (Qin et al., 2017).
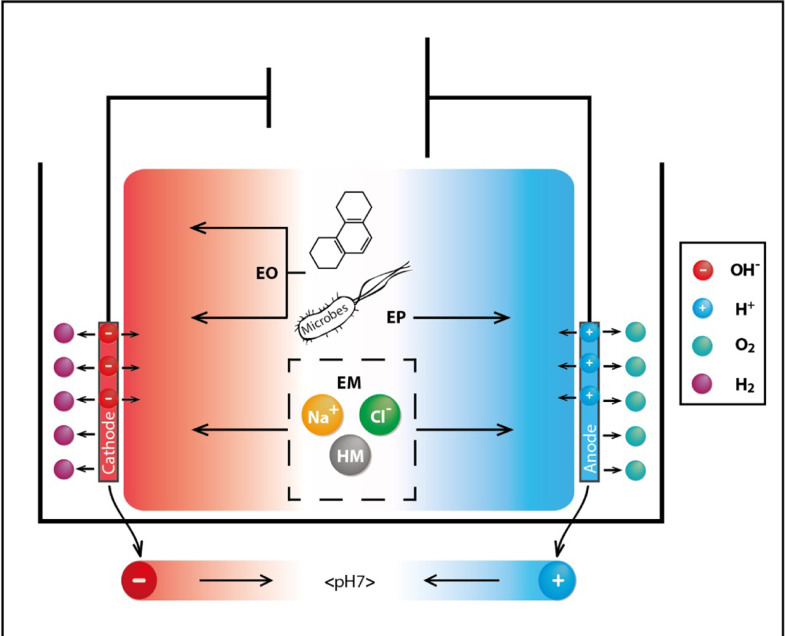
Low molecular weight (LMW) PAHs are more water-soluble and therefore more readily biodegradable than high molecular weight (HMW) ones. Some bacterial species have been reported to produce emulsifier substances called biosurfactants which can adeptly increase the solubility of these contaminants resulting in increased bioavailability and biodegradation of slightly soluble PAHs. Thermophilic strain Aeribacillus pallidus SL-1 evaluated for the biodegradation of crude oil and PAHs at 60 °C was found to produce SL-bioemulsifier which improved the solubility of PAHs (Tao et al., 2020). Recently, electro-bioremediation has emerged as a promising method for in situ remediation with ability to enhance the removal efficiency. Application of electric potential gradient among electrodes located in a contaminated site forms the basis of this method (Refer to Fig. 5). Electro-bioremediation used for field-scale remediation of a coking plant site resulted in 29.3% and 44.4% increase in degradation of the total and 4–6-ring PAHs, compared to bioremediation alone. Also, the total toxicity equivalent concentrations of total PAHs and 4-, 5- and 6-ring PAHs decreased 49.0%, 63.7%, 48.2% and 30.1%, respectively (F. Li et al., 2020).
Fig. 5.
Working of an electro-bioremediation setting (Acuña et al., 2012). The effect of electrokinetic phenomena on porous soil. Hydroxide ions and hydrogen gas are generated at the cathode and hydrogen ions and oxygen gas at the anode. pH gradient generated throughout the affected subsurface facilitates electrokinetic migration of soil constituents. Microbes and PAHs move to the cathode by electroosmosis (EO). Electronegative microbes move to the anode electrophoretically (EP). Whereas electromigration (EM) is responsible in the movement of ions and heavy metals (HM).
7. Conclusions and future perspectives
There has been great progress in the field of bioremediation, although the complete utilization of microbes for bioremediation of sites looks like a possibility of near to midterm future. Despite the high energy and expenditure required, conventional methods of treatment of environmental contaminants are mostly still in use. There is obvious need for bioremediation to take leaps in success to make it more suitable options compared to conventional methods. Some species have been found to be performing biotransformation rather than complete mineralization of toxic wastes which ultimately leads to production of metabolites of low, equal and sometimes even of higher toxicity than the initial toxic compound. Recent studies have made it clear that mixed consortia utilization can be more advantageous compared to pure cultures for complete mineralization. Co-metabolism using co-substrate can be a good method to improve the efficiency of biodegradation. The recent rise in the field of bioinformatics and the use of statistics can pave a way for easier optimization of culture conditions of microorganisms achieving optimum efficiency of biodegradation. There is still a lot of scope in this field to be covered, for example there are still many species for which biodegradation pathways are yet to be elucidated. Elucidation of these pathways can provide genetic engineering to improve the potential of biodegradation using microorganisms drastically.
Credit author statement
Paramdeep Kaur and Deepanshu Monga: Software, Data curation, Writing- Original draft preparation, Visualization, Investigation. Baljinder Singh: Conceptualization, Methodology, Supervision, Editing, Validation
Declaration of Competing Interest
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
References
- Abdel-Shafy H.I., Mansour M.S. A review on polycyclic aromatic hydrocarbons: source, environmental impact, effect on human health and remediation. Egypt. J. Pet. 2016;25(1):107–123. [Google Scholar]
- Abe F.R., Soares A.M., de Oliveira D.P., Gravato C. Toxicity of dyes to zebrafish at the biochemical level: cellular energy allocation and neurotoxicity. Environ. Pollut. 2018;235:255–262. doi: 10.1016/j.envpol.2017.12.020. [DOI] [PubMed] [Google Scholar]
- Acuña A.J., Pucci O.H., Pucci G.N. InTech New Tech Oil Gas Ind. 2012. Electrobioremediation of hydrocarbon contaminated soil from Patagonia Argentina; pp. 29–48. [Google Scholar]
- Agrawal S., Tipre D., Patel B., Dave S. Optimization of triazo Acid Black 210 dye degradation by Providencia sp. SRS82 and elucidation of degradation pathway. Process Biochem. 2014;49(1):110–119. [Google Scholar]
- Andeer P., Strand S.E., Stahl D.A. High-sensitivity stable-isotope probing by a quantitative terminal restriction fragment length polymorphism protocol. Appl. Environ. Microbiol. 2012;78(1):163–169. doi: 10.1128/AEM.05973-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asad S., Amoozegar M., Pourbabaee A.A., Sarbolouki M., Dastgheib S. Decolorization of textile azo dyes by newly isolated halophilic and halotolerant bacteria. Bioresour. Technol. 2007;98(11):2082–2088. doi: 10.1016/j.biortech.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Bae J.-.S., Freeman H.S., Kim S.D. Influences of new azo dyes to the aquatic ecosystem. Fibers Polym. 2006;7(1):30–35. [Google Scholar]
- Basak B., Bhunia B., Dutta S., Dey A. Enhanced biodegradation of 4-chlorophenol by Candida tropicalis PHB5 via optimization of physicochemical parameters using Taguchi orthogonal array approach. Int. Biodeterior. Biodegradation. 2013;78:17–23. [Google Scholar]
- Bidgoli S.A., Karimi M., Asami Z., Baher H., Zavarhei M.D. Association between testicular Aryl hydrocarbon Receptor levels and idiopathic male infertility: a case–control study in Iran. Sci. Total Environ. 2011;409(18):3267–3273. doi: 10.1016/j.scitotenv.2011.03.024. [DOI] [PubMed] [Google Scholar]
- Binks P.R., French C.E., Nicklin S., Bruce N.C. Degradation of pentaerythritol tetranitrate by Enterobacter cloacae PB2. Appl. Environ. Microbiol. 1996;62(4):1214–1219. doi: 10.1128/aem.62.4.1214-1219.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binks P.R., Nicklin S., Bruce N.C. Degradation of hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine (RDX) by Stenotrophomonas maltophilia PB1. Appl. Environ. Microbiol. 1995;61(4):1318–1322. doi: 10.1128/aem.61.4.1318-1322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan B., Lamar R., Hammel K. Fluorene Oxidation In Vivo by Phanerochaete chrysosporium and In Vitro during Manganese Peroxidase-Dependent Lipid Peroxidation. Appl. Environ. Microbiol. 1996;62(5):1788–1792. doi: 10.1128/aem.62.5.1788-1792.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaglia S., Broman E., Brindefalk B., Hedlund E., Hjorth T., Rolff C., Nascimento F.J., Udekwu K., Gunnarsson J.S. Activated carbon stimulates microbial diversity and PAH biodegradation under anaerobic conditions in oil-polluted sediments. Chemosphere. 2020;248 doi: 10.1016/j.chemosphere.2020.126023. [DOI] [PubMed] [Google Scholar]
- Bumpus J.A., Tatarko M. Biodegradation of 2, 4, 6-trinitrotoluene by Phanerochaete chrysosporium: identification of initial degradation products and the discovery of a TNT metabolite that inhibits lignin peroxidases. Curr. Microbiol. 1994;28(3):185–190. [Google Scholar]
- Cai S., Cai T., Liu S., Yang Q., He J., Chen L., Hu J. Biodegradation of N-methylpyrrolidone by Paracoccus sp. NMD-4 and its degradation pathway. Int. Biodeterior. Biodegrad. 2014;93:70–77. [Google Scholar]
- Cao X., Song H.-l., Yu C.-y., Li X.-n. Simultaneous degradation of toxic refractory organic pesticide and bioelectricity generation using a soil microbial fuel cell. Bioresour. Technol. 2015;189:87–93. doi: 10.1016/j.biortech.2015.03.148. [DOI] [PubMed] [Google Scholar]
- Caracciolo A.B., Topp E., Grenni P. Pharmaceuticals in the environment: biodegradation and effects on natural microbial communities. A review. J. Pharm. Biomed. Anal. 2015;106:25–36. doi: 10.1016/j.jpba.2014.11.040. [DOI] [PubMed] [Google Scholar]
- Castellet-Rovira F., Lucas D., Villagrasa M., Rodríguez-Mozaz S., Barceló D., Sarrà M. Stropharia rugosoannulata and Gymnopilus luteofolius: promising fungal species for pharmaceutical biodegradation in contaminated water. J. Environ. Manage. 2018;207:396–404. doi: 10.1016/j.jenvman.2017.07.052. [DOI] [PubMed] [Google Scholar]
- Cerniglia C. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation. 1992;3(2–3):351–368. [Google Scholar]
- Chaurasia A.K., Adhya T.K., Apte S.K. Engineering bacteria for bioremediation of persistent organochlorine pesticide lindane (γ-hexachlorocyclohexane) Bioresour. Technol. 2013;149:439–445. doi: 10.1016/j.biortech.2013.09.084. [DOI] [PubMed] [Google Scholar]
- Chequer F.M.D., Lizier T.M., de Felício R., Zanoni M.V.B., Debonsi H.M., Lopes N.P., Marcos R., de Oliveira D.P. Analyses of the genotoxic and mutagenic potential of the products formed after the biotransformation of the azo dye Disperse Red 1. Toxicol. in Vitro. 2011;25(8):2054–2063. doi: 10.1016/j.tiv.2011.05.033. [DOI] [PubMed] [Google Scholar]
- Chien C.C., Kao C.M., Chen D.Y., Chen S.C., Chen C.C. Biotransformation of trinitrotoluene (TNT) by Pseudomonas spp. isolated from a TNT-contaminated environment. Environ. Toxicol. Chem. 2014;33(5):1059–1063. doi: 10.1002/etc.2553. [DOI] [PubMed] [Google Scholar]
- Christodoulatos C., Bhaumik S., Brodman B.W. Anaerobic biodegradation of nitroglycerin. Water Res. 1997;31(6):1462–1470. [Google Scholar]
- Chung K.-.T. Azo dyes and human health: a review. J. Environ. Sci. Health. 2016;34(4):233–261. doi: 10.1080/10590501.2016.1236602. [DOI] [PubMed] [Google Scholar]
- Cleinmensen S., Jensen J.C., Jensen N.J., Meyer O., Olsen P., Würtzen G. Toxicological studies on malachite green: a triphenylmethane dye. Arch. Toxicol. 1984;56(1):43–45. doi: 10.1007/BF00316351. [DOI] [PubMed] [Google Scholar]
- Corsini E., Sokooti M., Galli C., Moretto A., Colosio C. Pesticide induced immunotoxicity in humans: a comprehensive review of the existing evidence. Toxicology. 2013;307:123–135. doi: 10.1016/j.tox.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Dat N.-.D., Chang M.B. Review on characteristics of PAHs in atmosphere, anthropogenic sources and control technologies. Sci. Total Environ. 2017;609:682–693. doi: 10.1016/j.scitotenv.2017.07.204. [DOI] [PubMed] [Google Scholar]
- de Souza M., Newcombe D., Alvey S., Crowley D., Hay A., Sadowsky M., Wackett L. Molecular basis of a bacterial consortium: interspecies catabolism of Atrazine. Appl. Environ. Microbiol. 1998;64(1):178–184. doi: 10.1128/aem.64.1.178-184.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drwal E., Rak A., Gregoraszczuk E.L. polycyclic aromatic hydrocarbons (PAHs)—Action on placental function and health risks in future life of newborns. Toxicology. 2019;411:133–142. doi: 10.1016/j.tox.2018.10.003. [DOI] [PubMed] [Google Scholar]
- Du L.-.N., Wang S., Li G., Wang B., Jia X.-.M., Zhao Y.-.H., Chen Y.-.L. Biodegradation of malachite green by Pseudomonas sp. strain DY1 under aerobic condition: characteristics, degradation products, enzyme analysis and phytotoxicity. Ecotoxicology. 2011;20(2):438–446. doi: 10.1007/s10646-011-0595-3. [DOI] [PubMed] [Google Scholar]
- Eaton R., Chapman P. Bacterial metabolism of naphthalene: construction and use of recombinant bacteria to study ring cleavage of 1,2-dihydroxynaphthalene and subsequent reactions. J. Bacteriol. 1992;174(23):7542–7554. doi: 10.1128/jb.174.23.7542-7554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhusseiny S.M., Amin H.M., Shebl R.I. Modulation of laccase transcriptome during biodegradation of naphthalene by white rot fungus Pleurotus ostreatus. Int. Microbiol. 2019;22(2):217–225. doi: 10.1007/s10123-018-00041-5. [DOI] [PubMed] [Google Scholar]
- Enayatizamir N., Tabandeh F., Rodríguez-Couto S., Yakhchali B., Alikhani H.A., Mohammadi L. Biodegradation pathway and detoxification of the diazo dye Reactive Black 5 by Phanerochaete chrysosporium. Bioresour. Technol. 2011;102(22):10359–10362. doi: 10.1016/j.biortech.2011.08.130. [DOI] [PubMed] [Google Scholar]
- Esteve-Núñez A., Caballero A., Ramos J.L. Biological degradation of 2, 4, 6-trinitrotoluene. Microbiol. Mol. Biol. Rev. 2001;65(3):335–352. doi: 10.1128/MMBR.65.3.335-352.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eum J., Kwak J., Kim H.J., Ki S., Lee K., Raslan A.A., Park O.K., Chowdhury M.A.U., Her S., Kee Y. 3D visualization of developmental toxicity of 2, 4, 6-trinitrotoluene in zebrafish embryogenesis using light-sheet microscopy. Int. J. Mol. Sci. 2016;17(11):1925. doi: 10.3390/ijms17111925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng F., Ge J., Li Y., He S., Zhong J., Liu X., Yu X. Enhanced degradation of chlorpyrifos in rice (Oryza sativa L.) by five strains of endophytic bacteria and their plant growth promotional ability. Chemosphere. 2017;184:505–513. doi: 10.1016/j.chemosphere.2017.05.178. [DOI] [PubMed] [Google Scholar]
- Fernando E., Keshavarz T., Kyazze G. Complete degradation of the azo dye Acid Orange-7 and bioelectricity generation in an integrated microbial fuel cell, aerobic two-stage bioreactor system in continuous flow mode at ambient temperature. Bioresour. Technol. 2014;156:155–162. doi: 10.1016/j.biortech.2014.01.036. [DOI] [PubMed] [Google Scholar]
- Gao P., da Silva E., Hou L., Denslow N.D., Xiang P., Ma L.Q. Human exposure to polycyclic aromatic hydrocarbons: metabolomics perspective. Environ. Int. 2018;119:466–477. doi: 10.1016/j.envint.2018.07.017. [DOI] [PubMed] [Google Scholar]
- Geiger E., Hornek-Gausterer R., Saçan M.T. Single and mixture toxicity of pharmaceuticals and chlorophenols to freshwater algae Chlorella vulgaris. Ecotoxicol. Environ. Saf. 2016;129:189–198. doi: 10.1016/j.ecoenv.2016.03.032. [DOI] [PubMed] [Google Scholar]
- Gibson D., Parales R. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr. Opin. Biotechnol. 2000;11(3):236–243. doi: 10.1016/s0958-1669(00)00090-2. [DOI] [PubMed] [Google Scholar]
- Gómez-Oliván L.M., Galar-Martínez M., Islas-Flores H., García-Medina S., SanJuan-Reyes N. DNA damage and oxidative stress induced by acetylsalicylic acid in Daphnia magna. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2014;164:21–26. doi: 10.1016/j.cbpc.2014.04.004. [DOI] [PubMed] [Google Scholar]
- Guo Y., Shen O., Han J., Duan H., Yang S., Zhu Z., Tong J., Zhang J. Circadian rhythm genes mediate fenvalerate-induced inhibition of testosterone synthesis in mouse Leydig cells. J. Toxicol. Environ. Health Part A. 2017;80(23–24):1314–1320. doi: 10.1080/15287394.2017.1384148. [DOI] [PubMed] [Google Scholar]
- Govarthanan M., Khalifa A.Y., Kamala-Kannan S., Srinivasan P., Selvankumar T., Selvam K., Kim W. Significance of allochthonous brackish water Halomonas sp. on biodegradation of low and high molecular weight polycyclic aromatic hydrocarbons. Chemosphere. 2020;243 doi: 10.1016/j.chemosphere.2019.125389. [DOI] [PubMed] [Google Scholar]
- Govindwar S.P., Kurade M.B., Tamboli D.P., Kabra A.N., Kim P.J., Waghmode T.R. Decolorization and degradation of xenobiotic azo dye Reactive Yellow-84A and textile effluent by Galactomyces geotrichum. Chemosphere. 2014;109:234–238. doi: 10.1016/j.chemosphere.2014.02.009. [DOI] [PubMed] [Google Scholar]
- Guillén-Jiménez F.d.M., Cristiani-Urbina E., Cancino-Díaz J.C., Flores-Moreno J.L., Barragán-Huerta B.E. Lindane biodegradation by the Fusarium verticillioides AT-100 strain, isolated from Agave tequilana leaves: kinetic study and identification of metabolites. Int. Biodeterior. Biodegrad. 2012;74:36–47. [Google Scholar]
- Guntupalli S., Thunuguntla V., Chalasani L.M., Rao C., Bondili J.S. Degradation and metabolite profiling of benz (a) anthracene, dibenz (a, h) anthracene and indeno [1, 2, 3-cd] pyrene by Aspergillus terricola. Polycycl. Aromat. Compd. 2019;39(1):84–92. [Google Scholar]
- Gupta J., Rathour R., Singh R., Thakur I.S. Production and characterization of extracellular polymeric substances (EPS) generated by a carbofuran degrading strain Cupriavidus sp. ISTL7. Bioresour. Technol. 2019;282:417–424. doi: 10.1016/j.biortech.2019.03.054. [DOI] [PubMed] [Google Scholar]
- Hadibarata T., Adnan L.A., Yusoff A.R.M., Yuniarto A., Zubir M.M.F.A., Khudhair A.B., Teh Z.C., Naser M.A. Microbial decolorization of an azo dye reactive black 5 using white-rot fungus Pleurotus eryngii F032. Water Air Soil Pollut. 2013;224(6):1–9. [Google Scholar]
- Halasz A., Spain J., Paquet L., Beaulieu C., Hawari J. Insights into the formation and degradation mechanisms of methylenedinitramine during the incubation of RDX with anaerobic sludge. Environ. Sci. Technol. 2002;36(4):633–638. doi: 10.1021/es011071g. [DOI] [PubMed] [Google Scholar]
- Hamadache M., Benkortbi O., Hanini S., Amrane A., Khaouane L., Moussa C.S. A quantitative structure activity relationship for acute oral toxicity of pesticides on rats: validation, domain of application and prediction. J. Hazard. Mater. 2016;303:28–40. doi: 10.1016/j.jhazmat.2015.09.021. [DOI] [PubMed] [Google Scholar]
- Hampel M., Alonso E., Aparicio I., Bron J.E., Santos J.L., Taggart J.B., Leaver M.J. Potential physiological effects of pharmaceutical compounds in Atlantic salmon (Salmo salar) implied by transcriptomic analysis. Environ. Sci. Pollut. Res. 2010;17(4):917–933. doi: 10.1007/s11356-009-0282-6. [DOI] [PubMed] [Google Scholar]
- Hawari J., Halasz A., Sheremata T., Beaudet S., Groom C., Paquet L., et al. Characterization of metabolites during biodegradation of hexahydro-1,3,5-Trinitro-1,3,5-Triazine (RDX) with municipal anaerobic sludge. Appl. Environ. Microbiol. 2000;66(6):2652–2657. doi: 10.1128/aem.66.6.2652-2657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homem V., Santos L. Degradation and removal methods of antibiotics from aqueous matrices–a review. J. Environ. Manage. 2011;92(10):2304–2347. doi: 10.1016/j.jenvman.2011.05.023. [DOI] [PubMed] [Google Scholar]
- Huang Y., Guo J., Lv N., Li S., Wu Y., Bai R., Shen J., Chen G., Zhang D. Associations of urinary polycyclic aromatic hydrocarbons with age at natural menopause in US women aged 35–65, NHANES 2003–2012. Environ. Pollut. 2018;243:1878–1886. doi: 10.1016/j.envpol.2018.09.109. [DOI] [PubMed] [Google Scholar]
- Hussar E., Richards S., Lin Z.-.Q., Dixon R.P., Johnson K.A. Human health risk assessment of 16 priority polycyclic aromatic hydrocarbons in soils of Chattanooga, Tennessee, USA. Water Air Soil Pollut. 2012;223(9):5535–5548. doi: 10.1007/s11270-012-1265-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilamathi R., Jayapriya J. Microbial fuel cells for dye decolorization. Environ. Chem. Lett. 2018;16(1):239–250. [Google Scholar]
- Jadhav S.S., David M. Biodegradation of flubendiamide by a newly isolated Chryseobacterium sp. strain SSJ1. 3 Biotech. 2016;6(1):31. doi: 10.1007/s13205-015-0347-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain K., Shah V., Chapla D., Madamwar D. Decolorization and degradation of azo dye–Reactive Violet 5R by an acclimatized indigenous bacterial mixed cultures-SB4 isolated from anthropogenic dye contaminated soil. J. Hazard. Mater. 2012;213:378–386. doi: 10.1016/j.jhazmat.2012.02.010. [DOI] [PubMed] [Google Scholar]
- Jin J., Yao J., Zhang Q., Liu J. Biodegradation of pyrene by pseudomonas sp. JPN2 and its initial degrading mechanism study by combining the catabolic nahAc gene and structure-based analyses. Chemosphere. 2016;164:379–386. doi: 10.1016/j.chemosphere.2016.08.113. [DOI] [PubMed] [Google Scholar]
- Kabeer F.A., John N., Abdulla M.H. Biodegradation of malachite green by a newly isolated Bacillus vietnamensis sp. MSB17 from continental slope of the Eastern Arabian Sea: enzyme analysis, degradation pathway and toxicity studies. Bioremediat. J. 2019;23(4):334–342. [Google Scholar]
- Kadri T., Cuprys A., Rouissi T., Brar S.K. Environmental Contaminants: Ecological Implications and Management. Springer; 2019. Microbial degradation of polyaromatic hydrocarbons; pp. 101–117. [Google Scholar]
- Kagawa N., Nagao T. Neurodevelopmental toxicity in the mouse neocortex following prenatal exposure to acetamiprid. J. Appl. Toxicol. 2018;38(12):1521–1528. doi: 10.1002/jat.3692. [DOI] [PubMed] [Google Scholar]
- Karami M.A., Amin M.M., Bina B., Sadani M., Mirzaei N., Teimouri F., Akrami A. Enhanced aerobic biodegradation of soil contaminated with explosives (TNT and PETN) by rhamnolipid. Eurasian J. Anal. Chem. 2017;12:641–652. [Google Scholar]
- Khan M.I., Lee J., Yoo K., Kim S., Park J. Improved TNT detoxification by starch addition in a nitrogen-fixing Methylophilus-dominant aerobic microbial consortium. J. Hazard. Mater. 2015;300:873–881. doi: 10.1016/j.jhazmat.2015.08.032. [DOI] [PubMed] [Google Scholar]
- Kong L., Zhu S., Zhu L., Xie H., Su K., Yan T., Wang J., Wang J., Wang F., Sun F. Biodegradation of organochlorine pesticide endosulfan by bacterial strain Alcaligenes faecalis JBW4. J. Environ. Sci. 2013;25(11):2257–2264. doi: 10.1016/s1001-0742(12)60288-5. [DOI] [PubMed] [Google Scholar]
- Krzmarzick M.J., Khatiwada R., Olivares C.I., Abrell L., Sierra-Alvarez R., Chorover J., Field J.A. Biotransformation and degradation of the insensitive munitions compound, 3-nitro-1, 2, 4-triazol-5-one, by soil bacterial communities. Environ. Sci. Technol. 2015;49(9):5681–5688. doi: 10.1021/acs.est.5b00511. [DOI] [PubMed] [Google Scholar]
- Kumar N., Sinha S., Mehrotra T., Singh R., Tandon S., Thakur I.S. Biodecolorization of azo dye acid black 24 by Bacillus pseudomycoides: process optimization using box Behnken design model and toxicity assessment. Bioresour. Technol. Rep. 2019;8 [Google Scholar]
- Kurade M.B., Waghmode T.R., Jadhav M.U., Jeon B.-.H., Govindwar S.P. Vol. 5. 2015. Bacterial–yeast consortium as an effective biocatalyst for biodegradation of sulphonated azo dye Reactive Red 198; pp. 23046–23056. (RSC Adv.). [Google Scholar]
- Lade H., Kadam A., Paul D., Govindwar S. Biodegradation and detoxification of textile azo dyes by bacterial consortium under sequential microaerophilic/aerobic processes. EXCLI J. 2015;14:158. doi: 10.17179/excli2014-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Guo S., Wu B., Wang S. Pilot-scale electro-bioremediation of heavily PAH-contaminated soil from an abandoned coking plant site. Chemosphere. 2020;244 doi: 10.1016/j.chemosphere.2019.125467. [DOI] [PubMed] [Google Scholar]
- Liang L., Song X., Kong J., Shen C., Huang T., Hu Z. Anaerobic biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by a facultative anaerobe Pseudomonas sp. JP1. Biodegradation. 2014;25(6):825–833. doi: 10.1007/s10532-014-9702-5. [DOI] [PubMed] [Google Scholar]
- Lin B., Lyu J., Lyu X.-j., Yu H.-q., Hu Z., Lam J.C., Lam P.K. Characterization of cefalexin degradation capabilities of two Pseudomonas strains isolated from activated sludge. J. Hazard. Mater. 2015;282:158–164. doi: 10.1016/j.jhazmat.2014.06.080. [DOI] [PubMed] [Google Scholar]
- Liu J., Cai Y., Lu G., Dan X., Wu D., Yan Z. Interaction of erythromycin and ketoconazole on the neurological, biochemical and behavioral responses in crucian carp. Environ. Toxicol. Pharmacol. 2017;55:14–19. doi: 10.1016/j.etap.2017.08.002. [DOI] [PubMed] [Google Scholar]
- Liu W., You Y., Sun D., Wang S., Zhu J., Liu C. Decolorization and detoxification of water-insoluble Sudan dye by Shewanella putrefaciens CN32 co-cultured with Bacillus circulans BWL1061. Ecotoxicol. Environ. Saf. 2018;166:11–17. doi: 10.1016/j.ecoenv.2018.09.055. [DOI] [PubMed] [Google Scholar]
- Logan B.E. Exoelectrogenic bacteria that power microbial fuel cells. Nat. Rev. Microbiol. 2009;7(5):375–381. doi: 10.1038/nrmicro2113. [DOI] [PubMed] [Google Scholar]
- Lovley D.R. Syntrophy goes electric: direct interspecies electron transfer. Annu. Rev. Microbiol. 2017;71:643–664. doi: 10.1146/annurev-micro-030117-020420. [DOI] [PubMed] [Google Scholar]
- Ma Y.-.L., Lu W., Wan L.-.L., Luo N. Elucidation of fluoranthene degradative characteristics in a newly isolated Achromobacter xylosoxidans DN002. Appl. Biochem. Biotechnol. 2015;175(3):1294–1305. doi: 10.1007/s12010-014-1347-7. [DOI] [PubMed] [Google Scholar]
- Mache S., Eickenhorst P., Vitzthum K., Klapp B.F., Groneberg D.A. Cognitive-enhancing substance use at German universities: frequency, reasons and gender differences. Wiener Medizinische Wochenschrift. 2012;162(11):262–271. doi: 10.1007/s10354-012-0115-y. [DOI] [PubMed] [Google Scholar]
- Mandelbaum R., Wackett L., Allan D. Mineralization of the s-triazine ring of atrazine by stable bacterial mixed cultures. Appl. Environ. Microbiol. 1993;59(6):1695–1701. doi: 10.1128/aem.59.6.1695-1701.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manickam N., Bajaj A., Saini H.S., Shanker R. Surfactant mediated enhanced biodegradation of hexachlorocyclohexane (HCH) isomers by Sphingomonas sp. NM05. Biodegradation. 2012;23(5):673–682. doi: 10.1007/s10532-012-9543-z. [DOI] [PubMed] [Google Scholar]
- Mansour H.B., Ayed-Ajmi Y., Mosrati R., Corroler D., Ghedira K., Barillier D., Chekir-Ghedira L. Acid violet 7 and its biodegradation products induce chromosome aberrations, lipid peroxidation, and cholinesterase inhibition in mouse bone. marrow. 2010;17(7):1371–1378. doi: 10.1007/s11356-010-0323-1. [DOI] [PubMed] [Google Scholar]
- Marco-Urrea E., Pérez-Trujillo M., Vicent T., Caminal G. Ability of white-rot fungi to remove selected pharmaceuticals and identification of degradation products of ibuprofen by Trametes versicolor. Chemosphere. 2009;74(6):765–772. doi: 10.1016/j.chemosphere.2008.10.040. [DOI] [PubMed] [Google Scholar]
- Martinez B., Tomkins J., Wackett L., Wing R., Sadowsky M. Complete Nucleotide Sequence and Organization of the Atrazine Catabolic Plasmid pADP-1 from Pseudomonas sp . Strain ADP. J. Bacteriol. 2001;183(19):5684–5697. doi: 10.1128/JB.183.19.5684-5697.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias F.T., Fockink D.H., Disner G.R., Prodocimo V., Ribas J.L.C., Ramos L.P., Cestari M.M., Silva de Assis H.C. Effects of low concentrations of ibuprofen on freshwater fish Rhamdia quelen. Environ Toxicol Pharmacol. 2018;59:105–113. doi: 10.1016/j.etap.2018.03.008. [DOI] [PubMed] [Google Scholar]
- Melvin S.D., Cameron M.C., Lanctôt C.M. Individual and mixture toxicity of pharmaceuticals naproxen, carbamazepine, and sulfamethoxazole to Australian striped marsh frog tadpoles (Limnodynastes peronii) J Toxicol Environ Health A. 2014;77(6):337–345. doi: 10.1080/15287394.2013.865107. [DOI] [PubMed] [Google Scholar]
- Meriç S., Selçuk H., Belgiorno V. Acute toxicity removal in textile finishing wastewater by Fenton's oxidation, ozone and coagulation–flocculation processes. Water Res. 2005;39(6):1147–1153. doi: 10.1016/j.watres.2004.12.021. [DOI] [PubMed] [Google Scholar]
- Mimeault C., Woodhouse A., Miao X.-.S., Metcalfe C., Moon T., Trudeau V. The human lipid regulator, gemfibrozil bioconcentrates and reduces testosterone in the goldfish, Carassius auratus. Aquatic. Toxicol. 2005;73(1):44–54. doi: 10.1016/j.aquatox.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Moorthy B., Chu C., Carlin D.J. Polycyclic aromatic hydrocarbons: from metabolism to lung cancer. Toxicol. Sci. 2015;145(1):5–15. doi: 10.1093/toxsci/kfv040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira I.S., Ribeiro A.R., Afonso C.M., Tiritan M.E., Castro P.M. Enantioselective biodegradation of fluoxetine by the bacterial strain Labrys portucalensis F11. Chemosphere. 2014;111:103–111. doi: 10.1016/j.chemosphere.2014.03.022. [DOI] [PubMed] [Google Scholar]
- Mueller E.J., Loida P.J., Sligar S.G. Cytochrome P450. Springer; Boston, MA: 1995. Twenty-five years of P450 cam research; pp. 83–124. [Google Scholar]
- Mukherjee T., Das M. Degradation of malachite green by Enterobacter asburiae strain XJUHX-4TM. CLEAN–Soil, Air, Water. 2014;42(6):849–856. [Google Scholar]
- Pan L.-j., Li J., Li C.-x., Yu G.-w., Wang Y. Study of ciprofloxacin biodegradation by a Thermus sp. isolated from pharmaceutical sludge. J. Hazard. Mater. 2018;343:59–67. doi: 10.1016/j.jhazmat.2017.09.009. [DOI] [PubMed] [Google Scholar]
- Patel J.G., Kumar J.N., Kumar R.N., Khan S.R. Biodegradation capability and enzymatic variation of potentially hazardous polycyclic aromatic hydrocarbons—Anthracene and pyrene by Anabaena fertilissima. Polycycl. Aromat. Compd. 2016;36(1):72–87. [Google Scholar]
- Perreault N.N., Crocker F.H., Indest K.J., Hawari J. Involvement of cytochrome c CymA in the anaerobic metabolism of RDX by Shewanella oneidensis MR-1. Can. J. Microbiol. 2012;58(2):124–131. doi: 10.1139/w11-116. [DOI] [PubMed] [Google Scholar]
- Popa C., Favier L., Dinica R., Semrany S., Djelal H., Amrane A., Bahrim G. Potential of newly isolated wild Streptomyces strains as agents for the biodegradation of a recalcitrant pharmaceutical, carbamazepine. Environ. Technol. 2014;35(24):3082–3091. doi: 10.1080/09593330.2014.931468. [DOI] [PubMed] [Google Scholar]
- Prasad S.S., Aikat K. Study of bio-degradation and bio-decolourization of azo dye by Enterobacter sp. SXCR. Environ. Technol. 2014;35(8):956–965. doi: 10.1080/09593330.2013.856957. [DOI] [PubMed] [Google Scholar]
- Qi S., Niu X., hui Wang D., Wang C., Zhu L., Xue X., Zhang Z., Wu L. Flumethrin at sublethal concentrations induces stresses in adult honey bees (Apis mellifera L.) Sci. Total Environ. 2020;700 doi: 10.1016/j.scitotenv.2019.134500. [DOI] [PubMed] [Google Scholar]
- Qin W., Fan F., Zhu Y., Huang X., Ding A., Liu X., Dou J. Anaerobic biodegradation of benzo (a) pyrene by a novel Cellulosimicrobium cellulans CWS2 isolated from polycyclic aromatic hydrocarbon-contaminated soil. Braz. J. Microbiol. 2018;49:258–268. doi: 10.1016/j.bjm.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W., Fan F., Zhu Y., Wang Y., Liu X., Ding A., Dou J. Comparative proteomic analysis and characterization of benzo (a) pyrene removal by Microbacterium sp. strain M. CSW3 under denitrifying conditions. Bioprocess Biosyst. Eng. 2017;40(12):1825–1838. doi: 10.1007/s00449-017-1836-5. [DOI] [PubMed] [Google Scholar]
- Qu Y., Cao X., Ma Q., Shi S., Tan L., Li X., Zhou H., Zhang X., Zhou J. Aerobic decolorization and degradation of Acid Red B by a newly isolated Pichia sp. TCL. J. Hazard. Mater. 2012;223:31–38. doi: 10.1016/j.jhazmat.2012.04.034. [DOI] [PubMed] [Google Scholar]
- Rana R.S., Singh P., Kandari V., Singh R., Dobhal R., Gupta S. A review on characterization and bioremediation of pharmaceutical industries’ wastewater: an Indian perspective. Appl Water Sci. 2017;7(1):1–12. [Google Scholar]
- Rathour R., Gupta J., Tyagi B., Kumari T., Thakur I.S. Biodegradation of pyrene in soil microcosm by Shewanella sp. ISTPL2, a psychrophilic, alkalophilic and halophilic bacterium. Bioresour. Technol. Rep. 2018;4:129–136. [Google Scholar]
- Reddy G., Chandra S.A., Lish J.W., Quails C.W., Jr Toxicity of 2, 4, 6-trinitrotoiuene (TNT) in hispid cotton rats (Sigmodon hispidus): hematological, biochemical, and pathological effects. Int. J. Toxicol. 2000;19(3):169–177. [Google Scholar]
- Reis P.J., Reis A.C., Ricken B., Kolvenbach B.A., Manaia C.M., Corvini P.F., Nunes O.C. Biodegradation of sulfamethoxazole and other sulfonamides by Achromobacter denitrificans PR1. J. Hazard. Mater. 2014;280:741–749. doi: 10.1016/j.jhazmat.2014.08.039. [DOI] [PubMed] [Google Scholar]
- Rivera-Utrilla J., Sánchez-Polo M., Ferro-García M.Á., Prados-Joya G., Ocampo-Pérez R. Pharmaceuticals as emerging contaminants and their removal from water. A review. Chemosphere. 2013;93(7):1268–1287. doi: 10.1016/j.chemosphere.2013.07.059. [DOI] [PubMed] [Google Scholar]
- Saez J.M., Benimeli C.S., Amoroso M.J. Lindane removal by pure and mixed cultures of immobilized actinobacteria. Chemosphere. 2012;89(8):982–987. doi: 10.1016/j.chemosphere.2012.06.057. [DOI] [PubMed] [Google Scholar]
- Salgado R., Oehmen A., Carvalho G., Noronha J., Reis M. Biodegradation of clofibric acid and identification of its metabolites. J. Hazard. Mater. 2012;241:182–189. doi: 10.1016/j.jhazmat.2012.09.029. [DOI] [PubMed] [Google Scholar]
- Sariwati A., Purnomo A.S., Kamei I. Abilities of Co-cultures of Brown-Rot Fungus Fomitopsis pinicola and Bacillus subtilis on Biodegradation of DDT. Curr. Microbiol. 2017;74(9):1068–1075. doi: 10.1007/s00284-017-1286-y. [DOI] [PubMed] [Google Scholar]
- Schwarzenbach R.P., Escher B.I., Fenner K., Hofstetter T.B., Johnson C.A., Von Gunten U., Wehrli B. The challenge of micropollutants in aquatic systems. Science. 2006;313(5790):1072–1077. doi: 10.1126/science.1127291. [DOI] [PubMed] [Google Scholar]
- Selvi A., Salam J.A., Das N. Biodegradation of cefdinir by a novel yeast strain, Ustilago sp. SMN03 isolated from pharmaceutical wastewater. World. J. Microbiol. Biotechnol. 2014;30(11):2839–2850. doi: 10.1007/s11274-014-1710-4. [DOI] [PubMed] [Google Scholar]
- Sharif A., Ashraf M., Javeed A., Anjum A.A., Akhtar M.F., Akhtar B., Saleem A. Oxidative stress responses in Wistar rats on subacute exposure to pharmaceutical wastewater. Environ. Sci. Pollut. Res. 2016;23(23):24158–24165. doi: 10.1007/s11356-016-7717-7. [DOI] [PubMed] [Google Scholar]
- Sharma A., Gangola S., Khati P., Kumar G., Srivastava A. Novel pathway of cypermethrin biodegradation in a Bacillus sp. strain SG2 isolated from cypermethrin-contaminated agriculture field. 3 Biotech. 2016;6(1):45. doi: 10.1007/s13205-016-0372-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shourian M., Noghabi K.A., Zahiri H.S., Bagheri T., Karbalaei R., Mollaei M., Rad I., Ahadi S., Raheb J., Abbasi H. Efficient phenol degradation by a newly characterized Pseudomonas sp. SA01 isolated from pharmaceutical wastewaters. Desalination. 2009;246(1–3):577–594. [Google Scholar]
- Sidhu G.K., Singh S., Kumar V., Dhanjal D.S., Datta S., Singh J. Toxicity, monitoring and biodegradation of organophosphate pesticides: a review. Crit. Rev. Environ. Sci. Technol. 2019;49(13):1135–1187. [Google Scholar]
- Singh B., Kaur J., Singh K. Microbial remediation of explosive waste. Crit. Rev. Microbiol. 2012;38(2):152–167. doi: 10.3109/1040841X.2011.640979. [DOI] [PubMed] [Google Scholar]
- Singh B., Singh K. Bacilli and Agrobiotechnology. Springer; 2016. Bacillus: as bioremediator agent of major environmental pollutants; pp. 35–55. [Google Scholar]
- Singh B., Singh K. Microbial degradation of herbicides. Crit. Rev. Microbiol. 2016;42(2):245–261. doi: 10.3109/1040841X.2014.929564. [DOI] [PubMed] [Google Scholar]
- Singh R.L., Singh P.K., Singh R.P. Enzymatic decolorization and degradation of azo dyes–A review. Int. Biodeterior. Biodegradation. 2015;104:21–31. [Google Scholar]
- Sunarko B., Sulistinah N. Paper presented at the IOP Conference Series: Earth and Environmental Science. 2019. Acrylonitrile degradation by whole cells of Corynebacterium sp. D5, isolated from polluted industrial waste. [Google Scholar]
- Talwar M., Mulla S., Ninnekar H. Biodegradation of organophosphate pesticide quinalphos by Ochrobactrum sp. strain HZM. J. Appl. Microbiol. 2014;117(5):1283–1292. doi: 10.1111/jam.12627. [DOI] [PubMed] [Google Scholar]
- Tan L., He M., Song L., Fu X., Shi S. Aerobic decolorization, degradation and detoxification of azo dyes by a newly isolated salt-tolerant yeast Scheffersomyces spartinae TLHS-SF1. Bioresour. Technol. 2016;203:287–294. doi: 10.1016/j.biortech.2015.12.058. [DOI] [PubMed] [Google Scholar]
- Tao W., Lin J., Wang W., Huang H., Li S. Biodegradation of aliphatic and polycyclic aromatic hydrocarbons by the thermophilic bioemulsifier-producing Aeribacillus pallidus strain SL-1. Ecotoxicol. Environ. Saf. 2020;189 doi: 10.1016/j.ecoenv.2019.109994. [DOI] [PubMed] [Google Scholar]
- Tian J., Yu C., Xue Y., Zhao R., Wang J., Chen L. Performance of trichlorfon degradation by a novel Bacillus tequilensis strain PA F-3 and its proposed biodegradation pathway. Biodegradation. 2016;27(4):265–276. doi: 10.1007/s10532-016-9771-8. [DOI] [PubMed] [Google Scholar]
- Tien C.-.J., Lin M.-.C., Chiu W.-.H., Chen C.S. Biodegradation of carbamate pesticides by natural river biofilms in different seasons and their effects on biofilm community structure. Environ. Pollut. 2013;179:95–104. doi: 10.1016/j.envpol.2013.04.009. [DOI] [PubMed] [Google Scholar]
- Torres-Farradá G., Manzano-León A.M., Rineau F., Leal M.R., Thijs S., Jambon I., Put J., Czech J., Rivera G.G., Carleer R. Biodegradation of polycyclic aromatic hydrocarbons by native Ganoderma sp. strains: identification of metabolites and proposed degradation pathways. Appl. Microbiol. Biotechnol. 2019;103(17):7203–7215. doi: 10.1007/s00253-019-09968-9. [DOI] [PubMed] [Google Scholar]
- Unnikrishnan S., Khan M.H., Ramalingam K. Dye-tolerant marine Acinetobacter baumannii-mediated biodegradation of reactive red. Water Sci. Eng. 2018;11(4):265–275. [Google Scholar]
- Velayutham K., Madhava A.K., Pushparaj M., Thanarasu A., Devaraj T., Periyasamy K., Subramanian S. Biodegradation of Remazol Brilliant Blue R using isolated bacterial culture (Staphylococcus sp. K2204) Environ. Technol. 2018;39(22):2900–2907. doi: 10.1080/09593330.2017.1369579. [DOI] [PubMed] [Google Scholar]
- Velázquez-Fernández J.B., Martínez-Rizo A.B., Ramírez-Sandoval M., Domínguez-Ojeda D. Biodegradation and bioremediation of organic pesticides. Pestic.-Recent Trend. Pestic. Residue assay. 2012:253–272. [Google Scholar]
- Verma J.P., Jaiswal D.K., Sagar R. Pesticide relevance and their microbial degradation: a-state-of-art. Rev. Environ. Sci. Bio/Technol. 2014;13(4):429–466. [Google Scholar]
- Verma Y. Acute toxicity assessment of textile dyes and textile and dye industrial effluents using Daphnia magna bioassay. Toxicol. Ind. Health. 2008;24(7):491–500. doi: 10.1177/0748233708095769. [DOI] [PubMed] [Google Scholar]
- Vikrant K., Giri B.S., Raza N., Roy K., Kim K.-.H., Rai B.N., Singh R.S. Recent advancements in bioremediation of dye: current status and challenges. Bioresour. Technol. 2018;253:355–367. doi: 10.1016/j.biortech.2018.01.029. [DOI] [PubMed] [Google Scholar]
- Vorbeck C., Lenke H., Fischer P., Spain J.C., Knackmuss H.-.J. Initial reductive reactions in aerobic microbial metabolism of 2, 4, 6-trinitrotoluene. Appl. Environ. Microbiol. 1998;64(1):246–252. doi: 10.1128/aem.64.1.246-252.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahla A.Q., Iqbal S., Anwar S., Firdous S., Mueller J.A. Optimizing the metribuzin degrading potential of a novel bacterial consortium based on Taguchi design of experiment. J. Hazard. Mater. 2019;366:1–9. doi: 10.1016/j.jhazmat.2018.11.054. [DOI] [PubMed] [Google Scholar]
- Wang H., Jin M., Mao W., Chen C., Fu L., Li Z., Du S., Liu H. Photosynthetic toxicity of non-steroidal anti-inflammatory drugs (NSAIDs) on green algae Scenedesmus obliquus. Sci. Total Environ. 2020;707 doi: 10.1016/j.scitotenv.2019.136176. [DOI] [PubMed] [Google Scholar]
- Wang J.a., Gao F., Liu Z., Qiao M., Niu X., Zhang K.-.Q., Huang X. Pathway and molecular mechanisms for malachite green biodegradation in Exiguobacterium sp. MG2. PLoS One. 2012;7(12):e51808. doi: 10.1371/journal.pone.0051808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zheng Y., Zhao B., Zhang Y., Liu Z., Xu J., Chen Y., Yang Z., Wang F., Wang H. Human metabolic responses to chronic environmental polycyclic aromatic hydrocarbon exposure by a metabolomic approach. J. Proteome Res. 2015;14(6):2583–2593. doi: 10.1021/acs.jproteome.5b00134. [DOI] [PubMed] [Google Scholar]
- Woźniak-Karczewska M., Čvančarová M., Chrzanowski Ł., Kolvenbach B., Corvini P.F.-X., Cichocka D. Isolation of two Ochrobactrum sp. strains capable of degrading the nootropic drug—Piracetam. N Biotechnol. 2018;43:37–43. doi: 10.1016/j.nbt.2017.07.006. [DOI] [PubMed] [Google Scholar]
- Wu F., Guo C., Liu S., Liang X., Lu G., Dang Z. Pyrene degradation by Mycobacterium gilvum: metabolites and proteins involved. Water Air Soil Pollut. 2019;230(3):1–13. [Google Scholar]
- Yan Z., Zhang Y., Wu H., Yang M., Zhang H., Hao Z., Jiang H. Vol. 7. 2017. Isolation and characterization of a bacterial strain Hydrogenophaga sp. PYR1 for anaerobic pyrene and benzo [a] pyrene biodegradation; pp. 46690–46698. (RSC Adv.). [Google Scholar]
- Yang L., Yan K., Zeng D., Lai X., Chen X., Fang Q., Guo H., Wu T., Zhang X. Association of polycyclic aromatic hydrocarbons metabolites and risk of diabetes in coke oven workers. Environ. Pollut. 2017;223:305–310. doi: 10.1016/j.envpol.2017.01.027. [DOI] [PubMed] [Google Scholar]
- Yang X., Wei H., Zhu C., Geng B. Biodegradation of atrazine by the novel Citricoccus sp. strain TT3. Ecotoxicol. Environ. Saf. 2018;147:144–150. doi: 10.1016/j.ecoenv.2017.08.046. [DOI] [PubMed] [Google Scholar]
- Ye Z., Zhao Q., Zhang M., Gao Y. Acute toxicity evaluation of explosive wastewater by bacterial bioluminescence assays using a freshwater luminescent bacterium, Vibrio qinghaiensis sp. Nov. J. Hazard. Mater. 2011;186(2–3):1351–1354. doi: 10.1016/j.jhazmat.2010.12.013. [DOI] [PubMed] [Google Scholar]
- Zamani H., Grakoee S.R., Rakhshaee R. Microbial degradation of Paclitaxel using Citrobacter amalonaticus Rashtia isolated from pharmaceutical wastewater: kinetic and thermodynamic study. World. J. Microbiol. Biotechnol. 2016;32(8):1–10. doi: 10.1007/s11274-016-2087-3. [DOI] [PubMed] [Google Scholar]
- Zhang C., Lu J., Wu J., Luo Y. Removal of phenanthrene from coastal waters by green tide algae Ulva prolifera. Sci. Total Environ. 2017;609:1322–1328. doi: 10.1016/j.scitotenv.2017.07.187. [DOI] [PubMed] [Google Scholar]
- Zhang L., Yin X., Li S.F.Y. Bio-electrochemical degradation of paracetamol in a microbial fuel cell-Fenton system. Chem. Eng. J. 2015;276:185–192. [Google Scholar]
- Zhang W., Deng Y., Chen L., Zhang L., Wang Z., Liu R., Diao J., Zhou Z. Effect of triadimefon and its metabolite on adult amphibians Xenopus laevis. Chemosphere. 2020;243 doi: 10.1016/j.chemosphere.2019.125288. [DOI] [PubMed] [Google Scholar]
- Zhang X., Gao Y., Zang P., Zhao Y., He Z., Zhu H., Song S., Zhang L. Study on the simultaneous degradation of five pesticides by Paenibacillus polymyxa from Panax ginseng and the characteristics of their products. Ecotoxicol. Environ. Saf. 2019;168:415–422. doi: 10.1016/j.ecoenv.2018.10.093. [DOI] [PubMed] [Google Scholar]
- Ziganshin A.M., Naumova R.P., Pannier A.J., Gerlach R. Influence of pH on 2, 4, 6-trinitrotoluene degradation by Yarrowia lipolytica. Chemosphere. 2010;79(4):426–433. doi: 10.1016/j.chemosphere.2010.01.051. [DOI] [PubMed] [Google Scholar]