Summary
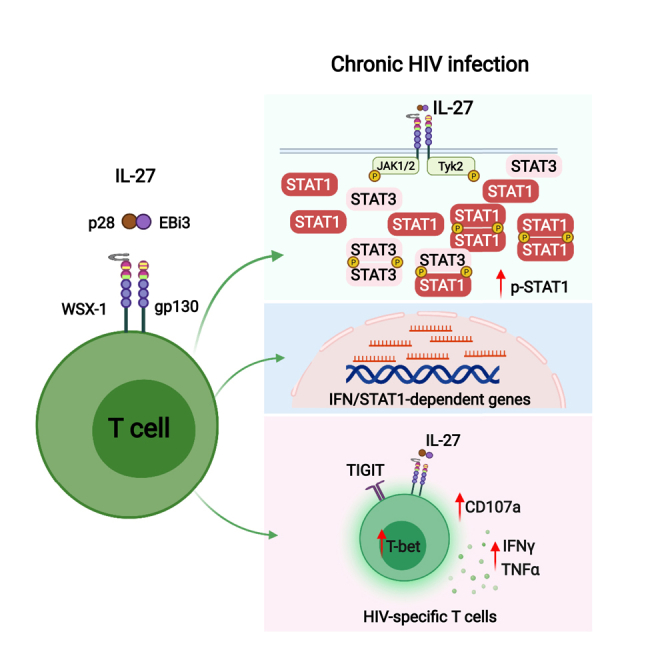
HIV-specific T cells have diminished effector function and fail to control/eliminate the virus. IL-27, a member of the IL-6/IL-12 cytokine superfamily has been shown to inhibit HIV replication. However, whether or not IL-27 can enhance HIV-specific T cell function is largely unknown. In the present manuscript, we investigated the role of IL-27 signaling in human T cells by evaluating the global transcriptional changes related to the function of HIV-specific T cells. We found that T cells from people living with HIV (PLWH), expressed higher levels of STAT1 leading to enhanced STAT1 activation upon IL-27 stimulation. Observed IL-27 induced transcriptional changes were associated with IFN/STAT1-dependent pathways in CD4 and CD8 T cells. Importantly, IL-27 dependent modulation of T-bet expression promoted IFNγ secretion by TIGIT+HIVGag-specific T cells. This new immunomodulatory effect of IL-27 on HIV-specific T cell function suggests its potential therapeutic use in cure strategies.
Subject areas: Immune response, Components of the immune system, Transcriptomics
Graphical abstract
Highlights
-
•
IL-27 induced transcriptional changes associated with IFN/STAT1-dependent pathways
-
•
HIV infection alters IL-27 signaling in T cells by enhancing STAT1 phosphorylation
-
•
IL-27 upregulates T-bet expression and enhances TIGIT+ HIVGag-specific T cell function
Immune response; Components of the immune system; Transcriptomics
Introduction
Persistent immune activation remains the hallmark of HIV infection even in the context of successfully suppressed viral replication by combination antiretroviral therapy (cART) (Catalfamo et al., 2008; Doisne et al., 2004; Klatt et al., 2013). The main feature of chronic T cell immune activation is an “activated/exhausted phenotype” characterized by the expression of one or co-expression of several immunomodulatory receptors including PD1, CTLA4, LAG3, TIGIT, TIM3, and CD160 among others. Particularly, exhausted HIV-specific T cells have diminished effector function and fail to control/eliminate the virus (Chew et al., 2016; Fromentin et al., 2016; Hatano et al., 2013; Kaufmann et al., 2007; Peretz et al., 2012; Petrovas et al., 2006; Pombo et al., 2015; Tian et al., 2015; Trautmann et al., 2006; Wykes and Lewin, 2018). Yet, CD8 T cells play a central role in controlling HIV/SIV replication during acute and chronic infection (Betts et al., 2006; Hersperger et al., 2011; Jin et al., 1999; Koup et al., 1994; Lifson et al., 2001; McDermott and Koup, 2012; Migueles and Connors, 2015; Schmitz et al., 1999). In contrast, CD4 T cells are the main targets of HIV infection and their role in T cell mediated immunity against HIV is compromised (Buggert et al., 2016, 2018; Kaufmann et al., 2007; Morou et al., 2014, 2019; Niessl and Kaufmann, 2018; Soghoian and Streeck, 2010). CD4 T cells are the main cell type harboring the HIV/SIV reservoirs in tissues and recent evidence determined that latently HIV infected CD4 T cells express checkpoint receptors promoting viral persistence (Chen et al., 2020; Churchill et al., 2016; Fromentin et al., 2016; Wykes and Lewin, 2018). Therefore, HIV cure strategies will require combination therapies targeting the viral reservoir and enhancement of HIV-specific T cell effector function.
IL-27, a relatively new member of the IL-6/IL-12 cytokine superfamily, has been a focus of intensive investigation because of its immunomodulatory functions (Hall et al., 2012; Hunter and Kastelein, 2012; Kastelein et al., 2007). IL-27 is formed by the IL-27p28 and Epstein Barr-Virus-induced gene 3 (EBI3) chains (Devergne et al., 1996; Kastelein et al., 2007; Pflanz et al., 2002). In humans, IL-27p28 is only secreted bound to EBI3, in contrast to the mouse IL-27 where there is an independent secretion of IL-27p28, suggesting intrinsic species-specific differences in its biology (Crabe et al., 2009; Muller et al., 2019; Pflanz et al., 2002). IL-27 signals through a heterodimer receptor composed of IL-27Rα (WSX1) and gp130 (shared by IL-6 receptor), and activates Janus Kinase (JAK)-Signal Transducer and Activator of Transcription (STAT1 and STAT3), and the mitogen-activated protein kinase (MAPK) signaling pathways (Hibbert et al., 2003; Kastelein et al., 2007; Lucas et al., 2003; Pflanz et al., 2004; Takeda et al., 2003; Villarino et al., 2003). IL-27 plays a crucial role in immunity, balancing protective and inflammatory responses (Hirahara et al., 2012; Hunter and Kastelein, 2012; Villarino et al., 2005). IL-27 induces expression of T-bet and IL-12Rbeta-2, promoting Th1 responses (Hibbert et al., 2003; Kamiya et al., 2004; Lucas et al., 2003; Owaki et al., 2005; Takeda et al., 2003). In contrast, the anti-inflammatory properties include inhibition of Th17 differentiation, upregulation of PD-L1 expression, IL-10 secretion, and differentiation of T regulatory cells (Batten et al., 2008; Hirahara et al., 2012; Peters et al., 2015; Stumhofer et al., 2007; Zhu et al., 2015).
In the context of HIV infection, the role of IL-27 is not well defined. IL-27 has antiviral properties, and in vitro inhibits HIV replication in CD4 T cells, monocyte-derived macrophages, and dendritic cells (Chen et al., 2013; Dai et al., 2013; Poudyal et al., 2019). Comparison of plasma levels of IL-27 between people living with HIV (PLWH) (untreated, and successfully suppressed viremia with cART), and healthy age matched controls showed no differences between the study groups (Swaminathan et al., 2014). In addition, plasma levels of IL-27 showed a weak positive association with serologic biomarkers of inflammation including D-dimer and sCD163 levels (Swaminathan et al., 2014). In contrast, two small studies reported contradictory changes of plasma levels of IL-27 during HIV infection (Ruiz-Riol et al., 2017; Zheng et al., 2017). These data suggest that plasma levels of IL-27 may not reflect the in vivo functions of this cytokine in the context of HIV infection.
The immunomodulatory functions of IL-27 on HIV-specific T cell responses remains largely unknown. We previously reported that HIV driven T cell immune activation alters cytokine signaling in a STAT1 dependent manner, particularly those cytokines associated with CD4 T cell depletion (IL-7) and HIV viral replication and reservoirs (Type I IFNs) impacting T cell immune activation and homeostasis (Catalfamo et al., 2008, 2011; Le Saout et al., 2014). In the present manuscript, we evaluated IL-27 signaling in the context of HIV infection to determine whether IL-27 can enhance HIV-specific T cell responses. We report that IL-27 signaling induces global transcriptional changes resembling with IFN/STAT1 signaling pathway activation, and also results in enhanced T-bet expression and cytokine secretion by TIGIT+ HIV-specific T cells. These data suggest that IL-27 can be considered a potential target in a cytokine-base therapy to treat HIV infection because of its dual effects including antiviral properties and enhancing HIV-specific T cell responses.
Results
In vitro stimulation of IL-27 enhanced STAT1 activation in T cell subsets from PLWH
IL-27 signals through the JAK-STAT pathway activating primarily STAT1 and STAT3 (Hall et al., 2012; Hirahara et al., 2015; Kastelein et al., 2007). We previously reported that T cells from PLWH have an increased expression of STAT1 that leads to an enhanced responsiveness to Type I IFN (Le Saout et al., 2014). These data propose a model in which the relative availability of STATs can regulate cytokine signaling in the context of HIV infection. In this study, we hypothesized that HIV infection promotes enhanced STAT1 activation by IL-27 stimulation.
We first evaluated the expression levels of the IL-27Rα (WSX1) and gp130 chains in human CD4 and CD8 T cells from healthy volunteers and PLWH (Figures S1A and S1B, Table S1 and S2 Panel 1). Similar to previous reports, surface expression levels of the IL-27Rα chain measured by flow cytometry was low in both groups (Schneider et al., 2011). The weak expression was observed in T cells from fresh and frozen peripheral blood mononuclear cells (PBMCs) (Figure S1A). There was no statistically significant difference of IL-27Rα expression in CD4 and CD8 T cells from the healthy volunteer and PLWH groups (Figure S1A). In contrast, the expression of gp130 was significantly increased in CD4 but not CD8 T cells from the PLWH group when compared to the healthy control group (Figure S1B). Of note is that expression of IL-27Rα and gp130 was higher in the CD4 compared to CD8 T cells from both study groups (Figure 1B). The expression of a functional receptor was further evaluated by blockade of the receptor leading to inhibition of STAT1 and STAT3 activation measured by detection of the phosphorylated forms, p-STAT1 and p-STAT3 (Figure S1C, Table S2 Panel 2 and Panel 3). IL-27Rα mAb efficiently blocked activation of STAT1 (p-STAT1) induced by 5 and 10 ng/mL of rhIL-27 (Figure S1C).
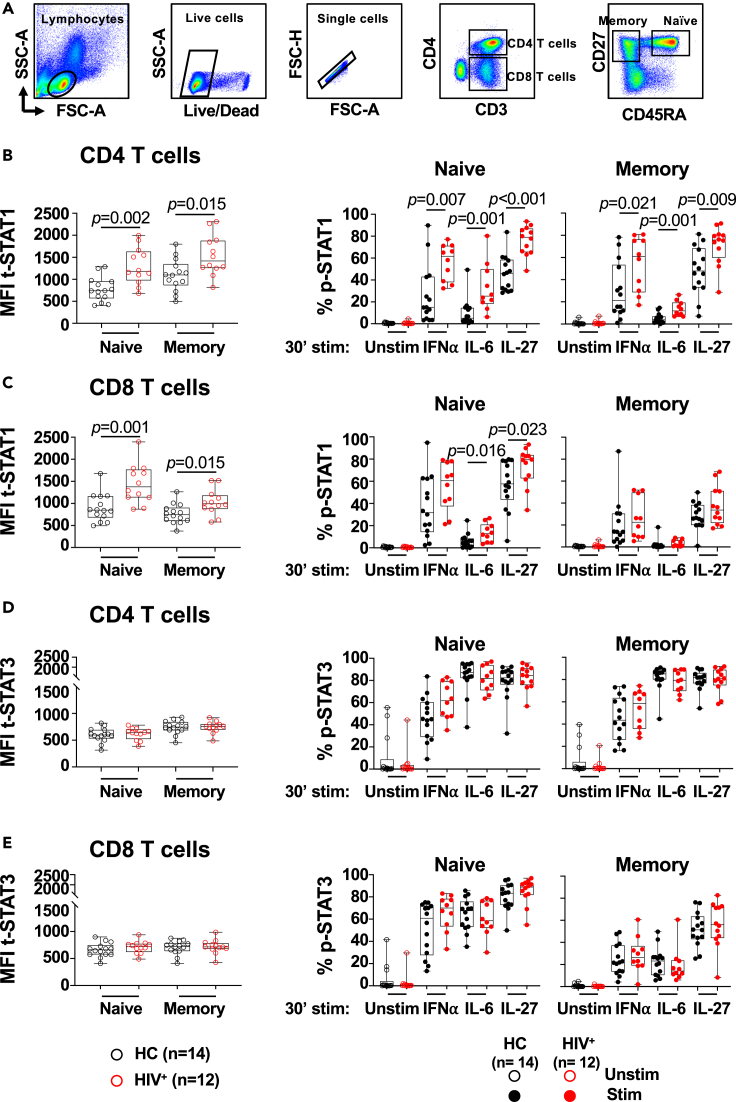
Figure 1.
In vitro IL-27 stimulation leads to enhanced STAT1 activation in naive and memory phenotype T cells from PLWH
PBMCs from PLWH (HIV+, n = 12) and healthy controls (HC, n = 14) were thaw, rested overnight and stimulated with 25 ng/mL of rhIL-27, 100 U/mL of rhIFN⍺, and 100 ng/mL of rhIL-6, for 30 min. (A) Gating strategy of naive (CD45RA+CD27+) and memory (CD45RA−CD27+) phenotype CD4 and CD8 T cells. Median Fluorescence Intensity (MFI) of intracellular expression of t-STAT1 and frequency of phosphorylated STAT1 (p-STAT1) in: (B) naive and memory phenotype CD4 T cell subsets; (C) naive and memory phenotype CD8 T cell subsets. Median Fluorescence Intensity (MFI) of intracellular expression of t-STAT3 and frequency of phosphorylated STAT3 (p-STAT3) in: (D) naive and memory phenotype CD4 T cell subsets; and (E) naive and memory phenotype CD8 T cell subsets. The graph is represented by box and whisker showing the median value with first and third quartiles in the box, with whiskers extending to the minimum and maximum values. Comparisons between the study groups PLWH and healthy controls were performed using Mann–Whitney test. p value <0.05 was considered significant.
Furthermore, stimulation of PBMCs from PLWH (n = 10) stimulated with IL-27 (10 ng/mL) in the presence of an anti-IL-27Rα mAb led to inhibition of STAT1 and STAT3 phosphorylation in CD4 and CD8 T cells (Figure S1D). As previously reported gp130 mAb alone was not sufficient to inhibit STAT1 and STAT3 activation mediated by IL-27 (Petes et al., 2018). However, anti-gp130 mAb effciently blocked STAT3 activation induced by IL-6 stimulation which signals through gp130 and activates STAT3 (Figure S1E) (Jones and Jenkins, 2018; Kastelein et al., 2007). In addition, combination of IL-27Rα and gp130 mAbs synergized in the inhibition both STAT1 and STAT3 activation induced by IL-27 (Figure S1D). Altogether these data suggest T cells express a functional IL-27 receptor and IL-27Rα is required for STAT1 and STAT3 activation.
Having established that T cells express a functional IL-27 receptor, we next determined the ability of in vitro IL-27 stimulation to activate STAT1 and STAT3 in CD4 and CD8 T cell subsets from PLWH with suppressed HIV replication by cART (n = 12, Table S1) and healthy volunteers (n = 14). We evaluated the levels of expression of the transcription factors and found a significantly higher expression levels of STAT1 (t-STAT1) by the naive (CD45RA+CD27+) and memory (CD45RA−CD27+) CD4 T cell subsets from PLWH when compared with healthy volunteers (Figures 1A and 1B, Table S2 Panel 3). A similar observation was noted in the CD8 T cell subsets (Figure 1C). Memory CD8 T cells showed lower expression than their naive counterparts as previously reported (Le Saout et al., 2014). The expression of STAT3 (t-STAT3) was similar between the study groups, and between the CD4 and CD8 T cell subsets (Figures 1D and 1E respectively).
We next evaluated whether the increased expression of STAT1 influences IL-27 signaling by measuring the activation of STAT1 and STAT3 (phosphorylated forms, p-STAT1 and p-STAT3) upon IL-27 stimulation at a concentration 25 ng/mL that induced maximum activation of STAT1 (Figure S1C). We used in vitro stimulation with IFN⍺ and IL-6 as controls for STAT1 and STAT3 activation respectively. In PLWH, IL-27 led to an increased p-STAT1 in naive and memory phenotype CD4 T cells and naive phenotype CD8 T cells when compared to healthy controls (Figures 1B and 1C). Consistent with the lower STAT1 expression, the memory CD8 T cell subset showed lower STAT1 activation than naive CD8 T cells, and while an enhanced trend was observed in PLWH did not reach statistical significance (Figure 1C). Consistent with previous observations, higher STAT1 expression led to enhanced responsiveness to IFN⍺ in the naive and memory phenotype CD4 T cell subsets, and a trend was observed in the CD8 T cell subsets but did not reach statistically significance (Le Saout et al., 2014).
Interestingly, naive CD4 T cells from PLWH, showed an enhanced STAT1 activation upon in vitro IL-6 stimulation. This effect was weak although significant in the memory CD4 and naive CD8 T cell subsets (Figures 1B and 1C respectively).
In contrast to STAT1, the activation of STAT3 by IL-27 stimulation was similar between PLWH and healthy controls in both CD4 and CD8 T cell subsets (Figures 1D and 1E).
These data suggest that HIV infection differentially impacts cytokine signaling in T cell subsets. Particularly, the increased expression of STAT1 led to an enhanced IL-27-dependent STAT1 activation in T cell subsets.
IL-27 signaling induces global transcriptional changes that overlap with that of IFNα and IL-6 signaling
The distinct activation of STAT1 by IL-27 stimulation in PLWH led us to hypothesize that infection alters the transcriptome of IL-27 signaling. To investigate these downstream effects, we performed RNAseq analysis of sorted naive (CD45RA+CD27+) and memory (CD45RA−CD27+) CD4 and CD8 T cells stimulated with IL-27 (100 ng/mL) for 90 min to assure maximum early transcriptonal changes induced by IL-27 signaling (Figure S1C, Table S1 and S2 Panel 4). Because HIV infection alters cytokine signaling (Figure 1), we used T cell subsets from healthy controls stimulated with IFNα and IL-6 as controls to evaluate the STAT1 and STAT3 regulated genes respectively.
The analysis of the global transcriptional changes at an early time point (90 min) showed that sets of differentially expressed genes (DEGs) responsive to IL-27 stimulation in naive and memory CD4 and CD8 T cells were also responsive to IFNα and IL-6 stimulation in both study groups PLWH and healthy controls (Figures 2A and 2B). In contrast, the number of genes specifically responsive only to IL-27 stimulation in CD4 and CD8 T cell subsets from PLWH was higher compared to healthy controls (Figures 2A and 2B).
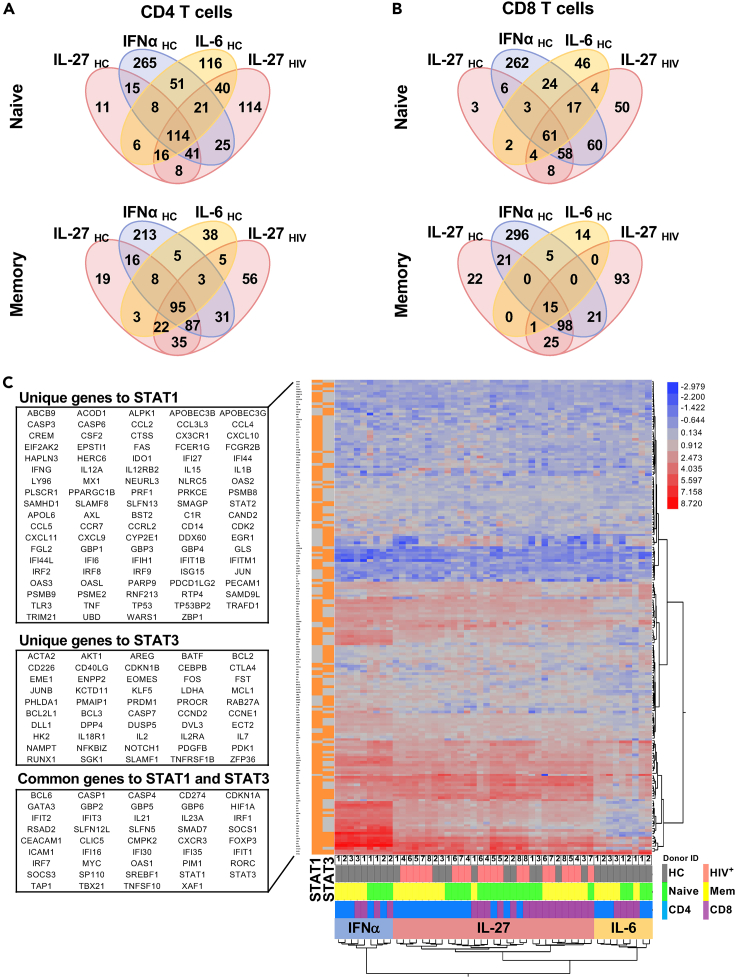
Figure 2.
IL-27 signaling induced a cluster of STAT1-dependent genes
RNAseq analysis of sorted naive (CD45RA+CD27+) and memory (CD45RA−CD27+) T cells from PLWH (HIV+, n = 5) and healthy controls (HC, n = 3) stimulated in vitro with 100 ng/mL of rhIL-27, and cells from healthy controls stimulated with 100 U/mL of rhIFN⍺ and 100 ng/mL of rhIL-6 for 90 min as controls.
(A) Venn diagrams represent overlapped differentially expressed gene transcripts (DEGs) regulated by IL-27 in CD4 T cell subsets from PLWH and HC compared to the DEGs induced by IFN⍺ and IL-6 stimulation in T cells from healthy controls.
(B) Venn diagrams represent overlapped DEGs regulated by IL-27 in CD8 T cell subsets from PLWH and HC compared to the DEGs induced by IFN⍺ and IL-6 stimulation in T cell from healthy controls.
(C) Ingenuity Pathway Analysis (IPA) was used to subset the RNA-seq data to include only target genes predicted by IPA to be regulated via STAT1 and/or STAT3 activity, as indicated in the left column, orange for genes downstream, gray for genes not downstream of the corresponding upstream regulator. The 2-way clustered heatmap of gene expression (log2 of stimulated/unstimulated ratio) for IL-27, IFNα and IL-6 stimulated T cell subsets. The color bar indicates: HC group (gray), HIV group (pink), CD4 T cells (blue), CD8 T cells (purple), naive subset (green), Memory (Mem, yellow). The lists (DEGs) were selected based on >2-fold change (| Log2 FC | > 1) and FDR <0.05.
IL-27 stimulation of naive CD4 and CD8 T cell subsets from PLWH showed an increase in the number of upregulated genes and minor changes in the set of downregulated genes (Figure S2). In contrast, in the memory CD4 and CD8 T cells from PLWH, there were a higher number of downregulated genes by IL-27 stimulation, particularly in the memory CD8 T cell subset (Figure S2). These data suggest potential differences in the biological effects of IL-27 stimulation in CD4 and CD8 T cell subsets. Moreover, the gene sets derived from canonical pathways in IL-27 stimulation showed genes associated with IFN signaling, pattern recognition receptors, and antiviral responses in CD4 and CD8 T cell subsets (Table S3). In addition, in both CD4 and CD8 T cell subsets from healthy controls and HIV infected, IPA predicted differences in cytokine signaling pathways between the healthy control and HIV infected groups including the JAK/STAT signaling, Prolactin signaling, Th1 pathway, STAT3 pathway, IL-23 and IL-9 signaling pathway (Table S3).
In CD4 and CD8 memory T cell subsets, the top 20 affected categories of diseases and functions predicted by IPA within DEGs of T cells stimulated with IL-27 showed increased antiviral responses, cell quantity and decreased DEGs associated with viral production, viral replication and viral life cycle (Table S4). In addition in both groups, cell viability, cell survival, cell death of immune cells, quantity of lymphocytes, migration of cells, differentiation of T lymphocytes annotations among other were predicted to be increased suggesting wide direct effects on T cells by IL-27 stimulation and the potential regulation of other cells. Of note is that in PLWH group, the molecules involved in each disease and function annotation was higher than in the healthy control group (Table S4).
These data suggest that in the context of HIV infection IL-27 stimulation in T cells induced early-phase transcriptional changes associated with antiviral responses and IFN-dependent signaling. In addition, IL-27 downregulated a set of DEGs in the memory CD8 T cell subset linked to cytokine signaling pathways.
IL-27 induced global transcripts enriched in IFN/STAT1-inducible genes
IL-27 signals through STAT1 and STAT3 transcription factors, and it has been shown that STAT1 provides the specificity of the IL-27 transcriptional profile that distinguishes it from other STAT3-dependent cytokines such as IL-6 (Hirahara et al., 2015).
Using the observed gene expression signature of the three cytokines (IFNα, IL-6 and IL-27) across STAT1 and STAT3 induced genes predicted by IPA, we characterized the IL-27 response by similarity to that of IFNα and IL-6 simultaneously and evaluate the relative contribution of these transcription factors in human T cells (Figure 2C and Tables S5 and S6).
The two-way hierarchical clustering of the main patterns of gene expression associated with the upstream regulators (UR)-STAT1 and (UR)-STAT3 (predicted by IPA) in the culture conditions shows that the overall response to IL-27 signaling clustered separately from IFNα and IL-6 signaling, but closer by correlation to IL-6 than to IFNα (Figure 2C). A set of genes mainly downstream of (UR)-STAT1 were clustered at the bottom of the heatmap were upregulated by IFNα and IL-27 stimulation, and in less degree by IL-6 signaling (STAT1, left column, Figure 2C). Most of these genes downstream of the (UR)-STAT3 were observed to be less upregulated or downregulated (STAT3 left column, Figure 2C; Tables S5 and S6).
In addition, other upstream regulators predicted to be active were associated primarily with IFN/STAT1 regulated genes including Interferon alpha, IFNG, IFNL1, IFNA2, PRL, Interferon alpha, and STAT1 (Table 1). These data suggest that IL-27 signaling induces a transcriptional gene profile that was enriched in a cluster of IFN/STAT1-dependent genes.
Table 1.
Top 20 IPA-predicted activated upstream regulators in memory CD4 and CD8 T cells from HC and PLWH stimulated in vitro with IL-27.
| Memory CD4 T cells |
Memory CD8 T cells |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HC |
HIV+ |
HC |
HIV+ |
||||||||
| Upstream regulator | Activation Z score | p value of overlap | Upstream Regulator | Activation Z score | p value of overlap | Upstream regulator | Activation Z score | p value of overlap | Upstream regulator | Activation Z score | p value of overlap |
| STAT1 | 6.82 | 8.75 × 10−68 | Interferon alpha | 5.86 | 2.98 × 10−72 | STAT1 | 6.22 | 1.01 × 10−61 | STAT1 | 6.08 | 2.37 × 10−65 |
| Interferon alpha | 5.19 | 7.66 × 10−65 | STAT1 | 6.74 | 4.06 × 10−72 | Interferon alpha | 4.69 | 6.94 × 10−57 | Interferon alpha | 4.87 | 2.94 × 10−56 |
| IFNA2 | 6.35 | 4.93 × 10−61 | IFNL1 | 6.67 | 6.56 × 10−68 | IFNG | 7.49 | 2.16E-50 | IFNG | 7.05 | 2.00 × 10−53 |
| IFNL1 | 6.30 | 3.03E-60 | IFNA2 | 6.72 | 1.65 × 10−67 | IFNL1 | 5.47 | 5.79 × 10−47 | IFNL1 | 5.81 | 4.26E-50 |
| IFNG | 8.19 | 5.04 × 10−59 | IFNG | 8.58 | 2.29 × 10−66 | IFNA2 | 5.49 | 4.27 × 10−46 | IFNA2 | 5.66 | 1.18 × 10−44 |
| NONO | 6.24 | 4.59E-50 | NONO | 6.69 | 4.62 × 10−57 | PRL | 5.35 | 3.83 × 10−37 | NONO | 5.94 | 9.08 × 10−41 |
| PRL | 6.13 | 6.24 × 10−46 | PRL | 6.36 | 2.02 × 10−48 | NONO | 5.43 | 7.27 × 10−36 | RC3H1 | −4.70 | 6.20 × 10−33 |
| MAPK1 | −5.67 | 1.02 × 10−38 | MAPK1 | −6.24 | 2.60 × 10−44 | RC3H1 | −4.49 | 5.19 × 10−33 | PRL | 5.35 | 7.68 × 10−33 |
| IRGM | −4.77 | 8.30 × 10−38 | IRGM | −5.41 | 7.87 × 10−42 | MAPK1 | −4.98 | 1.04 × 10−32 | MAPK1 | −5.32 | 2.63 × 10−32 |
| IRF3 | 5.74 | 2.56 × 10−37 | IRF3 | 5.74 | 2.54 × 10−39 | Irgm1 | −4.57 | 3.90 × 10−31 | Irgm1 | −4.33 | 4.95 × 10−29 |
| STAT3 | 1.13 | 1.69 × 10−35 | Irgm1 | −5.13 | 3.67 × 10−38 | STAT3 | 1.03 | 8.52 × 10−31 | IRF3 | 4.95 | 8.03 × 10−29 |
| Irgm1 | −5.16 | 1.89 × 10−35 | STAT3 | 1.40 | 1.73 × 10−37 | IRF3 | 4.76 | 6.24E-30 | Ifnar | 4.32 | 7.41 × 10−28 |
| TCR | 2.00 | 4.22 × 10−35 | TRIM24 | −5.54 | 3.20 × 10−37 | Ifnar | 4.21 | 8.25E-30 | IRGM | −4.57 | 8.02 × 10−28 |
| TRIM24 | −5.51 | 3.10 × 10−33 | RC3H1 | −5.01 | 1.63 × 10−36 | TRIM24 | −4.94 | 2.11 × 10−29 | TLR9 | 4.32 | 6.84 × 10−27 |
| RC3H1 | −5.19 | 3.26 × 10−33 | TCR | 2.91 | 1.31 × 10−34 | IRGM | −4.46 | 5.98 × 10−29 | TLR4 | 3.66 | 1.58 × 10−26 |
| PNPT1 | −4.76 | 4.28 × 10−32 | IL1RN | −5.47 | 3.04 × 10−34 | TLR9 | 4.83 | 9.86 × 10−29 | STAT3 | 0.42 | 4.24 × 10−26 |
| IL1RN | −4.68 | 2.05 × 10−31 | Ifnar | 4.93 | 6.64 × 10−34 | TCR | 2.91 | 2.90 × 10−26 | TRIM24 | −4.94 | 9.18 × 10−26 |
| TLR3 | 4.22 | 6.24 × 10−31 | PNPT1 | −4.76 | 1.39E-30 | TLR3 | 4.27 | 1.19 × 10−25 | TCR | 2.19 | 3.05 × 10−25 |
| TLR9 | 4.26 | 4.08E-30 | CNOT7 | −2.62 | 6.69E-30 | IRF7 | 4.38 | 2.57 × 10−24 | IRF7 | 4.10 | 1.79 × 10−24 |
| Ifnar | 5.08 | 1.49 × 10−29 | RNY3 | 4.36 | 1.34 × 10−29 | IFNB1 | 4.22 | 1.39 × 10−23 | RNY3 | 3.87 | 4.52 × 10−23 |
Lists of differentially expressed gene transcripts (DEGs) selected by criteria: | log2(FC) | > 1 and adjusted p values (<0.05) were generated for each condition and used as input in IPA software. Activation Z-score are based on a model that assigns random regulation directions for predicted upstream regulators. The p value of overlap was used to rank the significance associated for each Upstream Regulator. The pvalue indicates the significance of the overlap between the genes targeted by the upstream regulator in the database and the experimental dataset.
IL-27 upregulates CD69 and T-bet expression in CD4 and CD8 T cells
IL-27 plays a critical role in balancing protective and inflammatory responses in tissues, and regulates expression of checkpoint receptors (Chihara et al., 2018; DeLong et al., 2019; Hunter and Kastelein, 2012; Villarino et al., 2005; Yoshimura et al., 2006). In addition, IL-27 signaling in a STAT1 dependent fashion, promotes T-bet expression and Th1 differentiation (Hibbert et al., 2003; Lucas et al., 2003; Pflanz et al., 2002).
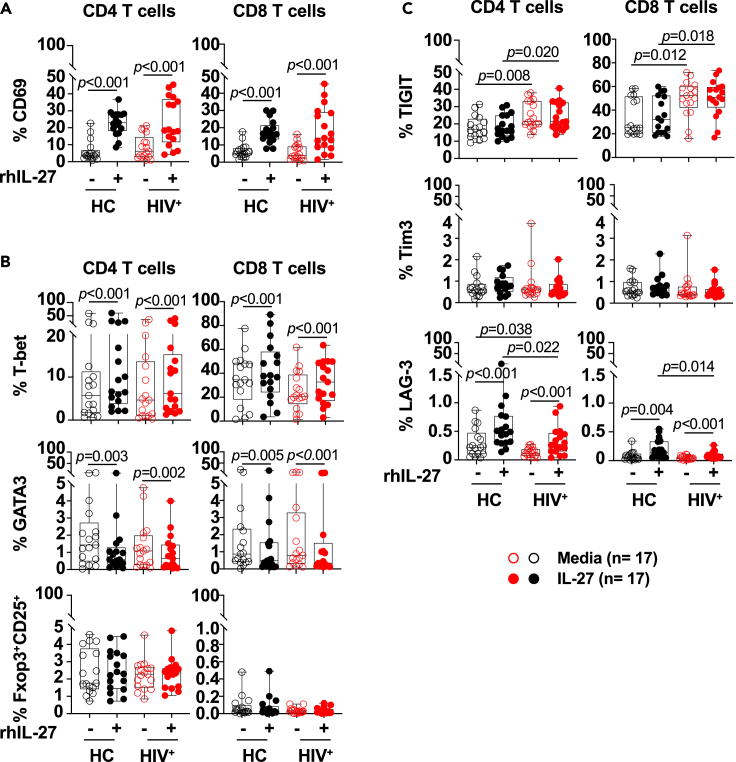
We investigated the core gene signature expression of Th1, Th2, Th17 and regulatory T cells (Treg) in the CD4 and CD8 T cell subsets (Figure S3A). T cells stimulated with IL-27 showed upregulation of Th1 master regulator TBX21 and down regulation of GATA3 and RORC master regulators associated with Th2 and Th17 differentiation respectively. In contrast, no changes were observed in the expression of FOXP3 (Figure S3A). In addition, CD69 an activation and tissue retention marker, was predicted to be activated by the upstream regulator IL27 (Kumar et al., 2017). Upregulation of CD69 protein expression was observed after overnight in vitro stimulation with IL-27 (Figure S3C).
We next investigated the impact of in vitro IL-27 stimulation in the protein expression of transcription factors associated with T helper differentiation and CD69 expression. PBMCs from PLWH (n = 17, Table S1) and healthy controls (n = 17) cultured overnight in the presence or absence of IL-27 (100 ng/mL) showed a significant increase of CD69 expression (Figure 3A, Table S2 Panel 5). The increased expression of T-bet induced by IL-27 was associated with a concomitant dowregulation of GATA3 expression in both groups (Figure 3B). In contrast, no effect was observed on the frequency of Treg (Foxp3+CD25+) (Figure 3B). In addition, the expression of GZA and GZB was not modulated by IL-27 stimulation (Figure S3D). These results suggest that IL-27 signaling in T cells from both heathy controls and in the context chronic HIV infection may favor Th1 function.
Figure 3.
In vitro IL-27 stimulation increased expression of CD69 and T-bet in CD4 and CD8 T cells
PBMCs from PLWH (HIV+, n = 17) and healthy controls (HC, n = 17) were cultured in the absence (opened symbols) or presence (closed symbols) of rhIL-27 (100 ng/mL) overnight. Flow cytometry analysis was performed for the expression of: (A) CD69; (B) T-bet, GATA3, and Foxp3+CD25+; (C) TIGIT, Tim3 and LAG3 in CD4 and CD8 T cells. Expressions of the markers are expressed as frequencies of the CD4 and CD8 T cells. The graphs are represented by box and whisker showing the median value with first and third quartiles in the box, with whiskers extending to the minimum and maximum values. Comparisons between the media and IL-27 stimulation were performed by non-parametric Wilcoxon test. Comparisons between the groups were performed by non-parametric Mann–Whitney test. p value <0.05 was considered significant.
In the context of HIV infection, chronic immune activation leads to accumulation of activated/exhausted T cells characterized by expression of checkpoint receptors (Chen et al., 2020; Wykes and Lewin, 2018). Because of the important implications in modulating HIV-specific T cell responses we analyzed the effect of IL-27 stimulation on the expression of checkpoint receptors and molecules involved in regulating T cell function (Figures 3C and S3B). IL-27 stimulation increased the expression of LAG3 although the frequency of T cells expressing LAG3 was very low, and no changes were noted in the expression of Tim3 (Figure 3C). In addition in PLWH, the frequency of cells expressing TIGIT was higher than healthy controls and its expression was not modulated by IL-27 in both CD4 and CD8 T cells (Figure 3C).
Altogether these data suggest that IL-27 signaling does not modulate TIGIT expression. The upregulation of T-bet expression can potentially regulate IFNγ production by HIV-specific T cells.
IL-27 enhanced cytokine secretion by TIGIT+ HIVGag-specific T cells
In the setting of HIV infection, chronic immune activation leads to dysfunctional HIV-specific T cells (Chew et al., 2016; Kaufmann et al., 2007; Tian et al., 2015; Trautmann et al., 2006). The data above suggest that IL27 stimulation have the potential to enhance Th1 function and we investigated whether this effect occurs in those virus-specific T cells expressing TIGIT.
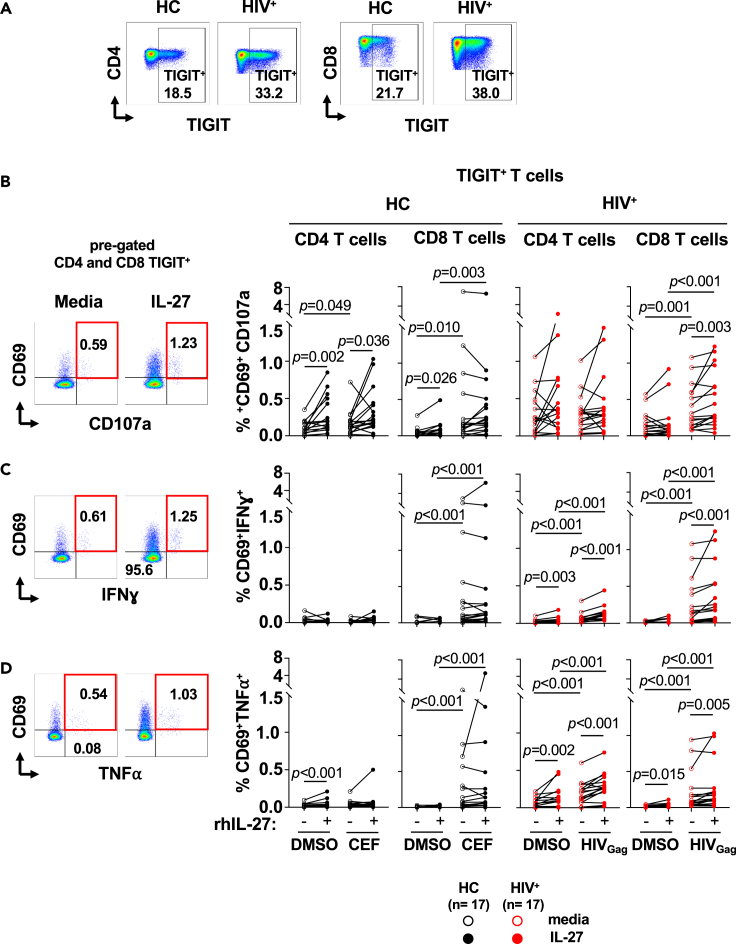
PBMCs from healthy controls (n = 17) and PLWH (n = 17) were cultured overnight in presence or absence of IL-27 followed by stimulation with CEF and HIVGag peptide pools respectively (Figure 4). IL-27 stimulation promoted cytokine secretion in TIGIT+ (CD69+IFNɣ+ and CD69+TNF⍺+) CD4 and CD8 T cells from PLWH (Figures 4C and 4D, respectively). IL-27 also increased the proportion of CD69+CD107a+ TIGIT+ CD8 T cells from PLWH (Figure 4B). These effects were not observed in the CEF-specific T cells from healthy controls (Figure 4). In addition, the frequency of TIGIT− antigen-specific CD4 and CD8 T cells were lower than TIGIT+ T cells in both groups (Figure S4).
Figure 4.
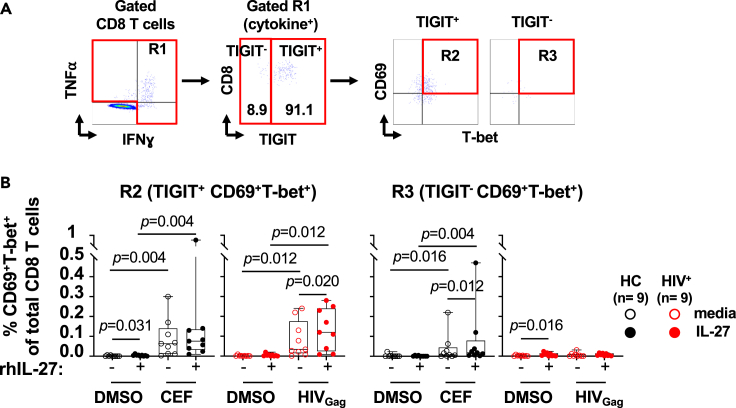
IL-27 enhances cytokine secretion of TIGIT+ HIVGag-specific T cells
PBMCs from PLWH (HIV+, n = 17) were cultured in the absence (red opened symbols) or presence (red closed symbols) of rhIL-27 (100 ng/mL) overnight follow by stimulation with HIVGag peptide pool. DMSO was used as control. PBMCs from and healthy controls (HC, n = 17) were stimulated in the absence (black opened symbols) or presence (black closed symbols) of rhIL-27 (100 ng/mL) overnight follow by stimulation with CEF (CMV, EBV and Influenza pool of peptides), and DMSO as control. Secretion of cytokines by TIGIT+ T cells was analyzed by flow cytometry. (A) Flow cytometry gating strategy for TIGIT+ CD4 and CD8 T cells. TIGIT+ CD4 and CD8 T cells were analyzed for expression of: (B) CD69+CD107a+; (C) CD69+IFNɣ+; and (D) CD69+TNF⍺+. The graphs are represented by box and whisker showing the median value with first and third quartiles in the box, with whiskers extending to the minimum and maximum values of the frequencies of T cells secreting cytokines. Comparisons between the culture conditions were performed by Wilcoxon test. Comparisons between PLWH and healthy controls were performed by Mann–Whitney test. p value <0.05 was considered significant.
The enhanced IL-27 induced IFNγ secretion by CD8 T cells was not associated with activation of HIV replication in the overnight cultures (Figure S5). In these culture conditions we did not detect HIVp24 production. HIVp24 was detected after 4 days culture of PBMCs stimulated with CD3 and CD28 mAbs used as positive control (Figure S5, Levinger et al., 2021).
The enhanced function mediated by IL-27 stimulation was associated with increased expression of CD69 and T-bet on the cytokine secreting TIGIT+ HIVGag-specific CD8 T cells. In contrast in the healthy controls, IL-27 upregulated the expression of CD69 and T-bet in the TIGIT− CEF-specific CD8 T cells (Figure 5). These data show that in PLWH, IL-27 stimulation enhances Th1 cytokine secretion and cytotoxic potential of TIGIT+ HIVGag -specific T cells.
Figure 5.
Enhanced TIGIT+HIVGag-specific CD8 T cell function is driven by IL-27 dependent upregulation of T-bet
PBMCs from PLWH (HIV+, n = 9) were cultured in the absence (red opened symbols) or presence (red closed symbols) of rhIL-27 (100 ng/mL) overnight follow by stimulation with HIVGag peptide pool. DMSO was used as control. PBMCs from and healthy controls (HC, n = 9) were cultured in the absence (black opened symbols) or presence (black closed symbols) of rhIL-27 (100 ng/mL) overnight follow by stimulation with were stimulated with CEF (CMV, EBV and Influenza pool of peptides), and DMSO was used as control. IL-27 upregulation of T-bet and CD69 was measured by flow cytometry. (A) Gating strategy for TIGIT+and TIGIT−virus-specific T cells secreting IFNγ+TNFα+.
(B) Expression of CD69+T-bet+ in cytokine secreting CD8 T cells. TIGIT+CD69+T-bet+ or TIGIT− CD69+T-bet+ cytokine secreting T cells is expressed as frequency of total CD8 T cells. The graph is represented by box and whisker showing the median value with first and third quartiles in the box, with whiskers extending to the minimum and maximum values. Comparisons between culture conditions were performed using non-parametric Wilcoxon test. p value <0.05 was considered significant.
Discussion
The immune therapeutic approaches in cure strategies for HIV infection require targeting the viral reservoir and enhancing HIV-specific T cell responses. In the present manuscript, we evaluated the immunomodulatory effects of IL-27, a cytokine that belongs to the IL-6/IL-12 superfamily and has been shown to inhibit HIV replication in vitro (Chen et al., 2013; Dai et al., 2013; Swaminathan et al., 2013). We determined the global transcriptional changes induced by IL-27 stimulation in human CD4 and CD8 T cells. More importantly, we showed that IL-27 enhances HIVGag-specific T cell function, particularly in T cells that express the checkpoint receptor TIGIT. These data and the previously reported antiviral properties of IL-27 against HIV highlight the potential dual therapeutic use of this cytokine by both targeting HIV viral replication and enhancing T cell mediated immunity against HIV.
IL-27 signals through the heterodimeric receptor composed by IL-27Rα and gp130 (Fabbi et al., 2017; Hall et al., 2012; Pflanz et al., 2004; Saito et al., 1992). A study had reported increased mRNA expression levels of IL-27RA in HIV viremic chronically infected individuals when compared to healthy controls. In contrast, HIV controllers (viremic and elite) expressed lower levels compared to healthy controls (Ruiz-Riol et al., 2017). In addition, the IL-27RA mRNA expression levels showed a positive association with plasma levels of IL-27 (Ruiz-Riol et al., 2017). In the present study, we have not observed increased surface protein expression levels of IL-27Rα in PLWH. Despite the low expression of IL-27Rα, blockade of the receptor efficiently inhibited IL-27 signaling. The enhanced activation of STAT1 in PLWH compared to the healthy individuals suggest that the levels of STAT1 can contribute in modulating IL-27 signaling as previously reported in the signaling of other cytokines (Le Saout et al., 2014, 2017).
IL-27 has been shown to be modulated by interferons in myeloid cells (Iyer et al., 2010; Liu et al., 2007; Molle et al., 2007; Pirhonen et al., 2007; Tan et al., 2018). IL-27p28 expression is induced by IFNγ in TLR4 stimulated murine macrophages (Liu et al., 2007). In human macrophages and dendritic cells, Type I IFNs and TLR stimulation or live viruses induced transcription of IL-27 (Greenwell-Wild et al., 2009; Pirhonen et al., 2007; Remoli et al., 2007). In the present study, IL-27 overnight stimulation apparently did not lead to a measurable p24 levels in the supernatants of the cultures as indication of HIV reactivation and potential production of Type I IFNs. However, we cannot exclude viral reactivation because of the limitation in the sensitivity of the assay and future studies should address the role of IL-27 in the context of HIV reactivation.
IL-27 signaling uses STAT1 and STAT3 transcription factors promoting expression of T-bet, and IL-12Rβ2 and IFNγ (Hibbert et al., 2003; Pflanz et al., 2002). CD4 and CD8 T cell subsets from PLWH show differential expression and activation of STAT1 upon IL-27 stimulation suggesting that HIV infection alters cytokine signaling similarly to that reported with IFNα and IL-7 (Le Saout et al., 2014, 2017). In contrast, no changes in STAT3 expression and activation are observed upon IL-27 stimulation.
The genes induced by IL-27 also responded to IFNα and IL-6 stimulation, cytokines that use primarily STAT1, STAT2, and STAT3 respectively. Particularly, IL-27 stimulation caused gene expression changes associated with IFN/STAT1 signaling pathway activation, and antiviral responses (Chen et al., 2013; Dai et al., 2013; Fakruddin et al., 2007; Imamichi et al., 2008). Similar to these results, a gene regulation prediction analysis in murine T cells reported the important role of STAT3 driving the observed pattern of transcriptome. They compared the global transcriptional changes induced by IL-27 and IL-6 signaling and showed that STAT1 cannot compensate STAT3 deficiency, and provide the specificity of the transcriptional response associated to IL-27 signaling (Hirahara et al., 2015). In addition, dysregulation of genes associated with cytokine signaling pathways were also observed including JAK/STAT signaling, Prolactin signaling, Th1 pathway, STAT3 pathway, IL-23 and IL-9 signaling pathway, and the functional implications of these pathways requires further investigation.
In the present study, we provide new insights of the transcriptional profile induced by IL-27 compared to IFNα, a cytokine with antiviral properties. We found STAT1 regulated genes responding to IL-27 stimulation in a pattern of expression changes similar to that observed in IFNα stimulated cells. These sets of genes are associated with host anti-viral responses and T cell function. These observations were also noted in T cells from PLWH suggesting that IL-27 may promote anti-viral responses even in the context of HIV infection. In addition, studies using STAT1 deficient mice had shown an indispensable role of STAT1 driving upregulation of T-bet and IL-12Rbeta2 and promoting Th1 differentiation in naive CD4 T cells. In contrast, STAT3 was involved in driving proliferation of CD4 naive T cells (Kamiya et al., 2004). Activation of other pathways by IL-27 including of the p38 MAPK/T-bet and ICAM1/LFA1/ERK1/2 has been shown to be involved in driving Th1 differentiation, although whether these pathways play a role in the setting of HIV infection requires further investigation (Hall et al., 2012; Hibbert et al., 2003; Owaki et al., 2006; Pflanz et al., 2002).
The immunomodulatory functions of IL-27 in the context of viral infection are not well defined. In a murine infection model by the JHM strain of mouse hepatitis virus (JHMV), IL-27 exerted immunomodulatory functions on CD8 effector T cells limiting immunopathology in the brain (de Aquino et al., 2014). In contrast, during chronic lymphocytic choriomeningitis virus (LCMV) clone 13 infection, IL-27 was critical in modulating plasmacytoid dendritic cell and CD4 T cell responses (Harker et al., 2013, 2018). In this setting, IL-27 also enhanced the expansion of self-renewing virus-specific CXCR5+ CD8 T cells, and this effect was STAT1 and IRF1 dependent (Huang et al., 2019). This evidence and the present findings suggest that IL-27 can play a role in enhancing immunity in the context of chronic infection. In addition, while we observed CD107a surface expression enhanced by IL-27 stimulation, future studies should address the cytotoxic function of the virus-specific CD8 T cells to kill HIV infected CD4 T cells.
In the setting of chronic HIV infection, we showed that in PLWH, IL-27 enhanced Th1 responses through modulation of STAT1/T-bet dependent genes suggesting a potential enhancement of T cell function at sites of HIV tissue reservoirs. These data suggest that IL-27 can be a therapeutic candidate to enhance HIV T cell mediated immunity in cure strategies.
Limitations of the study
In the present manuscript, we studied global transcriptional changes induced by IL-27 signaling. IL-27 signaling showed regulation of antiviral pathways that should be explored in future studies and particularly the impact of IL-27 in the viral reservoir. Moreover, we provide insights in the role of IL-27 in modulating HIVGag specific T cells function however in vivo studies should be performed to better dissect the role of IL-27 in T cell function and viral reservoir and its potential therapeutic candidacy to enhance HIV T cell mediated immunity in cure strategies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-human monoclonal anti-CD3 (UCHT1; BV711) | BD Biosciences | Cat# 563725; RRID:AB_2744392 |
| Mouse anti-human monoclonal anti-CD3 (SP34-2; BV650) | BD Biosciences | Cat# 563916; RRID:AB_2738486 |
| Mouse anti-human monoclonal anti-CD3 (OKT3; FITC) | Biolegend | Cat# 317306; RRID:AB_571907 |
| Mouse anti-human monoclonal anti-CD4 (SK3; BUV805) | BD Biosciences | Cat# 612887; RRID:AB_2870176 |
| Mouse anti-human monoclonal anti-CD4 (L200; PE) | BD Biosciences | Cat# 550630; RRID:AB_393790 |
| Mouse anti-human monoclonal anti-CD107a (H4A3; PE-Cy5) | BD Biosciences | Cat# 555802; RRID:AB_396136 |
| Mouse anti-human monoclonal anti-CD25 (M-A251; BV711) | Biolegend | Cat# 356138; RRID:AB_2632781 |
| Mouse anti-human monoclonal anti-CD27 (L128; BUV737) | BD Biosciences | Cat# 612829; RRID:AB_2870151 |
| Mouse anti-human monoclonal anti-CD27 (M-T271; APC-H7) | BD Biosciences | Cat# 560222; RRID:AB_1645474 |
| Mouse anti-human monoclonal anti-CD45RA (HI100; BV510) | Biolegend | Cat# 304141; RRID:AB_2561384 |
| Mouse anti-human monoclonal anti-CD45RA (HI100; BV605) | BD Biosciences | Cat# 562886; RRID:AB_273786 |
| Mouse anti-human monoclonal anti-CD69 (FN50; BV785) | BD Biosciences | Cat# 563834; RRID:AB_2738441 |
| Mouse anti-human monoclonal anti-CD8 (RPA-T8; BUV395) | BD Biosciences | Cat# 563795; RRID:AB_2722501 |
| Mouse anti-human monoclonal anti-CD8 (RPA-T8; APC) | BD Biosciences | Cat# 555369; RRID:AB_398595 |
| Mouse anti-human monoclonal anti-FOXP3 (259D; PE) | Biolegend | Cat# 320208; RRID:AB_492982 |
| Mouse anti-human monoclonal anti-GATA3 (16E10A23; Alexa fluor 647) | Biolegend | Cat# 653810; RRID:AB_2563217 |
| Mouse anti-human monoclonal anti-gp130 (2E1B02; PE-Cy7) | Biolegend | Cat# 362007; RRID:AB_2876682 |
| Mouse anti-human monoclonal anti-granzyme A (CB9; alexa fluor 700) | Biolegend | Cat# 507210; RRID:AB_961343 |
| Mouse anti-human monoclonal anti-granzyme B (GB11; V510) | BD Biosciences | Cat# 563388; RRID:AB_2738174 |
| Mouse anti-human monoclonal anti-IFNɣ (4S.B3; V570) | Biolegend | Cat# 502534; RRID:AB_2563880 |
| Mouse anti-human monoclonal anti-IL-27R⍺ (191106; PE) | R&D Systems | Cat# FAB14791P; RRID:AB_10718687 |
| Mouse anti-human monoclonal anti-LAG-3 (11C3C65; PE-Dazzle 594) | Biolegend | Cat# 369332; RRID:AB_2734422 |
| Mouse anti-human monoclonal anti-STAT1 (1/STAT1; alexa fluor 647) | BD Biosciences | Cat# 558560; RRID:AB_647143 |
| Mouse anti-human monoclonal anti-STAT3 (M59-50; PerCP-Cy5.5) | BD Biosciences | Cat# 564133; RRID:AB_2738614 |
| Mouse anti-human monoclonal anti-(pY701) STAT1 (4a; PE) | BD Biosciences | Cat# 612564; RRID:AB_399855 |
| Rabbit anti-human monoclonal anti-(pY705) STAT3 (D3A7; alexa fluor 488) | Cell Signaling | Cat# 4323S; RRID:AB_561299 |
| Mouse anti-human monoclonal anti-T-bet (4B10; V421) | Biolegend | Cat# 644816; RRID:AB_10959653 |
| Mouse anti-human monoclonal anti-TIGIT (A15153G; PE-Cy7) | Biolegend | Cat# 372714; RRID:AB_2632929 |
| Mouse anti-human monoclonal anti-TIM-3 (F38-2E2; APC-Cy7) | Biolegend | Cat# 345026; RRID:AB_2565717 |
| Mouse anti-human monoclonal anti-TNF⍺ (4MAb11; alexa fluor 488) | BD Biosciences | Cat# 557,722; RRID:AB_396831 |
| Mouse anti-human monoclonal anti-STAT3 (4G4B45; alexa fluor 594) | Biolegend | Cat# 678003; RRID:AB_2566584 |
| Rabbit anti-human monoclonal anti-(pY701) STAT1 (58D6; PE) | Cell Signaling | Cat# 8062; RRID:AB_10859888 |
| Mouse anti-human monoclonal anti-IL-27 R alpha (191106) | R&D Systems | Cat# MAB1479; RRID:AB_2249005 |
| Mouse anti-human monoclonal anti-gp130 (28126) | R&D Systems | Cat# MAB228; RRID:AB_2233737 |
| Mouse IgG isotype control antibody (20116) | R&D Systems | Cat# MAB004; RRID: AB_357346 |
| monoclonal anti-HIV-1 p24 (39/5.4A) | Zeptometrix Corporation | Cat# 0801080; RRID: AB_2895201 |
| Monoclonal anti-HIV-1/2 p24 | Capricorn Products LLC | Cat# HIV-018-48303; RRID: AB_2895202 |
| Chemicals, peptides, and recombinant proteins | ||
| X-VIVO 15 serum-free hematopoietic cell medium | Lonza | Cat# 04-418Q |
| Benzonase Nuclease | Millipore Sigma | Cat# 70,664-3 |
| LIVE/DEAD fixable blue dead cell stain kit | Invitrogen | Cat# L23105 |
| Recombinant human IFN-Alpha | pbl Assay Science | Cat# 11200-2 |
| Recombinant uman IL-6 | Tonbo Biosciences | Cat# 21-8069 |
| Recombinant human IL-6 | PeproTech | Cat# 200-06 |
| Recombinant human IL-27 | PeproTech | Cat# 200-38 |
| 16% paraformaldehyde aqueous solution | Electron Microscopy Sciences | Cat# 15710 |
| Human IgG | Millipore Sigma | Cat# I4506 |
| HIVGag peptide pool | NIH AIDS Reagent Program | Cat# 12425 |
| CEF peptide pool | NIH AIDS Reagent Program | Cat# 9809 |
| Dimethyl sulfoxide (DMSO) | Millipore Sigma | Cat# D2650 |
| Brefeldin A | Millipore Sigma | Cat# B6542 |
| TRIzol | Invitrogen | Cat# 15596026 |
| T cell TransAct | Miltenyi Biotec | 130-111-160 |
| HIV type 1 p24 | Zeptometrix | Cat# 0801002 |
| Deposited data | ||
| Raw data | This paper | GSE189997https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE189997 |
| Software and algorithms | ||
| GraphPad prism | GraphPad | https://www.graphpad.com, RRID:SCR_002798 |
| CLC genomics workbench V20 | QIAGEN | www.qiagenbioinformatics.com |
| IPA (Ingenuity pathway analysis) | QIAGEN | https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis |
| JMP pro 15 | SAS Institute Inc | https://www.jmp.com |
| Other | ||
| Ultrasensitive HIV-1 p24 detection ELISA assay | https://doi.org/10.1038/s41598-021-03072-7 | |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Marta Catalfamo (mc2151@georgetown.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Patient and healthy volunteers
The human study was conducted according to the principles expressed in the Declaration of Helsinki. Participants were studied under a MedStar Georgetown University Hospital and a NIAID Institutional Review Board approved HIV clinical research studies CR00000926 and 91-I-0140 respectively. PLWH and healthy controls provided written informed consent for the collection of samples and subsequent analysis. Healthy volunteers were obtained from the MedStar Georgetown University Hospital, and the NIH Blood Bank. Healthy volunteers have a median age of 47 (IQR: 34–57) and gender distribution was 54% male and 46% female. The characteristics of PLWH group used in this study are described in Table S1.
Method details
Flow cytometry
Staining of IL-27Ra/WSX-1 and gp130
Surface staining of the receptor was performed in PBMCs from PLWH (n = 10 frozen and n = 5 fresh). Characteristics of the study patiecipatnts is presented in Table S1. PBMCs from healthy individuals were used as controls (n = 10, frozen). PBMCs were thawed with X-Vivo media (Lonza, MD) containing Benzonase nuclease (50 U/mL; Millipore Sigma, MO) and rested overnight. Fresh PBMCs were isolated by ficoll and stained. Cells were incubated with human IgG (10 μg/mL, Sigma, MO) to block Fc receptors followed by a cocktail of mAbs for evaluating the expression of receptor of IL-27 (Table S2, Panel 1).
Detection of phosphorylated STAT1 and STAT3
PBMCs were thawed with X-Vivo media (Lonza, MD) containing Benzonase nuclease (50 U/mL; Millipore Sigma, MO) and rested overnight. PBMCs were labeled with Live/Dead (Invitrogen, CA), adjusted to a cell concentration of 2 × 106 cells/mL, and stimulated for 30 minutes at 37°C and 5% CO2. Cells were stimulated with rhIL-27 (PeproTech, NJ) at 5 ng/mL, 10 ng/mL, 25 ng/mL and 100 ng/mL and phosphorylated STAT1 was measured by flow cytometry (Figure S1C, Table S2 Panel 2). In other studies, cells were stimulated with rhIL-27 (25 ng/mL, PeproTech, NJ), rhIFN-α (100 U/mL, PBL Biomedical Laboratories, NJ), rhIL-6 (100 ng/mL; Tonbo Biosciences, CA and PeproTech, NJ). Unstimulated cells cultured in media culture were used as control.
For detetction of expression and STAT1 and STAT3 activation by flow cytometry, after stimulation, cells were fixed with 4% paraformaldehyde followed by a permeabilization step with pre-cooled methanol and acetone 1:1 volume mix for 30 minutes on ice. Cells were washed and incubated with human IgG (10 μg/mL; Sigma, MO) for 10 minutes to block Fc receptors before staining with a cocktail of mAbs (Table S2 Panel 2 and Panel 3). Cells were acquired in a Flow Cytometer Symphony and expression of total STAT1 (t-STAT1), total STAT3 (t-STAT3) and phosphorylated forms p-STAT1, p-STAT3 in T cell subsets were analyzed with FlowJo.
Inhibition of STAT1 and STAT3 phosphorylation
Inhibition of IL-27 induced STAT1 and STAT3 phosphorylation were performed using increasing concentration of anti-IL-27Rα mAb 5 μg/mL, 10 μg/mL and 20 μg/mL (clone:191,106, R&D Systems, MN), and anti-gp130 mAb (10 μg/mL, clone:28126, R&D Systems, MN) was used to blocked IL-6 induced STAT3 phpophorylation (Figure S1C and S1F respectively, Table S2 Panel 2 and Panel 3).
For the blockade experiments, PBMCs were pre-incubated with anti-IL-27Rα mAbs (10 μg/mL, clone:191106, R&D Systems, MN) or anti-gp130 mAbs (10 μg/mL, clone:28126, R&D Systems, MN), or a combination of both mAbs for 20 minutes on ice. Cells incubated with non-specific mouse IgG (10μg/mL, clone:20,116, R&D Systems, MN) was used as control. Cell were then stimulated with media and rhIL-27 (10 ng/mL; PeproTech, NJ). Staining for phosphorylated STAT1 and STAT3 was performed as describe above with the panel of mAb described in Table S2, Panel 2 and Panel 3.
Cytokine secretion assay
To study the effect of IL-27 in T cells a time course stimulation assay was performed to evaluate the upregulation of CD69 expression in T cells incubated with rhIL-27 at concentration: 0.5ng/mL, 5ng/mL, 25ng/mL and 100ng/mL and media as control for overnight or 6 hours (Figure S3C). PBMCs were thawed with X-Vivo media (Lonza, MD) containing Benzonase nuclease (50 U/mL; Millipore Sigma, MO) and cultured in the absence or presence of rhIL-27 (100 ng/mL). After overnight culture, PBMCs from HIV infected participants (n = 17, Table S1) were stimulated with HIVGag peptide pool (2 μg/mL, NIH AIDS Reagent Program), and PBMCs from healthy donors (n = 17) were stimulated with the CMV, EBV and Influenza (CEF) peptide pools (5μg/mL, NIH AIDS Reagent Program). DMSO was used as control in the unstimulated culture condition. After 2 hours of stimulation, Brefeldin A (10 μg/mL, Calbiochem, CA) was added and cultured for additional 4 hours. Cells were harvested and stained with Live/Dead staining (Invitrogen, CA). Cells were incubated with human IgG (10 μg/mL, Sigma, MO) to block Fc receptors followed by a cocktail of mAbs as described in Table S2, Panel 5.
T cell sorting and mRNA-Seq analysis
T cell subsets were sorted based on surface expression of CD45RA and CD27. Naïve (CD45RA+CD27+) and Memory (CD45RA−CD27+) CD4 and CD8 T cell subsets from PLWH (n = 5) and healthy controls (n = 3) were sorted in BD FACS Aria based on surface staining with antibody cocktail of mAbs (Table S2, Panel 4). Sorted cells were spun down and rested 3 hours for subsequent in vitro simulation for 90 minutes at 37°C and 5% CO2 with rhIL-27 (100 ng/mL; PeproTech, NJ) or media as unstimulated control. Cells from healthy control were also stimulated with rhIFN-α (100 U/mL, PBL Biomedical Laboratories, N), rhIL-6 (100 ng/mL, PeproTech, NJ) as control. After stimulation, the cells were washed with cold PBS, resuspended in TRIzol (Invitrogen, CA) and stored at −80°C.
RNA-sequencing and bioinformatic analysis
mRNA sequencing
Total RNA extraction from approx. 50,000 sorted cells of each sample was performed using Qiagen RNeasy Plus Universal mini kit (Qiagen, Hilden, Germany). Extracted RNA samples were quantified using Qubit 2.0 Fluorometer (Life Technologies, CA) and RNA integrity was analyzed using Agilent TapeStation 4200 (Agilent Technologies, CA). cDNA was converted by using SMARTer kit (Clontech, CA), and sequencing libraries were prepared using Illumina Nextera XT Library Preparation Kit (NEB, MA). The samples were sequenced using a 2 × 150 bp Paired End (PE) configuration on Illumina HiSeq instrument (Illumina, CA).
Illumina's bcl2fastq2.17 software was used to de-multiplex the reads into individual fastq files for each sample. All original RNA sequencing data were deposited in the NCBI's Gene Expression Omnibus database (GSE189997 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE189997).
Reads were mapped to human genome GRCh38, Ensemble version 91, using CLC Genomics Workbench v20 (QIAGEN) and “total exon reads” counted for each gene. Gene expression was measured as the count of exon-mapped after normalization using the transcripts per million (TPM) method. Differential gene expression was calculated from log2 of TPM using mixed-effects ANOVA with JMP/Genomics (SAS Institute) software, taking human subject as a random effect, and reported as “Log2 fold change” (the base 2 log of the expression ratio, stimulated/unstimulated) for each cell type and cytokine treatment. P-values adjusted to consider multiple testing were calculated using the BH False Discovery Rate (FDR) procedure.
Lists of Differentially Expressed Genes (DEGs) for knowledge base searches and Venn diagrams were defined by choosing those with the following criteria: the absolute value of Log2 fold change (L2FC) greater than 1 and FDR less than 0.05.
mRNAseq analysis
Venn diagrams were generated using Bioinformatics & Evolutionary Genomics web tools (http://bioinformatics.psb.ugent.be/webtools/Venn/). IPA (QIAGEN) software was used for canonical pathways and upstream regulator searches with lists of DEGs identified in this study. Heatmaps were analyzed for the core gene expression of Th1, Th2, Th17 and Treg and immune checkpoint receptors (Hollbacher et al., 2020; Kim et al., 2020; Ramesh et al., 2014; Utzschneider et al., 2020).
Heatmap (Hierarchical clustering analysis) clustered downstream genes of upstream regulators (UR)-STAT1 and STAT3 among over all 49 sample lists. Upstream Regulator (UR) bioinformatic analysis was computed in IPA (Qiagen) by searching with all 12 gene lists (4 cell subsets, 3 cytokines stimulation compared to unstimulated for each individual), then using the comparison analysis method to aggregate the results.
Detection of p24 by ultrasensitive planar array p24 Gag ELISA
PBMCs from PLWH were cultured with media in presence or abscence of rhIL-27 (100 ng/mL; PeproTech, NJ) for overnight or for three days. As control, PBMCs were stimulated with CD3/CD28 mAbs (T cell TransAct catalog # 130-111-160, Miltenyi Biotec, Germany) overnight and during three and four days. Supernatants were colleted for p24 detection using an ultra-sensitive ELISA (SP-X technology) as described by (Levinger et al., 2021). Briefly, plates coated with 1 μg/mL capture antibody (HIV-018-48,303, Capricorn Products LLC, ME) were incubated with 50 μL of each culture condition in triplicates for 2 hours on a shaker. Plates were washed four times and patted dry to remove excess wash buffer (ELISA wash buffer, Quanterix, MA). Before addition of the biotinylated anti-p24 detection antibody (1 μg/mL, clone: 39/5.4A, Zeptometrix Corporation, NY) a blockade step was performed by using 5% non-fat milk for 30 minutes. Detection of p24 was performed by incubation with streptavidin-HRP (Quanterix, MA) for 30 minutes on a shaker. The plate was washed six times and patted dry after each step. After adding mixed 25μL Stable Peroxide (Quanterix, MA) and 25μL SuperSignal Luminol (Quanterix, MA) for each well, the plate was immediately read on the SP-X Imager (Quanterix, MA). P24 was calculated based on a standard curve using recombinant p24 protein (Catalog# 0801002, ZeptoMetrix Corporation, NY) prepared in X-Vivo media (Lonza, MD) at concentrations: 100, 20, 4, 0.8, 0.16, 0.032. 0.0064 and 0 pg/mL.
Quantification and statistical analysis
Comparison between groups in the flow cytometry experiments were performed using non-parametric Mann-Whitney test, and Wilcoxon test for comparisons between culture conditions. p value <0.05 was considered significant.
Acknowledgments
The CEF and HIVGag peptide pools were obtained through the NIH-AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institute of Health. The graphical abstract was created with Biorender.com. This work was supported by Leidos Biomedical Research, Inc. and has been funded in whole or in part with federal funds from the National Cancer Institute, NIH, under Contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the US Government. MC is also supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number NIH R01AI145549-02 and by the District of Columbia Center for AIDS Research, an NIH funded program (P30AI117970) which is supported by the following NIH Co-Funding and Participating Institutes and Centers: NIAID, NCI, NICHD, NHLBI, NIDA, NIMH, NIA, NIDDK, NIMHD, NIDCR, NINR, FIC and OAR. TGM is supported by the Intramural Research Program of the NIH. The authors thank Dr. Hui Chen for the critical review of the manuscript and Mr. Ziang Zhu for helping with sample collection.
Author contributions
MC conceptualized and designed the study. JC designed and performed the experiments. JC, TGM, and MC analyzed and interpreted the data and wrote the manuscript. CL and AB perfomed the p24 detetction assay. PK, JK and BAG were involved in recruitment of participants of the study. All authors critically reviewed the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: January 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103588.
Supplemental information
Lists of differentially expressed gene (DEGs) induced by IFNα, IL-6, and IL-27 stimulation selected by | log2(FC) | > 1 compared with unstimulated condition and Adjusted p-value < 0. 05 were used as input in the upstream regulator analysis (IPA). DEGs predicted by IPA to be regulated via STAT1 activityare listed
Lists of differentially expressed genes (DEGs) induced by IFNα, IL-6, and IL-27 stimulation selected by | log2(FC) | >1 and adjusted p-value < 0. 05 was used as input in the upstream regulator analysis (IPA). DEGs predicted by IPA to be regulated via STAT3 activity are listed
Data and code availability
RNAseq data have been deposited at in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE189997 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE189997). GSE accession number is listed in the key resources table.
This paper does not report original code.
Data reported in this paper will be shared by the lead contact upon request. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Batten M., Kljavin N.M., Li J., Walter M.J., de Sauvage F.J., Ghilardi N. Cutting edge: IL-27 is a potent inducer of IL-10 but not FoxP3 in murine T cells. J. Immunol. 2008;180:2752–2756. doi: 10.4049/jimmunol.180.5.2752. [DOI] [PubMed] [Google Scholar]
- Betts M.R., Nason M.C., West S.M., De Rosa S.C., Migueles S.A., Abraham J., Lederman M.M., Benito J.M., Goepfert P.A., Connors M., et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggert M., Frederiksen J., Lund O., Betts M.R., Biague A., Nielsen M., Tauriainen J., Norrgren H., Medstrand P., Karlsson A.C., et al. CD4+ T cells with an activated and exhausted phenotype distinguish immunodeficiency during aviremic HIV-2 infection. AIDS. 2016;30:2415–2426. doi: 10.1097/QAD.0000000000001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggert M., Nguyen S., McLane L.M., Steblyanko M., Anikeeva N., Paquin-Proulx D., Del Rio Estrada P.M., Ablanedo-Terrazas Y., Noyan K., Reuter M.A., et al. Limited immune surveillance in lymphoid tissue by cytolytic CD4+ T cells during health and HIV disease. PLoS Pathog. 2018;14:e1006973. doi: 10.1371/journal.ppat.1006973. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Catalfamo M., Di Mascio M., Hu Z., Srinivasula S., Thaker V., Adelsberger J., Rupert A., Baseler M., Tagaya Y., Roby G., et al. HIV infection-associated immune activation occurs by two distinct pathways that differentially affect CD4 and CD8 T cells. Proc. Natl. Acad. Sci. U S A. 2008;105:19851–19856. doi: 10.1073/pnas.0810032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalfamo M., Wilhelm C., Tcheung L., Proschan M., Friesen T., Park J.H., Adelsberger J., Baseler M., Maldarelli F., Davey R., et al. CD4 and CD8 T cell immune activation during chronic HIV infection: roles of homeostasis, HIV, type I IFN, and IL-7. J. Immunol. 2011;186:2106–2116. doi: 10.4049/jimmunol.1002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Moussa M., Catalfamo M. The role of immunomodulatory receptors in the pathogenesis of HIV infection: a therapeutic opportunity for HIV cure? Front Immunol. 2020;11:1223. doi: 10.3389/fimmu.2020.01223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Swaminathan S., Yang D., Dai L., Sui H., Yang J., Hornung R.L., Wang Y., Huang da W., Hu X., et al. Interleukin-27 is a potent inhibitor of cis HIV-1 replication in monocyte-derived dendritic cells via a type I interferon-independent pathway. PLoS One. 2013;8:e59194. doi: 10.1371/journal.pone.0059194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew G.M., Fujita T., Webb G.M., Burwitz B.J., Wu H.L., Reed J.S., Hammond K.B., Clayton K.L., Ishii N., Abdel-Mohsen M., et al. TIGIT marks exhausted T cells, correlates with disease progression, and serves as a target for immune restoration in HIV and SIV infection. PLoS Pathog. 2016;12:e1005349. doi: 10.1371/journal.ppat.1005349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihara N., Madi A., Kondo T., Zhang H., Acharya N., Singer M., Nyman J., Marjanovic N.D., Kowalczyk M.S., Wang C., et al. Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature. 2018;558:454–459. doi: 10.1038/s41586-018-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill M.J., Deeks S.G., Margolis D.M., Siliciano R.F., Swanstrom R. HIV reservoirs: what, where and how to target them. Nat. Rev. Microbiol. 2016;14:55–60. doi: 10.1038/nrmicro.2015.5. [DOI] [PubMed] [Google Scholar]
- Crabe S., Guay-Giroux A., Tormo A.J., Duluc D., Lissilaa R., Guilhot F., Mavoungou-Bigouagou U., Lefouili F., Cognet I., Ferlin W., et al. The IL-27 p28 subunit binds cytokine-like factor 1 to form a cytokine regulating NK and T cell activities requiring IL-6R for signaling. J. Immunol. 2009;183:7692–7702. doi: 10.4049/jimmunol.0901464. [DOI] [PubMed] [Google Scholar]
- Dai L., Lidie K.B., Chen Q., Adelsberger J.W., Zheng X., Huang D., Yang J., Lempicki R.A., Rehman T., Dewar R.L., et al. IL-27 inhibits HIV-1 infection in human macrophages by down-regulating host factor SPTBN1 during monocyte to macrophage differentiation. J. Exp. Med. 2013;210:517–534. doi: 10.1084/jem.20120572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Aquino M.T., Kapil P., Hinton D.R., Phares T.W., Puntambekar S.S., Savarin C., Bergmann C.C., Stohlman S.A. IL-27 limits central nervous system viral clearance by promoting IL-10 and enhances demyelination. J. Immunol. 2014;193:285–294. doi: 10.4049/jimmunol.1400058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong J.H., O'Hara Hall A., Rausch M., Moodley D., Perry J., Park J., Phan A.T., Beiting D.P., Kedl R.M., Hill J.A., Hunter C.A. IL-27 and TCR stimulation promote T cell expression of multiple inhibitory receptors. Immunohorizons. 2019;3:13–25. doi: 10.4049/immunohorizons.1800083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devergne O., Hummel M., Koeppen H., Le Beau M.M., Nathanson E.C., Kieff E., Birkenbach M. A novel interleukin-12 p40-related protein induced by latent Epstein-Barr virus infection in B lymphocytes. J. Virol. 1996;70:1143–1153. doi: 10.1128/jvi.70.2.1143-1153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doisne J.M., Urrutia A., Lacabaratz-Porret C., Goujard C., Meyer L., Chaix M.L., Sinet M., Venet A. CD8+ T cells specific for EBV, cytomegalovirus, and influenza virus are activated during primary HIV infection. J. Immunol. 2004;173:2410–2418. doi: 10.4049/jimmunol.173.4.2410. [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbi M., Carbotti G., Ferrini S. Dual roles of IL-27 in cancer biology and immunotherapy. Mediators Inflamm. 2017;2017 doi: 10.1155/2017/3958069. 3958069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakruddin J.M., Lempicki R.A., Gorelick R.J., Yang J., Adelsberger J.W., Garcia-Pineres A.J., Pinto L.A., Lane H.C., Imamichi T. Noninfectious papilloma virus-like particles inhibit HIV-1 replication: implications for immune control of HIV-1 infection by IL-27. Blood. 2007;109:1841–1849. doi: 10.1182/blood-2006-02-001578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromentin R., Bakeman W., Lawani M.B., Khoury G., Hartogensis W., DaFonseca S., Killian M., Epling L., Hoh R., Sinclair E., et al. CD4+ T cells expressing PD-1, TIGIT and LAG-3 contribute to HIV persistence during ART. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005761. e1005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwell-Wild T., Vazquez N., Jin W., Rangel Z., Munson P.J., Wahl S.M. Interleukin-27 inhibition of HIV-1 involves an intermediate induction of type I interferon. Blood. 2009;114:1864–1874. doi: 10.1182/blood-2009-03-211540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A.O., Silver J.S., Hunter C.A. The immunobiology of IL-27. Adv. Immunol. 2012;115:1–44. doi: 10.1016/B978-0-12-394299-9.00001-1. [DOI] [PubMed] [Google Scholar]
- Harker J.A., Dolgoter A., Zuniga E.I. Cell-intrinsic IL-27 and gp130 cytokine receptor signaling regulates virus-specific CD4(+) T cell responses and viral control during chronic infection. Immunity. 2013;39:548–559. doi: 10.1016/j.immuni.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker J.A., Wong K.A., Dallari S., Bao P., Dolgoter A., Jo Y., Wehrens E.J., Macal M., Zuniga E.I. Interleukin-27R signaling mediates early viral containment and impacts innate and adaptive immunity after chronic lymphocytic choriomeningitis virus infection. J. Virol. 2018;92:e02196. doi: 10.1128/JVI.02196-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano H., Jain V., Hunt P.W., Lee T.H., Sinclair E., Do T.D., Hoh R., Martin J.N., McCune J.M., Hecht F., et al. Cell-based measures of viral persistence are associated with immune activation and programmed cell death protein 1 (PD-1)-expressing CD4+ T cells. J.Infect. Dis. 2013;208:50–56. doi: 10.1093/infdis/jis630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersperger A.R., Migueles S.A., Betts M.R., Connors M. Qualitative features of the HIV-specific CD8+ T-cell response associated with immunologic control. Curr.Opin. HIV AIDS. 2011;6:169–173. doi: 10.1097/COH.0b013e3283454c39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbert L., Pflanz S., De Waal Malefyt R., Kastelein R.A. IL-27 and IFN-alpha signal via Stat1 and Stat3 and induce T-Bet and IL-12Rbeta2 in naive T cells. J. Interferon Cytokine Res. 2003;23:513–522. doi: 10.1089/10799900360708632. [DOI] [PubMed] [Google Scholar]
- Hirahara K., Ghoreschi K., Yang X.P., Takahashi H., Laurence A., Vahedi G., Sciume G., Hall A.O., Dupont C.D., Francisco L.M., et al. Interleukin-27 priming of T cells controls IL-17 production in trans via induction of the ligand PD-L1. Immunity. 2012;36:1017–1030. doi: 10.1016/j.immuni.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirahara K., Onodera A., Villarino A.V., Bonelli M., Sciume G., Laurence A., Sun H.W., Brooks S.R., Vahedi G., Shih H.Y., et al. Asymmetric action of STAT transcription factors drives transcriptional outputs and cytokine specificity. Immunity. 2015;42:877–889. doi: 10.1016/j.immuni.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollbacher B., Duhen T., Motley S., Klicznik M.M., Gratz I.K., Campbell D.J. Transcriptomic profiling of human effector and regulatory T cell subsets identifies predictive population signatures. Immunohorizons. 2020;4:585–596. doi: 10.4049/immunohorizons.2000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Zak J., Pratumchai I., Shaabani N., Vartabedian V.F., Nguyen N., Wu T., Xiao C., Teijaro J.R. IL-27 promotes the expansion of self-renewing CD8(+) T cells in persistent viral infection. J. Exp. Med. 2019;216:1791–1808. doi: 10.1084/jem.20190173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C.A., Kastelein R. Interleukin-27: balancing protective and pathological immunity. Immunity. 2012;37:960–969. doi: 10.1016/j.immuni.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamichi T., Yang J., Huang D.W., Brann T.W., Fullmer B.A., Adelsberger J.W., Lempicki R.A., Baseler M.W., Lane H.C. IL-27, a novel anti-HIV cytokine, activates multiple interferon-inducible genes in macrophages. AIDS. 2008;22:39–45. doi: 10.1097/QAD.0b013e3282f3356c. [DOI] [PubMed] [Google Scholar]
- Iyer S.S., Ghaffari A.A., Cheng G. Lipopolysaccharide-mediated IL-10 transcriptional regulation requires sequential induction of type I IFNs and IL-27 in macrophages. J. Immunol. 2010;185:6599–6607. doi: 10.4049/jimmunol.1002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Bauer D.E., Tuttleton S.E., Lewin S., Gettie A., Blanchard J., Irwin C.E., Safrit J.T., Mittler J., Weinberger L., et al. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S.A., Jenkins B.J. Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nat. Rev. 2018;18:773–789. doi: 10.1038/s41577-018-0066-7. [DOI] [PubMed] [Google Scholar]
- Kamiya S., Owaki T., Morishima N., Fukai F., Mizuguchi J., Yoshimoto T. An indispensable role for STAT1 in IL-27-induced T-bet expression but not proliferation of naive CD4+ T cells. J. Immunol. 2004;173:3871–3877. doi: 10.4049/jimmunol.173.6.3871. [DOI] [PubMed] [Google Scholar]
- Kastelein R.A., Hunter C.A., Cua D.J. Discovery and biology of IL-23 and IL-27: related but functionally distinct regulators of inflammation. Annu. Rev. Immunol. 2007;25:221–242. doi: 10.1146/annurev.immunol.22.012703.104758. [DOI] [PubMed] [Google Scholar]
- Kaufmann D.E., Kavanagh D.G., Pereyra F., Zaunders J.J., Mackey E.W., Miura T., Palmer S., Brockman M., Rathod A., Piechocka-Trocha A., et al. Upregulation of CTLA-4 by HIV-specific CD4+ T cells correlates with disease progression and defines a reversible immune dysfunction. Nat. Immunol. 2007;8:1246–1254. doi: 10.1038/ni1515. [DOI] [PubMed] [Google Scholar]
- Kim K., Park S., Park S.Y., Kim G., Park S.M., Cho J.W., Kim D.H., Park Y.M., Koh Y.W., Kim H.R., et al. Single-cell transcriptome analysis reveals TOX as a promoting factor for T cell exhaustion and a predictor for anti-PD-1 responses in human cancer. Genome Med. 2020;12:22. doi: 10.1186/s13073-020-00722-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt N.R., Chomont N., Douek D.C., Deeks S.G. Immune activation and HIV persistence: implications for curative approaches to HIV infection. Immunol. Rev. 2013;254:326–342. doi: 10.1111/imr.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koup R.A., Safrit J.T., Cao Y., Andrews C.A., McLeod G., Borkowsky W., Farthing C., Ho D.D. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar B.V., Ma W., Miron M., Granot T., Guyer R.S., Carpenter D.J., Senda T., Sun X., Ho S.H., Lerner H., et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 2017;20:2921–2934. doi: 10.1016/j.celrep.2017.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Saout C., Hasley R.B., Imamichi H., Tcheung L., Hu Z., Luckey M.A., Park J.H., Durum S.K., Smith M., Rupert A.W., et al. Chronic exposure to type-I IFN under lymphopenic conditions alters CD4 T cell homeostasis. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1003976. e1003976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Saout C., Luckey M.A., Villarino A.V., Smith M., Hasley R.B., Myers T.G., Imamichi H., Park J.H., O'Shea J.J., Lane H.C., Catalfamo M. IL-7-dependent STAT1 activation limits homeostatic CD4+ T cell expansion. JCI Insight. 2017;2:e96228. doi: 10.1172/jci.insight.96228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinger C., Howard J., Cheng J., Tang P., Joshi A., Catalfamo M., Bosque A. An ultrasensitive planar array P24 Gag ELISA to detect HIV-1 in diverse biological matrixes. Sci. Rep. 2021;11:23682. doi: 10.1038/s41598-021-03072-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifson J.D., Rossio J.L., Piatak M., Jr., Parks T., Li L., Kiser R., Coalter V., Fisher B., Flynn B.M., Czajak S., et al. Role of CD8(+) lymphocytes in control of simian immunodeficiency virus infection and resistance to rechallenge after transient early antiretroviral treatment. J. Virol. 2001;75:10187–10199. doi: 10.1128/JVI.75.21.10187-10199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Guan X., Ma X. Regulation of IL-27 p28 gene expression in macrophages through MyD88- and interferon-gamma-mediated pathways. J. Exp. Med. 2007;204:141–152. doi: 10.1084/jem.20061440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas S., Ghilardi N., Li J., de Sauvage F.J. IL-27 regulates IL-12 responsiveness of naive CD4+ T cells through Stat1-dependent and -independent mechanisms. Proc. Natl. Acad. Sci. U S A. 2003;100:15047–15052. doi: 10.1073/pnas.2536517100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott A.B., Koup R.A. CD8(+) T cells in preventing HIV infection and disease. AIDS. 2012;26:1281–1292. doi: 10.1097/QAD.0b013e328353bcaf. [DOI] [PubMed] [Google Scholar]
- Migueles S.A., Connors M. Success and failure of the cellular immune response against HIV-1. Nat. Immunol. 2015;16:563–570. doi: 10.1038/ni.3161. [DOI] [PubMed] [Google Scholar]
- Molle C., Nguyen M., Flamand V., Renneson J., Trottein F., De Wit D., Willems F., Goldman M., Goriely S. IL-27 synthesis induced by TLR ligation critically depends on IFN regulatory factor 3. J. Immunol. 2007;178:7607–7615. doi: 10.4049/jimmunol.178.12.7607. [DOI] [PubMed] [Google Scholar]
- Morou A., Brunet-Ratnasingham E., Dube M., Charlebois R., Mercier E., Darko S., Brassard N., Nganou-Makamdop K., Arumugam S., Gendron-Lepage G., et al. Altered differentiation is central to HIV-specific CD4(+) T cell dysfunction in progressive disease. Nat. Immunol. 2019;20:1059–1070. doi: 10.1038/s41590-019-0418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morou A., Palmer B.E., Kaufmann D.E. Distinctive features of CD4+ T cell dysfunction in chronic viral infections. Curr.Opin. HIV AIDS. 2014;9:446–451. doi: 10.1097/COH.0000000000000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S.I., Friedl A., Aschenbrenner I., Esser-von Bieren J., Zacharias M., Devergne O., Feige M.J. A folding switch regulates interleukin 27 biogenesis and secretion of its alpha-subunit as a cytokine. Proc. Natl. Acad. Sci. U S A. 2019;116:1585–1590. doi: 10.1073/pnas.1816698116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessl J., Kaufmann D.E. Harnessing T follicular helper cell responses for HIV vaccine development. Viruses. 2018;10:336. doi: 10.3390/v10060336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owaki T., Asakawa M., Fukai F., Mizuguchi J., Yoshimoto T. IL-27 induces Th1 differentiation via p38 MAPK/T-bet- and intercellular adhesion molecule-1/LFA-1/ERK1/2-dependent pathways. J. Immunol. 2006;177:7579–7587. doi: 10.4049/jimmunol.177.11.7579. [DOI] [PubMed] [Google Scholar]
- Owaki T., Asakawa M., Morishima N., Hata K., Fukai F., Matsui M., Mizuguchi J., Yoshimoto T. A role for IL-27 in early regulation of Th1 differentiation. J. Immunol. 2005;175:2191–2200. doi: 10.4049/jimmunol.175.4.2191. [DOI] [PubMed] [Google Scholar]
- Peretz Y., He Z., Shi Y., Yassine-Diab B., Goulet J.P., Bordi R., Filali-Mouhim A., Loubert J.B., El-Far M., Dupuy F.P., et al. CD160 and PD-1 co-expression on HIV-specific CD8 T cells defines a subset with advanced dysfunction. PLoS Pathog. 2012;8:e1002840. doi: 10.1371/journal.ppat.1002840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A., Fowler K.D., Chalmin F., Merkler D., Kuchroo V.K., Pot C. IL-27 induces Th17 differentiation in the absence of STAT1 signaling. J. Immunol. 2015;195:4144–4153. doi: 10.4049/jimmunol.1302246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes C., Mariani M.K., Yang Y., Grandvaux N., Gee K. Interleukin (IL)-6 inhibits IL-27- and IL-30-mediated inflammatory responses in human monocytes. Front Immunol. 2018;9:256. doi: 10.3389/fimmu.2018.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovas C., Casazza J.P., Brenchley J.M., Price D.A., Gostick E., Adams W.C., Precopio M.L., Schacker T., Roederer M., Douek D.C., Koup R.A. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. J. Exp. Med. 2006;203:2281–2292. doi: 10.1084/jem.20061496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflanz S., Hibbert L., Mattson J., Rosales R., Vaisberg E., Bazan J.F., Phillips J.H., McClanahan T.K., de Waal Malefyt R., Kastelein R.A. WSX-1 and glycoprotein 130 constitute a signal-transducing receptor for IL-27. J. Immunol. 2004;172:2225–2231. doi: 10.4049/jimmunol.172.4.2225. [DOI] [PubMed] [Google Scholar]
- Pflanz S., Timans J.C., Cheung J., Rosales R., Kanzler H., Gilbert J., Hibbert L., Churakova T., Travis M., Vaisberg E., et al. IL-27, a heterodimeric cytokine composed of EBI3 and p28 protein, induces proliferation of naive CD4+ T cells. Immunity. 2002;16:779–790. doi: 10.1016/s1074-7613(02)00324-2. [DOI] [PubMed] [Google Scholar]
- Pirhonen J., Siren J., Julkunen I., Matikainen S. IFN-alpha regulates Toll-like receptor-mediated IL-27 gene expression in human macrophages. J. Leukoc. Biol. 2007;82:1185–1192. doi: 10.1189/jlb.0307157. [DOI] [PubMed] [Google Scholar]
- Pombo C., Wherry E.J., Gostick E., Price D.A., Betts M.R. Elevated expression of CD160 and 2B4 defines a cytolytic HIV-specific CD8+ T-cell population in elite controllers. J. Infect. Dis. 2015;212:1376–1386. doi: 10.1093/infdis/jiv226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poudyal D., Yang J., Chen Q., Goswami S., Adelsberger J.W., Das S., Herman A., Hornung R.L., Andresson T., Imamichi T. IL-27 posttranslationally regulates Y-box binding protein-1 to inhibit HIV-1 replication in human CD4+ T cells. AIDS. 2019;33:1819–1830. doi: 10.1097/QAD.0000000000002288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh R., Kozhaya L., McKevitt K., Djuretic I.M., Carlson T.J., Quintero M.A., McCauley J.L., Abreu M.T., Unutmaz D., Sundrud M.S. Pro-inflammatory human Th17 cells selectively express P-glycoprotein and are refractory to glucocorticoids. J. Exp. Med. 2014;211:89–104. doi: 10.1084/jem.20130301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remoli M.E., Gafa V., Giacomini E., Severa M., Lande R., Coccia E.M. IFN-beta modulates the response to TLR stimulation in human DC: involvement of IFN regulatory factor-1 (IRF-1) in IL-27 gene expression. Eur. J. Immunol. 2007;37:3499–3508. doi: 10.1002/eji.200737566. [DOI] [PubMed] [Google Scholar]
- Ruiz-Riol M., Berdnik D., Llano A., Mothe B., Galvez C., Perez-Alvarez S., Oriol-Tordera B., Olvera A., Silva-Arrieta S., Meulbroek M., et al. Identification of interleukin-27 (IL-27)/IL-27 receptor subunit alpha as a critical immune Axis for in vivo HIV control. J. Virol. 2017;91:e00441. doi: 10.1128/JVI.00441-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M., Yoshida K., Hibi M., Taga T., Kishimoto T. Molecular cloning of a murine IL-6 receptor-associated signal transducer, gp130, and its regulated expression in vivo. J. Immunol. 1992;148:4066–4071. [PubMed] [Google Scholar]
- Schmitz J.E., Kuroda M.J., Santra S., Sasseville V.G., Simon M.A., Lifton M.A., Racz P., Tenner-Racz K., Dalesandro M., Scallon B.J., et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- Schneider R., Yaneva T., Beauseigle D., El-Khoury L., Arbour N. IL-27 increases the proliferation and effector functions of human naive CD8+ T lymphocytes and promotes their development into Tc1 cells. Eur. J. Immunol. 2011;41:47–59. doi: 10.1002/eji.201040804. [DOI] [PubMed] [Google Scholar]
- Soghoian D.Z., Streeck H. Cytolytic CD4(+) T cells in viral immunity. Expert Rev. Vaccin. 2010;9:1453–1463. doi: 10.1586/erv.10.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumhofer J.S., Silver J.S., Laurence A., Porrett P.M., Harris T.H., Turka L.A., Ernst M., Saris C.J., O'Shea J.J., Hunter C.A. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat. Immunol. 2007;8:1363–1371. doi: 10.1038/ni1537. [DOI] [PubMed] [Google Scholar]
- Swaminathan S., Hu X., Zheng X., Kriga Y., Shetty J., Zhao Y., Stephens R., Tran B., Baseler M.W., Yang J., et al. Interleukin-27 treated human macrophages induce the expression of novel microRNAs which may mediate anti-viral properties. Biochem.Biophys. Res. Commun. 2013;434:228–234. doi: 10.1016/j.bbrc.2013.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S., Hu Z., Rupert A.W., Higgins J.M., Dewar R.L., Stevens R., Chen Q., Rehm C.A., Metcalf J.A., Baseler M.W., et al. Plasma interleukin-27 (IL-27) levels are not modulated in patients with chronic HIV-1 infection. PLoS One. 2014;9:e98989. doi: 10.1371/journal.pone.0098989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A., Hamano S., Yamanaka A., Hanada T., Ishibashi T., Mak T.W., Yoshimura A., Yoshida H. Cutting edge: role of IL-27/WSX-1 signaling for induction of T-bet through activation of STAT1 during initial Th1 commitment. J. Immunol. 2003;170:4886–4890. doi: 10.4049/jimmunol.170.10.4886. [DOI] [PubMed] [Google Scholar]
- Tan G., Xiao Q., Song H., Ma F., Xu F., Peng D., Li N., Wang X., Niu J., Gao P., et al. Type I IFN augments IL-27-dependent TRIM25 expression to inhibit HBV replication. Cell Mol. Immunol. 2018;15:272–281. doi: 10.1038/cmi.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Zhang A., Qiu C., Wang W., Yang Y., Qiu C., Liu A., Zhu L., Yuan S., Hu H., et al. The upregulation of LAG-3 on T cells defines a subpopulation with functional exhaustion and correlates with disease progression in HIV-infected subjects. J. Immunol. 2015;194:3873–3882. doi: 10.4049/jimmunol.1402176. [DOI] [PubMed] [Google Scholar]
- Trautmann L., Janbazian L., Chomont N., Said E.A., Gimmig S., Bessette B., Boulassel M.R., Delwart E., Sepulveda H., Balderas R.S., et al. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat. Med. 2006;12:1198–1202. doi: 10.1038/nm1482. [DOI] [PubMed] [Google Scholar]
- Utzschneider D.T., Gabriel S.S., Chisanga D., Gloury R., Gubser P.M., Vasanthakumar A., Shi W., Kallies A. Early precursor T cells establish and propagate T cell exhaustion in chronic infection. Nat. Immunol. 2020;21:1256–1266. doi: 10.1038/s41590-020-0760-z. [DOI] [PubMed] [Google Scholar]
- Villarino A., Hibbert L., Lieberman L., Wilson E., Mak T., Yoshida H., Kastelein R.A., Saris C., Hunter C.A. The IL-27R (WSX-1) is required to suppress T cell hyperactivity during infection. Immunity. 2003;19:645–655. doi: 10.1016/s1074-7613(03)00300-5. [DOI] [PubMed] [Google Scholar]
- Villarino A.V., Larkin J., 3rd, Saris C.J., Caton A.J., Lucas S., Wong T., de Sauvage F.J., Hunter C.A. Positive and negative regulation of the IL-27 receptor during lymphoid cell activation. J. Immunol. 2005;174:7684–7691. doi: 10.4049/jimmunol.174.12.7684. [DOI] [PubMed] [Google Scholar]
- Wykes M.N., Lewin S.R. Immune checkpoint blockade in infectious diseases. Nat. Rev. Immunol. 2018;18:91–104. doi: 10.1038/nri.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura T., Takeda A., Hamano S., Miyazaki Y., Kinjyo I., Ishibashi T., Yoshimura A., Yoshida H. Two-sided roles of IL-27: induction of Th1 differentiation on naive CD4+ T cells versus suppression of proinflammatory cytokine production including IL-23-induced IL-17 on activated CD4+ T cells partially through STAT3-dependent mechanism. J. Immunol. 2006;177:5377–5385. doi: 10.4049/jimmunol.177.8.5377. [DOI] [PubMed] [Google Scholar]
- Zheng Y.H., Xiao S.L., He B., He Y., Zhou H.Y., Chen Z., Zheng L.W., He M., Wang H.Y., Lin Y.H., Cao J. The role of IL-27 and its receptor in the pathogenesis of HIV/AIDS and anti-viral immune response. Curr. HIV Res. 2017;15:279–284. doi: 10.2174/1570162X15666170517130339. [DOI] [PubMed] [Google Scholar]
- Zhu C., Sakuishi K., Xiao S., Sun Z., Zaghouani S., Gu G., Wang C., Tan D.J., Wu C., Rangachari M., et al. An IL-27/NFIL3 signalling axis drives Tim-3 and IL-10 expression and T-cell dysfunction. Nat. Commun. 2015;6:6072. doi: 10.1038/ncomms7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lists of differentially expressed gene (DEGs) induced by IFNα, IL-6, and IL-27 stimulation selected by | log2(FC) | > 1 compared with unstimulated condition and Adjusted p-value < 0. 05 were used as input in the upstream regulator analysis (IPA). DEGs predicted by IPA to be regulated via STAT1 activityare listed
Lists of differentially expressed genes (DEGs) induced by IFNα, IL-6, and IL-27 stimulation selected by | log2(FC) | >1 and adjusted p-value < 0. 05 was used as input in the upstream regulator analysis (IPA). DEGs predicted by IPA to be regulated via STAT3 activity are listed
Data Availability Statement
RNAseq data have been deposited at in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE189997 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE189997). GSE accession number is listed in the key resources table.
This paper does not report original code.
Data reported in this paper will be shared by the lead contact upon request. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.