Abstract
A rapid and sensitive PCR assay for the detection of Candida albicans DNA in serum was established. DNA from human serum samples was purified using the QIAamp blood kit, which proved to be a fast and simple method for isolating minute amounts of Candida DNA from clinical specimens for diagnosis of invasive candidiasis. Universal primer sequences used in the PCR assay are derived from the internal transcribed spacer rRNA gene of fungi, whereas the biotinylated hybridization probe used in a DNA enzyme immunoassay (DEIA) binds specifically to C. albicans DNA. The sensitivity of this PCR-DEIA method is very high; the detection limit for genomic Candida DNA is one C. albicans genome per assay. Blood from uninfected and infected persons, ranging from healthy volunteers, patients with mucocutaneous infections, and patients at risk to develop a systemic Candida infection to patients with an established systemic candidiasis, was analyzed for the presence of C. albicans to diagnose fungal infection. Candida DNA could not be detected in sera of 16 culture-negative controls and from 11 nonsystemic candidal infections by PCR or DEIA. Blood cultures from patients at risk were all negative for Candida, whereas all blood cultures from systemic candidiasis patients were positive. However, Candida DNA could be detected by PCR and DEIA in the serum from three out of nine patients who were at risk for a systemic infection and in the serum of all seven patients who had already developed an invasive Candida infection. PCR is more sensitive than blood culture, since some of the patients at risk for invasive yeast infection, whose blood cultures were all negative for Candida, tested positive in the PCR amplification. These results indicate the potential value of PCR for detecting C. albicans in serum samples and for identifying patients at risk for invasive candidiasis.
Candida species are common human commensals that can cause a wide spectrum of disease. A major concern is a disseminated infection, which occurs with increased prevalence in postoperative and immunocompromised patients. Candida albicans, the major cause of invasive candidiasis, has become one of the pathogens most frequently isolated from the blood of postoperative and immunocompromised patients during the last decade (9, 12). The hematogenous spread of yeasts occurs frequently in these patients, leading to life-threatening disseminated infections and contributing significantly to mortality. Bloodstream infections due to Candida have risen to become the fourth-most-frequent cause of septicemia, with an attributable mortality rate of about 50% (22). To reduce mortality rates for patients with invasive candidiasis, early initiation of antifungal drug therapy is critical.
However, diagnosis remains difficult, since the only sign of infection may be a prolonged fever that is refractory to antibacterial treatment. Laboratory tests have been developed to detect circulating Candida antigens for rapid diagnosis of disseminated candidiasis (7). Detection of circulating antigens lacks sensitivity and, to some extent, specificity, so diagnosis can be delayed; in most cases, it is obtained only at autopsy. Existing diagnostic methods using antigen or antibody detection lack sensitivity and specificity (21, 27, 28). Antibody production in immunocompromised patients can be variable (32), complicating the diagnosis. Although two or more blood cultures are often used to identify disseminated disease, standard blood culturing methods can require two to three days or even longer for detection. Moreover, fungal blood culture, which is the “gold standard” in diagnosis, can remain negative despite widespread dissemination of Candida in internal organs (6).
Hence, a more rapid, specific, and sensitive test for the timely and accurate diagnosis of deep-seated Candida infections in both immunocompetent and immunocompromised patients is necessary. The development of DNA-based methods for detection of Candida (11) provides an alternative and potentially more sensitive means for diagnosing disseminated candidiasis. The detection of candidal DNA has already been conducted with amplification of the small subunit rRNA gene (19), lanosterol demethylase gene (16), 5.8S rRNA gene, including the adjacent nontranscribed spacer region (8), and the noncoding internal transcribed spacer (ITS) region of rRNA genes (2, 18). These assays worked well for cultured Candida cells or when blood was spiked with Candida cells and purified candidal DNA.
PCR has also been applied for the diagnosis of systemic candidiasis (10, 15). However, detection of C. albicans DNA recovered from clinical specimens lacked sensitivity, even if the blood culture was positive (1). Sensitivity could be improved to 10 cells per sample (10) or 3 cells per 0.1 ml of blood (15), but this required the use of Southern blotting coupled with radioactively labeled probes for detection. To increase the sensitivity of methods that do not involve radioactivity, the amplified product was bound to a streptavidin-coated microtiter plate using a biotinylated capture probe, and the amplicon was analyzed by an enzyme immunoassay (2, 5, 26). Recently, DNA from several microorganisms, e.g., Aspergillus in invasive aspergillosis (30), C. albicans (3), Legionella pneumophila (17), Coxiella burnetii (31), and human herpesvirus type 6 (20), was PCR amplified from serum of patients.
In this study, we describe a rapid and sensitive method for the detection of C. albicans DNA in serum samples, based on PCR amplification of the candidal 5.8S rRNA gene and the noncoding ITS region by using fungus-specific universal primers. A nonisotopic, C. albicans-specific biotin-labeled oligonucleotide probe was used in a DNA enzyme immunoassay. The QIAamp blood kit (Qiagen, Hilden, Germany) provides a fast pretreatment procedure for extracting DNA from serum samples in order to introduce a simple, specific, and more sensitive tool than blood culture and to improve diagnosis and management of invasive candidiasis.
MATERIALS AND METHODS
Yeast isolates.
Twelve yeast strains from the fungus collection of the Hygiene Institute, University of Heidelberg, Heidelberg, Germany, were used to test the universal fungi PCR system. These strains included C. albicans (HD 1447/95), C. tropicalis (HD 107/95), C. parapsilosis (HD 1173/95), C. krusei (HD 102/95), C. guilliermondii (HD 4941/92), C. pelliculosa (CBS-S110), C. rugosa (HD 529/95), C. glabrata (HD 126/95), C. lipolytica (CBS-S6124), C. lusitaniae (CBS 15.595), C. kefyr (CBS-S656), and C. neoformans (ATCC-3; HD 4544).
Clinical samples.
Swabs, stool specimens, blood, and sera were obtained from healthy volunteers or from patients at the University Hospital of Jakarta, Indonesia. Swabs taken from vagina and stool samples were collected in a sterile tube and inoculated on Sabouraud dextrose agar for 4 days. Blood cultures were incubated overnight, and sera were separated by centrifugation. The clot was cut into small pieces and inoculated on Sabouraud dextrose agar. Colonies were identified morphologically by the germ tube formation test and rice cream agar and biochemically by the sugar assimilation and fermentation test. Serum samples were obtained by centrifugation of clotted whole blood and were stored at −20°C until use. Healthy individuals and patients comprised four groups. Healthy volunteers without clinical evidence of mucocutaneous lesions and patients with vaginitis, who had tested negative for Candida in direct examinations and in cultures from smear, represented negative controls (group 1). Group 2 comprised 11 patients with mucocutaneous Candida infection. Group 3 consisted of 9 patients with predisposing factors for invasive candidiasis, such as underlying disease or surgery, who were admitted to the intensive care unit for more than 2 weeks. They were treated with antibiotics for a minimum of 2 weeks and developed fever in spite of prolonged antibiotic therapy. All patients in this group had already been tested three to five times by blood culture, which was always negative. Group 4 included patients with invasive candidiasis. These patients received antibiotic therapy for a minimum of 2 weeks and had at least three positive Candida blood cultures.
Extraction of Candida DNA.
Candida strains were grown on Sabouraud dextrose agar for 24 h. Cells were harvested, and genomic DNA was extracted (25). A thick suspension of C. albicans (200 μl) was mixed with 300 μl of lysis buffer (10 mM Tris-HCl, 10 mM EDTA, 50 mM NaCl, 0.2% sodium dodecyl sulfate). Proteinase K was then added to a final concentration of 20 mg/ml, incubated for 30 min at 56°C, boiled for 4 min, and subsequently kept on ice for 5 min. Extracted DNA was purified by phenol-chloroform extraction and ethanol precipitated (23), and the concentration was measured at 260 nm in a spectrophotometer. About 3 ng of purified DNA of all yeast strains was added to the PCR mixture for universal fungal PCR.
When sera from patients were tested, 200 μl of serum was treated with a QIAamp blood kit as recommended by the manufacturer. Proteinase K was added to 200 μl of serum and vortexed for 15 s. Subsequently, 200 μl of lysis buffer was added, vortexed again, and incubated for 10 min at 70°C. Then 225 μl of absolute ethanol was added, transferred to a Qiagen column, and spun down for 1 min at 6,000 × g. Afterwards, 500 μl of washing buffer was pipetted onto the column and spun down for 1 min at 6,000 × g. The washing procedure was repeated once, and buffer was centrifuged for 3 min at 13,000 × g. Finally, the column was placed in a microcentrifuge tube, and 200 μl of elution buffer was applied to the column and incubated for 5 min at 70°C. The eluted fraction was applied one more time to the column and spun down for 1 min at 6,000 × g, and 30 μl was assayed in PCR.
PCR.
PCR was performed according to standard procedures (24), and all clinical samples were assayed three times in independent experiments. Oligonucleotide primers were derived from rRNA genes of fungi and can be used for universal fungi PCR (29). Forward primer ITS3 (5′-GCA TCG ATG AAG AAC GCA GC-3′) corresponds to the 5.8S rRNA gene, and reverse primer ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′) corresponds to the 28S rRNA gene of fungi. The biotinylated probe used for hybridization (5′-ATT GCT TGC GGC GGT AAC GTC C-3′) was designed to bind specifically to the ITS2 region of C. albicans, located between the 5.8S rRNA gene and the 28S rRNA gene (5). Primers and the biotinylated probe were purchased from TIB MolBiol (Berlin, Germany). PCR was performed in a total volume of 100 μl containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin, a 200 μM concentration of each deoxynucleoside triphosphate, a 0.5 μM concentration of each primer, 2.5 U of Taq DNA polymerase (AmpliTaq; Perkin-Elmer), and 30 μl of extracted specimens. Samples were placed in a Perkin-Elmer GeneAmp 2400 DNA thermal cycler. After an initial step of 5 min at 94°C, 35 cycles were performed for 1 min at 94°C, 1 min at 57°C, and 1 min at 72°C. Finally, an additional extension was achieved for 7 min at 72°C, and samples were cooled to 4°C and kept at this temperature until further processing. For positive and negative controls, 30 μl containing 3 ng of purified Candida DNA or 30 μl of distilled water, respectively, was added to 200 μl of negative serum and processed for DNA extraction with the QIAamp blood kit. For visualization, 10 μl of the amplified product was electrophoresed for 30 min at 80 V in a vertical 8% polyacrylamide gel in TBE buffer (0.089 M Tris-HCl, 0.089 M boric acid, 0.002 M EDTA [pH 8.4]), stained for 15 min in 0.5 μg of ethidium bromide/ml, and photographed under ultraviolet illumination.
To avoid sample contamination, we used the precautions suggested by Kwok and Higuchi (13). Cross-contamination by aerosols was reduced by physical separation of laboratory rooms used for reagent preparation, sample processing, and DNA amplification. Other precautions included UV irradiation for microcentrifuge tubes, racks, surfaces of laboratory benches, and instruments. Such laboratory procedures as autoclaving of buffers and distilled water, use of fresh lots of previously aliquoted reagents, combined use of positive-displacement pipetters and aerosol-resistant pipette tips, frequent changing of gloves, premixing of reagents, addition of DNA as the last step, and testing of negative controls, including omission of either the primer or the DNA template during PCR, were used. Appropriate negative controls which contained all of the reagents except the template DNA were always included for each set of amplification. In all experiments the negative controls always tested negative.
DEIA.
PCR-amplified DNA was hybridized and detected using the GEN-ETI-K DNA enzyme immunoassay (DEIA) kit (Sorin Biomedica, Düsseldorf, Germany) as recommended by the manufacturer. Briefly, 100 μl of the biotinylated C. albicans probe (1 ng/μl) was added to each well of a streptavidin-coated microtitration plate and incubated for 18 h at 4°C. The microtitration plate was washed five times with washing buffer, and 100 μl of hybridization buffer was applied to each well. Meanwhile, amplicons were denatured for 15 min at 100°C and put on ice immediately. Then 20 μl of the denatured amplicons was pipetted into the wells and incubated for 1 h at 37°C. After fivefold washing at high stringency, 100 μl of anti-double-stranded DNA antibody was pipetted into each well and kept at room temperature for 30 min. The wells were washed five times, and then 100 μl of an enzyme tracer, protein A conjugated with horseradish peroxidase, was added to the wells and incubated for 30 min at room temperature. Again, all wells were washed, and 100 μl of a chromogen substrate solution for horseradish peroxidase was added and kept at room temperature for 30 min. Finally, 200 μl of blocking solution was applied to each well, and the absorbance was determined at 450 nm using a microtitration plate reader (Dynatech MR 5000; Dynex, Denkendorf, Germany). Optical density (OD) was determined after subtraction of the absorbance of the reagent blank. The cutoff value of this test is 0.2 OD unit above the mean OD of negative controls.
Sensitivity.
Sensitivities of the PCR assay and the DNA enzyme immunoassay were tested by spiking 200 μl of serum with serial dilutions of either C. albicans cells ranging from 106 to 1 cell or of C. albicans DNA at concentrations ranging from 40 ng to 4 fg of genomic DNA. The concentration of DNA from all samples was extracted using the QIAamp blood kit, then PCR amplified, separated in an 8% polyacrylamide gel, stained with ethidium bromide, and photographed under UV illumination. Amplicons were hybridized with the C. albicans-specific probe using the DNA enzyme immunoassay.
Direct sequencing of PCR products.
PCR products were phenol-chloroform extracted and precipitated with ethanol. DNA was dissolved in bidistilled water to a final concentration of 20 ng/μl. The PCR products were automatically sequenced with a 373A extended DNA sequencer using the DyeDeoxy Terminator Taq-cycle sequencing technique (Applied Biosystems, Weiterstadt, Germany). Each sequencing reaction was performed in a volume of 20 μl containing 100 ng of the PCR product, 50 pmol of the sequencing primer ITS3 or ITS4, and 10.5 μl of the DyeDeoxy Terminator reaction mixture. The cycle sequencing reaction was incubated for 28 cycles in an automated GeneE temperature cycler (Techne, Cambridge, United Kingdom) under cycling conditions of 96°C for 30 s and 60°C for 4 min per cycle. Electrophoresis of the samples was carried out on a polyacrylamide gel.
RESULTS
PCR amplification of Candida rDNA and detection.
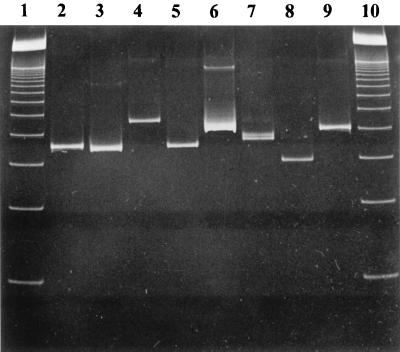
C. albicans and 11 other Candida species were used to test the applicability of the PCR system for the detection of Candida DNA. Of each species, about 3 ng of genomic DNA was used for PCR amplification. All species were amplified using universal fungal primers ITS3 and ITS4, most of them yielding PCR products differing from the 340-bp C. albicans amplicon. The amplified products of some Candida species are shown in Fig. 1. The specificity of the C. albicans capture probe used in our assay was tested by hybridization of the amplicons of all Candida species to the biotinylated capture probe specific for C. albicans. A positive hybridization result in the DEIA could be shown only for the C. albicans-derived amplicon; the results for all other Candida species were negative.
FIG. 1.
Ethidium bromide-stained polyacrylamide gel of PCR products obtained with primers ITS3 and ITS4 and DNAs from different Candida species. Lanes: 1, molecular weight marker (100-bp ladder); 2, C. albicans; 3, C. parapsilosis; 4, C. glabrata; 5, C. krusei; 6, C. pelliculosa; 7, C. tropicalis; 8, C. rugosa; 9, C. guilliermondii; 10, molecular weight marker (100-bp ladder).
Sensitivity for DNA detection from Candida cells and Candida DNA in serum.
To analyze the sensitivity of the PCR assay and the DEIA, serum from a noninfected healthy individual was spiked with serial dilutions of C. albicans cells, ranging from 106 to 1 cell per assay, or with serial dilutions of C. albicans genomic DNA, with concentrations ranging from 106 to 10−1 genomes (40 ng to 4 fg of purified C. albicans genomic DNA) per assay. The DNA was extracted by subjecting 200 μl of spiked serum to the QIAamp blood kit extraction procedure. After PCR amplification of genomic DNA, as few as 10 C. albicans genomes (400 fg of purified C. albicans genomic DNA) could be visualized in gel electrophoresis (Table 1). When the PCR amplicons were further analyzed in the DEIA by hybridization to the biotinylated C. albicans capture probe, as few as one C. albicans genome (40 fg of C. albicans genomic DNA) could be detected after hybridization in the DEIA (Table 1). Thus a tenfold increase in sensitivity was achieved in the DEIA.
TABLE 1.
Sensitivity of PCR and DEIA for Candida in seruma
| Assay | Result obtained with C. albicans genome amt ofb:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 106 | 105 | 104 | 103 | 102 | 101 | 1 | 0.1 | |
| PCRc | + | + | + | + | + | + | − | − |
| DEIAd | + | + | + | + | + | + | + | − |
+, positive; −, negative.
C. albicans genomic DNA was serially diluted, added to 200 μl of serum, and extracted using the QIAamp blood kit.
Measured as visible bands in ethidium bromide-stained gel monitored under ultraviolet illumination.
Hybridization of amplicons to the biotinylated C. albicans probe measured with DEIA.
Detection of C. albicans DNA in clinical samples.
Clinical samples from healthy volunteers or patients were tested by Candida culture and PCR followed by DEIA hybridization. For group 1, clinical samples from negative controls were analyzed for the presence of C. albicans or Candida DNA in serum (Table 2). Healthy volunteers 1 to 13 were physically examined and found to be negative for superficial Candida infection. Three patients with vaginitis were negative for Candida by direct examination and culture. C. albicans could be cultured from the stool of volunteer no. 3 due to an intestinal colonization with C. albicans. Furthermore, a Candida-specific 45-kDa band was detected in a Western blot by testing the serum of this individual (data not shown). All sera from group 1 were negative by PCR and hybridization, and all ODs of the hybridization were below the cutoff value (Table 2). For group 2, clinical samples from patients with mucocutaneous candidiasis were cultured, and C. albicans could be identified in all samples (Table 2). In contrast to the positive cultures from skin, nails, and swabs, Candida DNA could not be detected in sera of these patients either by PCR or by DEIA hybridization (Table 2). All serum samples from patients with proven mucocutaneous candidiasis were negative by PCR and DEIA.
TABLE 2.
Results of assay for Candida with culture from clinical specimens (swabs, stool, skin scrabs, and nail scrabs) and PCR with serum of negative control persons (group 1) and patients with mucocutaneous C. albicans infections (group 2)a
| Patient no. | Sex | Disease | Results with:
|
||
|---|---|---|---|---|---|
| Candida culture (swabs, etc.) | PCR (serum) | DEIA (OD) | |||
| Group 1 | |||||
| 1 | m | − | ND | − | − (0.042) |
| 2 | m | − | ND | − | − (0.023) |
| 3 | m | − | + | − | − (0.035) |
| 4 | m | − | ND | − | − (0.043) |
| 5 | m | − | ND | − | − (0.043) |
| 6 | m | − | ND | − | − (0.035) |
| 7 | m | − | ND | − | − (0.018) |
| 8 | m | − | ND | − | − (0.012) |
| 9 | m | − | ND | − | − (0.033) |
| 10 | m | − | ND | − | − (0.013) |
| 11 | m | − | ND | − | − (0.044) |
| 12 | m | − | ND | − | − (0.056) |
| 13 | m | − | ND | − | − (0.024) |
| 14 | f | Vaginitis | − | − | − (0.031) |
| 15 | f | Vaginitis | − | − | − (0.054) |
| 16 | f | Vaginitis | − | − | − (0.010) |
| Group 2 | |||||
| 17 | f | Vaginitis | + | − | − (0.044) |
| 18 | f | Vaginitis | + | − | − (0.160) |
| 19 | f | Vaginitis | + | − | − (0.042) |
| 20 | f | Vaginitis | + | − | − (0.163) |
| 21 | f | Vaginitis | + | − | − (0.079) |
| 22 | m | Skin candidiasis | + | − | − (0.089) |
| 23 | m | Onychomycosis | + | − | − (0.042) |
| 24 | m | Skin candidiasis | + | − | − (0.062) |
| 25 | f | Vaginitis | + | − | − (0.111) |
| 26 | f | Vaginitis | + | − | − (0.038) |
| 27 | f | Vaginitis | + | − | − (0.101) |
m, male; f, female; +, positive; −, negative; ND, not done. Candida culture refers to all Candida species.
Group 3 consisted of patients with predisposing factors for invasive candidiasis, e.g., underlying disease or surgery, who had been admitted to an intensive care unit for more than 2 weeks. Blood and serum samples of these patients were analyzed for the presence of Candida cells and Candida DNA, respectively. Candida blood cultures of all patients from group 3 were repeatedly negative. However, despite negative blood cultures, three out of nine serum samples of these patients (patients 28a, 31, and 34) were positive for Candida by PCR (Table 3). Any band by electrophoresis between approximately 300 and 400 bp is regarded as a positive result, since PCR is not genus specific. The specificity of each band must be determined either by hybridization with specific probes or by DNA sequencing. The amplified PCR products were further analyzed by DEIA hybridization using a C. albicans-specific probe. The PCR products from patients 28a and 31 could be verified as C. albicans in the DEIA (Table 3) and by direct sequencing of the PCR products. The DNA sequences of both amplicons are identical to the DNA sequence in the database (data not shown), thus confirming the specificity of the PCR-DEIA. Systemic candidiasis infection of patient 28a was proven 7 days later by a positive blood culture (patient 28b; Table 3), and clinical symptoms were in full agreement with those expected for a systemic Candida infection by that time. The amplicon from patient 34 tested negative in the DEIA hybridization, suggesting the presence of DNA from a Candida species other than C. albicans. Since no yeast could be grown in blood culture, direct DNA sequencing of this PCR product was performed and revealed the DNA sequence of the corresponding rRNA gene of C. krusei. C. albicans and C. krusei show only minor differences in the rRNA gene that is amplified during PCR; thus the PCR products from both species are about the same size. Interestingly, no PCR product could be amplified from the serum of patient 32, but C. albicans DNA could be detected in the hybridization assay. PCR followed by hybridization is about 10 times more sensitive than PCR without hybridization.
TABLE 3.
Results of Candida culture of blood and PCR assay of serum of patients at risk (group 3) and patients with systemic candidiasis (group 4)a
| Patient no. | Sex | Disease or condition | Result obtained with:
|
||
|---|---|---|---|---|---|
| Candida blood culture | PCR (serum) | DEIA (OD) | |||
| Group 3 | |||||
| 28a | f | Pancreatic malignancy | − | + | + (0.420) |
| 29 | f | Surgery | − | − | − (0.025) |
| 30 | m | Guillain-Barré syndrome | − | − | − (0.123) |
| 31 | m | Asthma | − | + | + (0.777) |
| 32 | m | Surgery | − | − | + (0.260) |
| 33 | m | Surgery | − | − | − (0.046) |
| 34 | m | Surgery | − | + | − (0.034) |
| 35 | m | Surgery | − | − | − (0.055) |
| 36 | m | Nephrectomy | − | − | − (0.035) |
| Group 4 | |||||
| 28b | f | Pancreatic malignancy | + | + | + (2.496) |
| 37 | m | Burn | + | + | + (2.290) |
| 38 | m | Pneumonia | + | + | + (1.490) |
| 39a | m | Surgery | + | + | + (2.324) |
| 39b | m | 39a after therapy | − | − | − (0.024) |
| 40 | m | Burn | + | + | + (0.355) |
| 41 | f | Myelopathy | + | + | + (0.583) |
| 42 | m | Pneumonia | + | + | + (2.429) |
m, male; f, female; +, positive; −, negative. Candida culture refers to all Candida species.
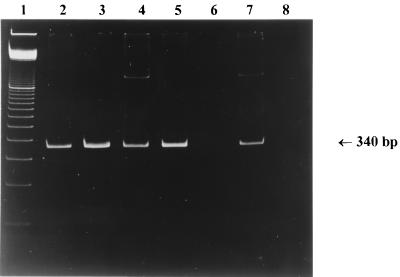
All patients with invasive candidiasis infections in group 4 tested positive for Candida by blood culture and PCR assay of serum. The PCR results of patients 37, 38, 39a, 39b, and 40, who were suffering from systemic candidiasis, are shown in Fig. 2. All PCR products from this group could be verified as C. albicans in the DEIA (Table 3). Most sera exerted strong positive signals in PCR and high ODs after hybridization. Patient 28b, who was initially blood culture negative but who was identified as being at risk for an invasive candidiasis (patient 28a), was now also included in group 4, since clinical symptoms and a positive blood culture by that time were in full agreement with a systemic Candida infection. Interestingly, patient 39a, who was positive by PCR and hybridization, became negative for Candida by blood culture and negative by PCR in serum, 3 days after therapy (patient 39b) with fluconazole was initiated (Table 3; Fig. 2).
FIG. 2.
Ethidium bromide-stained polyacrylamide gel of PCR products obtained with DNA extracted from serum samples of systemic candidiasis patients (group 4). Lanes: 1, molecular weight marker (100-bp ladder); 2, C. albicans DNA; 3, patient 37; 4, patient 38; 5, patient 39a (before therapy); 6, patient 39b (after therapy); 7, patient 40; 8, negative control. The size of the C. albicans PCR product is indicated by an arrow.
DISCUSSION
Given the fulminant and rapidly fatal outcome of invasive candidiasis, early and fast detection of Candida species and initiation of antifungal drug therapy are critical. Existing diagnostic methods using Candida blood culture, antigen, or antibody detection lack sensitivity and specificity (7, 21, 27). DNA-based diagnostic tests not only are sensitive and specific but also have the potential to decrease the time taken for the laboratory identification of pathogens that are growing slowly or difficult to culture. Therefore, earlier detection and identification would facilitate a prompt and appropriate treatment.
The presence of DNAs from several pathogens in blood of infected patients has been demonstrated, and recently, DNAs from several bacteria, fungi, and viruses were amplified from serum samples of patients (2, 17, 20, 31). However, the procedures used to extract the genomic DNAs of these microorganisms were rather time consuming and complicated. Difficult and labor-intensive methods for extraction of Candida DNA from blood and serum, such as mechanical disruption for serum spiked with Candida cells followed by protein digestion and DNA purification with phenol-chloroform, were performed (10). Burnie et al. (2) prepared the serum samples by addition of lysis buffer and chaotropic agents, such as guanidium thiocyanate, followed by DNA precipitation with sodium acetate and isopropanol. Chryssanthou et al. (3) used proteinase K and sodium dodecyl sulfate for protein denaturation and extracted DNA from serum with phenol-chloroform. The QIAamp procedure used in our study provides a standardized method and circumvents rather complicated extraction methods by application of ready-to-use columns for purification of genomic DNA. No hazardous reagents are needed anymore, and DNA extraction can be performed in about half an hour. Recently a variety of DNA extraction procedures have been applied to serum samples and intensively investigated (4). QIAamp is about two times more expensive than other extraction procedures, but the QIAamp method allows the processing of three to four times more samples per unit of time than other methods and does not use organic solvents. QIAamp was used for DNA purification of other fungal pathogens (14), which provided a sensitive method for a high yield of fungal DNA. Dixon et al. (4) recommended this method as a first choice for DNA extraction. In serum, DNA is supposedly present in a free form and can easily and effectively be purified using the QIAamp blood kit.
PCR was recently applied to the diagnosis of systemic candidiasis (10, 15). We chose universal fungal primers to amplify high-copy-number rRNA genes, the haploid genome harboring about 40 to 80 repeat copies. Most amplicons could be separated and differentiated from that of C. albicans by size in gel electrophoresis. The advantages of PCR are the relatively short processing time and its high sensitivity and specificity. The amplification feature of the PCR assay makes it ideal for detecting low levels of yeasts from minimal serum volumes.
The hybridization assay was different from previously reported ones in terms of labeling DNA with digoxigenin (2, 5, 26). This assay does not require DNA prelabeling prior to hybridization but uses a routine method, a common enzyme immunoassay. Radioactively labeled oligonucleotides (10, 15) are impractical for routine laboratory use because of their toxicity, expense, and short half-life. Although detection of Candida species other than C. albicans was not attempted in the present study, the specificity of other Candida species-specific probes introduced by Fujita et al. (5) indicates their potential for detection and differentiation of other Candida species in serum.
Using PCR amplification of the multicopy rRNA gene followed by the DEIA, we were able to detect as few as one C. albicans genome (40 fg of C. albicans genomic DNA) per 30 μl of serum. Burnie et al. (2) were able to detect 10 CFU using serum samples, whereas Holmes et al. (8) calculated a sensitivity of 15 CFU per ml of blood. The sensitivity was similar also to that obtained by Miyakawa (15), who reported detection of 3 cells per 0.1 ml of blood.
Serum samples from persons with vaginitis or with no symptoms at all showed negative results in PCR and hybridization. The results for these groups were similar to the negative results obtained by Kan et al. (10), who analyzed sera from healthy volunteers and from patients with active oral thrush but with no evidence of disseminated candidiasis. From the stool of a healthy volunteer, C. albicans could be isolated, but the serum was negative both by PCR and by hybridization. It can be concluded that the PCR-DEIA system will not react in the case of intestinal colonization.
No Candida DNA could be detected in sera from mucocutaneous patients either by PCR or by hybridization. A study by Burnie et al. (2) revealed positive PCR results for serum from 3 out of 16 patients colonized by C. albicans, but clinical pictures of the patients were not available. To evaluate clinical applicability of the PCR-DEIA, serum samples from patients at risk and from patients with systemic candidiasis were analyzed. Candida DNA could be detected in serum samples of some patients at risk of developing systemic candidiasis and in all patients with invasive candidiasis. However, despite negative blood cultures from at-risk patients, three out of nine serum samples of these patients were positive for Candida by PCR. Even for autopsy-verified candidiasis, a negative outcome of blood cultures is possible due to either the use of suboptimal culture systems or the fact that insufficient numbers of yeast cells were introduced into the bottles. Patient 28a, who was at risk for invasive candidiasis due to underlying disease, tested negative by blood culture but positive by the PCR assay and hybridization, thus indicating a systemic infection with C. albicans. The results for this patient demonstrate the high sensitivity of PCR and the low sensitivity of blood culture. The amplicon from patient 34 tested negative in the DEIA hybridization, suggesting the presence of DNA from a Candida species other than C. albicans. Direct sequencing of this PCR product revealed the DNA sequence of the corresponding rRNA gene of C. krusei. C. albicans and C. krusei show only minor differences in the rRNA gene that is amplified during PCR; thus the PCR products from both species are about the same size, and cross-reaction in hybridization has not been observed (5). Interestingly, no PCR product could be amplified from the serum of patient 32, but C. albicans DNA could be detected in the hybridization assay, demonstrating the higher sensitivity when PCR and DEIA are combined. Two samples were available from this patient, and both samples were tested three times. PCR was always negative; i.e., no band could be visualized in the gel. However, the DEIA results were always positive, and these samples were regarded as positive for C. albicans.
Assay for detection of C. albicans DNA always yielded positive results for patients with invasive candidiasis, and Candida could be cultured from blood. All PCR products from this group could be verified as C. albicans in the DEIA. Patient 28a, who was initially blood culture negative but who was identified as being at risk for invasive candidiasis, turned out to test positive by blood culture and by the PCR-DEIA 7 days later (patient 28b). Interestingly, patient 39, who was strongly positive by PCR and hybridization, became negative for Candida by blood culture and PCR from serum 3 days after therapy with fluconazole was initiated, thus indicating the response to therapy.
We describe a PCR-based method for the rapid detection and identification of C. albicans from serum. An extraction method that is simpler than previously described procedures for the recovery of Candida DNA from serum was applied, followed by PCR amplification and a microtitration plate enzyme immunosorbent assay. This method could be achieved within 6 h and has the potential for automatic processing.
ACKNOWLEDGMENTS
We thank the Deutscher Akademischer Austauschdienst (DAAD) for providing a scholarship to R.W., R. Kappe, University of Heidelberg, for providing the Candida strains, U. Bahr for his help in sequencing, and H. K. Geiss for critically reading the manuscript. We also thank G. P. Sibabiat and R. Sitompul for providing some of the clinical samples.
REFERENCES
- 1.Burgener-Kairuz P, Zuber J P, Jaunin P, Buchman T G, Bille J, Rossier M. Rapid detection and identification of Candida albicans and Torulopsis (Candida) glabrata in clinical specimens by species-specific nested PCR amplification of a cytochrome P-450 lanosterol-α-demethylase (L1A1) gene fragment. J Clin Microbiol. 1994;32:1902–1907. doi: 10.1128/jcm.32.8.1902-1907.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burnie J P, Golbang N, Mathews R C. Semiquantitative polymerase chain reaction enzyme immunoassay for diagnosis of disseminated candidiasis. Eur J Clin Microbiol Infect Dis. 1997;16:346–350. doi: 10.1007/BF01726361. [DOI] [PubMed] [Google Scholar]
- 3.Chryssanthou E, Anderson B, Petrint B, Lofdahl S, Tollemar J. Detection of Candida albicans DNA in serum by polymerase chain reaction. Scand J Infect Dis. 1994;26:479–485. doi: 10.3109/00365549409008623. [DOI] [PubMed] [Google Scholar]
- 4.Dixon S C, Horti J, Guo Y, Reed E, Figg W D. Methods for extracting and amplifying genomic DNA isolated from frozen serum. Nat Biotechnol. 1998;16:91–94. doi: 10.1038/nbt0198-91. [DOI] [PubMed] [Google Scholar]
- 5.Fujita S-I, Lasker B A, Lott T J, Reiss E, Morrison C J. Microtitration plate enzyme immunoassay to detect PCR-amplified DNA from Candida species in blood. J Clin Microbiol. 1995;33:962–967. doi: 10.1128/jcm.33.4.962-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geha D J, Roberts G D. Laboratory detection of fungemia. Clin Lab Med. 1994;14:83–97. [PubMed] [Google Scholar]
- 7.Herent P, Stynen D, Hernando F, Fruit J, Poulain D. Retrospective evaluation of two latex agglutination tests for detection of circulating antigens during invasive candidosis. J Clin Microbiol. 1992;30:2158–2164. doi: 10.1128/jcm.30.8.2158-2164.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holmes A R, Cannon R D, Sheperd M G, Jenkinson H F. Detection of C. albicans and other yeasts in blood by PCR. J Clin Microbiol. 1994;32:228–231. doi: 10.1128/jcm.32.1.228-231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holzman D C. NIH grant for Candida and AIDS research. Mol Med Today. 1995;7:300. doi: 10.1016/s1357-4310(95)80020-4. [DOI] [PubMed] [Google Scholar]
- 10.Kan V L. Polymerase chain reaction for the diagnosis of candidemia. J Infect Dis. 1993;168:779–783. doi: 10.1093/infdis/168.3.779. [DOI] [PubMed] [Google Scholar]
- 11.Kappe R, Okeke C N, Fauser C, Maiwald M, Sonntag H-G. Molecular probes for the detection of pathogenic fungi in the presence of human tissue. J Med Microbiol. 1998;47:811–820. doi: 10.1099/00222615-47-9-811. [DOI] [PubMed] [Google Scholar]
- 12.Komshian S V, Uwaydah A K, Sobel J D, Crane L R. Fungemia caused by Candida species and Torulopsis glabrata in the hospitalized patient: frequency, characteristics, and evaluation of factors influencing outcome. Rev Infect Dis. 1989;3:379–390. doi: 10.1093/clinids/11.3.379. [DOI] [PubMed] [Google Scholar]
- 13.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature (London) 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 14.Loeffler J, Hebart H, Schumacher U, Reitze H, Einsele H. Comparison of different methods for extraction of DNA of fungal pathogens from cultures and blood. J Clin Microbiol. 1997;35:3311–3312. doi: 10.1128/jcm.35.12.3311-3312.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyakawa Y, Mabuchi T, Fukazawa Y. New method for detection of Candida albicans in human blood by polymerase chain reaction. J Clin Microbiol. 1993;31:3344–3347. doi: 10.1128/jcm.31.12.3344-3347.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morace G, Sanguinetti M, Posteraro B, Cascio G L, Fadda G. Identification of various medically important Candida species in clinical specimens by PCR-restriction enzyme analysis. J Clin Microbiol. 1997;35:667–672. doi: 10.1128/jcm.35.3.667-672.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murdoch D R, Walford E J, Jennings L C. Use of the polymerase chain reaction to detect Legionella DNA in urine and serum samples from patients with pneumonia. Clin Infect Dis. 1996;23:475–480. doi: 10.1093/clinids/23.3.475. [DOI] [PubMed] [Google Scholar]
- 18.Nho S, Anderson M J, Moore C B, Denning D W. Species differentiation by internally transcribed PCR and HhaI digestion of fluconazole-resistant Candida krusei, Candida inconspicua, and Candida norvegensis strains. J Clin Microbiol. 1997;35:1036–1039. doi: 10.1128/jcm.35.4.1036-1039.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niesters H G M, Goessens W H F, Meis J M F G, Quint W G V. Rapid, polymerase chain reaction-based identification assays for Candida species. J Clin Microbiol. 1993;3:904–910. doi: 10.1128/jcm.31.4.904-910.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osiowy C, Prud'Homme I, Monette M, Zou S. Detection of human herpesvirus 6 DNA in serum by a microplate PCR-hybridization assay. J Clin Microbiol. 1998;36:68–72. doi: 10.1128/jcm.36.1.68-72.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Philips P, Dowd A, Jewesson P, Radigan G, Tweedale M G, Clarke A, Geere I, Kelly M. Nonvalue of antigen detection immunoassays for diagnosis of candidemia. J Clin Microbiol. 1990;10:2320–2326. doi: 10.1128/jcm.28.10.2320-2326.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reiss E, Morrison C J. Nonculture methods for diagnosis of disseminated candidiasis. Clin Microbiol Rev. 1993;6:311–323. doi: 10.1128/cmr.6.4.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rigsby W S, Torres Bauza L J, Wills J W, Townes T M. DNA content, kinetic complexity, and the ploidy question in Candida albicans. Mol Cell Biol. 1982;2:853–862. doi: 10.1128/mcb.2.7.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Shin J H, Nolte F S, Morrison C J. Rapid identification of Candida species in blood cultures by a clinically useful PCR method. J Clin Microbiol. 1997;35:1454–1459. doi: 10.1128/jcm.35.6.1454-1459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Deventer A J M, van Vliet H J A, Hop W C J, Goessens W H F. Diagnostic value of anti-Candida enolase antibodies. J Clin Microbiol. 1994;1:17–23. doi: 10.1128/jcm.32.1.17-23.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiss C, Kappe R, Sonntag H-G. Western blot analysis of the immune response to Candida albicans antigens in 391 long term intensive care patients. Mycoses. 1997;40:153–157. doi: 10.1111/j.1439-0507.1997.tb00207.x. [DOI] [PubMed] [Google Scholar]
- 29.White T J, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. San Diego, Calif: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 30.Yamakami Y, Hashimoto A, Tokimatsu I, Nasu M. PCR detection of DNA specific for Aspergillus species in serum of patients with invasive aspergillosis. J Clin Microbiol. 1996;34:2464–2468. doi: 10.1128/jcm.34.10.2464-2468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang G Q, Nguyen S V, To H, Ogawa M, Hotta A, Yamaguchi T, Kim H J, Fukushi H, Hirai K. Clinical evaluation of a new PCR assay for detection of Coxiella burnetii in human serum samples. J Clin Microbiol. 1998;36:77–80. doi: 10.1128/jcm.36.1.77-80.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zöller L, Krämer I, Kappe R, Sonntag H-G. Enzyme immunoassays for invasive Candida infections: reactivity of somatic antigens of Candida albicans. J Clin Microbiol. 1991;29:1860–1867. doi: 10.1128/jcm.29.9.1860-1867.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]