Abstract
The lysosome represents a central degradative compartment of eukaryote cells, yet little is known about the biogenesis and function of this organelle in parasitic protists. Whereas the mannose 6-phosphate (M6P)-dependent system is dominant for lysosomal targeting in metazoans, oligosaccharide-independent sorting has been reported in other eukaryotes. In this study, we investigated the phagolysosomal proteome of the human parasite Trichomonas vaginalis, its protein targeting and the involvement of lysosomes in hydrolase secretion. The organelles were purified using Percoll and OptiPrep gradient centrifugation and a novel purification protocol based on the phagocytosis of lactoferrin-covered magnetic nanoparticles. The analysis resulted in a lysosomal proteome of 462 proteins, which were sorted into 21 classes. Hydrolases represented the largest functional class and included proteases, lipases, phosphatases, and glycosidases. Identification of a large set of proteins involved in vesicular trafficking (80) and turnover of actin cytoskeleton rearrangement (29) indicate a dynamic phagolysosomal compartment. Several cysteine proteases such as TvCP2 were previously shown to be secreted. Our experiments showed that secretion of TvCP2 was strongly inhibited by chloroquine, which increases intralysosomal pH, thus indicating that TvCP2 secretion occurs through lysosomes rather than the classical secretory pathway. Unexpectedly, we identified divergent homologues of the M6P receptor TvMPR in the phagolysosomal proteome, although T. vaginalis lacks enzymes for M6P formation. To test whether oligosaccharides are involved in lysosomal targeting, we selected the lysosome-resident cysteine protease CLCP, which possesses two glycosylation sites. Mutation of any of the sites redirected CLCP to the secretory pathway. Similarly, the introduction of glycosylation sites to secreted β-amylase redirected this protein to lysosomes. Thus, unlike other parasitic protists, T. vaginalis seems to utilize glycosylation as a recognition marker for lysosomal hydrolases. Our findings provide the first insight into the complexity of T. vaginalis phagolysosomes, their biogenesis, and role in the unconventional secretion of cysteine peptidases.
Keywords: Trichomonas vaginalis, phagolysosome, proteome, cysteine peptidase, glycosylation, mannose 6-phosphate receptor
Abbreviations: AP, acid phosphatase; BA, β-amylase; BBS, borate buffered saline; BFA, Brefeldin A; bHX, β-hexosaminidase; CD-MPR, cation-dependent mannose 6-phosphate receptor; CI-MPR, cation-independent mannose 6-phosphate receptor; CLCP, cathepsin L-like cysteine peptidase; CLQ, chloroquine; CP, cysteine protease; CPBF1, cysteine protease-binding protein family 1; DMAP, DNA methyltransferase-associated protein; EDC, 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride; ER, endoplasmic reticulum; FITC, fluorescein isothiocyanate; GGA, Golgi-localized, γ-ear-containing, ADP ribosylation factor-binding protein; GlcNAc-PT, N-acetylglucosamine-1-phosphotransferase; HA, hemagglutinin; LAMP, lysosome-associated membrane glycoprotein; LERP, lysosomal enzyme receptor protein; LF, lactoferrin; LFQ MS, label-free quantitative mass spectrometry; LGF, large granule fraction; LTS, lysosomal targeting sequence; M6P, mannose 6-phosphate; mBA, mutated β-amylase; mCLCP, mutated cathepsin L-like cysteine peptidase; MES, 2-[n-morpholino]-ethanesulfonic acid buffer; MPR, mannose 6-phopshate receptor; MPR300, mannose 6-phopshate receptor 300; MRH, mannose 6-phosphate receptor homology; MS, mass spectrometry; nanoLC-MS, nano reverse-phase liquid chromatography mass spectrometry; NHS, N-hydroxysuccinimide; PBS, phosphate-buffered saline; s-LTS, lysosomal targeting sequence of soluble protein; SDC, sodium deoxycholate; SNAP, soluble N-ethylmaleimide-sensitive-factor attachment protein; SNARE, SNAP receptor; ST, sucrose-tris buffer; t-LTS, lysosomal targeting sequence of transmembrane protein; TBSR, Trichomonas beta-sandwich repeat protein; TCA, trichloracetic acid; TEAB, triethylammonium bicarbonate; TGN, trans Golgi network; TLCK, tosyl-L-lysyl-chloromethane hydrochloride; TMD, transmembrane domain; TSP, tetraspanin; TYM, tryptone-yeast extract-maltose medium; UCE, uncovering enzyme, N-acetylglucosamine-1-phosphodiester α-N-acetylglucosaminidase; vATPase, vacuolar ATPase
Graphical Abstract
Highlights
-
•
Trichomonas vaginalis phagolysosome consist of over 460 proteins.
-
•
Lysosomes are involved in secretion of virulence factors such as TvCP2.
-
•
N-glycosylation is required for lysosomal protein targeting.
-
•
T. vaginalis possesses homologs of mannose 6-phosphate receptor.
In Brief
Lysosomes represent a central degradative compartment of eukaryotes, yet little is known about biogenesis and function of this organelle in the parasitic protist Trichomonas vaginalis. We analyzed the phagolysosomal proteome that consists of over 460 proteins including important virulence factors. We demonstrated that glycosylation is involved in lysosomal protein targeting in T. vaginalis, which is unprecedented in parasitic protists. In addition to the classical secretory pathway, lysosomes are involved in unconventional protein secretion.
Trichomonas vaginalis is a flagellated parasitic protist that causes the most common nonviral sexually transmitted disease, with 276 million new infections annually (1). In women, the parasite can cause vaginitis and increase the risk of HIV transmission, preterm delivery, low birth weight, and cervical cancer. Most infected men are asymptomatic, but long-term infections increase the risk of prostate cancer development (2, 3, 4, 5). In the vaginal mucosa, T. vaginalis parasites actively phagocytose host cells such as epithelial cells, lymphocytes, erythrocytes, as well as cell debris and microbes, including yeast and bacteria (6, 7, 8, 9). Moreover, T. vaginalis secretes a large number of biologically active molecules, such as adhesins for cytoadherence, cytotoxic cysteine proteases (CPs), amylases and glycosidases to metabolize available glycogen (4, 10), and peptidoglycan hydrolases that are active against the bacterial cell wall (11). T. vaginalis secretes proteins through the classical secretory pathway (10) or unconventionally via exosomes, which are derived from the endolysosomal pathway (12). Vesicles with engulfed material fuse with lysosomes to form phagolysosomes, which are acidic organelles specializing in the breakdown of a broad range of biomolecules (13, 14, 15, 16). In addition, lysosomes participate in various other cellular processes, such as autophagy (17, 18), secretion, and degradation of misfolded proteins within the secretory pathway (19).
The biogenesis of lysosomes depends on the delivery of newly synthesized proteins from the trans-Golgi network (TGN) via the transport vesicles that deliver cargo within the cell and through the endosomal pathway that imports proteins from the plasma membrane (20, 21). Lysosomes are supplied with over 60 hydrolases (15, 22), as well as other proteins such as acidifying vacuolar ATPases (vATPases), lysosome-associated membrane glycoproteins (LAMPs), and over 50 lysosomal channel proteins and transporters (14, 15).
In metazoans, the sorting of most lysosomal hydrolases depends on the mannose 6-phosphate (M6P) pathway (23). Soluble lysosomal proteins are glycosylated on asparagine residues within the sequence Asn-X-[Ser/Thr] (soluble lysosomal targeting sequence, s-LTS) in the endoplasmic reticulum (ER) (23, 24). Then, in the Golgi body, N-acetylglucosamine-1-phosphotransferase (GlcNAc-PT) and N-acetylglucosamine-1-phosphodiester α-N-acetylglucosaminidase (“uncovering enzyme,” UCE) form M6P for interaction with M6P receptors (MPRs). Two MPRs, a cation-dependent (CD-MPR) and cation-independent MPR (CI-MPR), have been identified (22, 23, 25). Other lysosomal sorting receptors have also been described, including LIMP-2 (mammals), VSR (plants, algae, alveolates), and sortilin/Vps10, which were studied in mammals and yeast; however, sortilin homologues have been identified in members of all eukaryotic groups (23). Novel receptors for the delivery of CPs to lysosomes (CP-binding protein family 1) were identified in Entamoeba histolytica (26).
Receptors involved in protein delivery to lysosomes consist of a luminal domain that binds cargo in the Golgi body, at least one transmembrane domain, and a C-terminal tail that contains a lysosomal targeting sequence (LTS) facing the cytosol. LTSs of transmembrane proteins and receptors (t-LTS) are most frequently dileucine-based ([DE]xxxL[LI], DxxLL) or tyrosine-based (YxxØ) (24) and regulate endosomal/lysosomal sorting and internalization from the plasma membrane (27). T-LTSs are bound by cytosolic Golgi-localized, γ-ear-containing, ADP ribosylation factor-binding proteins (GGAs) that mediate sorting at the TGN, which is further facilitated by adaptor proteins. Cargo is released into acidified endosomes, and receptors are recycled. However, GGAs are known to be opisthokont-specific innovations while adaptor protein complexes that recognize the same LTSs are found across eukaryotes and are known to be involved in trafficking throughout the endolysosomal system (23, 25, 28, 29, 30, 31).
Little is known about the lysosomal proteome, its biogenesis, and function in T. vaginalis. The genome of this protist encodes 447 proteases; thus, a high number of these proteins can be expected in lysosomes (32). Indeed, some cysteine proteases involved in T. vaginalis pathogenesis, such as TvCP4, have been shown to reside partially in lysosomes (33, 34, 35). The secretome of T. vaginalis revealed active secretion of almost 90 proteins, including over 20 hydrolases (10). Moreover, the genome of T. vaginalis encodes an extensive complement of at least 73 adaptin subunit genes implying their role in the endolysosomal system (32). However, these data did not clarify the role of lysosomes in secretion, nor has lysosomal protein targeting been studied in this organism.
Considering the highly complex lysosomal content originating from external and internal sources and the high sensitivity of mass spectrometry (MS), the major challenge is to distinguish the actual lysosomal proteome from contaminants. Therefore, we used three different methods to isolate phagolysosomes and lysosome-enriched samples, which were analyzed by MS to estimate the phagolysosomal proteome. To uncover the mode of T. vaginalis lysosomal targeting, we evaluated the role of specific glycosylation. Furthermore, we demonstrated that lysosomes are involved in the unconventional secretion of a papain-like cysteine peptidase. These studies provide the first insights into the phagolysosomal composition and the endolysosomal route of T. vaginalis.
Experimental Procedures
Cell Cultivation
The T. vaginalis strains T1 (36) and TV17-48 (37) were cultured axenically in tryptone-yeast extract-maltose medium (TYM; pH 6.2) with 10% (v/v) heat-inactivated horse serum. The culture medium was supplemented with 200 μg/ml Geneticin G418 (single transfectants) or with both 200 μg/ml G418 and 40 μg/ml puromycin (double transfectants) for transfectant selection (10). Cultures were grown at 37 °C.
Gene Cloning and T. vaginalis Transfection
The genes encoding Rab7a (TVAG_159730), acid phosphatase (AP, TVAG_169070), Trichomonas beta-sandwich repeat protein 5 (TBSR-5, TVAG_340570), β-hexosaminidase (bHX, TVAG_110660), MPR-2 (TVAG_351790), MPR-3 (TVAG_498650), β-amylase 2 (BA2, TVAG_080000) and mutated BA2 (mBA2), and the cysteine peptidases TvCP2 (TVAG_057000), CLCP (TVAG_485880), and mutated CLCP (mCLCP) were cloned into the vector pTagVag-HA-Neo (38), enabling the expression of these proteins with a C-terminal haemagglutinin (HA) tag. The gene encoding BA1 (TVAG_436700) was cloned into the vector pTagVag-V5-Pur (10), fusing the protein to a V5 tag at the C-terminus. Genes were expressed with their respective native promotor (300 bp upstream of coding sequence). The plasmids were transfected into trichomonads by nucleofection using the Human T Cell Nucleofector Kit (Lonza). Approximately 4 ml of cells in the logarithmic growth phase (1.5 × 106 cells/ml) was harvested and resuspended gently in 82 μl Nucleofector Solution and 18 μl Supplement 1. Then, 10 μg of plasmid was added, and the mixture was incubated at room temperature for 10 min. The manufacturer's program U.033 was used for transfection (39). For double transfection, the cells were consecutively transfected with pTagVag-HA-Neo and pTagVag-V5-Pur. All primers used for gene amplification and cloning are listed in Table S1.
Isolation of Lysosomal Fractions
Lysosomes were isolated by three approaches. (i) For density gradient centrifugation in Percoll, we used a modified procedure based on the protocol by Bradley et al. (40). Briefly, 1 l of TV17-48 expressing HA-tagged TvRab7a (TV17-48-Rab7a) (1.5 × 106 cells/ml) was harvested by centrifugation (1200g, 10 min, 4 °C) and washed twice with phosphate-buffered saline (PBS) and once with sucrose-tris (ST) buffer (500 mM sucrose, 20 mM Tris, 1 mM KCl, pH 7.2). The cells were resuspended in 7.5 ml of ST buffer with 10 μg/ml leupeptin and 50 μg/ml tosyl-L-lysyl-chloromethane hydrochloride (TLCK). The cells were disintegrated by sonication on ice, and unbroken cells were removed by centrifugation at 800g for 10 min at 4 °C. The supernatant was centrifuged at 14,000g for 20 min at 4 °C. The top white layer of the pellet was separated from a brown layer of hydrogenosomes, gently resuspended in 7.5 ml of ST buffer, and centrifuged again under the same conditions. The white pellet was collected and resuspended in 2 ml of ST buffer with protease inhibitors as described above (large granule fraction, LGF) and centrifuged in a 45% Percoll gradient at 30,000 rpm for 30 min at 4 °C using a Beckman Optima XPN-90 ultracentrifuge and a Ti70 rotor. Deceleration was set to 9 to avoid disruption of the gradient. The upper lysosomal fraction was collected and centrifuged again in a 45% Percoll gradient under the same conditions. The final lysosomal fraction was washed twice with ST buffer at 14,000g for 20 min at 4 °C, resuspended in 500 μl ST buffer with protease inhibitors, and frozen before further processing.
(ii) Density gradient centrifugation using OptiPrep. The LGF of TV17-48-Rab7a was obtained as above and resuspended in 800 μl of a buffer consisting of 0.25 M sucrose, 1 mM EDTA, and 10 mM Tris-HCl, pH 7.4. The suspension was loaded into a 5% to 50% continuous OptiPrep gradient and centrifuged at 200,000g for 2 h at 4 °C using a Beckman Optima XPN-90 ultracentrifuge with an SW 41 Ti rotor. Acceleration was set to 4 and deceleration to 9. Three fractions corresponding to three visible bands were collected, washed, and frozen.
(iii) Phagolysosomes were isolated using lactoferrin (Lf)-covered Dynabeads. Lf was coupled with fluorescein isothiocyanate (FITC) in one volume PBS and two volumes borate buffered saline (BBS, pH 9) at a final concentration of 100 ng FITC/1 μg Lf. Then, the PBS-BBS solution was exchanged for 25 mM 2-[n-morpholino]-ethanesulfonic acid (MES) buffer (pH 6) using Amicon Ultra-4 30K Centrifugal Filters (Millipore). Magnetic Dynabeads MyOne Carboxylic Acid (Invitrogen) of 1 μm in diameter were coupled with the FITC-Lf in MES buffer according to Invitrogen's two-step protocol using 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS). Subsequently, FITC-Lf-coupled Dynabeads were incubated with 300 μl of 50 mM Tris, pH 7.4, for 15 min to quench nonreacted activated carboxylic acid groups. The FITC-Lf-coupled Dynabeads were washed four times with Tris for 5 min and resuspended in 100 μl PBS. For the isolation of phagolysosomes, 1 l of TV17-48-Rab7a cells (1.5 × 106 cells/ml) was harvested. The cells were resuspended in 20 ml TYM medium and incubated with 100 μl of FITC-Lf-coupled Dynabeads in a 150 ml tissue culture flask for 1 h at 37 °C. After incubation, the cells were gently disintegrated by sonication on ice. Lysis was evaluated under a microscope at 30-s intervals. Leupeptin and TLCK were added as before to avoid protein degradation. The lysosomes containing Dynabeads were washed three times with PBS by applying a magnet for 15 min after each wash and then filtering them through a 3 μm pore size filter (Whatman, GE Healthcare). Then, the lysosomes were treated with 0.1% Triton X-100 to release the Dynabeads. A magnet was applied, and the supernatant containing the lysosomal content was transferred to a new tube and frozen.
Immunoblot Analysis
The presence of marker proteins in cellular fractions was tested by immunoblotting using mouse monoclonal anti(α)-HA antibody (Exbio) against HA-tagged TvRab7a (lysosomes) and rat polyclonal α-OsmC antibody (hydrogenosomes) (41). Rat polyclonal α-soluble protein disulphide-isomerase (sPDI) antibody (endoplasmic reticulum) was raised against a recombinant PDI produced in Escherichia coli BL21 DE. The sPDI gene (TVAG_267400) was cloned into the bacterial expression vector pET42b, and the His-tagged protein was isolated by Ni-NTA agarose affinity chromatography (Qiagen). Anti-mouse or anti-rat polyclonal antibodies coupled with peroxidase were used as secondary antibodies (Sigma-Aldrich). Visualization was performed with chemiluminescence (Immobilon Forte, Merck), and images were obtained using the Amersham Imager 600 (GE Healthcare), and signals quantified using ImageJ/Fiji software (42).
Fluorescence Microscopy
T. vaginalis cells were incubated with 2.5 μM LysoTracker Deep Red (Thermo Fisher Scientific) for 5 min or with 5% FITC-Lf (v/v) for 30 min at 37 °C. Then, the cells were washed in PBS, fixed with 2% formaldehyde, and processed as described (43). Transfectants were stained using mouse monoclonal α-HA antibody (Exbio) and the secondary donkey α-mouse antibody conjugated to Alexa Fluor 488 or 594 (Life Technologies), and rabbit polyclonal α-V5 antibody (Abcam) and Alexa Fluor 594 donkey α-rabbit (Life Technologies). To observe T. vaginalis phagocytosis, TV17-48 cells expressing HA-tagged TvRab7a were incubated with FITC-Lf coupled Dynabeads for 40 min at 37 °C, washed in PBS, and processed as before with TvRab7a labeled with Alexa Fluor 594. Slides were observed with a Leica TCS SP8 confocal laser scanning microscope (Leica Microsystems). Images were processed using Huygens Professional version 19.10 (Scientific Volume Imaging) and further processed using ImageJ/Fiji software (42) and the Imaris 9.7.2 Package for Cell Biologists (Bitplane AG). Voxel-based colocalization was performed using ImarisColoc. Costes's automatic thresholding was applied to the images in the Z stack, and Pearson's correlation coefficient (PCC) in colocalized volume was calculated.
Protein Preparation
Protein samples were precipitated with trichloroacetic acid (TCA) for 1 h at −20 °C (44), pelleted at 21,000g for 5 min at 4 °C, washed twice with cold acetone, dried for 5 min at 95 °C, and stored at −80 °C. LFQ MS analysis was performed as described previously (10). Briefly, proteins were dissolved in 100 mM TEAB with 1% sodium deoxycholate (SDC). Samples were digested with trypsin, and detergent was removed by liquid–liquid extraction (45).
Mass Spectrometry Data Acquisition
Tryptic peptides were injected on a nano reverse-phase liquid chromatograph (UltiMate 3000 RSLC, Thermo Scientific) coupled with MS (nanoLC-MS) using an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific) as described previously (10). Briefly, peptides were loaded onto an Acclaim PepMap300 trap column (300 μm × 5 mm) packed with C18 (5 μm, 300 Å) in loading buffer (0.1% trifluoroacetic acid in 2% acetonitrile) for 4 min at a flow rate of 15 μl/min and then separated on an EASY-Spray column (75 μm × 50 cm) packed with C18 (2 μm, 100 Å, Thermo Scientific) at a flow rate of 300 nl/min. Mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile) were used to establish a 60-min gradient from 4% to 35% B. Eluted peptides were ionized by electrospray. The full MS spectrum (350–1400 m/z range) was acquired at a resolution of 120,000 at m/z 200 and a maximum ion accumulation time of 100 ms. Dynamic exclusion was set to 60 s. Higher-energy collisional dissociation (HCD) MS/MS spectra were acquired in iontrap in rapid mode, and the normalized collision energy was set to 30% with a maximum ion accumulation time of 35 ms. An isolation width of 1.6 m/z units was used for MS 2.
Analysis of MS Data
Raw data were processed using MaxQuant version 1.6.2.0 (Max-Planck-Institute of Biochemistry) (46). Searches were performed using the T. vaginalis database from EUpathDB (47) (release 2020-05-27, 60,330 entries). Trypsin cleaving specificity was used to generate the peptides, and two missed cleavages were allowed. The protein modifications were set as follows: cysteine (unimod nr: 39) as static and methionine oxidation (unimod: 1384) and protein N-terminus acetylation (unimod: 1) as variable. The false discovery rates for peptides and proteins were set to 1%. Search mass tolerances were used in MaxQuant default settings for Orbitrap and Iontrap. The precursor ion mass tolerance in the initial search was 20 ppm, the tolerance in the main search was 4.5 ppm, and the fragment ion mass tolerance was 0.5 Da.
Experimental Design and Statistical Rationale
Three different isolation methods were used, three independent biological experiments were performed for each method, and each biological sample was analyzed by MS in three technical replicates. The lysosomal proteome identifications were filtered using following criteria: (i) the protein identification was supported by at least three peptides, (ii) the protein was identified in at least two out of three biological replicates, and (iii) the protein was identified by all three methods for organelle isolation.
Bioinformatics
For each protein identified in the lysosomal proteome, targeting to the secretory pathway was predicted using the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP-4.1/), the TargetP 2.0 server (http://www.cbs.dtu.dk/services/TargetP/), and the SecretomeP 2.0 server (http://www.cbs.dtu.dk/services/SecretomeP/). Transmembrane domains were predicted using the TMHMM server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). Conserved domains were predicted using the Pfam 34.0 database (http://pfam.xfam.org, 19,179 entries). Identified proteins were annotated and sorted based on TrichDB annotation (https://trichdb.org/trichdb/), Pfam database, consensus of BLASTp top hits against the refseq database for the vesicle-trafficking specific proteins, and molecular function gene ontology (http://geneontology.org/docs/ontology-documentation/). Putative lysosomal targeting motifs were detected using the Protein Motif Pattern tool (https://trichdb.org/trichdb/app/search/transcript/GenesByMotifSearch). For sequence alignments, Clustal Omega 1.2.4 was used (48).
Site-Directed Mutagenesis
Q5 Site-Directed Mutagenesis Kit (New England Biolabs, Inc) was used alongside its high-efficiency 5-alpha competent E. coli to introduce mutations into the CLCP gene. NEBaseChanger was used to design primers (Table S1), and mutagenesis was performed according to the manufacturer's protocol.
Monitoring of Protein Secretion
T. vaginalis T1 cells (50 ml, approximately 7.5 × 107 cells) were harvested by centrifugation and washed three times with TYM without horse serum. The cells were resuspended in 2 ml TYM without horse serum and incubated for 60 min at 37 °C. The cells were removed by centrifugation at 1200g for 10 min at 4 °C, and then the supernatant was centrifuged at 10,000g for 10 min at 4 °C to remove cell debris, followed by filtration through a 0.2 μm pore size filter (Whatman, GE Healthcare). Proteins in the supernatant were precipitated with TCA as described previously (44) and analyzed by immunoblotting as above. Alternatively, cells were incubated with 1 μM chloroquine (Sigma-Aldrich) or 50 μg/ml brefeldin A (Sigma-Aldrich) for 15 min or 45 min prior to protein isolation from the supernatant.
Results
Lysosomal Localization of TvRab7a
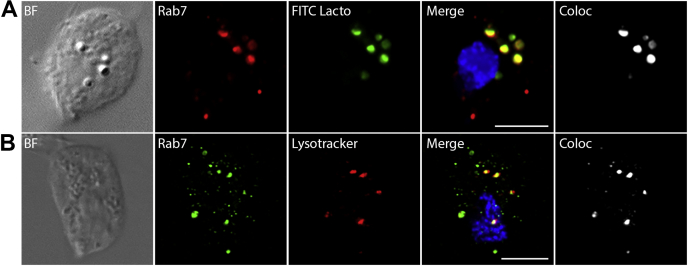
Before the separation of T. vaginalis phagolysosomes, we selected Rab7 as a potential phagolysosomal protein marker. There are three Rab7 paralogues in the T. vaginalis genome, of which we selected TvRab7a (49), which displayed the highest pairwise protein sequence identity with human Rab7 (45.5%). To verify its lysosomal localization in T. vaginalis, FITC-Lf and LysoTracker Deep Red were used as lysosomal markers. Extracellular Lf is known to be imported by trichomonads via receptor-mediated endocytosis; thus, TV17-48-Rab7a cells were incubated with FITC-Lf prior to slide preparation to allow for FITC-Lf internalization. FITC-Lf appeared in a few small but mainly larger vesicles corresponding to endosomes and lysosomes (Fig. 1A). TvRab7a and LysoTracker appeared in numerous small- to larger-sized vesicles scattered throughout the cell (Fig. 1B). Colocalization of TvRab7a with FITC-Lf was found mainly in the larger vesicles (Fig. 1A). Similarly, LysoTracker colocalized with TvRab7a mainly in larger vesicles, which further supports that these vesicles represent lysosomes (Fig. 1B).
Fig. 1.
Lysosomal localization of TvRab7a in T. vaginalis TV17-48. TvRab7a (Rab7) was visualized using mouse monoclonal α-HA antibody and (A) Alexa Fluor 594 (red) or (B) Alexa Fluor 488 (green) donkey α-mouse antibody. FITC-lactoferrin (A, green) or LysoTracker (B, red) was used as a lysosomal marker. BF, brightfield; Coloc, colocalization channel (Imaris Coloc software). Scale bar = 5 μm.
Proof of Principle of Lysosomal Isolation Methods
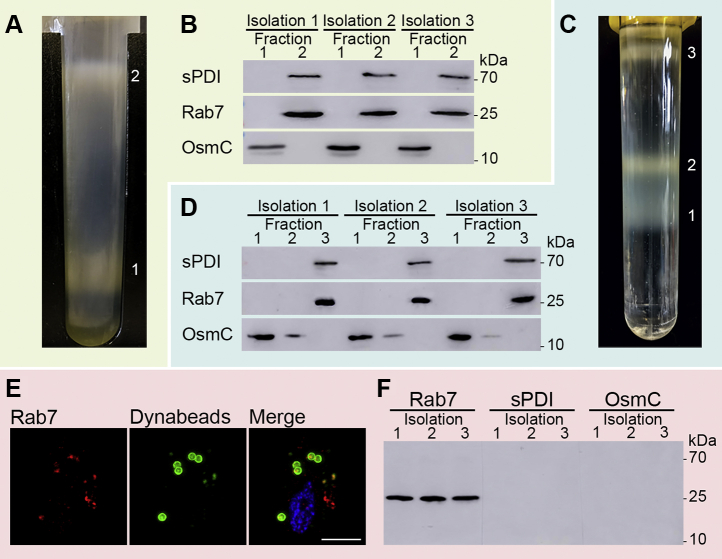
Three different methods were used for the isolation of lysosomes/phagolysosomes. First, we employed 45% Percoll gradient centrifugation as previously described (40), which resulted in the separation of a brown hydrogenosomal fraction (lower band) and a white upper lysosome-enriched fraction (Fig. 2A). The lysosomal fraction was subsequently repurified on a second 45% Percoll gradient. Using the OsmC protein as a marker (41), immunoblot analyses confirmed a clear separation of hydrogenosomes from the lysosomal fraction with TvRab7a; however, the lysosomal fraction also contained a weaker sPDI signal, suggesting the presence of ER-derived vesicles (Fig. 2B). Next, we employed an OptiPrep density gradient, which appeared to be more favorable to the isolation of T. vaginalis hydrogenosomes compared with the Percoll gradient (50). Separation of the LGF using OptiPrep resulted in the formation of three visible bands (Fig. 2C), and immunoblot analyses revealed that lysosomes were present in the uppermost band with a strong TvRab7a signal and a low sPDI signal (Fig. 2D). Hydrogenosomes detected by OsmC were clearly separate from the lysosomal fraction. Finally, we employed a method based on T. vaginalis phagocytic activity. TV17-48-Rab7a cells were incubated with magnetic beads coupled with Lf that were ingested by phagocytosis. Fluorescence microscopy confirmed the uptake of beads into phagolysosomes colocalizing with TvRab7a (Fig. 2E). Phagolysosomes containing beads were then separated by a magnet. This approach resulted in the separation of a phagolysosomal fraction with the TvRab7a marker that was free of OsmC and sPDI (Fig. 2F). Lysosomal fractions obtained by all three methods were submitted for MS.
Fig. 2.
The isolation of lysosomes.A, the Percoll gradient yielded two bands corresponding to hydrogenosomes (1) and lysosomes (2). B, Western blot of Percoll fractions from three independent isolations. HA-tagged TvRab7a was enriched in fraction 2, where the ER marker sPDI was also present. Hydrogenosomal OsmC was detected only in fraction 1. C, the OptiPrep gradient showed three fractions. D, Western blot of OptiPrep fractions. E, immunofluorescence microscopy of T. vaginalis TV17-48 with endocytosed Dynabeads. The beads covered with FITC-lactoferrin (green) colocalize with mouse monoclonal α-HA and Alexa Fluor 594 donkey α-mouse-labeled TvRab7a (red) in phagolysosomes. F, Western blot of three isolations by Dynabeads. Scale bar = 5 μm.
Proteomic Analysis
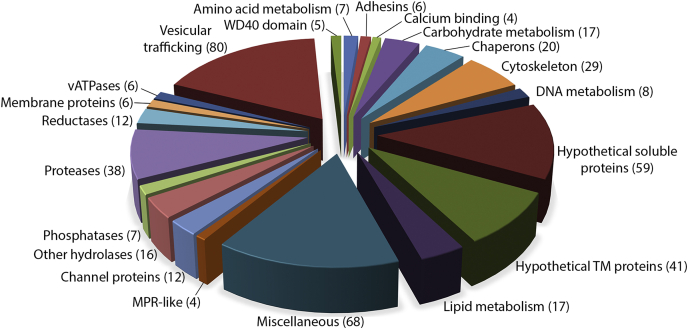
The total number of proteins identified by MS was 3442 in the Percoll purified fraction, 4055 using the OptiPrep gradient, and 1351 in the fraction obtained with magnetic beads (Table S2-S4). The initial dataset was filtered using two parameters: the protein must be identified in at least two out of three biological experiments, and the protein identification must be supported by at least three peptides. Therefore, we obtained 1758 (Percoll), 2748 (OptiPrep), and 748 (Dynabeads) proteins. These datasets overlapped in 550 proteins of which 88 proteins were considered contaminants based on their annotation and known cellular localization, and the remaining 462 proteins were considered to represent phagolysosomal proteins. These proteins were sorted into 21 functional groups considering TrichDB annotations, Pfam domain identifications, and molecular function gene ontology (Fig. 3, Table S5). Seventy-six percent (351 proteins) appeared to be soluble proteins, and the remaining 24% (111 proteins) possessed one or more transmembrane domains (TMDs), with the majority (75 proteins) possessing a single TMD. Hydrolases, including proteases, phosphatases, lipases, and glycosidases, represented the largest functional class in the phagolysosomal proteome. Cysteine proteases formed the largest protease set (13 proteins). They included TvCP2, TvCP3, TvCP4, and TvCPT, which were previously shown to be secreted by T. vaginalis (51); the legumain-like protease TvLEGU-1 (52); ten metallopeptidases (53); and eight serine peptidases (10, 54). The other typical lysosomal hydrolase was represented by a β-hexosaminidase that was present as two paralogues. In the category of carbohydrate metabolism, we identified 17 proteins, including a putative glycogen debranching enzyme (TVAG_150430) and three paralogues of 4-α-glucanotransferase. Another class of typical lysosomal proteins are the vATPases, of which we identified six, with two of them possessing seven TMDs. One vATPase (TVAG_075320) has also been found on the surface (54). Among the channel proteins, we found three ABC transporters, a class of proteins known to couple ATP hydrolysis with the transport of substrates across membranes. Interestingly, we identified one Piezo channel (TVAG_223490) with 37 TMDs. In Dictyostelium, this group of proteins is proposed to act as a pressure sensor to regulate cell movement (55).
Fig. 3.
Lysosomal proteome. The 462 proteins were sorted into 21 groups with the help of TrichDB annotations, Pfam motif identifications, and molecular function gene ontology.
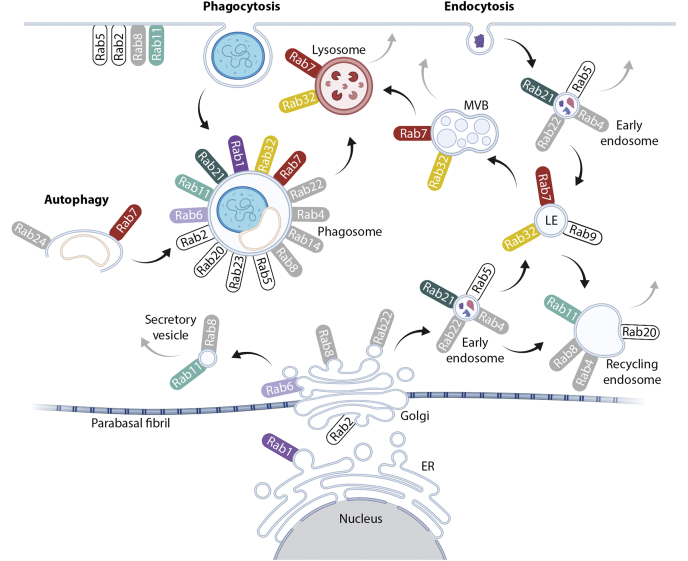
In the vesicular trafficking group, we identified 43 Rab proteins, including multiple paralogues in most categories. Of these, six Rabs were assigned to known categories based on orthology searches (Fig. 4, Table S5). As expected, we identified Rab7a, which controls transport to late endosomes and lysosomes and is required for phagosomal maturation (56). We also identified three paralogues of Rab32 that are known to promote the fusion of phagosomes with lysosomes (57) and Rab 6, which has been shown to play a role in targeting material to lysosome-related organelles in mammalian systems (58). Furthermore, we identified Rabs 21 and notably Rab1. The metazoan-specific paralogue of Rab1, Rab35 is well-established to play a role in regulating phagosomal maturation (59, 60). Finally, we identified Rab11, which although not directly lysosomal is a well-known marker of the recycling endosome, an upstream endocytic compartment (61). Five Rabs were present with less supported classifications, including Rab4, Rab8, Rab14, Rab22, and Rab24, and interestingly, 18 Rabs with no clear classification. An overview of the identified Rab proteins is given in Figure 4. In addition to Rabs, we identified components of the retromer complex (Vps35), of the lysosome-associated HOPS complex (Vps16, 33, 39), of the late endosomal/multivesicular body associated ESCRT complexes (SNF7), and several other endosomally associated Vps proteins (e.g., Vps9). We also identified SNARE proteins, notably syntaxin 16, Vta1, and several Vamp7 homologues, all of which mediate endolysosomal trafficking and cargo delivery. Additional proteins associated with endocytic vesicle formation included a single enthoprotin and three dynamins. Finally, we found clathrin and six subunits of adaptor complexes including a near complete AP4 complex (missing only the sigma subunit) and mu subunits for AP1, AP2, and AP4. It is the mu subunit that binds the YXXØ motif in proteins during lysosomal trafficking (62).
Fig. 4.
Schematic of Rabs in the endomembrane system. Rabs displayed in color were identified in the lysosomal proteome and annotated with high confidence. Rabs in gray were found, but their phylogenetic classifications were weakly supported. White Rabs have been previously described to be present in the respective organelle but were not found in our lysosomal proteome. Rab7 (TvRab7a) was used in this study as lysosomal marker. Gray arrows indicate the transport of cargo to the cell surface and secretion. ER, endoplasmic reticulum; MVB, multivesicular body.
Another striking category was the cytoskeleton, for which we identified 29 proteins. These proteins included two actins and other actin-interacting proteins such as two actinins, three profilins, two gelsolins, and one twinfilin. The identification of proteins involved in vesicular trafficking and proteins associated with the cytoskeleton in the lysosomal proteome is in line with the established model of endolysosomal trafficking and organellar movement along the cytoskeleton (63, 64, 65).
A total of 55 proteins in the phagolysosomal proteome overlap with the surface proteome (54) (Fig. 5), of which 45 are TM proteins. These proteins include two putative adhesins of the BspA family, and in our phagolysosomal proteome, we identified three more BspA members, the tetraspanin protein TSP1 (TVAG_019180), which is associated with multivesicular bodies (12), and five paralogues of the Trichomonas β-sandwich repeat protein (TBSR). A relatively small number of TM proteins overlapping with the surface proteome possess an internalization motif in the cytosolic tail; a single protein contains the classical NPxY signal (TBSR-5, TVAG_340570), five proteins harbor the [DE]xxxL[LI] motif, and 16 proteins possess the YxxØ signal (Table S5). The phagolysosomal proteome and the T. vaginalis secretome overlapped with only 26 proteins, eight of which are hydrolases, including β-hexosaminidase, an acid phosphatase, a serine and two cysteine peptidases, two phospholipases, and a glucosaminidase (Fig. 5) (10). A relatively low level of overlap between phagolysosomal proteome and previously reported proteomes of the secreted proteins, and the surface proteome supports the successful separation of the phagolysosomal fraction. However, it also indicates an expected engulfment of some surface proteins and their transport to the lysosomes, and conversely, possible participation of lysosomes in unconventional protein secretion.
Fig. 5.
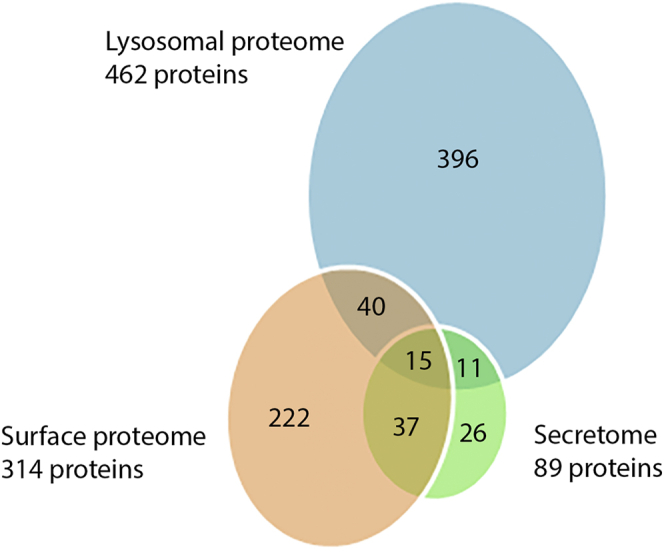
Overlap of the lysosomal proteome with the secretome (10) and the surface proteome (54). Fifteen proteins were found in all three proteomic studies. The lysosomal proteome and surface proteome had 55 overlapping proteins, and the lysosomal proteome and secretome had 26 overlapping proteins.
Experimental Cellular Localization of Selected Proteins
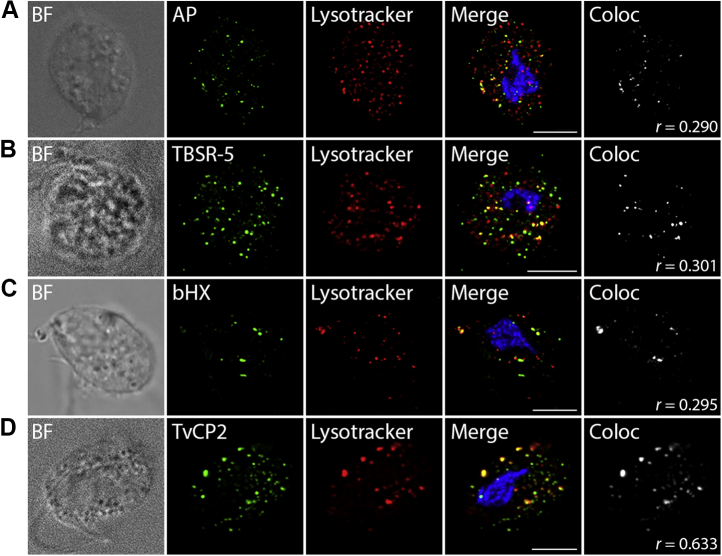
Acid phosphatase (AP, TVAG_169070), TBSR-5 (TVAG_340570), β-hexosaminidase (bHX, TVAG_110660), TvCP2 (TVAG_057000), and CLCP (TVAG_485880) were selected for experimental verification of their cellular localization. AP and bHX are known to reside in lysosomes in other organisms and may serve as lysosomal markers (66), TBSR-5 was selected as an example of a membrane protein with C-terminal NPIY signal for endocytic internalization (Table S5), TvCP2 is known to be secreted as an important virulence factor (67), while CLCP represents a new resident cysteine protease that was not identified in other T. vaginalis proteomes, thus far. The corresponding genes were expressed in T. vaginalis under the control of their respective native promotors and with a C-terminal HA-tag and investigated by confocal microscopy. Colocalization with LysoTracker supported their lysosomal localization in large vesicles, whereas limited colocalization was observed in smaller vesicles (Figs. 6 and 7A WT). The PCC in colocalized volume ranged from low (PCC r = 0.290, AP) to moderate (PCC r = 0.633, TvCP2) values (Figs. 6 and 7C).
Fig. 6.
Immunofluorescence microscopy of selected proteins. The proteins were visualized using mouse monoclonal α-HA antibody and Alexa Fluor 488 donkey α-mouse (green). LysoTracker (red) was used as a lysosomal marker. A, acid phosphatase, (B) Trichomonas β-sandwich repeat protein 5, (C) β-hexosaminidase, (D) Trichomonas vaginalis cysteine protease 2. BF, brightfield; Coloc, colocalization channel (ImarisColoc software). Scale bar = 5 μm.
Fig. 7.
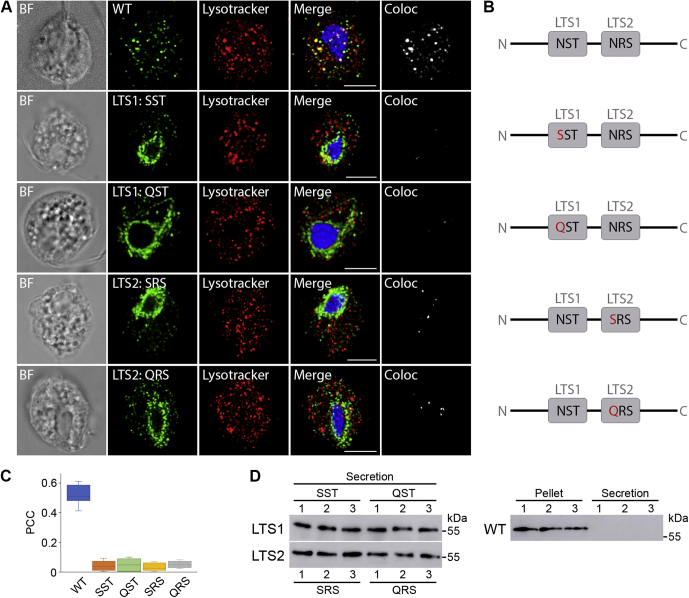
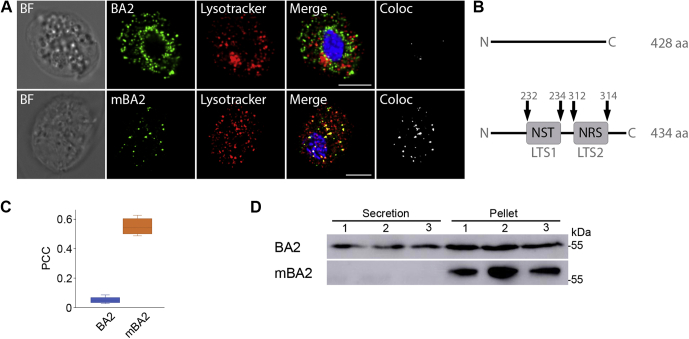
Effect of LTS mutations on CLCP localization.A, localization of wild-type CLCP (WT) and CLCP with mutated glycosylation motifs LTS1 and LTS2 (mCLCP) in T. vaginalis T1. WT CLCP and mCLCP versions were visualized using mouse monoclonal α-HA antibodies and Alexa Fluor 488 donkey α-mouse antibodies (green). BF, brightfield; Coloc, colocalization channel (ImarisColoc software). Scale bar = 5 μm. B, schematic illustration of the amino acid substitutions (in red) introduced into glycosylation motifs LTS1 and LTS2 of CLCP. C, boxplot chart of Pearson's correlation coefficient (PCC) in colocalized volume for WT CLCP and mCLCP versions with LysoTracker. WT CLCP and LysoTracker colocalized with PCC r = 0.512 ± 0.1 (n = 16), whereas the PCCs of the mCLCP versions were below r = 0.1 (n = 12 cells per each version). Error bars represent standard deviation. D, secretion of mutated and WT CLCP. CLCP with mutated LTS1 (SST, QST) and LTS2 (SRS, QRS) was secreted and detected in the extracellular supernatant after 1 h incubation at 37 °C. WT CLCP was not secreted and was associated only with cells (Pellet). Each experiment was performed with biological triplicates.
Mannose 6-phosphate-like Receptors
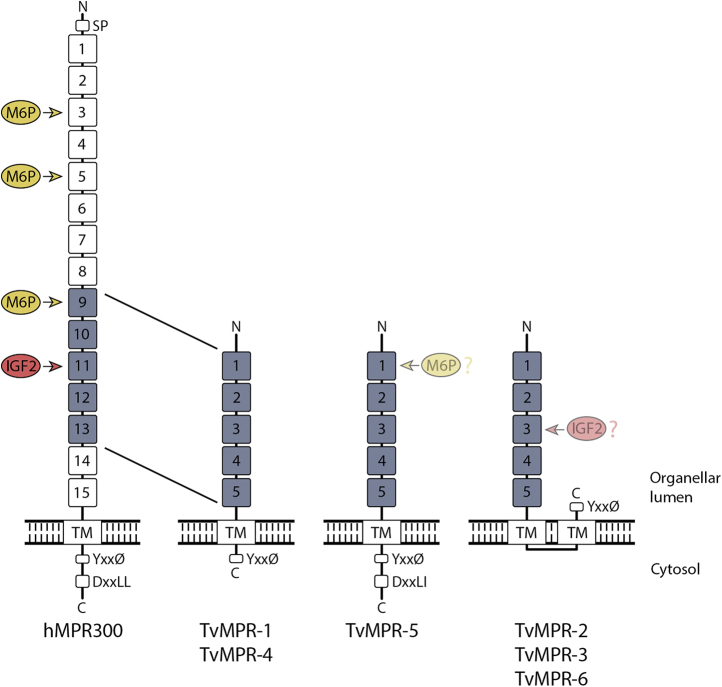
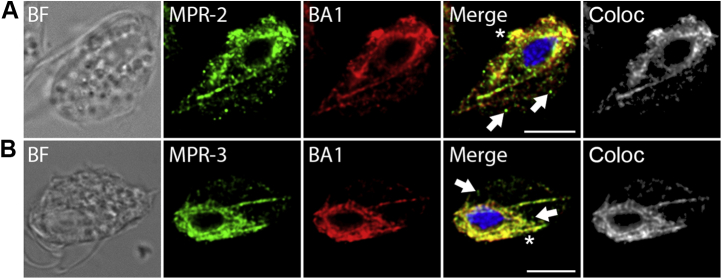
Four transmembrane proteins identified in phagolysosomes appear to be homologues of mammalian cation-independent mannose 6-phosphate receptor 300 (MPR300). These proteins were named TvMPR-1 (TVAG_177320), TvMPR-2 (TVAG_351790), TvMPR-3 (TVAG_498650), and TvMPR-4 (TVAG_166760). In addition, we identified two more paralogues, TvMPR-5 (TVAG_160180) and TvMPR-6 (TVAG_445110), in the T. vaginalis genome, which are between 882 and 991 amino acids long. Their N-terminal domains range in length between 809 and 924 amino acid residues, and their C-terminal tails are between eight and 49 amino acids long. Human MPR300 (hMRP300) consists of 15 characteristic mannose 6-phosphate receptor homology (MRH) domains in the N-terminus (Fig. 8), of which MRH3, 5, and 9 were implicated in binding M6P and MRH11 has been implicated in binding insulin-like growth factor II (IGF2) (23, 68). Each MRH domain consists of approximately 150 amino acid residues with eight conserved cysteine residues. Interestingly, T. vaginalis MPRs consist of only five MRH domains, which aligned with MRH9 to MRH13 of hMPR300 (Fig. S1). Although the protein sequence similarity of T. vaginalis MRH domains to MRH of hMPR300 appeared to be rather low (7.2% to 35.2%), most of the conserved cysteine residues are present. TvMPR structure is similar to the Drosophila lysosomal enzyme receptor protein (LERP), which also possesses five MRH domains (Fig. S1). Moreover, similar to LERP, TvMPR proteins lack most residues in hMPR300 that are essential for the binding of M6P and IGF2. All TvMPR proteins possess either the YxxØ or DxxL[LI] sorting motif for recognition by adaptor proteins (Figs. 8 and S1). However, only three of them, TvMPR-1, TvMPR-4, and TvMPR-5, were predicted to be type I glycoproteins with a single TMD, similar to hMPR300 and LERP, which is consistent with the cytosolic topology of their sorting signal (Fig. 8). The other three TvMPR proteins were predicted to possess two TMDs, resulting in the C-terminal YxxØ motif most likely being in the same compartment as the N-terminal domain. Although we found TvMPR proteins in the phagolysosomal proteome and previously identified them in the secretome and surface proteome, the expression of HA-tagged TvMPR-2 and TvMPR-3 revealed that they localize predominantly to the structures of the ER and Golgi body (Fig. 9). V5-tagged β-amylase 1 was used as a marker for the ER/Golgi body (10) that was coexpressed in double T. vaginalis transfectants and colocalized with TvMPR-2 and TvMPR-3 (Fig. 9).
Fig. 8.
Domain composition of human MPR300 (hMPR300) and T. vaginalis homologues (TvMPR). hMPR300 consists of 15 repeats, of which five are homologous to TvMPRs. Detected C-terminal signal sequence motifs DxxLL and YxxØ (Ø, hydrophobic amino acid residue), which are involved in the internalization and trafficking of membrane proteins is indicated. M6P and IGF2 binding sites are absent in TvMPRs; only TvMPR-5 possesses three M6P binding residues, and TvMPR-1 and TvMPR-6 contain a single residue. TvMPR-2 contains the isoleucine that is essential for IGF2 binding. No N-terminal signal peptide was predicted in TvMPRs. SP, signal peptide; TM, transmembrane.
Fig. 9.
Cellular localization of TvMPR. Immunofluorescence microscopy of (A) TvMPR-2 and (B) TvMPR-3. The proteins were visualized using mouse monoclonal α-HA antibody and Alexa Fluor 488 donkey α-mouse (green) in T. vaginalis T1. BA1 was used as an ER marker and was stained with rabbit polyclonal α-V5 antibody and Alexa Fluor 594 donkey α-rabbit (red). TvMPR-2 and TvMPR-3 strongly colocalized with BA1 in the ER and possibly Golgi body (asterisk). Some TvMPR-2 and TvMPR-3 appeared in small vesicles not colocalizing with BA1 (arrows). BA1, β-amylase 1; BF, brightfield; Coloc, colocalization channel (ImarisColoc software); TvMPR, Trichomonas vaginalis mannose 6-phosphate receptor. Scale bar = 5 μm.
Glycosylation-Dependent Targeting to Lysosomes
The observed localization of TvMPR proteins is consistent with the model of the M6P-dependent signaling pathway that is initiated in the ER, where lysosomal cargo is glycosylated on the asparagine of the s-LTS motif Asn-X-[Ser/Thr] and subsequently modified in the Golgi body to be recognized by MPR. However, as TvMPR proteins lack most residues required for M6P-dependent recognition, M6P signalling is unlikely. Moreover, the generation of M6P on N-linked glycans requires the consecutive activity of GlcNAc-PT and UCE. Thus, we searched for corresponding homologues in the T. vaginalis database using the human α/β GlcNAc-PT (Q3T906) and UCE (Q9UK23) amino acid sequences as queries. We identified the protein TVAG_166090 with only limited homology to one of four characteristic GlcNAc-PT stealth domains (69), and we did not identify any UCE homologues, which further supports the absence of M6P signalling. Thus, we hypothesized that another M6P-independent feature, such as a peptide structure or glycans, may serve as a signal. To test whether glycosylation is involved in the protein targeting of T. vaginalis lysosomes, we selected phagolysosome-resident CLCP. This protein is composed of 452 amino acid residues and possesses two putative s-LTSs, Asn-Ser-Thr at position 188–190 (LTS1) and Asn-Arg-Ser at position 268–270 (LTS2). A series of CLCPs with mutated s-LTSs (mCLCP) was prepared in which asparagine was replaced by serine or glutamine (Fig. 7B). Whereas wild-type CLCP was observed in lysosomes with a PCC r = 0.512 ± 0.1 (n = 16) (Fig. 7C), all mCLCP variants accumulated predominantly around the nucleus (Fig. 7A). Some mCLCP variants were observed in vesicles scattered throughout the cell, in which cases the mCLCP signal exceptionally colocalized with LysoTracker. The PCC of mCLCP was below r = 0.1 (Fig. 7C). Next, we were interested in whether mCLCP is arrested in the ER or secreted into the cell environment. Thus, the cells were incubated for 1 h at 37 °C in TYM, and secreted mCLCP was detected by immunoblotting. Whereas wild-type CLCP was not secreted, which is consistent with its absence in the secretome (10), all mutated s-LTSs versions of mCLCP were secreted (Fig. 7D).
To further test the ability of the CLCP LTS1 and LTS2 sequences to target a protein to lysosomes, we introduced both motifs into BA2 (mBA2), a nonlysosomal protein that does not possess any glycosylation sites and expressed the protein with a C-terminal HA tag. LTS1 was introduced at positions 232–234 and LTS2 at positions 312–314, thereby maintaining the same distance of 78 amino acids between the signals as in CLCP (Fig. 10B). The active sites of BA2 are located at positions 133 and 330 and thus were not altered by the insertions. While wild-type BA2 did not localize to lysosomes (PCC r = 0.051 ± 0.029, n = 29, Fig. 10) but labeled ER structures surrounding the nucleus as demonstrated previously (10), cells expressing mBA2 showed significant colocalization with LysoTracker in lysosomes (Fig. 10A) with a PCC r = 0.556 ± 0.07 (n = 22, Fig. 10C), whereas no accumulation in the ER/Golgi was observed. Next, we were interested in whether the insertion of LTS1 and LTS2 into BA2 influences protein secretion. Therefore, the cells were incubated in TYM as before, and the conditioned medium was analyzed by immunoblotting. Whereas wild-type BA2 was secreted, mBA2 was associated exclusively with cells, and no secretion was observed (Fig. 10D). These experiments demonstrated that the tandem N-glycosylation sites were decisive for protein sorting, they represent the s-LTS of CLCP, and they are essential for protein delivery to lysosomes. The screening of the lysosomal proteome for tandem N-glycosylation sites that have a similar distance to one another as in CLCP (78 amino acids ± 20 amino acid range) revealed 27% of soluble proteins with such a motif (Table S5). Interestingly, within the soluble proteases, 44% contain this motif. These results suggest that the tandem glycosylation sites might be a general motif for lysosomal delivery in T. vaginalis; however, such an assumption needs experimental verification in future. Moreover, the distance between the glycosylation sites in different proteins might not be conserved because the steric position of glycans that are recognized by receptors is likely dependent on the protein conformation. Further studies are needed to establish which and how lysosomal receptor recognizes this s-LTS for which TvMPR proteins are the best candidates.
Fig. 10.
Effect of LTS on BA2 localization.A, localization of wild-type and mutated BA2 (mBA2) in T. vaginalis T1 transformants. Both were visualized using mouse monoclonal α-HA antibodies and Alexa Fluor 488 donkey α-mouse antibodies (green). LysoTracker (red) was used as endolysosomal marker. BF, brightfield; Coloc, colocalization channel (ImarisColoc software). Scale bar = 5 μm. B, schematic illustration of LTS1 and LTS2 insertions introduced into BA2. C, boxplot chart of Pearson's correlation coefficient (PCC) in colocalized volume for BA2 and mBA2 with LysoTracker. BA2 and LysoTracker did not colocalize with a PCC r = 0.051 ± 0.029 (n = 29), whereas the PCC of mBA2 was r = 0.556 ± 0.07 (n = 22). Error bars represent standard deviation. D, secretion of BA2 and mBA2. BA2 is secreted and can be detected in the extracellular supernatant and in cells (pellet) after 1 h incubation at 37 °C, whereas mBA2 was not secreted. Each experiment was performed with biological triplicates.
Lysosomes Are Involved in Unconventional Secretion of CP
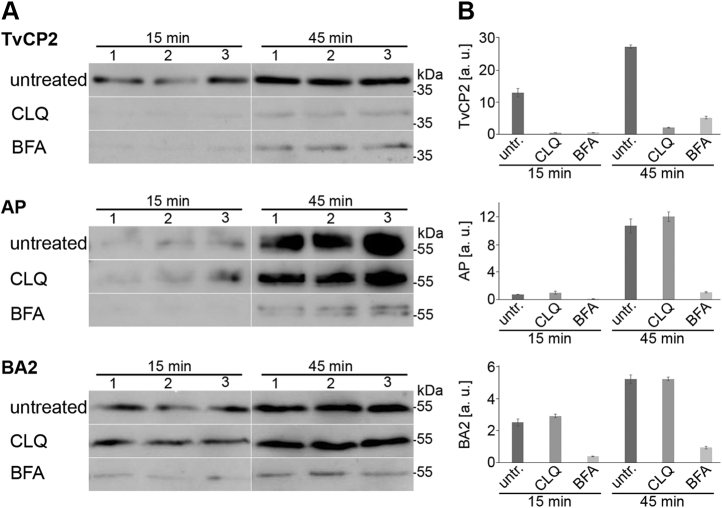
A few proteins were observed in the phagolysosomal proteome as well as in the secretome. This phenomenon prompted us to test whether lysosomes are involved in protein secretion in addition to the classical ER-Golgi secretory pathway. We selected the two proteins TvCP2 and AP that were found in the phagolysosome and secretome and BA2 as a control for classical secretion (10). The proteins were expressed with an HA-tag and transfected trichomonads were incubated in TYM medium with either 1 μM chloroquine (CLQ) or 50 μg/ml brefeldin A (BFA). CLQ blocks the endolysosomal pathway by increasing lysosomal pH (70, 71), while BFA inhibits classical secretion via stimulation of retrograde transport from the Golgi body to the ER (10, 72). The secretion of TvCP2 was strongly decreased by both CLQ and BFA, while the secretion of BA2 and AP was inhibited only by BFA (Fig. 11). These results indicated that lysosomes were involved in the unconventional secretion of TvCP2, as opposed to BA2 and AP, which are secreted through the classical secretory pathway, and AP is eventually delivered to lysosomes via endocytosis (Fig. 11).
Fig. 11.
pH-dependent secretion of TvCP2.A, Western blot of secreted TvCP2, acid phosphatase (AP), and β-amylase 2 (BA2) after 15 min and 45 min incubation at 37 °C. Both chloroquine (CLQ) and brefeldin A (BFA) inhibited secretion of TvCP2 compared with the untreated control. Secretion of AP and BA2 was inhibited upon incubation with BFA compared with the untreated controls, but no change in secretion was observed upon treatment with CLQ. Each experiment was performed with biological triplicates. B, quantification of western blots using densitometry that shows the relative amounts of secreted TvCP2, AP, and BA2 from three biological replicates. Error bars indicate standard deviation. a. u., arbitrary unit; untr, untreated.
Discussion
Lysosomes play a key role in the pathogenicity and virulence of parasitic protists (73). However, little is known about their biogenesis, lysosomal protein targeting, and composition outside of classical mammalian and yeast models. In this paper, we characterized the proteome of T. vaginalis phagolysosomes, showed that glycosylation is involved in lysosomal targeting of CLCP and that lysosomes are involved in protease secretion. Unexpectedly, we identified homologues of the M6P receptor, TvMPRs. While this protein has bioinformatically been identified across eukaryotes, it is inconsistently conserved and to our knowledge only functionally characterized in metazoans and fungi (31).
To separate the T. vaginalis lysosome-enriched fraction, we initially employed a method based on differential and gradient centrifugation using two different isoosmotic gradient media, Percoll and OptiPrep. MS analysis identified 1758 (Percoll) and 2748 (OptiPrep) reproducibly enriched proteins in these preparations, which is comparable to the 3703 proteins identified in the macrophage lysosome-enriched fraction using a similar approach (74). Although these methods are widely applied, lysosomes tend to copurify with other organelles, typically the endoplasmic reticulum, Golgi, and mitochondria (74, 75), or may contain proteins of other compartments due to autophagy (18). Indeed, we observed the presence of the PDI ER marker in samples of the lysosome-enriched fractions using immunoblotting, and contaminants were apparent from the annotations of identified proteins. Therefore, we developed a new experimental protocol for phagolysosome purification based on the ingestion of magnetic nanobeads by T. vaginalis. To increase the specific binding of the nanobeads, we covered the beads with Lf, which binds to trichomonad receptors on the surface (76). This analysis identified 748 proteins and resulted in a set of 462 phagolysosomal proteins when combined with the previous proteomic datasets. The obtained size of the trichomonad phagolysosomal proteome is close to the approximately 450 lysosomal proteins identified in the lysosomes of mouse embryonic fibroblasts isolated using magnetite solution (77).
Comparisons of the phagolysosomal proteome with previous proteomic studies of T. vaginalis surface membrane proteins (54), secretome (10), and exosomes (12) allow us to estimate the purity of our preparation and detect proteins with multiple localizations. Indeed, there was a very low overlap with these compartments that corroborated efficient organellar separation. The highest number of shared proteins (55 proteins) was found with the surface proteome. However, multiple localizations are expected for membrane proteins, and this is consistent with the endocytic pathways of some lysosomal proteins that are expressed on the plasma membrane as receptors, adhesins, or proteases and subsequently internalized via endocytosis/phagocytosis and transported to lysosomes. These shared proteins included the leucine-rich repeat protein BspA (78), β-hexosaminidase, and the surface adhesin TvAD1, which is also associated with exosomes (79). Moreover, there were 19 conserved membrane proteins of unknown function, which provide interesting targets for future studies.
Proteases have been repeatedly implicated in T. vaginalis virulence (33, 80); however, their cellular localization is mostly unknown. A comparison of the lysosomal proteome that included 38 peptidases with previous studies allowed us to distinguish three groups. The first and the major group includes primary lysosomal resident peptidases (34 proteins) that have not been detected in other cellular compartments. This group included seven new CPs, such as CLCP, in addition to four CPs (TvCP3, TvCP4, TvCPT, and TvLEGU-1) that have been previously recognized in trichomonad cellular lysates (81). TvLEGU-1 has been observed in lysosomes, the Golgi complex, and on the trichomonad surface using immunofluorescence and immunoelectron microscopy (52), whereas it has been absent from the secretome and the surface proteome (10, 54). However, the expression of TvLEGU-1 seems to be strongly dependent on the iron level, which may explain the observed differences in cellular localization (52). The first group further included seven serine peptidases and nine metallopeptidases of different families (M1, 8, 16, 20, 24, 28, 67) that had not been characterized thus far. The second group consists of two soluble CPs that are secreted (10) including TvCP2, an important virulence factor (67), whereas leishmanolysin-like metallopeptidase (GP-63 like) and subtilisin-like serine peptidase (SUB1) were two membrane proteases of the third group, which were shared with the surface proteome (54). Previous cellular localization of SUB1 using fluorescence microscopy suggested its presence in intracellular vesicles and on the plasma membrane, which supports our proteomic data (82). In pathogenic fungi, subtilisin proteases have been shown to be important for host invasion (83, 84); however, their role in T. vaginalis has not been established. Another hydrolase group that we found in the lysosomal proteome was phospholipase B, of which we identified eight paralogues, two of which are secreted (TVAG_215920, TVAG_333010). Phospholipase B belongs to the group of acyl esterases and represents an important lysosomal component to hydrolyze glycerophospholipids (85). Secreted phospholipases have been shown to kill bacteria (86) and display antiviral activity (87), however, have not been studied in trichomonads. Finally, β-hexosaminidase is a typical lysosomal hydrolase that we identified two copies of. Interestingly, one copy (TVAG_110660) possesses a single TMD and was shared between the secretome and the surface proteome. Its cell membrane localization and previously determined N-acetyl-β-D-glucosaminidase activity in T. vaginalis (88) suggest that β-hexosaminidase may participate in T. vaginalis virulence via host mucin degradation. Interestingly, in E. histolytica, β-hexosaminidase is transported to the phagolysosome by a unique cysteine protease binding protein family 8 (CPBF8) receptor that is not present in T. vaginalis (26).
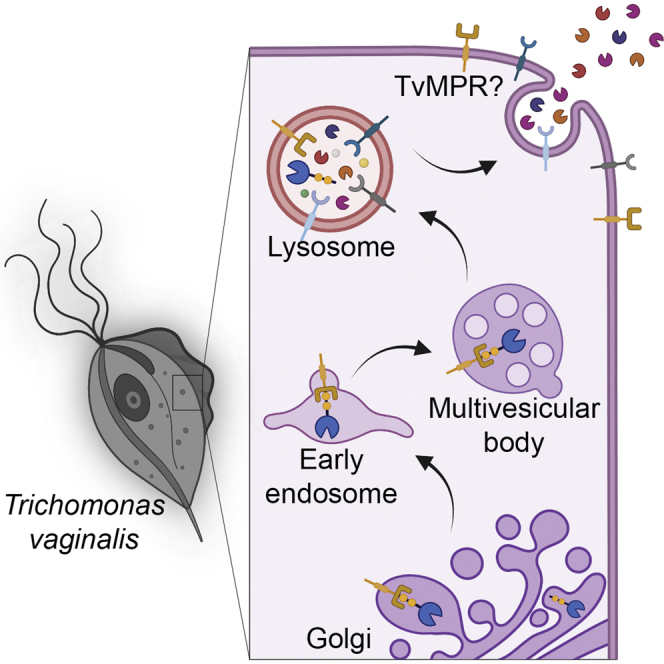
The model of protein delivery to lysosome and secretion in T. vaginalis, which is based on our results, is provided in Figure 11. In metazoans, most lysosomal proteins are typically targeted via M6P-dependent pathways, whereas in other eukaryotic lineages, sorting of hydrolases was not reported to depend on oligosaccharides. Alternative receptors such as sortilins/Vps10 and plant vacuolar sorting receptors (VSRs) directly recognize the polypeptide motifs of cargo (23). Vsp10 is conserved in most eukaryotic lineages (31), and its function as a sorting receptor was shown in Giardia intestinalis (89) and Toxoplasma gondii (90). However, there is no obvious orthologue of Vsp10 in the T. vaginalis genome (31). Surprisingly, we identified four paralogues of the putative M6P receptor TvMPR in the lysosomal proteome and two other paralogues in the genome. These proteins contain the characteristic N-terminal domain repeats with typical cysteines (22) that are mostly conserved in TvMRPs. However, unlike the mammalian CI-MPR with 15 domains, TvMPRs contain only five domains, which resemble the structure of the MPR-like protein LERP in Drosophila (91). Moreover, TvMPR-1, TvMPR-4, and TvMPR-5 possess a single TMD and the endocytic sorting signal YxxØ or a dileucine motif (DXXLI]) in their C-termini, which further supports their role as receptors and is consistent with the presence of adaptin complex proteins, particularly the mu subunits that we identified. Interestingly, TvMPR-2, -3, and -6 were predicted to possess double TMDs at their C-termini, which may indicate a different topology of their N-terminal domains. Similar to Drosophila, TvMPRs lack most of the residues involved in M6P binding and the two enzymes (GlcNAc-PT and UCE) required for the formation of M6P recognition markers. Interestingly, LERP has been shown to rescue the missorting of lysosomal proteins in MPR-deficient mouse cells in an M6P-independent manner, although it is not known whether LERP recognizes a glycan or a peptide structure (91). Thus, the identification of LERP-like TvMPR prompted us to test whether glycans are involved in lysosomal targeting in T. vaginalis. Indeed, we demonstrated that two sites for N-glycosylation of the Nx[ST] motif within the lysosomal resident CLCP are essential for its lysosomal localization. Moreover, introducing the two Nx[ST] motifs into nonglycosylated BA2, which is secreted by the classical ER-Golgi pathway (10), redirected this protein from the secretory pathway to lysosomes (Fig. 12). However, glycosylation apparently needs a specific structure/conformation to provide the lysosomal signal. First, we demonstrated that both Nx[ST] motifs in CLCP are needed for correct targeting, as mutation of a single site mistargeted CLCP to the secretory pathway. Second, a number of other glycosylated proteins, such as BA1, that possess multiple Nx[ST] motifs are predominantly secreted (10) (Fig. 12). For future investigations of recognition markers, it also needs to be considered that T. vaginalis possesses a limited set of glycosyltransferases available for glycan synthesis in the ER, producing a simplified glycan structure consisting of two N-acetyl glucosamines and only five mannose residues, as opposed to GlcNAc2Man9Glu3 in the majority of eukaryotes (10, 92). Collectively, these results indicate that glycosylation is required for the recognition and targeting of lysosomal proteins in T. vaginalis; however, whether TvMPRs or other possible receptors are involved in this process and the precise structure of the lysosomal recognition marker need to be clarified in future studies.
Fig. 12.
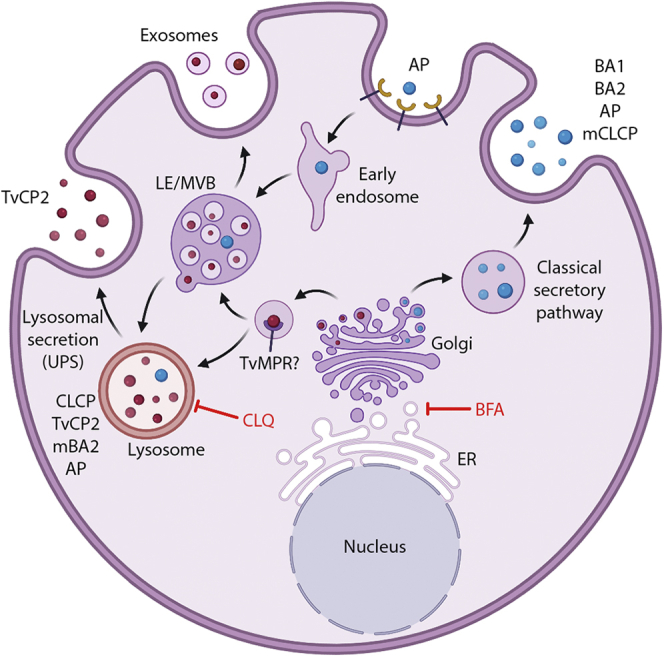
Hypothetical protein delivery to lysosomes and secretion in T. vaginalis. CLCP is a resident lysosomal peptidase, delivery of which is dependent on a glycosylation signal. Upon signal mutation (mCLCP), the protein is secreted. BA1, BA2, AP, and mCLCP are secreted via the classical ER-Golgi secretory pathway that is inhibited by brefeldin A but not chloroquine. They are delivered by secretory vesicles directly to the plasma membrane and released to the extracellular space. AP is eventually endocytosed and transported to lysosomes. Introduction of CLCP glycosylation signal into BA2 (mBA2) redirected this protein to lysosomes. TvCP2 is a lysosomal peptidase, which is secreted. Inhibition of acidification by chloroquine inhibited TvCP2 secretion, which indicates that this peptidase is secreted via the unconventional lysosomal pathway (unconventional protein secretion, UPS). AP, acid phosphatase; BA, β-amylase; BFA, brefeldin A; CLQ, chloroquine; ER, endoplasmic reticulum; LE, late endosome; MVB, multivesicular body; TvCP2, Trichomonas vaginalis cysteine protease 2; TvMPR, T. vaginalis mannose 6-phosphate receptor. Arrows indicate possible directions of the transport.
The secretion of TvCP2 has been shown to contribute to the host cell cytotoxicity caused by T. vaginalis. It has been suggested that this hydrolase is not a lysosomal protein and is secreted via secretory vesicles (67). However, the presence of TvCP2 in lysosomes, which was supported by immunofluorescence microscopy and the proteomic analysis in this study, suggested that TvCP2 might be secreted by an unconventional secretory pathway via lysosomes (Fig. 12) (93). The involvement of acidic organelles in TvCP2 secretion supported the treatment of T. vaginalis with the lysosomotropic amine chloroquine, which is known to increase the intralysosomal pH and inhibit lysosomal function (70, 71). The secretion of TvCP2 under chloroquine treatment was strongly inhibited, whereas no inhibition was observed for the secretion of nonlysosomal BA2. Secretion of both proteins was inhibited by brefeldin A, which impairs transport between the ER and Golgi (10, 72). Unlike TvCP2, secretion of lysosomal acid phosphatase (TVAG_169070) was insensitive to chloroquine, suggesting that this soluble protein is released via the secretory pathway as BA2 and possibly internalized via endocytosis. Interestingly, chloroquine was observed to have the opposite effect on lysosomal protein secretion in human cells that employed the M6P recognition marker. Treatment with chloroquine resulted in enhanced secretion of lysosomal proteins via the secretory pathway. In these cells, MPR proteins bind lysosomal cargo in the TGN and deliver the cargo through the intracellular pathway or via the plasma membrane to acidic vesicles, where the cargo dissociates and MPRs are recycled. Enhanced secretion of lysosomal proteins was explained by the inhibition of MPR recycling and consequently the decreased capacity to bind M6P (24, 70). The striking differences in the effect of chloroquine on lysosomal protein secretion and the absence of the M6P-dependent pathway in T. vaginalis suggest that the parasite employs specific unconventional mechanisms for the secretion of virulence factors such as TvCP2.
In conclusion, our proteomics study provided an extensive dataset of phagolysosmal proteins, including known and potentially novel virulence factors. Future investigations need to be performed to elucidate their functions and, in particular, to decipher the molecular mechanism of lysosomal sorting, which may provide an interesting target for the development of new antiparasitic strategies.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (94) partner repository with the dataset identifier PXD029152.
Supplemental data
This article contains supplemental data (10, 12, 54, 91).
Conflict of interest
The authors declare no competing interests.
Acknowledgments
We acknowledge the Imaging Methods Core Facility at BIOCEV, an institution supported by the MEYS CR (Large RI Project LM2018129 Czech-BioImaging), and the OMICS Proteomics of BIOCEV for mass spectrometry analysis. We also thank BioRender.com, which was used to create Figures 4, 12, and the graphical abstract. The authors wish to thank Michaela Marcinčiková for her excellent technical assistance, and Marie Olšinová for her help with fluorescence microscopy.
Funding and additional information
This work was supported by the Charles University Grant Agency, project number 18218 (N. Z.) and UNCE/SCI/012 (J. T.), the Czech Science Foundation (19-18773J) (J. T.), European Regional Development Funds project CePaViP (02.1.01/0.0/0.0/16_019/0000759) (J. T.), and H2020 Spreading Excellence and Widening Participation MICOBION (H2020 No 81022) (J. T.).
Author contributions
N. Z. and J. T. conceptualization; V. Ž., K. Z., J. D., and J. T. formal analysis; N. Z. and J. T. funding acquisition; N. Z., T. S., I. H., P. R., and K. H. investigation; N. Z. methodology; N. Z. and J. T. writing–original draft.
Supplemental data
Supplemental Table S1
Legends
References
- 1.World Health Organisation . WHO Press, World Health Organization; Geneva, Switzerland: 2012. Global Incidence and Prevalence of Selected Curable Sexually Transmitted Infections 2008. [Google Scholar]
- 2.Twu O., Dessí D., Vu A., Mercer F., Stevens G.C., De Miguel N., Rappelli P., Cocco A.R., Clubb R.T., Fiori P.L., Johnson P.J. Trichomonas vaginalis homolog of macrophage migration inhibitory factor induces prostate cell growth, invasiveness, and inflammatory responses. Proc. Natl. Acad. Sci. U. S. A. 2014;111:8179–8184. doi: 10.1073/pnas.1321884111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kissinger P. Trichomonas vaginalis: A review of epidemiologic, clinical and treatment issues. BMC Infect. Dis. 2015;15:307. doi: 10.1186/s12879-015-1055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrin D., Delgaty K., Bhatt R., Garber G. Clinical and microbiological aspects of Trichomonas vaginalis. Clin. Microbiol. Rev. 1998;11:300–317. doi: 10.1128/cmr.11.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kissinger P., Adamski A. Trichomoniasis and HIV interactions: A review. Sex. Transm. Infect. 2013;89:426–433. doi: 10.1136/sextrans-2012-051005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benchimol M., De Andrade Rosa I., Da Silva Fontes R., Burla Dias Â.J. Trichomonas adhere and phagocytose sperm cells: Adhesion seems to be a prominent stage during interaction. Parasitol. Res. 2008;102:597–604. doi: 10.1007/s00436-007-0793-3. [DOI] [PubMed] [Google Scholar]
- 7.Midlej V., Benchimol M. Trichomonas vaginalis kills and eats - Evidence for phagocytic activity as a cytopathic effect. Parasitology. 2010;137:65–76. doi: 10.1017/S0031182009991041. [DOI] [PubMed] [Google Scholar]
- 8.Pereira-Neves A., Benchimol M. Phagocytosis by Trichomonas vaginalis: New insights. Biol. Cell. 2007;99:87–101. doi: 10.1042/BC20060084. [DOI] [PubMed] [Google Scholar]
- 9.Rendón-Maldonado J.G., Espinosa-Cantellano M., González-Robles A., Martínez-Palomo A. Trichomonas vaginalis: In vitro phagocytosis of lactobacilli, vaginal epithelial cells, leukocytes, and erythrocytes. Exp. Parasitol. 1998;89:241–250. doi: 10.1006/expr.1998.4297. [DOI] [PubMed] [Google Scholar]
- 10.Štáfková J., Rada P., Meloni D., Zárský V., Smutná T., Zimmann N., Harant K., Pompach P., Hrdý I., Tachezy J. Dynamic secretome of Trichomonas vaginalis: Case study of β-amylases. Mol. Cell. Proteomics. 2018;17:304–320. doi: 10.1074/mcp.RA117.000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinheiro J., Biboy J., Vollmer W., Hirt R.P., Keown J.R., Artuyants A., Black M.M., Goldstone D.C., Simoes-Barbosa A. The protozoan Trichomonas vaginalis targets bacteria with laterally acquired NlpC/P60 peptidoglycan hydrolases. MBio. 2018;9 doi: 10.1128/mBio.01784-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Twu O., de Miguel N., Lustig G., Stevens G.C., Vashisht A.A., Wohlschlegel J.A., Johnson P.J. Trichomonas vaginalis exosomes deliver cargo to host cells and mediate host:parasite interactions. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyttinen J.M.T., Niittykoski M., Salminen A., Kaarniranta K. Maturation of autophagosomes and endosomes: A key role for Rab7. Biochim. Biophys. Acta Mol. Cell Res. 2013;1833:503–510. doi: 10.1016/j.bbamcr.2012.11.018. [DOI] [PubMed] [Google Scholar]
- 14.Ganley I.G. Autophagosome maturation and lysosomal fusion. Essays Biochem. 2013;55:65–78. doi: 10.1042/bse0550065. [DOI] [PubMed] [Google Scholar]
- 15.Xu H., Ren D. Lysosomal physiology. Annu. Rev. Physiol. 2015;77:57–80. doi: 10.1146/annurev-physiol-021014-071649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinchen J.M., Ravichandran K.S. Phagosome maturation: Going through the acid test. Nat. Rev. Mol. Cell Biol. 2008;9:781–795. doi: 10.1038/nrm2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu L., Chen Y., Tooze S.A. Autophagy pathway: Cellular and molecular mechanisms. Autophagy. 2018;14:207–215. doi: 10.1080/15548627.2017.1378838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benchimol M. Hydrogenosome autophagy: An ultrastructural and cytochemical study. Biol. Cell. 1999;91:165–174. doi: 10.1016/s0248-4900(99)80039-2. [DOI] [PubMed] [Google Scholar]
- 19.Sun Z., Brodsky J.L. Protein quality control in the secretory pathway. J. Cell Biol. 2019;218:3171–3187. doi: 10.1083/jcb.201906047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peden A.A., Oorschot V., Hesser B.A., Austin C.D., Scheller R.H., Klumperman J. Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J. Cell Biol. 2004;164:1065–1076. doi: 10.1083/jcb.200311064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janvier K., Bonifacino J.S. Role of the endocytic machinery in the sorting of lysosome-associated membrane proteins. Mol. Biol. Cell. 2005;16:4231–4242. doi: 10.1091/mbc.E05-03-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castonguay A.C., Lasanajak Y., Song X., Olson L.J., Cummings R.D., Smith D.F., Dahms N.M. The glycan-binding properties of the cation-independent mannose 6-phosphate receptor are evolutionary conserved in vertebrates. Glycobiology. 2012;22:983–996. doi: 10.1093/glycob/cws058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lousa C.D.M., Denecke J. Lysosomal and vacuolar sorting: Not so different after all. Biochem. Soc. Trans. 2016;44:891–897. doi: 10.1042/BST20160050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braulke T., Bonifacino J.S. Sorting of lysosomal proteins. Biochim. Biophys. Acta Mol. Cell Res. 2009;1793:605–614. doi: 10.1016/j.bbamcr.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 25.Coutinho M.F., Prata M.J., Alves S. Mannose-6-phosphate pathway: A review on its role in lysosomal function and dysfunction. Mol. Genet. Metab. 2012;105:542–550. doi: 10.1016/j.ymgme.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 26.Nakada-Tsukui K., Tsuboi K., Furukawa A., Yamada Y., Nozaki T. A novel class of cysteine protease receptors that mediate lysosomal transport. Cell. Microbiol. 2012;14:1299–1317. doi: 10.1111/j.1462-5822.2012.01800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonifacino J.S., Traub L.M. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 28.Puertollano R., Aguilar R.C., Gorshkova I., Crouch R.J., Bonifacino J.S. Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science. 2001;292:1712–1716. doi: 10.1126/science.1060750. [DOI] [PubMed] [Google Scholar]
- 29.Höning S., Sandoval I., Von Figura K. A di-leucine-based motif in the cytoplasmic tail of LIMP-II and tyrosinase mediates selective binding of AP-3. EMBO J. 1998;17:1304–1314. doi: 10.1093/emboj/17.5.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kyttälä A., Yliannala K., Schu P., Jalanko A., Luzio J.P. AP-1 and AP-3 facilitate lysosomal targeting of Batten disease protein CLN3 via its dileucine motif. J. Biol. Chem. 2005;280:10277–10283. doi: 10.1074/jbc.M411862200. [DOI] [PubMed] [Google Scholar]
- 31.Koumandou V.L., Klute M.J., Herman E.K., Nunez-Miguel R., Dacks J.B., Field M.C. Evolutionary reconstruction of the retromer complex and its function in Trypanosoma brucei. J. Cell Sci. 2011;124:1496–1509. doi: 10.1242/jcs.081596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carlton J.M., Hirt R.P., Silva J.C., Delcher A.L., Schatz M., Zhao Q., Wortman J.R., Bidwell S.L., Alsmark U.C.M., Besteiro S., Sicheritz-Ponten T., Noel C.J., Dacks J.B., Foster P.G., Simillion C., et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernández H.M., Marcet R., Sarracent J. Biological roles of cysteine proteinases in the pathogenesis of Trichomonas vaginalis. Parasite. 2014;21:54. doi: 10.1051/parasite/2014054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott D.A., North M.J., Coombs G.H. The pathway of secretion of proteinases in Trichomonas vaginalis. Int. J. Parasitol. 1995;25:657–666. doi: 10.1016/0020-7519(95)99370-x. [DOI] [PubMed] [Google Scholar]
- 35.Cárdenas-Guerra R.E., Arroyo R., Rosa de Andrade I., Benchimol M., Ortega-López J. The iron-induced cysteine proteinase TvCP4 plays a key role in Trichomonas vaginalis haemolysis. Microbes Infect. 2013;15:958–968. doi: 10.1016/j.micinf.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 36.Tai J.H., Su H.M., Tsai J., Shaio M.F., Wang C.C. The divergence of Trichomonas vaginalis virus RNAs among various isolates of Trichomonas vaginalis. Exp. Parasitol. 1993;76:278–286. doi: 10.1006/expr.1993.1033. [DOI] [PubMed] [Google Scholar]
- 37.Kulda J., Vojtechovska M., Tachezy J., Demes P., Kunzová E. Metronidazole resistance of Trichomonas vaginalis as a cause of treatment failure in trichomoniasis. A case report. Br. J. Vener. Dis. 1982;58:394–399. doi: 10.1136/sti.58.6.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hrdy I., Hirt R.P., Dolezal P., Bardonová L., Foster P.G., Tachezy J., Embley T.M. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004;432:618–622. doi: 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
- 39.Janssen B.D., Chen Y.P., Molgora B.M., Wang S.E., Simoes-Barbosa A., Johnson P.J. CRISPR/Cas9-mediated gene modification and gene knock out in the human-infective parasite Trichomonas vaginalis. Sci. Rep. 2018;8:270. doi: 10.1038/s41598-017-18442-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bradley P.J., Lahti C.J., Plumper E., Johnson P.J. Targeting and translocation of proteins into the hydrogenosome of the protist Trichomonas: Similarities with mitochondrial protein import. EMBO J. 1997;16:3484–3493. doi: 10.1093/emboj/16.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nývltová E., Smutná T., Tachezy J., Hrdý I. OsmC and incomplete glycine decarboxylase complex mediate reductive detoxification of peroxides in hydrogenosomes of Trichomonas vaginalis. Mol. Biochem. Parasitol. 2016;206:29–38. doi: 10.1016/j.molbiopara.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 42.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woessner D.J., Dawson S.C. The Giardia median body protein is a ventral disc protein that is critical for maintaining a domed disc conformation during attachment. Eukaryot. Cell. 2012;11:292–301. doi: 10.1128/EC.05262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Méchin V., Damerval C., Zivy M. Total protein extraction with TCA-acetone. Methods Mol. Biol. 2007;355:1–8. doi: 10.1385/1-59745-227-0:1. [DOI] [PubMed] [Google Scholar]
- 45.Masuda T., Tomita M., Ishihama Y. Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J.Proteome.Res. 2008;7:731–740. doi: 10.1021/pr700658q. [DOI] [PubMed] [Google Scholar]
- 46.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, Termed MaxLFQ. Mol. Cell. Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warrenfeltz S., Basenko E., Crouch K., Harb O., Kissinger J., Roos D., Shanmugasundram A., Silva-Franco F. EuPathDB: The eukaryotic pathogen genomics database resource. Methods Mol. Biol. 2018;1757:69–113. doi: 10.1007/978-1-4939-7737-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lal K., Field M.C., Carlton J.M., Warwicker J., Hirt R.P. Identification of a very large Rab GTPase family in the parasitic protozoan Trichomonas vaginalis. Mol. Biochem. Parasitol. 2005;143:226–235. doi: 10.1016/j.molbiopara.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 50.Beltrán N.C., Horváthová L., Jedelský P.L., Šedinová M., Rada P., Marcinčiková M., Hrdý I., Tachezy J. Iron-induced changes in the proteome of Trichomonas vaginalis hydrogenosomes. PLoS One. 2013;8 doi: 10.1371/journal.pone.0065148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sommer U., Costello C.E., Hayes G.R., Beach D.H., Gilbert R.O., Lucas J.J., Singh B.N. Identification of Trichomonas vaginalis cysteine proteases that induce apoptosis in human vaginal epithelial cells. J. Biol. Chem. 2005;280:23853–23860. doi: 10.1074/jbc.M501752200. [DOI] [PubMed] [Google Scholar]
- 52.Rendón-Gandarilla F.J., Ramón-Luing L.D.L.A., Ortega-López J., Rosa De Andrade I., Benchimol M., Arroyo R. The TvLEGU-1, a legumain-like cysteine proteinase, plays a key role in Trichomonas vaginalis cytoadherence. Biomed. Res. Int. 2013;2013 doi: 10.1155/2013/561979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma L., Meng Q., Cheng W., Sung Y., Tang P., Hu S., Yu J. Involvement of the GP63 protease in infection of Trichomonas vaginalis. Parasitol. Res. 2011;109:71–79. doi: 10.1007/s00436-010-2222-2. [DOI] [PubMed] [Google Scholar]
- 54.De Miguel N., Lustig G., Twu O., Chattopadhyay A., Wohlschlegel J.A., Johnson P.J. Proteome analysis of the surface of Trichomonas vaginalis reveals novel proteins and strain-dependent differential expression. Mol. Cell. Proteomics. 2010;9:1554–1566. doi: 10.1074/mcp.M000022-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Srivastava N., Traynor D., Piel M., Kabla A.J., Kay R.R. Pressure sensing through Piezo channels controls whether cells migrate with blebs or pseudopods. Proc. Natl. Acad. Sci. U. S. A. 2020;117:2506–2512. doi: 10.1073/pnas.1905730117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guerra F., Bucci C. Multiple roles of the small GTPase Rab7. Cells. 2016;5:34. doi: 10.3390/cells5030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu Z.Q., Rao C.L., Tang M.L., Zhang Y., Lu X.X., Chen J.G., Mao C., Deng L., Li Q., Mao X.H. Rab32 GTpase, as a direct target of miR-30b/c, controls the intracellular survival of Burkholderia pseudomallei by regulating phagosome maturation. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patwardhan A., Bardin S., Miserey-Lenkei S., Larue L., Goud B., Raposo G., Delevoye C. Routing of the RAB6 secretory pathway towards the lysosome related organelle of melanocytes. Nat. Commun. 2017;8 doi: 10.1038/ncomms15835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Egami Y., Fukuda M., Araki N. Rab35 regulates phagosome formation through recruitment of ACAP2 in macrophages during FcγR-mediated phagocytosis. J. Cell Sci. 2011;124:3557–3567. doi: 10.1242/jcs.083881. [DOI] [PubMed] [Google Scholar]
- 60.Verma K., Datta S. The Monomeric GTPase Rab35 regulates phagocytic cup formation and phagosomal maturation in Entamoeba histolytica. J. Biol. Chem. 2017;292:4960–4975. doi: 10.1074/jbc.M117.775007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gutierrez M.G. Functional role(s) of phagosomal Rab GTPases. Small GTPases. 2013;4:148–158. doi: 10.4161/sgtp.25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohno H., Stewart J., Fournier M.C., Bosshart H., Rhee I., Miyatake S., Saito T., Gallusser A., Kirchhausen T., Bonifacino J.S. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- 63.Caviston J.P., Holzbaur E.L.F. Microtubule motors at the intersection of trafficking and transport. Trends Cell Biol. 2006;16:530–537. doi: 10.1016/j.tcb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 64.Krendel M., Mooseker M.S. Myosins: Tails (and heads) of functional diversity. Physiology. 2005;20:239–251. doi: 10.1152/physiol.00014.2005. [DOI] [PubMed] [Google Scholar]
- 65.Bálint Š., Vilanova I.V., Álvarez Á.S., Lakadamyali M. Correlative live-cell and superresolution microscopy reveals cargo transport dynamics at microtubule intersections. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3375–3380. doi: 10.1073/pnas.1219206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lübke T., Lobel P., Sleat D.E. Proteomics of the lysosome. Biochim. Biophys. Acta. 2009;1793:625–635. doi: 10.1016/j.bbamcr.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miranda-Ozuna J.F.T., Rivera-Rivas L.A., Cárdenas-Guerra R.E., Hernández-García M.S., Rodríguez-Cruz S., González-Robles A., Chavez-Munguía B., Arroyo R. Glucose-restriction increases Trichomonas vaginalis cellular damage towards HeLa cells and proteolytic activity of cysteine proteinases (CPs), such as TvCP2. Parasitology. 2019;146:1156–1166. doi: 10.1017/S0031182019000209. [DOI] [PubMed] [Google Scholar]
- 68.Dahms N.M., Hancock M.K. P-type lectins. Biochim. Biophys. Acta. 2002;1572:317–340. doi: 10.1016/s0304-4165(02)00317-3. [DOI] [PubMed] [Google Scholar]
- 69.Liu L., Lee W.S., Doray B., Kornfeld S. Role of spacer-1 in the maturation and function of GlcNAc-1-phosphotransferase. FEBS Lett. 2017;591:47–55. doi: 10.1002/1873-3468.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gonzalez-Noriega A., Grubb J.H., Talkad V., Sly W.S. Chloroquine inhibits lysosomal enzyme pinocytosis and enhances lysosomal enzyme secretion by impairing receptor recycling. J. Cell Biol. 1980;85:839–852. doi: 10.1083/jcb.85.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fedele A.O., Proud C.G. Chloroquine and bafilomycin A mimic lysosomal storage disorders and impair mTORC1 signalling. Biosci. Rep. 2020;40 doi: 10.1042/BSR20200905. BSR20200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chardin P., McCormick F. Brefeldin A: The advantage of being uncompetitive. Cell. 1999;97:153–155. doi: 10.1016/s0092-8674(00)80724-2. [DOI] [PubMed] [Google Scholar]
- 73.Doyle P., Sajid M., O'Brien T., DuBois K., Engel J., Mackey Z., Reed S. Drugs targeting parasite lysosomes. Curr. Pharm. Des. 2008;14:889–900. doi: 10.2174/138161208784041060. [DOI] [PubMed] [Google Scholar]
- 74.Gao Y., Chen Y., Zhan S., Zhang W., Xiong F., Ge W. Comprehensive proteome analysis of lysosomes reveals the diverse function of macrophages in immune responses. Oncotarget. 2017;8:7420–7440. doi: 10.18632/oncotarget.14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schröder B.A., Wrocklage C., Hasilik A., Saftig P. The proteome of lysosomes. Proteomics. 2010;10:4053–4076. doi: 10.1002/pmic.201000196. [DOI] [PubMed] [Google Scholar]
- 76.Peterson K.M., Alderete J.F. Iron uptake and increased intracellular enzyme activity follow host lactoferrin binding by Trichomonas vaginalis receptors. J. Exp. Med. 1984;160:398–410. doi: 10.1084/jem.160.2.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mosen P., Sanner A., Singh J., Winter D. Targeted quantification of the lysosomal proteome in complex samples. Proteomes. 2021;9:1–17. doi: 10.3390/proteomes9010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Noël C.J., Diaz N., Sicheritz-Ponten T., Safarikova L., Tachezy J., Tang P., Fiori P.L., Hirt R.P. Trichomonas vaginalis vast BspA-like gene family: Evidence for functional diversity from structural organisation and transcriptomics. BMC Genomics. 2010;11:99. doi: 10.1186/1471-2164-11-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Molgora B.M., Rai A.K., Sweredoski M.J., Moradian A., Hess S., Johnson P.J. A novel Trichomonas vaginalis surface protein modulates parasite attachment via protein:Host cell proteoglycan interaction. MBio. 2021;12:1–17. doi: 10.1128/mBio.03374-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arroyo R., Cárdenas-Guerra R.E., Figueroa-Angulo E.E., Puente-Rivera J., Zamudio-Prieto O., Ortega-López J. Trichomonas vaginalis cysteine proteinases: Iron response in gene expression and proteolytic activity. Biomed. Res. Int. 2015;2015 doi: 10.1155/2015/946787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ramón-Luing L.A., Rendón-Gandarilla F.J., Cárdenas-Guerra R.E., Rodríguez-Cabrera N.A., Ortega-López J., Avila-González L., Angel-Ortiz C., Herrera-Sánchez C.N., Mendoza-García M., Arroyo R. Immunoproteomics of the active degradome to identify biomarkers for Trichomonas vaginalis. Proteomics. 2010;10:435–444. doi: 10.1002/pmic.200900479. [DOI] [PubMed] [Google Scholar]
- 82.Hernández-Romano P., Hernández R., Arroyo R., Alderete J.F., López-Villaseñor I. Identification and characterization of a surface-associated, subtilisin-like serine protease in Trichomonas vaginalis. Parasitology. 2010;137:1621–1635. doi: 10.1017/S003118201000051X. [DOI] [PubMed] [Google Scholar]
- 83.Hu G., St. Leger R.J. A phylogenomic approach to reconstructing the diversification of serine proteases in fungi. J. Evol. Biol. 2004;17:1204–1214. doi: 10.1111/j.1420-9101.2004.00786.x. [DOI] [PubMed] [Google Scholar]
- 84.Li J., Yu L., Yang J., Dong L., Tian B., Yu Z., Liang L., Zhang Y., Wang X., Zhang K. New insights into the evolution of subtilisin-like serine protease genes in Pezizomycotina. BMC Evol. Biol. 2010;10:1–14. doi: 10.1186/1471-2148-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shayman J.A., Tesmer J.J.G. Lysosomal phospholipase A2. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2019;1864:932–940. doi: 10.1016/j.bbalip.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harwig S.S.L., Tan L., Qu X.D., Cho Y., Eisenhauer P.B., Lehrer R.I. Bactericidal properties of murine intestinal phospholipase A2. J. Clin. Invest. 1995;95:603–610. doi: 10.1172/JCI117704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fenard D., Lambeau G., Valentin E., Lefebvre J.C., Lazdunski M., Doglio A. Secreted phospholipases A2, a new class of HIV inhibitors that block virus entry into host cells. J. Clin. Invest. 1999;104:611–618. doi: 10.1172/JCI6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sanon A., Tournaire-Arellano C., Younes El Hage S., Bories C., Caujolle R., Loiseau P.M. N-acetyl-β-d-hexosaminidase from Trichomonas vaginalis: Substrate specificity and activity of inhibitors. Biomed. Pharmacother. 2005;59:245–248. doi: 10.1016/j.biopha.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 89.Rivero M.R., Miras S.L., Feliziani C., Zamponi N., Quiroga R., Hayes S.F., Rópolo A.S., Touz M.C. Vacuolar protein sorting receptor in Giardia lamblia. PLoS One. 2012;7 doi: 10.1371/journal.pone.0043712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sloves P.J., Delhaye S., Mouveaux T., Werkmeister E., Slomianny C., Hovasse A., Dilezitoko Alayi T., Callebaut I., Gaji R.Y., Schaeffer-Reiss C., Van Dorsselear A., Carruthers V.B., Tomavo S. Toxoplasma sortilin-like receptor regulates protein transport and is essential for apical secretory organelle biogenesis and host infection. Cell Host Microbe. 2012;11:515–527. doi: 10.1016/j.chom.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 91.Dennes A., Cromme C., Suresh K., Kumar N.S., Eble J.A., Hahnenkamp A., Pohlmann R. The novel Drosophila lysosomal enzyme receptor protein mediates lysosomal sorting in mammalian cells and binds mammalian and Drosophila GGA adaptors. J. Biol. Chem. 2005;280:12849–12857. doi: 10.1074/jbc.M410626200. [DOI] [PubMed] [Google Scholar]
- 92.Samuelson J., Banerjee S., Magnelli P., Cui J., Kelleher D.J., Gilmore R., Robbins P.W. The diversity of dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets glycosyltranferases. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1548–1553. doi: 10.1073/pnas.0409460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee J., Ye Y. The roles of endo-lysosomes in unconventional protein secretion. Cells. 2018;7:198. doi: 10.3390/cells7110198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D., Inuganti A., Griss J., Mayer G., Eisenacher M., Al E. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1
Legends
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (94) partner repository with the dataset identifier PXD029152.