Abstract
A high-throughput, retrovirus-mediated mutagenesis method based on gene trapping in embryonic stem cells was used to identify a novel mouse gene. The human ortholog encodes a transmembrane protein containing five extracellular immunoglobulin-like domains that is structurally related to human NEPHRIN, a protein associated with congenital nephrotic syndrome. Northern analysis revealed wide expression in humans and mice, with highest expression in kidney. Based on similarity to NEPHRIN and abundant expression in kidney, this protein was designated NEPH1 and embryonic stem cells containing the retroviral insertion in the Neph1 locus were used to generate mutant mice. Analysis of kidney RNA from Neph1−/− mice showed that the retroviral insertion disrupted expression of Neph1 transcripts. Neph1−/− pups were represented at the expected normal Mendelian ratios at 1 to 3 days of age but at only 10% of the expected frequency at 10 to 12 days after birth, suggesting an early postnatal lethality. The Neph1−/− animals that survived beyond the first week of life were sickly and small but without edema, and all died between 3 and 8 weeks of age. Proteinuria ranging from 300 to 2,000 mg/dl was present in all Neph1−/− mice. Electron microscopy demonstrated NEPH1 expression in glomerular podocytes and revealed effacement of podocyte foot processes in Neph1−/− mice. These findings suggest that NEPH1, like NEPHRIN, may play an important role in maintaining the structure of the filtration barrier that prevents proteins from freely entering the glomerular urinary space.
NEPHRIN is a transmembrane protein of the immunoglobulin (Ig) superfamily that is expressed by epithelial podocytes of developing glomeruli (13, 17). Congenital nephrotic syndrome of the Finnish type results from mutations in NPHS1, the human gene encoding NEPHRIN, indicating a role for NEPHRIN in maintaining the filtration barrier that prevents proteins from freely entering the glomerular urinary space (6, 17). Recent studies localized NEPHRIN to the slit diaphragms that form the junctions between podocyte foot processes interdigitating along the glomerular basement membrane. This and other studies suggest that NEPHRIN proteins extending toward each other from adjacent podocyte foot processes may interdigitate in a zipper-like structure to form the crucial filtration barrier in the slit diaphragm. The eight Ig-like domains of each NEPHRIN protein, which are of the C2 type of Ig domain known to be involved in cell-cell interactions, are thought to provide the homophilic interactions that bind these NEPHRIN proteins together (13, 17).
We use a high-throughput mutagenesis method based on gene trapping in embryonic stem (ES) cells that allows automated production of sequence tags from the trapped and mutated genes (20). These ES cell clones are stored in a library called Omnibank, and the sequence tag from the gene trapped in each clone, referred to as the Omnibank sequence tag or OST, is entered into a searchable database. A protein with Ig domains was identified within this database and was designated NEPH1 since (i) human and mouse sequences for this protein were homologous to NEPHRIN and (ii) additional studies showed that this protein was highly expressed in human and mouse kidney. To obtain insight into the function of NEPH1, particularly with respect to the role of NEPH1 in kidney, we used ES cells harboring the mutated Neph1 gene to generate mutant mice. We now report that mutation of the Neph1 locus results in perinatal lethality accompanied by proteinuria.
MATERIALS AND METHODS
Generation of Neph1 mutant mice.
An ES cell clone with a retroviral vector (see Fig. 3 for a description of the vector) inserted in the Neph1 locus was injected into C57BL/6-Tyrc-Brd host blastocysts (21). Chimeric mice were generated and bred to C57BL/6-Tyrc-Brd mice; the resulting Neph1+/− offspring were interbred to produce Neph1−/− mice. Tail DNA was genotyped by quantitative dot blots (Bio-Rad) using a neomycin phosphotransferase gene fragment and a fragment containing exon 1 of the murine Csk gene (MMU05247) as probes to detect the virus integration and a single-copy endogenous gene, respectively.
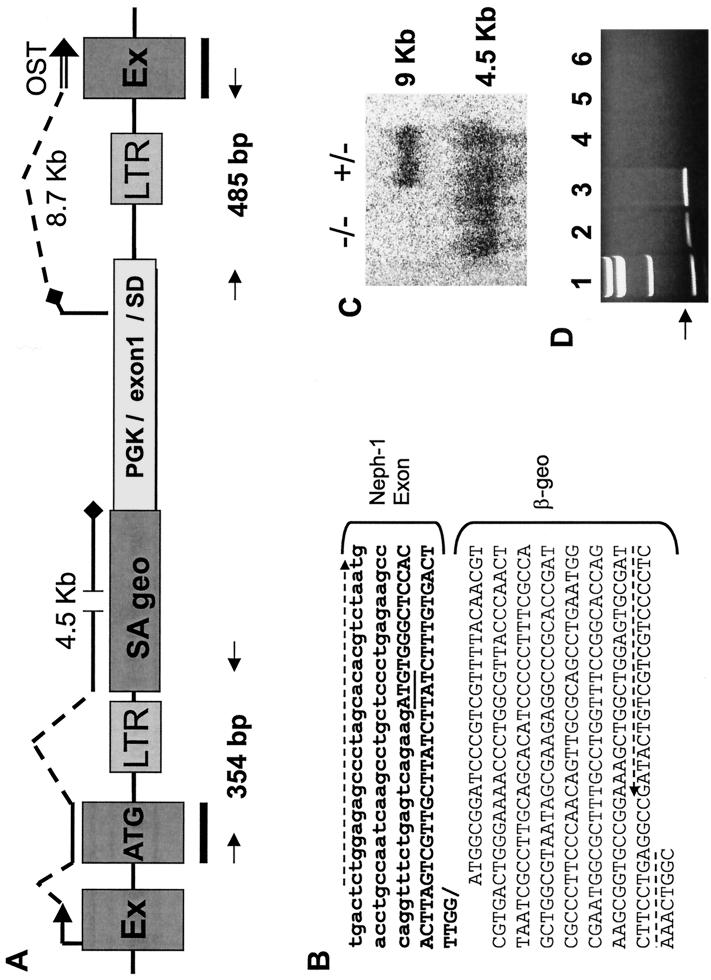
FIG. 3.
Retroviral insertion into the Neph1 locus. (A) Structure of the gene trap vector integration site in the Neph1 locus. The retroviral construct, which contains flanking viral long terminal repeats (LTRs), is inserted in the intron downstream of the ATG-containing Neph1 exon. Transcripts initiating at the Neph1 promoter (indicated by an arrow followed by broken and solid lines) encode a 4.5-kb message that utilizes the splice acceptor (SA) site upstream of the β-geo fusion gene. Transcripts initiating from the PGK promoter utilize the splice donor site in the inserted heterologous exon 1-splice donor (SD)-intron 1 cassette and splice into the downstream Neph1 exon (EX) to produce a roughly 8.7-kb RNA which is detectable only in ES cells and is absent in vivo (see text). The heterologous exon 1 is a noncoding exon that contains a natural in-frame stop codon (black diamond) that eliminates the likelihood of expression of a protein from the trapped Neph1 locus. A probe spanning the ATG-containing exons and downstream exons of Neph1 (black bars) was used to perform the Northern blot shown in Fig. 2 and 3C. The OST sequence is represented by a double line. The primers used to clone the Neph1–β-geo and the exon 1-Neph1 fusion transcript splice junctions are indicated by arrows and are expected to produce 354- and 485-bp bands, respectively; the sequence of the 5′ transcript is given in panel B. (B) Sequence of the hybrid transcript spanning the splice junction of the Neph1 ATG-containing exon and the β-geo-coding region. The ATG in the Neph1 coding region (boldface) is underlined, and the primers used for RT-PCR of this fragment are designated by broken arrows. (C) Northern blot of kidney RNA obtained from 1-day-old mice bearing either one copy (+/−) or two copies (−/−) of the trapped Neph1 locus. The blot, hybridized with a probe spanning the Neph1 ATG-containing and downstream exons (see panel A), shows RNA transcripts of 9 and 4.5 kb. (D) Ethidium bromide-stained gel showing RT-PCR products from the heterologous exon 1-Neph1 fusion transcripts. Lane 1 contains molecular-weight standards. The expected band size of 485 bp (see panel A) is clearly observed in the RNA samples from the ES cell line with the Neph1 mutation (lanes 2 and 3). This band is undetectable in kidney RNA from Neph1+/+ and Neph1+/− mice (lanes 4 and 6, respectively) and is negligible in kidney RNA from Neph1−/− mice (lane 5).
Cloning of Neph1 cDNAs.
Mouse total kidney RNA was used in 5′ and 3′ rapid amplification of cDNA ends (Clontech) employing primers derived from the OST sequence. The product (mouse cDNA; GenBank accession no. AYO17368) was cloned into the PCRII-TOPO vector (Invitrogen) and sequenced on an ABI 377 sequencer using BigDye terminators. The full-length human cDNA clone (GenBank accession no. AYO17369) was identified by screening a kidney cDNA library (Edge BioSystems, Gaithersburg, Md.) by PCR using two gene-specific primers (5′-GTGCTCCCCTGTGTGCTGCTCAACTACT-3′ and 5′-CAGAGAGCTCAGCATCTGTGATCTCCAGGT-3′) derived from the human genomic DNA sequence (GenBank accession no. AL139010). Positive clones were PCR amplified with Expand High Fidelity DNA Polymerase (Roche) using two different primer sets for the 5′ and 3′ cDNA ends. The 3′ cDNA end was amplified using a NEPH1-specific primer (5′-GTGCTCCCCTGTGTGCTGCTCAACTACT–3′) and a λGT10 vector-specific primer (5′-AGCAAGTTCAGCCTGGTTAAG-3′), while the 5′ cDNA end was amplified using a NEPH1-specific primer (5′-CAGAGAGCTCAGCATCTGTGATCTCCAGGT-3′) and a λGT10 primer (5′-CTTATGAGTATTTCTTCCAGGGTA-3′).
RNA analysis.
Human and mouse Northern blots containing 2 μg of poly(A)+ RNA in each lane were probed as recommended by the supplier (OriGene). Probing was performed with 32P-labeled fragments spanning bp 882 to 1225 of the human NEPH1 cDNA and from bp 40 to 403 of the murine Neph1 cDNA, respectively. Fusion transcripts consisting of a heterologous exon and downstream Neph1 exons (see Fig. 3 and Results) were reverse transcribed and PCR amplified (RT-PCR) using Expand Polymerase as recommended by the manufacturer (Gibco-BRL); 5′-CCAGAGTCTTCAGAGATCAAGTC-3′ was the 5′ primer, and 5′-GGCATTAAATGCTCTGCACGTGAGGTTGTAG-3′ was the 3′ primer. RNA quality in the samples was assessed using primers to the mouse β-actin cDNA (GenBank no. M12481).
Histopathology.
Whole-mount LacZ staining of kidneys was performed as described previously (10). After LacZ-stained samples were fixed in 4% paraformaldehyde and embedded in paraffin, 4-μm sections were cut and counterstained with nuclear fast red. Freshly dissected kidneys were fixed in paraformaldehyde, processed into paraffin blocks, and stained with hematoxylin and eosin for morphological assessment (8). Glycogen content and basement membranes were evaluated using the periodic acid-Schiff reaction (2).
To confirm that LacZ was expressed in glomerular epithelial cells, glutaraldehyde-fixed tissue was postfixed in buffered osmium and then dehydrated in graded ethanol solutions. Specimens were embedded in Araldite 502. Ultrathin sections were stained with lead citrate and aranyl acetate and were analyzed by electron microscopy.
Mouse care and study.
Mouse studies were performed according to federal guidelines (1). Mice were housed at 24°C on a fixed 12-h light/12-h dark cycle and had ad libitum access to water and rodent chow (product no. 5001; Purina, St. Louis, Mo.). Urinalysis was performed using urine dipsticks (Multistix; Bayer, Pittsburgh, Pa.).
RESULTS
Gene trapping and characterization of the Neph1 gene.
We use a retrovirus-mediated gene trap strategy that selects for integrations into gene introns. This approach ablates gene function because the vector introduces heterologous splice acceptor and donor sites that disrupt the normal splicing of the trapped gene (20). The gene trap vector also contains an internal phosphoglycerate kinase 1 (PGK) promoter that generates a fusion transcript incorporating the downstream exons of the trapped locus. Fusion transcripts from over 200,000 trapped ES cell clones have been sequenced and deposited into a searchable database. Because these sequences are exonic, they are informative with respect to the identity of the trapped gene. Searching this database identified an OST encoding a novel protein with homology to human NEPHRIN, a protein mutated in patients with congenital nephrotic syndrome of the Finnish type (6). Since the protein product of this novel gene is homologous to NEPHRIN and is highly expressed in kidney (see below), the gene was designated Neph1 (gene name standardized by the mouse nomenclature committee at the Jackson Laboratories).
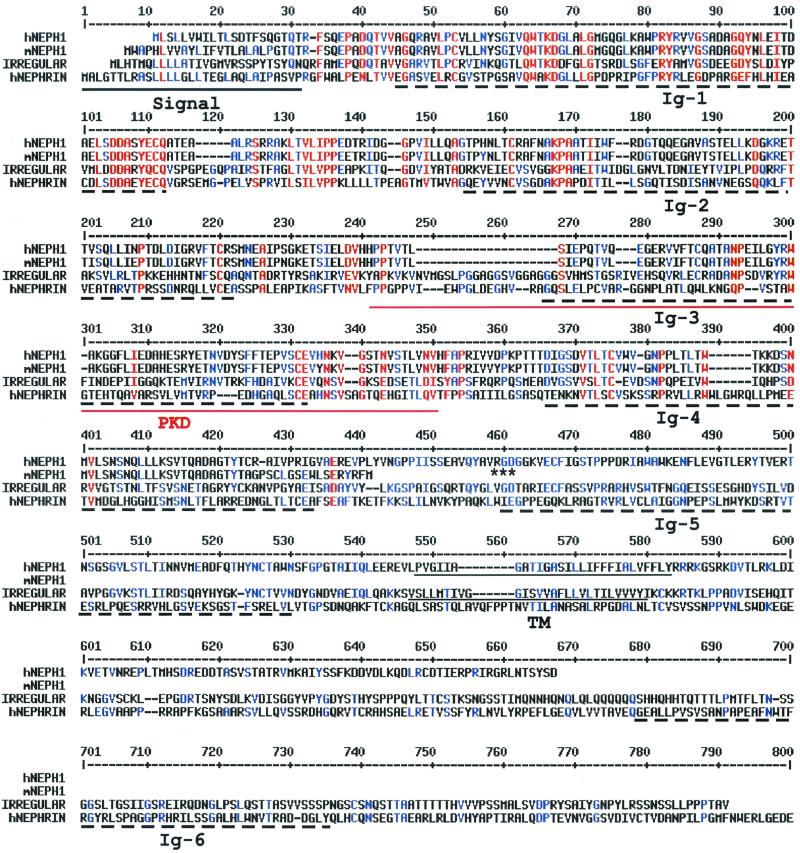
The human NEPH1 cDNA is predicted to encode a 67-kDa protein with five Ig domains and the integrin-recognition motif RGD found in proteins such as fibronectin (Fig. 1) (9). Ig3 is similar to the polycystic kidney disease (PKD) domains first identified in the polycystin-1 protein (18). These PKD domains, which have Ig-like folds, are evolutionarily distinct from members of the Ig superfamily; their function in polycystin-1 is unclear at present (4). Human NEPH1 also includes two hydrophobic domains: the presumptive signal sequence at the N terminus and a transmembrane region. A homology search revealed that NEPH1 is related to the Drosophila melanogaster protein IRREGULAR CHIASM (12) in addition to NEPHRIN (6) (25.6 and 18% identity, respectively). Despite their low overall identity, these proteins are structurally related due to conservation of Ig domains (Fig. 1). The mouse cDNA clone appears to be truncated just past the Ig4 domain (Fig. 1); because we failed to detect short Neph1 transcripts in mouse tissues (not shown), it is possible that this cDNA is a partial clone. Northern analyses performed on poly(A)+ RNA from multiple human and mouse tissues identified a single 9-kb transcript, which was widely expressed, with highest levels of expression in kidney (Fig. 2).
FIG. 1.
Alignment of NEPH1 with homologous proteins. The human NEPH1 (hNEPH1; GenBank accession no. AYO17369), a truncated mouse NEPH1 (mNEPH1; GenBank accession no. AYO17368), the D. melanogaster irregular chiasmata (IRREGULAR; GenBank accession no. L11040), and the human NEPHRIN (hNEPHRIN; GenBank accession no. AF190637) proteins were aligned using the MultiAlin version 5.4.1 program (http://www.toulouse.inra.ft/multalin). The signal and transmembrane (TM) domains in the hNEPH1, mNEPH1, and IRREGULAR proteins, identified by DNAStar Protean V. 4.0, are underlined. Ig-like domains (Ig) are marked by broken lines, and the PKD domain is doubly underlined; these domains were identified using the Pfam search engine (http://pfam.wustl.edu). The RGD sequence of NEPH1 is marked by asterisks. The hNEPHRIN protein sequence extends an additional 459 amino acid residues beyond what is shown here.
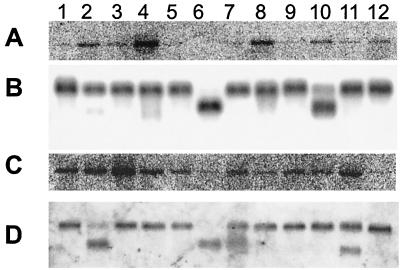
FIG. 2.
Neph1 expression in human and mouse tissues. (A) Northern blot showing a 9-kb Neph1 transcript in adult human brain (lane 1), colon (lane 2), heart (lane 3), kidney (lane 4), liver (lane 5), lung (lane 6), muscle (lane 7), placenta (lane 8), small intestine (lane 9), spleen (lane 10), stomach (lane 11), and testis (lane 12). (B) The same blot as in panel A hybridized with a probe to the human β-actin gene for RNA loading control. (C) Northern blot showing a 9-kb Neph1 transcript in adult mouse brain (lane 1), heart (lane 2), kidney (lane 3), liver (lane 4), lung (lane 5), muscle (lane 6), skin (lane 7), small intestine (lane 8), spleen (lane 9), stomach (lane 10), testis (lane 11), and thymus (lane 12). (D) The same blot as in panel C hybridized with a human β-actin probe.
Since NEPH1 is highly expressed in kidney and is homologous to NEPHRIN, we chose to disrupt the Neph1 gene in mice. ES cells expressing the NEPH1 OST were injected into blastocysts, and the resulting chimeric mice were bred to generate Neph1+/− and Neph1−/− mice (see Materials and Methods). To fully characterize the insertion site of the gene trap vector, we performed RT-PCR on Neph1+/− kidney RNA using primers to the 5′ end of the Neph1 cDNA and to the promoterless β-galactosidase–neomycin phosphotransferase (β-geo) fusion gene (5) introduced by the gene trap vector. These primers generated a 354-bp product from the predicted Neph1–β-geo fusion transcript (shown schematically in Fig. 3A). Sequence analysis of the PCR product confirmed that the ATG-containing Neph1 exon spliced into the splice acceptor site in the β-geo gene in the vector to produce a fusion transcript (Fig. 3B). Thus, the vector inserted in the intron just 3′ to the ATG-containing exon of Neph1.
To verify that the gene trap event ablated expression of normal Neph1 transcripts in Neph1+/− and Neph1−/− mice, we used a mouse cDNA fragment that spans the two Neph1 exons flanking the retrovirus integration site to probe a kidney RNA Northern blot (probe is shown in Fig. 3A). This probe should hybridize to mRNA transcripts originating from both the native mouse Neph1 promoter and the PGK promoter. When used to hybridize to Neph1+/− kidney RNA, this probe recognized the 9-kb native Neph1 transcript and a 4.5-kb transcript representing the fusion of Neph1 upstream exons (0.3 kb) to β-geo (4.2 kb) (Fig. 3C). When used to hybridize to Neph1−/− kidney RNA, this probe identified the 4.5-kb transcript but not the 9-kb native transcript, confirming the loss of normal Neph1 RNA due to retroviral insertion. The predicted 8.7-kb heterologous exon 1-Neph1 downstream fusion transcript, which must be present in the trapped ES cell line because it is the source of the OST sequence (Fig. 3A), should also hybridize to the probe in both Neph1−/− and Neph1+/− kidney samples. However, it would only be detectable in the Neph1−/− sample, as it is similar in size to the 9-kb native transcript present in the Neph1+/− sample and is therefore not distinguishable. Interestingly, it is absent from the Neph1−/− sample, suggesting that its expression is regulated differently in vivo and in ES cells in vitro. To address this possibility, we performed RT-PCR using primers derived from the heterologous and downstream Neph1 exons (Fig. 3A). The expected 485-bp RT-PCR product was present in RNA derived from ES cells but was negligible in RNA from Neph1+/− and Neph1−/− kidneys (Fig. 3D); all samples exhibited robust control PCR products using primers that amplified the mouse β-actin gene (not shown). A likely explanation for this discrepancy is that gene transcription initiating from a heterologous promoter internal to a retrovirus may be silenced in vivo (7, 16). This appears to be a general phenomenon because we confirmed the in vivo loss of both endogenous and 3′ fusion transcripts in dozens of other Omnibank gene-trapped mouse lines (B. Zambrowicz, unpublished observations).
Expression pattern of the Neph1 gene.
To identify cell types expressing NEPH1 in various mouse tissues, we exploited the activity of the β-geo protein expressed as part of the fusion transcript transcribed by the Neph1 promoter (Fig. 3A). In newborn Neph1+/− mice, kidneys showed focal LacZ staining in glomerular epithelium, mesangium, and proximal tubules (Fig. 4A) and in blastemal cells and collecting ducts (not shown). Staining was also noted in leptomeninges of the brain and in smooth muscle of liver, lung, and heart (not shown). In the adult, LacZ staining in kidney was limited to juxtamedullary glomeruli and proximal tubules (Fig. 4B and C). Some weak staining was also observed in ovarian follicular cells and rare cells of the red pulp in the spleen (not shown). In adult brain, β-geo was expressed by some Purkinje cells; by neurons of the small molecular layer and granule layer; by CA1 and CA4 in the hippocampus; and in the basal ganglia, brain stem, and cortex (not shown).
FIG. 4.
Neph1 expression as indicated by LacZ staining (blue) of kidneys from newborn (A) and adult (B and C) heterozygous mice. (A) Staining in the glomeruli (g, arrow) and proximal tubules (t, arrowhead) of a 1-day-old newborn pup. (B) Staining in the proximal tubules (t, arrowhead) of an adult kidney. (C) Staining in a juxtamedullary glomerulus (g, arrow) of an adult mouse kidney.
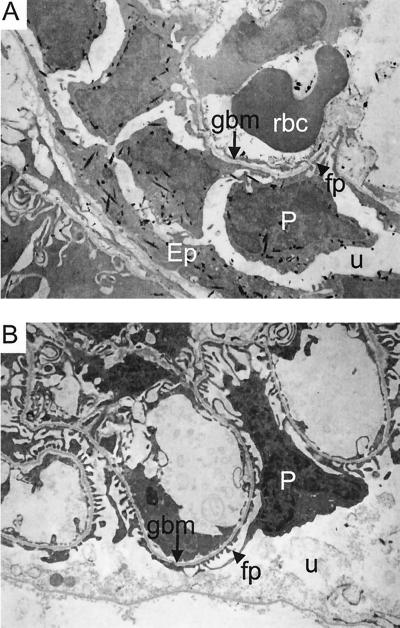
To confirm that the Neph1 promoter is active in glomerular podocytes, LacZ-stained kidney tissue from newborn Neph1+/+ and Neph1−/− mice was analyzed by electron microscopy. LacZ crystals were abundant in the cytoplasm of glomerular podocytes from Neph1−/− kidney (Fig. 5A) and were also present in tubular epithelial, vascular smooth muscle, and endothelial cells. Interestingly, foot processes of Neph1−/− podocytes were effaced along the glomerular basement membrane, suggesting heavy proteinuria (see below). In contrast, LacZ crystals were not detected in Neph1+/+ kidney and podocyte foot processes were not effaced (Fig. 5B).
FIG. 5.
Electron micrographs of LacZ-stained Neph1−/− kidney. (A) Electron micrograph depicting LacZ stain crystals (black specks) indicating expression of Neph1–β-geo fusion transcripts in the cytoplasm of podocytes (P) and parietal epithelium (Ep) of a Neph1−/− newborn mouse. The glomerular basement membrane (gbm) is marked with an arrow, and a podocyte foot process (fp) is depicted by an arrowhead. There is significant effacement of the podocyte foot processes in Neph1−/− mutant animals (see text and compare to panel B). The urinary space (u) and a red blood cell (rbc) are also noted. (B) Electron micrograph depicting no LacZ stain crystals in a Neph1+/+ kidney. The podocyte (P), glomerular basement membrane (gbm), foot processes (fp), and urinary space (u) are marked as in panel A.
Consequences of Neph1 gene disruption.
To determine the effects of loss of normal Neph1 transcripts, we intercrossed Neph1+/− mice to generate over 236 progeny. When these mice were genotyped at 10 to 12 days after birth, only 6 Neph1−/− mice were identified out of the 59 Neph1−/− pups expected. These six Neph1−/− pups were sickly and small but without edema; most were failing to thrive by 1 week of age and all six died between 3 and 8 weeks of age. Genotyping of an additional 48 pups at 1 to 3 days of age identified 13 (27%) as Neph1−/−. This expected Mendelian frequency at birth suggests that most Neph1−/− pups die in the 1- to 12-day postnatal period and not during embryogenesis.
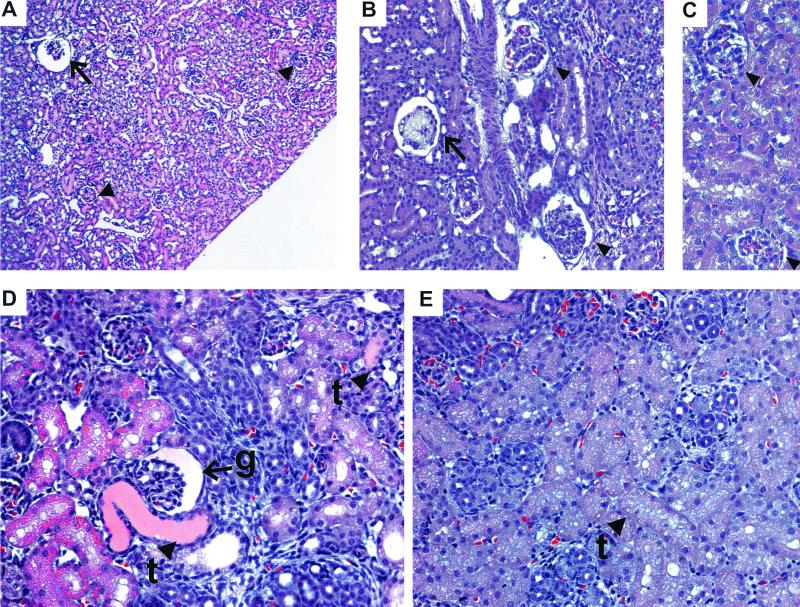
Urine from eight newborn (1 to 3 days of age) mice and one 3-week-old Neph1−/− mouse exhibited heavy proteinuria (five with 2,000 mg/dl; four with 300 mg/dl). Urine from 22 Neph1+/− and 11 Neph1+/+ age-matched littermates contained either trace protein or 30 mg of protein/dl. In general, kidneys from Neph1−/− pups appeared normal by gross examination. However, histopathological examination revealed a variety of glomerular and tubular abnormalities (Fig. 6). Three-week-old Neph1−/− kidneys exhibited dilated juxtamedullary glomeruli (Fig. 6A), diffuse mesangial hypercellularity, and increased mesangial matrix, which was more marked focally in the juxtamedullary region, and occasional mesangiolysis (Fig. 6B). Kidneys from newborn Neph1−/− mice showed occasional cystic glomeruli, proximal tubules focally dilated and lined by epithelial cells that in some cases contained vacuoles and/or protein droplets, and protein-filled tubules (Fig. 6D). At necropsy, there was no gross or microscopic evidence of edema in kidney or any other tissue examined. Neph1−/− neonates had less milk in their stomachs than their littermates, suggesting poor feeding. However, histological examination failed to demonstrate abnormalities in any organ other than the kidney (data not shown).
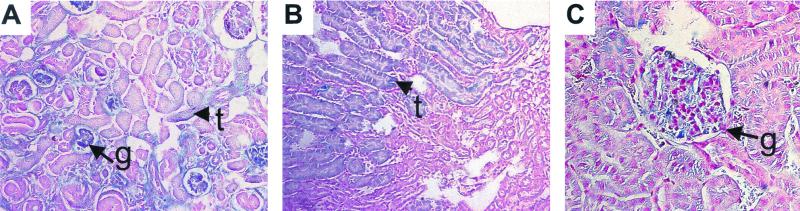
FIG. 6.
Kidney abnormalities in Neph1−/− mice. (A) Dilated (cystic) juxtamedullary glomerulus (arrow) in a 3-week-old Neph1−/− mouse. Cortical glomeruli (arrowheads) look normal. (B) Mesangiolysis (arrow) and mesangial hypercellularity and increased matrix (arrowheads) in glomeruli from 3-week-old Neph1−/− kidney. (C) Normal glomeruli (arrowheads) from a control 3-week-old littermate. (D) Protein-filled tubules (t, arrowheads) and cystic glomerulus (g, arrow) from a newborn Neph1−/− mouse. (E) Control newborn mouse kidney showing normal tubules (t, arrowhead).
DISCUSSION
NEPH1 and NEPHRIN share a number of characteristics: (i) both proteins are expressed in glomerular podocytes; (ii) both contain multiple extracellular Ig domains of the C2 type; (iii) both contain domains near their single transmembrane region, which should allow interaction with integrins or other membrane-bound proteins; (iv) absence of either protein results in effacement of podocyte foot processes (11; this study); and (v) absence of either protein results in proteinuria in the newborn period (11; this study). The histology of Neph1−/− kidneys is consistent with urinary protein loss, since kidneys from newborn Neph1−/− mice showed effacement of podocyte foot processes and occasional protein-filled tubules. In addition, 3-week-old Neph1−/− mouse kidneys exhibited diffuse mesangial hypercellularity and increased mesangial matrix, which were more marked in juxtamedullary glomeruli, a pattern noted in some forms of nephrotic syndrome as an intermediate step in the progression of the renal lesion to glomerular scarring. These findings suggest that NEPH1 may play an important role in maintaining the structure of the protein filtration barrier in the slit diaphragm, similar to the role proposed for NEPHRIN (17).
Despite the similarities between NEPH1 and NEPHRIN, recent studies of mice with disruption of the Nphs1 gene encoding murine NEPHRIN shed light on the relative contribution of these two proteins in maintaining the filtration function of the slit diaphragm. Nphs1−/− mice are born with the expected frequency of 25% but rapidly develop heavy proteinuria and edema; death occurs within 24 h (17). The widespread enlargement of Bowman's spaces, dilated tubules, effacement of podocyte foot processes, and absence of slit diaphragms in kidneys from newborn Nphs1−/− mice constitute a more abnormal histological phenotype than that noted in kidneys from Neph1−/− newborn mice, consistent with the more severe clinical phenotype of Nphs1−/− mice. This suggests that NEPHRIN plays a more crucial role than NEPH1 in maintaining the integrity of the podocyte slit diaphragm.
Similar to NEPH1, NEPHRIN is expressed in brain and other extrarenal tissues (17) and their homolog IRREGULAR CHIASM is expressed in specialized neural tissues of D. melanogaster (12). IRREGULAR CHIASM is known to participate in cell-cell interactions by mechanisms that include both homophilic and heterophilic interactions (15, 19). These data suggest a possible role for NEPH1 in cell-cell interactions in each of the tissues where it is expressed and raise the possibility that NEPH1 may interact with other NEPH1 proteins or in a heterophilic interaction with NEPHRIN, in the slit diaphragm between podocytes.
Congenital nephrotic syndrome caused by the absence of functional NEPHRIN is rapidly fatal to mice but is less grave in humans (11). This suggests that lack of functional NEPH1, which is rapidly fatal to mice, might also be less grave in humans and might present as nephrotic syndrome. The human NEPH1 gene apparently maps to 1q21-25, since a genomic clone containing NEPH1 exon sequence maps to this interval (GenBank accession no. AL139010). Although familial forms of nephrotic syndrome secondary to focal segmental glomerulosclerosis also map to the long arm of chromosome 1, most of these families carry mutations in the NPHS2 gene encoding the glomerular protein PODOCIN (3, 14). However, it may be advantageous to screen the NEPH1 locus for inactivating mutations in familial forms of nephrotic syndrome that do not exhibit mutations in NPHS2.
ACKNOWLEDGMENTS
We thank June Wingert and David Tran for expert technical assistance; Kate Holt, Lance Ishimoto, and Peter Sefarian for reviewing the manuscript; and Jim Piggott for helpful discussions.
REFERENCES
- 1.Anonymous. Guide for the care and use of laboratory animals. Bethesda, Md: National Institutes of Health; 1985. [Google Scholar]
- 2.Bancroft J, Stevens A. Theory and practice of histological techniques. 2nd ed. New York, N.Y: Churchill Livingstone; 1982. [Google Scholar]
- 3.Boute N, Gribouval O, Roselli S, Benessy F, Lee H, Fuchshuber A, Dahan K, Gubler M C, Niaudet P, Antignac C. NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat Genet. 2000;24:349–354. doi: 10.1038/74166. . (Erratum, 25:125.) [DOI] [PubMed] [Google Scholar]
- 4.Bycroft M, Bateman A, Clarke J, Hamill S J, Sandford R, Thomas R L, Chothia C. The structure of a PKD domain from polycystin-1: implications for polycystic kidney disease. EMBO J. 1999;18:297–305. doi: 10.1093/emboj/18.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 6.Kestila M, Lenkkeri U, Mannikko M, Lamerdin J, McCready P, Putaala H, Ruotsalainen V, Morita T, Nissinen M, Herva R, Kashtan C E, Peltonen L, Holmberg C, Olsen A, Tryggvason K. Positionally cloned gene for a novel glomerular protein—nephrin—is mutated in congenital nephrotic syndrome. Mol Cell. 1998;1:575–582. doi: 10.1016/s1097-2765(00)80057-x. [DOI] [PubMed] [Google Scholar]
- 7.Kuriyama S, Sakamoto T, Kikukawa M, Nakatani T, Toyokawa Y, Tsujinoue H, Ikenaka K, Fukui H, Tsujii T. Expression of a retrovirally transduced gene under control of an internal housekeeping gene promoter does not persist due to methylation and is restored partially by 5-azacytidine treatment. Gene Ther. 1998;5:1299–1305. doi: 10.1038/sj.gt.3300738. [DOI] [PubMed] [Google Scholar]
- 8.Luna L. Manual of histologic staining methods of the Armed Forces Institute of Pathology. 3rd ed. New York, N.Y: Blakiston Division, McGraw-Hill; 1968. [Google Scholar]
- 9.Pierschbacher M D, Ruoslahti E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature. 1984;309:30–33. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 10.Puri M C, Rossant J, Alitalo K, Bernstein A, Partanen J. The receptor tyrosine kinase TIE is required for integrity and survival of vascular endothelial cells. EMBO J. 1995;14:5884–5891. doi: 10.1002/j.1460-2075.1995.tb00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Putaala H, Soininen R, Kilpelainen P, Wartiovaara J, Tryggvason K. The murine nephrin gene is specifically expressed in kidney, brain and pancreas: inactivation of the gene leads to massive proteinuria and neonatal death. Hum Mol Genet. 2001;10:1–8. doi: 10.1093/hmg/10.1.1. [DOI] [PubMed] [Google Scholar]
- 12.Ramos R G, Igloi G L, Lichte B, Baumann U, Maier D, Schneider T, Brandstatter J H, Frohlich A, Fischbach K F. The irregular chiasm C-roughest locus of Drosophila, which affects axonal projections and programmed cell death, encodes a novel immunoglobulin-like protein. Genes Dev. 1993;7:2533–2547. doi: 10.1101/gad.7.12b.2533. [DOI] [PubMed] [Google Scholar]
- 13.Ruotsalainen V, Ljungberg P, Wartiovaara J, Lenkkeri U, Kestila M, Jalanko H, Holmberg C, Tryggvason K. Nephrin is specifically located at the slit diaphragm of glomerular podocytes. Proc Natl Acad Sci USA. 1999;96:7962–7967. doi: 10.1073/pnas.96.14.7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salomon R, Gubler M C, Niaudet P. Genetics of the nephrotic syndrome. Curr Opin Pediatr. 2000;12:129–134. doi: 10.1097/00008480-200004000-00008. [DOI] [PubMed] [Google Scholar]
- 15.Schneider T, Reiter C, Eule E, Bader B, Lichte B, Nie Z, Schimansky T, Ramos R G, Fischbach K F. Restricted expression of the irreC-rst protein is required for normal axonal projections of columnar visual neurons. Neuron. 1995;15:259–271. doi: 10.1016/0896-6273(95)90032-2. [DOI] [PubMed] [Google Scholar]
- 16.Stewart C L, Schuetze S, Vanek M, Wagner E F. Expression of retroviral vectors in transgenic mice obtained by embryo infection. EMBO J. 1987;6:383–388. doi: 10.1002/j.1460-2075.1987.tb04766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tryggvason K. Unraveling the mechanisms of glomerular ultrafiltration: nephrin, a key component of the slit diaphragm. J Am Soc Nephrol. 1999;10:2440–2445. doi: 10.1681/ASN.V10112440. [DOI] [PubMed] [Google Scholar]
- 18.Van Adelsberg J S, Frank D. The PKD1 gene produces a developmentally regulated protein in mesenchyme and vasculature. Nat Med. 1995;1:359–364. doi: 10.1038/nm0495-359. [DOI] [PubMed] [Google Scholar]
- 19.Venugopala Reddy G, Reiter C, Shanbhag S, Fischbach K F, Rodrigues V. Irregular chiasm-C-roughest, a member of the immunoglobulin superfamily, affects sense organ spacing on the Drosophila antenna by influencing the positioning of founder cells on the disc ectoderm. Dev Genes Evol. 1999;209:581–591. doi: 10.1007/s004270050292. [DOI] [PubMed] [Google Scholar]
- 20.Zambrowicz B P, Friedrich G A, Buxton E C, Lilleberg S L, Person C, Sands A T. Disruption and sequence identification of 2,000 genes in mouse embryonic stem cells. Nature. 1998;392:608–611. doi: 10.1038/33423. [DOI] [PubMed] [Google Scholar]
- 21.Zheng B, Sage M, Cai W W, Thompson D M, Tavsanli B C, Cheah Y C, Bradley A. Engineering a mouse balancer chromosome. Nat Genet. 1999;22:375–378. doi: 10.1038/11949. [DOI] [PubMed] [Google Scholar]