Abstract
Four dietary polyphenols, theaflavin (TF1), theaflavin-3-gallate (TF2A), theaflavin-3′-gallate (TF2B) and theaflavin-3,3′-digallate (TF3), have been isolated from black tea, and their effects on oxidative phosphorylation and superoxide production in a model system (E. coli) have been examined. The esterified theaflavins were all potent inhibitors of the membrane-bound ATP synthase, inhibiting at least 90% of the activity, with IC50 values in the range of 10-20 μM. ATP-driven proton translocation was inhibited in a similar fashion, as was the purified F1-ATPase, indicating that the primary site of inhibition was in the F1 sector. Computer modeling studies supported this interpretation. All four theaflavins were also inhibitory towards the electron transport chain, whether through Complex I (NDH-1) or the alternative NADH dehydrogenase (NDH-2). Inhibition of NDH-1 by TF3 appeared to be competitive with respect to NADH, and this was supported by computer modeling studies. Rates of superoxide production during NADH oxidation by each dehydrogenase were measured. Superoxide production was completely eliminated in the presence of about 15 μM TF3, suggesting that inhibition of the respiratory chain by theaflavins does not contribute to superoxide production.
Keywords: Theaflavin, Superoxide, ATP synthase, Complex I, NDH-1, NDH-2, Oxidative phosphorylation, Inhibitor, Respiratory chain
Introduction
Tea, a widely consumed beverage in the world, has long been considered to be beneficial to health (for reviews see [1-3]). Tea is distinguished by high levels of polyphenols, which are thought to be primarily responsible for any benefits to health. (For a comprehensive review see [4]). Green tea polyphenols consist primarily of catechins, mainly including (−) epicatechin (EC), (−) epicatechin gallate (ECG), (−) epigallocatechin (EGC), (−) epigallocatechin gallate (EGCG), (+) gallocatechin, and (+) catechin [5]. During the production of black tea, the catechins are enzymatically oxidized to produce theaflavins, which are orange or orange-red in color and possess a benzotropolone skeleton. The major theaflavins in black tea are theaflavin (TF1), theaflavin-3-gallate (TF2A), theaflavin-3′-gallate (TF2B), and theaflavin-3,3′-digallate (TF3). Theaflavins contribute importantly to properties of black tea including its color, ‘mouthfeel’ and extent of tea cream formation [6]. The oxidation process also forms larger, and less well characterized, polymers, referred to as thearubigins. The chemistry of these compounds has been reviewed recently [7]. The chemical structures of the theaflavins and EGCG are shown in Fig. 1.
Figure 1.
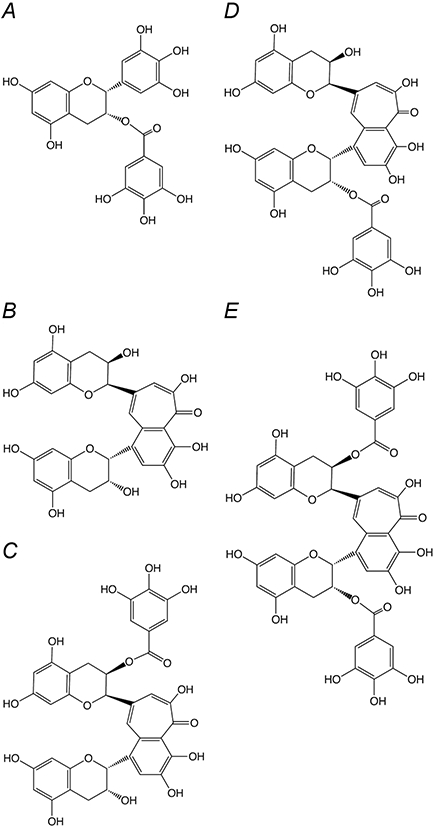
Chemical Structure of Polyphenols used in this Study. (A) (−) epigallocatechin gallate (EGCG) (B) Theaflavin (TF1) (C) Theaflavin-3-gallate (TF2A) (D) Theaflavin-3′-gallate (TF2B) (E) Theaflavin-3,3′-digallate (TF3)
The potential health benefits associated with tea consumption have been partially attributed to the antioxidative property of tea polyphenols [8]. Reaction mechanisms and products of reactions with peroxyl radicals [8, 9], hydrogen peroxide [10, 11] and superoxide [12, 13] have been investigated. The antioxidant potential of theaflavins in particular has been examined in vitro [14, 15]. In mammals, most superoxide is produced in the mitochondria, and principally from the electron transport chain [16]. Dioxygen can be reduced to superoxide at both Complex I and Complex III, and this preferentially occurs when key components of the electron transport chain are reduced. Many, but not all, inhibitors of the electron transport chain cause an increase in the rate of superoxide production [17]. Inhibitors of the ATP synthase can have the same effect, since when ATP synthesis is inhibited, the proton motive force builds up, and the electron transport chain slows down [18].
Several plant polyphenols, including tea catechins, have been shown to be inhibitors of the mitochondrial ATP synthase [19-21]. In the lab of Walker, the bovine F1 sector of the ATP synthase was co-crystallized with three different polyphenols: resveratrol, piceatannol, and quercetin [22, 23]. All three were bound inside the enzyme along side the rotary shaft of the gamma subunit, and not at the nucleotide binding sites. The ATP synthase from E. coli, known to be a valuable model for the mitochondrial enzyme, was also found to be inhibited by many similar compounds. Treatment with piceatannol, morin, silymarin, baicalein, silibinin, rimantadin, amantidin, and epicatechin resulted in complete inhibition, while resveratrol, quercetin, quercetrin, quercetin-3-β-D-glucoside, hesperidin, chrysin, kaempferol, diosmin, apigenin, genistein, and rutin were partially inhibitory, in the range of 40–80% [24, 25].
In this paper we have extended previous work by showing that polyphenols from black tea, the theaflavins, can also inhibit the ATP synthase from E. coli, and that they are more potent than is EGCG. In similar fashion, these compounds are also inhibitory towards Complex I (NDH-1) of the electron transport chain. No evidence of increased production of superoxide was found in the presence of these inhibitors.
Materials and methods
Chemicals
EGCG, NADH, deamino-NADH, capsaicin, hexamine ruthenium chloride, and other chemicals were from Sigma-Aldrich (St. Louis, MO USA). ATP was from Roche (Indianapolis, IN USA). ACMA (9-amino 3-chloro 2-methoxy acridine) and MitoSOX were purchased from Invitrogen (Carlsbad, CA USA). Theaflavins were prepared as described previously [15]
Strains used
The wild type strain was 1100 [26]. BA14 is a derivative of strain 1100 with a deletion of the nuo genes, and so lacks all NDH-1 activity[27]. MWC215 contains a knockout of the ndh gene, and so lacks all NDH-2 activity [28]. It was generously provided by Dr. Bob Gennis (U. Illinois, Urbana IL USA). Strain DK8 with plasmid pFV2 [29] was used for purification of F1.
Purification and preparations
1100 cells (wild type) or BA14 cells were grown in LB medium containing 1% tryptone, 0.5% yeast extract and 0.5 % NaCl at 37 °C. DK8/pFV2 cells were grown in LB medium supplemented with ampicillin at 0.05 g/L. MWC215 cells were grown in rich medium containing 3% tryptone, 1.5% yeast extract, 0.15% NaCl, and 1% (v/v) glycerol at 37 °C, supplemented with chloramphenicol at 0.05 g/L. Membrane vesicles for assays of ATP synthase and purified F1 were isolated as described by Volkov, et al [30]. F1-ATPase was purified essentially as described in Wise [31], except that the final purification of the ATPase was achieved by Ni-NTA chromatography. The enzyme was eluted at 180 mM imidazole. Membrane vesicles for assays of NDH-1 and NDH-2 were prepared at pH 6.0 as described previously [27].
Functional assays
The rate of ATP hydrolysis was measured by quantifying the release of inorganic phosphate using a colorimetric assay. After the equilibration of 200 μg of membrane vesicles from wild type strain 1100 or 40 μg of purified F1 with test compounds in 500 μl buffer containing 5 mM MgCl2, 50mM Tris (pH 7.5) for 5 min at room temperature, the assay was started by adding 5 mM ATP and 4 mM MgCl2. The reaction was stopped after 3 min by addition of 250 μl of 10% SDS and 250 μl of color reagent [32]. The total phosphate was measured spectrophotometrically at 700 nm after 10 min. Phosphate at zero time point was determined by adding 10% SDS to the ATPase cocktail before adding the protein.
ATP-dependent ACMA fluorescence quenching was measured in 2 ml of buffer containing 50 mM MOPS and 10 mM MgCl2 (pH 7.3). After the equilibration of 200 μg of membrane vesicles from wild type strain 1100 with tested compounds for 5 min, the reaction was initiated by addition of 0.1 mM of ATP and 1 μM of the dye under constant stirring at 37 °C, and stopped by addition of 3 μM FCCP . Fluorescence was excited at 410 nm and monitored at 490 nm using a Perkin-Elmer LS-5B luminescence spectrometer [29].
NADH oxidase activity in membrane vesicles was measured using a Beckman Coulter DU800 spectrophotometer by following NADH oxidation at 340 nm (ε=6.22 mM−1 cm−1) at 30 °C. For the NADH oxidase activity of NDH-1 from strain 1100 and NDH-2 from strain BA14, the reaction medium (1 ml) contained 50 mM MOPS (pH 7.3), 10 mM MgCl2, and 200 μg of membrane vesicles. For the NADH- HAR (hexamine ruthenium) reductase activity, the reaction medium (1 ml) contained 50 mM MOPS (pH 7.3), 10 mM MgCl2, 200 μg of membrane vesicles from strain MWC215, 10 mM KCN, and 1 mM HAR. The reaction was started by adding 75 μM deamino-NADH (for strain 1100) or 250 μM NADH (for strain BA14 or MWC215) after the equilibration of membrane vesicles with the test compounds for 5 min. The plotted concentrations represent the final concentration in the assay chamber.
Inhibition kinetics
The modes of inhibition and inhibition constants were determined by means of Dixon plots [33] and Cornish-Bowden plots [34], in which the reciprocal velocity, 1/v or s/v was plotted against the inhibitor concentration, i, at three values of s, the substrate concentration. Other details of the assays were indicated in the Figure legends.
Detection of superoxide with MitoSOX
Generation of superoxide by membrane vesicles from strain MWC215 or BA14 was estimated using the MitoSOX™ Red superoxide indicator in opaque-walled 96-well tissue culture plates. The reaction medium (0.2 ml) in each well contained 50 mM MOPS (pH 7.3), 10 mM MgCl2, 200 μg of membrane vesicles. The reaction was started by adding 5 μM MitoSOX™ reagent and 1.25 mM NADH after the incubation of membrane vesicles with tested compounds for 5 min. SOD (150 units) was added in the control experiments. The plates protected from light were incubated for 15 min at 30 °C. The increase in fluorescence was monitored on a Molecular Devices Gemini XPS plate reader (Sunnyvale, CA) at 510/580 nm.
Under all assay conditions described above, without the addition of membrane vesicles, the five polyphenols tested had no relevant effects on the absorption or fluorescence of the indicators used.
Molecular docking
ChemDraw ultra 8.0 software [Chemical Structure Drawing Standard; Cambridge Soft corporation, USA (2003)] was used for construction of compounds which were converted to 3D structures using Chem3D ultra 8.0 software and the constructed 3D structures were energetically minimized by using VEGA 2.3.2 [35, 36]. Bovine F1-ATPase (Protein Data Bank code: 1efr [37], resolution: 3.10 Å, co-crystallized with efrapeptin) and the hydrophilic domain of respiratory complex I from Thermus thermophilus (pdb code: 3i9v [38], resolution: 3.10 Å) were obtained from the Protein Data Bank (http://www.rcsb.org). The files were edited by using Chimera software [39]. All the water and inhibitors were removed. AutoDock 4.2 was employed for all docking calculations [40]. Polar hydrogen atoms and Gasteiger charges were added to the protein (receptor) model prior to calculation of grid maps using a grid box with a npts (number of points in xyz) of 62–62–62 Å box. For F1-ATPase docking, the box spacing was 1.0 Å and grid center was designated at dimensions (x, y, z): 107.703, 74.005 and 73.815. For complex I docking, the box spacing was 0.7 Å and grid center was designated at dimensions (x, y, z): −29.238, 8.223 and 37.853. Ten runs were generated by using Lamarckian genetic algorithm searches. Default settings were used with a population size of 150, a maximum number of 2.5 × 106 energy evaluations, and a maximum number of 2.7 × 104 generations. A mutation rate of 0.02 and a crossover rate of 0.8 were chosen. The best ligand–receptor structure for each of the docking simulation was chosen based on the lowest energy.
Results
Inhibition of the ATP synthase
The four major theaflavin compounds: theaflavin (TF1), theaflavin-3-gallate (TF2A), theaflavin-3′-gallate (TF2B), and theaflavin-3, 3′-digallate (TF3) were tested as inhibitors of the membrane-bound F1Fo-ATPase, along with (−) epigallocatechin gallate (EGCG). Several catechins, including EGCG have been previously shown to be inhibitors of the rat liver mitochondrial ATPase [20], but the theaflavins are somewhat larger than EGCG (see Fig. 1), and therefore might not be capable of binding to the same sites. Membrane vesicles were prepared from wild type cells, and ATP hydrolysis was measured in the presence of increasing amounts of the polyphenols. The results are summarized in Table 1. The theaflavins with gallate esters (TF2A, TF2B, TF3) all inhibited to an extent of 90-95 % with IC50 values in the range 10-20 μM. TF1 and EGCG were somewhat less potent, with IC50 values of about 60 and 30 μM respectively. Complete inhibition data are shown in Supplementary Fig. 1. Previous work has indicated that the sites of inhibition by polyphenols are in the F1 sector of the ATP synthase [22]. Since the theaflavins are twice as large as many other polyphenols, it was possible that they might bind to other regions of the enzyme. For that reason, isolated F1-ATPase was examined as well. These parallel results are also presented in Table 1 and Supplementary Fig. 1. In each case, a very similar pattern of results was obtained, except that the IC50 values were about 10-fold lower using the isolated F1-ATPase.
Table 1.
Inhibition of ATPase Activity by Theaflavins
| Membrane-bound ATPase Activitya | F1-ATPase Activityb | |||
|---|---|---|---|---|
| Inhibitor | IC50 | Maximum inhibition | IC50 | Maximum inhibition |
| (μM)c | (%)c | (μM)c | (%)c | |
| TF1 | 60 | 85 | 4.0 | 85 |
| TF2A | 20 | 95 | 3.0 | 90 |
| TF2B | 15 | 95 | 1.5 | 95 |
| TF3 | 10 | 90 | 0.7 | 90 |
| EGCG | 30 | 95 | 4.5 | 90 |
Membrane-bound ATPase activity was about 0.7 μmoles ATP/min/mg protein
F1-ATPase activity was about 1.8 μmoles ATP/min/mg
IC50 concentrations and maximum inhibition % were determined from two replicates of 5-6 concentrations of inhibitor. The data are displayed in Supplementary Figure1.
Proton translocation requires the coupling of ATP hydrolysis to the proton translocation apparatus of the membrane-bound Fo sector. This can be detected by monitoring the fluorescence quenching of an acridine dye, ACMA. In this assay, the rate and extent of fluorescence quenching is indicative of the generation of a proton gradient across the membrane. Each of the four theaflavins was tested as an inhibitor of ATP-driven proton translocation, using membrane vesicles, and the results are presented in Fig. 2. In each case, the inhibition of proton translocation seemed to follow that of the inhibition of ATP hydrolysis (Table 1), and so there was no evidence for additional inhibitory binding sites in the membrane sector of the enzyme. The binding site for several polyphenols in the F1-ATPase has been well-established by the co-crystallization of the bovine enzyme with resveratrol, piceatannol, and quercetin, each binding to the same position inside the enzyme near the membrane-distal end [22]. Considering the size of the theaflavin polyphenols, especially the digallate ester (TF3), it seemed unlikely that they could bind to the same site as the smaller flavonol quercetin. Computer modeling and docking studies were carried out with each of the theaflavins, along with EGCG, to see where they might bind in the F1-ATPase. Using the bovine F1-ATPase structure (Protein Data Bank file 1efr) [37], stripped of ligands as the template, quercetin was docked to a position very close to the actual binding site discovered by co-crystallization studies (see Fig. 3A). In contrast, TF3 was docked to a totally different site, closer to the membrane, and overlapping with the actual binding site of efrapeptin [37] (see Fig. 3B).
Figure 2.
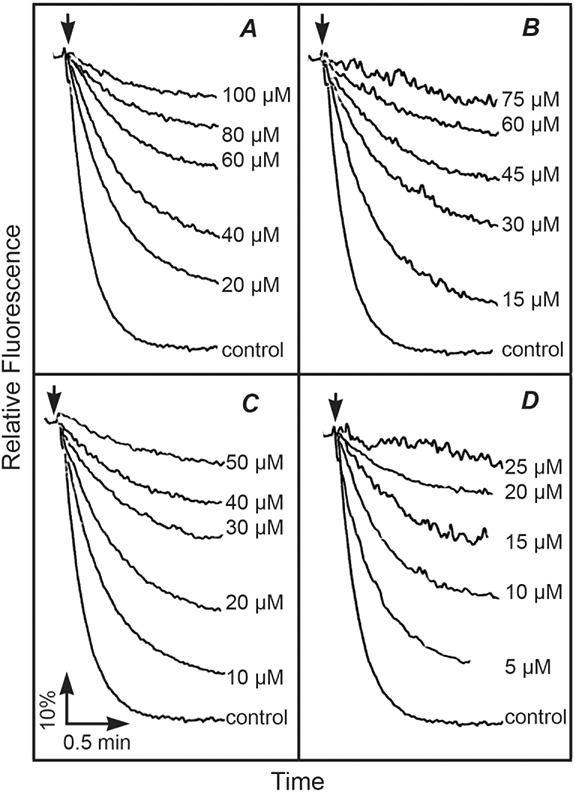
Inhibition of ATP-driven Proton Translocation by Polyphenols. Proton translocation by membrane vesicles is indicated by the quenching of the fluorescence of ACMA upon the addition of ATP. Membranes without the addition of polyphenols are indicated by CK. In each case, the fluorescence quenching could be eliminated by the addition of FCCP (carbonyl cyanide p-(trifluoromethoxy)phenylhydrazone), a protonophore (results not shown). (A) TF1, (B) TF2A, (C) TF2B, (D) TF3. The final concentrations of inhibitors are indicated in the panels. The results shown are typical from 2 assays of 2 different preparations of membranes.
Figure 3.
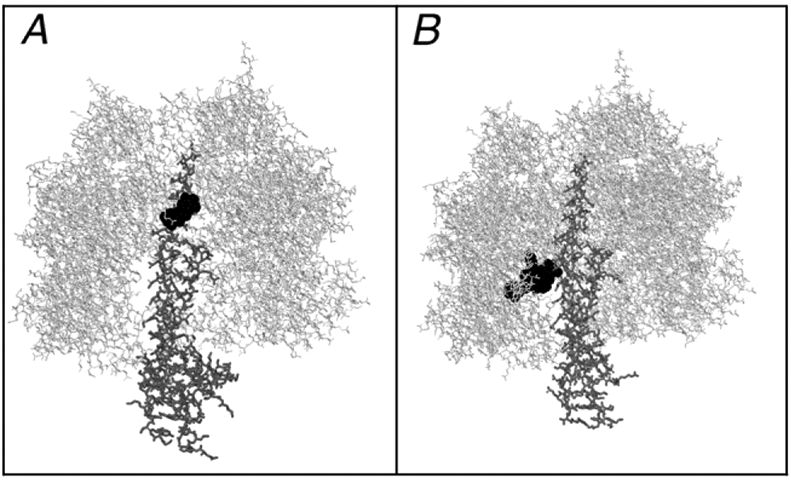
Comparison of Actual and Predicted Inhibitor Binding Sites in Bovine F1-ATPase. F1 is shown in gray wireframe, the gamma subunit is in thick wireframe, and the inhibitors are shown in black spacefilling. (A) The model shown is from Protein Data Bank file 2jj2. (B) The results from a docking study of TF3 and the F1-ATPase from Protein Data Bank file 1efr are shown. In both cases the inhibitors are bound to the gamma subunit, but the TF3 is docked much lower, and below the nucleotide binding sites. The TF3 site in panel B is accessible from the bottom of the structure, as shown, rather than from the top, as in panel A.
Inhibition of Electron Transport
Since the polyphenol resveratrol has previously been shown to inhibit the mitochondrial respiratory chain [41, 42], the theaflavins were also tested as inhibitors of the electron transport chain in membrane vesicles. E. coli has two distinct NADH dehydrogenases in its plasma membrane [28, 43]. Both use the oxidation of NADH to reduce ubiquinone, which is re-oxidized by one of two quinol oxidases, bd and bo [44]. NDH-1 is homologous to the mitochondrial Complex I. It is a multi-subunit, proton translocating enzyme, and is similar to the ATP synthase, in that it has two large sectors, one embedded in the membrane, and another peripheral to it. The peripheral sector, binds NADH, while the membrane sector is involved with ion translocation. In contrast, NDH-2 is a single polypeptide that is monotopically bound to the membrane and does not translocate ions. In E. coli, they can be distinguished by the use of strains in which one enzyme is knocked out genetically, or by the use of deamino-NADH, which only NDH-1 is able to use. Each NADH dehydrogenase was tested with the four theaflavins and EGCG, and the results of the inhibition are presented in Table 2. Complete inhibition data are shown in Supplementary Fig. 2. For NDH-1 the IC50 values for the 5 polyphenols range from about 10-50 μM, and the inhibition reaches 90-95%. In contrast, the theaflavins inhibit NDH-2 with lower IC50 values in the range of 1-4 μM, while the extent of inhibition is 75-95%. However, the IC50 for EGCG is about the same with respect to both NADH dehydrogenases. Clearly, the inhibitors have different targets in these two respiratory chains.
Table 2.
Inhibition of NADH Oxidase Activity by Theaflavins
| NDH-1 Deamino-NADH oxidasea | NDH-2 NADH oxidaseb | |||
|---|---|---|---|---|
| Inhibitor | IC50 | Maximum inhibition | IC50 | Maximum inhibition |
| (μM)c | (%)c | (μM)c | (%)c | |
| TF1 | 45 | 95 | 3.0 | 75 |
| TF2A | 15 | 95 | 3.5 | 95 |
| TF2B | 8 | 95 | 2.5 | 90 |
| TF3 | 7 | 90 | 1.5 | 95 |
| EGCG | 20 | 90 | 20 | 65 |
NDH-1 activity using deamino-NADH was about 0.5 μmoles deamino-NADH/min/mg protein
NDH-2 activity using NADH was about 0.25 μmoles NADH/min/mg protein
IC50 concentrations and maximum inhibition % were determined from two replicates of 5-6 concentrations of inhibitor. The data are displayed in Supplementary Figure2.
Next, the modes of inhibition by TF3 were examined, by varying the amounts of NADH and TF3, and these results are presented in Fig. 4. Both Dixon [33] plots (1/v vs. s) and Cornish-Bowden [34] plots (s/v vs. s) are shown, and the assays were done with both a wild type strain, using deamino-NADH, and with a mutant strain lacking NDH-1. The results are consistent with competitive inhibition, since the lines are parallel in the Cornish-Bowden plot, and intersect in the upper left quadrant in the Dixon plot. The Ki can be estimated to be about 0.4 μM from the wild type strain, and about 1 μM from the mutant strain. In contrast, NDH-2 shows uncompetitive behavior under inhibition by TF3, as shown in Fig. 5. The Dixon plots are parallel lines, and the Cornish-Bowden lines intersect in the upper left quadrant. From these plots the Ki′ can be estimated to be 0.8 μM.
Figure 4.
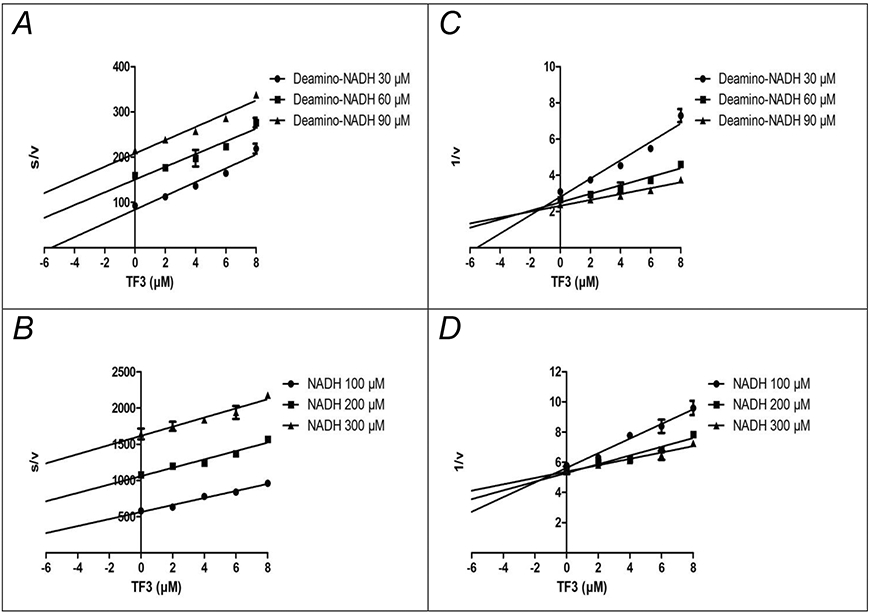
Inhibition of NDH-1 by TF3 is Competitive with Respect to NADH. NADH oxidase activity was measured with deamino-NADH using wild type membranes (1100) (panels A, C), and with NADH using strain MWC215, which lacks NDH-2 activity (panels B, D). Assays were carried out with 3 different concentrations of deamino-NADH or NADH, with increasing amounts of TF3 added. The results are plotted according to Cornish-Bowden (panels A, B) or Dixon (panels C, D). The units of s/v are μM TF3 (μmoles NADH/min/mg protein) −1, and the units of 1/v are (μmoles NADH/min/mg protein) −1. Each point shown represents the mean of two measurements.
Figure 5.
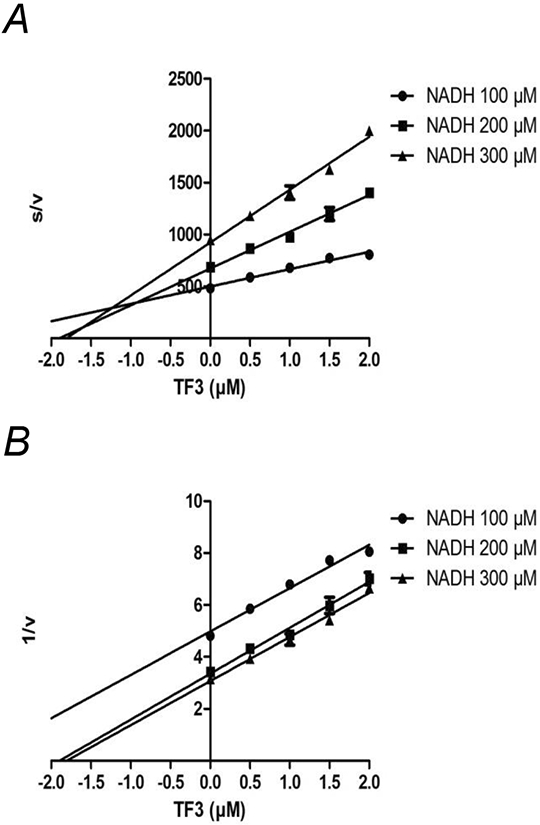
Inhibition of NDH-2 by TF3 is Uncompetitive with Respect to NADH. NADH-oxidase activity was measured with NADH using strain BA14, which lacks NDH-1 activity. Assays were carried out with 3 different concentrations of NADH, with increasing amounts of TF3 added. The results are plotted according to Cornish-Bowden (panels A) or Dixon (panels B). The units of s/v are μM TF3 (μmoles NADH/min/mg protein) −1, and the units of 1/v are (μmoles NADH/min/mg protein) −1. Each point shown represents the mean of two measurements.
To support the finding that the NDH-1 was competitively inhibited by TF3, computer modeling studies were carried out using the peripheral sector of the Thermus thermophilus Complex I (Protein Data Bank file 3i9v) [38]. Results showed that the highest affinity binding occurred on the surface of the enzyme at a site that partially overlapped with the NADH binding site. In addition, the activity of NDH-1 was assayed by the use of an artificial electron acceptor, hexamine ruthenium (HAR), which is thought to draw off electrons before they reach the membrane sector. In this case, using TF3 as the inhibitor, the results showed uncompetitive inhibition (results not shown), where the Dixon plots have parallel lines, and the Cornish-Bowden lines intersect in the upper left quadrant. The Ki′ was estimated to be 0.16 μM. This appeared to be in conflict with the earlier finding that TF3 inhibits competitively at the NADH site.
Modulation of the Rate of Superoxide Production
Inhibitors of electron transport typically cause an increase in superoxide production during the consumption of NADH. Since TF3 was found to be a rather potent inhibitor of both NDH-1 and NDH-2, rates of superoxide production were measured in membrane vesicles with increasing levels of TF3. The results, presented in Fig. 6 showed that NADH oxidation by NDH-1 caused higher rates of superoxide production than by NDH-2. Addition of superoxide dismutase reduced the fluorescence, and so indicated the amount that was due to superoxide. When TF3 was added at levels in the range of the IC50 value, superoxide did not increase, but rather decreased. This shows that while TF3 is an inhibitor of the electron transport chain in E. coli membranes, it does not cause an increase in superoxide production.
Figure 6.
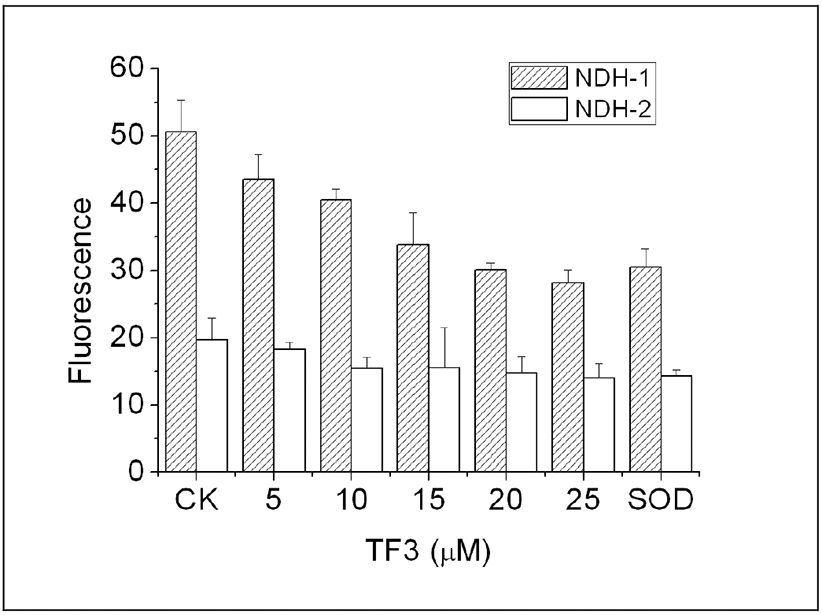
The Effect of Increasing Levels of TF3 on Superoxide Production by NDH-1 and NDH-2. Superoxide was measured by the increase in fluorescence of the Mitosox reagent, in the absence of NADH (CK) and in the presence of superoxide dismutase (SOD), which reflect the background fluorescence. To differentiate between NDH-1 and NDH-2, strains MWC215 was used for NDH-1 (shaded bars) and strain BA14 was used for NDH-2 (open bars). The results shown are the mean of three measurements with the error bars indicating the standard deviation.
Discussion
Flavonoid polyphenols are dietary compounds from plants that are known to interact directly with enzymes. The ATP synthase was previously found to be inhibited by several simple polyphenols, including resveratrol, quercetin, and EGCG [19, 20, 23-25]. In this report theaflavins, a class of polyphenols found exclusively in black tea, were also shown to be inhibitors of the ATP synthase. The four theaflavins tested varied in size according to the presence of 0, 1, or 2 gallate esters. The apparent affinities, as indicated by IC50 values, had a four-fold range of values, and correlated well with overall size. The importance of the gallate group is also suggested by the inhibitory properties of EGCG. It is smaller than TF1, but contains both a gallate ester and a galloyl group, and has an IC50 value that is similar to that of TF1. Furthermore, in another inhibition study of the E. coli ATP synthase [25], epicatechin, lacking both the galloyl and gallate groups, was found to have a substantially larger IC50 of 4 mM. Similar to the results here, rat liver mitochondrial ATP synthase was found to be inhibited by EGCG with an IC50 of 17 μM [20].
The extents of inhibition also correlated with the size of the polyphenols. That is probably related to the fact that the ATP synthase is a rotary motor, and many of its inhibitors appear to be mechanical inhibitors. For example, the polyphenols that have been-co-crystallized with the enzyme, such as quercetin or resveratrol are found to bind between the shaft of the rotary gamma subunit and the stationary alpha and beta subunits, which bind the nucleotides [22]. Since the theaflavins are somewhat larger than quercetin, and much larger than resveratrol, it could be expected that they would not bind to the same site, even though they are related chemically. Indeed, the computer modeling studies suggested a different binding site, one which overlaps with a previously identified site for the antibiotic efrapeptin, from crystallographic studies. This site is accessible from the opposite end of the enzyme, and it is a region that undergoes large conformational changes during rotary catalysis. It is possible that even with full occupancy, some of the smaller polyphenols fail to inhibit completely because they do not totally block rotation, but only hinder it.
Assays carried out with the isolated F1-ATPase demonstrated that the site of inhibition was located in the F1 sector of the enzyme, rather than Fo. The lower IC50 values obtained relative to the membrane-bound enzyme might reflect complexities in the activity of the enzyme. The rate of ATP hydrolysis by the membrane-bound enzyme is quite sensitive to pH [45] and other conditions [46, 47] that cause it to be released from the membrane sector. The results shown do not preclude additional bindings sites in the membrane sector. Other inhibitors, such as the antibiotic oligomycin, are known to act there. However, the assays of proton translocation showed no evidence for additional inhibitor sites in the membrane sector. While it cannot be ruled out that lower affinity sites exist, the polarity of the theaflavins, especially those with gallate esters, might be too great to allow binding in the lipid phase.
Two different enzymes allow NADH to initiate electron transport in E. coli, and both were found to be inhibited by all 5 polyphenols tested. In the case of NDH-1, the IC50 values followed the same pattern as with the ATP synthase: affinity correlated with size, except that TF1 had a larger IC50 than did EGCG. This again indicates the likely importance of the gallate groups in binding. The computer docking of TF3 suggested important interactions of the benzotropolone group and one of the gallates (3′) with the enzyme, supporting that conclusion and the relatively low IC50 for TF2B. Overall, the polyphenols had relatively high extents of inhibition (90-95%), which is consistent with a competitive mode of inhibition, as was found for TF3, in which substrate binding is precluded. The inhibition of NDH-2 followed the pattern of larger theaflavins having lower IC50 values, but in this case, EGCG had the highest IC50 value of all by a factor of 5-10. This suggests importance of the benzotropolone group, in conjunction with at least one gallate. Overall, the extents of inhibition were somewhat lower than for NDH-1, with values in the range of 65-95%. Since the mode of inhibition by TF3 was determined to be uncompetitive, it suggests that for some of the inhibitors, the enzyme-inhibitor-NADH complexes retained some activity.
It was not expected that TF3 would be a competitive inhibitor with respect to NADH, since it does not resemble NADH structurally. On the other hand, it also does not closely resemble ubiquinone, so there was no basis to predict that it would bind in the membrane sector. The finding of its competitive mode of inhibition of NADH oxidase activity was in conflict with the finding that it was uncompetitive with respect to NADH using the HAR reductase assay. The former result requires TF3 binding in the absence of NADH, while the later requires binding in the presence of NADH. An important consideration is that the mechanism of the HAR reaction is unknown. A recent paper by Birrell [48] suggests that HAR might draw off electrons through the protein from one of the Fe-S centers that is close enough to the surface. Given such uncertainties about this artificial electron acceptor, we favor the interpretation that the inhibition is competitive with respect to NADH. Supporting that view are the findings by computer modeling that a potential binding site exists that is partially overlapping with the entrance to the NADH binding site. In this way the binding of TF3 could block the binding of NADH, without significant structural similarity to it.
Many inhibitors of the electron transport chain are significant contributors to superoxide production in the mitochondrion, because they tend to keep particular electron carriers in a reduced state. For example, some Complex I inhibitors are known to increase the rate of superoxide production [17, 49]. Superoxide has been suggested to be formed at various sites in Complex I, including at the flavin, at an Fe-S center, or at a ubisemiquinone site [50-54]. The mode of the inhibition by TF3 is consistent with its lack of stimulation of superoxide production. A competitive inhibitor will tend to prevent reduction of the electron carriers, rather than to keep them in a reduced state. In addition, theaflavins are known to be superoxide scavengers [14]. In this study, TF3 caused a decrease in superoxide production while it inhibited the electron transport chain in E. coli membranes at levels in the range of the IC50 value.
The bioavailability of catechin compounds in the human body is thought to be low [55, 56], notwithstanding the possibility of higher bioavailability for some catechins such as EGCG [57], or in some tissues such as mouth and esophagus [58]. Thus, given the micromolar IC50 values measured here, and previous studies on the health benefits of theaflavins as antioxidants in vivo, we hypothesized that theaflavins had dual effects on cells in a dose-dependent way. At low doses, theaflavins play an important role as antioxidants, and the physiological consequences of the inhibition of ATP synthase or Complex I by theaflavins might be limited. At high dosage, theaflavins might have some effect on the electron transport chain. These results suggest the possibility of theaflavins used as drugs targeting Complex I or ATP synthase of some pathogens such as Mycobacterium tuberculosis, or in some diseases such as obesity [59]. In addition, the results presented here suggest that there are modes of inhibition of these enzymes that might be common to a variety of polyphenols, or related compounds, found in the human diet.
Supplementary Material
Acknowledgments
This work was supported by grant GM40508 from the National Institutes of Health, USA, and grant N-1378 from the Welch Foundation (S.B.V.), and the China Scholarship Council and the National Science Foundation for Young Researchers of China (Grant No.30901002 ) to Y.T. S.B.V was also supported as a Guang Biao Professor at Zhejiang University by the K. P. Chao Hi-Tech Foundation for Scholars and Scientists. From the Department of Biological Sciences at Southern Methodist University we thank Jessica De Leon Rangel and Shaotong Zhu for excellent advice and technical assistance, and Susan Pandey and Dr. Pia Vogel for help with F1-ATPase preparations.
List of Abbreviations:
- ACMA
9-amino 3-chloro 2-methoxy acridine
- EC
(-) epicatechin
- ECG
(-) epicatechin gallate
- EGC
(-) epigallocatechin
- EGCG
(-) epigallocatechin gallate
- HAR
hexamine ruthenium
- IC50
concentration of inhibitor required to achieve 50% of the maximal level of inhibition
- NDH-1
NADH dehydrogenase 1, or Complex I
- NDH-2
alternative NADH dehydrogenase
- TF1
theaflavin
- TF2A
theaflavin-3-gallate
- TF2B
theaflavin-3′-gallate
- TF3
theaflavin-3,3′-digallate
References
- [1].McKay DL, Blumberg JB. The role of tea in human health: An update. J Am Coll Nutr. 2002; 21:1–13. [DOI] [PubMed] [Google Scholar]
- [2].Cabrera C, Artacho R, Gimenez R. Beneficial effects of green tea-A review. J Am Coll Nutr. 2006; 25:79–99. [DOI] [PubMed] [Google Scholar]
- [3].Khan N, Mukhtar H. Tea polyphenols for health promotion. Life Sci 2007; 81:519–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sajilata MG, Bajaj PR, Singhal RS. Tea polyphenols as nutraceuticals. Compr Rev Food Sci Food Saf 2008; 7:229–54. [DOI] [PubMed] [Google Scholar]
- [5].Lin YL, Tsai SH, Lin-Shiau SY, Ho CT, Lin JK. Theaflavin-3,3′-digallate from black tea blocks the nitric oxide synthase by down-regulating the activation of NF-kappaB in macrophages. Eur J Pharmacol 1999; 367:379–88. [DOI] [PubMed] [Google Scholar]
- [6].Yang ZY, Tu YY, Xia HL, Jie GL, Chen XM, He PM. Suppression of free-radicals and protection against H2O2-induced oxidative damage in HPF-1 cell by oxidized phenolic compounds present in black tea. Food Chem 2007; 105:1349–56. [Google Scholar]
- [7].Drynan JW, Clifford MN, Obuchowicz J, Kuhnert N. The chemistry of low molecular weight black tea polyphenols. Nat Prod Rep 2010; 27:417–62. [DOI] [PubMed] [Google Scholar]
- [8].Kondo K, Kurihara M, Miyata N, Suzuki T, Toyoda M. Scavenging mechanisms of (−)-epigallocatechin gallate and (−)-epicatechin gallate on peroxyl radicals and formation of superoxide during the inhibitory action. Free Radic Biol Med 1999; 27:855–63. [DOI] [PubMed] [Google Scholar]
- [9].Tejero I, González-García N, González-Lafont À, Lluch JM. Tunneling in green tea: Understanding the antioxidant activity of catechol-containing compounds. A variational transition-state theory study. J Am Chem Soc 2007; 129:5846–54. [DOI] [PubMed] [Google Scholar]
- [10].Polovka M, Brezová V, Stasko A. Antioxidant properties of tea investigated by EPR spectroscopy. Biophys Chem 2003; 106:39–56. [DOI] [PubMed] [Google Scholar]
- [11].Sang S, Tian S, Jhoo J.-w, Wang H, Stark RE, Rosen RT, Yang CS, Ho C-T. Chemical studies of the antioxidant mechanism of theaflavins: radical reaction products of theaflavin 3,3′-digallate with hydrogen peroxide. Tetrahedron Lett 2003; 44:5583–87. [Google Scholar]
- [12].Guo Q, Zhao B, Shen S, Hou J, Hu J, Xin W. ESR study on the structure-antioxidant activity relationship of tea catechins and their epimers. Biochim Biophys Acta 1999; 1427:13–23. [DOI] [PubMed] [Google Scholar]
- [13].Severino JF, Goodman BA, Kay CWM, Stolze K, Tunega D, Reichenauer TG, Pirker KF. Free radicals generated during oxidation of green tea polyphenols: Electron paramagnetic resonance spectroscopy combined with density functional theory calculations. Free Radic Biol Med 2009; 46:1076–88. [DOI] [PubMed] [Google Scholar]
- [14].Jovanovic SV, Hara Y, Steenken S, Simic MG. Antioxidant Potential of Theaflavins. A Pulse Radiolysis Study. J Am Chem Soc 1997; 119:5337–43. [Google Scholar]
- [15].Yang Z, Jie G, Dong F, Xu Y, Watanabe N, Tu Y. Radical-scavenging abilities and antioxidant properties of theaflavins and their gallate esters in H2O2-mediated oxidative damage system in the HPF-1 cells. Toxicol In Vitro 2008; 22:1250–56. [DOI] [PubMed] [Google Scholar]
- [16].Brand MD. The sites and topology of mitochondrial superoxide production. Exp Gerontol 2010; 45:466–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fato R, Bergamini C, Bortolus M, Maniero AL, Leoni S, Ohnishi T, Lenaz G. Differential effects of mitochondrial Complex I inhibitors on production of reactive oxygen species. Biochim Biophys Acta 2009; 1787:384–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Johnson KM, Cleary J, Fierke CA, Opipari AW Jr.; Glick GD. Mechanistic basis for therapeutic targeting of the mitochondrial F1Fo-ATPase. ACS Chem Biol 2006; 1:304–8. [DOI] [PubMed] [Google Scholar]
- [19].Zheng J, Ramirez VD. Piceatannol, a stilbene phytochemical, inhibits mitochondrial FoF1-ATPase activity by targeting the F1 complex. Biochem Biophys Res Commun 1999; 261:499–503. [DOI] [PubMed] [Google Scholar]
- [20].Zheng J, Ramirez VD. Inhibition of mitochondrial proton FoF1-ATPase/ATP synthase by polyphenolic phytochemicals. Br J Pharmacol 2000; 130:1115–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kipp JL, Ramirez VD. Effect of estradiol, diethylstilbestrol, and resveratrol on FoF1-ATPase activity from mitochondrial preparations of rat heart, liver, and brain. Endocrine 2001; 15:165–75. [DOI] [PubMed] [Google Scholar]
- [22].Gledhill JR, Montgomery MG, Leslie AGW, Walker JE. Mechanism of inhibition of bovine F1-ATPase by resveratrol and related polyphenols. Proc Natl Acad Sci U.S.A 2007;104:13632–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gledhill JR, Walker JE. Inhibition sites in F1-ATPase from bovine heart mitochondria. Biochem J 2005;386:591–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dadi PK, Ahmad M, Ahmad Z. Inhibition of ATPase activity of Escherichia coli ATP synthase by polyphenols. Int J Biol Macromol 2009; 45:72–9. [DOI] [PubMed] [Google Scholar]
- [25].Chinnam N, Dadi PK, Sabri SA, Ahmad M, Kabir MA, Ahmad Z. Dietary bioflavonoids inhibit Escherichia coli ATP synthase in a differential manner. Int J Biol Macromol 2010; 46:478–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Humbert R, Brusilow WS, Gunsalus RP, Klionsky DJ, Simoni RD. Escherichia coli mutants defective in the uncH gene. J Bacteriol 1983; 153:416–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Amarneh B, Vik SB. Mutagenesis of subunit N of the Escherichia coli Complex I. Identification of the initiation codon and the sensitivity of mutants to decylubiquinone. Biochemistry 2003; 42:4800–08. [DOI] [PubMed] [Google Scholar]
- [28].Calhoun MW, Gennis RB. Demonstration of separate genetic loci encoding distinct membrane-bound respiratory NADH dehydrogenases in Escherichia coli. J Bacteriol 1993; 175:3013–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ishmukhametov RR, Galkin MA, Vik SB. Ultrafast purification and reconstitution of His-tagged cysteine-less Escherichia coli F1Fo ATP synthase. Biochim Biophys Acta 2005; 1706:110–6. [DOI] [PubMed] [Google Scholar]
- [30].Volkov OA, Zaida TM, Voeller P, Lill H, Wise JG, Vogel PD. De-novo modeling and ESR validation of the cyanobacterial FoF1-ATP synthase subunit bb’ left-handed coiled coil. Biochim Biophys Acta 2009; 1787:183–90. [DOI] [PubMed] [Google Scholar]
- [31].Wise JG. Site-directed mutagenesis of the conserved β subunit tyrosine 331 of Escherichia coli ATP synthase yields catalytically active enzymes. J Biol Chem 1990; 265:10403–9. [PubMed] [Google Scholar]
- [32].Taussky HH, Shorr E. A microcolorimetric method for the determination of inorganic phosphorus. J Biol Chem 1953; 202:675–85. [PubMed] [Google Scholar]
- [33].Dixon M The determination of enzyme inhibitor constants. Biochem J 1953; 55:170–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cornish-Bowden A A simple graphical method for determining the inhibition constants of mixed, uncompetitive and non-competitive inhibitors. Biochem J 1974. ; 137:143–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pedretti A, Villa L, Vistoli G. VEGA: a versatile program to convert, handle and visualize molecular structure on Windows-based PCs. J Mol Graph Model 2002; 21:47–9. [DOI] [PubMed] [Google Scholar]
- [36].Pedretti A, Villa L, Vistoli G. VEGA – An open platform to develop chemo-bio-informatics applications, using plug-in architecture and script programming. J Comput Aided Mol Des 2004; 18:167–73. [DOI] [PubMed] [Google Scholar]
- [37].Abrahams JP, Buchanan SK, Van Raaij MJ, Fearnley IM, Leslie AG, Walker JE. The structure of bovine F1-ATPase complexed with the peptide antibiotic efrapeptin. Proc Natl Acad Sci U.S.A 1996; 93:9420–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Berrisford JM, Sazanov LA. Structural basis for the mechanism of respiratory complex I. J Biol Chem 2009; 284:29773–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera-A visualization system for exploratory research and analysis. J Comput Chem 2004; 25:1605–12. [DOI] [PubMed] [Google Scholar]
- [40].Morris GM, Goodsell DS, Huey R, Olson AJ. Distributed automated docking of flexible ligands to proteins: parallel applications of AutoDock 2.4. J Comput Aided Mol Des 1996; 10:293–304. [DOI] [PubMed] [Google Scholar]
- [41].Fang N, Casida JE. Anticancer action of cube insecticide: correlation for rotenoid constituents between inhibition of NADH:ubiquinone oxidoreductase and induced ornithine decarboxylase activities. Proc Natl Acad Sci U.S.A 1998; 95:3380–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zini R, Morin C, Bertelli A, Bertelli AA, Tillement JP. Effects of resveratrol on the rat brain respiratory chain. Drugs Exp Clin Res 1999; 25:87–97. [PubMed] [Google Scholar]
- [43].Matsushita K, Ohnishi T, Kaback HR. NADH-ubiquinone oxidoreductases of the Escherichia coli aerobic respiratory chain. Biochemistry 1987; 26:7732–7. [DOI] [PubMed] [Google Scholar]
- [44].Calhoun MW, Oden KL, Gennis RB, de Mattos MJ, Neijssel OM. Energetic efficiency of Escherichia coli: effects of mutations in components of the aerobic respiratory chain. J Bacteriol 1993; 175:3020–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Al-Shawi MK, Parsonage D, Senior AE. Directed mutagenesis of the strongly conserved aspartate 242 in the β-subunit of Escherichia coli proton-ATPase. J Biol Chem 1988; 263:19633–9. [PubMed] [Google Scholar]
- [46].Tuttas-Dörschug R, Hanstein WG. Coupling factor 1 from Escherichia coli lacking subunits δ and ε: preparation and specific binding to depleted membranes, mediated by subunits δ or ε. Biochemistry 1989; 28:5107–13. [DOI] [PubMed] [Google Scholar]
- [47].Vogel G, Steinhart R. ATPase of Escherichia coli: purification, dissociation, and reconstitution of the active complex from the isolated subunits. Biochemistry 1976; 15:208–16. [DOI] [PubMed] [Google Scholar]
- [48].Birrell JA, Yakovlev G, Hirst J. Reactions of the flavin mononucleotide in complex I: a combined mechanism describes NADH oxidation coupled to the reduction of APAD(+), ferricyanide, or molecular oxygen. Biochemistry 2009; 48:12005–13. [DOI] [PubMed] [Google Scholar]
- [49].Lambert AJ, Brand MD. Inhibitors of the quinone-binding site allow rapid superoxide production from mitochondrial NADH:ubiquinone oxidoreductase (Complex I). J Biol Chem 2004; 279:39414–20. [DOI] [PubMed] [Google Scholar]
- [50].Galkin A, Brandt U. Superoxide radical formation by pure complex I (NADH:ubiquinone oxidoreductase) from Yarrowia lipolytica. J Biol Chem 2005; 280:30129–35. [DOI] [PubMed] [Google Scholar]
- [51].Genova ML, Ventura B, Giuliano G, Bovina C, Formiggini G, Parenti Castelli G, Lenaz G The site of production of superoxide radical in mitochondrial Complex I is not a bound ubisemiquinone but presumably iron-sulfur cluster N2. FEBS Lett 2001; 505:364–8. [DOI] [PubMed] [Google Scholar]
- [52].Grivennikova VG, Vinogradov AD. Generation of superoxide by the mitochondrial Complex I. Biochim Biophys Acta 2006; 1757:553–61. [DOI] [PubMed] [Google Scholar]
- [53].Kussmaul L, Hirst J. The mechanism of superoxide production by NADH:ubiquinone oxidoreductase (complex I) from bovine heart mitochondria. Proc Natl Acad Sci U.S.A 2006; 103:7607–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ohnishi ST, Shinzawa-Itoh K, Ohta K, Yoshikawa S, Ohnishi T. New insights into the superoxide generation sites in bovine heart NADH-ubiquinone oxidoreductase (Complex I): The significance of protein-associated ubiquinone and the dynamic shifting of generation sites between semiflavin and semiquinone radicals. Biochim Biophys Acta 2010; 1797: 1901–9. [DOI] [PubMed] [Google Scholar]
- [55].Lafay S, Gil-Izquierdo A. Bioavailability of phenolic acids. Phytochem Rev 2008; 7:301–11. [Google Scholar]
- [56].Manach C, Williamson G, Morand C, Scalbert A, Remesy C. Bioavailability and bioefficacy of polyphenols in humans. I. Review of 97 bioavailability studies. Am J Clin Nutr 2005; 81:230S–42S. [DOI] [PubMed] [Google Scholar]
- [57].Del Rio D, Calani L, Cordero C, Salvatore S. Pellegrini N; Brighenti F. Bioavailability and catabolism of green tea flavan-3-ols in humans. Nutrition 2010; 26:1110–6. [DOI] [PubMed] [Google Scholar]
- [58].Yang CS, Kim S, Yang GY, Lee MJ, Liao J, Chung JY, Ho CT. Inhibition of carcinogenesis by tea: bioavailability of tea polyphenols and mechanisms of actions. Proc Soc Exp Biol Med 1999; 220:213–7. [DOI] [PubMed] [Google Scholar]
- [59].Hong S, Pedersen PL. ATP synthase and the actions of inhibitors utilized to study its roles in human health, disease, and other scientific areas. Microbiol Mol Biol R 2008; 72: 590–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


