Dear editor,
Recent published study reported that angiotensin-converting enzyme 2 (ACE2) of various mammals may contribute to cross-species transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).1 In fact, there is strong evidence that SARS-CoV-2 is a zoonotic spillover from bats into human populations through bridging hosts.2 Therefore, ongoing investigation on possible intermediate hosts would facilitate understanding the mechanisms of cross-species transmission of SARS-CoV-2 with potential benefits for the control of the coronavirus disease 2019 (COVID-19).
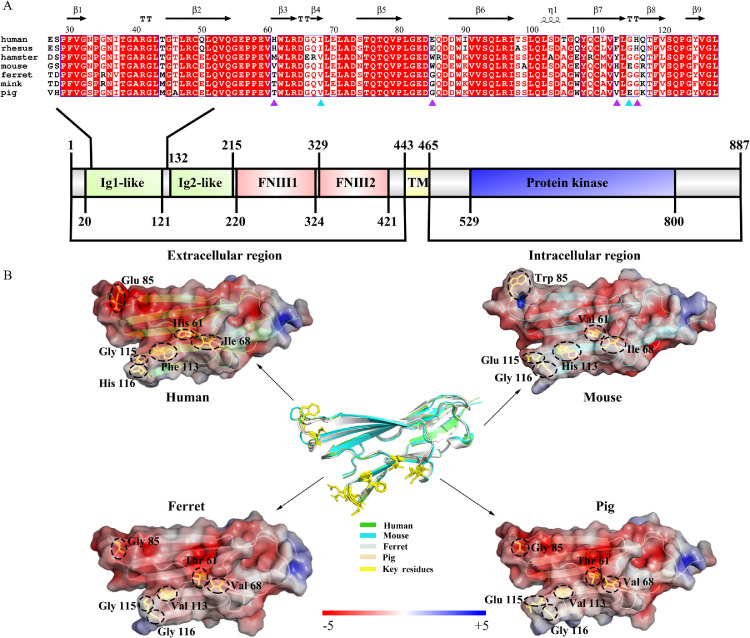
Notably, the receptor is the crucial host restrict factor in SARS-CoV-2 infection. ACE2 is proved to serve as a primary factor for the cellular entry of SARS-CoV-2, which may determine the host tropism.3 , 4 However, the low-abundance expression of ACE2 in human lung and trachea indicated that additional cellular mediators may exist that promote cellular uptake of SARS-CoV-2. Recently research demonstrated that the tyrosine-protein kinase receptor UFO (AXL) is a novel candidate receptor, especially in the respiratory system.5 Since pigs as important livestock species and the huge size of their global population, it is of great significance to reveal the putative susceptibility of swine to SARS-CoV-2.6 Although previous data suggested that porcine ACE2 could bind SARS-CoV-2 spike (S),3 the interaction between porcine AXL and S that predict infection capacity is still unclear. Here, the amino acid (aa) sequences of AXLs in different species containing the residues interfacing with the N-terminal domain (NTD) of S were aligned to screen the critical sites by ESPript 3.0 (https://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). According the structure of human AXL-S NTD complex, P57–58, E59, H61, I68, E70, E85, F113, and G115 are located in the interface and interact with S NTD. As illustrated in Fig. 1 A, the key residues of porcine AXL were consistent with those of mink and ferret, which are efficiently infected with SARS-CoV-2, and showed weaker affinity characteristics than human AXL. Among these sites, T61H and G85E substitutions may reduce the binding affinities, while V68I and E115G mutations would not disrupt the formation of hydrogen bonds.5 Specifically, G116H substitution existing in hamster, mouse, mink, ferret, and pig AXLs result in a new interaction on the interface, which may have little effect on interfacial binding properties. The distribution of electrostatic potential also confirmed that the more identical pattern in AXL-S interface between pig and ferret, which appears to be significantly different from mouse (Fig. 1B). In consequence, the local structural difference of AXLs among different species suggested that porcine AXL may exhibit the similar binding affinity with S NTD to those of mink and ferret.
Fig. 1.
Alignment and surface potential analysis of crucial amino acids in AXL proteins. (A) Comparative analysis of the residues of AXLs at the interface binding to spike of SARS-CoV-2 from human (GenBank accession no. NP_068713), rhesus (GenBank accession no. EHH30062.1), hamster (GenBank accession no. XP_040590606), mouse (GenBank accession no. NP_033491), ferret (GenBank accession no. XP_004776133), mink (GenBank accession no. XP_044113292.1), and pig (GenBank accession no. NP_001121930). The ALX residues at position 68 and 115 are marked in cyan triangles, position 61, 85, 113, and 116 are labeled with purple triangles. (B) Surface potential diagram of interface zone of AXLs. The structural superposition of the AXL region 27–128 from human (green, PDB code 4RA0), mouse (cyan, Uniprot entry Q0093), ferret (gray), and pig (wheat) is in the center. The AXL structures of pig and ferret were modeled using the homology models of human AXL (PDB code 4RA0 and 5VXZ, respectively) as the templates by SWISS-MODEL (https://swissmodel.expasy.org/). The six key differential residues of AXL interacting with spike protein of SARS-CoV-2 are represented by yellow sticks in the structural superposition, the details of which are further circled by dash line in the surface electrostatic potential maps of each AXL. The electrostatic potential color range is -/+5.
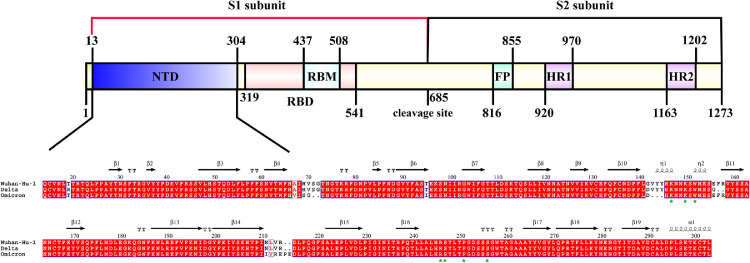
Interestingly, the experimental infection results in two independent studies did not support that pig is susceptible to SARS-CoV-2 infection,7 , 8 in contrast to previous ACE2 results and the above AXL analysis associated with the spike-binding peculiarity.1 , 3 Nevertheless, several porcine cell lines are identified to be permissive for SARS-CoV-2 infection. Owing to the limited pig breeds and numbers, age and health condition should been taken into account in future study,7 which may combine the pre-existing illnesses to develop typical COVID-19 symptoms.9 In addition, SARS-CoV-2 possesses the largest RNA genome, and evolves quickly to generate genetic diversity. The newly emerging variants omicron and delta have stronger ACE2 binding affinity and higher contiguous than these predecessors, with the mild clinical manifestations. To address the impact of the certain changes in S NTD of different SARS-CoV-2 strains, comparative analysis of aa sequences was performed. The result in Fig. 2 uncovered that the key residues K147, K150, W152, R246, S247, P251, and S256 in the AXL-S NTD complexes were unaltered, while multiple residue deletions and substitutes occurred in the same positions between two variants. More importantly, whether these aa differences would have an influence on the ability of SARS-CoV-2 to cause COVID-19 related diseases in pigs is urgently needed to be explored. Once the SARS-CoV-2 variants enhance the infectivity and transmission in pigs, the outcome for evolution of zoonotic potential would become more unpredictable and complicated when these variants are introduced into swine herds.
Fig. 2.
Sequence alignment of spike protein from three SARS-CoV-2 strains including Wuhan-Hu-1 (GenBank accession no.YP_009724390), delta variant (GenBank accession no. QYM88683), and omicron variant (GenBank accession no. UFS23237). The important residues in NTD of S interacting with AXL are marked with green stars.
In summary, alignment of AXL from different species and S NTD of three representative SARS-CoV-2 strains would be conducive to quest for the potential recipient hosts in zoonotic transmission. Considering pig as the dominant livestock and the idea human biomedical model,10 a specific focus on interaction between porcine AXL and S of SARS-CoV-2 variants may also help prevent the risk of next pandemic and food supply stability.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 32002289) and the Science and Technology Project of Jiangxi Province (Nos. 2018ACB21027, 20212ACB205004, and 20203BBF63020). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
References
- 1.Li R., Qiao S., Zhang G. Analysis of angiotensin-converting enzyme 2 (ACE2) from different species sheds some light on cross-species receptor usage of a novel coronavirus 2019-nCoV. J Infect. 2020;80(4):469–496. doi: 10.1016/j.jinf.2020.02.013. PubMed PMID: 32092392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruiz-Aravena M., McKee C., Gamble A., Lunn T., Morris A., Snedden C.E., et al. Ecology, evolution and spillover of coronaviruses from bats. Nat Rev Microbiol. 2021:1–16. doi: 10.1038/s41579-021-00652-2. https://www.nature.com/articles/s41579-021-00652-2#Sec2 PubMed PMID: 34799704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. PubMed PMID: 32015507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ren W., Zhu Y., Wang Y., Shi H., Yu Y., Hu G., et al. Comparative analysis reveals the species-specific genetic determinants of ACE2 required for SARS-CoV-2 entry. PLoS Pathog. 2021;17(3) doi: 10.1371/journal.ppat.1009392. PubMed PMID: 33760889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang S., Qiu Z., Hou Y., Deng X., Xu W., Zheng T., et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31(2):126–140. doi: 10.1038/s41422-020-00460-y. PubMed PMID: 33420426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muñoz-Fontela C., Dowling W.E., Funnell S.G.P., Gsell P.S., Riveros-Balta A.X., Albrecht R.A., et al. Animal models for COVID-19. Nature. 2020;586(7830):509–515. doi: 10.1038/s41586-020-2787-6. PubMed PMID: 32967005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlottau K., Rissmann M., Graaf A., Schön J., Sehl J., Wylezich C., et al. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. Lancet Microbe. 2020;1(5):e218–ee25. doi: 10.1016/S2666-5247(20)30089-6. PubMed PMID: 32838346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368(6494):1016–1020. doi: 10.1126/science.abb7015. PubMed PMID: 32269068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du X., Guo Z., Fan W., Hai T., Gao F., Li P., et al. Establishment of a humanized swine model for COVID-19. Cell Discov. 2021;7(1):70. doi: 10.1038/s41421-021-00313-x. PubMed PMID: 34404772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lunney J.K., Van Goor A., Walker K.E., Hailstock T., Franklin J., Dai C. Importance of the pig as a human biomedical model. Sci Transl Med. 2021;13(621):eabd5758. doi: 10.1126/scitranslmed.abd5758. PubMed PMID: 34818055. [DOI] [PubMed] [Google Scholar]