Abstract
Cardiac magnetic resonance enables comprehensive cardiac evaluation; however, intense time and labor requirements for data acquisition and processing have discouraged many clinicians from using it. We have developed an alternative image-processing algorithm that requires minimal user interaction: an ultrafast algorithm that computes left ventricular ejection fraction (LVEF) by using temporal intensity variation in cine balanced steady-state free precession (bSSFP) short-axis images, with or without contrast medium. We evaluated the algorithm's performance against an expert observer's analysis for segmenting the LV cavity in 65 study participants (LVEF range, 12%–70%). In 12 instances, contrast medium was administered before cine imaging. Bland-Altman analysis revealed quantitative effects of LV basal, midcavity, and apical morphologic variation on the algorithm's accuracy.
Total computation time for the LV stack was <2.5 seconds. The algorithm accurately delineated endocardial boundaries in 1,132 of 1,216 slices (93%). When contours in the extreme basal and apical slices were not adequate, they were replaced with manually drawn contours. The Bland-Altman mean differences were <1.2 mL (0.8%) for end-diastolic volume, <5 mL (6%) for end-systolic volume, and <3% for LVEF. Standard deviation of the difference was ≤4.1% of LV volume for all sections except the midcavity in end-systole (8.3% of end-systolic volume).
We conclude that temporal intensity variation–based ultrafast LVEF computation is clinically accurate across a range of LV shapes and wall motions and is suitable for postcontrast cine SSFP imaging. Our algorithm enables real-time processing of cine bSSFP images on a commercial scanner console within 3 seconds in an unobtrusive automated process.
Keywords: Algorithms; automation; cardiac volume/physiology; image processing, computer-assisted/methods; magnetic resonance imaging, cine/methods; predictive value of tests; reference values; reproducibility of results; ventricular dysfunction, left/diagnosis
Cardiovascular disease is the leading cause of death worldwide.1 Impaired myocardial contractile function caused by diminished perfusion or cardiomyopathies can manifest itself as reduced left ventricular ejection fraction (LVEF) and lead to heart failure.2 Cardiac magnetic resonance (CMR), the gold standard for estimating LVEF, enables clinicians to characterize pathologic tissue and evaluate myocardial morphology, perfusion, viability, and global and regional function with excellent spatial and contrast resolution. However, the intense time and labor required for data acquisition and processing have discouraged many clinicians from using CMR.
In standard CMR evaluation, LVEF is estimated by analyzing a stack of cine balanced steady-state free precession (bSSFP) short-axis (SA) slices during a time-consuming process. Images of the left ventricle (LV) are acquired on a scanner console, and the data are transferred to a remote postprocessing workstation. To determine global LV function, a CMR expert must draw myocardial contours on a stack of SA slices for the entire LV. Increasingly, at CMR sites, the cine slices are being acquired after a contrast agent has been administered, to save time in patients who are undergoing angiography or myocardial viability evaluation.
Fluidly integrating data acquisition and postprocessing necessitates a rapid, clinically accurate computational algorithm that is optimally robust for capturing varying LV shapes and wall motions, as well as in the presence of contrast medium. However, typical LV segmentation algorithms rely on the substantial contrast difference between the myocardium and the blood pool in precontrast images. In myocardial viability imaging, advantageous workflow involves acquiring bSSFP images after contrast administration. In these instances, the blood-to-myocardial contrast ratio is substantially diminished, and segmentation algorithms that rely solely on spatial contrast may fail. To achieve clinical accuracy in the presence of LV variations, an optimal algorithm should be driven predominantly by data rather than by training-based models. In addition, its use should require minimal input from experts, and it should have minimal memory requirements and low computational costs.
We prospectively compared the clinical performance of our ultrafast, data-driven LV segmentation algorithm against that of a manual LV segmentation process, and we report our findings.
Participants and Methods
The study included 65 participants: 49 patients from our CMR clinical schedule who were being evaluated for LV dysfunction (35 men and 14 women; mean age, 51 ± 9 yr; age range, 17–83 yr) and 16 asymptomatic volunteers (8 men and 8 women; mean age, 38 ± 5 yr; age range, 27–54 yr). Our Institutional Ethics Committee approved this study, and all participants gave written informed consent.
Two thirds of the patients were evaluated for myocardial viability, and the others for dilated or hypertrophic cardiomyopathy, LV noncompaction, or LV aneurysm. In 12 patients evaluated for myocardial viability, images were acquired <10 min after 0.2 mmol/kg of gadolinium was administered as a contrast agent.
Image Acquisition
Images were acquired with use of an Achieva 1.5T scanner (Philips Healthcare), equipped with a 32-element phased-array surface coil and vector-cardiographic (VCG) gating. All study participants held their breath for 10 to 12 heartbeats while non-VCG–gated scout images of the thoracic cavity were acquired along the 3 orthogonal planes. Then, during similar breath-holds, images of VCG-gated 2-chamber views, 4-chamber views, and 10 to 13 contiguous SA slices covering the LV from apex to base were acquired.
The repetition time was 3 to 3.2 ms; the echo time, 1.5 to 1.6 ms; and the flip angle, 55°. The acquired voxel size was 1.25–1.5 × 1.25–1.5 × 8 mm3, the sensitivity-encoding acceleration factor was 2, and the temporal resolution was 40 to 50 ms. Images were stored in Digital Imaging and Communications in Medicine (DICOM) standard format.
Manual Image Analysis
The DICOM data were transferred to an on-site View-Forum postprocessing workstation (Philips Healthcare). A board-certified cardiac radiologist (BYCC), who had 7 years of clinical CMR experience and was blinded to the algorithmic results, reviewed the SA cine images and manually drew the LV myocardial contours, including the papillary muscles (PMs) and trabeculae in the LV volume. The global LV functional indices of end-diastolic volume (EDV), end-systolic volume (ESV), and LVEF were then calculated as references for the algorithm's accuracy. By visually inspecting the cine loop of the midcavity slice, the reviewer defined end-diastole (ED) and end-systole (ES) as the phases with the largest and smallest cavity areas, respectively, and the basal slice as that in which at least 50% of the ED blood volume was surrounded by ventricular myocardium.3 To study anatomic effects on our algorithm's accuracy, we annotated each slice as basal (from the mitral valve annulus to the tips of the PMs at ED), midcavity (the length of the PMs), or apical (beyond the PMs to the apex). To ensure accurate volumetric and LVEF measurements, we included the slice containing the LV outflow tract with the basal slice if it encompassed any LV volume.
The algorithm-drawn contours were then given to the reviewer. For consistent quantitative comparison, he inspected the resultant contours for proper shape (in the extreme basal slice) and for size (LV volume inscribed by the extreme apical LV contour), and he redrew those that were clinically unacceptable.
In-Line Image Analysis by Algorithm
The LVEF computation program, written in MATLAB R2009a (MathWorks, Inc.), runs as an unobtrusive background extension of the acquisition protocol on the console of a 1.5T Achieva clinical scanner (Intel Xeon, 3.2 GHz, 6 GB RAM). The major processing steps are LV localization and LV segmentation.
Localization. Immediately after images are written to the database, the in-line processing tool automatically launches. From the displayed diastolic frame of each SA slice, the user selects and inputs the most basal and apical slices (Fig. 1). The algorithm identifies the LV midcavity slice halfway between the specified slices.
Fig. 1.
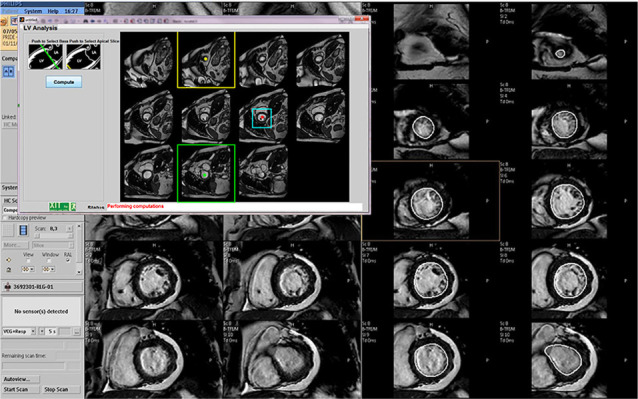
For in-line computation of ejection fraction on the cardiac magnetic resonance scanner console, the user clicks inside the left ventricle during end-diastole and, in the pop-up window, specifies the basal (green outline) and apical (yellow outline) slices. The algorithm computes the region of interest (ROI) for the midcavity slice (cyan outline), then processes one short-axis ROI slice at a time to extract endocardial contours for all slices and phases. The main console window displays the resultant contoured slices (the 2 columns on the right) next to the corresponding original images (the 2 columns on the left).
Otsu's method4 is used on an 8-bit histogram of the midcavity image (Fig. 2A) to determine an intensity threshold for identifying bright-signal regions of blood, fat, and fluid (Fig. 2B). Hollow and irregularly shaped regions are eliminated on the basis of solidity (area/convex area) and eccentricity (ratio of the distance between the foci of the fitted ellipse and its major axis length) of the convex hull of the regions (Fig. 2C). The remaining bright regions are ranked according to the combined score (N/D), where N is the region's area normalized to the image area, and D is the region's distance from the intersection point of the midcavity slice and the 3-dimensional (3D) line connecting the user-specified basal and apical points. The highest-ranked region is labeled the region of interest (ROI) (Fig. 2D). The algorithm then processes the data, slice by slice, toward the base and apex.
Fig. 2.
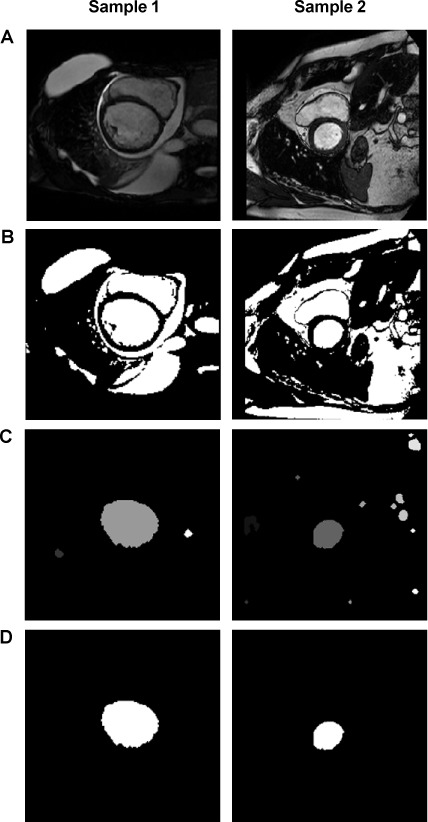
Images represent the algorithmic steps for left ventricular localization in a patient with significant pericardial effusion and a dilated left ventricle (Sample 1) and a patient with substantial epicardial fat (Sample 2). A) An 8-bit histogram is used to determine an intensity threshold to identify B) bright-signal regions. C) Concave and irregularly shaped regions have been eliminated, and the remaining regions have been ranked according to combined spatial proximity and area. D) In midcavity slices, the left ventricle is the highest-ranked region of interest; 3-dimensional geometric continuity constraints are used for the remaining slices.5
Segmentation. After the center of the LV cavity has been identified, the algorithm for LV segmentation proceeds as follows:
Individual pixels along 8 radial lines that intersect at the center of an identified LV region (Fig. 3A) are analyzed to create time-intensity profiles, which show the variation in intensity over time (Fig. 3B). The mean value of the individual time-intensity (TI) profiles is then subtracted from the value at each time point to generate zero-clamped TI profiles (Fig. 3C).
Some pixel locations are occupied entirely by blood or myocardium throughout the cardiac cycle (Fig. 3B). However, some pixels such as those adjoining the endocardium may be occupied by blood during early and late diastole but by myocardium during systole. These transitory pixels have zero-clamped TI profiles that show only 2 zero crossovers during the cardiac cycle, the first going from a positive to a negative value (Fig. 3C). The pixels transition from blood-only to partial volumes (PVs) of blood and myocardium, and then to myocardium-only during the ED-to-ES cycle, and in reverse during the ES-to-ED cycle.
Transitory pixels in the cine phases between the crossover time points (Fig. 3C) are chiefly myocardium-only and are classified as blood-myocardium PV pixels. The lower and higher signal intensities corresponding to half the peak pixel population are recorded in a histogram (Fig. 3D). Gaussian distribution of transitory pixels determines the myocardial upper-intensity threshold (Fig. 3B).
After the ROI threshold is established at the upper intensity limit, the LV is localized among the blood, fat, and fluids by eliminating the regions touching the ROI edges and by using solidity and eccentricity criteria and 3D geometric constraints on the LV from adjoining slices (Fig. 2).5
A convex hull is fitted to the blood pool (Fig. 4A). To prevent concave portions during curve-fitting, salient points of the convex hull are determined by curvature and spatial proximity to the adjacent corner.
A piecewise closed Bezier curve of 2nd-order geometric continuity is fitted through the salient points of the convex hull (Fig. 4B) to delineate endocardial contours for a given slice (Fig. 4C–D).6
Fig. 3.
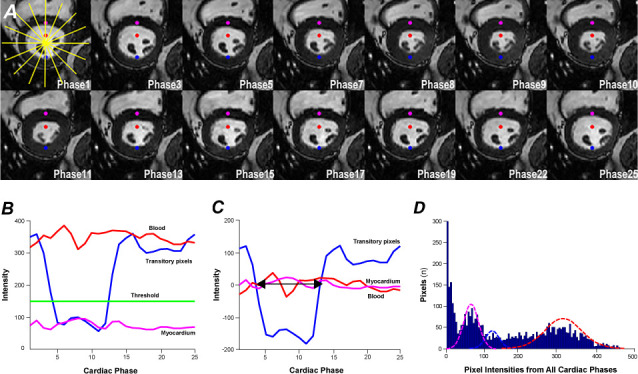
Images show steps for classifying partial-volume (PV) pixels. A) On individual pixels along 8 radial lines centered on the identified left ventricular (LV) region, B) time-intensity (TI) profiles are created. In A, the red dot shows the blood-only pixels. In B, the red line indicates their TI profile; the magenta line, that of myocardium-only pixels (magenta dot in A); and the blue line, that of transitory pixels (blue dot in A). The blue line shows signal changes as the pixels cycle from blood to PV blood-myocardium, myocardium, PV myocardium-blood, and blood again. C) Zero-clamped TI profiles of the pixels depicted in A are shown; the mean intensity during the cardiac cycle is subtracted from intensity at each time point. The transitory pixels between the crossover time points are PV (blood-myocardium) pixels (between arrows in C). D) Histogram shows overlaid distributions of blood pool (red), myocardium (magenta), and PV myocardium-blood (blue). Gaussian distribution of transitory pixels determines the myocardial upper-intensity threshold (green line in B).
Fig. 4.
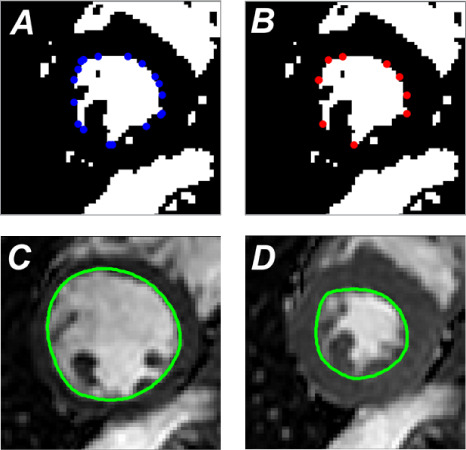
Images illustrate algorithmic delineation of endocardial contours. The convex hull is fitted to the resultant images after the myocardial upper-intensity threshold is applied (blue dots correspond to corners of the polygon). A) The curvature and the spatial proximity to the adjacent corner are used on blue points to select salient points of the convex hull. B) A piecewise closed Bezier curve of 2nd-order geometric continuity is fitted through the salient points. Corresponding endocardial contours (green outline) are shown for C) end-diastole and D) end-systole.
Basal slices are automatically rejected if the LV segmented area increases abruptly, thus indicating shape discontinuity. After all slices are processed, the cardiac phase halfway between the zero-crossover time points is used as the ES phase for automatic LVEF reporting.
Statistical Analysis
Bland-Altman analysis was used to compare the EDV, ESV, and LVEF results with those derived manually.7 The bias (mean difference ± SD) was compared with previously obtained corresponding values for inter- and intraobserver variability.8 Contours unacceptable to the CMR expert were redrawn for analytic consistency. To evaluate morphologic effects of PMs and trabeculae on variations between the respective results, the SA slices were trifurcated into basal, midcavity, and apical sections. Each was analyzed for volume (mL) and difference as a percentage of the manually traced volume for the entire LV.
Results
We evaluated a total of 1,216 slices: 494 ED and 433 ES from the 49 patients, and 158 ED and 131 ES from the 16 volunteers. The computation time per slice was 80 to 120 ms, and the total time per participant was 2 ± 0.2 sec. Algorithmic contours were redrawn in 27 ED and 29 ES extreme basal slices, as well as in 13 ED and 15 ES extreme apical slices, to exclude the outflow tract from LV volume and include extremely small apical LV sections (volume, <2 mL). The algorithm's relative performance was also evaluated by region: basal (188 ED and 113 ES slices), midcavity (294 ED, 297 ES), and apical (170 ED, 154 ES).
Examples of comparative manual and computed images are shown in Figure 5, and Figure 6 shows examples of adjusted contours.
Fig. 5.
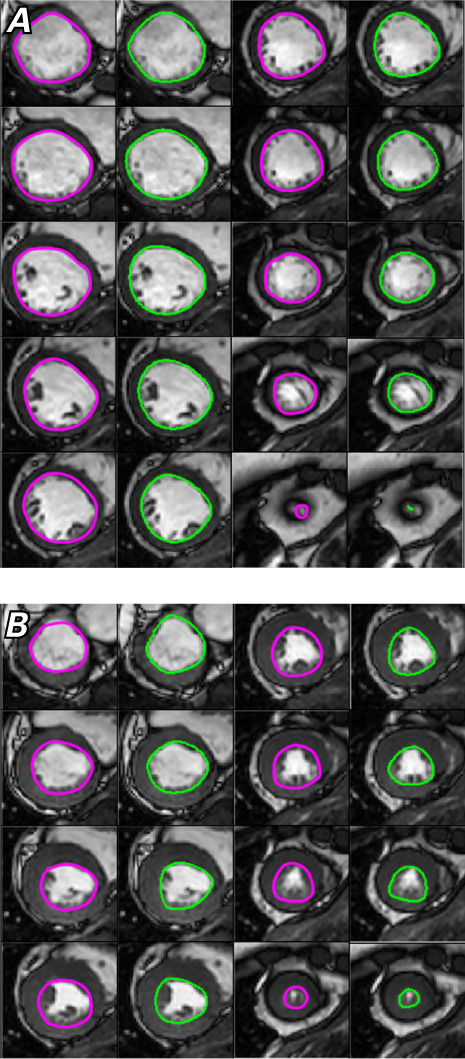
Images show manual (magenta) and computed (green) left ventricular contours in A) end-diastole and B) end-systole.
Fig. 6.
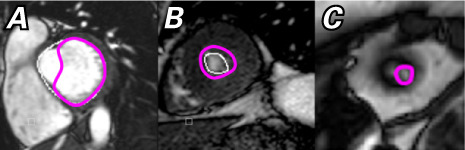
Images show manual adjustments (magenta) to the algorithmic contours (white) for A) a basal slice with outflow tract and B), C) extreme apical slices.
Table I shows the distribution of the manually traced LV volumes. Myocardial hyperenhancement was seen in 30% of the patients. The diagnoses were dilated cardiomyopathy in 25%; systolic dysfunction in 35%; and hypertrophic cardiomyopathy, LV noncompaction, or LV aneurysm in the rest.
TABLE I.
Manually Traced Left Ventricular Volumetric Indices in 16 Volunteers and 49 Patients
| End-Diastolic Volume (mL) | End-Systolic Volume (mL) | Ejection Fraction (%) | ||||
|---|---|---|---|---|---|---|
|
|
|
|
||||
| Distribution | Volunteers | Patients | Volunteers | Patients | Volunteers | Patients |
| Minimum | 118 | 75 | 48 | 30 | 49 | 12 |
| 1st Quartile | 152 | 143 | 61 | 66 | 53 | 37 |
| Median | 185 | 175 | 75 | 81 | 56 | 53 |
| 3rd Quartile | 203 | 211 | 93 | 127 | 62 | 59 |
| Maximum | 271 | 456 | 121 | 387 | 70 | 66 |
We calculated the Bland-Altman bias and the SD of the difference between the manual and algorithmic contours for the apical, midcavity, and basal regions (Table II) and by participant type in the presence or absence of contrast administration (Table III).
TABLE II.
Bland-Altman Analysis Comparing Manual and Algorithmic Contours by Slice Location
| End-Diastolic Volume (mL, %) | End-Systolic Volume (mL, %) | Ejection Fraction (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||
| Apical | Mid | Basal | Total | Apical | Mid | Basal | Total | Actual | Computed* | |
| Bias | 0.2, 0 | −0.3, 0 | −1.5, −0.4 | −1.2, −0.8 | 1.5, 1.3 | 4.5, 6 | −0.3, −0.2 | 4, 5.2 | −3 | −5.4 |
| SDD | 3.9, 1.6 | 8, 4 | 5.1, 2.8 | 14.7, 7.4 | 3.9, 3.8 | 7.8, 8.3 | 3.6, 4.1 | 12.3, 13.5 | 4.4 | 5 |
Mid = midcavity; SDD = standard deviation of the difference
*Computed after area was set to zero to reject unacceptable contours
Data are presented as mL and percent error (percentage of total left ventricular volume calculated manually). Bias is the mean difference.
TABLE III.
Bland-Altman Analysis Comparing Manual and Algorithmic Contours by Participant Type and Contrast Status
| Volunteers | Patients, No Contrast | Patients, Contrast | |||||||
|---|---|---|---|---|---|---|---|---|---|
| EDV (mL, %) | ESV (mL, %) | EF (%) | EDV (mL, %) | ESV (mL, %) | EF (%) | EDV (mL, %) | ESV (mL, %) | EF (%) | |
| Bias | 3, 1.7 | 4.3, 5.9 | −2 | −2.7, −1.2 | 5.7, 7.1 | −4.1 | −4.8, −3.1 | −1.5, −1.8 | −0.8 |
| SDD | 2.8, 1.4 | 6.7, 9.3 | 3.8 | 12.7, 7 | 11.5, 12.3 | 4 | 25.9, 11.8 | 18.4, 19.3 | 5.5 |
EDV = end-diastolic volume; EF = ejection fraction; ESV = end-systolic volume; SDD = standard deviation of the difference
Data are presented as mL and percent error (percentage of total left ventricular volume calculated manually). Bias is the mean difference.
The algorithm underestimated ESV by 7% and, consequently, overestimated LVEF by 4% in patients not given contrast medium (Fig. 7). Figure 8 shows representative algorithmic contours for postcontrast data.
Fig. 7.
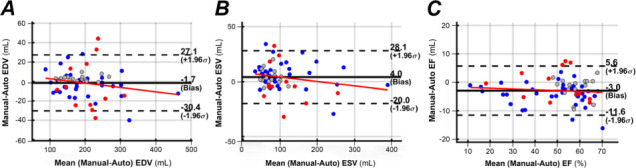
Bland-Altman plots show analyses of variability between manual and algorithmic computations for left ventricular A) end-diastolic volume (EDV), B) end-systolic volume (ESV), and C) ejection fraction (EF) in volunteers (gray circles), patients with no contrast administered (blue circles), and patients with contrast administered (red circles).
Fig. 8.
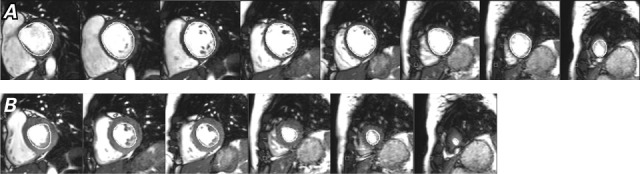
Postcontrast images show left ventricular endocardial contours during A) end-diastole and B) end-systole.
Percentages for EDV, ESV, and LVEF bias were comparable for patients with and without contrast. However, in postcontrast cases, the SD increased by 69% in EDV and by 57% in ESV. The ESV was underestimated chiefly in the midcavity, where PMs blended with endocardium.
Discussion
Our algorithm worked successfully when LV volumes ranged from 75 mL to 456 mL at ED and when LVEF ranged from 12% to 70%. Results were accurate in 13 patients with LVEF <36% and in 12 whose images were acquired after contrast administration. Thus, determining myocardial intensity thresholds by using TI profiles is a robust method in patients who have reduced wall motion or diminished blood-to-myocardial contrast. The manual and algorithmic endocardial contours were qualitatively similar in ED and were close in ES. Quantitatively, the respective volumes were comparable. Bias was within typical inter- and intraobserver variability (0.05 LVEF) among experienced clinicians.9–12
Manually delineating myocardial boundaries within cine bSSFP images takes time and is susceptible to human error. Developing an automated algorithm to perform this task has challenged investigators for nearly 2 decades.13–15 Segmentation methods proposed for extracting LV functional indices have ranged from simple signal intensity–based approaches to sophisticated model-based algorithms.10,11,16–43 In general, LV volume analysis involves 1) basic signal processing to classify intensity and compute energy derived for features in the image (for example, edges), and 2) advanced mathematical modeling that uses shape-motion constraints to minimize the computed energy. However, myocardium and the fine structures projecting from it have similar signal intensities, and experts include these structures in the endocardial contour to estimate LV function and analyze wall motion. Computationally intensive models can account for these geometric considerations but are sensitive to initial placement and assumptions underlying spatiotemporal behavior. Left ventricular dilation or cavity obliteration in hypertrophic cardiomyopathy violates these assumptions, often necessitating contour correction. Previously, in-line algorithms with delays of 10 to 15 seconds per slice were implemented fully44 or partially45 in CMR reconstruction. In unsupervised approaches, LV coverage in ED starts from the most basal slice, and in ES, the most basal slice is presumptively one slice below that in ED.44 Instead of relying on the contrast-diminished intensity difference between the blood pool and myocardium alone, we use a priori knowledge of intensity variation in pixels near the endocardium—a minimally assumptive, data-driven approach.
When PMs were well isolated from the endocardium during ED, the SD for our algorithmic volume estimates in all LV sections was ≤4%, with minimal bias (< −0.4%). Accuracy was lowest for the midcavity in ES, when the PMs blended with the myocardium (bias, 6%; SD, 8.3%). A remaining challenge is to extract acceptable endocardial contours when the outflow tract intrudes into the basal slices, especially when a CMR expert draws the semilunar contours.
Initial Efficacy of the Algorithm's Main Design Elements
The distinctive signal intensity of pixels adjoining the endocardium enables better threshold determination in ES phases than does global signal intensity, especially in postcontrast images. Although using intensity alone cannot isolate PMs from endocardium during complete blending at ES, geometric constraint to avoid sharp concave indentations achieves endocardial contours similar to manual contours by forcing the contour through most convex boundary points.
Our algorithm's low computational cost enables real-time LVEF estimation, generation of LV time-volume curves, and further wall-motion analysis. Optimizing the process in C or C++ can increase speed. Before and after replacing unacceptable basal contours, we observed a 1.8% change in bias and a 1.7% change in SD for LVEF, indicating that underestimated EDV and ESV cause further overestimation of LVEF. In future versions, contours can be written as an editable overlay for rapid correction or removal.
The algorithm requires minimal user interaction—a CMR technician need only specify the basal and apical slices on the scanner console. This user-driven approach enables LVEF estimation, even from the stack of SA slices that span the ventricles as well as the atria, as is often done when evaluating congenital heart disease.
The analysis protocol seamlessly retrieves, processes, and saves the images to the patient's database, displaying the computed LVEF. The algorithm loads into memory only the first cardiac phase of the central SA slice (treated as ED) to localize the LV and constrain the ROI,12 thus minimizing memory competition with routine scanner operation. Of note, we found no effects on the spectrometer or reconstruction control of the host, and data acquisition was uninterrupted. In our clinic, we have already developed other in-line postprocessing tools, such as an M-mode display to enable viewing of delayed septal-lateral wall motion.46
Study Limitations
This study had limitations. First, the inclusion of all consenting participants created a study population with heterogeneous clinical indications, so we plan to test the algorithm specifically in patients who have atrial fibrillation, hypertrophic cardiomyopathy, or dilated cardiomyopathy. Second, MATLAB precludes users from editing contours on the scanner itself. Finally, we relied on previously reported inter- and intraobserver variability in EDV, ESV, and LVEF when determining clinically acceptable bias. Although there is no reason to believe that these numbers will differ significantly, repeated contouring by our expert reviewer and additional contouring by another reviewer would have provided observer variability specific to our study population.
Conclusion
Our cost-effective algorithm for LV segmentation enables clinically accurate computation of global LV functional indices (EDV, ESV, and LVEF), with or without contrast administration. The real-time process is completed in <3 seconds on a CMR scanner console as an automated, unobtrusive extension of data acquisition.
Acknowledgment
The Department of Scientific Publications at the Texas Heart Institute provided editorial support.
Footnotes
Conflict of interest disclosures: Dr. Pednekar is employed by Philips Healthcare. Dr. Muthupillai has an institutional research agreement with Philips Healthcare.
Funding/support: This study was partially supported by funding from Philips Healthcare.
References
- 1.Alwan A, World Health Organization Geneva: World Health Organization; 2011. Global status report on noncommunicable diseases 2010. [Google Scholar]
- 2.Solomon SD, Anavekar N, Skali H, McMurray JJV, Swedberg K, Yusuf S et al. Influence of ejection fraction on cardiovascular outcomes in a broad spectrum of heart failure patients. Circulation . 2005;112(24):3738–44. doi: 10.1161/CIRCULATIONAHA.105.561423. [DOI] [PubMed] [Google Scholar]
- 3.Alfakih K, Plein S, Thiele H, Jones T, Ridgway JP, Sivananthan MU. Normal human left and right ventricular dimensions for MRI as assessed by turbo gradient echo and steady-state free precession imaging sequences. J Magn Reson Imaging . 2003;17(3):323–9. doi: 10.1002/jmri.10262. [DOI] [PubMed] [Google Scholar]
- 4.Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern . 1979;9(1):62–6. [Google Scholar]
- 5.Pednekar AS, Muthupillai R, Lenge VV, Kakadiaris IA, Flamm SD. Automatic identification of the left ventricle in cardiac cine-MR images: dual-contrast cluster analysis and scout-geometry approaches. J Magn Reson Imaging . 2006;23(5):641–51. doi: 10.1002/jmri.20552. [DOI] [PubMed] [Google Scholar]
- 6.Farin GE. 3rd ed. San Diego: Academic Press Professional, Inc; 1993. Curves and surfaces for computer-aided geometric design: a practical guide. [Google Scholar]
- 7.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet . 1986;1(8476):307–10. [PubMed] [Google Scholar]
- 8.Sardanelli F, Quarenghi M, Di Leo G, Boccaccini L, Schiavi A. Segmentation of cardiac cine MR images of left and right ventricles: interactive semiautomated methods and manual contouring by two readers with different education and experience. J Magn Reson Imaging . 2008;27(4):785–92. doi: 10.1002/jmri.21292. [DOI] [PubMed] [Google Scholar]
- 9.Codella NCF, Cham MD, Wong R, Chu C, Min JK, Prince MR et al. Rapid and accurate left ventricular chamber quantification using a novel CMR segmentation algorithm: a clinical validation study. J Magn Reson Imaging . 2010;31(4):845–53. doi: 10.1002/jmri.22080. [DOI] [PubMed] [Google Scholar]
- 10.Lu YL, Connelly KA, Dick AJ, Wright GA, Radau PE. Automatic functional analysis of left ventricle in cardiac cine MRI. Quant Imaging Med Surg . 2013;3(4):200–9. doi: 10.3978/j.issn.2223-4292.2013.08.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Brien SP, Ghita O, Whelan PF. A novel model-based 3D +time left ventricular segmentation technique. IEEE Trans Med Imaging . 2011;30(2):461–74. doi: 10.1109/TMI.2010.2086465. [DOI] [PubMed] [Google Scholar]
- 12.Pednekar AS, Muthupillai R, Cheong B, Flamm SD. Automatic computation of left ventricular ejection fraction from spatiotemporal information in cine-SSFP cardiac MR images. J Magn Reson Imaging . 2008;28(1):39–50. doi: 10.1002/jmri.21363. [DOI] [PubMed] [Google Scholar]
- 13.Petitjean C, Dacher JN. A review of segmentation methods in short axis cardiac MR images. Med Image Anal . 2011;15(2):169–84. doi: 10.1016/j.media.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Suh DY, Mersereau RM, Eisener RL, Pettigrew RI. Knowledge-based boundary detection applied to cardiac magnetic resonance image sequences. Int Conf Acoustics, Speech, and Signal Processing. Vol. 4. IEEE; 1990. pp. 2341–4. p. Available from: https://ieeexplore.ieee.org/document/116053. [Google Scholar]
- 15.Suinesiaputra A, Cowan BR, Al-Agamy AO, Elattar MA, Ayache N, Fahmy AS et al. A collaborative resource to build consensus for automated left ventricular segmentation of cardiac MR images. Med Image Anal . 2014;18(1):50–62. doi: 10.1016/j.media.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cocosco CA, Niessen WJ, Netsch T, Vonken EJPA, Lund G, Stork A, Viergever MA. Automatic image-driven segmentation of the ventricles in cardiac cine MRI. J Magn Reson Imaging . 2008;28(2):366–74. doi: 10.1002/jmri.21451. [DOI] [PubMed] [Google Scholar]
- 17.Codella NCF, Weinsaft JW, Cham MD, Janik M, Prince MR, Wang Y. Left ventricle: automated segmentation by using myocardial effusion threshold reduction and intravoxel computation at MR imaging. Radiology . 2008;248(3):1004–12. doi: 10.1148/radiol.2482072016. [DOI] [PubMed] [Google Scholar]
- 18.Cordero-Grande L, Vegas-Sanchez-Ferrero G, Casasecade-la-Higuera P, San-Roman-Calvar JA, Revilla-Orodea A, Martin-Fernandez M, Alberola-Lopez C. Unsupervised 4D myocardium segmentation with a Markov Random Field based deformable model. Med Image Anal . 2011;15(3):283–301. doi: 10.1016/j.media.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Goshtasby A, Turner DA. Segmentation of cardiac cine MR images for extraction of right and left ventricular chambers. IEEE Trans Med Imaging . 1995;14(1):56–64. doi: 10.1109/42.370402. [DOI] [PubMed] [Google Scholar]
- 20.Gupta A, O'Donnell T, Singh A. Segmentation and tracking of cine cardiac MR and CT images using a 3-D deformation model. Computers in Cardiology. IEEE; 1994. pp. 661–4. p. Available from: https://ieeexplore.ieee.org/document/470099. [Google Scholar]
- 21.Jolly MP. Automatic segmentation of the left ventricle in cardiac MR and CT images. Int J Comput Vis . 2006;70(2):151–63. [Google Scholar]
- 22.Jolly MP. Combining edge, region, and shape information to segment the left ventricle in cardiac MR images. In: Niessen WJ, Viergever MA, editors. Medical Image Computing and ComputerAssisted Intervention—MICCAI. Berlin: Springer; Lecture Notes in Computer Science (vol. 2208); 2001. p. 482–90. Available from: https://link.springer.com/chapter/10.1007/3-540-45468-3_58. [Google Scholar]
- 23.Jolly MP, Xue H, Grady L, Guehring J. Combining registration and minimum surfaces for the segmentation of the left ventricle in cardiac cine MR images. Med Image Comput Assist Interv . 2009;12(Pt 2):910–8. doi: 10.1007/978-3-642-04271-3_110. [DOI] [PubMed] [Google Scholar]
- 24.Kaus MR, von Berg J, Weese J, Niessen W, Pekar V. Automated segmentation of the left ventricle in cardiac MRI. Med Image Anal . 2004;8(3):245–54. doi: 10.1016/j.media.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 25.Lalande A, Legrand L, Walker PM, Guy F, Cottin Y, Roy S, Brunotte F. Automatic detection of left ventricular contours from cardiac cine magnetic resonance imaging using fuzzy logic. Invest Radiol . 1999;34(3):211–7. doi: 10.1097/00004424-199903000-00010. [DOI] [PubMed] [Google Scholar]
- 26.Lee HY, Codella NCF, Cham MD, Weinsaft JW, Wang Y. Automatic left ventricle segmentation using iterative thresholding and an active contour model with adaptation on short-axis cardiac MRI. IEEE Trans Biomed Eng . 2010;57(4):905–13. doi: 10.1109/TBME.2009.2014545. [DOI] [PubMed] [Google Scholar]
- 27.Lee HY, Codella N, Cham M, Prince M, Weinsaft J, Wang Y. Left ventricle segmentation using graph searching on intensity and gradient and a priori knowledge (lvGIGA) for short-axis cardiac magnetic resonance imaging. J Magn Reson Imaging . 2008;28(6):1393–401. doi: 10.1002/jmri.21586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lelieveldt BP, van der Geest RJ, Reiber JH, Bosch JG, Mitchell SC, Sonka M. Time-continuous segmentation of cardiac image sequences using active appearance motion models. In: Insana MF, Leahy RM, editors. Information Processing in Medical Imaging. Berlin: Springer-Verlag; Lecture Notes in Computer Science (vol. 2082); 2001. p. 446–52. Available from: https://link.springer.com/chapter/10.1007/3-540-45729-1_47. [Google Scholar]
- 29.Lorenzo-Valdes M, Sanchez-Ortiz GI, Elkington AG, Mohiaddin RH, Rueckert D. Segmentation of 4D cardiac MR images using a probabilistic atlas. Med Image Anal . 2004;8(3):255–65. doi: 10.1016/j.media.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Lotjonen J. Segmentation of MR images using deformable models: application to cardiac images. Int J Bioelectromagnetism . 2001;3(2):49–58. [Google Scholar]
- 31.Lotjonen J, Kivisto S, Koikkalainen J, Smutek D, Lauerma K. Statistical shape model of atria, ventricles and epicardium from short- and long-axis MR images. Med Image Anal . 2004;8(3):371–86. doi: 10.1016/j.media.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 32.Makowski P, Sorensen TS, Therkildsen SV, Materka A, Stodkilde-Jorgensen H, Pedersen EM. Two-phase active contour method for semiautomatic segmentation of the heart and blood vessels from MRI images for 3D visualization. Comput Med Imaging Graph . 2002;26(1):9–17. doi: 10.1016/s0895-6111(01)00026-x. [DOI] [PubMed] [Google Scholar]
- 33.Merrifield R, Keegan J, Firmin D, Yang GZ. Dual contrast TrueFISP imaging for left ventricular segmentation. Magn Reson Med . 2001;46(5):939–45. doi: 10.1002/mrm.1280. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell SC, Lelieveldt BP, van der Geest RJ, Bosch HG, Reiber JH, Sonka M. Multistage hybrid active appearance model matching: segmentation of left and right ventricles in cardiac MR images. IEEE Trans Med Imaging . 2001;20(5):415–23. doi: 10.1109/42.925294. [DOI] [PubMed] [Google Scholar]
- 35.Paragios N. A level set approach for shape-driven segmentation and tracking of the left ventricle. IEEE Trans Med Imaging . 2003;22(6):773–6. doi: 10.1109/TMI.2003.814785. [DOI] [PubMed] [Google Scholar]
- 36.Bi-temporal 3D active appearance models with applications to unsupervised ejection fraction estimation. In: Fitzpatrick JM, Reinhardt JM, editors. Proc SPIE Medical Imaging 2005: Image Processing; 2005. Available from: https://neurophotonics.spiedigitallibrary.org/conference-proceedings-of-spie/5747/0000/Bi-temporal-3D-active-appearance-models-with-applications-to-unsupervised/10.1117/12.594930.short. [Google Scholar]
- 37.Suh DY, Eisner RL, Mersereau RM, Pettigrew RI. Knowledge-based system for boundary detection of four-dimensional cardiac magnetic resonance image sequences. IEEE Trans Med Imaging . 1993;12(1):65–72. doi: 10.1109/42.222668. [DOI] [PubMed] [Google Scholar]
- 38.Sun H, Frangi AF, Wang H, Sukno FM, Tobon-Gomez C, Yushkevich PA. Automatic cardiac MRI segmentation using a biventricular deformable medial model. Med Image Comput Assist Interv . 2010;13(Pt 1):468–75. doi: 10.1007/978-3-642-15705-9_57. [DOI] [PubMed] [Google Scholar]
- 39.Thedens DR, Skorton DJ, Fleagle SR. Methods of graph searching for border detection in image sequences with applications to cardiac magnetic resonance imaging. IEEE Trans Med Imaging . 1995;14(1):42–55. doi: 10.1109/42.370401. [DOI] [PubMed] [Google Scholar]
- 40.Uzumcu M, van der Geest RJ, Swingen C, Reiber JHC, Lelieveldt BPF. Time continuous tracking and segmentation of cardiovascular magnetic resonance images using multidimensional dynamic programming. Invest Radiol . 2006;41(1):52–62. doi: 10.1097/01.rli.0000194070.88432.24. [DOI] [PubMed] [Google Scholar]
- 41.Cardiac LV segmentation using a 3D active shape model driven by fuzzy inference. In: Ellis RE, Peters TM, editors. Lecture Notes in Computer Science; Proc 6th Int Medical Image Computing and Computer-Assisted Intervention - MICCAI 2003; Berlin: Springer; 2003. pp. 533–40. p. [Google Scholar]
- 42.van Assen HC, Danilouchkine MG, Dirksen MS, Reiber JHC, Lelieveldt BPF. A 3-D active shape model driven by fuzzy inference: application to cardiac CT and MR. IEEE Trans Inf Technol Biomed . 2008;12(5):595–605. doi: 10.1109/TITB.2008.926477. [DOI] [PubMed] [Google Scholar]
- 43.van der Geest RJ, Buller VG, Jansen E, Lamb HJ, Baur LH, van der Wall EE, et al. Comparison between manual and semiautomated analysis of left ventricular volume parameters from short-axis MR images. J Comput Assist Tomogr . 1997;21(5):756–65. doi: 10.1097/00004728-199709000-00019. [DOI] [PubMed] [Google Scholar]
- 44.Theisen D, Sandner TA, Bauner K, Hayes C, Rist C, Reiser MF, Wintersperger BJ. Unsupervised fully automated inline analysis of global left ventricular function in CINE MR imaging. Invest Radiol . 2009;44(8):463–8. doi: 10.1097/RLI.0b013e3181aaf429. [DOI] [PubMed] [Google Scholar]
- 45.Li B, Liu Y, Occleshaw CJ, Cowan BR, Young AA. Inline automated tracking for ventricular function with magnetic resonance imaging. JACC Cardiovasc Imaging . 2010;3(8):860–6. doi: 10.1016/j.jcmg.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 46.Pednekar A, Arena C, Swaab J, Cheong B, Muthupillai R. Ultrafast in-line interactive m-mode tool for quantification of left-ventricular (LV) septo-lateral wall motion (SLWM) from high-temporal resolution (6–12ms) cardiac cine steady-state free precession (SSFP) images. J Cardiovasc Magn Reson . 2011;13(Suppl 1):P37. [Google Scholar]


